Abstract
Cervical cancer is acknowledged as the most prevalent gynecological tumor and a severe public issue that threatens female health, resulting from its high incidence and fatality rate. Surging evidence have shown that circular RNAs (circRNAs) play significant roles in the initiation and progression of various malignancies. Although circAMOTL1 has been testified to execute oncogenic properties in breast cancer and prostate cancer, literature on its function and regulatory mechanism in cervical cancer development is still scanty. Using a bioinformatics analysis, we found circ_0004214 was a circular form of AMOTL1. Through qRT-PCR analysis, circAMOTL1 and its host gene AMOTL1 were both upregulated in cervical cancer tissues and closely correlated with poor prognosis of cervical cancer. Gain- or loss-of-function assays and in vivo experiments demonstrated that AMOTL1 promoted cervical cancer cell growth both in vitro and in vivo. Mechanically, circAMOTL1 served as a competing endogenous RNA (ceRNA) to prompt the expression of AMOTL1 through sponging miR-485-5p. Rescue assays revealed that miR-485-5p/AMOTL1 axis was involved in circ_AMOTL1-mediated cervical cancer progression. Our findings provide a better understanding of the molecular mechanism underlying circAMOTL1 in cervical cancer and indicated circAMOTL1/miR-485-5p/AMOTL1 as a promising novel therapeutic strategy for the treatment of this disease.
Keywords: circAmotl1, cervical cancer, Amotl1, miR-485-5p
Introduction
Cervical cancer is listed as the most prevalent gynecological tumor and the fourth leading reason for mortality in female malignancies worldwide.1 On the basis of global statistics, cervical cancer has become a severe public issue that threatens female health, leading to approximately 527,600 new cases and 265,700 mortalities annually and showing an increasing trend.2 Although encouraging improvements have been achieved in therapeutic options, treatment effectiveness is limited, and the 5-year survival rate of cervical cancer patients remains only 40%–50%.3 Considering that, it is necessary to elucidate the indistinct mechanism of cervical cancer and figure out promising potent strategies for the remedy of this disease.
It is well known that circular RNAs (circRNAs) are a novel category of non-coding RNAs, which are extensively characterized by lack of 5′ caps or 3′ poly(A) tails.4 Mounting documentation has proven that circRNAs are implicated in the initiation and progression of multiple malignant tumors, including cervical cancer. For instance, circRNA8924 promotes cervical cancer cell proliferation, migration, and invasion by competitively binding to the miR-518d-5p/519-5p family and modulating the expression of CBX8.5 circRNA ciRS-7 correlates with advance disease and poor prognosis, and its downregulation inhibits cell proliferation while inducing cell apoptosis in non-small-cell lung cancer.6 circZMYM2 regulated JMJD2C in pancreatic cancer by endogenously competing with miR-335-5p.7 It has been reported that circAMOTL1 (hsa_circ_0004214) exerts a crucial role in breast cancer.8 However, whether circAMOTL1 participates in cervical cancer development remains to be further explored.
Angiomotin-like 1 (AMOTL1) is a member of the motin family, which plays a core role in modulating the migration and polarity of endothelial cells, together with angiomotin (Amot) and angiomotin-like 2 (Amotl2).9, 10, 11 It is confirmed by several researchers that AMOTL1 is a pivotal effector in the Hippo pathway by negatively regulating YAP expression so as to elicit oncogenic property in breast cancer.12, 13, 14 Besides, AMOTL1 has been documented to facilitate vasculogenic mimicry and cell motility in cervical cancer progression.15 Nevertheless, the interplay between circAMOTL1 and its host gene AMOTL1 in cervical cancer is still unclear.
The purpose of this study was to investigate the participation of circAMOTL1 and AMOTL1 in cervical cancer and expound the detailed regulatory mechanism of circAMOTL1 on AMOTL1 expression, which may contribute to enriching the therapeutic strategy for cervical cancer.
Results
Existence of circAMOTL1 in Cervical Cancer Cells
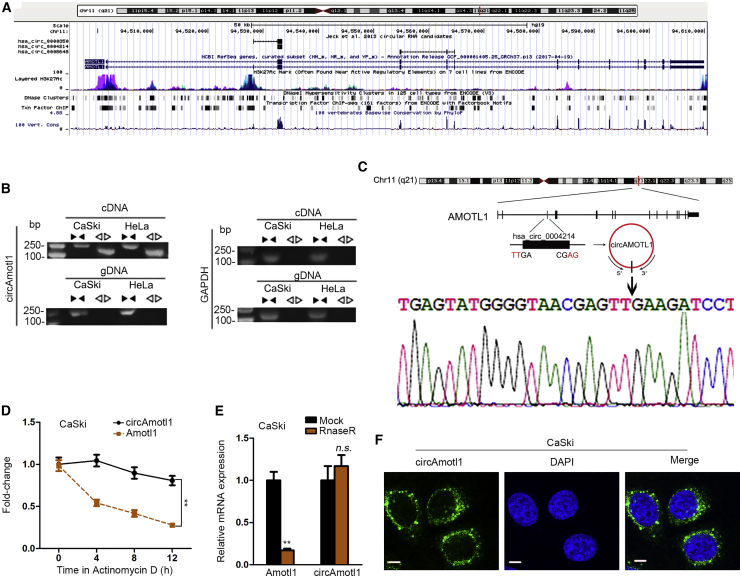
Searching the University of California Santa Cruz (UCSC) database, we discovered that circAMOTL1 was located in chromosome 11 and AMOTL1 was its host gene (Figure 1A). By employing the circRNADisease database, we subsequently found circAMOTL1 to be upregulated in cervical cancer. In view of that, we intended to evaluate the involvement of circAMOTL1 in cervical cancer development. Our data manifested the existence of circAMOTL1 in cervical cancer by evaluation of cDNA and genomic DNA from two randomly selected cervical cancer cell lines CaSki and HeLa (Figure 1B). Besides, Sanger sequencing was adopted to sequence the amplified product and corroborated the circularized junction of the circAMOTL1 (Figure 1C). We observed that circAMOTL1 exhibited more resistance to transcription inhibitor actinomycin D (ActD) in comparison with AMOTL1 (Figure 1D). Concordantly, results of RNase R exonuclease treatment certified the circular nature of the circAMOTL1 (Figure 1E). FISH assay was conducted to assess the subcellular localization of circAMOTL1 in CaSki cells, and we found that circAMOTL1 was principally localized in the cytoplasm (Figure 1F). On the whole, our findings validate the circular characterization of circAMOTL1 and the relationship between circAMOTL1 and cervical cancer.
Figure 1.
Existence of circAMOTL1 in Cervical Cancer Cells
(A) Data from the UCSC website revealed that AMOTL1 was the host gene for circAMOTL1. (B) The qPCR analysis was applied to validate the expression of circAMOTL1in cervical cancer cell lines. (C) The “head-to-tail” splicing sites of circAMOTL1 detected by Sanger sequencing. (D) The expression level of circAMOTL1 and linear AMOTL1 in CaSki cells treated with Actinomycin D in different time points. (E) The expression level of circAMOTL11 and AMOTL1 mRNA in CaSki cells treated with or without Rnase R was measured by qRT-PCR. (F) The localization of circAMOTL1 in CaSki cell was evaluated by FISH assay. Scale bar, 200 μm. **p < 0.01. n.s.: no significance.
circAMOTL1 and Its Host Gene AMOTL1 Indicate Poor Prognosis of Cervical Cancer
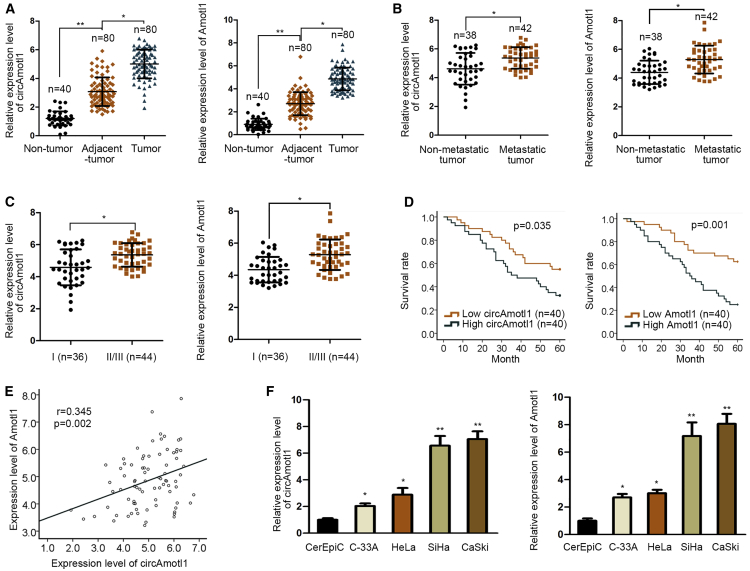
In order to probe the role of circAMOTL1 and AMOTL1 in cervical cancer, we estimated circAMOTL1 and AMOTL1 expression in clinical tissues and cell lines of cervical cancer. The qPCR analysis revealed that both circAMOTL1 and AMOTL1 were upregulated in cervical cancer tissues compared with that in adjacent normal tissues (Figure 2A). Additionally, in contrast with non-metastatic tumor tissues, circAMOTL1and AMOTL1 were abundant in metastatic tumor tissues (Figure 2B). We unveiled that the expression of circAMOTL1 and its host gene AMOTL1 were prominently elevated attributable to the increase of the clinical classification (Figure 2C). In agreement with the above results, cervical cancer patients with high levels of circAMOTL1 and AMOTL1 had worse survival rates than those with low expression of circAMOTL1 and AMOTL1 (Figure 2D), and results of Pearson’s correlation coefficient analysis unraveled the positive association between circAMOTL1 and AMOTL1 (Figure 2E). There were five circular forms of AMOTL1 that had the same exons. Through qRT-PCR examination and northern blot analysis, we determined that four circular forms of AMOTL1 had no significant expression change in the normal cervical epithelial cell line and four CC cell lines (Figures S1A and S1B). However, circ_0004214 (corcAmotl1) and its host gene Amotl1 were upregulated in cervical cancer cells compared with that in CerEpiC cell (Figure 2F). By and large, upregulation of circAMOTL1 and AMOTL1 correlates with poor outcomes of cervical cancer.
Figure 2.
circAMOTL1 and Its Host Gene AMOTL1 Indicate Poor Prognosis of Cervical Cancer
(A) The non-cancer, para-cancer, and tumor tissues were subjected to qRT-PCR for examination of circAMOTL1 and AMOTL1 expression. (B) The higher expression level of circAMOTL1 and AMOTL1 mRNA in patient samples with metastasis. (C) The high expression level of circAMOTL1 and AMOTL1 mRNA in samples obtained from patients in stage II/III. (D) Kaplan-Meier curve was generated to analyze the overall survival of cervical cancer patients with high or low level of circ_ or. (E) The relationship between circ and was analyzed by Pearson’s analysis. (F) qRT-PCR was implemented to detect the expression levels of circAMOTL1 and AMOTL1 in normal cervical epithelial cells (CerEpiC) and cervical cancer cell lines (C-33A, HeLa, Siha, and CaSki). *p < 0.05, **p < 0.01.
circAMOTL1 Boosts Cell Proliferation and Migration in Cervical Cancer
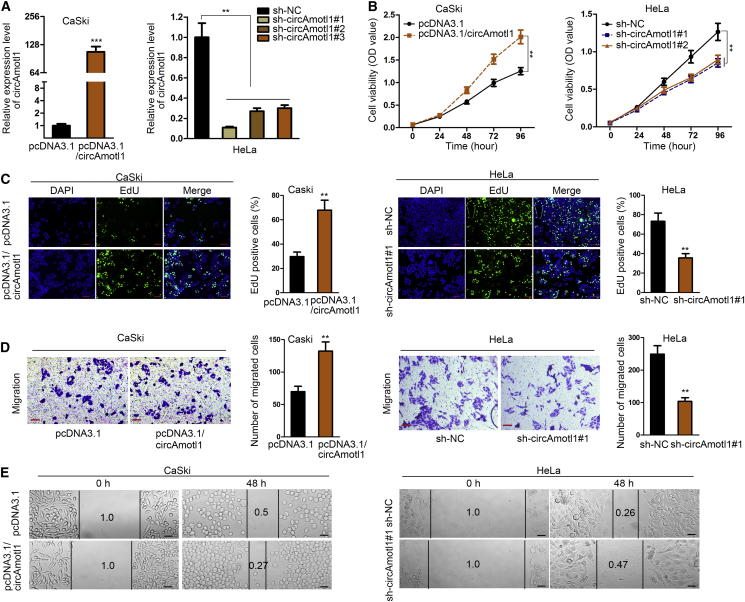
Consequent upon the findings mentioned, we subsequently explored the effect of circAMOTL1 on the proliferative and migratory capability of cervical cancer cells. First, circAMOTL1 was overexpressed in CaSki cells, and the expression of circAMOTL1 in HeLa cells was notably depleted after transfection (Figure 3A). The CCK-8 assay demonstrated that ectopic expression of circAMOTL1 promoted CaSki cell viability, whereas circAMOTL1 downregulation restrained cell growth of cervical cancer (Figure 3B). The EdU assay expounded that circAMOTL1 overexpression resulted in an overt increase of EdU-positive cells, and silencing of circAMOTL1 produced the opposite consequence, further validating that circAMOTL1 contributed to the promotion of cell proliferation in cervical cancer (Figure 3C). Moreover, we verified that cell migration was suppressed due to upregulation of circAMOTL1 and vice versa (Figure 3D). This result was confirmed by wound healing assay (Figure 3E). To summarize, we illuminate that circAMOTL1 induces the proliferation and migration of cervical cancer cells.
Figure 3.
circAMOTL1 Boosts Cell Proliferation and Migration in Cervical Cancer
(A) qPCR was utilized to determine the abundance of circAMOTL1 after transfection. (B–C) Increased or decreased cell viability was assessed in circAmotl1-silenced CaSki cell or circAmotl1-overexpressed HeLa cell by CCK-8 assay. (D–E) EdU assay assessed the proliferative ability of circAmotl1-silenced CaSki cell or circAmotl1-overexpressed HeLa cell. **p < 0.01, ***p < 0.001.
circAMOTL1 Modulates the Expression of AMOTL1 through Sponging miR-485-5p
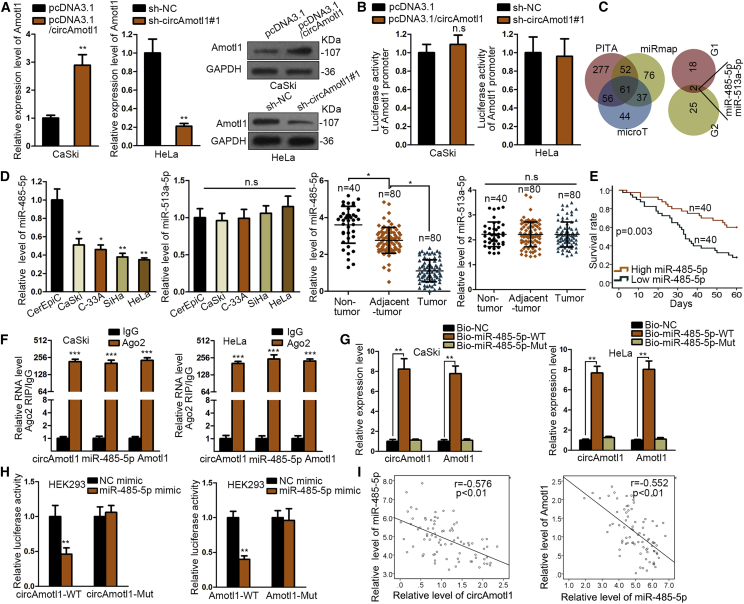
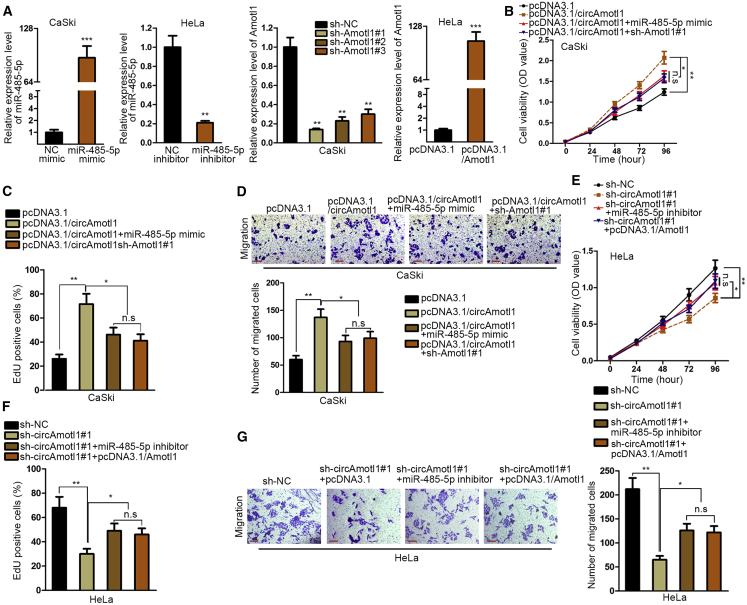
To interrogate the regulation of circAMOTL1 on AMOTL1, we examined the expression level of AMOTL1 after circAMOTL1 was affected and discovered that circAMOTL1 overexpression enhanced AMOTL1 expression at mRNA and protein levels, whereas circAMOTL1 inhibition decreased the mRNA and protein expression of AMOTL1 (Figure 4A). In view of the fact that the luciferase activity of the AMOTL1 promoter had no response to upregulation or knockdown of circAMOTL1, we reasoned that circAMOTL1 may function as a competing endogenous RNA (ceRNA) to modulate AMOTL1 (Figure 4B). We performed bioinformatics analysis with the assistance of three databases (PITA, miRmap, and microT) and uncovered that there were 61 joint predictive binding miRNAs of AMOTL1. In addition, only miR-485-5p and miR-513a-5p can combine with both circAMOTL1 and AMOTL1 (Figure 4C). As a result, we intended to identify the miRNA targeting AMOTL1 through detection of its expression in cervical cancer tissues and cells. Our data illustrated that miR-485-5p was dramatically declined in cervical cancer cells, and the level of miR-485-5p was lower in tumor samples than in normal and adjacent cancer tissues, but there was no alteration of miR-513a-5p expression in tumor samples and cells (Figure 4D). Kaplan-Meier analysis revealed that survival times of cervical cancer patients in low miR-485-5p expression were much shorter than those in high miR-485-5p level (Figure 4E). RNA FISH assay revealed that circAmtol1 and miR-485-5p were co-localized in CaSki cells (Figure S1C). We observed that circAMOTL1, miR-485-5p, and AMOTL1 were enriched by antibody against Ago2 in contrast with immunoglobulin G (IgG) antibody (Figure 4F). Meanwhile, only wild-type (WT) miR-485-5p can pull down circAMOTL1 and AMOTL1 compared with mutant miR-485-5p and miR-NC (NC) (Figure 4G). Consistently, luciferase activity of circAMOTL1-WT and AMOTL1-WT presented a significant reduction under ectopic expression of miR-485-5p, whereas there was no change in circAMOTL1-Mut and AMOTL1-Mut (Figure 4H). Moreover, it was indicated by Pearson’s analysis that circAMOTL1 was negatively correlated with miR-485-5p, and there was a negative relationship between miR-485-5p and AMOTL1 (Figure 4I). Taken together, we conclude that circAMOTL1 regulates AMOTL1 by acting as a sponge of miR-485-5p. As previously reported, circAMOTL1 can interact with c-myc to regulate tumorigenesis. Here, we detected whether circAMOTL1 can regulate c-myc expression in cervical cancer cells transfected with circAMOTL1 expression vector or sh-circAMOTL1#1. Both mRNA and protein levels of c-myc were not significantly changed (Figures S2A and S2B).
Figure 4.
circAMOTL1 Modulates the Expression of AMOTL1 through Sponging miR-485-5p
(A) The regulation of circAMOTL1 on AMOTL1expression was estimated by qPCR analysis and western blot. (B) The luciferase reporter assay was applied to evaluate whether circAMOTL1affected the transcription level of AMOTL1. (C) Results of bioinformatics analysis using PITA, miRmap, and microT databases. (D) Candidate miRNAs were screened through their expression in cervical cancer cells and tissues measured by qPCR. (E) The correlation between miR-485-5p expression and survival rate was revealed by Kaplan-Meier analysis. (F) Ago2-RIP assay demonstrated the enrichment of circAMOTL1, miR-485-5p and AMOTL1 in anti-Ago2 group compared to the negative control anti-IgG. (G) circAMOTL1 and AMOTL1 were efficiently pulled down by Bio-miR-485-5p-WT but not Bio-NC or Bio-miR-485-5p-MUT. (H) The luciferase activity of reporter vector containing circAMOTL1-WT was decreased after co-transfection with miR-485-5p mimics but that of circAMOTL1-MUT was not significantly changed. (I) Pearson’s analysis exposed the association between miR-485-5p, circ, and *p < 0.05, **p < 0.01, ***p < 0.001. n.s., no significance.
circAMOTL1 Exerts Oncogenic Activities in Cervical Cancer Progression via the miR-485-5p/AMOTL1 Axis
To investigate whether circAMOTL1 executed function through targeting miR-485-5p and AMOTL1, we conducted rescue experiments. First, miR-485-5p was overexpressed in CaSki cells and knocked down in HeLa cells, whereas AMOTL1 was silenced in CaSki cells and upregulated in HeLa cells. Also, the efficiency of transfection was verified by qPCR assay (Figure 5A). Afterward, CCK-8 and EdU assays delineated that overexpression of miR-485-5p or repression of AMOTL1 renovated the increased proliferation of cervical cancer cells caused by circAMOTL1 upregulation (Figures 5B and 5C). Finally, transwell assay corroborated that the rebound of cell migration promoted by circAMOTL1 overexpression occurred with miR-485-5p ectopic expression or silencing of AMOTL1 (Figure 5D). Inversely, miR-485-5p knockdown or AMOTL1 upregulation reversed the restraining influence of circAMOTL1 depletion on cell proliferation and migration in cervical cancer (Figures 5E–5G). Namely, circAMOTL1 facilitates the tumorigenesis of cervical cancer by targeting miR-485-5p/AMOTL1.
Figure 5.
circAMOTL1 Exerts Oncogenic Activities in Cervical Cancer Progression via the miR-485-5p/AMOTL1 Axis
(A) The increased or decreased level of miR-485-5p or Amotl1 expression was measured after transfection with indicted plasmids. (B–G) CCK-8, EdU assay and transwell migration assay detected that the cell proliferation and migration affected by the downregulation or overexpression of circAmotl1 was recovered after co-transfection with miR-485-5p mimic or inhibitor or sh-Amotl1 or pcDNA3.1-Amotl1. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., no significance.
circAMOTL1 Accelerates the Deterioration of Cervical Cancer In Vivo
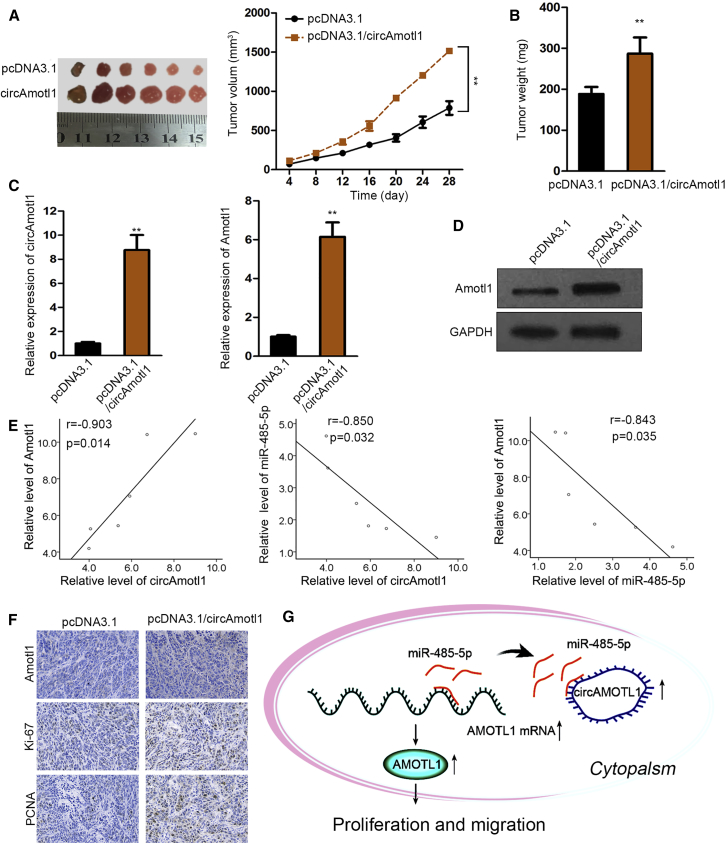
Animal experiments were carried out to further prove the carcinogenic function of circAMOTL1 in vivo. circAMOTL1 stable overexpression C-33A cells were hypodermically inoculated into nude mice. We manifested that elevated tumor sizes and weights were observed owing to upregulation of circAMOTL1 (Figures 6A and 6B). It was suggested by qPCR analysis and western blot that circAMOTL1 and AMOTL1 exhibited higher expression in tumors from the circAMOTL1upregulation group than in xenografts of the control group (Figures 6C and 6D). Furthermore, circAMOTL1 expression was positively associated with the level of AMOTL1 in neoplasms, whereas there was a negative association between miR-485-5p and circAMOTL1, as well as miR-485-5p and AMOTL1, in xenografts (Figure 6E). Similarly, IHC assay indicated that overexpressing circAMOTL1 contributed to a diminished level of AMOTL1 and the increased expression of Ki-67 and Proliferating Cell Nuclear Antigen (PCNA) in tumor tissues (Figure 6F). In a word, it is confirmed that circAMOTL1 promotes cervical cancer development in vivo.
Figure 6.
circAMOTL1 Accelerates the Deterioration of Cervical Cancer In Vivo
(A) Tumors derived from circAmotl1-overexpressed cells were bigger than those derive from cells transfected with empty vector. (B) Tumor weight in circAmotl1-overexpressed group were more than that in empty vector group. (C) The expression level of circAmotl1 and Amotl1 mRNA was higher in circAmotl1-overexpressed group. (D) Protein level of Amotl1 was higher in circAmotl1-overexpressed group than that in empty vector group. (E) The expression correlation between circAmotl1, miR-485-5p and Amotl1 in tumor samples. (F) The expression of Amotl1, Ki-67 and PCNA were assessed and found to be higher in circAmotl1-overexpressed group than that in empty vector group. (G) circAmotl1 promotes tumor progression in cervical cancer by sponging miR-485-5p to upregulate Amotl1. *p < 0.05, **p < 0.01.
Discussion
Cervical cancer is identified as a main contributor of cancer-associated deaths worldwide, constituting a large proportion of global female mortalities.16 Due to its high incidence and fatality rate, probing the potential molecular mechanisms underlying cervical cancer tumorigenesis has great significance for the improvement of therapeutic effects.
circRNAs, originating from back splicing or exon skipping of precursor mRNA templates, has been validated to exert important functions in human cancer by regulation of cell processes, such as cell proliferation, migration, invasion, and apoptosis.17, 18, 19 For instance, circRNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492.20 circRNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge.21 circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma.22 It has been reported that circAMOTL1 acts as an oncogene in breast cancer and prostate cancer,8,23 but the exploration on the function of circAMOTL1 in cervical cancer remains deficient. By application of the circRNADisease database, we found that circAMOTL1 was upregulated in cervical cancer, and then our results proved the circular characterization of circAMOTL1 and that high expression of circAMOTL1 predicted poor prognosis of cervical cancer. In addition, we unveiled that circAMOTL1 facilitated the proliferative and migratory ability of cervical cancer cells.
AMOTL1 belongs to the family of angiomotins (Amots) and is known as an adaptor protein, localizing in the nuclear, cytoplasmic, or membranous fraction in a cell context-dependent manner.24 Growing evidence has certified that AMOTL1 controls biological activities, including migration, cell polarity, tight junction formation, and angiogenesis.11,25,26 Furthermore, previous investigations have demonstrated that AMOTL1 exhibits cancerigenic activities in malignancies, such as breast cancer and cervical cancer.12,15 However, the specific molecular mechanism of AMOTL1 in cervical cancer needs to be illuminated. With the aid of the UCSC database, we verified that AMOTL1 was the host gene of circAMOTL1, implying the underlying administration of circAMOTL1 on AMOTL1. Further experiments manifested that AMOTL1 was obviously upregulated in cervical cancer tissues, which had close correlation with poor outcomes of cervical cancer patients. Moreover, circAMOTL1 turned out to modulate AMOTL1 expression at mRNA and protein levels, whereas it had no effect on luciferase activity of AMOTL1.
An increasing number of mechanistic studies have suggested that circAMOTL1 can protect mRNAs from miRNA-mediated degradation by serving as sponges for miRNAs.27, 28, 29 Therefore, we reasoned that circAMOTL1 may function as a ceRNA to regulate AMOTL1. By bioinformatics analysis, miR-485-5p was predicted to interact with circAMOTL1 and target AMOTL1, and our findings revealed that circAMOTL1 upregulated the expression of AMOTL1 through sponging for miR-485-5p. Additionally, circAMOTL1 exerted oncogenic activities in cervical cancer progression via the miR-485-5p/AMOTL1 axis, and in vivo assays further testified the pro-cancer role of circAMOTL1 in cervical cancer. A previous study reported that circ_AMOTL1 promoted tumorigenesis via interacting with c-myc.8 In our current study, c-myc expression was found to be unchanged by the silencing or overexpression of circ_AMOTL1. We may do further mechanism investigation about the circ_AMOTL1/c-myc axis in the future. In conclusion, this study exposed that circAMOTL1 prompted the expression of its host gene AMOTL1 to accelerate cervical cancer deterioration as miR-485-5p sponge (Figure 6G), which offered clues for clinical therapy of patients with cervical cancer and indicated circAMOTL1 as a novel target.
Materials and Methods
Clinical Samples
Forty normal specimens from healthy volunteers and 80 pairs of tumor samples and matched adjacent non-tumor tissues from cervical cancer patients were acquired from The First Affiliated Hospital of Wenzhou Medical University. All patients enrolled in this study signed written informed consent. Procedures of this study were in line with the Declaration of Helsinki and approved by the Institutional Review Board of The First Affiliated Hospital of Wenzhou Medical University. As soon as surgical resection was completed, tissues were immediately frozen with liquid nitrogen and stored at −80°C for subsequent use.
Cell Culture
Human cervical cancer cell lines (C-33A, HeLa, SiHa, and CaSki) were bought from American Type Culture Collection (ATCC, Bethesda, MD, USA) and cultivated in DMEM medium (GIBCO, Carlsbad, CA, USA) mixed with 10% fetal bovine serum (FBS; GIBCO) and 100 U/mL penicillin sodium. Human CerEpiCs were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and maintained in Cervical Epithelial Cell Growth Supplement (CerEpiCGS; ScienCell Research Laboratories), a complete medium designed for CerEpiC in vitro. All cells were cultured at 37°C under a moist incubator with 5% CO2.
Cell Transfection
Short hairpin RNA (shRNA) targeting circAMOTL1 (sh-circAMOTL1#1/2/3) or AMOTL1 (sh-AMOTL1#1/2/3) and NC shRNA (sh-NC) were used for silencing of circAMOTL1 or AMOTL1.
The sequences for sh-NC or circAMOTL1-specific shRNAs were listed as follows: sh-NC: 5′-CCGCAACGAGTTCAACTCGTTACCCTCGAGGTAACGAGTTGAACTCGTTTTTTTG-3′; sh-circAMOTL1#1: 5′-CCGCGGTAACGAGTTGAAGATCCTCCTCGAGGAGGATCTTCAACTCGTTACCTTTTTG-3′; sh-circAMOTL1#2: 5′-CCGCGTATGGGGTAACGAGTTGAAGCTCGAGCTTCAACTCGTTACCCCATACTTTTTG-3′; sh-circAMOTL1#3: 5′-CCGCCTCGTTACCTTGAAGATCCTCCTCGAGGAGGATCTTCAAGGTAACGAGTTTTTG-3′. For overexpression of circAMOTL1 or AMOTL1, circAMOTL1 or AMOTL1 sequences were cloned into pcDNA3.1(+) CircRNA Mini Vector (YBscience, Shanghai, China), and empty vector served as NC. miR-485-5p mimic, miR-485-5p inhibitor, and NC (NC mimic and NC inhibitor) were procured from Gene Pharma. All vectors were transfected into cells with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions.
qPCR
Total RNA extraction was carried out with TRIzol reagent (Life Technologies, Grand Island, NY, USA) in light of the manufacturer’s instructions. Transcriptor First Strand cDNA Synthesis Kit (Takara, Tokyo, Japan) was adopted to synthesize cDNA. Subsequently, qPCR was accomplished with SYBR PremixEx Taq (Takara) under ABI 7500 real-time PCR system (Thermo, Rockford, IL, USA) in accordance with the manufacturer’s protocol. The gene expression was depicted using the 2−ρρCt method, and each sample was detected in triplicate. GAPDH and U6 served as internal controls, respectively. The sequences of primers were as follows: circAMOTL1 (forward): 5′-GATGGTCAAGCCCTACCCTG-3′ and (reverse): 5′-CCCTGATGCTACTGGTTGCC-3′; miR-485-5p (forward): 5′-AGAGGCTGGCCGTGAT-3′ and (reverse): 5′-GAACATGTCTGCGTATCTC-3′; miR-513a-5p (forward): 5′-TTCACAGGGAGGTGTCA-3′ and (reverse): 5′-GAACATGTCTGCGTATCTC-3′; AMOTL1 (forward): 5′-GTCTACCACCAAGCGAGAATCG-3′ and (reverse): 5′-CTGCTGGATAGTTGCCTGTTAGC-3′; U6 (forward): 5′-CTCGCTTCGGCAGCACATA-3′ and (reverse): 5′-CGAATTTGCGTGTCATCCT-3′; GAPDH (forward): 5′-GGAGCGAGATCCCTCCAAAAT-3′ and (reverse): 5′-GGCTGTTGTCATACTTCTCATGG-3′.
CCK-8 Assay
CCK-8; Beyotime Institute of Biotechnology, Haimen, China) was applied for the evaluation of cell viability. Transfected cells (103/well) were plated to 96-well plates and then incubated for 0, 24, 48, 72, or 96 h prior to the supplement of 10 μL CCK-8 solution. After another 4 h of incubation at 37°C, the absorbance at 450 nm was detected under a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
EdU Incorporation Assay
According to product manuals of the EdU labeling/detection kit (RiboBio, Guangzhou, China), cells were treated with 50 μM EdU labeling solution, fixed with 4% paraformaldehyde, followed by incubation with glycine, washed by PBS, stained with anti-EdU working solution, and then permeabilized with PBS containing 0.5% Triton X-100. DAPI (5 μg/mL) was applied to stain the cell nucleus, and cells were observed under fluorescent microscopy.
Western Blot
After transfection, cells were lysed with RIPA assay lysate blend with protease inhibitor. Protein concentration was determined by BCA method, and equal protein extracts were detached by electrophoresing on SDS-PAGE, transferred to PVDF membrane, and blocked in 5% skim milk powder for 1 h. Then, PVDF membranes were probed by primary antibody against AMOTL1 (Abcam, Cambridge, MA, USA) at 4°C overnight and incubated with appropriate secondary antibody at room temperature for 2 h. Finally, the enhanced chemiluminescence kit (Pierce, Thermo) was employed to visualize the bands, and the gray value of bands was analyzed by Scion Image analysis software (Scion Corporation, Frederick, MD, USA) with GAPDH (Abcam) as the endogenous control.
Wound Healing Assay
Cells were plated in six-well plates and grown to confluence. Subsequently, the scratch wound was obtained with a pipette tip, and the generated floating cells were washed off using PBS. Cells were incubated at 37°C for 48 h, and images were captured with a microscope (Nikon, Tokyo, Japan) at 0 and 48 h after the scratch wound was made.
Cell Migration Assay
Transwell assay was utilized to estimate cell migration using transwell chambers (8-μm pore size; Corning, NY, USA). Cells resuspended in serum-free culture medium were added to the upper chamber, and the lower chamber was supplemented with the medium containing 10% FBS. After 24 h of incubation, cells on the bottom surface were fixed in 4% paraformaldehyde, stained by 0.1% crystal violet, photographed, and counted by using a microscope.
FISH
FISH kit (Guangzhou Biosense Bioscience, Guangzhou, China) was adopted to identify the subcellular localization of circAmolt1 following the manufacturer’s recommendations. Harvested cells were fixed by 4% paraformaldehyde, followed by permeabilization with 0.5% Triton X-100, and treated with hybridization solution. Afterward, cells were incubated with the probe bond to the backspliced junction of circAmolt1 overnight at 37°C, and cell nuclei were stained using DAPI at room temperature. The laser scanning confocal microscope (LSM700; Carl Zeiss, Germany) was used to capture fluorescence images.
RNA Pull-Down Assay
According to the manufacturer’s recommendations, WT miR-485-5p, mutant miR-485-5p, and NC miR-NC were biotinylated with Pierce RNA 3′ End Desthiobiotinylation kit (Pierce; Thermo Fisher Scientific, Waltham, MA, USA). Cell lysates were obtained using lysis buffer (Sigma-Aldrich; Merck, Darmstadt, Germany) and incubated with the streptavidin magnetic beads (Sigma-Aldrich) for 2 h at 4°C. Lastly, precipitated RNAs were eluted from magnetic beads by elution buffer (Sigma-Aldrich) and subjected to qPCR analysis.
Luciferase Reporter Assay
The WT or mutant of circAMOTL1 or AMOTL1 was integrated into a pmir-GLO Dual-luciferase vector to construct the circAMOTL1-WT, circAMOTL1-Mut, AMOTL1-WT, or AMOTL1-Mut reporter vector. HEK293 cells were then co-transfected with reporter vectors and miR-485-5p mimic or NC mimic using Lipofectamine 2000. 48 h later, the luciferase activity was determined with the dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Tumor Xenograft Assay
Procedures of animal experiments were carried out with the approval of the Animal Care and Use Committee of The First Affiliated Hospital of Wenzhou Medical University. The 4-week-old BALB/c nude mice were injected subcutaneously with 5 × 106 pcDNA3.1/circAMOTL1 stably transfected HeLa cells or the empty vector transfected HeLa cells, respectively. Volumes of tumors were monitored every 4 days. After 4 weeks, the mice were euthanized and tumor weights were measured.
RIP Assay
RNA immunoprecipitation (RIP) experiment was carried out using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) in line with the manufacturer’s protocol. Cell extracts were acquired with RNA lysis buffer and then blended with RIP buffer and magnetic beads coated with anti-Ago2 antibody (Abcam) or NC IgG (Millipore). Subsequently, protein A/G beads were utilized to enrich RNA-protein complexes, and precipitated RNAs were eluted and detected by qPCR assay.
IHC Assay
The slides were dewaxed with xylene and dehydrated using a graded alcohol series. Tissue sections were rinsed three times by PBS and treated with 3% hydrogen peroxide. After washing with PBS three times, antigen retrieval was carried out in citrate-buffered solution (pH 6.0) with pressure cooking. Tissues went through overnight incubation with the following primary antibodies: anti-AMOTL1 (Abcam), anti-Ki67 (Abcam), and anti-PCNA (Abcam) at 4°C, followed by incubation with appropriate secondary antibody at room temperature for 1 h. Lastly, sections were treated with 3′-diaminobenzidine tetrahydrochloride (DAB) and counterstained with hematoxylin. Images were photographed under a microscope.
Statistical Analysis
GraphPad Prism version 5 (La Jolla, CA, USA) was employed for statistical analysis, and all data were displayed as mean ± SEM. Overall survival analysis was conducted with Kaplan-Meier plots and log rank tests. Differences between groups were identified by Student’s t test or one-way ANOVA test. Correlations were analyzed by Pearson’s correlation coefficient analysis. p < 0.05 was defined as the criterion for statistical significance.
Author Contributions
J.L., Q.Z., and F.L. designed the experiments. R.O., L. Zhao, and L. Zhu performed the experiments. R.O. collected the clinical samples and analyzed the data. F.H., T.L., Y.R., and X.L. prepared figures and Y.X. drafted the manuscript. R.O. supervised laboratorial processes. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We are sincerely grateful for the vigorous support of Natural Science Foundation of China (NSFC; funding numbers 81571395, 81671408, 81771531, 81701634, and 81871129).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.09.022.
Supplemental Information
References
- 1.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Wright J.D., Chen L., Tergas A.I., Burke W.M., Hou J.Y., Neugut A.I., Ananth C.V., Hershman D.L. Population-level trends in relative survival for cervical cancer. Am. J. Obstet. Gynecol. 2015;213:670.e1–670.e7. doi: 10.1016/j.ajog.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Wang D., Long Z., Liu J., Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. Cell. Physiol. Biochem. 2018;48:173–184. doi: 10.1159/000491716. [DOI] [PubMed] [Google Scholar]
- 6.Yan B., Zhang W., Mao X.W., Jiang L.Y. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8712–8721. doi: 10.26355/eurrev_201812_16636. [DOI] [PubMed] [Google Scholar]
- 7.An Y., Cai H., Zhang Y., Liu S., Duan Y., Sun D., Chen X., He X. circZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell. Physiol. Biochem. 2018;51:2224–2236. doi: 10.1159/000495868. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratt A., Birot O., Sinha I., Veitonmäki N., Aase K., Ernkvist M., Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J. Biol. Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- 10.Moleirinho S., Guerrant W., Kissil J.L. The Angiomotins—from discovery to function. FEBS Lett. 2014;588:2693–2703. doi: 10.1016/j.febslet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y., Vertuani S., Nyström S., Audebert S., Meijer I., Tegnebratt T., Borg J.P., Uhlén P., Majumdar A., Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ. Res. 2009;105:260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]
- 12.Couderc C., Boin A., Fuhrmann L., Vincent-Salomon A., Mandati V., Kieffer Y., Mechta-Grigoriou F., Del Maestro L., Chavrier P., Vallerand D. AMOTL1 Promotes Breast Cancer Progression and Is Antagonized by Merlin. Neoplasia. 2016;18:10–24. doi: 10.1016/j.neo.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong W. Angiomotin’g YAP into the nucleus for cell proliferation and cancer development. Sci. Signal. 2013;6:pe27. doi: 10.1126/scisignal.2004573. [DOI] [PubMed] [Google Scholar]
- 14.Jiang W.G., Watkins G., Douglas-Jones A., Holmgren L., Mansel R.E. Angiomotin and angiomotin like proteins, their expression and correlation with angiogenesis and clinical outcome in human breast cancer. BMC Cancer. 2006;6:16. doi: 10.1186/1471-2407-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan H.Y., Li Q.Q., Zhang Y., Tian W., Li Y.N., Liu M., Li X., Tang H. MiR-124 represses vasculogenic mimicry and cell motility by targeting amotL1 in cervical cancer cells. Cancer Lett. 2014;355:148–158. doi: 10.1016/j.canlet.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 17.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng H., Chen T., Li C., Xu C., Ding C., Chen J., Ju S., Zhang Z., Liang Z., Cui Z., Zhao J. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag. Res. 2019;11:443–454. doi: 10.2147/CMAR.S189338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Fan S., Song E. Noncoding RNAs: New Players in Cancers. Adv. Exp. Med. Biol. 2016;927:1–47. doi: 10.1007/978-981-10-1498-7_1. [DOI] [PubMed] [Google Scholar]
- 20.Yan L., Zheng M., Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag. Res. 2019;11:1033–1041. doi: 10.2147/CMAR.S186857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y., Yu L.L., Zhang B., Liu C.F., Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz. J. Med. Biol. Res. 2018;51:e7811. doi: 10.1590/1414-431X20187811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Luo P., Jing W., Zhou H., Liang C., Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. 2018;11:2853–2863. doi: 10.2147/OTT.S158008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z., Qu C.B., Zhang Y., Zhang W.F., Wang D.D., Gao C.C., Ma L., Chen J.S., Liu K.L., Zheng B. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516–2532. doi: 10.1038/s41388-018-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang T., Zhou Y., Zhang J., Cheng A.S.L., Yu J., To K.F., Kang W. The physiological role of Motin family and its dysregulation in tumorigenesis. J. Transl. Med. 2018;16:98. doi: 10.1186/s12967-018-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagné V., Moreau J., Plourde M., Lapointe M., Lord M., Gagnon E., Fernandes M.J. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil. Cytoskeleton. 2009;66:754–768. doi: 10.1002/cm.20405. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y., Zhang Y., Barutello G., Chiu K., Arigoni M., Giampietro C., Cavallo F., Holmgren L. Angiomotin like-1 is a novel component of the N-cadherin complex affecting endothelial/pericyte interaction in normal and tumor angiogenesis. Sci. Rep. 2016;6:30622. doi: 10.1038/srep30622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guil S., Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem. Sci. 2015;40:248–256. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Cai H., Hu B., Ji L., Ruan X., Zheng Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am. J. Transl. Res. 2018;10:1690–1702. [PMC free article] [PubMed] [Google Scholar]
- 29.Rong D., Lu C., Zhang B., Fu K., Zhao S., Tang W., Cao H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer. 2019;18:25. doi: 10.1186/s12943-019-0958-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.