Abstract
Triple-negative breast cancer (TNBC) cells, which do not express genes for estrogen receptor (ER), progesterone receptor (PR), and Her2/neu, develop highly aggressive and metastatic tumors resistant to chemo- and hormonal therapies. We found that expression of glutathione peroxidase-1 (Gpx1) is silenced in the non-TNBC cells but significantly maintained in the TNBC cell lines. Such Gpx1 expression plays a vital role in the metastasis of TNBC cells by regulating cell adhesion. Transcriptomic and signaling pathway analyses demonstrate that depletion of Gpx1 essentially impairs cell adhesion/spreading by down-regulating FAK/c-Src activation. Mechanistically, Gpx1 interacts with FAK kinase and prevents the kinase inactivation by H2O2, not lipid hydroperoxide. As a result, depletion of Gpx1 suppresses lung metastasis of TNBC cells in vivo. Overall, our study identifies that Gpx1 is a redox safeguard of FAK kinase and its inhibition may provide an effective way to control the metastasis of deadly malignant TNBC.
Keywords: Glutathione peroxidase, Triple-negative breast cancer, Metastasis, Adhesion, Focal adhesion kinase
Highlights
-
•
Gpx1 is expressed in the triple-negative, not luminal-type, breast cancer cells.
-
•
Gpx1 regulates the adhesion of triple-negative breast cancer cells by interacting with FAK1 kinase.
-
•
FAK1 is an oxidation-sensitive kinase.
-
•
Gpx1 regulates the metastasis of triple-negative breast cancer cells in vivo.
1. Introduction
Breast cancer (BC) is the second leading cause of cancer death among women and is classified into four major molecular subtypes: Luminal A, Luminal B, Basal-like/triple-negative, and HER2 types [1,2]. Treatment response of primary BC patients is generally good and a five-year survival rate is over 90% according to the American Cancer Society. However, the triple-negative/basal-like breast cancer (TNBC) with little or no estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor (HER2) expression shows highly metastatic and recurrent behaviors [[3], [4], [5]]. Therefore, a targeted eradication of TNBC is critical for increasing a relapse-free survival of BC patients [6].
Molecular mechanism of metastasis is complicated and yet to be fully understood because it involves the serial processes: local tumor cell invasion, entry into the vasculature, the exit of cancer cells from the circulation, and colonization at the distal sites [7,8]. A critical step in the processes is the colonization by adhesion of the circulating cancer cells to the extracellular matrix (ECM) in the second metastasized tissues. Cell adhesion is mainly regulated by focal adhesion kinase (FAK), a non-receptor tyrosine kinase concentrated in focal adhesions [[9], [10], [11]]. The well-characterized regulatory mechanism of FAK activation is the autophosphorylation on FAK Y397, which creates a high-affinity binding site for the Src kinase. Upon c-Src binding to FAK, the activated c-Src phosphorylates both Y576 and Y577 on the kinase domain of FAK, which is required for full activation of FAK [[12], [13], [14]]. FAK activity was shown to be upregulated in cancer cell lines and tumor tissues obtained from BC patients with metastasis [15]. In particular, recent analyses of TCGA database for invasive BC cohort have indicated that the FAK expression is higher in the TNBC patients, which is correlated with a poor survival of TNBC patients [16,17]. Related to the cancer cell adhesion, several studies have shown that certain carcinoma cells exposed to oxidative stress exhibit a decreased attachment to basal lamina [18,19]. However, the molecular mechanism underlying the relationship between oxidative stress and cell adhesion remains to be clarified.
Cancer cells harbor the intrinsic oxidative stress and thus upregulate antioxidant enzymes including glutathione peroxidase (Gpx) [20]. Gpx is a selenium-containing enzyme that protects cells against oxidative stress by eliminating hydrogen peroxide and organic hydroperoxides using reducing equivalents from NADPH via the glutathione-glutathione reductase system [21]. To date, numerous studies have attempted to seek the role of Gpx in various cancers [22]. In the BC, the allelic variations were found to affect the Gpx1 activity [23]. An immunohistochemical analysis of tumor tissue samples from 60 BC patients with invasive ductal carcinoma has indicated that high Gpx1 expression is associated with high mortality and shorter survival [24]. The Gpx1 expression was shown to be silenced by promoter methylation in several BC cell lines, such as MCF7, and in some of primary tumor tissues [25]. Nonetheless, the cellular function of Gpx1 beyond a general antioxidant scavenger has not been explored in the BC cells.
In this study, we scrutinized the molecular function of Gpx1 in the TNBC cell lines. We demonstrated that the depletion of Gpx1 reduces the proliferation and adhesion of TNBC cells by increasing an oxidative inactivation of FAK kinase and further inhibits the metastasis of TNBC cells to the lung in vivo. Overall, our study provides the evidence that Gpx1 regulates the TNBC metastasis specifically through the FAK-dependent cell adhesion.
2. Materials and methods
2.1. Antibodies and reagents
Polyclonal anti-Gpx1 antibody (ab#59546) and anti-Gpx4 antibody (ab#125066) were purchased from Abcam. Antibodies against FAK-pY925 (3284), FAK-pY577/576 (3281), FAK-pY397 (8556), FAK (3285), Src-pY416 (2101), Src (2107) were purchased from Cell Signaling Technology. Mouse anti-tubulin (T5168) was purchased from Sigma. Horseradish peroxidase (HRP)-conjugated secondary rabbit IgG (31460) and mouse IgG (31250) antibodies were purchased from Pierce. Alexa Fluor 488-phalloidin was purchased from Invitrogen. RNeasy mini Kit (74104) was obtained from Qiagen. Oligo-dT primer (ab-1247) was purchased from Thermo. SYBR Green PCR Kit (06924204001) was purchased from Roche.
A siRNA specific to human Gpx1 (5′-GCA ACC AGU UUG GGC AUC A-3′) and Gpx4 (5′-ACG AAG AGA UCA AAG AGU U-3′) were selected from a SMART pool (Dharmacon). The control siRNA against firefly luciferase was purchased from Dharmacon.
The following pre-designed primers for real-time qPCR were purchased from Qiagen.
| Gene symbol | Cat.no | Gene symbol | Cat.no |
|---|---|---|---|
| PFN2 | QT10667386 | LAMC1 | QT00072142 |
| ARPC5 | QT00085176 | CDC42 | QT00066528 |
| CRK | QT00066402 | GNAI2 | QT00013419 |
| SHC1 | QT00060445 | MYL12A | QT00044373 |
| LIMA1 | QT00095564 | ACTN1 | QT00083678 |
2.2. Cell culture
MCF7, MDA-MB-468, BT549, Hs578T, and T47D cells were purchased from ATCC and cultured in the high-glucose RPMI 1640 (Hyclone) supplemented with 10% fetal bovine serum (Hyclone), 100 units/ml penicillin, and 100 units/ml streptomycin. MDA-MB-231 cells were cultured and passaged in DMEM (Hyclone) supplemented with 10% fetal bovine serum (Hyclone), 100 units/ml penicillin, and 100 units/ml streptomycin. These cells were maintained at 37 °C with an atmosphere of 5% CO2.
2.3. Immunoprecipitation, in vitro kinase assay, and immunoblotting
Cells were washed once with ice-cold phosphate-buffered saline (PBS) and lysed in a lysis buffer containing 20 mM HEPES (pH 7.0), 1%Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 2 mM EGTA, 5 mM Na3VO4, 5 mM NaF, 1 mM DTT, 1 mM AEBSF, aprotinin (5 μg/ml), and leupeptin (5 μg/ml). Cell lysates were clarified by centrifugation at 12,000×g for 10 min and then subjected to the protein determination by a Bradford assay. For immunoprecipitation, the clarified cell lysates (2 mg proteins) were pre-cleared with 20 μl of protein-A/G Sepharose 4 Fast Flow beads (Amersham Biosciences) for 1 h. The supernatant was incubated overnight with 1 μg of the appropriate antibody with rotation at 4 °C and then precipitated by mixing with 20 μl of protein-A/G beads for an additional 3 h. The beads were washed three times with 1 ml of the chilled lysis buffer and then subjected to either the in vitro kinase assay or immunoblotting. For in vitro FAK kinase assay, the precipitated immunocomplexes were incubated with an assay cocktail (20 mM HEPES pH 7.5, 20 mM MgCl2, 20 mM glycerophosphate, and 200 μM Na3VO4, 10 μCi γ-32[P]-ATP) in a 30 μl of reaction volume at 30 °C for 20 min. The reaction was stopped by the addition of 5 × SDS sample buffer. The samples were boiled and then separated on a denaturing gel. The gel was vacuum-dried and subjected to autoradiography. For immunoblotting, cell lysates or immunoprecipitates were mixed with an SDS sample buffer and boiled for 5 min. The samples were separated on a denaturing gel and then electrophoretically transferred onto nitrocellulose membranes. The blots were probed with the primary antibodies and the immune-reactive bands were detected with HRP-conjugated secondary antibodies and enhanced chemiluminescence system (YonginFrontier, Seoul, Korea). For loading control, the blots were stripped with a stringent buffer (62.5 mM Tris-HCl and 2% SDS) for 30 min at 60 °C and then re-probed.
2.4. RNA-seq and data analyses
After depleting ribosomal RNAs (rRNAs), sequencing libraries were constructed by the Illumina Qiagen RNeasy mini Kit and then subjected to sequencing by an Illumina HiSeq 2500 system (101bp paired-end reads, DNA Link Inc., Seoul, Korea). Raw sequencing reads were checked for quality control using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter sequences were trimmed bysickle (v1.33) (https://github.com/najoshi/sickle). Resulting reads were then aligned to the human genome (hg19) using MapSplice (v2.1.8) with the default options. RNA abundance was estimated using RSEM (v1.2.12). Differentially expressed genes (DEGs) were identified for each cell line using Limma Voom R package with multiple testing correction of Benjamini-Hochberg method at the corrected p value threshold < 10−5. Common DEGs from both cell lines were analyzed for functional enrichment in KEGG pathways using the DAVID bioinformatics resources (v6.8) (p-value < 0.05). The protein-protein interaction (PPI) network was constructed for genes involved in cell adhesion process using the STRING plug-in available for Cytoscape (v1.7.2). We included molecular interactions from textmining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. Minimum required interaction scores were >0.4 (medium confidence).
2.5. RT-PCR
Total RNA was isolated from cells using RNeasy kit and subjected to the reverse transcription for the synthesis of complimentary DNA (cDNA) with oligo-dT primer. The level of gene expression was measured qualitatively or quantitatively by PCR. The forward and reverse PCR primers for Gpx1 were 5′-AAG GTA CTA CTT ATC GAG AAT-GTG-3′ and 5′-GTC AGG CTC GAT GTC AAT GGT CTG-3′, respectively. The forward and reverse PCR primers for GAPDH were 5′-TGG ACT CCA CGA ACT CA-3′ and 5′-GGA AGG TTG TCA TCA ATG GAA-3, respectively. The quantitative real time PCR (qPCR) was performed in triplicate using SYBR Green PCR kit on a Lightcycler96 (Roche).
2.6. Migration, invasion, clonogenic assays
MDA-MB-231 and Hs578T cells were transfected with control and Gpx1-specific siRNA for 36 h and transferred onto fresh culture devices for cell assays. For migration assay, the transfected cells were serum starved for additional 12 h in media containing 0.5% fetal bovine serum. The bottom of the upper chamber was coated with gelatin B (1 mg/ml) and air-dried for 20 min before the cell seeding. The transfected cells (5 × 104 cells per well) were transferred onto the polycarbonate upper chamber (8 μm pore size) in a 24-well Trans-well culture chamber (Costar) and then stimulated with 20% fetal bovine serum in the lower chamber for 24 h. The upper chambers were fixed with methanol and stained with 0.6% hematoxylin (DAKO), following the removal of the non-migrated cells from the top. The stained cells were photographed and counted. The number of migratory cells was averaged from triplicate wells. The invasion assay was similarly performed except the inner surface of upper chambers was coated with 40 μg Matrigel (BD Biosciences) followed by coating the bottom surface with gelatin B (1 mg/ml). For clonogenic assay, the transfected cells (3 × 103 cells per well) were transferred onto a 6-well culture plate and cultured for 10 days. The cells were washed once with PBS and then stained with crystal violet. The blue-stained colonies were manually counted.
2.7. Real-time cell analysis
The siRNA-transfected cells were collected by trypsinization and seeded onto 16-well E-plates at 3000 cells/well. Following seeding, cells were monitored every 5 min by the xCELLigence system (Roche) for adhesion and spreading. RTCA Software 2.0 (Roche) was used to calculate the cell index (CI) values.
2.8. Methylation-specific PCR
The genomic DNA was extracted from the cells using Laird's lysis buffer (1% SDS, 50 mM Tris (pH 8.0), 100 mM EDTA, 100 mM NaCl, and 10 μg/ml Proteinase K). DNA (500 ng) was sheared into single-strand DNA and treated with sodium bisulfite using EZ DNA Methylation kit (Zymo Research) according to the manufacturer's protocol. Methylation status of Gpx1 promoter was analyzed using the methylation and unmethylation primers. The sequence of primers are as follows: forward primer (5′-CTA ACC GAA CAA CAC ACA TAA CG-3′) and reverse primer (5′-GAG GCG GGA TTT TTA GGT TC-3′) for methylation; forward primer (5′-ACC AAA CAA CAC ACA TAA CAC A-3′) and reverse primer (5′-ATG AGG TGG GAT TTT TAG GTT T-3′) for unmethylation.
2.9. Glutathione peroxidase assay
A standard glutathione-dependent peroxidase assay was carried out in a 200 μl reaction mixture containing 250 μM NADPH, 1 mM GSH, 1 U yeast GR, 20 μg cell proteins, 1 mM EDTA, and 50 mM HEPES buffer (pH 7.0). After pre-incubation for 3 min, the reaction was initiated by adding 1.2 mM H2O2. The reduction of UV absorbance at 340 nm was monitored for 3 min at 30 °C in an UV/VIS spectrophotometer (Hewlett Packard, USA). The initial rate of reaction was calculated using the linear portion of the curve and expressed as the amount of NADPH oxidized per minute per mg protein.
2.10. Immunofluorescence staining
Cells were transfected with siRNAs, grown on a cover glass for 36 h. Subsequently, cells were fixed with 3.7% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min. For F-actin staining, cells were incubated with Alexa Fluor 488-phalloidin (1:500 dilution) for 1 h at room temperature. The nuclei were further stained with 4’,6- diamidino-2-phenylindole (DAPI) for 10 min. Images were taken using a Zeiss LSM 880 confocal microscope.
2.11. Proximity ligation assay
MDA-MB-231 cells were grown on a cover glass for 36 h and then stimulated with FBS for 2 h. After stimulation, cells were rinsed twice with cold PBS and then fixed with 3.7% paraformaldehyde for 10 min. The fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and incubated with a drop of blocking buffer for 30 min at 37 °C. The plus and minus antibodies (1:1000 and 1:500 for FAK and Gpx1 antibodies, respectively) were diluted in blocking buffer containing 1 × Duolink assay reagent (Sigma-Aldrich) and incubated with the samples overnight at 4 °C. The polymerase chain reaction was performed according to the manufacturer's instructions. After polymerase chain reaction, the F-actin filaments and nuclei were stained with Alexa Fluor 488-phalloidin (1:500; Invitrogen) and DAPI for 1 h and 10 min, respectively, at room temperature. Images were taken using a Zeiss LSM 880 confocal microscope.
2.12. Measurement of reactive oxygen species
Intracellular reactive oxygen species (ROS) level was assessed with an oxidation-sensitive fluorescent dye 5,6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCF-DA, Invitrogen). The cells were immediately washed once with phenol red-free media and then incubated with the Hank's Balanced Salt Solution (HBSS) containing 5 μM CM-H2DCF-DA for 5 min. The dichlorofluorescein (DCF) fluorescence images were taken in an inverted Axiovert 200 fluorescence microscope (Zeiss). The lipid hydroperoxide (LOOH) level was measured with C11-BODIPY581/591 ratiometric fluorescent probe. Cells were transfected with the indicated siRNAs for 36 h, rinsed with ice-cold PBS, and fixed with 3.7% formaldehyde. The fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and stained with 1 μM C11-BODIPY for 30 min at 37 °C. The green (oxidized) and red (reduced) fluorescence images were simultaneously acquired using a Zeiss LSM 880 confocal microscope.
2.13. Production of inducible stable cell line
MDA-MB-231 cells were transfected with a lentiviral pTRIPZ vector encoding Gpx1-specific shRNAmir under Tet-on inducible promoter (V3THS_413713; GE healthcare). The transfected cells were cultured and passaged in the complete media containing 1.5 μg/ml puromycin for two weeks. The selected cells were trypsinized, diluted, and re-plated onto a 96-well culture plate in a single cell level. Each cell was cultured until a visible clone is obtained and then expanded on 6-well culture plates. The individual clones were tested for the Gpx1 knockdown after treatment of doxycycline (5 μg/ml) for 24 h. The knockdown level of Gpx1 protein was evaluated by immunoblot analysis.
2.14. Lung metastasis of MDA-MB-231 cells in mice
The mouse experiments were approved by the Institutional Animal Care and Use Committee of Ewha Womans University and conformed to Guide for Care and Use of Laboratory Animals published by the US National Institutes of Health (The National Academies Press, 8th Edition, 2011). MDA-MB-231 cells stably harboring inducible Gpx1 shRNA were treated with 5 μg/ml doxycycline for 24 h, trypsinized, and then re-suspended in Hank's balanced salt solution (HBSS). The suspended cells were injected intravenously into 4-week-old female Balb/c nu/nu mice (1 × 106 cells per mouse). After 10 days, the mice were euthanized and received the intra-tracheal injection of 7.5% India Ink. India ink-injected lungs were washed once with and then placed overnight in Fekte's solution (300 ml 70% ethanol, 30 ml 37% formaldehyde, 5 ml glacial acetic acid). Pulmonary metastases were measured by counting white tumor nodules on the blue background.
For histopathological analysis, the lungs were fixed in formalin, paraffin-embedded, and sectioned with a rotary microtome (Leica RM2255). The tissue sections (5 μm in thickness) were stained with hematoxylin and eosin (H&E). Images were taken with a phase-contrast microscope (Zeiss).
2.15. Statistics
All experiments were repeated at least three times. Unless stated otherwise, the quantified data were analyzed with Student's t-test to determine the statistical significance (p value). A P < 0.05 was considered to be statistically significant.
3. Results
3.1. Gpx1 is essential for the cellular functions of TNBC cells
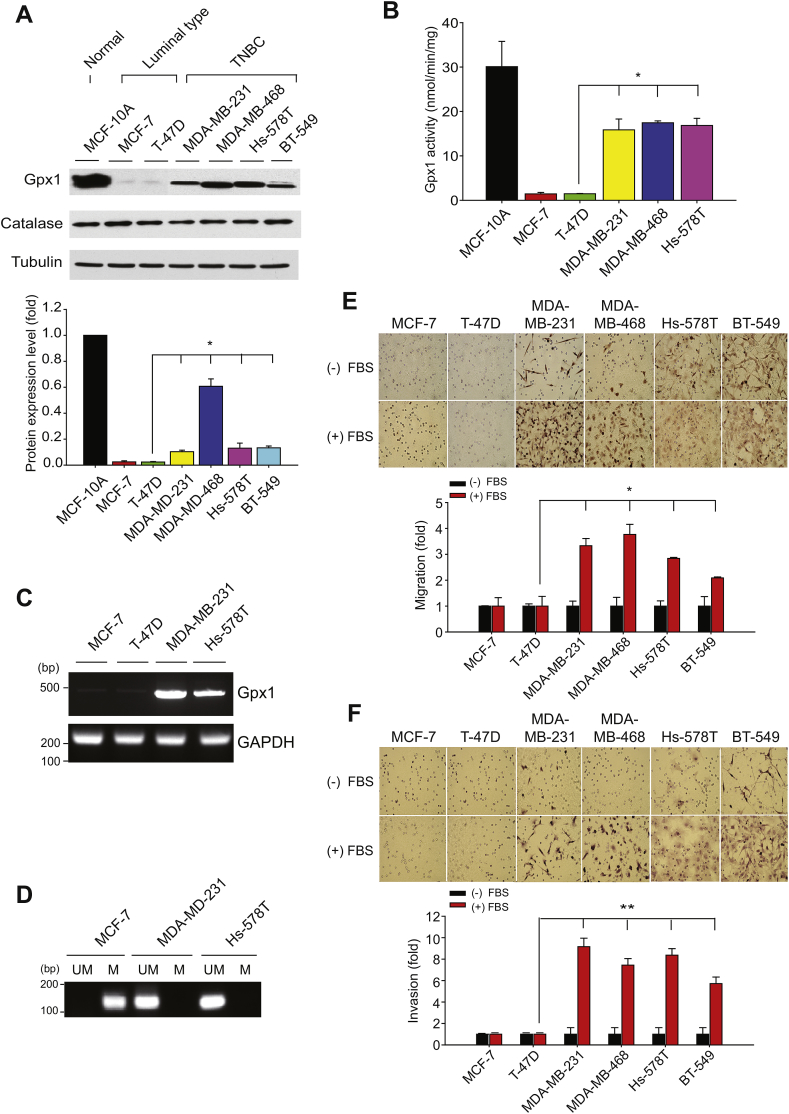
Through an immunoblot analysis of the Gpx1 expression in a panel of BC cell lines, we found that the Gpx1 expression was high in a normal breast epithelial cell line MCF10A, absent in the luminal-type BC cells, and again appeared in the basal-type/TNBC cells (Fig. 1A). Since the level of a peroxisomal enzyme catalase was the same among the BC and normal breast epithelial cells, we thought that the dynamic change of Gpx1 expression is a specific phenomenon in the BC cells. To strengthen this finding, we examined the cellular Gpx1 activity and mRNA level (Fig. 1B and C), which was well corresponded to the Gpx1 protein level. In addition, as previously reported [25], the Gpx1 expression in the luminal-type BC cells was appeared to be silenced by promoter methylation (Fig. 1D). Moreover, the in vitro cell assays showed that the migratory and invasive activities of TNBC cells were much stronger than those of the luminal-type BC cells (Fig. 1E and F), which interestingly corresponded with the cellular level of Gpx1 expression.
Fig. 1.
Gpx1 expression is preserved in the TNBC cells.
(A) Level of Gpx1 protein expression in human normal breast epithelial cell and breast cancer cells. Data in the graph are means ± SD of the relative fold change of band intensity versus that in MCF10A (n = 3, *P < 0.005).
(B) H2O2 -reducing peroxidase activity of Gpx1 in the soluble cell extracts. The activity assay was performed in the reaction mixture by adding 20 μg proteins. Data in the graph are means ± SD of the specific activities based on initial velocities of NADPH oxidation at 340 nm (n = 3, *P < 0.0001).
(C) RT-PCR analysis of Gpx1 mRNA expression among BC cells. GAPDH is an endogenous control gene.
(D) CpG methylation of Gpx1 promoter. The methylation-specific PCR was performed with DNA samples isolated in the indicated cells. The PCR product is 173 base pairs in size. UM, unmethylated; M, methylated. In C and D, representative images of agarose gels are shown (n = 3). The 100-base pair (bp) size markers are indicated.
(E and F) The six different BC cells were subjected to the migration (E) and invasion (F) assays using transwell plates. Cells were plated on the upper chamber and stimulated with 20% fetal bovine serum (FBS) in the lower chamber. Representative images are shown. Data in the graph are means ± SD of fold increase of the number of migrating or invading cells (n = 3, *P < 0.05, **P < 0.001).
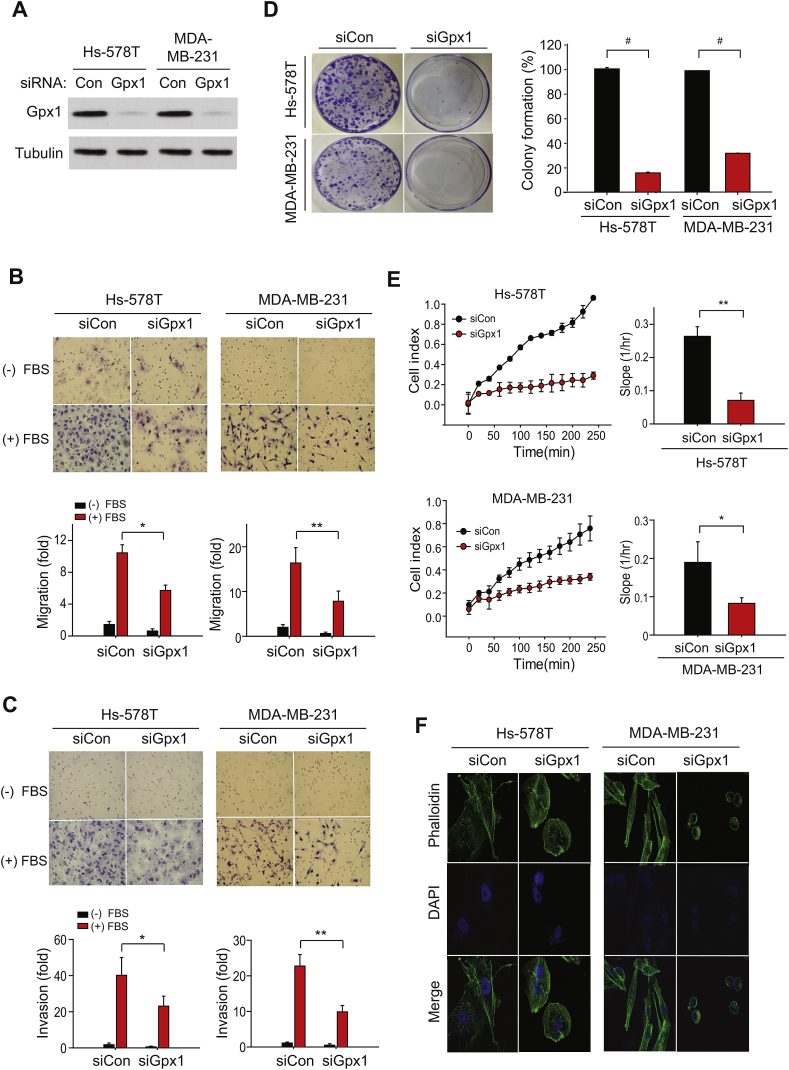
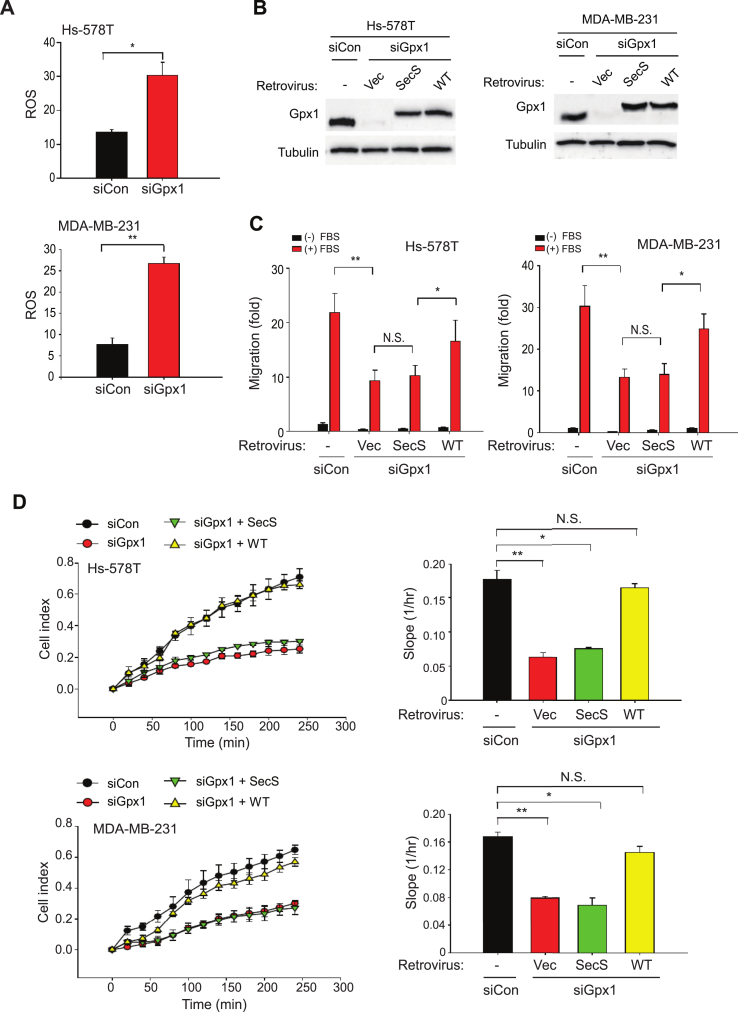
To study the importance of the Gpx1 expression in the TNBC cells, we selected two representative TNBC cells, MDA-MB-231 and Hs578T, and then performed the in vitro cell assays for migration and invasion. To achieve the goal, the Gpx1 expression was transiently knocked down using the specific small-interfering RNA (siRNA) (Fig. 2A). The Gpx1 knockdown resulted in the strong reduction of protein level and peroxidase activity, but did not cause a cytotoxic death (Supplementary Fig. S1A - C). Subsequent transwell assays showed that the Gpx1 depletion reduced the migratory and invasive activities of MDA-MB-231 and Hs578T cells (Fig. 2B and C). However, the Gpx1 depletion did not change the metabolic activities, such as ATP levels and oxygen consumption rates, in both TNBC cells (Supplementary Fig. S1D), suggesting a specific signaling, not metabolic, role of Gpx1 in TNBC cells. When the long-term effect of the Gpx1 depletion was assessed by clonogenic assay, the Gpx1 depletion drastically ablated the colony formation in the MDA-MB-231 and Hs578T cells (Fig. 2D), indicating that Gpx1 might also play a role in the cell proliferation/survival. To delineate how Gpx1 regulates various cellular functions, we traced the cell behavior using a real-time cell analysis. The MDA-MB-231 and Hs578T cells were detached and seeded on a specialized microtiter plate equipped with gold microelectrodes, which measure electrical impedance that reflects the attachment and spreading of cells. The cell index was immediately increased after seeding of control siRNA-transfected MDA-MB-231 and Hs578T cells, whereas it was seriously retarded in the Gpx1-depleted cells (Fig. 2E). This result indicates that Gpx1 depletion caused a defective adhesion and thereby spreading, which consequently influenced the overall cellular functions. More significantly, the phalloidin staining against F-actin filaments visualized the defective spreading in the Gpx1-depleted TNBC cells (Fig. 2F). We then addressed whether Gpx1 regulates the cellular functions via its peroxidase activity. The Gpx1 depletion increased the level of cellular reactive oxygen species (ROS) in both MDA-MB-231 and Hs578T cells (Fig. 3A). In contrast, the level of lipid hydroperoxide (LOOH) as another substrate of Gpx1 was undetected in both control and Gpx1-depleted TNBC cells (Supplementary Fig. S2), which indicates that Gpx1 mainly controls the level of H2O2, not LOOH, in the TNBC cells. In addition, we observed that the LOOH level was markedly increased by the depletion of Gpx4, which catalyzes a reduction reaction of phospholipid hydroperoxide. Then, we prepared an inactive mutant (SecS) of Gpx1 by substituting the active-site selenocysteine to serine residue (Fig. 3B). As a result, WT restored the migratory activities of TNBC cells that had been dampened by the Gpx1 depletion, whereas the inactive mutant did not (Fig. 3C). Furthermore, the adhesion and spreading of TNBC cells were also rescued by the Gpx1 WT, not the inactive mutant (Fig. 3D). Collectively, the data implicate that the adhesion and spreading of TNBC cells on the extracellular matrix are redox-dependent as being regulated by Gpx1 peroxidase activity.
Fig. 2.
Absence of Gpx1 impairs the adhesion function of TNBC cells.
(A) Knockdown of Gpx1 expression in the Hs-578T and MDA-MB-231 cells by transfection of Gpx1-specific siRNAs for 36 h. A representative immunoblot is shown.
(B–D). The migratory (B), invasive (C), and colony-forming (D) activities of the TNBC cells transfected with control or Gpx1 siRNA (siCon or siGpx1). Cellular function assay was performed as described in the Methods. Data in the graphs are means ± SD of percent or fold increase of the number of colonies or migrated/invaded cells, respecitvely (n = 3, *P < 0.001, **P < 0.0005, #P < 0.00001).
(E) Real-time measurement of adhesion of the siRNA-transfected TNBC cells using the xCELLigence system. Real-time traces of electrical impedance are shown (Left panel). Data in the graph are means ± SD of the slopes of cell index increments based on electrical impedance (n = 3, *P < 0.001, **P < 0.0005).
(F) Analysis of F-actin filaments with phalloidin-conjugated probes in the siRNA-transfected TNBC cells. The representative images were taken by confocal microscopy (n = 3). F-actin filaments and nuclei were stained with Alexa Fluor 488-phalloidin (green) and DAPI (blue), respectively. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Gpx1 peroxidase activity is required for regulating the functions of TNBC cells.
(A) Level of intracellular ROS in the TNBC cells transfected with control or Gpx1 siRNA (siCon or siGpx1). Data in the graph are means ± SD of the relative DCF fluorescence averaged from 60 cells. (n = 3, *P < 0.001, **P < 0.0005).
(B and C) The siRNA-transfected TNBC cells were infected with retrovirus encoding Gpx1 WT or Sec47S (SecS) mutant. The expression level of exogenous Gpx1 with a Myc epitope tag was validated by immunoblotting (B). Data in the graphs (C) are means ± SD of number of migrated cells (n = 3, *P < 0.05, **P < 0.001). N.S., not significant.
(D) Real-time measurement of ECM-mediated adhesion of TNBC cells using the xCELLigence system. Real-time traces of electrical impedance are shown (Left panels). Data in the graph are means ± SD of the slopes of cell index increments based on electrical impedance (n = 3, *P < 0.001, **P < 0.0005). N.S., not significant.
3.2. FAK/c-Src activation is defective in the absence of Gpx1
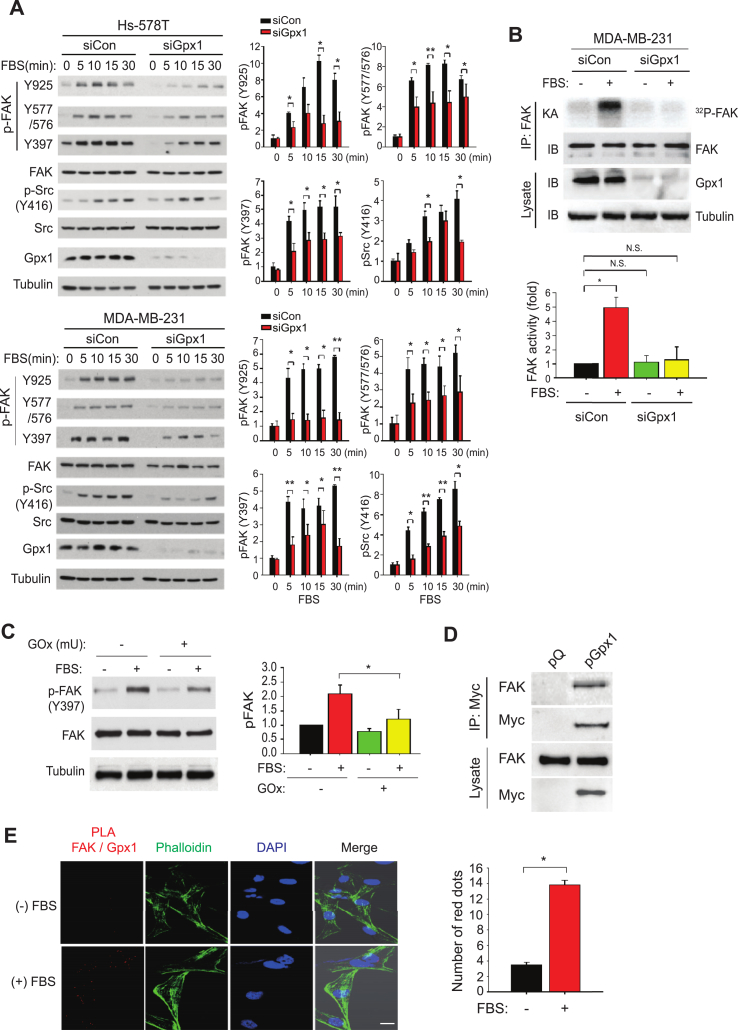
To dissect the adhesion signaling pathways dependent on Gpx1, we performed the phosphorylation pathway analysis in the TNBC cells stimulated with serum as a source of growth factors. As a result, we found that the serum-induced activation of FAK and c-Src kinases were concomitantly depressed by the Gpx1 depletion (Fig. 4A). Specifically, the Gpx1 depletion inhibited the FAK activation by reducing the autophosphorylation on the Y397 residue critical for full activation as well as the c-Src-dependent phosphorylation on the other Tyr residues [26]. Although depletion of Gpx4 increased the cellular LOOH level as shown above, the same condition had no effect on the serum-induced FAK activation (Supplementary Fig. S3), which suggests that the FAK activation is specific to Gpx1. In addition, the Gpx1 depletion also inhibited the activation of MAP kinase ERK1/2 and Akt compared to the control cells (Supplementary Fig. S4). Given that FAK/c-Src signaling complex is key for cell adhesion [27], we pointed the redox sensitivity of the FAK kinase. The in vitro kinase assay demonstrated that the Gpx1 depletion strongly inhibited the induction of FAK kinase activity in response to the serum stimulation (Fig. 4B). To directly address whether H2O2 targets the FAK kinase, we treated the MDA-MB-231 cells with glucose oxidase that extracellularly produces H2O2 using the medium glucose as a substrate. Unexpectedly, treatment with glucose oxidase alone sufficiently impaired the auto-phosphorylation of FAK on Y397 induced by serum (Fig. 4C), which represents that FAK kinase is oxidation-sensitive. Subsequently, we performed co-immunoprecipitation experiment using a myc epitope-tagged Gpx1. The expressed Gpx1 interacted with the endogenous FAK kinase in the serum-deprived condition (Fig. 4D). The endogenous interaction between FAK and Gpx1 was further examined by a proximity ligation assay (PLA) capable of detecting the protein-protein interactions in situ at the level of single molecular events. The interaction-based PLA signals consistently appeared without stimulation and strongly increased by serum stimulation (Fig. 4E). Moreover, the PLA signals were perfectly merged with the F-actin filaments. Therefore, the data suggest that Gpx1 functions as a redox safeguard that preserves FAK activation.
Fig. 4.
Gpx1 positively regulates the FAK and Src activations.
(A) Serum-induced activation of intracellular signaling molecules in the TNBC cells transfected with control or Gpx1 siRNA (siCon or siGpx1). The transfected cells were deprived of serum for 12 h and then stimulated with 20% fetal bovine serum for indicated times. The signaling activation was analyzed by immunoblotting with phospho-specific antibodies. Data are means ± SD of the relative intensities of phospho-specific bands normalized by corresponding protein bands (n = 3, *P < 0.05, **P < 0.001). Representative immunoblots are shown.
(B) Serum-induced FAK activation in the MDA-MB-231 cells. The siRNA-transfected MDA-MB-231 cells were serum-starved for 12 h and then stimulated with or without 20% fetal bovine serum for 30 min. The in vitro kinase assay (KA) was performed using immunoprecipited (IP) FAK enzymes as described in the Methods. Representative immunoblot (IB) and autoradiogram (KA) are shown. Data in the graph are means ± SD of fold increase of the intensities of the radioactive bands normalized by corresponding FAK bands (n = 3, *P < 0.001). N.S., not significant.
(C) H2O2-mediated inhibition of FAK activation in response to serum stimulation. MDA-MB-231 cells were pretreated with or without glucose oxidase (GOx, 100 mU) for 30 min and then stimulated with 20% fetal bovine serum for 30 min. The FAK activation was measured by immunoblotting auto-phosphorylation of FAK (pY397). Data are means ± SD of the relative intensities of phospho-FAK bands normalized by corresponding FAK bands (n = 3, *P < 0.05). Representative immunoblots are shown.
(D) Direct interaction between FAK and Gpx1 proteins. HEK293 cells were transfected with the empty vector (pQ) or vector encoding mouse Gpx1 with a Myc epitope (pGpx1). The immunoprecipitated mouse Gpx1 proteins were subjected to the immunoblotting. Representative immunoblots are shown (n = 3). (E) The in situ proximity analysis of endogenous FAK and Gpx1 interaction in the MDA-MB-231 cells. The control and serum-stimulated MDA-MB-231 cells were subjected to the proximity ligation assay (PLA) followed by phalloidin staining. The merged images show the co-localization of PLA red dots in the actin filaments. Data in the graph are means ± SD of number of the PLA red dots (n = 3, *P < 0.00005). Scale bar, 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Transcriptomic analysis reveals Gpx1-dependent regulation of cell adhesion genes in TNBC cells
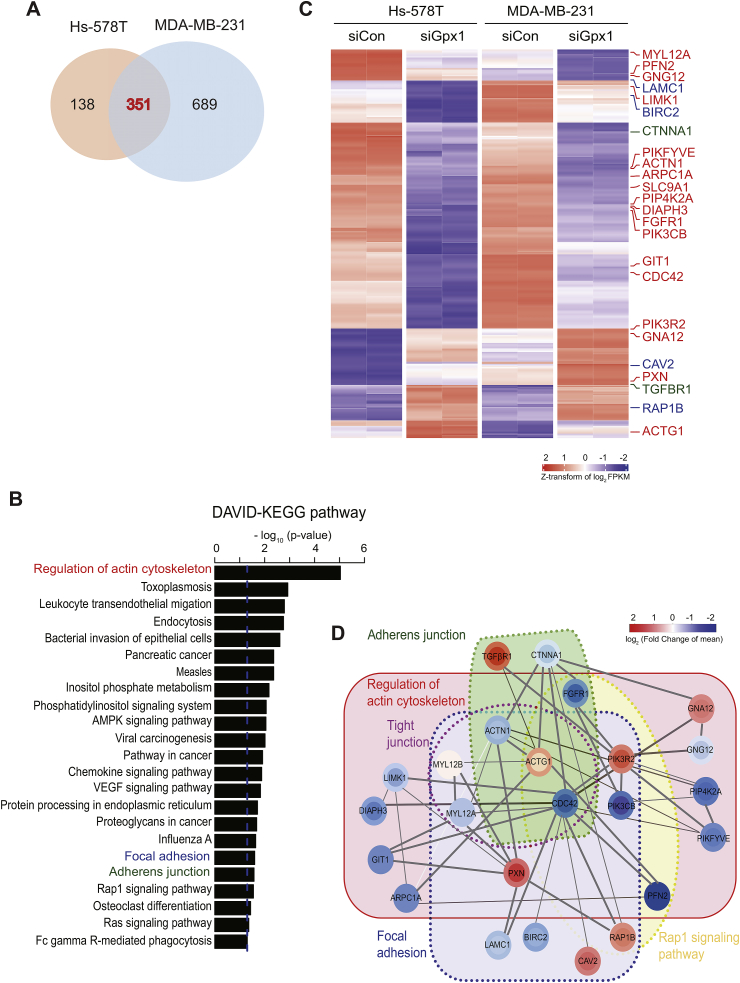
To obtain an unbiased picture on the roles of Gpx1 in the TNBC cells, we performed whole mRNA sequencing for the control versus Gpx1-depleted MDA-MB-231 and Hs578T cells in duplicates. The computational pipelines identified 351 common genes that were differentially expressed in both cells (Fig. 5A and Supplementary Table S1). Then, we performed a functional enrichment analysis for 351 differentially-expressed genes (DEGs) using DAVID bioinformatics resources with the KEGG pathways [28]. As a result, the analysis revealed that the signaling pathways involved in the adhesion, phosphatidylinositol, and migration were enriched (Fig. 5B). In particular, 24 out of the top 30 genes with the lowest p-value in the DEG analysis were associated with the three pathways including actin cytoskeleton, focal adhesion, and adherens junction, which hence strongly suggest that cell migration and adhesion are the most prominent features characterized by Gpx1 depletion in the TNBC cells (Fig. 5C). The representative genes include actin, cdc42, and laminin, whose differential expression was then verified by RT-PCR (Supplementary Fig. S5A). In addition, since the cellular proliferation processes, including phosphatidylinositol and Ras signaling, were enriched, we confirmed experimentally that key genes, including CRK1 and Shc1, were down-regulated by the Gpx1 depletion (Supplementary Fig. S5B). To understand the relationship among the featured cellular processes, we built a protein-protein interaction network using STRING [29]. The network analysis constituted the densely inter-connected network modules where CDC42, ACTN1, ACTG1, PIK3R2, and PXN genes were the network hubs, possibly suggesting their crosstalk in the regulation of adhesion signaling (Fig. 5D). Overall, our transcriptome and subsequent network analyses demonstrated that the Gpx1 depletion cooperatively directs the expression of gene sets involving not only cell adhesion but also proliferation signaling and some metabolic pathways.
Fig. 5.
Transcriptome analysis highlights the role of Gpx1 in TNBC cell adhesion.
(A) Venn diagram depicting the number of differentially-expressed genes (DEGs) identified from control and Gpx1-depleted TNBC cells. False discovery rate (FDR) < 10−5 was used to obtain 489 and 1040 DEGs for Hs578T and MDA-MB-231 cells, respectively. Subsequent analyses were performed for 351 common DEGs including 99 upregulated genes and 252 downregulated genes.
(B) KEGG pathway terms significantly enriched for the DEGs (P < 0.05).
(C) Expression heatmap for 351 DEGs in control and Gpx1-depleted cells. Experimental duplicates were separated by dashed lines. Genes involved in the cell adhesion process were identified by colors: regulation of actin cytoskeleton (red), focal adhesion (blue), and adherens junction (green).
(D) STRING network analysis depicting the functional protein-protein interaction networks. Color of each node represents the log2 (FoldChange) in Hs578T (inner circle) and MDA-MB-231 (outer circle) cells, where red and blue colors indicate up- and down-regulation on Gpx1 depletion, respectively. Genes belonging to the same KEGG pathway were grouped together to show the network modules. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Absence of Gpx1 reduces lung metastasis of TNBC cells in vivo
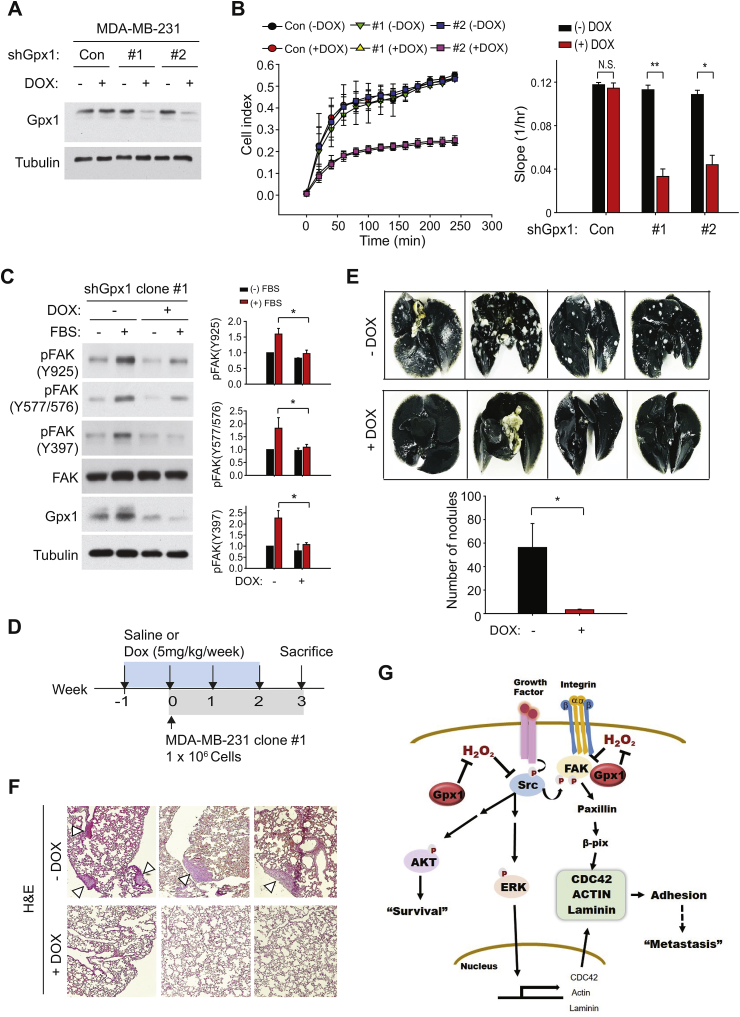
Lastly, the biological significance of the Gpx1-mediated regulation of TNBC cell adhesion was assessed in vivo through a lung metastasis assay. To this end, we produced a stable MDA-MB-231 cell line harboring the microRNA-adapted small-hairpin RNA (shRNAmir) specific to Gpx1, whose expression is inducible by tetracycline. The induction of Gpx1 depletion was confirmed in the two independent clones of stable MDA-MB-231 cells after treatment of doxycycline (Dox) for 24 h (Fig. 6A). The real-time cell analysis showed that the Dox-induced Gpx1 depletion markedly reduced the adhesion and spreading of the stable MDA-MB-231 clones (Fig. 6B). The following immunoblot analysis demonstrated that the Dox-induced Gpx1 depletion consistently reduced the phosphorylation-dependent activation of FAK kinase (Fig. 6C). Then, we performed the in vivo metastasis assay by intravenously injecting the stable cells into the athymic nu/nu mice (Fig. 6D). After either saline or Dox was administered for four weeks, the lungs were stained with blue India ink and excised for analyzing macroscopic metastases. The number of metastatic white nodules was markedly decreased by the Dox-induced Gpx1 depletion compared to the non-induced control samples (Fig. 6E). The histological analysis again revealed that the hyperplastic tumor lobules disappeared almost completely in the lung tissues from Dox-treated mice compared to those from the saline-treated control mice (Fig. 6F). Hence, the Gpx1-dependent cell adhesion is crucial for the metastatic growth of TNBC cells.
Fig. 6.
Deletion of Gpx1 suppresses the metastasis of TNBC cells in vivo.
(A) Generation of the inducible MDA-MD-231 cell lines harboring small-hairpin Gpx1 (shGpx1) sequence. Two clones of MDA-MD-231 stable cell lines (#1 and #2) were established using the shGpx1-encoding lentiviral vector as described in the Methods. A control cell line contains the empty pTRIPZ vector. Depletion of Gpx1 was induced by treatment of doxycycline (DOX, 5 μg/ml) for 24 h. A representative immunoblot for Gpx1 level is shown.
(B) Real-time measurement of adhesion of MDA-MD-231 stable cells using the xCELLigence system. Real-time traces of electrical impedance are shown (Left panel). Data in the graph are means ± SD of the slopes of cell index increments based on electrical impedance (n = 3, *P < 0.005, **P < 0.001). N.S., not significant.
(C) Serum-induced FAK activation in the MDA-MB-231 stable cells. MDA-MB-231 stable cells were serum-starved for 12 h in the presence or absence of doxycycline and then stimulated with or without 20% fetal bovine serum for 30 min. Representative immunoblots are shown. Data in the graph are means ± SD of the relative intensities of phospho-FAK bands normalized by corresponding FAK bands (n = 3, *P < 0.05).
(D) Scheme for injection schedules of DOX and MDA-MB-231 stable cells in the nude mice. (E) Tumor nodules formed by MDA-MB-231 stable cells in the mouse lungs. Lung was stained with India ink. Representative photographs are shown. Data in the graph are means ± SD of the number of white nodules (n = 4, *P < 0.05).
(F) HE images of lung tissue sections showing adenomatous hyperplasia lesions (arrowheads).
(G) Proposed model for the Gpx1-mediated adhesion signaling in the TNBC cells.
4. Discussion
Metastasis is considered to be an insurmountable blockade in cancer therapy because the metastatic cancer cells are rare in the circulation and become refractory to the existing chemo- and radio-therapies when colonized in the secondary organs [30]. Among the metastasis processes, the cancer cell-ECM adhesion is the most critical step for colonization and re-growth. Therefore, targeting integrin-FAK signaling has been thought to be an efficient way to inhibit the metastatic tumor growth [31,32]. In terms of the antioxidant system related to the BC, the role of Gpx1 in BC metastasis is largely unknown. In this study, we report the evidence that FAK is a redox-sensitive tyrosine kinase, whose activity is preserved by antioxidant enzyme Gpx1 in the TNBC cells. In the absence of Gpx1, the activation of FAK kinase is impaired and hence the BC cell adhesion/spreading signaling is abrogated. Moreover, the absence of Gpx1 also causes the transcriptional repression of actin cytoskeleton-related genes, including actin, cdc42, and laminin, which all cooperate for cell adhesion/spreading (Fig. 6G). Our in-depth signaling analysis further indicates that the activation of c-Src kinase, which is required for full activation of FAK, was also dampened by the Gpx1 depletion. It is somewhat explained by a previous study showing that a Src family member Lyn kinase is redox-sensitive [33]. Such strong evidence for the cellular function of Gpx1 in the TNBC cells encouraged us to look into the clinical relevance and significance of our study. A local clinical study in Brazil has showed that high Gpx1 expression is significantly correlated with a shorter survival of BC patients and particularly with a high mortality of the patients received with chemo/radiotherapy [24]. However, a correlation between the overall survival of BC patients and the Gpx1 expression was not seen in the large dataset in the Kaplan-Meier plotter (3951 patients). We surmised that the present bulk sequencing data does not reflect the dynamics of Gpx1 expression that we observed among the normal breast epithelial and BC cell lines. Thus, the data analysis of BC samples along with normal matches may be necessary for defining the clinical significance of Gpx1. Moreover, we pointed that the Gpx1 plays a pro-metastatic function via FAK in the TNBC cells. In fact, it has been shown that the FAK expression is low in normal breast epithelial cells but induced in the specific sites of invasive BC tissues [34]. Therefore, we thought that the functional significance of Gpx1 in the invasive BC may be associated with the FAK expression.
Our study also address an intriguing subject that two Gpx isoforms, Gpx1 and Gpx4, may play the differential roles in the cellular system. The in vitro peroxidase activities of Gpx1 and Gpx4 are somewhat different in terms of substrate preference by utilizing H2O2 and phospholipid hydroperoxides, respectively [22]. Moreover, Gpx4 has recently been received attentions as a modulator of ferroptotic cancer cell death [35]. At least in the TNBC cells, we showed that the depletion of Gpx1 increased the level of cellular H2O2, not LOOH, and resulted in the inactivation of FAK kinase. However, the depletion of Gpx4 actually increased the LOOH level in the TNBC cells, but the same condition did not alter the FAK activation. These results represent that both isoforms may have a different substrate specificity and thus exert different biological functions in the TNBC cells, i.e. cell adhesion versus ferroptotic cell death.
Since ER, PR and HER2 are usually overexpressed in BC cells, the BC patients are treated with anti-cancer drugs targeting the hormone receptors [[36], [37], [38]]. However, TNBCs are unlikely to respond to anti-estrogen therapy or HER2 antagonists [5]. Thus, the development of alternative therapy for TNBC treatment is urgently needed for curing aggressive and metastatic BCs. Although studies have indicated that the cellular antioxidant enzymes are abundant in various cancer types, the non-specific targeting of ROS metabolism was entirely failed [39]. The lesson from previous redox-cancer studies was that the antioxidant enzymes play the localized and target-specific role in the cancer cells [[40], [41], [42]]. One of the most significant finding in this study is that the Gpx1 expression is resumed in the TNBC cells and interacted with FAK. If cancer cells face on oxidative microenvironment created by residential immune cells while migrating inside the secondary metastatic tissues, the functional integrity of focal adhesion complex may be an essential factor for successful colonization. In this context, it is logical that the restoration of Gpx1 expression is necessary for preserving FAK activation. Hence, our study suggests that the specific targeting of Gpx1 for anti-BC treatment may require the additional subtyping of metastatic BC based on the correlated expression of Gpx1 and FAK as a part of personalized medicine.
Author contribution
E.K.L. designed and performed experiments; A.L., Y.J., and S.L. performed and interpreted bioinformatics analysis; N.K., J.I.Y., and S.Y. K. provided expertise and feedback; S.W.K. conceived experiments, interpreted data, supervised the project; E.K.L., S.L., and S.W.K. wrote the manuscript.
Data availability
RNA-Seq data: Gene Expression Omnibus GSE126675 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126675).
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgements
We thank Prof. Eok-Soo Oh and Dr. Eun-Woo Lee for sharing antibodies and the lab members for technical assistance. This study was supported by grants from the National Research Foundation of Korea (2018R1A2B3006323 and 2017M3A9B6073098) and the National R&D Program for Cancer Control (1420280).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101391.
Contributor Information
Sanghyuk Lee, Email: sanghyuk@ewha.ac.kr.
Sang Won Kang, Email: kangsw@ewha.ac.kr.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., Fluge O., Pergamenschikov A., Williams C., Zhu S.X., Lonning P.E., Borresen-Dale A.L., Brown P.O., Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Tavassoli F.A. Classification of metaplastic carcinomas of the breast. Pathol. Annu. 1992;27 Pt 2:89–119. [PubMed] [Google Scholar]
- 3.Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 4.Fornier M., Fumoleau P. The paradox of triple negative breast cancer: novel approaches to treatment. Breast J. 2012;18:41–51. doi: 10.1111/j.1524-4741.2011.01175.x. [DOI] [PubMed] [Google Scholar]
- 5.Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 6.Wahba H.A., El-Hadaad H.A. Current approaches in treatment of triple-negative breast cancer. Canc. Biol. Med. 2015;12:106–116. doi: 10.7497/j.issn.2095-3941.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fidler I.J. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 8.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 9.Weiner T.M., Liu E.T., Craven R.J., Cance W.G. Expression of focal adhesion kinase gene and invasive cancer. Lancet. 1993;342:1024–1025. doi: 10.1016/0140-6736(93)92881-s. [DOI] [PubMed] [Google Scholar]
- 10.Luo M., Fan H., Nagy T., Wei H., Wang C., Liu S., Wicha M.S., Guan J.L. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Cancer Res. 2009;69:466–474. doi: 10.1158/0008-5472.CAN-08-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitra S.K., Hanson D.A., Schlaepfer D.D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 12.Guan J.L., Shalloway D. Regulation of focal adhesion-associated protein tyrosine kinase by both cellular adhesion and oncogenic transformation. Nature. 1992;358:690–692. doi: 10.1038/358690a0. [DOI] [PubMed] [Google Scholar]
- 13.Parsons J.T. Focal adhesion kinase: the first ten years. J. Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 14.Schlaepfer D.D., Hauck C.R., Sieg D.J. Signaling through focal adhesion kinase. Prog. Biophys. Mol. Biol. 1999;71:435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 15.Cance W.G., Harris J.E., Iacocca M.V., Roche E., Yang X., Chang J., Simkins S., Xu L. Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: correlation with preinvasive and invasive phenotypes. Clin. Cancer Res. 2000;6:2417–2423. [PubMed] [Google Scholar]
- 16.Rigiracciolo D.C., Santolla M.F., Lappano R., Vivacqua A., Cirillo F., Galli G.R., Talia M., Muglia L., Pellegrino M., Nohata N., Di Martino M.T., Maggiolini M. Focal adhesion kinase (FAK) activation by estrogens involves GPER in triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. : CR (Clim. Res.) 2019;38:58. doi: 10.1186/s13046-019-1056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golubovskaya V.M., Ylagan L., Miller A., Hughes M., Wilson J., Wang D., Brese E., Bshara W., Edge S., Morrison C., Cance W.G. High focal adhesion kinase expression in breast carcinoma is associated with lymphovascular invasion and triple-negative phenotype. BMC Canc. 2014;14:769. doi: 10.1186/1471-2407-14-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafer Z.T., Grassian A.R., Song L., Jiang Z., Gerhart-Hines Z., Irie H.Y., Gao S., Puigserver P., Brugge J.S. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felding-Habermann B., O'Toole T.E., Smith J.W., Fransvea E., Ruggeri Z.M., Ginsberg M.H., Hughes P.E., Pampori N., Shattil S.J., Saven A., Mueller B.M. Integrin activation controls metastasis in human breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1853–1858. doi: 10.1073/pnas.98.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Autreaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 21.Flohe L., Toppo S., Cozza G., Ursini F. A comparison of thiol peroxidase mechanisms. Antioxidants Redox Signal. 2011;15:763–780. doi: 10.1089/ars.2010.3397. [DOI] [PubMed] [Google Scholar]
- 22.Lubos E., Loscalzo J., Handy D.E. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxidants Redox Signal. 2011;15:1957–1997. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bera S., Weinberg F., Ekoue D.N., Ansenberger-Fricano K., Mao M., Bonini M.G., Diamond A.M. Natural allelic variations in glutathione peroxidase-1 affect its subcellular localization and function. Cancer Res. 2014;74:5118–5126. doi: 10.1158/0008-5472.CAN-14-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jardim B.V., Moschetta M.G., Leonel C., Gelaleti G.B., Regiani V.R., Ferreira L.C., Lopes J.R., Zuccari D.A. Glutathione and glutathione peroxidase expression in breast cancer: an immunohistochemical and molecular study. Oncol. Rep. 2013;30:1119–1128. doi: 10.3892/or.2013.2540. [DOI] [PubMed] [Google Scholar]
- 25.Kulak M.V., Cyr A.R., Woodfield G.W., Bogachek M., Spanheimer P.M., Li T., Price D.H., Domann F.E., Weigel R.J. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene. 2013;32:4043–4051. doi: 10.1038/onc.2012.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaller M.D., Hildebrand J.D., Shannon J.D., Fox J.W., Vines R.R., Parsons J.T. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 1994;14:1680–1688. doi: 10.1128/mcb.14.3.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huveneers S., Danen E.H. Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehlen P., Puisieux A. Metastasis: a question of life or death. Nat. Rev. Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 31.Yilmaz M., Christofori G. Mechanisms of motility in metastasizing cells. Mol. Cancer Res. : MCR. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 32.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo S.K., Starnes T.W., Deng Q., Huttenlocher A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature. 2011;480:109–112. doi: 10.1038/nature10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watermann D.O., Gabriel B., Jager M., Orlowska-Volk M., Hasenburg A., zur Hausen A., Gitsch G., Stickeler E. Specific induction of pp125 focal adhesion kinase in human breast cancer. Br. J. Canc. 2005;93:694–698. doi: 10.1038/sj.bjc.6602744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassannia B., Vandenabeele P., Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Piccart-Gebhart M.J., Procter M., Leyland-Jones B., Goldhirsch A., Untch M., Smith I., Gianni L., Baselga J., Bell R., Jackisch C., Cameron D., Dowsett M., Barrios C.H., Steger G., Huang C.S., Andersson M., Inbar M., Lichinitser M., Lang I., Nitz U., Iwata H., Thomssen C., Lohrisch C., Suter T.M., Ruschoff J., Suto T., Greatorex V., Ward C., Straehle C., McFadden E., Dolci M.S., Gelber R.D. Herceptin adjuvant trial study, T., trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 37.Johnston S., Pippen J., Jr., Pivot X., Lichinitser M., Sadeghi S., Dieras V., Gomez H.L., Romieu G., Manikhas A., Kennedy M.J., Press M.F., Maltzman J., Florance A., O'Rourke L., Oliva C., Stein S., Pegram M. Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer. J. Clin. Oncol. : Off. J. Am. Soc. Clin. Oncol. 2009;27:5538–5546. doi: 10.1200/JCO.2009.23.3734. [DOI] [PubMed] [Google Scholar]
- 38.Cortes J., Saura C., Bellet M., Munoz-Couselo E., Ramirez-Merino N., Calvo V., Perez J., Vidal M. HER2 and hormone receptor-positive breast cancer--blocking the right target. Nat. Rev. Clin. Oncol. 2011;8:307–311. doi: 10.1038/nrclinonc.2010.185. [DOI] [PubMed] [Google Scholar]
- 39.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 40.Kang D.H., Lee D.J., Lee S., Lee S.Y., Jun Y., Kim Y., Kim Y., Lee J.S., Lee D.K., Lee S., Jho E.H., Yu D.Y., Kang S.W. Interaction of tankyrase and peroxiredoxin II is indispensable for the survival of colorectal cancer cells. Nat. Commun. 2017;8:40. doi: 10.1038/s41467-017-00054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris I.S., Treloar A.E., Inoue S., Sasaki M., Gorrini C., Lee K.C., Yung K.Y., Brenner D., Knobbe-Thomsen C.B., Cox M.A., Elia A., Berger T., Cescon D.W., Adeoye A., Brustle A., Molyneux S.D., Mason J.M., Li W.Y., Yamamoto K., Wakeham A., Berman H.K., Khokha R., Done S.J., Kavanagh T.J., Lam C.W., Mak T.W. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 42.Woo H.A., Yim S.H., Shin D.H., Kang D., Yu D.Y., Rhee S.G. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data: Gene Expression Omnibus GSE126675 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126675).