Drug repurposing is a quick and economical strategy for developing new therapies with approved drugs. H5N1 is a highly pathogenic avian influenza virus subtype that can cause severe acute lung injury (ALI) and a high mortality rate due to limited treatments. The use of RNA interference (RNAi) is a reliable approach to identify essential genes in diseases. In most genomewide RNAi screenings, virus replication is the readout of interference. Since H5N1 virus infection could induce significant cell death and the percentage of cell death is associated with virus lethality, we designed a genomewide RNAi screening method to identify repurposable drugs against H5N1 virus with cell death as the readout. We discovered that the neurological drug ifenprodil and the anticancer drug flavopiridol could effectively ameliorate murine ALI after influenza A H5N1 virus infection, suggesting that they might be novel remedies for H5N1 virus-induced ALI in addition to the traditional indications.
KEYWORDS: H5N1, lung injury, ifenprodil, flavopiridol, drug repurposing, genomewide RNAi, H5N1
ABSTRACT
Due to the limitations of effective treatments, avian influenza A H5N1 virus is the most lethal influenza virus strain that causes severe acute lung injury (ALI). To develop effective drugs ameliorating H5N1-induced ALI, we explore an RNA interference (RNAi) screening method to monitor changes in cell death induced by H5N1 infection. We performed RNAi screening on 19,424 genes in A549 lung epithelial cells and examined cell death induced by H5N1 infection. These screens identified 1,137 host genes for which knockdown altered cell viability by over 20%. DrugBank searches of these 1,137 host genes identified 146 validated druggable target genes with 372 drug candidates. We obtained 104 commercially available drugs with 65 validated target genes and examined their improvement of cell viability following H5N1 infection. We identified 28 drugs that could significantly recover cell viability following H5N1 infection and tested 10 in an H5N1-induced-ALI mouse model. The neurological drug ifenprodil and the anticancer drug flavopiridol markedly decreased leukocyte infiltration and lung injury scores in infected mouse lungs, significantly ameliorated edema in infected mouse lung tissues, and significantly improved the survival of H5N1-infected mice. Ifenprodil is an antagonist of the N-methyl-d-aspartate (NMDA) receptor, which is linked to inflammation and lung injury. Flavopiridol is an inhibitor of cyclin-dependent kinase 4 (CDK4), which is linked to leukocyte migration and lung injury. These results suggest that ifenprodil and flavopiridol represent novel remedies against potential H5N1 epidemics in addition to their proven indications. Furthermore, our strategy for identifying repurposable drugs could be a general approach for other diseases.
IMPORTANCE Drug repurposing is a quick and economical strategy for developing new therapies with approved drugs. H5N1 is a highly pathogenic avian influenza virus subtype that can cause severe acute lung injury (ALI) and a high mortality rate due to limited treatments. The use of RNA interference (RNAi) is a reliable approach to identify essential genes in diseases. In most genomewide RNAi screenings, virus replication is the readout of interference. Since H5N1 virus infection could induce significant cell death and the percentage of cell death is associated with virus lethality, we designed a genomewide RNAi screening method to identify repurposable drugs against H5N1 virus with cell death as the readout. We discovered that the neurological drug ifenprodil and the anticancer drug flavopiridol could effectively ameliorate murine ALI after influenza A H5N1 virus infection, suggesting that they might be novel remedies for H5N1 virus-induced ALI in addition to the traditional indications.
INTRODUCTION
Influenza A virus is the most common type among the viruses that cause influenza epidemics, and influenza A viruses can be divided into various subtypes based on the hemagglutinin and neuraminidase proteins on the viral surface (1). H5N1 is a highly pathogenic avian influenza virus subtype that can cause severe acute lung injury (ALI) in human subjects (2). Infection with H5N1 virus is associated with a 52.79% mortality rate (https://www.who.int/influenza/human_animal_interface/2019_02_12_tableH5N1.pdf?ua=1). However, effective treatments are limited. The use of corticoids is controversial, and extracorporeal membrane oxygenation (ECMO), artificial liver support system (ALSS), and mechanical ventilation are often expensive (3). Thus, there is an urgent and continuous need for novel treatments.
Drug repurposing is a quick and economical strategy to identify new therapeutic uses of approved drugs outside their original scope and is estimated to be the source of approximately 30% of newly approved drugs every year (4). An example of successful drug repurposing is the use of the chemotherapeutic agent azidothymidine as a therapy for HIV in the 1980s (4). Thus, the exploration of an efficient method for drug repurposing is necessary.
RNA interference (RNAi) represents a reliable approach to identify genes important in diseases such as acute myeloid leukemia, epithelial ovarian cancer, and schistosomiasis (5–7). Genomewide RNAi screening has been used to identify host genes involved in the replication of the moderately pathogenic influenza H1N1 virus (8–15). Data mining using public databases such as the DrugBank database and the Therapeutic Target Database (TTD) has been increasingly used to identify candidate drugs that could potentially interact with known validated target genes (16, 17). We combined RNAi screening and DrugBank and TTD target gene searches to develop an effective method for identifying repurposable drugs.
In the current study, we used genomewide RNAi screening to identify host genes that influence host cell viability following H5N1 virus infection. The results showed <10% overlap in the candidate host gene targets between H5N1 and the less lethal H1N1 strain. Next, we screened 1,137 critical host genes in RNAi screens using the DrugBank database and identified 372 approved or clinically investigated drugs (with 146 target genes). Validation experiments performed with 104 commercially available drugs (and 65 target genes) identified 28 candidate drugs that significantly increased the viability of H5N1-infected A549 cells. The two most effective drugs among these candidates in a mouse model of H5N1-induced ALI were ifenprodil, an agent with neurological indications, and flavopiridol, an antineoplastic agent.
RESULTS
Genomewide RNAi screening of target host genes.
A total of 19,424 human genes were screened by culturing with RNAi; 6,540 effective genes were selected for a second round of genomewide RNAi screening. The viability of H5N1-infected A549 cells was affected by a total of 2,957 genes (Fig. 1A and B; see also Fig. S1 and Table S1 in the supplemental material). Among these host genes, 1,137 and 337 RNAi treatments altered cell viability with H5N1 infection by more than 20% and 30%, respectively, in both screenings (Fig. 1B). The Metacore database indicated that the most significant Gene Ontology (GO) terms for biological process clusters for the 337 genes correspond to immune responses, the cell cycle, and stress responses (Fig. 1C). On the basis of enrichment of the GO term cellular component (CC) as well as on protein-protein interaction (PPI) analysis using the STRING database (https://string-db.org/), knockdown of the genes related to spliceosomes ameliorated cell death; in contrast, knockdown of the genes related to mitochondrial respiratory chain complex I aggravated cell death (Fig. S2). Among the top 10 functional enrichment pathways, 4 were related to the immune response and 3 to apoptosis (Fig. 1D). Our results obtained through the RNAi platform show that cell viability may be influenced by over a thousand host genes and that those genes might represent potential drug targets against H5N1 virus.
FIG 1.
Genomewide RNAi screening to identify essential host factors for H5N1 infection. (A) Rank-ordered distribution of cell viability based on 6,540 human genes in the second round of genomewide RNAi screening. (B) Counts of genes associated with changes in cell viability of more than 10%, 20%, or 30% in two genomewide RNAi screens. (C) GO term (biological process) enrichment of 337 genes based on Metacore analysis. Enrichment was visualized using the Enrichment Map application in Cytoscape. Biological process terms are presented as nodes, while the node color indicates the significance of terms and node size reflects the object count enriched in the terms. The connection between terms is based on shared objects. (D) Functional enrichment of the top 10 significant pathways for 337 genes (false discovery rate < 0.05). IAP, inhibitor of apoptosis; SCF, Skp, Cullin, F-box-containing complex; G-CSF, granulocyte colony-stimulating factor; NALP3, NACHT, LRR, and PYD domain-containing protein 3; BAD, BCL2-associated agonist of cell death; TLR, Toll-like receptor; IL-17, interleukin-17.
Genomewide RNAi screen for host genes associated with H5N1 virus infection. (A) Flow chart of the genomewide RNAi screen of 19,424 genes to identify those important in influenza A H5N1 virus infection. (B) Heat map of cell viability related to 337 genes whose knockdown altered cell viability by >30% in each of two siRNA screens. The two columns of the heat map represent the first and second rounds of genomewide RNAi screening. Download FIG S1, PDF file, 0.4 MB (425KB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional enrichment of essential host genes associated with H5N1 infection. (A) Enriched Gene Ontology cellular component terms associated with 337 genes, as analyzed using Metacore. (B) Interactions among clusters of spliceosomes. The three kinds of colored circles (dark red, medium red, and light red) indicate that the cell viability increased more than 30%, 20%, and 10%, respectively. (C) Interactions among the clusters of mitochondrial respiratory chain complex I. The three kinds of colored circles (dark blue, medium blue, and light blue) indicate that the cell viability decreased more than 30%, 20%, and 10%, respectively. Download FIG S2, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Host genes identified in the genomewide RNAi screen. Download Table S1, XLSX file, 0.3 MB (268.1KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selection of candidate drugs and screening in H5N1-infected A549 cells.
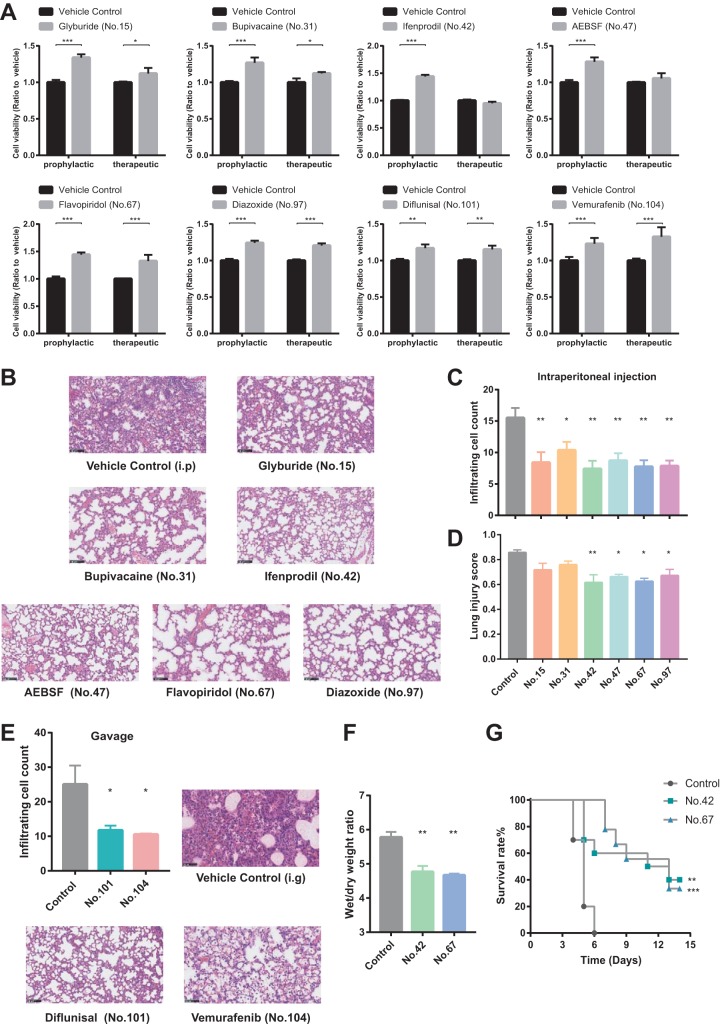
By searching the DrugBank database, we identified 146 drug target genes (372 drug candidates) among 1,137 host genes that altered cell viability by more than 20% in the RNAi screen after H5N1 virus infection. We further evaluated the prophylactic and therapeutic effects of 104 commercially available drugs (with 65 validated target genes) by examining their ameliorative effects in a cell viability assay in H5N1-infected A549 cells (Fig. S3A and B and Table S2) and found 28 drugs that significantly enhanced cell viability with low cytotoxicity (Fig. 2A and Fig. S3C; Table S3).
FIG 2.
Drugs effective against influenza A H5N1 virus infection in vitro and in vivo. (A) Viability of A549 cells infected with H5N1 at a multiplicity of infection of 3.0 and treated with drugs prophylactically (3 h before infection) or therapeutically (3 h after infection). Viability was determined at 48 h after infection and is expressed relative to vehicle-treated control cells. (B) Representative images of lung pathology in hematoxylin-and-eosin-stained sections from H5N1-infected C57BL/6 mice (n = 3 to 5) treated with drugs or vehicle by intraperitoneal injection. Scale bar = 100 μm. Lung tissues were removed at 3 days after infection. i.p, intraperitoneal. (C and D) Numbers of infiltrating cells (C) and lung injury scores (D) per microscopic field (means ± standard errors of the means [SEM]) are shown in the bar graph (n = 100 fields per treatment). (E) Representative images of lung pathology in hematoxylin-and-eosin-stained sections from H5N1-infected C57BL/6 mice (n = 3 to 5) treated with drugs or vehicle by intragastric administration. Lung tissues were removed at 3 days after infection. The numbers of infiltrating cells per microscopic field (means ± SEM) are shown in the bar graph (n = 100 fields per treatment). Scale bar = 100 μm. i.g, intragastric. (F) Wet-to-dry-weight ratios of lung tissue from H5N1-infected C57BL/6 mice treated by intraperitoneal injection with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle. In vitro and in vivo experiments were repeated three times. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed t test). (G) Kaplan-Meier survival curves of H5N1-infected C57BL/6 mice treated with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle by intraperitoneal injection. **, P < 0.01; ***, P < 0.001 (log rank test). Detailed information about effective drug concentrations in vitro and drug dosages in vivo are shown in Table S3.
In vitro validation of drugs against H5N1 virus infection. (A) Flow chart of drug screening based on 1,137 genes and 104 drug candidates. (B) Flow chart of the in vitro validation of drug efficacy against H5N1 infection. (C) Viability of A549 cells infected with H5N1 at a multiplicity of infection of 3.0 and treated with drugs prophylactically (3 h before infection) or therapeutically (3 h after infection). Viability was assessed at 48 h after infection and is expressed relative to that of vehicle-treated control cells. In vitro experiments were repeated three times. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed t test). Download FIG S3, PDF file, 0.3 MB (405.9KB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Drug information. Download Table S2, XLSX file, 0.01 MB (13.4KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro and in vivo drug information. Download Table S3, XLSX file, 0.01 MB (11.2KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Drug candidates that ameliorated ALI and improved survival rates in H5N1-infected mice.
Among the 28 drugs with which we observed a significant recovery of cell viability following H5N1 infection, 8 were glucocorticoids, which, at low doses, are used in conventional treatments against influenza virus-induced ALI (18, 19). Therefore, we focused on the other 20 nonhormonal drug candidates and tested the top 10 nonhormonal drug candidates that enhanced the viability of H5N1-infected A549 cells against ALI in C57BL/6 mice infected with H5N1 (Fig. S4A; see also Table S3). The following 8 of those 10 drugs significantly decreased the number of infiltrating cells in mouse lung tissues: glyburide (no. 15), bupivacaine (no. 31), ifenprodil (no. 42), 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF) (no. 47), flavopiridol (no. 67), diazoxide (no. 97), diflunisal (no. 101), and vemurafenib (no. 104) (Fig. 2B, C, and E). The following 4 of those 8 drugs significantly reduced lung injury scores in animals: ifenprodil, AEBSF, flavopiridol, and diazoxide (Fig. 2B and D; see also Fig. S4B). Both ifenprodil treatment and flavopiridol treatment ameliorated lung edema by decreasing the wet-to-dry-weight ratio in H5N1-infected mice with induced ALI (Fig. 2F; see also Fig. S4C and D). We measured the impact of these 2 drugs on major subsets of leukocytes during the response to H5N1 virus infection and found that ifenprodil or flavopiridol significantly decreased the levels of neutrophils, natural killer (NK) cells, and T cells in H5N1 virus-infected mouse lung tissues (Fig. S5). Notably, both ifenprodil treatment and flavopiridol treatment significantly increased the survival rate of H5N1 virus-infected mice (Fig. 2G). The weight loss of mice administered ifenprodil or flavopiridol was recovered at the second week after H5N1 virus infection (Fig. S4E). Taken together, these results indicate that 8 of the top 10 nonhormonal drugs can ameliorate ALI in H5N1-infected mice, with ifenprodil and flavopiridol being the most effective.
In vivo validation of drugs against H5N1 infection. (A) Flow chart of the in vivo validation of drug efficacy. (B) Lung injury scores of tissue from H5N1-infected C57BL/6 mice (n = 3 to 5) treated with drug candidates by intragastric administration. Lung tissues were obtained at 3 days after infection. Lung injury scores per microscopic field are shown in the bar graph. n = 100 fields were analyzed for each group. (C and D) Wet-to-dry-weight ratios of lung tissue from H5N1-infected C57BL/6 mice treated with drugs by intraperitoneal injection (C) or intragastric administration (D). Lung tissues were obtained at 3 days after infection. Compound identities: no. 15, glyburide; no. 31, bupivacaine; no. 47, AEBSF; no. 97, diazoxide; no. 101, diflunisal; no. 104, vemurafenib. The data shown represent means ± standard errors of the means (SEMs). In vivo experiments were repeated three times. *, P < 0.05; **, P < 0.01 (two-tailed test). (E) Body weight changes in H5N1-infected C57BL/6 mice treated with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle by intraperitoneal injection. Download FIG S4, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ifenprodil and flavopiridol impact inflammatory cell infiltration. (A) Representative images of lung pathology by immunofluorescence staining from H5N1-infected C57BL/6 mice (n = 3 to 5) treated with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle by intraperitoneal injection. Lung tissues were removed at 3 days after infection. The blue areas indicate cell nuclei stained with DAPI, the red areas indicate CD45 protein (leukocyte common antigen), and the green areas indicate marker proteins. Scale bar, 100 μm. The numbers of neutrophil cells (CD45+ MPO+), T cells (CD45+ CD3e+), and NK cells (CD45+ NCR1+) per microscopic field are shown in the bar graph (means ± SEM, n = 100 fields per treatment). (B and C) The numbers of macrophage cells (B) and dendritic cells (C) per microscopic field are shown in the bar graph (means ± SEM, n = 100 fields per treatment). *, P < 0.05; **, P < 0.01 (two-tailed t test). Download FIG S5, PDF file, 2.2 MB (2.2MB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ifenprodil and flavopiridol attenuated H5N1-induced cytokine storm in mice.
RNA sequencing (RNA-Seq) analysis of the lung tissue showed that ifenprodil and flavopiridol alleviated cytokine storm in H5N1-infected mice (Fig. 3A and B; see also Fig. S6A and B). We obtained blood samples from H5N1-infected mice and performed multiplex analyses to measure the levels of 23 cytokines/chemokines in plasma from mice administered ifenprodil or flavopiridol. Overall, ifenprodil treatment and flavopiridol treatment significantly decreased the measured levels of 8 and 12 cytokines/chemokines, respectively, in the plasma of H5N1-infected mice (Fig. 3C). The other six drugs decreased the mRNA levels of several cytokines and cytokine receptors in mouse blood (Fig. S6C to H). Our results suggest that these drugs, especially ifenprodil and flavopiridol, can attenuate the excessive inflammation caused by H5N1 virus.
FIG 3.
Ifenprodil and flavopiridol alleviated H5N1-induced cytokine storm in mouse lungs. (A and B) Heat maps of relative cytokine mRNA levels in lung tissues of H5N1-infected C57BL/6 mice treated with ifenprodil (no. 42) (A), flavopiridol (no. 67) (B), or vehicle. Lung tissues were obtained at 3 days after infection, and total RNA was isolated for RNA sequencing. The columns of the heat maps represent each experimental mouse. The relative expression levels of cytokine mRNAs were normalized by Z-score transformation. (C) The levels of cytokines and chemokines in plasma from H5N1-infected C57BL/6 mice treated with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle. Blood samples were obtained at 3 days after infection. *, P < 0.05; **, P < 0.01 (Mann-Whitney U test). GM-CSF, granulocyte-macrophage colony-stimulating factor; TNF-α, tumor necrosis factor alpha; IFN-γ, gamma interferon; RANTES, regulated on activation normally T-cell expressed and secreted; MCP-1, metaphase chromosome protein 1; MIP-1α, macrophage inflammatory protein 1 alpha.
Drugs antagonize H5N1-induced upregulation of cytokines. (A and B) Relative mRNA levels of cytokines and chemokines in H5N1-infected C57BL/6 mice (n = 3 to 5) treated with ifenprodil (no. 42) (A), flavopiridol (no. 67) (B), or vehicle. Lung tissue samples were collected at 72 h after infection, and total RNA was isolated for RNA sequencing. #, P < 0.1; *, P < 0.05; **, P < 0.01 (two-tailed t test). (C to H) Relative mRNA levels of cytokines and cytokine receptors in H5N1-infected C57BL/6 mice (n = 3 to 5) treated with glyburide (no. 15) (C), bupivacaine (no. 31) (D), AEBSF (no. 47) (E), diazoxide (no. 97) (F), diflunisal (no. 101) (G), vemurafenib (no. 104) (H), or vehicle. Blood samples were collected at 72 h after infection, and total RNA was isolated for RNA sequencing. #, P < 0.1; *, P < 0.05; **, P < 0.01 (two-tailed t test). Download FIG S6, PDF file, 0.8 MB (810.7KB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ifenprodil-targeted NMDA receptor genes and flavopiridol-targeted cyclin-dependent kinase 4 (CDK4) genes are linked to lung injury.
Ifenprodil is a selective N-methyl-d-aspartate (NMDA) receptor antagonist (20) and has been clinically used to treat neuronal injury and neurological disorders induced by overstimulation of the NMDA receptor (21). Our RNAi screening data revealed that knockdown of the NMDA receptor gene (GRIN2B) could alter the viability of H5N1-infected cells (Table S1). We obtained RNA sequencing data from lung tissues of H5N1-infected mice treated with ifenprodil and analyzed biological processes and pathways influenced by ifenprodil compared to vehicle control mice. We found that ifenprodil influenced the immune response and neurophysiological processes in the mouse lung (Fig. 4A). Nine of the top 10 pathways of the ifenprodil treatment group were linked to the immune response (Fig. 4C). Dozens of genes in most of the top 10 pathways were reported to be related to lung diseases as well as to traditional neuropathic indications of ifenprodil (Fig. 4C; see also Table S4 in the supplemental material). Although previous reports indicated that NMDA receptors are expressed in lung tissues, that NMDA receptor signaling is linked to inflammation, and that overstimulation of the NMDA receptor can trigger lung injury (22, 23), our study revealed for the first time that ifenprodil is effective in avian influenza A H5N1 virus infection and lung injury.
FIG 4.
Functional processes and pathways influenced by ifenprodil and flavopiridol in H5N1-infected mice. (A and B) GO term (biological process) enrichment of DEGs associated with ifenprodil (no. 42) treatment (A) or flavopiridol (no. 67) treatment (B) in H5N1-infected mice, as analyzed by Metacore. Enrichment was visualized using the Enrichment Map application in Cytoscape. Biological processes are presented as nodes, while the node color indicates the significance of the biological processes, and the node size reflects the object count enriched in the biological processes. The connection between biological processes is based on shared objects. (C and D) Heat maps of the percentage of objects related to the traditional indication or to lung diseases in the top 10 enriched pathways associated with ifenprodil (no. 42) treatment (C) or flavopiridol (no. 67) treatment (D). LN, lung neoplasms; LI, lung diseases (interstitial); LO, lung diseases (obstructive); PF, pulmonary fibrosis; T, traditional indication; SLE, systemic lupus erythematosus; NETosis, neutrophil extracellular trap-associated cell death; PR, progesterone receptor; ERK, extracellular signal-regulated kinase; PI3K, phosphatidylinositol 3-kinase; MAPK, mitogen-activated protein kinase; COPD, chronic obstructive pulmonary disease. Detailed information about the enriched pathways and objects is provided in Table S4.
Traditional/lung disease-related objects in the top 10 pathways for ifenprodil (no. 42) and flavopiridol (no. 67) administration. Download Table S4, XLSX file, 0.06 MB (62.5KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Flavopiridol is an inhibitor of cyclin-dependent kinases (CDKs) (including CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, CDK8, and CDK9) and has been investigated for the treatment of several types of cancers, including leukemia, liver cancer, and renal cancer, in clinical phase 2 trials (24–26). Previous research found that CDK4 inhibition resulted in the improvement of lung injury by blocking leukocyte adhesion and migration and that flavopiridol inhibited avian influenza H7N9 virus replication in A549 cells but that its efficiency in vivo could not be confirmed (27). Our RNAi screening results showed that the viability of H5N1 virus-infected cells was increased due to CDK4 gene knockdown (Table S1). Using RNA sequencing data for biological process and pathway analysis, we found that flavopiridol treatment could alter the immune response as well as cell development and differentiation in H5N1 virus-infected mouse lung tissues (Fig. 4B). Among the top 10 pathways of the flavopiridol treatment group, 5 are related to the immune response of lung disease and 2 are involved in cancer (Fig. 4D). We found that dozens of genes in most of the top 10 pathways were linked to lung diseases as well as to the known antineoplastic function of flavopiridol (Fig. 4D; see also Table S4). Our study revealed for the first time that flavopiridol is effective in lung injury.
DISCUSSION
In this study, we established an efficient whole-genome RNAi-based platform for repurposing drugs for H5N1 infection in A549 cells. Compared to primary epithelial cells, A549 cells are widely used in large-scale experiments for the study of influenza A virus based on convenience and economic considerations (11, 13, 15). We identified potential gene targets via a genomewide RNAi screen, in which we searched for drugs that would maximize the viability of H5N1-infected cells. Most genomewide RNAi screens use replication of the moderately pathogenic influenza H1N1 virus as the readout of interference (8–15). Since the highly pathogenic avian influenza H5N1 virus can cause significant cell death (28) and since cell viability is highly correlated to virus lethality, we designed our genomewide RNAi screening platform to use cell death as the readout in order to identify repurposable drugs for treatment of H5N1 virus infection. Of the 28 effective drugs identified in vitro, 8 were glucocorticoids, which are already used to treat ALI in patients (18, 19). We showed that 8 of the top 10 nonhormone drugs effectively ameliorated ALI in H5N1-infected mice. We identified the neurological drug ifenprodil and the anticancer drug flavopiridol as potential novel effective remedies for avian influenza A H5N1 virus-infected lung injury.
The mechanisms by which ifenprodil and flavopiridol ameliorate lung injury and improve the survival of H5N1-infected mice may operate via an influence on the host immune response. Previous studies reported that limiting the infiltration of leukocytes such as NK cells and neutrophils could alleviate influenza A virus-induced ALI (29, 30). We found that both ifenprodil and flavopiridol significantly decreased the number of infiltrating cells in H5N1-infected mouse lung tissues. In addition, hypercytokinemia was reported in the blood samples of patients with lethal avian influenza A virus infection, and plasma cytokine and chemokine levels were linked to disease fatal outcomes (31, 32). In our study, both ifenprodil and flavopiridol alleviated cytokine storm, as they significantly decreased the RNA and protein levels of some cytokines and chemokines in lung tissues and blood samples from mice with H5N1 virus infection. In addition, we obtained H5N1-infected mouse lung tissues for RNA sequencing and conducted bioinformatic analysis, which showed that the genes influenced by ifenprodil or flavopiridol administration were highly clustered in GO functional terms and pathways related to the immune response. From these collective results, we speculated that the impact of ifenprodil and flavopiridol on the immune response contributes to the amelioration of H5N1-induced ALI and to improvement in mouse survival. However, the efficacy of ifenprodil and flavopiridol for the treatment of H5N1-induced ALI also needs clinical evaluation.
In our study, we evaluated the efficiency in H5N1-infected A549 cells of only 104 commercially available drugs from 372 drug candidates identified using our genomewide RNAi screening method; the efficiencies of the rest of the 268 drugs need to be further tested. Furthermore, we examined the efficiencies of only 10 of the 28 in vitro-effective drugs in an animal model; the rest of the drug candidates also need to be tested in vivo.
Our report provides an economical, quick, and highly effective method (compared with traditional drug development strategies) for identifying novel remedies against avian influenza virus-induced lung injury. This approach could be generalized for identifying other contexts in which drugs can be repurposed.
MATERIALS AND METHODS
Cells and virus.
A549 human lung adenocarcinoma epithelial cells (ATCC, Rockville, MD, USA) were maintained in Ham’s F12 nutrient medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U ml−1 penicillin, and 100 U ml−1 streptomycin. Influenza virus A/Jilin/9/2004 (H5N1) was propagated by inoculation into specific-pathogen-free embryonated fowl eggs (10 to 11 days old) via the allantoic route. All in vivo and in vitro experiments involving live virus were performed in biosafety level 3 facilities.
Genomewide siRNA screen.
A genomewide small interfering RNA (siRNA) library targeting 19,424 human genes (with three siRNAs targeting each gene) and negative-control (NC) siRNAs was purchased from RiboBio (Guangzhou, China). A549 cells were plated into 96-well plates and transfected with siRNA (100 nM) using Lipofectamine RNAiMAX reagent (Invitrogen, CA, USA). At 24 h after transfection, A549 cells were infected with H5N1 influenza virus (multiplicity of infection [MOI], 3.0) or administered an equal volume of allantoic fluid (AF). At 48 h after infection, cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H (MTS) assay (catalog no. G3582, Promega, WI, USA). Cell viability values were normalized by calculating the ratio with respect to the viability of cells transfected with NC siRNA in the same plates as follows:
where siRNAH5N1 represents the H5N1-infected siRNA group, siRNAAF represents the AF-treated siRNA group, NCH5N1 represents the H5N1-infected NC group, and NCAF represents the AF-treated NC group. A two-tailed P value of <0.05 was considered statistically significant. Genes associated with normalized cell viabilities that were increased or decreased by >10% were taken into the second round of screening. Detailed information about the host genes identified in the genomewide RNAi screen is provided in Table S1 in the supplemental material.
Gene enrichment and network analyses.
The top hits in the siRNA screen were functionally grouped according to Gene Ontology (GO) terms and functional pathways as clustered by Metacore (Clarivate Analytics) software. The criteria used to identify “disease-related genes” followed the standards of Metacore (Clarivate Analytics) software. Cytoscape (version 3.6.1) with the Enrichment Map application (version 3.1.0) was used for visualization. Interactions among the genes were assessed using the STRING database (https://string-db.org/).
Drugs.
A total of 1,137 genes were searched as drug targets in the DrugBank database (www.DrugBank.ca/), and 104 drugs were ultimately selected as candidates. All drugs were purchased from Selleck Chemicals (Houston, TX, USA), and detailed information about the drugs is provided in Table S2.
In vitro experiments.
Drugs were added at 1, 10, and 100 μM prophylactically (3 h before infection) or therapeutically (3 h after infection) to A549 cells infected with H5N1 influenza virus, and cell viabilities were measured by MTS assay at 48 h after infection. We performed two rounds of screens to confirm drug efficacy. Drug cytotoxicity was measured by assessing the viability of drug-treated A549 cells that were not infected with H5N1. Negative-control cultures were treated with dimethyl sulfoxide (DMSO). Drugs that significantly improved the viability of H5N1-infected A549 cells and that showed low cytotoxicity in the two screens were considered effective for treating H5N1 infection in vitro. Drug information and the effective concentrations are listed in Table S3.
In vivo experiments.
Wild-type C57BL/6 mice (6 to 8 weeks old; catalog no. 5653791, RRID MGI:5653791) were purchased from Vital River (Beijing, China) and divided into two groups (5 mice per group) based on whether drug was administered by intraperitoneal (i.p.) injection or by gavage. A control group was administered vehicle. Mice were intratracheally instilled with live H5N1 virus (106 50% tissue culture infectious doses [TCID50]), and they received drug or vehicle at 4 time points: 24 h and 3 h before and 24 h and 48 h after infection. Drug administration methods and concentrations are listed in Table S3. Mice were sacrificed at 3 days after viral infection, and bilateral lungs were collected to assess lung injury and pulmonary edema (33). The animal experiments in this work were approved by the Ethics Committee of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (ACUC-A02-2017-014) and adhered to the Chinese National Guidelines for the Care of Laboratory Animals and the Institutional Animal Care.
Immunofluorescence staining.
Lung tissue was fixed with 4% formaldehyde for at least 24 h and was then embedded in paraffin. Next, antigen retrieval was performed. For immunofluorescence staining, sections were blocked in 1% bovine serum albumin (BSA) and subsequently incubated overnight with the following primary antibodies in phosphate-buffered saline (PBS) at the indicated dilutions: anti-CD45 (rat; Abcam), 1:200; anti-CD11b (rabbit; Abclonal), 1:100; anti-ITGAX (CD11c) (rabbit; Abclonal), 1:100; anti-NCR1 (rabbit; Abclonal), 1:100; anti-CD3e (rabbit; Abclonal), 1:100; antimyeloperoxidase (anti-MPO) (rabbit; CST), 1:500. Secondary antibodies conjugated to Alexa Fluor 488, Cy3, or 4′,6-diamidino-2-phenylindole (DAPI) were used at a dilution of 1:200. Whole-section images were scanned with a Nikon (Tokyo, Japan) Eclipse C1 fluorescence microscope and Nikon DS-U3 imaging system to identify leukocyte subsets.
Measurement of cytokines and chemokines.
Three groups of five C57BL/6 mice (6 to 8 weeks old) were used in this study. Mice were infected with live H5N1 virus (106 TCID50) and administered vehicle, ifenprodil (20 mg/kg/body weight, intraperitoneally), or flavopiridol (5 mg/kg, intraperitoneally) at 4 time points: 24 h and 3 h before and 24 h and 48 h after infection. Mouse plasma samples were collected 72 h after infection. The levels of cytokines and chemokines in the plasma were measured with a Bio-Plex mouse cytokine 23-plex panel in a Bio-Plex protein array system (Bio-Rad Laboratories).
Measurement of the survival rate and body weight changes.
Wild-type C57BL/6 mice were divided into three groups (9 to 10 mice per group). Mice were administered vehicle, ifenprodil (20 mg/kg, intraperitoneally), or flavopiridol (5 mg/kg, intraperitoneally) at 24 h and 3 h before and 24 h and 48 h after infection (106 TCID50 of H5N1 virus). The survival and body weight changes of the mice in each group were assessed daily for 14 days. The log rank test was used to compare the Kaplan-Meier survival curves.
RNA sequencing and data analysis.
Lung tissue and blood from mice treated with drug or vehicle were obtained at 3 days after H5N1 infection. Total RNA was isolated from lung tissue and blood using TRIzol (Invitrogen, CA, USA). High-throughput strand-specific RNA sequencing was performed using an Illumina HiSeq 2500 platform (Berry Genomics, Beijing, China). Following quality control of the RNA sequencing reads by FastQC (version 0.11.2), the reads were mapped to the mouse genome (version mmc10; http://hgdownload.cse.ucsc.edu/downloads.html) with Bowtie2 (version 2.1.0) and TopHat2 (version 2.0.11). We then assembled the transcription units to calculate the number of mapped fragments per kilobase per million (FPKM) and analyzed the differentially expressed genes (DEGs) between samples using Cufflinks (version 2.2.1), Cuffmerge (version 2.2.1), and Cuffdiff (version 2.2.1). The functional pathways of DEGs between animals administered drug or vehicle were analyzed using Metacore.
Statistical analysis.
Differences between two groups were assessed for significance using unpaired t tests or the Mann-Whitney U test. Differences among three or more groups were assessed using analysis of variance (ANOVA). The log rank test was used for Kaplan-Meier survival analysis. Statistical analysis was performed using GraphPad Prism 7.0. Differences associated with a two-tailed P value of <0.05 and a false-discovery-rate value (q value) of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81788101), the Ministry of Science and Technology of China (2015CB553406), the National Natural Science Foundation of China (81490531), and the CAMS Innovation Fund for Medical Sciences (2017-I2M-1-009).
We declare that we have no competing interests.
C. Jiang conceived the project. C. Jiang and N. Jin designed the study and analyzed the data. C. Zhang performed the biosafety level 3 experiments employing the genomewide RNAi screen, with assistance from Q. Zhang, Q. Liu, H. Lu, and X. Li. C. Zhang and Y. Qin performed cell viability and the biosafety level 3 experiments in mice. Y. Zhang and D. Shang analyzed the pathology results. C. Zhang, Y. Zhang, and D. Shang screened the candidate drugs. F. Huang led the bioinformatic analysis with assistance from C. Zhang. C. Zhou helped analyze the data. F. Huang, C. Zhang, Y. Zhang, and C. Jiang wrote the manuscript.
REFERENCES
- 1.Medina RA, Garcia-Sastre A. 2011. Influenza A viruses: new research developments. Nat Rev Microbiol 9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H, Gao Z, Feng Z, Shu Y, Xiang N, Zhou L, Huai Y, Feng L, Peng Z, Li Z, Xu C, Li J, Hu C, Li Q, Xu X, Liu X, Liu Z, Xu L, Chen Y, Luo H, Wei L, Zhang X, Xin J, Guo J, Wang Q, Yuan Z, Zhou L, Zhang K, Zhang W, Yang J, Zhong X, Xia S, Li L, Cheng J, Ma E, He P, Lee SS, Wang Y, Uyeki TM, Yang W. 2008. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS One 3:e2985. doi: 10.1371/journal.pone.0002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alessandri F, Pugliese F, Ranieri VM. 2018. The role of rescue therapies in the treatment of severe ARDS. Respir Care 63:92–101. doi: 10.4187/respcare.05752. [DOI] [PubMed] [Google Scholar]
- 4.Nosengo N. 2016. Can you teach old drugs new tricks? Nature 534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 5.Guidi A, Mansour NR, Paveley RA, Carruthers IM, Besnard J, Hopkins AL, Gilbert IH, Bickle QD. 2015. Application of RNAi to genomic drug target validation in schistosomes. PLoS Negl Trop Dis 9:e0003801. doi: 10.1371/journal.pntd.0003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kodama M, Kodama T, Newberg JY, Katayama H, Kobayashi M, Hanash SM, Yoshihara K, Wei Z, Tien JC, Rangel R, Hashimoto K, Mabuchi S, Sawada K, Kimura T, Copeland NG, Jenkins NA. 2017. In vivo loss-of-function screens identify KPNB1 as a new druggable oncogene in epithelial ovarian cancer. Proc Natl Acad Sci U S A 114:E7301–E7310. doi: 10.1073/pnas.1705441114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 11.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su WC, Chen YC, Tseng CH, Hsu PW, Tung KF, Jeng KS, Lai MM. 2013. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc Natl Acad Sci U S A 110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward SE, Kim HS, Komurov K, Mendiratta S, Tsai PL, Schmolke M, Satterly N, Manicassamy B, Forst CV, Roth MG, Garcia-Sastre A, Blazewska KM, McKenna CE, Fontoura BM, White MA. 2012. Host modulators of H1N1 cytopathogenicity. PLoS One 7:e39284. doi: 10.1371/journal.pone.0039284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, Kawaoka Y. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16:795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Ji ZL, Chen YZ. 2002. TTD: Therapeutic Target Database. Nucleic Acids Res 30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. 2008. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meduri GU, Bridges L, Shih MC, Marik PE, Siemieniuk RAC, Kocak M. 2016. Prolonged glucocorticoid treatment is associated with improved ARDS outcomes: analysis of individual patients’ data from four randomized trials and trial-level meta-analysis of the updated literature. Intensive Care Med 42:829–840. doi: 10.1007/s00134-015-4095-4. [DOI] [PubMed] [Google Scholar]
- 19.Anonymous. 2017. Erratum: clinical practice guideline of acute respiratory distress syndrome. Tuberc Respir Dis (Seoul) 80:95. doi: 10.4046/trd.2017.80.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H. 2016. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534:63–68. doi: 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gogas KR. 2006. Glutamate-based therapeutic approaches: NR2B receptor antagonists. Curr Opin Pharmacol 6:68–74. doi: 10.1016/j.coph.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Said SI, Berisha HI, Pakbaz H. 1996. Excitotoxicity in the lung: N-methyl-d-aspartate-induced, nitric oxide-dependent, pulmonary edema is attenuated by vasoactive intestinal peptide and by inhibitors of poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A 93:4688–4692. doi: 10.1073/pnas.93.10.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Luo Z, Liu S, Li L, Deng X, Huang F, Shang L, Jian C, Yue S. 2009. Glutamate mediates hyperoxia-induced newborn rat lung injury through N-methyl-d-aspartate receptors. Am J Respir Cell Mol Biol 40:260–267. doi: 10.1165/rcmb.2008-0135OC. [DOI] [PubMed] [Google Scholar]
- 24.Ang C, O'Reilly EM, Carvajal RD, Capanu M, Gonen M, Doyle L, Ghossein R, Schwartz L, Jacobs G, Ma J, Schwartz GK, Abou-Alfa GK. 2012. A nonrandomized, phase II study of sequential irinotecan and flavopiridol in patients with advanced hepatocellular carcinoma. Gastrointest Cancer Res 5:185–189. [PMC free article] [PubMed] [Google Scholar]
- 25.Stadler WM, Vogelzang NJ, Amato R, Sosman J, Taber D, Liebowitz D, Vokes EE. 2000. Flavopiridol, a novel cyclin-dependent kinase inhibitor, in metastatic renal cancer: a University of Chicago phase II consortium study. J Clin Oncol 18:371–375. doi: 10.1200/JCO.2000.18.2.371. [DOI] [PubMed] [Google Scholar]
- 26.Karp JE, Garrett-Mayer E, Estey EH, Rudek MA, Smith BD, Greer JM, Drye DM, Mackey K, Dorcy KS, Gore SD, Levis MJ, McDevitt MA, Carraway HE, Pratz KW, Gladstone DE, Showel MM, Othus M, Doyle LA, Wright JJ, Pagel JM. 2012. Randomized phase II study of two schedules of flavopiridol given as timed sequential therapy with cytosine arabinoside and mitoxantrone for adults with newly diagnosed, poor-risk acute myelogenous leukemia. Haematologica 97:1736–1742. doi: 10.3324/haematol.2012.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perwitasari O, Yan X, O'Donnell J, Johnson S, Tripp RA. 2015. Repurposing kinase inhibitors as antiviral agents to control influenza A virus replication. Assay Drug Dev Technol 13:638–649. doi: 10.1089/adt.2015.0003.drrr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Li C, Shu Y, Ju X, Zou Z, Wang H, Rao S, Guo F, Liu H, Nan W, Zhao Y, Yan Y, Tang J, Zhao C, Yang P, Liu K, Wang S, Lu H, Li X, Tan L, Gao R, Song J, Gao X, Tian X, Qin Y, Xu KF, Li D, Jin N, Jiang C. 2012. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal 5:ra16. doi: 10.1126/scisignal.2001931. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Li F, Sun R, Gao X, Wei H, Tian Z. 2014. Klebsiella pneumoniae alleviates influenza-induced acute lung injury via limiting NK cell expansion. J Immunol 193:1133–1141. doi: 10.4049/jimmunol.1303303. [DOI] [PubMed] [Google Scholar]
- 30.Narasaraju T, Yang E, Samy RP, Ng HH, Poh WP, Liew AA, Phoon MC, van Rooijen N, Chow VT. 2011. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol 179:199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, Huang F, Liu J, Chen Y, Wang W, Cao B, Zou Z, Liu S, Pan J, Bao C, Zeng M, Xiao H, Gao H, Yang S, Zhao Y, Liu Q, Zhou H, Zhu J, Liu X, Liang W, Yang Y, Zheng S, Yang J, Diao H, Su K, Shao L, Cao H, Wu Y, Zhao M, Tan S, Li H, Xu X, Wang C, Zhang J, Wang L, Wang J, Xu J, Li D, Zhong N, Cao X, Gao GF, Li L, Jiang C. 2015. The serum profile of hypercytokinemia factors identified in H7N9-infected patients can predict fatal outcomes. Sci Rep 5:10942. doi: 10.1038/srep10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, Ju X, Liang Z, Liu Q, Zhao Y, Guo F, Bai T, Han Z, Zhu J, Zhou H, Huang F, Li C, Lu H, Li N, Li D, Jin N, Penninger JM, Jiang C. 2014. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun 5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomewide RNAi screen for host genes associated with H5N1 virus infection. (A) Flow chart of the genomewide RNAi screen of 19,424 genes to identify those important in influenza A H5N1 virus infection. (B) Heat map of cell viability related to 337 genes whose knockdown altered cell viability by >30% in each of two siRNA screens. The two columns of the heat map represent the first and second rounds of genomewide RNAi screening. Download FIG S1, PDF file, 0.4 MB (425KB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional enrichment of essential host genes associated with H5N1 infection. (A) Enriched Gene Ontology cellular component terms associated with 337 genes, as analyzed using Metacore. (B) Interactions among clusters of spliceosomes. The three kinds of colored circles (dark red, medium red, and light red) indicate that the cell viability increased more than 30%, 20%, and 10%, respectively. (C) Interactions among the clusters of mitochondrial respiratory chain complex I. The three kinds of colored circles (dark blue, medium blue, and light blue) indicate that the cell viability decreased more than 30%, 20%, and 10%, respectively. Download FIG S2, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Host genes identified in the genomewide RNAi screen. Download Table S1, XLSX file, 0.3 MB (268.1KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro validation of drugs against H5N1 virus infection. (A) Flow chart of drug screening based on 1,137 genes and 104 drug candidates. (B) Flow chart of the in vitro validation of drug efficacy against H5N1 infection. (C) Viability of A549 cells infected with H5N1 at a multiplicity of infection of 3.0 and treated with drugs prophylactically (3 h before infection) or therapeutically (3 h after infection). Viability was assessed at 48 h after infection and is expressed relative to that of vehicle-treated control cells. In vitro experiments were repeated three times. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed t test). Download FIG S3, PDF file, 0.3 MB (405.9KB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Drug information. Download Table S2, XLSX file, 0.01 MB (13.4KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro and in vivo drug information. Download Table S3, XLSX file, 0.01 MB (11.2KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vivo validation of drugs against H5N1 infection. (A) Flow chart of the in vivo validation of drug efficacy. (B) Lung injury scores of tissue from H5N1-infected C57BL/6 mice (n = 3 to 5) treated with drug candidates by intragastric administration. Lung tissues were obtained at 3 days after infection. Lung injury scores per microscopic field are shown in the bar graph. n = 100 fields were analyzed for each group. (C and D) Wet-to-dry-weight ratios of lung tissue from H5N1-infected C57BL/6 mice treated with drugs by intraperitoneal injection (C) or intragastric administration (D). Lung tissues were obtained at 3 days after infection. Compound identities: no. 15, glyburide; no. 31, bupivacaine; no. 47, AEBSF; no. 97, diazoxide; no. 101, diflunisal; no. 104, vemurafenib. The data shown represent means ± standard errors of the means (SEMs). In vivo experiments were repeated three times. *, P < 0.05; **, P < 0.01 (two-tailed test). (E) Body weight changes in H5N1-infected C57BL/6 mice treated with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle by intraperitoneal injection. Download FIG S4, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ifenprodil and flavopiridol impact inflammatory cell infiltration. (A) Representative images of lung pathology by immunofluorescence staining from H5N1-infected C57BL/6 mice (n = 3 to 5) treated with ifenprodil (no. 42), flavopiridol (no. 67), or vehicle by intraperitoneal injection. Lung tissues were removed at 3 days after infection. The blue areas indicate cell nuclei stained with DAPI, the red areas indicate CD45 protein (leukocyte common antigen), and the green areas indicate marker proteins. Scale bar, 100 μm. The numbers of neutrophil cells (CD45+ MPO+), T cells (CD45+ CD3e+), and NK cells (CD45+ NCR1+) per microscopic field are shown in the bar graph (means ± SEM, n = 100 fields per treatment). (B and C) The numbers of macrophage cells (B) and dendritic cells (C) per microscopic field are shown in the bar graph (means ± SEM, n = 100 fields per treatment). *, P < 0.05; **, P < 0.01 (two-tailed t test). Download FIG S5, PDF file, 2.2 MB (2.2MB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Drugs antagonize H5N1-induced upregulation of cytokines. (A and B) Relative mRNA levels of cytokines and chemokines in H5N1-infected C57BL/6 mice (n = 3 to 5) treated with ifenprodil (no. 42) (A), flavopiridol (no. 67) (B), or vehicle. Lung tissue samples were collected at 72 h after infection, and total RNA was isolated for RNA sequencing. #, P < 0.1; *, P < 0.05; **, P < 0.01 (two-tailed t test). (C to H) Relative mRNA levels of cytokines and cytokine receptors in H5N1-infected C57BL/6 mice (n = 3 to 5) treated with glyburide (no. 15) (C), bupivacaine (no. 31) (D), AEBSF (no. 47) (E), diazoxide (no. 97) (F), diflunisal (no. 101) (G), vemurafenib (no. 104) (H), or vehicle. Blood samples were collected at 72 h after infection, and total RNA was isolated for RNA sequencing. #, P < 0.1; *, P < 0.05; **, P < 0.01 (two-tailed t test). Download FIG S6, PDF file, 0.8 MB (810.7KB, pdf) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Traditional/lung disease-related objects in the top 10 pathways for ifenprodil (no. 42) and flavopiridol (no. 67) administration. Download Table S4, XLSX file, 0.06 MB (62.5KB, xlsx) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.