Abstract
Sugarcane is one of the most sustainable energy crops among cultivated crops presenting the highest tonnage of cultivated plants. Its high productivity of sugar, bioethanol and bioelectricity make it a promising green alternative to petroleum. Furthermore, the myriad of products that can be derived from sugarcane biomass has been driving breeding programs towards varieties with a higher yield of fiber and a more vigorous and sustainable performance: the energy cane. Here we provide an overview of the energy cane including plant description, breeding efforts, types, and end-uses. In addition, we describe recently published genomic resources for the development of this crop, discuss current knowledge of cell wall metabolism, bioinformatic tools and databases available for the community.
Keywords: Biomass, Bioenergy, Biofuels, Bioproducts, Genomics, Grasses
1. Introduction
Sugarcane (Saccharum spp.) is a perennial, tropical or subtropical non-cereal C4 grass; the major crop for food and bioenergy production and the highest tonnage crop in the world [1]. Mainly grown for sugar production in the tropical and subtropical regions of the world, sugarcane has one of the highest solar energy conversion efficiency and is a crop with one of the highest biomass yields [2], [3], [4]. Due to the high yields of both sugar and lignocellulosic biomass it is considered an important feedstock for substituting fossil fuel energy, either as natural biomass or transformed into liquid or gaseous forms [5]. An additional advantage is that this semi-perennial grass allows harvesting for several years (4 to 5 years) without the need for replanting, which reduces the cost of bioenergy production [6]. The development of conversion processes that use all plant carbon in a Biorefineries approach [7], [8] has stimulated the development of plants with several co-products for different applications, including sugars, biofuels and bioelectricity. In this scenario, breeders are designing crosses towards the development and improvement of energy cane, a new type of cane containing a high yield of fiber.
Over the last century, there was a great effort in breeding programs and conventional agricultural research to increase the yield of sugarcane and sugar to reach the current levels [9]. In addition, research groups around the world, have generated a large amount of molecular and physiological data on sugarcane. In addition, sugarcane geneticists have invested significant effort to explore and dissect the complex genome of cane using a wide range of genomic tools. These combined factors make data integration a key step to achieve a broader understanding of cane physiology and its interaction with the environment and climate changes as well as to help the design of more productive varieties.
In this mini review, we focus on bioenergy and energy cane and provide an overview of the current genomic resources and databases for the development of this crop. It is important to note that the theoretical potential of sugarcane dry biomass production is 177 t/(ha yr) or a fresh weight cane yield of 381 t/(ha yr) [10]. Worldwide sugarcane yield averages around 39 and 84 t/(ha yr), respectively. There is great interest and opportunity to decrease this yield gap.
2. The Saccharum complex
Sugarcane is the world’s leading biomass crop, produced in over 100 countries [1]. Modern sugarcane cultivars are polyploid interspecific hybrids, typically with 10–13 sets of their 10 basic chromosomes, 80–85% of Saccharum officinarum (2n = 80), 10–15% of S. spontaneum (2n = 40–128) and ~5% with recombined chromosomes between these two ancestors [11], [12]. Gene duplications as a result of polyploidization alter the transcriptional landscape [13] and provide additional flexibility to adapt and evolve new patterns of gene expression for homo(eo)logous gene copies [14]. This flexibility has been suggested to be an important mechanism allowing the diversification of adaptive traits [15], [16] through neofunctionalization of duplicated genes [17] and tissue-specific expression [18].
The high productivity cane makes this crop an excellent source of sugar, bioethanol and bioelectricity [19] and a promising green alternative to petroleum [20], [21], [22] with vast potential to mitigate climate change without affecting food security [23]. Additionally, the myriad of products that can be derived from sugarcane biomass [24], such cellulosic bioethanol, further enhance opportunities for sugarcane in a portfolio of technologies needed to transition to a low carbon ‘bioeconomy’. In this scenario, hybrids obtained through the cross-breeding of commercial varieties of sugarcane with ancestral species, such as S. spontaneum, have allowed the production of genotypes characterized by high fiber content, moderate brix levels, fine stalks and higher tillering rate – the energy cane [25], [26], [27].
3. The energy cane
Energy cane is an ideal type of sugarcane with high yield of fiber, more vigorous and rustic, i.e. these plants are less demanding in soil, climate, water and nutrients, more resistant to pests and diseases, which brings a series of economic and environmental advantages [25], [28], [29]. Efforts to develop this crop include interspecific hybridization between modern sugarcane varieties and other closely related wild species, such as S. spontaneum, which has the greatest potential as a source of genetic variation for a number of important traits for bioenergy production and low-input adaptability [30].
Initially, Tew and Cobil [31] classified energy cane hybrids into two categories: one (Type I) defined as a cane closer to the conventional sugarcane regarding sucrose content but higher fiber content; and other (Type II) with only marginal content of sugar but with fiber content higher than Type I, used exclusively for biomass production. More recently, Kumar et al. [32] proposed the classification of cane varieties considering variation in sucrose and fiber content. According to the authors, cane type I includes traditional commercial sugarcane varieties, with high sugar and commercial yield (13% sucrose and 12% fiber content); cane type II also comprises varieties with high sugar and commercial yield, but with an increase in fiber content (13% sucrose content and >14% fiber content); cane type III (energy cane) includes varieties for multiple use purposes, focusing on high biomass production (sucrose content <12% and >22% fiber content); and cane type IV (energy cane) embraces varieties for energy cogeneration purposes (sucrose content <5% and fiber >22%).
Compared to sugarcane commercial varieties, energy canes have higher ratooning ability and number of tillers [27], [29], [30], [33], which combined are very important in defining total biomass yield (Fig. 1). Because this crop is “vegetative propagation based”, this characteristic is also important in overcoming one of the most significant economic constraints in the cane cultivation: clonal multiplication [27]. In addition, there is also a profound difference regarding the root systems; energy cane produced an abundant and vigorous root system, surpassing the conventional cane in lateral extension, depth and volume. This trait, shared with the S. spontaneum progenitor, which is considered an invasive weed in some countries [34], allows its cultivation in marginal lands because it gives grater rusticity, helps mitigate soil erosion, boost permanent carbon sequestration and extends the crop life cycle up to 10 years; this is an important attribute due to the high cost of replanting sugarcane [27], [30].
Fig. 1.
An energy cane RB hybrid and a sugarcane variety (SP791011) at six months after planting, under field conditions at the experimental site ‘Estação de Floração e Cruzamento da Serra do Ouro’ (lat 9° 13′ S, long 35° 50′ W, alt 450 m asl) in Alagoas, Brazil.
The energy cane breeding initiatives began in Puerto Rico, with a pioneering commercial project established to conduct an integrated exploration of sugarcane as a biomass feedstock for multiple products, instead of only sugar [35]. The growing interest in bioenergy in recent decades pushed several sugarcane breeding programs world-wide to also produce energy cane commercial varieties. In the United States, the Cultivar L 79-1002 (‘CP 52-68’ x Tainan, S. spontaneum clone) was developed by the Louisiana State University Agricultural Center in cooperation with the USDA-ARS and the American Sugarcane League, Inc. This cultivar has high biomass yield and fiber content, on average of 257 g kg−1 [36]. The breeding program in Barbados has vigorous canes with exceptional fiber content (>30%), which are suitable for energy cogeneration [37]. Mauritius developed a similar program, aiming the increasing of biomass and fiber yield [38]. Other genetic improvement initiatives have been conducted in Australia [39], [40], Colombia [41], Japan [42], [43], [44], and Thailand [45]. Further details about breeding programs using S. spontaneum as an integral part of their activities can be found in Matsuoka et al. [27] and da Silva [30].
In Brazil, energy cane hyrids were obtained by Canavialis, a private sugarcane breeding company [27]. In 2011, an initiative was launched to create the first biorefinery in South America to produce cellulosic ethanol from sugarcane residues and energy cane genotypes, the Brazilian Group GranBio, an innovation industry for ethanol generation through biomass conversion [46]. New varieties have been developed by RIDESA (Inter-University Network for the Development of Sugarcane Industry) at the experimental site ‘Estação de Floração e Cruzamento da Serra do Ouro’ (lat 9° 13′ S, long 35° 50′ W, alt 450 m asl), Federal University of Alagoas (UFAL), Brazil. This institution maintains an important collection of sugarcane germplasm, which holds modern hybrids and a myriad of Saccharum, Erianthus and Miscanthus accessions. In a study conducted by UFAL/RIDESA researchers, six energy cane clones were selected which presented an overall average of 24.7% higher yield of dry biomass/ha as compared to the standard sugarcane variety (RB0442) (Table 1). Plants were grown under field conditions at São Miguel dos Campos – Alagoas – Brazil; and traits were measured at 13 months after planting and in the second harvest year.
Table 1.
Comparison of yield related traits between six RB energy cane clones and a sugarcane standard variety (RB0442).
| Value |
Traits |
||||||
|---|---|---|---|---|---|---|---|
| FB | DB | Fiber (%) | TFH | TRS | TSH | ||
| Top 6 RB energy cane clones | Min | 131.8 | 43.2 | 20.2 | 26.6 | 51 | 10.6 |
| Average | 137.6 | 46.4 | 22.7 | 31.2 | 67.8 | 12.7 | |
| Max | 153.3 | 50.1 | 24.1 | 33.9 | 96.2 | 15.1 | |
| RB0442 | Average | 126.5 | 37.2 | 13.8 | 17.4 | 122.1 | 18 |
| RB clones vs RB0442 | % | 8.8 | 24.7 | 64.5 | 79.3 | −44.5 | −29.4 |
| CV | % | 20.94 | 21.12 | 16.97 | 25.00 | 29.01 | 28.49 |
CV = coefficient of variation; FB = fresh biomass (t/ha); DB = dry biomass (t/ha); TFH = fiber (t/ha); TRS = Total Recoverable Sugar (%); TSH = sugar (t/ha)
There are three major problems in the selection of energy cane clones: high incidence of smut disease (Sporisorium scitamineum), high flowering rates and low unit stem mass. Thus, the major challenge of genetic breeding for the commercial cultivation and consolidation of energy cane cultivars is to use more effective strategies to overcome these three problems. In addition, there are other technological (agro-industrial) bottlenecks, such as: (i) in mechanized harvesting, development of harvesting machines for high biomass and high fiber cultivars; and (ii) in industrial processing, improve the efficiency of grinding for broth extraction (Geraldo Barbosa, UFAL, personal communication).
4. Sugarcane genomic resources
As mentioned before, modern sugarcane cultivars are polyploid interspecific hybrids and have a large (~10 Gb) and complex genome. Nevertheless, opportunities to accelerate breeding progress and enrich the knowledge of the fundamental biology of this important crop drive efforts to explore and dissect its complex genome using different genomic tools and to develop a high-quality reference genome.
After over a decade of multiple parallel genome sequencing initiatives [47], [48], Garsmeur et al. published the first mosaic monoploid genome reference of the modern cultivar R570 [49]. This study relies on 4535 bacterial artificial chromosomes (BACs) sequences that were colinear to the gene-rich portion of the sorghum genome (used as a reference). The final assembly consisted of 382 Mb of high-quality sequence in 3965 contigs, organized as a single tiling path, representing the single copy sugarcane gene space, and includes 25,316 predicted protein-coding gene models.
The next milestone was the publication of the first allele-defined genome reference of a tetraploid S. spontaneum genotype (AP85-441) [50]. To this end, authors took advantage of multiple sequencing technologies including high-throughput chromatin conformation capture (Hi-C) to assemble 32 pseudo-chromosomes (2.9 Gbp) comprising 8 homologous groups of 4 members each, bearing 35,525 genes with alleles defined. Subsequently, Nascimento et al. [51] published a new S. spontaneum gene space reference (from accession US851008), including 39,234 genes.
Recently, our group completed the assembly (4.26 Gb) of the Brazilian modern cultivar SP80-3280 [52], which includes the complete sequence of 373,869 genes and their upstream regions which may be further explored to identify regulatory promoter elements. This is the largest genomic data set available for the sugarcane researchers’ community and includes putative homo(eo)logs (mostly 2–5 copies) for a large fraction of the SP80-3280 gene space [52].
To date, most of the sugarcane RNA-seq initiatives are based on de novo transcriptome assembly [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69]. Nevertheless, a few studies also consider the closely related species Sorghum bicolor [70], the S. officinarum gene indices (SoGI) v3.0 [71] or the previously de novo assembled transcriptome [72] as a reference for transcriptome analysis. Further transcriptome studies are expected to take advantage of the multiple reference genomes now available.
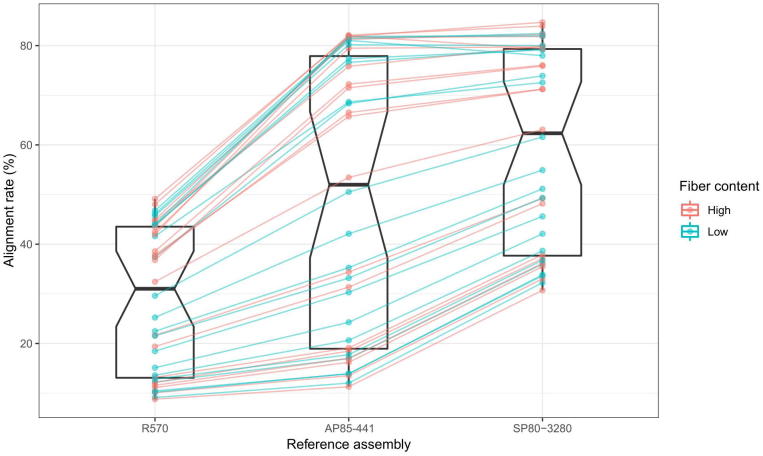
To investigate the value of the three largest public assemblies as a genomic resource, we used RNA-seq data from Kasirajan et al. [71]. These authors created RNA-seq libraries from top and bottom internodes of 20 different genotypes, including commercial cultivars and introgression lines derived from crosses with wild S. spontaneum relatives and Erianthus. We used HISAT2 [73] with default parameters to align these RNA-seq reads against the monoploid R570 hybrid assembly [49], the AP85-441 tetraploid S. spontaneum reference [50] and the SP80-3280 hybrid gene space assembly [52].
For the vast majority of sequenced samples, the SP80-3280 assembly resulted in higher alignment rates than those against the monoploid reference and the S. spontaneum assembly (Fig. 2). Only in the very high end of alignment rates did the AP85-441 assembly perform better for three of the 40 samples. These results show that these genome assemblies, and in particular the SP80-3280, can be used as a reference for downstream genomic studies. Combining both de novo and reference-guided transcriptome approaches, especially by taking advantage of multiple genome references, may allow the understanding of the extent to which homo(eo)logs resemble or differ from each other in their expression patterns, the spatiotemporal dynamics of these relationships, and how epistatic interactions between individual homo(eo)logs affect biological traits.
Fig. 2.
Alignment rates of RNA-seq libraries from the top and bottom internodes of 20 different genotypes [71], contrasting for fiber content, against the monoploid R570 hybrid assembly [49], the AP85-441 tetraploid S. spontaneum genome reference [50] and the SP80-3280 hybrid gene space assembly [52].
5. Linking genomic data to biomass improvement by exploring the plant cell wall metabolism
Plant biomass is composed mainly by secondary cell walls (SCW) and, consequently, achieving tailor-made biomass for bioenergy could benefit from a detailed understanding of SCW biosynthesis. The potential use of currently available sugarcane genomic resources for identifying classes of genes involved in SCW biosynthesis will help to shed light on these aspects. Table 2 indicates the number of cell wall-related genes found in different Saccharum genomic resources as well as transcription factors (TFs) from the two main families, NAC and MYB, involved in the gene regulatory network (GRN) controlling SCW biosynthesis.
Table 2.
Number of cell wall-related genes and NAC and MYB transcription factors identified in different Saccharum ssp. genomic databases: the R570 monoploid genome reference (R570) [49]; the allele-define genome reference of S. spontaneum (AP85-441) [50]; the SP80-3280 gene-space assembly; and the SAS (Sugarcane Assembled Sequences) from the SUCEST Project (SUCEST) [94].
| Gene class | R570 | AP85-441 | SP80-3280 | SUCEST |
|---|---|---|---|---|
| Cell Wall Differentiation | 83 | 386 | 742 | 175 |
| Cell Wall Growth/Extension | 54 | 219 | 434 | 58 |
| Lignin metabolism | 31 | 168 | 541 | 53 |
| Other Glycan Degradation | 117 | 456 | 870 | 177 |
| Phenylpropanoid biosynthesis | 58 | 311 | 511 | 123 |
| Polysaccharide biosynthesis | 40 | 220 | 475 | 112 |
| Structural proteins | 15 | 68 | 103 | 27 |
| Unknown | 1 | 7 | 7 | 5 |
| MYB | 178 | 999 | 613 | 120 |
| NAC | 101 | 412 | 884 | 63 |
The SCW-GRN has been elucidated in Arabidopsis and it is comprised by a three-layered structure. At the top level, TFs from the NAC family, named VASCULAR-RELATED NAC-DOMAIN (VNDs) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR (NSTs) genes, act as master switches activating cell differentiation, including programmed cell death to form tracheary elements, and SCW deposition in vessels and fibers [74], [75], [76]. They activate a second layer of master switches, comprised by TFs from the MYB family (AtMYB46 [77] and AtMYB83 [78] in Arabidopsis). These second level TFs activate biosynthetic genes of cellulose, hemicellulose (xylan) and lignin and a third layer of TFs, turning on SCW deposition. These downstream TFs activate other aspects of SCW deposition, with some redundancies, and includes transcriptional repressors of NAC master switches, such as AtMYB32 [79], establishing a negative feedback loop.
Besides this three-layer core structure and key players of GRN being conserved among vascular plants studied so far [80], [81], including grasses like rice, maize, brachypodium, sorghum, miscanthus and switchgrass, considerable divergence of transcription factors target diversities has been reported [82], [83], [84], [85], [86], [87], [88], [89], [90], [91]. Even among grasses some divergences of TF target repertoires may exist, such as in MYB SCW repressors orthologs [92]. Within the R570, SP80-3280, AP85 and SUCEST SAS (see below) databases (Table 2), we have found up to 884 and 999 NAC and MYB genes, respectively, which are potential targets for further exploration. For the NAC family, this number is 8-11x higher than in rice (105 genes) and Arabidopsis thaliana (75 genes) genomes [93], putting in perspective all the intricacy and diversity found in the Saccharum complex.
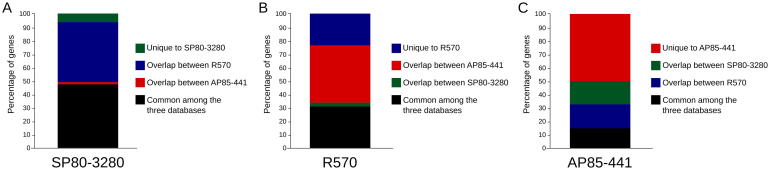
By comparing the overlap of genes among the three references, we can estimate how these datasets can complement each other. In the transcription factor MYB family, 15–50% of these genes is common to all three genotypes (Fig. 3), suggesting that all three databases have their particularities, given the fact that they are derived from different species (S. spontaneum) or varieties (R570 and SP80-3280). This is evident for AP85-441 (S. spontaneum) which reflects in approximately 50% of MYB genes not overlapping to sugarcane varieties (Fig. 3C).
Fig. 3.
Overlap of MYB genes among the three sugarcane genomic assemblies (datasets). All genes classified as MYBs (Table 2) were used for reciprocal blastp analysis among all three datasets. A, overlap of SP80-3280 with the other two databases; B, overlap of R570 with the other two databases; C, overlap of AP85-441 with the other two databases. Genes with coverage and identity >=90% were considered “overlapping genes”.
As lignin is one of the main causes of biomass recalcitrance, hampering lignocellulosic biofuels production [95], [96], [97], [98], [99], much effort has been made to understand its biosynthesis and polymerization and how to engineer it. Lignin is a phenolic polymer composed of three main units, p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S), crosslinked to hemicellulose providing strength and rigidity to the cell wall [100]. Although known for years [100], additional insight into the phenylpropanoid pathway and lignin synthesis have occurred recently. For example, the Caffeoyl-shikimate esterase (CSE) [101], the bifunctional phenylalanine/tyrosine ammonia lyase (PTAL) [102] and a bifunctional cytosolic ascorbate peroxidase functioning as C3H [103] are missing links in the phenylpropanoid pathway that have only recently been discovered. There are ~511 genes in the phenylpropanoid pathway of SP80-3280 (Table 2), whereas other species like Arabidopsis thaliana and the model C4 grass Setaria viridis have only 26 [104] and 56 [105], respectively. Gaps in our knowledge include how monolignols, the lignin monomers, are transported to the apoplast for polymerization [106]. To date, only one transporter has been characterized, AtABCG29, which is responsible for transporting the precursor of p-hydroxyphenyl (H-units), a minor component of lignin, but other routes should exist since loss-of-function mutations in AtABCG29 reduce H-units, but do not eliminate it [107].
The majority of studies about plant cell wall metabolism are derived from dicot model plants, particularly Arabidopsis. However, grasses and dicots diverged ~150 million years ago; therefore, considerable differences in their vascular and morpho-anatomic patterns and cell wall structure and composition have emerged. Grasses have several distinct features from eudicots cell walls [108], with different abundance of pectin, structural proteins and phenolic compounds and also hemicellulose structure and composition [109]. Eudicots have only traces of H-units and low levels of other phenolic compounds in their cell walls, whereas grasses have significant amounts of H-units and increased levels of hydroxycinammic acids [110], especially ferulic and p-coumaric acids esterified to arabinoxylan [108], [111], [112] and ferulate-monolignol conjugates incorporated to lignin [113]. Furthermore, the flavonoid tricin was discovered in monocot lignin [114], [115], acting as a nucleation site for lignification.
Grasses have arabinoxylan as the main hemicellulose, but eudicots do not have arabinosyl substitution in secondary wall xylan, which affects how lignin is crosslinked to hemicellulose [116]. Moreover, mixed-linkage glucan is a monocot-specific hemicellulose, due to the absence in eudicots of the genes responsible for its biosynthesis, cellulose synthase-like F and H (CslF e CslH) [117], [118], [119]. Also absent in eudicots, the bifunctional PTAL can use tyrosine as well as phenylalanine as substrate in the first step of phenylpropanoid pathway yielding 4-coumarate, thus bypassing the reaction catalyzed by cinnamate 4-hydroxylase (C4H), giving plasticity to the metabolism [102]. Furthermore, a transcription factor from the MYB family (BdSWAM1) was recently reported as SCW biosynthesis regulator in Brachypodium distachyon, although its clade is not found in the Brassicaceae family [120], which includes Arabidopsis. On the other hand, CSE, an essential enzyme in eudicot phenylpropanoid pathway whose down-regulation improves biomass saccharification, does not have a bona-fide ortholog identified in grasses so far [101], [121], [122]. Given all these differences, it is expected that considerable genetic divergence may be found and much of the knowledge from dicot cell wall cannot be extrapolated to grasses. Therefore, these differences can only be uncovered by studying grass functional genomics.
How SCW biosynthesis is connected to plant growth and biomass accumulation is still less understood. However, unknown factors linking these two processes may exist [123]. For example, one of these factors could be the transcriptional regulatory Mediator complex, since it has been reported to directly control lignin biosynthesis and its disruption rescues dwarfing phenotype in Arabidopsis lignin-deficient mutants [124], [125]. Such interactions are completely unknown for grasses and may be species-specific, raising the need to study crop plant omics [123]. Categorizing these hidden molecular hubs linking SCW biosynthesis to plant growth and other physiological processes are crucial to move forward in developing novel biotechnological strategies to improve plant biomass [123].
Identifying genes of interest, addressing grass specificities and finding the missing links to improve biomass accumulation and quality is a major challenge, especially in a complex genome species such as sugarcane. We expect that the sugarcane researchers can take advantage of genomic databases such as the ones described here to explore cell wall related-genes (Table 2), for example, thus helping to advance sugarcane functional genomics and giving new opportunities for molecular breeding to achieve and improve energy canes.
6. SUCEST-FUN Database: A platform for sugarcane data integration in a genomic context
In addition to the recent genome assemblies, plant genomic databases such as GRASSIUS [126], TropGENE [127], Phytozome [128], Plant TF database [129], MOROKOSHI [130], KBase [131], Gramene [132], PLAZA [133] and Plant GDB [134], are important foundations for molecular breeders to mine candidate genes and to facilitate molecular crop breeding.
Specially for sugarcane and energy cane breeders, the SUCEST-FUN Platform (http://sucest-fun.org/) [135] was developed to allow data analysis on five main aspects: i) gene annotation; ii) gene expression; iii) integration of public resources; iv) sequencing projects; and v) functional genomics. The database was initially based on 43,141 SAS (Sugarcane Assembled Sequences) from the SUCEST Project [94] and subsequently the 17,500 ORFeome genes generated using RNA-seq of sugarcane ancestral and hybrid varieties [69], which are useful for protein characterization, single nucleotide polymorphism analysis, splicing variants identification, evolutionary and comparative studies.
An important advantage of the SUCEST-FUN Database is the in-depth automatic and manual annotation conducted by our group and the definition of curated catalogs of transcription factors, cell wall genes, signal transduction genes (including kinases and phosphatases), KEGG metabolic pathways and enzymes, transposable elements, as well as orthologous gene analysis among grasses. For gene expression studies, the SUCEST-FUN Database supports three microarray platforms, including: (i) the Signal Transduction (SUCAST) array, composed by 1900 genes with 152 hybridizations; (ii) the RNA/Carbohydrate Metabolism and Signal Transduction (SUCAMET) array, composed by 4600 genes with more than 150 hybridizations; and (iii) the general regulatory function (CaneRegNet) array, composed by 14,522 genes, including sense and antisense probes with 122 hybridizations. These transcriptome studies used samples from multiple plant materials such as ancestral genotypes and commercial varieties; multiple tissues, such as leaves, internodes and roots; multiple conditions, such as field and greenhouse; and multiple treatments, such as drought stress, developmental and circadian stages, high CO2 [136], [137], [138], [139], [140]. In this scenario, these experiments, summarized in Table 3, are a valuable data source for co-expression analysis, a promising approach to unravel complex biological processes and regulatory networks, which can be extracted from available tools in the platform [141].
Table 3.
Public microarray data using the Signal Transduction (SUCAST [137]) array and general regulatory function (CaneRegNet [138]) array.
| Platform |
GEO accession number |
Experiment description | Number of hibridizations | Reference | |
|---|---|---|---|---|---|
| Platform | Series | ||||
| SUCAST | GPL3799 | GSE4966 | Phosphate starvation | 16 | [137]* |
| GSE4967 | Response to herbivory by Diatraea saccharalis | 8 | * | ||
| GSE4968 | ABA treatment | 16 | * | ||
| GSE4969 | MeJa treatment | 12 | * | ||
| GSE4970 | Response to N2-fixing endophytic bacteria association | 8 | * | ||
| GSE4971 | Drought response | 12 | * | ||
| GSE14732 | Sucrose content relate to drought and cell wall metabolism | 80 | * | ||
| CaneRegNet | GPL14862 | GSE33574 | Drought | 6 | [138] |
| GSE42725 | Circadian rhythms | 22 | [139] | ||
| GSE87826 | Sugarcane vs Leifsonia xyli subsp. xyli. | 4 | [142] | ||
| GSE124990 | SP80-3280 growth and maturation | 30 | [52] | ||
| Sugarcane Ancestral | 36 | [141] | |||
| GPL22278 | GPL22278 | Ethephon- and AVG-induced transcriptional changes | 24 | [143] | |
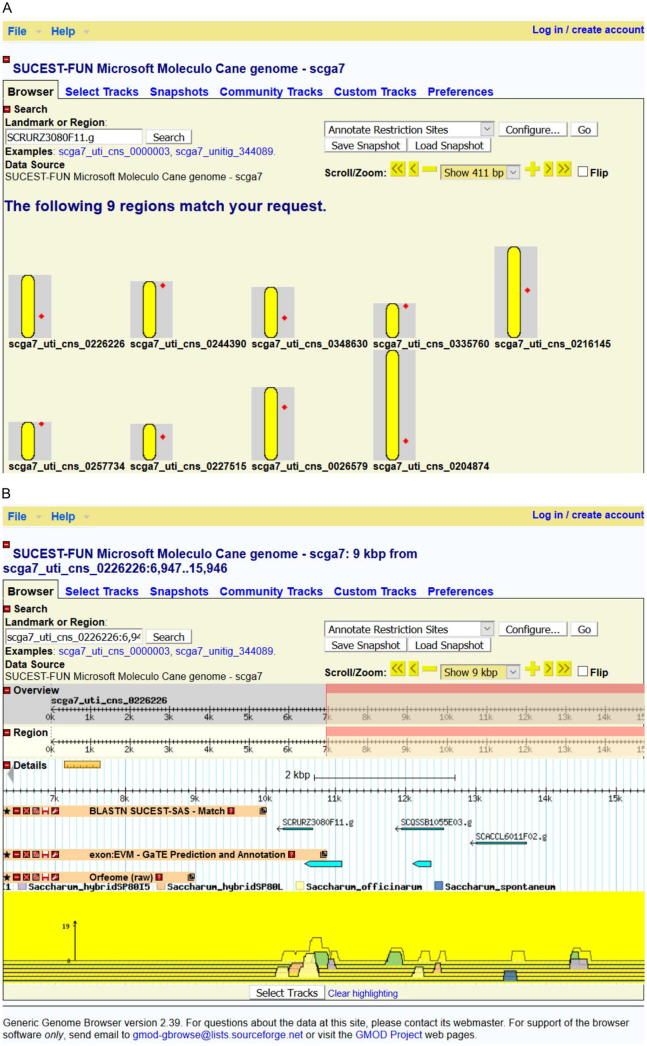
At the genome level, the annotation of public Sugarcane BAC sequences [47] and availability of a genome browser (available at http://sucest-fun.org/cgi-bin/cane_regnet/gbrowse2/gbrowse/microsoft_genome_moleculo_scga7/) with the gene space assembly of SP80-3280 polyploid cultivar [52] enables the survey of sequences and annotation in a global and dynamic way. For instance, we present one example of how we can explore this genomic tool in Fig. 4. Using the ‘SCRURZ3080F11.g’ SAS ID we searched for SP80-3280 contigs holding this transcript sequence (annotated as a MYB transcription factor). As a result, we found unique matches to nine different contigs, which may represent putative homo(eo)logs for this gene (Fig. 4A). In addition, we further present the genomic features of one of these contigs (scga7_uti_cns_0226226), such as SAS [94] matches, predicted genes [52] and RNA-seq [69] alignment results (Fig. 4B).
Fig. 4.
A view of the SUCEST-FUN genome browser, available at http://sucest-fun.org/cgi-bin/cane_regnet/gbrowse2/gbrowse/microsoft_genome_moleculo_scga7/. A: Screen shot of the result of searching for the ‘SCRURZ3080F11.g’ SAS (Sugarcane Assembled Sequence) derived from the SUCEST Project [94]. This SAS is annotated as a MYB transcription factor and has 9 matches (putative homo(eo)logs) in the SP80-3280 gene space. Yellow bars represent contigs and red diamonds indicate match position. B: Screen shot of the result of searching for the ‘scga7_uti_cns_0226226’ contig, which contains ‘SCRURZ3080F11.g’. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
7. Final considerations
Increasing plant yields is one of the greatest challenges of biotechnology. Computational tools that link genome sequences, their functions and possible attributes useful for breeding are greatly needed to speed up the process of improving sugarcane and the energy cane. Plant yields are directly impacted by lignocellulosic metabolism. We give an overview of the main genes involved and their regulators. Considering the size and complexity of cane genomes, the fact that many species have been used in breeding and the polyploidization that arose, datamining for genes of interest is a significant bioinformatics challenge for this crop. The SUCEST-FUN Platform comprises a robust infrastructure for storage, continuously updating, annotation, easy and controlled access and integration among the functional catalogs of the sugarcane transcriptome. Through various data-driven clustering analysis tools, crossings and enrichment analysis it allows for a systems biology approach. This will be an important resource considering progenies need to be analyzed in an integrated manner for multiple characteristics (technological, physiological, biochemical, genetic traits) for the construction of gene networks.
Author contributions
GMS conceived the topic and outline, wrote and critically revised the manuscript. ALD wrote the manuscript and consolidated authors’ contribution. MSC, JMS and GVSB provided energy cane breeding information and image. SSF wrote and critically revised the manuscript and identified cell wall-related genes and NAC and MYB transcription factors in different Saccharum genomic references. GRAM performed RNA-seq alignment to Saccharum genomic references. FTC curated the SUCEST-FUN genome browser. All authors reviewed the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was financially supported by State of São Paulo Foundation (FAPESP grant n° 2014/50921-8) under the BIOEN Program. GMS was a recipient of a CNPq Productivity Fellowship 304360/2014-7; ALD is a recipient of a FAPESP Fellowship 2017/02270-6; SSF is a recipient of a FAPESP Fellowship 2016/06917-1; GRAM is supported by the FAPESP grant 2015/22993-7; FTC was a recipient of a FAPESP Fellowship 2017/02842-0; MSC, JMS and GVSB were financially supported by RIDESA's partners sugar mills.
References
- 1.FAOSTAT. Production/Crops, Food and Agriculture Organization of the United Nations - Statistics Division 2018. http://www.fao.org/faostat/en/#home.
- 2.Henry R.J. Evaluation of plant biomass resources available for replacement of fossil oil. Plant Biotechnol J. 2010;8:288–293. doi: 10.1111/j.1467-7652.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrt C.S., Grof C.P.L., Furbank R.T. C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspectivefree access. J Integr Plant Biol. 2011;53:120–135. doi: 10.1111/j.1744-7909.2010.01023.x. [DOI] [PubMed] [Google Scholar]
- 4.Jakob K., Zhou F., Paterson A.H. Genetic improvement of C4 grasses as cellulosic biofuel feedstocks. Vitro Cell DevBiol-Plant. 2009;45:291–305. [Google Scholar]
- 5.Lam E., Shine J., Silva J.D., Lawton M., Bonos S., Calvino M. Improving sugarcane for biofuel: engineering for an even better feedstock. GCB Bioenergy. 2009;1:251–255. [Google Scholar]
- 6.Botha FC. Energy yield and cost in a sugarcane biomass system. Proceedings of the 2009 Conference of the Australian Society of Sugar Cane Technologists, 2009, p. 1–10.
- 7.Bonomi A, Cavalett O, da Cunha MP, Lima MAP. The Virtual Sugarcane Biorefinery Concept. In: Bonomi A, Cavalett O, Pereira da Cunha M, Lima MAP, editors. Virtual Biorefinery, Cham: Springer International Publishing; 2016, p. 5–11. https://doi.org/10.1007/978-3-319-26045-7_2.
- 8.Mariano A.P., Dias M.O.S., Junqueira T.L., Cunha M.P., Bonomi A., Filho R.M. Butanol production in a first-generation Brazilian sugarcane biorefinery: technical aspects and economics of greenfield projects. Bioresour Technol. 2013;135:316–323. doi: 10.1016/j.biortech.2012.09.109. [DOI] [PubMed] [Google Scholar]
- 9.Dal-Bianco M., Carneiro M.S., Hotta C.T., Chapola R.G., Hoffmann H.P., Garcia A.A.F. Sugarcane improvement: how far can we go? Curr Opin Biotechnol. 2012;23:265–270. doi: 10.1016/j.copbio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Waclawovsky A.J., Sato P.M., Lembke C.G., Moore P.H., Souza G.M. Sugarcane for bioenergy production: an assessment of yield and regulation of sucrose content. Plant Biotechnol J. 2010;8:263–276. doi: 10.1111/j.1467-7652.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- 11.D’Hont A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenetic and Genome Research. 2005;109:27–33. doi: 10.1159/000082378. [DOI] [PubMed] [Google Scholar]
- 12.Jannoo N., Grivet L., Seguin M., Paulet F., Domaingue R., Rao P.S. Molecular investigation of the genetic base of sugarcane cultivars. Theor Appl Genet. 1999;99:171–184. [Google Scholar]
- 13.Renny-Byfield S., Wendel J.F. Doubling down on genomes: polyploidy and crop plants. Am J Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- 14.Feldman M., Levy A.A., Fahima T., Korol A. Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot. 2012;63:5045–5059. doi: 10.1093/jxb/ers192. [DOI] [PubMed] [Google Scholar]
- 15.Van de Peer Y., Mizrachi E., Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- 16.Mudge S.R., Osabe K., Casu R.E., Bonnett G.D., Manners J.M., Birch R.G. Efficient silencing of reporter transgenes coupled to known functional promoters in sugarcane, a highly polyploid crop species. Planta. 2009;229:549–558. doi: 10.1007/s00425-008-0852-8. [DOI] [PubMed] [Google Scholar]
- 17.Kondrashov F.A. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B: Biol Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makova K.D. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 2003;13:1638–1645. doi: 10.1101/gr.1133803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldemberg J. Ethanol for a sustainable energy future. Science. 2007;315:808–810. doi: 10.1126/science.1137013. [DOI] [PubMed] [Google Scholar]
- 20.Souza G.M., Ballester M.V.R., de Brito Cruz C.H., Chum H., Dale B., Dale V.H. The role of bioenergy in a climate-changing world. Environ Dev. 2017;23:57–64. [Google Scholar]
- 21.Jaiswal D., De Souza A.P., Larsen S., LeBauer D.S., Miguez F.E., Sparovek G. Brazilian sugarcane ethanol as an expandable green alternative to crude oil use. Nat Clim Change. 2017;7:788–792. [Google Scholar]
- 22.Souza GM, Victoria RL, Joly CA, Verdade LM. Bioenergy & sustainability: bridging the gaps. Paris Cedex: Scientific Committee on Problems of the Environment (SCOPE); 2015.
- 23.Kline K.L., Msangi S., Dale V.H., Woods J., Souza G.M., Osseweijer P. Reconciling food security and bioenergy: priorities for action. GCB Bioenergy. 2017;9:557–576. [Google Scholar]
- 24.Souza G.M., Filho R.M. Industrial biotechnology and biomass: what next for Brazil’s future energy and chemicals? Ind Biotechnol. 2016;12:24–25. [Google Scholar]
- 25.Carvalho-Netto O.V., Bressiani J.A., Soriano H.L., Fiori C.S., Santos J.M., Barbosa G.V. The potential of the energy cane as the main biomass crop for the cellulosic industry. Chem Biol Technol Agric. 2014;1:20. [Google Scholar]
- 26.Kandel R, Yang X, Song J, Wang J. Potentials, Challenges, and genetic and genomic resources for sugarcane biomass improvement. Front Plant Sci 2018;9. https://doi.org/10.3389/fpls.2018.00151. [DOI] [PMC free article] [PubMed]
- 27.Matsuoka S, Kennedy AJ, Santos EGD dos, Tomazela A, L, Rubio LCS. Energy cane: its concept, development, characteristics, and prospects. Adv Botany 2014. https://doi.org/10.1155/2014/597275.
- 28.Goldemberg J., Coelho S.T., Guardabassi P. The sustainability of ethanol production from sugarcane. Energy Policy. 2008;36:2086–2097. [Google Scholar]
- 29.Alexander AG. The energy cane alternative. Amsterdam: Elsevier; 1985.
- 30.da Silva J.A. The importance of the wild cane saccharum spontaneum for bioenergy genetic breeding. Sugar Tech. 2017;19:229–240. [Google Scholar]
- 31.Tew TL, Cobill RM. Genetic improvement of sugarcane (Saccharum spp.) as an energy crop. In: Vermerris W, editor. Genetic improvement of bioenergy crops, New York, NY: Springer New York; 2008, p. 273–94. https://doi.org/10.1007/978-0-387-70805-8_9.
- 32.Kumar A., Kumar N., Baredar P., Shukla A. A review on biomass energy resources, potential, conversion and policy in India. Renew Sustain Energy Rev. 2015;45:530–539. [Google Scholar]
- 33.Wang L.-P., Jackson P.A., Lu X., Fan Y.-H., Foreman J.W., Chen X.-K. Evaluation of sugarcane × saccharum spontaneum progeny for biomass composition and yield components. Crop Sci. 2008;48:951–961. [Google Scholar]
- 34.Hammond B.W. Saccharum spontaneum (Gramineae) in panama: the physiology and ecology of invasion. J Sustainable For. 1999;8:23–38. [Google Scholar]
- 35.Samuels A., Alexander A.G., Rios C., Garcia M. The production of energy cane in Puerto Rico: the Hatillo project. Proc Cong Int Soc Sugar Cane Technol. 1984;3:14–17. [Google Scholar]
- 36.Bischoff K.P., Gravois K.A., Reagan T.E., Hoy J.W., Kimbeng C.A., LaBorde C.M. Registration of ‘L 79–1002’ sugarcane. J Plant Registrations. 2008;2:211. [Google Scholar]
- 37.Rao PS, Davis H, Simpson S. New sugarcane cultivars and year round sugar and ethanol production with bagassebased cogeneration in Barbados and Guiana. Proc 2007 Congr 2007;26:1169–76.
- 38.Santchurn D., Badaloo M.G.H., Zhou M., Labuschagne M.T. Contribution of sugarcane crop wild relatives in the creation of improved varieties in Mauritius. Plant Genet Resour: Characterization Utilization. 2019;17:151–163. [Google Scholar]
- 39.Jackson P. Utilizing wild germplasm in sugarcane breeding - progress and prospects. ISSCT Joint, Okinawa: 2018, p. 12.
- 40.Aitken K, Jingchuan L, Piperidis G, Qing C, Yuanhong F, Jackson P. Worldwide genetic diversity od Saccharum spontaneum and lavel of diversity captured in a sugarcane breeding program. ISSCT Joint, Okinawa: 2018, p. 44.
- 41.López J, Jaimes H, Franco M, Ocampo I, Barrios R, Salazar F, et al. Development of transgenic sugarcane associate with increasing biomass, sugar and stress tolerance in Colombia. ISSCT Joint, Okinawa: 2018, p. 54.
- 42.Terajima Y, Matsuoka M, Ujihara K, et al. The simultaneous production of sugar and biomass ethanol using high-biomass sugarcane derived from interspecific and intergeneric cross in Japan. Proceedings of the Biomass Asia Workshop, Tokyo: 2005.
- 43.Terajima Y, Sugimoto A, Fukuhara S, et al. The feature of root growth and activity of a high yielding interspecific hybrid between Saccharum hybrid and S. spontaneum L. Proceedings of the 25th Congress of the International Society of Sugar Cane Technologists, 2005, p. 255–8.
- 44.Sugimoto A, Terajima Y, Terauchi T, et al. Developing new types of sugarcane by hybridization between commercial sugarcane cultivars and wild relatives. Proceedings of the Symposium FAO RAP-NIAS, vol. 2012/1, Tsukuba: 2012, p. 11–24.
- 45.Tippayawat A., Terajima Y., Werapon P., Sansayawichai T., Irei S., Sugimoto A. Agronomic traits and root distribution of intergeneric F1 and BC1 hybrids between Saccharum spp. hybrid and Thai Erianthus Arundianaceus in north-east Thailand. ISSCT Joint, Okinawa. 2018:33. [Google Scholar]
- 46.de Morais LK, de Aguiar MS, de Albuquerque e Silva P, Câmara TMM, Cursi DE, Júnior ARF, et al. Breeding of Sugarcane. In: Cruz VMV, Dierig DA, editors. Industrial Crops, vol. 9, New York, NY: Springer New York; 2015, p. 29–42. https://doi.org/10.1007/978-1-4939-1447-0_2.
- 47.de Setta N., Monteiro-Vitorello C.B., Metcalfe C.J., Cruz G.M.Q., Del Bem L.E., Vicentini R. Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics. 2014;15:540. doi: 10.1186/1471-2164-15-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riaño-Pachón D.M., Mattiello L. Draft genome sequencing of the sugarcane hybrid SP80-3280. F1000Res. 2017;6:861. doi: 10.12688/f1000research.11859.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garsmeur O., Droc G., Antonise R., Grimwood J., Potier B., Aitken K. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Zhang X., Tang H., Zhang Q., Hua X., Ma X. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet. 2018;50:1565–1573. doi: 10.1038/s41588-018-0237-2. [DOI] [PubMed] [Google Scholar]
- 51.Nascimento L.C., Yanagui K., Jose J., Camargo E.L.O., Grassi M.C.B., Cunha C.P. Unraveling the complex genome of Saccharum spontaneum using Polyploid Gene Assembler. DNA Res. 2019;26:205–216. doi: 10.1093/dnares/dsz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Souza G.M., Van Sluys M.-A., Lembke C.G., Lee H., Margarido G.R.A., Hotta C.T. Assembly of the 373K gene space of the polyploid sugarcane genome reveals reservoirs of functional diversity in the world’s leading biomass crop. GigaScience. 2019 doi: 10.1093/gigascience/giz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardoso-Silva C.B., Costa E.A., Mancini M.C., Balsalobre T.W.A., Canesin L.E.C., Pinto L.R. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0088462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Que Y., Su Y., Guo J., Wu Q., Xu L. A Global view of transcriptome dynamics during sporisorium scitamineum challenge in sugarcane by RNA-seq. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0106476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vargas L., Santa Brígida A.B., Mota Filho J.P., de Carvalho T.G., Rojas C.A., Vaneechoutte D. Drought tolerance conferred to sugarcane by association with gluconacetobacter diazotrophicus: a transcriptomic view of hormone pathways. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0114744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vicentini R, Bottcher A, Brito M dos S, dos Santos AB, Creste S, Landell MG de A, et al. Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLOS ONE 2015;10:e0134909. https://doi.org/10.1371/journal.pone.0134909. [DOI] [PMC free article] [PubMed]
- 57.Mattiello L., Riaño-Pachón D.M., Martins M.C.M., da Cruz L.P., Bassi D., Marchiori P.E.R. Physiological and transcriptional analyses of developmental stages along sugarcane leaf. BMC Plant Biol. 2015 doi: 10.1186/s12870-015-0694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li M., Liang Z., Zeng Y., Jing Y., Wu K., Liang J. novo analysis of transcriptome reveals genes associated with leaf abscission in sugarcane (Saccharum officinarum L.) BMC Genomics. 2016 doi: 10.1186/s12864-016-2552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang D.-L., Gao Y.-J., Gui Y.-Y., Chen Z.-L., Qin C.-X., Wang M. Transcriptome of high-sucrose sugarcane variety GT35. Sugar Tech. 2016;18:520–528. [Google Scholar]
- 60.Santa Brigida A.B., Rojas C.A., Grativol C., de Armas E.M., Entenza J.O.P., Thiebaut F. Sugarcane transcriptome analysis in response to infection caused by Acidovorax avenae subsp. avenae. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0166473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong M., Cheng G., Peng L., Xu Q., Yang Y., Xu J. Transcriptome analysis of sugarcane response to the infection by sugarcane steak mosaic virus (SCSMV) Trop Plant Biol. 2017;10:45–55. [Google Scholar]
- 62.Hoang N.V., Furtado A., Mason P.J., Marquardt A., Kasirajan L., Thirugnanasambandam P.P. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genomics. 2017;18 doi: 10.1186/s12864-017-3757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belesini A.A., Carvalho F.M.S., Telles B.R., de Castro G.M., Giachetto P.F., Vantini J.S. novo transcriptome assembly of sugarcane leaves submitted to prolonged water-deficit stress. Genet Mol Res. 2017;16 doi: 10.4238/gmr16028845. [DOI] [PubMed] [Google Scholar]
- 64.Hoang N.V., Furtado A., O’Keeffe A.J., Botha F.C., Henry R.J. Association of gene expression with biomass content and composition in sugarcane. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0183417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu S., Wang J., Shang H., Huang Y., Yao W., Chen B. Transcriptomic characterization and potential marker development of contrasting sugarcane cultivars. Sci Rep. 2018;8 doi: 10.1038/s41598-018-19832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoang N.V., Furtado A., Thirugnanasambandam P.P., Botha F.C., Henry R.J. De novo assembly and characterizing of the culm-derived meta-transcriptome from the polyploid sugarcane genome based on coding transcripts. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNeil M.D., Bhuiyan S.A., Berkman P.J., Croft B.J., Aitken K.S. Analysis of the resistance mechanisms in sugarcane during Sporisorium scitamineum infection using RNA-seq and microscopy. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0197840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Telles BR, Carvalho FM de S, Vantini J da S, Belesini AA, de Castro GM, Giachetto PF, et al. Prolonged water deficit reveals new profile of sugarcane gene expression and metabolic pathway related to tolerance. Sugar Tech 2019;21:451–61. https://doi.org/10.1007/s12355-018-0674-3.
- 69.Nishiyama M.Y., Ferreira S.S., Tang P.-Z., Becker S., Pörtner-Taliana A., Souza G.M. Full-length enriched cDNA libraries and ORFeome analysis of sugarcane hybrid and ancestor genotypes. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0107351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wai C.M., Zhang J., Jones T.C., Nagai C., Ming R. Cell wall metabolism and hexose allocation contribute to biomass accumulation in high yielding extreme segregants of a Saccharum interspecific F2 population. BMC Genomics. 2017;18 doi: 10.1186/s12864-017-4158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kasirajan L., Hoang N.V., Furtado A., Botha F.C., Henry R.J. Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci Rep. 2018;8 doi: 10.1038/s41598-018-30033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaker P.D.C., Palhares A.C., Taniguti L.M., Peters L.P., Creste S., Aitken K.S. RNAseq transcriptional profiling following whip development in sugarcane smut disease. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0162237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.HISAT: a fast spliced aligner with low memory requirements n.d. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4655817/ (accessed May 29, 2019). [DOI] [PMC free article] [PubMed]
- 74.Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong R., Demura T., Ye Z.-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong R., Richardson E.A., Ye Z.-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarthy R.L., Zhong R., Ye Z.-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- 79.Wang H., Zhao Q., Chen F., Wang M., Dixon R.A. NAC domain function and transcriptional control of a secondary cell wall master switch: NST1 function and regulation. Plant J. 2011;68:1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x. [DOI] [PubMed] [Google Scholar]
- 80.Zhong R., Lee C., Ye Z.-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010;15:625–632. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 81.Nakano Y., Yamaguchi M., Endo H., Rejab N.A., Ohtani M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci. 2015;6:288. doi: 10.3389/fpls.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ambavaram M.M.R., Krishnan A., Trijatmiko K.R., Pereira A. Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 2011;155:916–931. doi: 10.1104/pp.110.168641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhong R., Lee C., McCarthy R.L., Reeves C.K., Jones E.G., Ye Z.-H. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011;52:1856–1871. doi: 10.1093/pcp/pcr123. [DOI] [PubMed] [Google Scholar]
- 84.Valdivia E.R., Herrera M.T., Gianzo C., Fidalgo J., Revilla G., Zarra I. Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J Exp Bot. 2013;64:1333–1343. doi: 10.1093/jxb/ers394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida K, Sakamoto S, Kawai T, Kobayashi Y, Sato K, Ichinose Y, et al. Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front Plant Sci 2013;4. https://doi.org/10.3389/fpls.2013.00383. [DOI] [PMC free article] [PubMed]
- 86.Zhong R., Yuan Y., Spiekerman J.J., Guley J.T., Egbosiuba J.C., Ye Z.-H. Functional characterization of NAC and MYB transcription factors involved in regulation of biomass production in switchgrass (Panicum virgatum) PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0134611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Noda S., Koshiba T., Hattori T., Yamaguchi M., Suzuki S., Umezawa T. The expression of a rice secondary wall-specific cellulose synthase gene, OsCesA7, is directly regulated by a rice transcription factor, OsMYB58/63. Planta. 2015;242:589–600. doi: 10.1007/s00425-015-2343-z. [DOI] [PubMed] [Google Scholar]
- 88.Scully E.D., Gries T., Sarath G., Palmer N.A., Baird L., Serapiglia M.J. Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J. 2016;85:378–395. doi: 10.1111/tpj.13112. [DOI] [PubMed] [Google Scholar]
- 89.Golfier P., Volkert C., He F., Rausch T., Wolf S. Regulation of secondary cell wall biosynthesis by a NAC transcription factor from Miscanthus. Plant Direct. 2017;1 doi: 10.1002/pld3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao Wenhan, Yang Yue, Yu Jingjuan. ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.) Plant Sci. 2018;266:83–94. doi: 10.1016/j.plantsci.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 91.Rao X., Chen X., Shen H., Ma Q., Li G., Tang Y. Gene regulatory networks for lignin biosynthesis in switchgrass (Panicum virgatum) Plant Biotechnol J. 2019;17:580–593. doi: 10.1111/pbi.13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Agarwal T., Grotewold E., Doseff A.I., Gray J. MYB31/MYB42 Syntelogs Exhibit Divergent Regulation of Phenylpropanoid Genes in Maize, Sorghum and Rice. Sci Rep. 2016;6:28502. doi: 10.1038/srep28502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ooka H. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 94.Vettore A.L. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003;13:2725–2735. doi: 10.1101/gr.1532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Himmel M.E., Ding S.-Y., Johnson D.K., Adney W.S., Nimlos M.R., Brady J.W. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 96.Chen F., Dixon R.A. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 97.Vanholme R., Morreel K., Ralph J., Boerjan W. Lignin engineering. Curr Opin Plant Biol. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 98.Zhao X., Zhang L., Liu D. Biomass recalcitrance. Part I: the chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels, Bioprod Biorefin. 2012;6:465–482. [Google Scholar]
- 99.Zeng Y., Zhao S., Yang S., Ding S.-Y. Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr Opin Biotechnol. 2014;27:38–45. doi: 10.1016/j.copbio.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 100.Bonawitz N.D., Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet. 2010;44:337–363. doi: 10.1146/annurev-genet-102209-163508. [DOI] [PubMed] [Google Scholar]
- 101.Vanholme R., Cesarino I., Rataj K., Xiao Y., Sundin L., Goeminne G. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in arabidopsis. Science. 2013;341:1103–1106. doi: 10.1126/science.1241602. [DOI] [PubMed] [Google Scholar]
- 102.Barros J., Serrani-Yarce J.C., Chen F., Baxter D., Venables B.J., Dixon R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat Plants. 2016;2:16050. doi: 10.1038/nplants.2016.50. [DOI] [PubMed] [Google Scholar]
- 103.Barros J., Escamilla-Trevino L., Song L., Rao X., Serrani-Yarce J.C., Palacios M.D. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat Commun. 2019;10:1994. doi: 10.1038/s41467-019-10082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fraser C.M., Chapple C. The Phenylpropanoid Pathway in Arabidopsis. The Arabidopsis Book. 2011;9 doi: 10.1199/tab.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferreira S.S., Simões M.S., Carvalho G.G. de Lima LGA, Svartman RM de A, Cesarino I. The lignin toolbox of the model grass Setaria viridis. Plant Mol Biol. 2019 doi: 10.1007/s11103-019-00897-9. [DOI] [PubMed] [Google Scholar]
- 106.Perkins M., Smith R.A., Samuels L. The transport of monomers during lignification in plants: anything goes but how? Curr Opin Biotechnol. 2019;56:69–74. doi: 10.1016/j.copbio.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 107.Alejandro S., Lee Y., Tohge T., Sudre D., Osorio S., Park J. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol. 2012;22:1207–1212. doi: 10.1016/j.cub.2012.04.064. [DOI] [PubMed] [Google Scholar]
- 108.Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 109.Carpita N.C. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- 110.Harris P.J., Trethewey J.A.K. The distribution of ester-linked ferulic acid in the cell walls of angiosperms. Phytochem Rev. 2010;9:19–33. [Google Scholar]
- 111.Ralph J., Grabber J.H., Hatfield R.D. Lignin-ferulate cross-links in grasses: active incorporation of ferulate polysaccharide esters into ryegrass lignins. Carbohydr Res. 1995;275:167–178. [Google Scholar]
- 112.Lam T.B.T., Kadoya K., Iiyama K. Bonding of hydroxycinnamic acids to lignin: ferulic and p-coumaric acids are predominantly linked at the benzyl position of lignin, not the β-position, in grass cell walls. Phytochemistry. 2001;57:987–992. doi: 10.1016/s0031-9422(01)00052-8. [DOI] [PubMed] [Google Scholar]
- 113.Karlen S.D., Zhang C., Peck M.L., Smith R.A., Padmakshan D., Helmich K.E. Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lan W., Lu F., Regner M., Zhu Y., Rencoret J., Ralph S.A. Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 2015;167:1284–1295. doi: 10.1104/pp.114.253757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eudes A., Dutta T., Deng K., Jacquet N., Sinha A., Benites V.T. SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0178160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Terrett O.M., Dupree P. Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr Opin Biotechnol. 2019;56:97–104. doi: 10.1016/j.copbio.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 117.Burton R.A., Wilson S.M., Hrmova M., Harvey A.J., Shirley N.J., Medhurst A. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 118.Yin Y., Huang J., Xu Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009;9:99. doi: 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Doblin M.S., Pettolino F.A., Wilson S.M., Campbell R., Burton R.A., Fincher G.B. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA. 2009;106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Handakumbura P.P., Brow K., Whitney I.P., Zhao K., Sanguinet K.A., Lee S.J. Secondary wall associated MYB1 is a positive regulator of secondary cell wall thickening in Brachypodium distachyon and is not found in the Brassicaceae. Plant J. 2018;96:532–545. doi: 10.1111/tpj.14047. [DOI] [PubMed] [Google Scholar]
- 121.Ha C.M., Escamilla-Trevino L., Yarce J.C.S., Kim H., Ralph J., Chen F. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 2016;86:363–375. doi: 10.1111/tpj.13177. [DOI] [PubMed] [Google Scholar]
- 122.Saleme M de LS, Cesarino I, Vargas L, Kim H, Vanholme R, Goeminne G, et al. Silencing CAFFEOYL SHIKIMATE ESTERASE Affects Lignification and Improves Saccharification in Poplar. Plant Physiol 2017;175:1040–57. https://doi.org/10.1104/pp.17.00920. [DOI] [PMC free article] [PubMed]
- 123.Ohtani M., Demura T. The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr Opin Biotechnol. 2019;56:82–87. doi: 10.1016/j.copbio.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 124.Bonawitz N.D., Kim J.I., Tobimatsu Y., Ciesielski P.N., Anderson N.A., Ximenes E. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature. 2014;509:376–380. doi: 10.1038/nature13084. [DOI] [PubMed] [Google Scholar]
- 125.Dolan W.L., Dilkes B.P., Stout J.M., Bonawitz N.D., Chapple C. Mediator complex subunits MED2, MED5, MED16, and MED23 genetically interact in the regulation of phenylpropanoid biosynthesis. Plant Cell. 2017;29:3269–3285. doi: 10.1105/tpc.17.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yilmaz A., Nishiyama M.Y., Fuentes B.G., Souza G.M., Janies D., Gray J. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–180. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ruiz M., Rouard M., Raboin L.M., Lartaud M., Lagoda P., Courtois B. TropGENE-DB, a multi-tropical crop information system. Nucleic Acids Res. 2004;32:D364–D367. doi: 10.1093/nar/gkh105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Makita Y., Shimada S., Kawashima M., Kondou-Kuriyama T., Toyoda T., Matsui M. MOROKOSHI: transcriptome database in sorghum bicolor. Plant Cell Physiol. 2015;56:e6. doi: 10.1093/pcp/pcu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ware D., Jaiswal P., Ni J., Pan X., Chang K., Clark K. Gramene: a resource for comparative grass genomics. Nucleic Acids Res. 2002;30:103–105. doi: 10.1093/nar/30.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., Van de Peer Y. 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018;46:D1190–D1196. doi: 10.1093/nar/gkx1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Duvick J., Fu A., Muppirala U., Sabharwal M., Wilkerson M.D., Lawrence C.J. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nishiyama M.Y., Vicente F.F.R., Lembke C.G., Sato P.M., Dal-Bianco M.L., Fandino R.A. The SUCEST-FUN regulatory network database: designing an energy grass. Int Sugar J. 2012;114:821–826. [Google Scholar]
- 136.Papini-Terzi FS, Rocha FR, Vêncio RZN, Oliveira KC, Felix J de M, Vicentini R, et al. Transcription profiling of signal transduction-related genes in sugarcane tissues. DNA Res 2005;12:27–38. https://doi.org/10.1093/dnares/12.1.27. [DOI] [PubMed]
- 137.Rocha F.R., Papini-Terzi F.S., Nishiyama M.Y., Vêncio R.Z., Vicentini R., Duarte R.D. Signal transduction-related responses to phytohormones and environmental challenges in sugarcane. BMC Genomics. 2007;8:71. doi: 10.1186/1471-2164-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lembke C.G., Nishiyama M.Y., Sato P.M., de Andrade R.F., Souza G.M. Identification of sense and antisense transcripts regulated by drought in sugarcane. Plant Mol Biol. 2012;79:461–477. doi: 10.1007/s11103-012-9922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hotta C.T., Nishiyama M.Y., Souza G.M. Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0071847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Souza A.P., Gaspar M., Da Silva E.A., Ulian E.C., Waclawovsky A.J., Nishiyama M.Y. Elevated CO2 increases photosynthesis, biomass and productivity, and modifies gene expression in sugarcane. Plant Cell Environ. 2008;31:1116–1127. doi: 10.1111/j.1365-3040.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- 141.Ferreira SS, Hotta CT, Poelking VG de C, Leite DCC, Buckeridge MS, Loureiro ME, et al. Co-expression network analysis reveals transcription factors associated to cell wall biosynthesis in sugarcane. Plant Mol Biol 2016;91:15–35. https://doi.org/10.1007/s11103-016-0434-2. [DOI] [PMC free article] [PubMed]
- 142.Cia M.C., de Carvalho G., Azevedo R.A., Monteiro-Vitorello C.B., Souza G.M., Nishiyama-Junior M.Y. Novel insights into the early stages of ratoon stunting disease of sugarcane inferred from transcript and protein analysis. Phytopathology. 2018;108:1455–1466. doi: 10.1094/PHYTO-04-18-0120-R. [DOI] [PubMed] [Google Scholar]
- 143.Cunha C.P., Roberto G.G., Vicentini R., Lembke C.G., Souza G.M., Ribeiro R.V. Ethylene-induced transcriptional and hormonal responses at the onset of sugarcane ripening. Sci Rep. 2017;7 doi: 10.1038/srep43364. [DOI] [PMC free article] [PubMed] [Google Scholar]