Abstract
ODF1 has been described as an exclusively expressed testicular protein and is located in the outer dense fibers along the sperm tail. ODF1 has been involved in the sperm motility and in the development of the flagellum, but the function of ODF1 is not already clear. Other ODF proteins, such as ODF2 have been characterized in other tissues like the basal body of the kidney primary cilium, but so far only the mRNA of ODF1 has been described in other tissues. These observations let us to hypothesize that the expression of the protein ODF1 could not be limited to the testis. Therefore, in the present work we proposed to evaluate if the ODF1 protein could also be present in tissues other than the testis. Here we demonstrated through western blot, immunofluorescence, and RT-PCR techniques that the protein and mRNA of ODF1 have been identified in the rat kidney. Finally, the presence of ODF1 in kidney has also been confirmed through proteomic analysis using mass spectrometry. The results derived from these different complementary approaches indicate that, to our knowledge and for the first time, ODF1 is demonstrated to be present in an additional organ different to testis. This results raise new questions about potential other functions and locations of the ODF1 protein.
Keywords: Cell biology, Proteins, Reproductive system, ODF1, Proteomic, Collecting duct
Cell biology; Proteins; Reproductive system; ODF1, Proteomic, Collecting duct
1. Introduction
The sperm tail contains the axoneme and certain accessory structures like the mitochondrial sheath, outer dense fibers (ODF) and fibrous sheath [1].These structures are assembled during spermatogenesis through a precise arrangement of microtubules, mitochondrial migration and Golgi transformation [2].
The ODF proteins are synthesized after the transcriptional activity has ended, in the latter part of spermiogenesis. This implies that these major integral proteins of the tail are translationally regulated and ODF mRNAs are long-live [3, 4].
The ODF were clearly defined by transmission electron microscopy and they are composed of more than a dozen different proteins of which only a few have been identified [5, 6, 7, 8, 9, 10, 11]. ODF seem to be important for the stability and the elastic recoil of the sperm tail, as well as for support of the flagellar beat [12, 13].Other authors suggest that the phosphorylation state of the ODF polypeptides may influence such functions and possibly affect sperm motility [5]. ODFs proteins are expressed together and their interaction have been described in the spermatogenesis and in the sperm tail [14].
ODF1(Outer dense fiber 1), also called HSPB10(Heat Shock Protein B10) is the major protein of the ODF in the mammalian sperm tail [10], and it has a molecular mass of 27 kDa. Analysis of ODF1 amino acid structure indicated the presence of a putative leucine zipper dimerization motif in the N-terminus and the presence of PCX repeats in the C-terminus [6]. The N-terminus motif mediates the interaction with the heavy chain of KCL3 [1], ODF2 [14], Spag4, and Spag5 [15].Our group in 2011 described that ODF1, undergoes maturational changes during epididymal transit like sulfhydryl oxidation. When the sperm cysteines were blocked by monobromobimane in rat [16] and human sperm [17] the spermatozoa lost the progressive motility, without affecting ATP production.
ODF1 therefore seems to be functionally involved in spermatid differentiation and formation of the sperm tail [18]. ODF1 deficiency in homozygous mice resulted in male infertility due to sperm decapitation. However heterozygous mice were fertile but showed reduced sperm motility [19].
ODFs proteins are expressed together in meiotic and post-meiotic germ cells, and ODF1 and ODF2 co-localize and strongly interact via their leucine zippers during the spermiogenesis [14, 20, 21, 22].Even though the ODF2 expression was firstly restricted to germ cells [21], many authors have described the ODF2 as a modulator of the spindle orientation in the appendage of the mother centriole in the kidney [23].
In spite that the mRNA of ODF1 has been described in other tissues [24, 25] so far there was no information in regard to the expression of the protein ODF1. Therefore, we hypothesized that the expression of the protein ODF1 could not limited to the testis and we proposed to evaluate if ODF1 could also be present in other tissues. In the present work, by different technique and proteomic analysis we have confirmed that the ODF1 protein is also expressed in the kidney, and specifically in the collecting ducts.
2. Materials and methods
2.1. Experimental animals and protocol
Adult male Wistar rats were obtained from our animal facility and maintained under the guidelines of local animal care committee. Animals were sacrificed in accordance to the local guidelines. The protocol was approved by the Institutional Committee for use of Laboratory Animal (ICULA- Res. 32/95, School of medicine, UNCuyo).
2.2. Tissue preparation
Testes, livers, kidneys(total fraction: cortex plus medulla), brains and lungs from three different animals were homogenized in chilled extraction buffer containing 300 mM Sucrose, 18 mM Trizma Base, 5 mM EGTA, pH 7.4 [26] with a tissue homogenizer Ultraturrax T25 (IKA-TRON- Staufen, Germany). The homogenates were centrifuged at 14,000 xg for 20 min at 4 °C (Eppendorf Model 5417R Hamburg, Germany). Both fractions (supernatant and pellet) were boiled for 5 min and the protein concentration was determined.
2.3. Isolation of cytoskeletal fraction in kidney
Kidney cortex (KC) and medulla (KM) were isolated and homogenized in chilled extraction buffer containing 0.1% Triton X-100, 30 mM imidazole, 10 mM EDTA, 2 mM MgCl2, 0.1 mM dithiothreitol, and Protease Inhibitor Cocktail (cat#P8340 Sigma Aldrich), pH 7.4. The homogenates were centrifuged at 35,000 xg for 10 min at 4 °C to separate the Triton-soluble supernatant (non-cytoskeleton: NC) protein fraction, from the Triton-insoluble pelleted fraction (cytoskeleton: C). The pellets were re-suspended in the extraction buffer to complete the volume of the original homogenate, resulting in similar protein concentration as in supernatants [27]. Both fractions (supernatant and pellet) were boiled for 5 min and the protein concentration was determined.
2.4. Protein determination and western blot analysis
By BCA method the total protein concentration were obtained [28] and were added with Laemmli sample buffer and were loaded onto 15% (w/v) acrylamide gel with 4% (w/v) stacking gel [29]. All Blue-Bio Rad (cat#161–0373) was used as molecular weight marker.
Proteins were transferred to a nitrocellulose membrane [30]. For immunoblotting, the membrane was incubated overnight in blocking buffer (3% (v/v) Teleostean gelatin fish in TBS-T, 1.92 mM Trizmabase, 0.1% (v/v) Tween 20) at 4 °C, incubated 1 h at rt (room temperature) with primary antibody anti-ODF1 against the N-Terminal (cat#sc-27907Santa Cruz, 1:1000) and middle fraction (cat#sc-23132 Santa Cruz,1:1000). The labeling was visualized with biotinylated anti-goat IgG antibodies (cat#B7014 Sigma Aldrich, 1:2500) and with extravidin-peroxidase (cat#E2886Sigma Aldrich, 1:750).
As loading control immunoblotting against α-tubulin (cat#69125MP-Biomedicals 1:7500) was carried on. The labeling was visualized with biotinylated anti mouse IgG antibodies (cat#B7014 Sigma Aldrich, 1:2500) and with extravidin-peroxidase (cat#E2886Sigma Aldrich, 1:750). All the antibodies were incubated for 1 h at rt and washed three times with TBS-T for 5 min. Extravidin-peroxidase was incubated for 45 min at rt and washed three times with TBS.
Bound antibodies were visualized by enhanced chemiluminescence (1 M Trizma base pH 8.5, 250 mM luminol, 90 mM cumaric acid, 3% (v/v) H2O2) and the images were captured by a camera model LAS 4.000 (Fujifilm Tokyo, Japan).
2.5. Relative quantitative Reverse Transcriptase polymerase chain reaction (RT-PCR)
Total RNA was extracted from testis, liver and kidney from two different animals, 100 mg of frozen tissues were homogenized mechanically in 1 ml Trizol reagent (cat#15596–026 Invitrogen) and RNA extracted with 0.2 ml chloroform per ml. Lysates were allowed to stand for 5 min at rt and centrifuged at 12,000 xg for 15 min at 4 °C. Upper aqueous phases containing RNA were transferred to fresh tubes with 0.5 ml isopropanol per ml Trizol to precipitate the RNA at rt for 10 min. RNA was pelleted out by centrifugation at 12,000 xg for 10 min at 4 °C. RNA pellet were washed with 1 ml of chilled 70% ethanol, centrifuged at 7,500 xg for 5 min at 4 °C and air dried. RNA pellets were solubilized in 25 μl UltraPure™ DNase⁄RNase-Free Distilled Water. Concentration and purity of the samples was spectroscopically (Nano drop lite Thermo Scientific). Two (2) micrograms of total RNA was randomly reverse transcribed with 200 units M-MLV enzyme Reverse Transcriptase (Cat#28025–013 Invitrogen). Twenty (20) μl of reaction mixture were added, following the manufacturer's instructions.
PCR was then performed using the reverse transcription products obtained as described above.
A primer designed using Primer3® software (www.ncbi.nlm.nih.gov/tools/primer-blast/) was used for the amplification by PCR at equimolar concentration.Odf1 primers (Invitrogen). Fw:GACCATAATGGCCGCACTG. Rv:CGATCTTGACACAACTGCCG. Product: 560 pb. As a positive control, β-Actin was used (Cat#B072-40 Promega). Fw:GGAACCGCTCATTGCC. Rv:ACCCACACTGTGCCCATCTA. Product: 289 pb.
The PCRs were carried out in a 25 μl reaction volume containing 2 μl of cDNA, 23 pmol of each primer, 200 WM dNTPs (cat#10297–018) 5 mM MgCl2, 1.5 U of Taq DNA polymerase (cat#11615–036) and 1X Taq DNA polymerase PCR buffer (Invitrogen). The cycling parameters were as follows: 95 °C, 5min; 35 cycles: 95 °C, 60s; 62 °C, 90s; 72 °C, 1min. Negative control included PCR reaction without template DNA. Amplified products were run on agarose gel (2% w/v), 100 pb DNA ladder (cat#15620-019Invitrogen) was used and dyed with SYBR Safe - DNA Gel Stain (Invitrogen). The images were captured by a camera LAS 4.000.Densitometry analysis was performed in Image J software(Version 1.46; NIH), and Odf1 mRNA expression was normalized to the β-Actin mRNA expression, the testicular expression of ODF1 that was considered as 100% [31].
2.6. Indirect immunofluorescence
Rats were anaesthetized with sodium pentobarbital (60 mg/kg IP). Then, kidneys were perfused through the abdominal aorta with ice-cold phosphate buffered saline (PBS) solution to rinse away all the blood. Then, the kidneys were fixed by retrograde perfusion with 40 ml of 4% paraformaldehyde in 9.4 mM Na2B4O7, 0.34 mM Na2SO3, 0.16 M H3BO3, pH 7.4.
The kidney, testis and liver were removed and placed in paraformaldehyde for 4 h at room temperature and processed by routine histological technique [32].
Paraffin sections (5–6 μm thickness) were dewaxed in xylol, rehydrated, and incubated with 50 mM of NH4Cl under UV light to quench the tissue auto-fluorescence. For antigen retrieval the slides were heated in buffer containing 10 mM Trizma base pH 8, 1 mM EDTA disodium salt, 0.05% (v/v) Tween 20. Endogenous peroxidase was blocked and the tissue was permeabilized with 10% (v/v) goat serum 2% (w/v) bovine serum albumin, 10% (v/v) Triton X100 in PBS for 30 min at rt. After washes, the sections were incubated with the antibody against ODF1 (cat#sc-279071:100). As a polyvalent biotinylated secondary antibody and the later fluorescent labeling (Extravidin-Fluorescein isothiocyanate) we used a Vectastain Kit R.T.U. 23 ® (Vector laboratories, USA). To identify collecting ducts a goat polyclonal antibody against aquaporine 2 (AQP2, cat#sc-9882Santa Cruz,1:1000) was used. The same polyvalent biotinylated antibody described above was used and was reveled with Extravidin-Texas Red. Hoechst was used for nuclear stain. Negative controls included tissues unexposed to primary antibodies Confocal laser microscopy was performed using an Olympus FV1000 (Japan).
2.7. Kidney proteomic analysis
2.7.1. Protein isolation
Proteins were obtained from the testis, kidney medulla and further cytoskeleton fraction isolation, as was described above, but without addition of protease inhibitor. Firstly, a proteomic analysis was carried out in testis and the peptides of ODF1 that were detected were compared with the ones isolated from kidney.
2.7.2. Mass spectrometry analysis
Protein digestion and Mass Spectrometry analysis were performed at the Proteomics Core Facility CEQUIBIEM, at the University of Buenos Aires/CONICET (National Research Council) as follows: Coomassie G250-stained SDS-PAGE gel excised protein bands were sequentially washed and faded with 50 mM ammonium bicarbonate, 25 mM ammonium bicarbonate 50% acetonitrile, and 100% acetonitrile; reduced and alkylated with 10 mM DTT and 20 mM iodoacetamide and in-gel digested with 100 ng Trypsin (Promega V5111) in 25 mM ammonium bicarbonate overnight at 37 °C.
Peptides were recovered by elution with 50% acetonitrile-0.5% trifluoroacetic acid, including brief sonication, and further concentrated by speed-vacuum drying. Samples were resuspended in 15 μl of water containing 0.1% Formic Acid.
After that, the peptides were purified and desalted with ZipTip C18 columns (Millipore). The digests were analysed by nanoLC-MS/MS in a Thermo Scientific QExactive Mass Spectrometer coupled to a nanoHPLC EASY-nLC 1000 (Thermo Scientific, Germany). For the LC-MS/MS analysis, approximately 1 μg of peptides was loaded onto the column and eluted for 120 min using a reverse phase column (C18, 2 μm, 100A, 50 μm × 150 mm) Easy-Spray Column PepMap RSLC (P/N ES801) suitable for separating protein complexes with a high degree of resolution. The flow rate used for the nano column was 300 nL min-1 and the solvent range from 7% B (5 min) to 35% (120 min). Solvent A was 0.1% formic acid in water whereas B was 0.1% formic acid in acetonitrile. The injection volume was 2μL. The MS equipment has a high collision dissociation cell (HCD) for fragmentation and an Orbitrap analyzer (Q-Exactive-Thermo Scientific, Germany). A voltage of 3,5 kV was used for Electro Spray Ionization (Thermo Scientific, EASY-SPRAY).
XCalibur 3.0.63 (Thermo Scientific, Germany) software was used for data acquisition and equipment configuration that allows peptide identification at the same time of their chromatographic separation. A Data dependent method was used: Full-scan mass spectra were acquired in the Orbitrap analyzer. The scanned mass range was 400–1800 m/z, at a resolution of 70000 at 400 m/z and the twelve most intense ions in each cycle, were sequentially isolated, fragmented by HCD and measured in the Orbitrap analyzer. Peptides with a charge of +1 or with unassigned charge state were excluded from fragmentation for MS2.
In a second stage a targeted approach was used, employing an m/z inclusion list with the m/z of ODF peptides which had been previously detected in testes samples (see Table 1).
Table 1.
List with the m/z of ODF peptides which had been previously detected in testes samples.
| Mass [m/z] | Formula [M] | Comments |
|---|---|---|
| 407.19978 | C47 H85 N18 O18 S | VCVSAERENR (C2 (∗Carb)) |
| 490.2293 | C38 H68 N12 O16 S | CIDEISSR (C1 (∗Carb)) |
| 589.32915 | C52 H90 N16 O13 S | LYCLROSLR (C3 (∗Carb)) |
| 603.78593 | C53 H85 N13 O15 S2 | KYSYMNICK (C8 (∗Carb)) |
| 610.29604 | C47 H84 N18 O18 S | VCVSAERENR (C2 (∗Carb)) |
| 621.28259 | C50 H82 N16 O19 S | ENRYDCLGSK (C6 (∗Carb)) |
| 660.81065 | C58 H91 N13 O20 S | EFSLPPCVDEK (C7 (∗Carb)) |
| 681.65093 | C82 H138 N27 O30 S2 | VCVSAERENRYDCLGSK (∗Carb); C13 (∗Carb)) |
| 724.83994 | C63 H99 N15 022 S | DVTYSYGLGSCVK (C11 (∗Carb)) |
| 917.42778 | C121 H187 N28 O41 S2 | EFSLPPCVDEKDVTYSYGLGSCVK (C7 (∗Carb); C22 (∗Carb)) |
| 1032.83522 | C129 H217 N38 O44 S3 | ILASSCCSSNILGSVNSCGFEPDQVKVR (C6 (∗Carb); C7 (∗Carb); C18 (∗Carb)) |
| 1123.99393782 | C94 H141 N23 O31 S5 | SCGLCDLYYPCCLCDYK (C2 (∗Carb); C5 (∗Carb); C11 (∗Carb); C12 (∗Carb); C14 (∗Carb)) |
| 410.70272 | C32 H59 N11 O12 S | VCVSAER (C2 (∗Carb)) |
| 654.13242 | C138 H237 N40 O45 S3 | ILASSCCSSNILGSVNVCGFEPDQVKVRVK (C6 (∗Carb); C7 (∗Carb); C6 (∗Carb); C7 (∗Carb)) |
| 831.66908 | C140 H239 N41 O46 S3 | ILASSCCSSNILGSVNVCGFEPDQVKVRVK (C6 (∗Cab), C7 (∗Cab), C18 (∗Cab) ILASSCCSSNILGSVNVCGFEPDQVKVRVK (C6 (∗Cab), C7 (∗Cab), C18 (∗Cab)) |
Carb: Carbamidomethyl.
2.8. Analysis of MS data
QExactive raw data was processed using Proteome Discoverer software (version 2.1.1.21 Thermo Scientific) and searched against Rattus norvegicus Uniprot database (http://www.uniprot.org/) and ODF sequence, with trypsin specificity and a maximum of two missed cleavage per peptide. Carbamidomethylation of cysteine residues was set as a fixed modification and oxidation of methionine was set as variable modification.
Proteome Discoverer searches were performed with a precursor mass tolerance of 10 ppm and product ion tolerance to 0.05 Da. Protein hits were filtered for high confidence peptide matches with a maximum protein and peptide false discovery rate of 1% calculated by employing a reverse database strategy, for medium confidence peptide matches a peptide false discovery rate of 5% was allowed.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [33] partner repository with the dataset identifier PXD014848.
3. Results
3.1. Detection of ODF1 in kidney by western –blot and RT-PCR
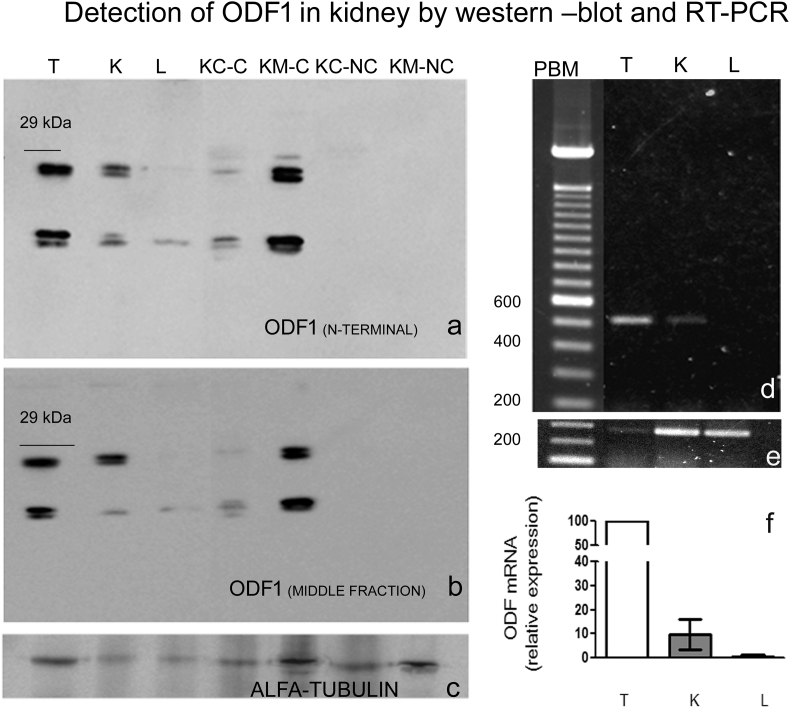
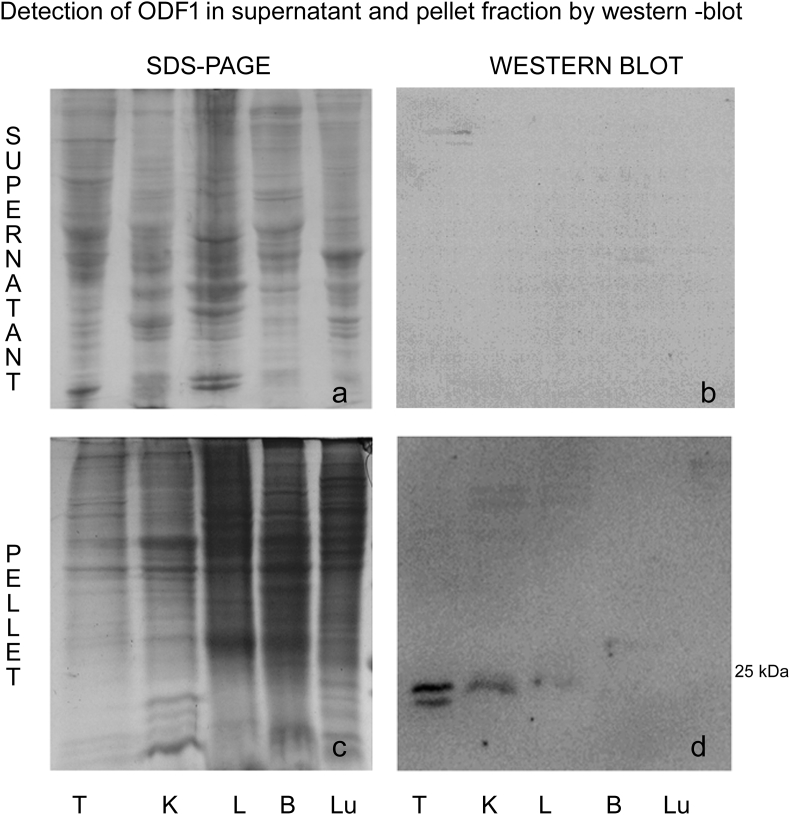
To determine possible new locations of ODF1, we performed western blot analysis in different tissue. We found that anti-ODF1 antibody against the N-terminal (Figure 1a) and middle fraction (Figure 1b) of the protein, recognized two or more proteins bands under 29 kDa, in the pellet fraction of the testis and kidney. No signal was observed in the liver (Figure 1a and Figure 2) brain and lung (Figure 2) pellet fraction. Due to the stabilization by disulfide bonds, ODF1 is an insoluble protein, so as we expected in the supernatant fraction obtained from testis, kidney, liver, brain and lung no positive band to anti-ODF1 was detected (Figure 2).
Figure 1.
ODF1 immunodetection by western blot, in proteins obtained from testis (T), kidney total fraction (K), liver (L), kidney cortex-cytoskeleton (KC–C) kidney medulla-cytoskeleton (KM-C), kidney cortex- non cytoskeleton (KC-NC) kidney medulla- non cytoskeleton (KM-NC). Different antibodies were used: in panel a: an antibody raised against a peptide mapped near the N-terminus and in panel ban antibody raised against a peptide mapping within an internal region, of ODF1 of human origin. In panel c an antibody against α-tubulin was used. On the left the approximate molecular weight was indicated. The results are representative from three separated experiments. In panel d: Odf1-mRNA expression detected by RT-PCR. In panel e: β actin-mRNA expression detected by RT-PCR. From left to right: PBM (pair base marker), T, K and L. The results are representative from two separated experiments. In panel f: optical density ratio of ODF1 relative to β-ACTIN and normalized to testis expression.
Figure 2.
Coomassie blue staining of protein profile observed in the supernatant (a) and the pellet (c), obtained of homogenates from testis (T), kidney (K), liver (L), brain (B) and lung (Lu). ODF1 immunodetection by western blot in the supernatant (b) and in the pellet (d) observed in homogenates from testis (T), kidney (K), liver (L), brain (B) and lung (Lu).On the right the approximate molecular weight was indicated. The results are representative from three separated experiments.
The strongest signal was observed in the testis and in the kidney medulla-cytoskeleton fraction (KM-C) and a weak signal was found in the kidney cortex-cytoskeleton fraction (KC–C). Conversely in the non-cytoskeleton fraction of the cortex (KC-NC) and the medulla (KM-NC) no peptide has been recognized by ODF1 antibodies. α-tubulin antibody was used as loading control (Figure 1c).
In order to detect the Odf1 mRNA we carried out the RT-PCR and we amplified the respective messenger in testis and kidney, but not in liver of the studied rats. As expected the highest expression was observed in the testis and a lower expression was also found in male kidney (Figure 1d and e).
3.2. Localization of ODF1 in kidney by immunofluorescence
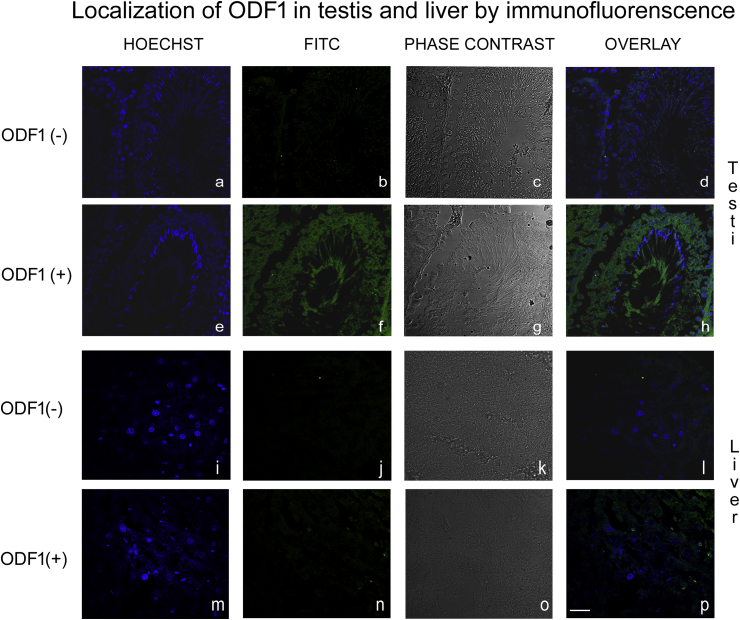
In order to characterize the location of ODF1 in kidney we applied immunofluorescence analysis. Testis cross-sections were used as a positive control of ODF1 antibodies, as well as liver cross-sections were used as a negative control due to the negative signal observed in western-blot.
Confocal images showed a positive signal to ODF1 in the epithelia of the seminiferous tubules (Figure 3f and h). In contrast liver confocal images did not display a positive reaction to ODF1-antibodies, only a weak signal and with different location was observed (Figure 3n and p).
Figure 3.
Confocal fluorescence micrographs of testis sections, incubated without (a, b, c and d) and with (e, f, g and h)) anti-ODF1.Confocal fluorescence micrographs of liver sections, incubated without (i, j, k and l) and with (m, n, o and p)) anti-ODF1. Blue fluorescence: Nuclei localization with Hoechst (a, e, i and m). Green fluorescence: anti-ODF1 labeled with FITC (b, f, j and n). Phase contrast (c, g, k and o). Overlay of blue and green fluorescence signal (d, h, l and p). Images shown are representative of three separate experiments. 40X Scale bar: 20 μm.
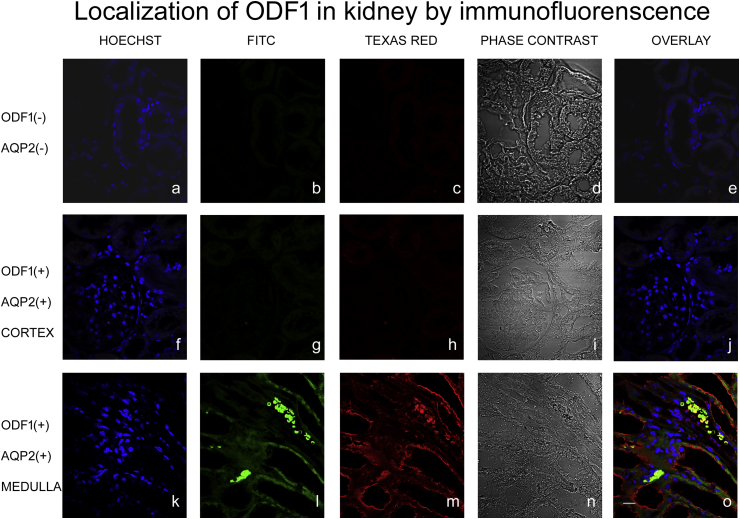
In images obtained from kidney-medulla a positive reaction to anti-ODF1 in the cytoplasm of the collecting ducts was observed (Figure 4l and o). The same tubules were also positive to AQP2, even though no co-localization was detected (Figure 4m and o). The kidney-cortex images analyzed did not reveal any positive signal to ODF1 neither to AQP2 (Figure 4g, h, and j).
Figure 4.
Confocal fluorescence micrographs of kidney sections, incubated without (a, b, c, d and e) and with (f, g, h, i, j, k, l, m, n and o) anti-ODF1and anti AQP2. Blue fluorescence: Nuclei localization (a, f and k). Green fluorescence: anti-ODF1 (b, g and l). Red fluorescence: anti AQP2 (c, h and m). Phase contrast (d, i and n). Overlay of blue, green and red fluorescence signal (e, j and o). Cortex (f, g, h, i and j) and medulla (k, l, m, n and o) cross-section are exposed. Images shown are representative of three separate experiments.60X Scale bar: 20 μm.
3.3. ODF1 proteomic detection in kidney
To confirm the presence of ODF1 in the kidney, we use the proteomic strategy. In the targeted analysis (priority MSMS list) against the rat data base, nine peptides with 99% of confidence (High) and one peptide with 95% of confidence (medium) were detected in testis as derived from 208 PSMs (peptide spectrum matches). In non-cytoskeleton fraction kidney medulla extracts, also 2 peptides were identified at 95% of confidence as derived from three PSMs detected. In cytoskeleton fraction kidney medulla extracts, also 3 peptides were identified at 95% of confidence as derived from four PSMs detected (Table 2 and Figure 5).
Table 2.
Peptide sequence that match with ODF1 (P21769) in testis and kidney.
| Tissue | Peptide Sequence | XCorr Sequest HT |
|---|---|---|
| Testis | EFSLPPCVDEKDVTYSYGLGSCVK | 5.16 |
| SCGLCDLYYPCCLCDYK | 3.96 | |
| ENRYDCLGSK | 3.92 | |
| DVTYSYGLGSCVK | 3.79 | |
| ILASSCCSSNILGSVNVCGFEPDQVKVR | 3,4 | |
| EFSLPPCVDEK | 2.84 | |
| CIDEISSR | 2.69 | |
| LYCLRPSLR | 2.22 | |
| VCVSAER | 2.1 | |
| Kidney medulla | EFSLPPCVDEKDVTYSYGLGSCVK (C) | 1.98 |
| VCVSAERENRYDCLGSK (NC and C) | 1.27 | |
| ENRYDCLGSK (C) | 0.9 | |
| DVTYSYGLGSCVK (NC) | 0.81 |
NC: peptide found in the non-cytoskeleton fraction of kidney medulla.
C: peptide found in the cytoskeleton fraction of kidney medulla.
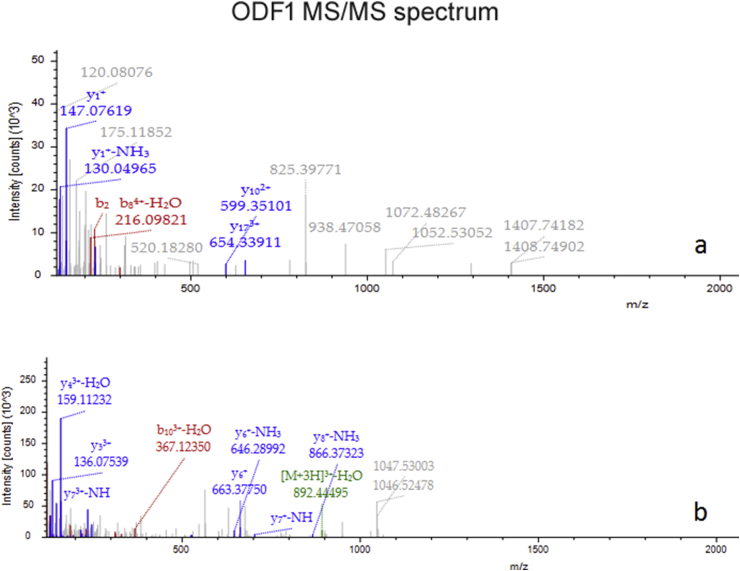
Figure 5.
ODF1 MS/MS spectrum. In (a) the spectrum of the peptide sequence: ILASSCCSSNILGSVNVCGFEPDQVKVRVK, C6-Carbamidomethyl (57.021146 Da), C7-Carbamidomethyl (57.02146 Da), C18-Carbamidomethyl (57.02146 Da). In (b) the spectrum of the peptide sequence EFSLPPCVDEKDVTYSYGLGSCVK, C22-Carbamidomethyl (57.021146 Da).
4. Discussion
ODF1 is a component of the outer dense fibers and so far had been described only as a testicular and sperm protein, but for the first time in the current work we also described using different complementary approaches its presence in male rat kidney, and specifically in the collecting ducts.
ODFs proteins are expressed together in meiotic and post-meiotic germ cells, and ODF1 and ODF2 co-localize and strongly interact via their leucine zippers during the spermiogenesis [14, 19, 20, 21]. A proteomic profiling of flagellar structures identified multiple forms of ODF proteins, in special ODF1 and ODF2 that could represent isoforms or post-translationally modified versions of either proteins [34]. The Odf2 gene encodes at least ten proteins by alternative splicing [35], and ODF2 have been localized in centrosomes, in photoreceptor primary cilia, and in basal bodies of ciliated cells in the respiratory epithelium and in the kidney ducts [36]. Despite many authors suggest that ODF1 is exclusively a testicular protein, specifically in the centrosome-derived head-tail coupling apparatus [37]and in the sperm tail [38], surprisingly in this article we describe the mRNA and the ODF1 protein expression in the kidney.
In these sense the Bgee and Uniprot KB (P21769), a database that provides a link between genes and phenotypes [38, 39], using RNA-seq as source of the information, suggests that the highest expression of Odf1 mRNA is in the testis and indicates that the mRNA corresponding to this protein also presents lower expression in skeletal muscle, brain, colon and male kidney too. But no information was reported regarding to the expression of the protein.
The experimental results presented in the current article demonstrated the expression of ODF1 in the kidney and particularly its location in medullar collecting ducts as confirmed by the co-expression with AQP2.
Even though the peptides found in the kidney have a 95% of confidence with ODF1, the detection of this protein by western blot and indirect immunofluorescence in kidney, and the detection of corresponding mRNA by RT-PCR, allowed us to validate its expression and the presence of ODF1 in the medulla kidney rat.
Many reports characterized to ODF1 as a phosphoprotein. At least, 6 phosphopeptides were described in the spermatozoa of fertile men using a phosphopeptide enrichment before proteomic analysis [39]. But when this technique was applied in other tissues, only in testis, ODF1 phosphopeptides were identified [40]. Phosphoproteome, using medulla kidney enriched samples, are necessary to determine the presence of phosphopeptide of ODF1 in kidney.
Kidney ducts and in special the collecting ducts develop a primary cilium. Spermatogenesis and ciliogenesis are processes that involve cell polarization and microtubule rearrangement [41]. They also express in common many proteins [39, 40], that are necessary in the development of cilium and flagellum and their impairment are involved in ciliopathies and infertility [41, 42].
5. Conclusion
ODF1 is a component of the outer dense fibers and had been described so far only as a testicular and sperm protein, but in the current work we also describe its expression in the kidney cells. This result is supported by 3 independent lines of evidence; by combining western blot, RT-PCR techniques and LC-MS/MS proteomic analysis. Using immunofluorescence, we also demonstrate a co-expression with AQP2. Taking into account that collecting ducts cells develop a primary cilium and that ODF1 is expressed in these cells we suggest that ODF1 is part of the cilium. This result provides further insights on the expression of ODF1 in the genesis of flagella and cilia in health and disease in different organs and therefore should stimulate further research towards a better understanding of cilia biology and perhaps new therapeutic targets.
Declarations
Author contribution statement
María Eugenia Cabrillana: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Victoria Bocanegra: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
María Ángeles Monclus, Estefania Saez Lancellotti: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Layla Simon, Abi Funes, Regina Colombo, Martín Ruiz Estrabón, Amanda Vincenti: Performed the experiments.
Rafael Oliva: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Miguel Fornes: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by the Research council of the Aconcagua University, Argentina [Grant number 065/17 to María Eugenia Cabrillana]; the National University of Cuyo, Argentina [Grant number 571/2015 to Miguel W Fornes]. University of Cuyo, Argentina [Grant number J071/2019 to María Eugenia Cabrillana].
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014848.
References
- 1.Bhullar B., Zhang Y., Junco A., Oko R., van der Hoorn F.A. Association of kinesin light chain with outer dense fibers in a microtubule-independent fashion. J. Biol. Chem. 2003;278:16159–16168. doi: 10.1074/jbc.M213126200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner R.M. Moving to the beat: a review of mammalian sperm motility regulation. Reprod. Fertil. Dev. 2006;18:25–38. doi: 10.1071/rd05120. [DOI] [PubMed] [Google Scholar]
- 3.Kierszenbaum A.L., Tres L.L. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch. Histol. Cytol. 2004;67:271–284. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- 4.Oko R.J., Clermont Y. Biogenesis of specialized cytoskeletal elements of rat spermatozoa. Ann. N. Y. Acad. Sci. 1991;637:203–223. doi: 10.1111/j.1749-6632.1991.tb27311.x. http://www.ncbi.nlm.nih.gov/pubmed/1785773 [DOI] [PubMed] [Google Scholar]
- 5.Oko R. Comparative analysis of proteins from the fibrous sheath and outer dense fibers of rat spermatozoa. Biol. Reprod. 1988;39:169–182. doi: 10.1095/biolreprod39.1.169. http://www.ncbi.nlm.nih.gov/pubmed/3207795 [DOI] [PubMed] [Google Scholar]
- 6.Petersen C., Füzesi L., Hoyer-Fender S. Outer dense fibre proteins from human sperm tail: molecular cloning and expression analyses of two cDNA transcripts encoding proteins of approximately 70 kDa. Mol. Hum. Reprod. 1999;5:627–635. doi: 10.1093/molehr/5.7.627. http://www.ncbi.nlm.nih.gov/pubmed/10381817 [DOI] [PubMed] [Google Scholar]
- 7.Vera J.C., Brito M., Zuvic T., Burzio L.O. Polypeptide composition of rat sperm outer dense fibers. A simple procedure to isolate the fibrillar complex. J. Biol. Chem. 1984;259:5970–5977. http://www.ncbi.nlm.nih.gov/pubmed/6715381 [PubMed] [Google Scholar]
- 8.Martínez-Heredia J., Estanyol J.M., Ballescà J.L., Oliva R. Proteomic identification of human sperm proteins. Proteomics. 2006;6:4356–4369. doi: 10.1002/pmic.200600094. [DOI] [PubMed] [Google Scholar]
- 9.de Mateo S., Martínez-Heredia J., Estanyol J.M., Domínguez-Fandos D., Domíguez-Fandos D., Vidal-Taboada J.M., Ballescà J.L., Oliva R. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics. 2007;7:4264–4277. doi: 10.1002/pmic.200700521. [DOI] [PubMed] [Google Scholar]
- 10.Amaral A., Castillo J., Estanyol J.M., Ballescà J.L., Ramalho-Santos J., Oliva R. Human sperm tail proteome suggests new endogenous metabolic pathways. Mol. Cell. Proteomics. 2013;12:330–342. doi: 10.1074/mcp.M112.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amaral A., Castillo J., Ramalho-Santos J., Oliva R. The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Hum. Reprod. Update. 2014;20:40–62. doi: 10.1093/humupd/dmt046. [DOI] [PubMed] [Google Scholar]
- 12.Baltz J.M., Williams P.O., Cone R.A. Dense fibers protect mammalian sperm against damage. Biol. Reprod. 1990;43:485–491. doi: 10.1095/biolreprod43.3.485. http://www.ncbi.nlm.nih.gov/pubmed/2271730 [DOI] [PubMed] [Google Scholar]
- 13.Lindemann C.B. Functional significance of the outer dense fibers of mammalian sperm examined by computer simulations with the geometric clutch model. Cell Motil. Cytoskeleton. 1996;34:258–270. doi: 10.1002/(SICI)1097-0169(1996)34:4<258::AID-CM1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Shao X., Tarnasky H.A., Schalles U., Oko R., van der Hoorn F.A. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. Leucine zippers mediate associations of Odf84 and Odf27. J. Biol. Chem. 1997;272:6105–6113. doi: 10.1074/jbc.272.10.6105. http://www.ncbi.nlm.nih.gov/pubmed/9045620 [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald C.J., Oko R.J., van der Hoorn F.A. Rat Spag5 associates in somatic cells with endoplasmic reticulum and microtubules but in spermatozoa with outer dense fibers. Mol. Reprod. Dev. 2006;73:92–100. doi: 10.1002/mrd.20388. [DOI] [PubMed] [Google Scholar]
- 16.Cabrillana M.E., Monclus M.A., Sáez Lancellotti T.E., V Boarelli P., Clementi M.A., Vincenti A.E., Yunes R.F.M., Fornés M.W. Characterization of flagellar cysteine-rich sperm proteins involved in motility, by the combination of cellular fractionation, fluorescence detection, and mass spectrometry analysis. Cytoskeleton (Hoboken) 2011;68:491–500. doi: 10.1002/cm.20525. [DOI] [PubMed] [Google Scholar]
- 17.Cabrillana M.E., Monclus M. de L.Á., Lancellotti T.E.S., Boarelli P.V., Vincenti A.E., Fornés M.M., Sanabria E.A., Fornés M.W. Thiols of flagellar proteins are essential for progressive motility in human spermatozoa. Reprod. Fertil. Dev. 2017;29:1435–1446. doi: 10.1071/RD16225. [DOI] [PubMed] [Google Scholar]
- 18.Yang K., Grzmil P., Meinhardt A., Hoyer-Fender S. Haplo-deficiency of ODF1/HSPB10 in mouse sperm causes relaxation of head-to-tail linkage. Reproduction. 2014;148:499–506. doi: 10.1530/REP-14-0370. [DOI] [PubMed] [Google Scholar]
- 19.Yang K., Meinhardt A., Zhang B., Grzmil P., Adham I.M., Hoyer-Fender S. The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Mol. Cell. Biol. 2012;32:216–225. doi: 10.1128/MCB.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao X., Murthy S., Demetrick D.J., van der Hoorn F.A. Human outer dense fiber gene, ODF2, localizes to chromosome 9q34. Cytogenet. Cell Genet. 1998;83:221–223. doi: 10.1159/000015183. [DOI] [PubMed] [Google Scholar]
- 21.Hoyer-Fender S., Petersen C., Brohmann H., Rhee K., Wolgemuth D.J. Mouse Odf2 cDNAs consist of evolutionary conserved as well as highly variable sequences and encode outer dense fiber proteins of the sperm tail. Mol. Reprod. Dev. 1998;51:167–175. doi: 10.1002/(SICI)1098-2795(199810)51:2<167::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 22.Turner K.J., Sharpe R.M., Gaughan J., Millar M.R., Foster P.M., Saunders P.T. Expression cloning of a rat testicular transcript abundant in germ cells, which contains two leucine zipper motifs. Biol. Reprod. 1997;57:1223–1232. doi: 10.1095/biolreprod57.5.1223. http://www.ncbi.nlm.nih.gov/pubmed/9369191 [DOI] [PubMed] [Google Scholar]
- 23.Tateishi K., Yamazaki Y., Nishida T., Watanabe S., Kunimoto K., Ishikawa H., Tsukita S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J. Cell Biol. 2013;203:417–425. doi: 10.1083/jcb.201303071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastian F., Parmentier G., Roux J., Moretti S., Laudet V., Robinson-Rechavi M. Bgee: integrating and comparing heterogeneous transcriptome data among species. In: Bairoch A., Cohen-Boulakia S., Froidevaux C., editors. Data Integr. Life Sci. Springer Berlin Heidelberg; Berlin, Heidelberg: 2008. pp. 124–131. [Google Scholar]
- 25.UniProt A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berrebi-Bertrand I., Maixent J.M. Immunodetection and enzymatic characterization of the alpha 3-isoform of Na,K-ATPase in dog heart. FEBS Lett. 1994;348:55–60. doi: 10.1016/0014-5793(94)00550-8. http://www.ncbi.nlm.nih.gov/pubmed/8026584 [DOI] [PubMed] [Google Scholar]
- 27.Ruete M.C., Carrizo L.C., Vallés P.G. Na+/K+ -ATPase stabilization by Hsp70 in the outer stripe of the outer medulla in rats during recovery from a low-protein diet. Cell Stress Chaperones. 2008;13:157–167. doi: 10.1007/s12192-008-0021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown R.E., Jarvis K.L., Hyland K.J. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal. Biochem. 1989;180:136–139. doi: 10.1016/0003-2697(89)90101-2. http://www.ncbi.nlm.nih.gov/pubmed/2817336 [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. http://www.ncbi.nlm.nih.gov/pubmed/5432063 [DOI] [PubMed] [Google Scholar]
- 30.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biotechnology. 1979;24:145–149. http://www.ncbi.nlm.nih.gov/pubmed/1422008 1992. [PubMed] [Google Scholar]
- 31.Gill-Sharma M.K., Choudhuri J., Ansari M.A., D’Souza S. Putative molecular mechanism underlying sperm chromatin remodelling is regulated by reproductive hormones. Clin. Epigenet. 2012;4:23. doi: 10.1186/1868-7083-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suvarna K.S., Bancroft J.D., Layton C. Elsevier Health Sciences; 2012. Bancroft’s Theory and Practice of Histological Techniques. [Google Scholar]
- 33.Yasset Perez-Riverol J.A.V., Csordas Attila, Bai Jingwen, Bernal-Llinares Manuel, Suresh Hewapathirana, Kundu Deepti J., Inuganti Avinash, Griss Johannes, Mayer Gerhard, Eisenacher Martin, Pérez Enrique, Uszkoreit Julian, Pfeuffer Julianus, Sachsenberg Timo, Şule The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;8 doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao W., Gerton G.L., Moss S.B. Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol. Cell. Proteomics. 2006;5:801–810. doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Chang J., Seo S.G., Lee K.H., Nagashima K., Bang J.K., Kim B.Y., Erikson R.L., Lee K.-W., Lee H.J., Park J.-E., Lee K.S. Essential role of Cenexin1, but not Odf2, in ciliogenesis. Cell Cycle. 2013;12:655–662. doi: 10.4161/cc.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa H., Kubo A., Tsukita S., Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 37.Higgy N.A., Pastoor T., Renz C., Tarnasky H.A., Van der Hoorn F.A. Testis-specific RT7 protein localizes to the sperm tail and associates with itself. Biol. Reprod. 1994;50:1357–1366. doi: 10.1095/biolreprod50.6.1357. http://www.ncbi.nlm.nih.gov/pubmed/7521678 [DOI] [PubMed] [Google Scholar]
- 38.Morales C.R., Oko R., Clermont Y. Molecular cloning and developmental expression of an mRNA encoding the 27 kDa outer dense fiber protein of rat spermatozoa. Mol. Reprod. Dev. 1994;37:229–240. doi: 10.1002/mrd.1080370215. [DOI] [PubMed] [Google Scholar]
- 39.Hetherington L., Schneider E.K., Scott C., DeKretser D., Muller C.H., Hondermarck H., Velkov T., Baker M.A. Deficiency in outer dense fiber 1 is a marker and potential driver of idiopathic male infertility. Mol. Cell. Proteomics. 2016;15:3685–3693. doi: 10.1074/mcp.M116.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundby A., Secher A., Lage K., Nordsborg N.B., Dmytriyev A., Lundby C., Olsen J.V. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 2012;3:876. doi: 10.1038/ncomms1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperry A.O. The dynamic cytoskeleton of the developing male germ cell. Biol. Cell. 2012;104:297–305. doi: 10.1111/boc.201100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Li W., Zhang Y., Zhang L., Teves M.E., Liu H., Strauss J.F., Pazour G.J., Foster J.A., Hess R.A., Zhang Z. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol. Biol. Cell. 2016 doi: 10.1091/mbc.E16-05-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]