Abstract

Monitoring the β-lactam antibiotic level has been an important task in food industry and clinical practice. Here, we report the development of a fluorescent PenP β-lactamase, PenP-E166Cf/N170Q, for efficient β-lactam antibiotic detection. It was constructed by covalently attaching fluorescein onto the active-site entrance of a thermostable E166Cf/N170Q mutant of a Bacillus licheniformis PenP β-lactamase. It gave a fluorescence turn-on signal toward various β-lactam antibiotics, where the fluorescence enhancement was attributed to the acyl–enzyme complex formed between PenP-E166Cf/N170Q and the β-lactam antibiotic. It demonstrated enhanced signal stability over its parental PenP-E166Cf because of the suppressed hydrolytic activity by the N170Q mutation. Compared with our previously constructed PenPC-E166Cf biosensor, PenP-E166Cf/N170Q was more thermostable and advanced in detecting β-lactams in terms of response time, signal stability, and detection limit. Positive fluorescence signals generated by E166Cf/N170Q in response to the penicillin-containing milk and mouse serum illustrated the feasibility of the biosensor for antibiotic detection in real samples. Taken together, our findings suggest the potential application of PenP-E166Cf/N170Q in biosensing β-lactam antibiotics.
1. Introduction
β-Lactam antibiotics, including penicillins and cephalosporins, are nowadays the most consumed antibiotics worldwide and have been extensively used in human and veterinary medicine.1−3 Surveillance of their level in food and biological samples has posed significant implications in food industry and clinical practice. Presence of β-lactam antibiotic residues in edible products from animal origin (e.g., milk and poultry meat), which has often been attributed to improper use and overuse of the antibiotics, can lead to adverse consequences including difficulties in fermentation in food industry, potential public health threats of antibiotic allergy in hypersensitive individuals, and emergence of antibiotic-resistant superbugs.4−8 Thus, it is necessary to stringently monitor the residual level of β-lactam antibiotics for food safety control. In addition, quantification of the antibiotic level in clinical samples (e.g., human plasma) can help evaluate the pharmacokinetics of the therapeutics, which is important information for the determination of effective dosing regimens in the infection treatment.9,10
Presently, numerous methods have been devised for detection of various types of antibiotics in food and biological samples. In general, they fall into three major categories: microbial inhibition tests, rapid tests, and analytical methods. Microbial inhibitory assays rely on the susceptibility of bacteria to the residual antibiotics in the samples.11−14 They are simple and cost-effective, but lengthy and lack of selectivity. Rapid tests (e.g., ELISA and surface plasmon resonance biosensor) are screening methods based on the recognition between antibiotics or receptor proteins specific to a particular type of antibiotics.11,15−20 They allow fast and sensitive but usually semi-quantitative determination for the antibiotic level. Analytical methods, including high-performance liquid chromatography and mass spectrometry (MS), provide a confirmatory assessment of the antibiotic’s authenticity and a precise qualitative measurement of the antibiotic level.21−25 However, these methods are usually laborious, requiring sophisticated equipment and tedious sample preparation steps. All these three categorical approaches have often been employed for practical β-lactam antibiotic detection.11,14,17−20 Microbial inhibition tests provide a nondiscriminatory detection of β-lactam antibiotics, whereas analytical analyses offer a discriminatory examination of the drug level. In parallel to these two approaches, rapid tests specific to β-lactam antibiotics, for example, Charm test and Parallux, are often adopted for fast screening.17,26−28 Furthermore, there has been a growing trend in developing innovative biosensors for simple, speedy, reliable, and efficient screening of antibiotic residues in food and clinical specimens.19,29−32
In the face of the growing demands for reliable, sensitive, and rapid detection system for β-lactam antibiotics, we previously developed a fluorescent β-lactamase-based biosensor, PenPC-E166Cf, as a reagentless platform for screening β-lactam antibiotics.29 β-lactamase is a bacterial enzyme that can hydrolyze the β-lactam ring of the antibiotics, conferring bacterial resistance to these drugs.33 Taking the advantage of the substrate specificity of β-lactamase to a broad spectrum of β-lactam antibiotics, in our previous study, a PenPC β-lactamase from Bacillus cereus was engineered into a biosensor by replacing the catalytically important glutamate residue at position 166 (E166) with a cysteine, followed by a conjugation of a fluorescein molecule at that site. Although PenPC-E166Cf demonstrated a dramatically reduced hydrolytic activity than the wild-type PenPC β-lactamase, its binding ability with β-lactams was greatly restored.29 For antibiotic detection, the active-site pocket of PenPC-E166Cf served as the recognition element of the biosensor by allowing specific binding of the antibiotic substrate to the biosensor. Upon substrate binding, fluorescein located in close proximity to the active site flipped out from a buried hydrophobic region to a more solvent-exposed environment.29 In this way, fluorescein participated as a signal transducer in which its movement away from the active-site cavity triggered a fluorescence enhancement of PenPC-E166Cf, thereby reporting the presence of antibiotics in the tested sample.34 This hypothesis of the biosensing mechanism of fluorescein-modified β-lactamase was confirmed by crystallographic analysis using an E166Cf derived from a thermostable PenP β-lactamase from Bacillus licheniforms.35 PenP β-lactamase shares 52% identity in the amino acid sequence with the PenPC enzyme.36,37 Both PenP and PenPC enzymes belong to class A β-lactamase, possessing conserved structural and functional elements essential for the same mechanistic action for hydrolyzing β-lactam antibiotics via the transient formation of an acyl–enzyme complex (Scheme 1).38,39 PenP-E166Cf was prepared by the same active-site-selective labeling approach as PenPC-E166Cf, and it exhibited a similar fluorescence profile for antibiotic detection as its PenPC counterpart.35 From our previous studies, both PenPC- and PenP-E166Cf biosensors were shown to enable a reliable and specific fluorescence detection of the β-lactam antibiotics in human plasma with low signal interference by the biological fluid.40,41
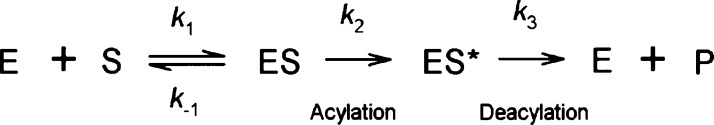
Scheme 1. Hydrolytic Reaction of β-Lactam Antibiotics by a Class A β-Lactamase.
Binding of β-lactamase (E) with the antibiotic substrate (S) leads to the formation of the Michaelis complex (ES). ES then undergoes an irreversible step of acylation to form an acyl–enzyme complex (ES*). Deacylation of ES* eventually takes place, leading to a release of the hydrolyzed product (P) from the active site of the enzyme.
With an aim of improving the biosensor performance of E166Cf, in this study, we have established a new PenP-E166Cf/N170Q system for β-lactam antibiotic detection. This biosensor is derived from PenP-E166Cf, in which an N170Q mutation was incorporated into the protein core to lower its rate of deacylation. We have postulated that PenP-E166Cf/N170Q would show more advanced biosensor features compared to PenPC- and PenP-E166Cf biosensors. First, it would exhibit enhanced protein stability than PenPC-E166Cf because of its thermostable PenP β-lactamase scaffold. Second, it would give a more stable fluorescence signal than the E166Cf biosensors because the degradation rate of the fluorescence signal contributing the acyl–enzyme complex would be reduced owing to its impaired catalytic activity by the N170Q mutation. Here, we report the development of PenP-E166Cf/N170Q as an improved version of the E166Cf sensor. Its thermostability and performance in β-lactam antibiotic detection were assessed and compared with those of the E166Cf biosensors.
2. Results and Discussion
2.1. Catalytic Impairment in E166C and E166C/N170Q Mutants
E166 is a catalytically important residue playing an indispensable role in deacylation of the β-lactamase hydrolysis.42 According to the kinetic results (Table 1), mutation of E166 caused a ∼600-fold decrease and a ∼40 000-fold decrease in the catalytic efficiency of PenP β-lactamase for CENTA and nitrocefin, respectively. When compared with the wild-type PenP enzyme, the E166C mutant displayed a ∼200-fold decrease in the kcat value and a 3-fold increase in the Km value for CENTA, whereas it showed a 4 × 105-fold reduction in kcat with a ∼10-fold drop in Km for nitrocefin. These data implied that E166C mutation posed a severe effect on the hydrolytic rate without significantly affecting the enzyme’s binding affinity with the substrate. In our study, we introduced an N170Q mutation onto the E166C mutant to further suppress the catalytic activity of that mutant. N170, a conserved residue among class A β-lactamases, is important for deacylation. It has been reported that N170Q mutation can render the enzyme defective in deacylation by eliminating the hydrolytic water molecule in the active site.42,43 According to our data, N170Q mutation in the E166C mutant resulted in a profound reduction in the catalytic efficiency of PenP β-lactamase. Catalytic efficiencies of E166C/N170Q were ∼1000-fold lower for CENTA and ∼1.3 × 105-fold lower for nitrocefin than those of the wild type. Compared with E166C, the E166C/N170Q mutant exhibited a 1.1-fold decrease in Km and a 1.9-fold decrease in kcat in the hydrolysis of CENTA, whereas it gave a 1.7-fold lower Km value and a 5-fold lower kcat value in the hydrolysis of nitrocefin. These findings reveal that N170Q mutation caused a slight increase in substrate-binding affinity and a further impairment in the catalytic activity of the E166C mutant.
Table 1. Kinetic Parameters of the PenP β-Lactamase Wild Type and Mutants for Various Antibiotics.
| wild type | E166C | E166C/N170Q | ||
|---|---|---|---|---|
| CENTA | Km (μM) | 34.1 ± 6.6 | 106.7 ± 22.9 | 96.0 ± 19.7 |
| kcat (s–1) | 640.7 ± 38.4 | 3.4 ± 0.25 | 1.8 ± 0.12 | |
| kcat/Km (μM–1 s–1) | 18.8 ± 3.8 | 0.032 ± 0.007 | 0.019 ± 0.004 | |
| nitrocefin | Km (μM) | 41.0a | 4.24 ± 0.23 | 2.52 ± 0.27 |
| kcat (s–1) | 1088a | (2.7 ± 0.05) × 10–3 | (0.54 ± 0.01) × 10–3 | |
| kcat/Km (μM–1 s–1) | 26.5a | (6.4 ± 0.36) × 10–4 | (2.1 ± 0.23) × 10–4 |
Kinetic constants of the non-MBP fused PenP β-lactamase wild type was cited from Escobar et al.44
2.2. Active-Site Labeling Approach for the Construction of PenP Biosensors
In this study, PenP β-lactamase mutants, which served as the biosensor scaffolds, were produced as a fusion protein with a maltose binding protein (MBP). To construct the fluorescent PenP β-lactamase-based biosensors, we adopted an active-site labeling approach to conjugate a fluorescent probe to MBP-PenP enzymes. Residue E166 of a wild-type PenP β-lactamase was first mutated to be a unique cysteine in the active-site pocket, and then a fluorescein molecule was specifically tethered to that location covalently via a thiol reaction between fluorescein-5-maleimide and the sulfhydryl group of the cysteine. Labeled β-lactamase E166C and E166C/N170Q mutants showed fluorescent bands on the protein gel under UV illumination (Figure S2). This indicates the successful labeling of these mutants. As evaluated by MS, these mutants were site-specifically conjugated with fluorescein in a 1:1 ratio to give PenP-E166Cf and PenP-E166Cf/N170Q, respectively, with a more than 90% labeling efficiency (Figure S3). CD analysis revealed that neither mutations nor modification with fluorescein-5-maleimide imposed a significant distortion on the overall secondary structure of the PenP enzyme (Figure S4).
2.3. Fluorescence Biosensing Mechanism of PenP Biosensors
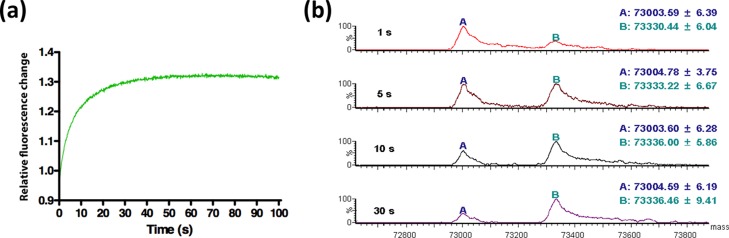
In our previous study, fluorescence signal generation of PenP-E166Cf was examined by fluorometric studies, mass spectrometric analysis, and crystallographic study.35 In PenP-E166Cf, fluorescein at position 166 is located at the entrance of the active-site cavity and situated on a flexible Ω-loop of PenP β-lactamase. In the apo structure, the rigid planar core of fluorescein protruded toward the active site, partially occupying the active-site pocket.35 In the presence of the β-lactam substrate, the entry of the substrate to the β-lactamase’s active site induced a change in the local environment of the fluorescein. Flexibility of the Ω-loop allowed a displacement of fluorescein from a buried hydrophobic region to a more solvent-exposed environment so as to provide space for the accommodation of the substrate in the active-site pocket. This drove a fluorescence enhancement of PenP-E166Cf, reporting the occupancy of the active site. With substrate hydrolysis by E166Cf, the release of the degraded products from the active site allowed the fluorescein to restore its original orientation, thereby leading to a recovery of the fluorescence signal to its basal level. In the current study, PenP-E166Cf/N170Q was shown to employ a similar biosensing mechanism as PenP-E166Cf. Upon the addition of penicillin G, the fluorescence intensity of PenP-E166Cf/N170Q was rapidly enhanced and became level-off in the first 40 s (Figure 1a). This fluorescence turn-on signal was characterized by a mass spectrometric analysis under the same experimental condition. From the mass spectra obtained within the initial 30 s of the reaction between PenP-E166Cf/N170Q and penicillin G, the disappearance of the enzyme (E) was accompanied by a progressive formation and accumulation of the acyl–enzyme complex (ES*) (Figure 1b). This indicated that ES* predominantly contributed to the fluorescence enhancement upon the addition of the β-lactam antibiotic.
Figure 1.
Formation of the acyl–enzyme complex between PenP-E166Cf/N170Q and penicillin G was monitored by (a) stopped-flow fluorescence measurement and (b) MS. (a) Initial fluorescence change of the PenP-E166Cf/N170Q after the addition of penicillin G was recorded for 100 s. (b) Reaction between PenP-E166Cf/N170Q and penicillin G was quenched at different time intervals and subjected to mass spectrometric analysis. Peak A refers to the free enzyme of PenP-E166Cf/N170Q (E), whereas peak B refers to the covalent acyl–enzyme complex formed between PenP-E166Cf/N170Q and penicillin G (ES*).
2.4. PenP-E166Cf/N170Q Displayed an Improved Biosensing Performance Over the E166Cf Biosensors
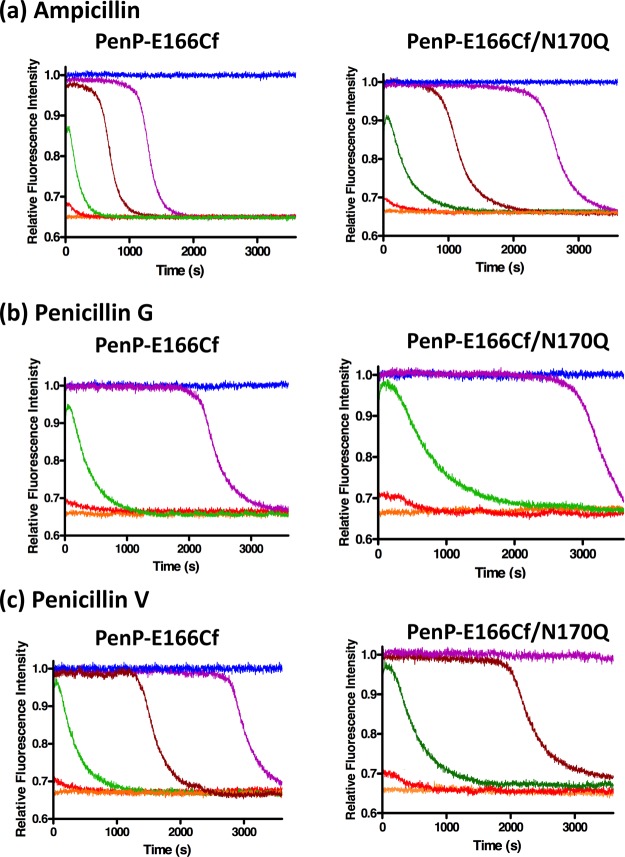
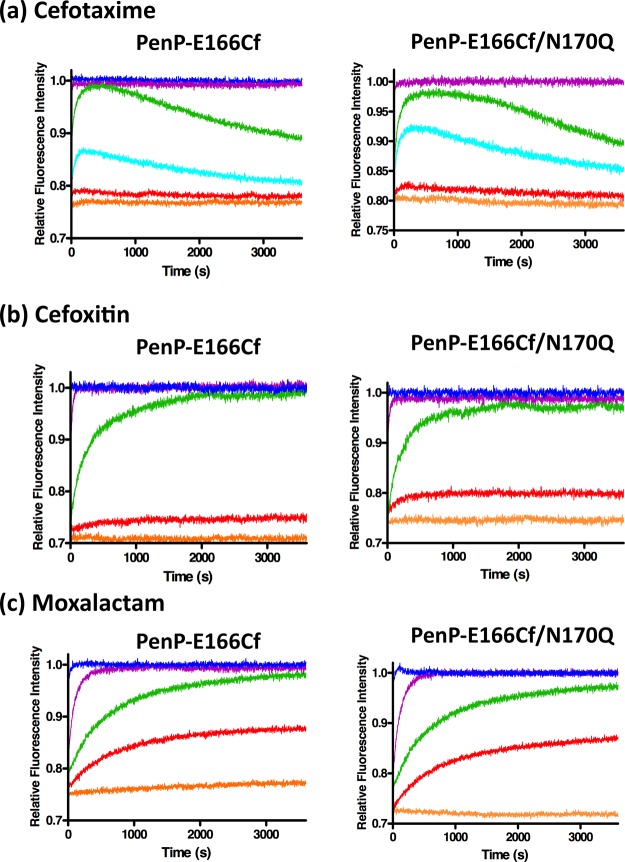
To evaluate the performance of the fluorescent biosensors for β-lactam detection, fluorescence measurements of PenP-E166Cf and PenP-E166Cf/N170Q were conducted with the addition of three penicillins (ampicillin, penicillin G, and penicillin V) (Figure 2) and three cephalosporins (cefotaxime, cefoxitin, and moxalactam) (Figure 3). In general, the two biosensors gave an enhanced fluorescence signal in an antibiotic concentration-dependent manner toward these β-lactam antibiotics. The rising phase of the fluorescence signal resulted from the binding of the biosensor with the antibiotic substrate. When the active site of a biosensor was saturated with the antibiotics, the signal reached its plateau level. With the substrate hydrolysis by the biosensor, the signal declined and returned to its basal level. The calibration curve for the detection of each antibiotic was prepared by plotting the maximum change of the fluorescence intensity against the concentration of the antibiotic (Figure S6). Parameters relating to the biosensor performance of PenP-E166Cf and PenP-E166Cf/N170Q are compared together with those of the previously reported PenPC-E166Cf and summarized in Table 2.
Figure 2.
Fluorescence spectra of 0.1 μM PenP-E166Cf and PenP-E166Cf/N170Q biosensors in 50 mM potassium phosphate (pH 7.0) with various concentrations of (a) ampicillin, (b) penicillin G, and (c) penicillin V: 0 (orange); 0.01 μM (red); 0.1 μM (green); 0.5 μM (brown); 1 μM (purple); and 10 μM (blue).
Figure 3.
Fluorescence spectra of 0.1 μM PenP-E166Cf and PenP-E166Cf/N170Q biosensors in 50 mM potassium phosphate (pH 7.0) with various concentrations of (a) cefotaxime, (b) cefoxitin, and (c) moxalactam: 0 (orange); 0.01 μM (red); 0.05 μM (cyan); 0.1 μM (green); 1 μM (purple); and 10 μM (blue). The figure of detection of cefotaxime by PenP-E166Cf (a; left panel) was adapted from our previously published data and reprinted in part with permission from Wong et al.35 Copyright 2011 BMC.
Table 2. Characterization of PenP-E166Cf, PenP-E166Cf/N170Q, and PenPC-E166Cf in Terms of Biosensor Performance and Thermostability.
| PenP-E166Cf | PenP-E166Cf/N170Q | PenPC-E166Cfa | |||
|---|---|---|---|---|---|
| biosensor performance | response time (s)b | ampicillin | NDg | ND | NDi |
| penicillin G | ND | ND | ND | ||
| penicillin V | ND | ND | NDi | ||
| cefotaxime | ND | ND | NAj | ||
| cefoxitin | 133 | 125 | 1000 | ||
| moxalactam | 688 | 600 | 1385 | ||
| maximum fluorescence change (%)c | ampicillin | 34 | 31.6 | 59.2i | |
| penicillin G | 35.6 | 34 | 60 | ||
| penicillin V | 30 | 33 | 56.3i | ||
| cefotaxime | 22.5 | 20 | NA | ||
| cefoxitin | 29.1 | 25 | 23.8 | ||
| moxalactam | 25.8 | 29.1 | 44 | ||
| duration of signal (s)d | ampicillin | 842 | 1947 | 1781i | |
| penicillin G | 1867 | 2353 | 1286 | ||
| penicillin V | 2474 | >3600; NMh | 1375i | ||
| cefotaxime | >3600; NM | >3600; NM | NA | ||
| cefoxitin | >3600; NM | >3600; NM | >3600; NM | ||
| moxalactam | >3600; NM | >3600; NM | >3600; NM | ||
| limit of detection (μM)e | ampicillin | 0.01 | 0.01 | 0.1i | |
| penicillin G | 0.01 | 0.01 | 0.1 | ||
| penicillin V | 0.01 | 0.01 | 0.1i | ||
| cefotaxime | 0.01 | 0.01 | NA | ||
| cefoxitin | 0.01 | 0.01 | 0.1 | ||
| moxalactam | 0.01 | 0.01 | 0.1 | ||
| thermostability | Tm (°C)f | 57 | 57 | 40 | |
| residual activity after 1 h incubation at 50 °C(%) | 100 | 100 | 0 |
Fluorescence data of PenPC-E166Cf in the presence of penicillin G, cefoxitin, and moxalactam are shown in Figure S5.
Response time: time taken by the biosensor to give a signal reaching a stable maximal level after the addition of a minimal saturating concentration of antibiotics. The lowest antibiotic concentration that saturates PenP biosensors and the PenPC biosensor were 1 and 10 μM, respectively.
Maximum fluorescence change (%) is determined in the presence of antibiotics at a saturating concentration.
Duration of the signal is assessed in the presence of a minimal saturating concentration of antibiotics (for PenP biosensors: 1 μM and for PenPC biosensor: 10 μM) and indicated by the time at which a stable maximal level of the fluorescence signal starts to decline.
Limit of detection: the lowest antibiotic level that can be detected by the biosensor.
Tm (oC): temperature at the relative ellipticity at 222 nm of a protein is dropped to 50% as observed by CD spectroscopy.
ND: not detectable.
NM: not measured.
values obtained by analyzing the results obtained from Chan et al.29
NA: not available.
At the saturating concentration of the β-lactam antibiotics, the maximum intensity change of PenP-based biosensors and PenPC-E166Cf was ∼25–36% and ∼24–60%, respectively (Table 2 and Figure S6). Thus, the amplitude of the signal given by the two PenP biosensors was similar, but lower than that generated by PenPC-E166Cf.
For the detection of penicillins, all the PenP and PenPC biosensors showed a rapid fluorescence enhancement (Figures 2, S5 and Table 2). Because the initiation of the fluorescence turn-on upon the addition of the penicillin-type antibiotic was too fast to be detected, the recorded fluorescence intensity had already reached the maximal level and differences in the response time were not detectable among the three biosensors. In contrast, for the detection of cephalosporins, the two PenP biosensors displayed a faster response than PenPC-E166Cf (Figures 3, S5 and Table 2).
Signal stability of the β-lactamase-based biosensors correlated to the susceptibility of the β-lactam analytes toward the hydrolysis by the biosensors and the stability of the acyl–enzyme complex. Therefore, it was envisaged that biosensors with inefficient deacylation can give rise to a stable fluorescence signal owing to the accumulation of the acyl–enzyme adduct. Mutation of the catalytic residue E166 significantly lowered the deacylation rate of the E166Cf biosensors. From our results, fluorescence signals attributed to PenP-based E166Cf biosensors were steadier than those from PenPC-E166Cf (Figures 2 and S5). In particular, PenP-E166Cf/N170Q showed a longer duration of signal than PenP-E166Cf (Table 2). This implied that a further impairment in deacylation of PenP-E166Cf by the introduction of N170Q mutation improved the stability of the PenP biosensors.
For the three tested antibiotics, PenP-E166Cf and PenP-E166Cf/N170Q offered a lower detection limit than PenPC-E166Cf. From our data, the lowest detection level for PenP-E166Cf and PenP-E166Cf/N170Q was 0.01 μM, whereas that for PenPC-E166Cf was 0.1 μM (Table 2 and Figure S6).
Overall, PenP-E166Cf/N170Q showed a similar biosensor performance as PenP-E166Cf in terms of the response time, maximum fluorescence change, and limit of detection, but it demonstrated higher signal stability than PenP-E166Cf. In addition, both PenP biosensors showed superiority over their PenPC counterpart with respect to the signal stability, response time, and detection limit.
2.5. PenP Biosensors Were More Thermostable than PenPC-E166Cf
Thermostability of the PenP and PenPC biosensors was compared in terms of the secondary structure and catalytic activity (Table 2). To assess the structural stability against thermal stress, the thermal denaturation curve of the proteins was determined by plotting the ellipticity change of the proteins at 222 nm as a function of temperature using circular dichroism (Figure 4a). The mid-point of the thermal denaturation curve corresponds to the temperature at which a protein denatures (melting temperature, Tm). MBP-PenP β-lactamase exhibited a Tm of 64 °C which was consistent with the reported Tm of PenP (63 °C),45 revealing that the MBP demonstrated no effect on the thermostability of the enzyme. PenP-E166C and PenP-E166C/N170Q showed the same Tm of 59 °C. Fluorescein-labeled mutants, PenP-E166Cf and PenPC-E166Cf/N170Q, displayed a similar value of Tm, which was lower than those of the unlabeled mutants, revealing that fluorescein labeling slightly destabilized the structure of the cysteine mutants. No matter the cysteine mutants were native or labeled, PenP mutants demonstrated a higher Tm than the PenPC-E166C mutant, illustrating a higher thermostability of the PenP enzyme than the PenPC β-lactamase.
Figure 4.
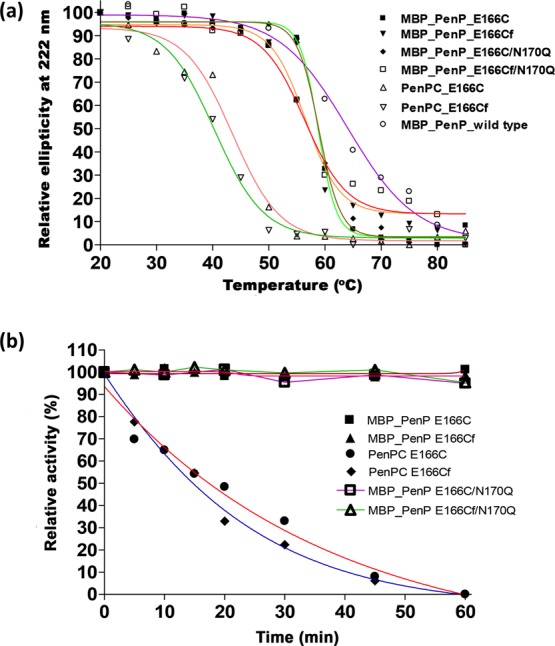
Thermostability of the PenP and PenPC biosensors. (a) Thermal denaturation curves of the biosensors determined by CD. Temperature at which the relative ellipticity at 222 nm is reduced to 50% is regarded as the melting temperature (Tm) of the protein. (b) Residual activities of PenP- and PenPC-based biosensors at 50 °C.
To examine the effect of heat stress on the activity of the β-lactamase-based biosensors, a heat inactivation study was performed by determining the residual activity of β-lactamase for hydrolyzing nitrocefin after 50 °C incubation at different time intervals (Figure 4b). Upon 1 h incubation at 50 °C, unlabeled and fluorescein-labeled PenP mutants showed no reduction in enzyme activity. This observation was in good agreement that the PenP enzymes had Tm higher than 50 °C, and thus these proteins can preserve their structural conformation and maintain their proper function in hydrolyzing the β-lactam substrate at 50 °C. Conversely, activities of the PenPC counterparts, which possessed a Tm value lower than 50 °C, dropped gradually with time and eventually ceased in 1 h. Taken together, PenP biosensors were more thermally stable than PenPC-E166Cf. Moreover, higher tolerance toward thermal stress probably facilitates the storage and durability during detection of these fluorescent biosensors.
2.6. Potential Applications of PenP-Based Biosensors
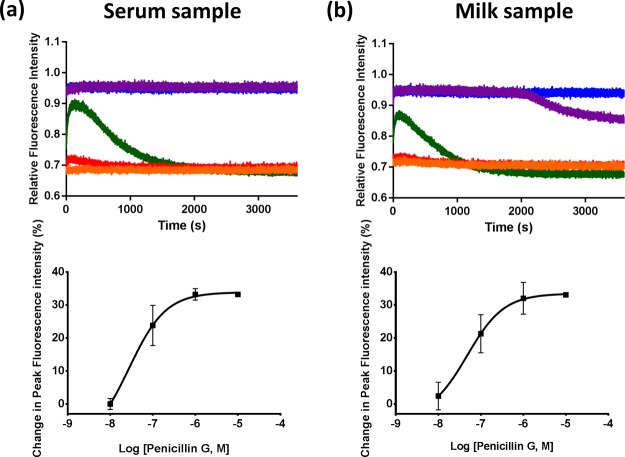
In our previous studies, PenPC-E166Cf and PenP-E166Cf have shown their applications in monitoring the pharmacokinetics of meropenem (a β-lactam antibiotic used in the current clinical setting) in human and animal plasma.40,41 In this study, PenP-E166Cf/N170Q gave fluorescence turn-on signals in the penicillin G containing mouse serum (Figure 5a). The change in its fluorescence intensity was in an antibiotic concentration-dependent fashion. This finding reveals its capability in determining the antibiotic level of the biological samples in clinical practice.
Figure 5.
Detection of penicillin G in the (a) mouse serum and (b) milk sample using PenP-E166Cf/N170Q. Upper panels: Time-resolved fluorescence traces of PenP-E166Cf/N170Q with mouse serum or milk containing various concentrations of penicillin G: 0 (orange); 0.01 μM (red); 0.1 μM (green); 1 μM (purple); and 10 μM (blue). Each point of the trace was the average value collected from three independent measurements. Lower panels: The corresponding calibration curve for the detection of penicillin G. Each point shows the mean ± S.E.M. of three measurements.
Apart from this, our current study also shows that PenP-E166Cf/N170Q allowed antibiotic detection in milk. PenP-E166Cf/N170Q displayed a fluorescence enhancement with the addition of the penicillin G-containing milk sample (Figure 5b). Such a fluorescence profile in milk is similar to those observed in potassium buffer (Figure 2b, right panel) and animal serum (Figure 5a), although there are some variations in parameters, for example, signal duration, which may be due to the batch variation of the biosensor and/or effect of the different compositions of the testing sample medium. Here, it is the first time we demonstrate the potential application of the β-lactamase-based biosensors for sensing antibiotics in milk.
Furthermore, PenP biosensors may become tools for investigating the structure–function relationship of β-lactamase. Nowadays, class A β-lactamases are the most prevalent β-lactamases worldwide,39 and understanding of their interaction with different substrates provides valuable information for the development of effective and potent therapeutics for combating these clinically problematic enzymes. As pointed out by our previous and current studies, fluorescence enhancement in the antibiotic detection of the E166C-based biosensors is mainly resulted from the accumulation of the acyl–enzyme complex (ES*).34,35 The fluorescence traces of these biosensors provide a real-time monitoring of ES*, thereby reflecting the enzymatic reactions occurred in the β-lactamases’ active site. Our current study showed that an incorporation of N170Q mutation onto E166Cf has led to changes in the fluorescence signal pattern in accordance to the mutational effects (e.g., an increased signal duration due to a reduction in the deacylation rate of PenP-E166Cf/N170Q). Therefore, we envision that via the introduction of various mutations of interest onto E166Cf and a comparison of the fluorescence signals between E166Cf and these derivatives, a fast screening of the mutations for their influences on catalytic function and substrate specificity might be easily achieved.
3. Conclusions
In this present study, a fluorescent biosensing molecule, PenP-E166Cf/N170Q, has been prepared by conjugating fluorescein onto the active site’s entrance of a thermostable and catalytically defective β-lactamase mutant. In the β-lactam antibiotic detection, PenP-E166Cf/N170Q demonstrated similar biosensing performance as its parental PenP-E166Cf, but with further impairment in deacylation, it gave a more stable signal than its E166Cf counterpart. Compared with our previously developed PenPC-E166Cf, this new biosensor showed higher stability against heat stress and also better detection performance in terms of response time, signal stability, and detection limit. In conclusion, PenP-E166Cf/N170Q illustrates a great potential to be a versatile tool for antibiotic detection in clinical practice and food industry, and its parental PenP-E166Cf may find its application as a screening platform for β-lactamase mutants’ functions.
4. Experimental Procedures
4.1. Chemicals
Ampicillin, cefotaxime, cefoxitin, CENTA, moxalactam, penicillin G, penicillin V, and kanamycin were purchased from Sigma (St. Louis, MO, USA). Nitrocefin was from Becton Dickinson Company (Cockeysville, Md). Fluorescein-5-maleimide was purchased from Molecular Probes Inc. (Eugene, OR, USA). The chemical structures of the fluorescein-5-maleimide and antibiotics used in this study are shown in Figure S1.
4.2. Overexpression and Purification of PenP β-Lactamase
The PenP β-lactamase wild type and mutants were produced in Escherichia coli with a fusion partner of an MBP at their N-terminus. Briefly, the PenP β-lactamase gene (encoding amino acid residue 29–293) from Bacillus licheniformis was subcloned into pMAL-c2X (New England Biolabs, Beverly, MA, USA). To avoid contamination with TEM-1 β-lactamase encoded by the ampicillin resistance marker of pMAL-c2X, the ampicillin resistance gene was replaced with a kanamycin resistance marker by PCR mutagenesis. The resultant MBP-PenP construct was then used as a template for the construction of PenP-E166C and PenP-E166C/N170Q mutants. These mutants were prepared by site-directed mutagenesis using the QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The mutagenic primers were as follows: for E166C mutation: forward: 5′–CGA TTC TGC CCA GAG TTA AAT GAA GTG AAT CCG GGT GAA–3′ and reverse: 5′–TTC ACC CGG ATT CAC TTC ATT TAA CTC TGG GCA GAA TCG–3′; and for E166C/N170Q double mutation: forward: 5′–CGA TTC TGC CCA GAG TTA CAG GAA GTG AAT CCG GGT GAA–3′ and reverse: 5′–TTC ACC CGG ATT CAC TTC CTG TAA CTC TGG GCA GAA TCG–3′. The authenticity of the MBP-fusion constructs was confirmed by DNA sequencing. The MBP-fusion constructs of the PenP wild type and mutants were transformed into E. coli BL21(DE3) for protein expression. The detailed procedure for the overexpression and purification of the PenP enzymes is mentioned in the Supporting Information.
4.3. Fluorescence Labeling
Approximately 2 mg of the β-lactamase cysteine mutant was reconstituted in 4 mL of 6 M guanidine hydrochloride. The protein solution was then incubated at room temperature for 30 min to unfold the mutant. The pH of the solution was then adjusted to 7.5 with 0.2 M sodium hydroxide. In parallel, a 20 mM stock solution of fluorescein-5-maleimide was prepared by dissolving it in dimethylformamide. A 10-fold molar excess of fluorescein-5-maleimide from the stock solution was then added into the protein solution. This mixture was then stirred at room temperature for 2–4 h in the dark to allow labeling of the mutant. Following the labeling reaction, excess label was then removed by dialyzing the labeling mixture against 1 L of 50 mM potassium phosphate buffer (pH 7.0) at 4 °C with a dialysis tubing of 12 kDa cut-off size. Upon the removal of guanidine hydrochloride from the reaction mixture by dialysis, enzymes were allowed to refold into their native structures. The refolded fluorophore-conjugated protein was stored at −20 °C.
4.4. β-Lactamase Activity Assay
The activity of the β-lactamase wild type and mutants was measured by the spectrophotometric method using a PerkinElmer Lambda Bio 20 UV/visible spectrometer. All assays were performed at 20 °C in 50 mM potassium phosphate buffer (pH 7.0). Initial velocities of substrate hydrolysis were monitored by an increase in absorbance at 405 nm for CENTA and 500 nm for nitrocefin for the first 5 min. The extinction coefficients of CENTA and nitrocefin are 6400 and 15 900 M–1 cm–1, respectively. Kinetic parameters (Km and kcat) were determined by fitting the initial rates of substrate hydrolysis to the Michaelis–Menten equation with nonlinear regression (Enzyme Kinetics, Trinity software).
4.5. Thermal Denaturation Study
Thermal denaturation of the MBP-β-lactamase was studied by monitoring the far-UV molar ellipticity at 222 nm at a scan rate of 50 nm/min in 5 mM potassium phosphate (pH 7.0). Data were recorded as a function of increasing temperature with a rate of 1 °C/min over the temperature range of 20–85 °C. The protein concentration of the samples was in the range of 150–200 μg/mL.
4.6. Thermal Inactivation Study
β-Lactamases were incubated at 50 °C in 50 mM potassium phosphate (pH 7.0). Samples were withdrawn at different time intervals and immediately cooled on ice. The residual activities of the enzymes in hydrolyzing nitrocefin at 20 °C were then measured in duplicate.
4.7. Fluorescence Measurements
Fluorescence measurements were performed on a PerkinElmer LS50B spectrofluorimeter at 20 °C in 50 mM potassium phosphate (pH 7.0). Excitation and emission wavelengths were at 494 and 515 nm, respectively. Both excitation and emission slit widths were 5 nm and scan speed was 250 nm/min.
4.8. Detection of Antibiotics in Serum
Penicillin G was first dissolved in phosphate-buffered saline (PBS) and then serially diluted to give stock solutions at different concentrations. After that, antibiotic-containing mouse serum samples were prepared by mixing an aliquot of 5 μL of penicillin G from the stock solution. For the control with the omission of antibiotics, PBS was added to the serum to substitute penicillin G. For the fluorescence detection of antibiotics, 1 μL of the serum sample was added to 0.1 μM of PenP-E166Cf/N170Q in 50 mM potassium phosphate buffer (pH 7.0) in a quartz cuvette. The reaction mixture was mixed manually, and its fluorescence signal was acquired by using a Cary Eclipse Fluorescence Spectrometer (Agilent, US). Excitation and emission wavelengths were 494 and 515 nm, respectively. Both excitation and emission slits were set at 5 nm. All measurements were conducted in triplicate at room temperature.
4.9. Detection of Antibiotics in Milk
The milk sample was purchased from the local supermarket in Hong Kong. To prepare antibiotic-containing milk, penicillin G was first dissolved in the fresh milk and then serially diluted with milk to give stock solution at various concentrations. Milk with the omission of penicillin G was served as the negative control for the detection. Prior to the analysis, 5 mL of the milk sample was added with 0.4 mL of 10% trichloroacetic acid (TCA; Sigma) in a sterilized centrifuge tube. The solution was vortexed briefly for 5 min. Then, it was subjected to centrifugation at 4000 rpm for 5 min. The clear supernatant was collected and filtered by using a 0.22 μm-syringe filter (PALL; acrodisc; hydrophilic polyethersulfone PES). Five microliters of the filtrate was subjected to the fluorescence assay. The condition and settings for the fluorescence measurement were the same as the aforementioned for fluorescence detection in mouse serum. All measurements were performed in triplicate.
Acknowledgments
This work was supported by the RGC GRF grant (grant 151019/14M) and RGC grants (C5031-14E, G-YBB3, G-YB RX, and K-BBX4).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b02211.
Experimental details; chemical structures of fluorescein-5-maleimide and β-lactam antibiotics used in this study; SDS-PAGE analysis of the PenP mutants before and after the labeling reaction; mass spectra of PenP-E166Cf and PenP-E166Cf/N170Q; CD spectra of wild-type PenP, cysteine mutants, and fluorescently labeled mutants; fluorescence spectra of PenPC-E166Cf in the presence of penicillin G, cefoxitin, and moxalactam; calibration curves for antibiotic detection; and references (PDF)
Author Contributions
§ These authors have contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Van Boeckel T. P.; Gandra S.; Ashok A.; Caudron Q.; Grenfell B. T.; Levin S. A.; Laxminarayan R. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. 10.1016/s1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- Van Boeckel T. P.; Brower C.; Gilbert M.; Grenfell B. T.; Levin S. A.; Robinson T. P.; Teillant A.; Laxminarayan R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 5649–5654. 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Critically Important Antimicrobials for Human Medicine, 5th revision, 2017.
- Kjeldgaard J.; Cohn M. T.; Casey P. G.; Hill C.; Ingmer H. Residual antibiotics disrupt meat fermentation and increase risk of infection. mBio 2012, 3, e00190 10.1128/mbio.00190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A.; Gaeta F.; Arribas Poves M. F.; Valluzzi R. L. Cross-reactivity among β-lactams. Curr. Allergy Asthma Rep. 2016, 16, 24. 10.1007/s11882-016-0594-9. [DOI] [PubMed] [Google Scholar]
- Marston H. D.; Dixon D. M.; Knisely J. M.; Palmore T. N.; Fauci A. S. Antimicrobial resistance. JAMA 2016, 316, 1193–1204. 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- Khan D. A.; Banerji A.; Bernstein J. A.; Bilgicer B.; Blumenthal K.; Castells M.; Ein D.; Lang D. M.; Phillips E. Cephalosporin allergy: Current understanding and future challenges. J. Allergy Clin. Immunol. Pract. 2019, 7, 2105–2114. 10.1016/j.jaip.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachi S.; Ferdous J.; Sikder M.; Hussani S. Antibiotic residues in milk: Past, present, and future. J. Adv. Vet. Anim. Res. 2019, 6, 315–332. 10.5455/javar.2019.f350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levison M. E.; Levison J. H. Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect. Dis. Clin. N. Am. 2009, 23, 791–815. 10.1016/j.idc.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch R.; Chatelle C.; Kling A.; Rebmann B.; Wirth S.; Schumann S.; Weber W.; Dincer C.; Urban G. Clinical on-site monitoring of β-lactam antibiotics for a personalized antibiotherapy. Sci. Rep. 2017, 7, 3127. 10.1038/s41598-017-03338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cháfer-Pericás C.; Maquieira Á.; Puchades R. Fast screening methods to detect antibiotic residues in food samples. TrAC, Trends Anal. Chem. 2010, 29, 1038–1049. 10.1016/j.trac.2010.06.004. [DOI] [Google Scholar]
- Virolainen N.; Karp M. Biosensors, antibiotics and food. Adv. Biochem. Eng. 2014, 145, 153–185. 10.1007/978-3-662-43619-6_5. [DOI] [PubMed] [Google Scholar]
- Pikkemaat M. G.; Rapallini M. L. B. A.; Zuidema T.; Elferink J. W. A.; Oostra-van Dijk S.; Driessen-van Lankveld W. D. M. Screening methods for the detection of antibiotic residues in slaughter animals: comparison of the European Union Four-Plate Test, the Nouws Antibiotic Test and the PremiTest (applied to muscle and kidney). Food Addit. Contam., Part A 2011, 28, 26–34. 10.1080/19440049.2010.535027. [DOI] [PubMed] [Google Scholar]
- Das S.; Kumar N.; Vishweswaraiah R. H.; Haldar L.; Gaare M.; Singh V. K.; Puniya A. K. Microbial based assay for specific detection of β-lactam group of antibiotics in milk. J. Food Sci. Technol. 2014, 51, 1161–1166. 10.1007/s13197-011-0609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller N.; Mueller-Seitz E.; Scholz O.; Hillen W.; Bergwerff A. A.; Petz M. A new strategy for the analysis of tetracycline residues in foodstuffs by a surface plasmon resonance biosensor. Eur. Food Res. Technol. 2007, 224, 285–292. 10.1007/s00217-006-0392-z. [DOI] [Google Scholar]
- Peng J.; Cheng G.; Huang L.; Wang Y.; Hao H.; Peng D.; Liu Z.; Yuan Z. Development of a direct ELISA based on carboxy-terminal of penicillin-binding protein BlaR for the detection of β-lactam antibiotics in foods. Anal. Bioanal. Chem. 2013, 405, 8925–8933. 10.1007/s00216-013-7311-5. [DOI] [PubMed] [Google Scholar]
- Ahmed S.; Ning J.; Cheng G.; Ahmad I.; Li J.; Mingyue L.; Qu W.; Iqbal M.; Shabbir M. A. B.; Yuan Z. Receptor-based screening assays for the detection of antibiotics residues – A review. Talanta 2017, 166, 176–186. 10.1016/j.talanta.2017.01.057. [DOI] [PubMed] [Google Scholar]
- Chen T.; Cheng G.; Ahmed S.; Wang Y.; Wang X.; Hao H.; Yuan Z. New methodologies in screening of antibiotic residues in animal-derived foods: Biosensors. Talanta 2017, 175, 435–442. 10.1016/j.talanta.2017.07.044. [DOI] [PubMed] [Google Scholar]
- Gaudin V. Advances in biosensor development for the screening of antibiotic residues in food products of animal origin – A comprehensive review. Biosens. Bioelectron. 2017, 90, 363–377. 10.1016/j.bios.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Durel L.; Gallina G.; Pellet T. Assessment of ceftiofur residues in cow milk using commercial screening test kits. Vet. Rec. Open 2019, 6, e000329 10.1136/vetreco-2018-000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgou E.; Christoforidou S.; Ioannidou M.; Psomas E.; Samouris G. Detection of β-lactams and chloramphenicol residues in raw milk-development and application of an HPLC-DAD method in comparison with microbial inhibition assays. Foods 2018, 7, 82. 10.3390/foods7060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier M.-C.; Tribut O.; Tattevin P.; Le Tulzo Y.; Michelet C.; Bentué-Ferrer D. Simultaneous determination of 12 β-lactam antibiotics in human plasma by high-performance liquid chromatography with UV detection: application to therapeutic drug monitoring. Antimicrob. Agents Chemother. 2011, 55, 4873–4879. 10.1128/aac.00533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.; Ryu H.-D.; Chung E. G.; Kim Y.; Lee J.-k. A review of analytical procedures for the simultaneous determination of medically important veterinary antibiotics in environmental water: Sample preparation, liquid chromatography, and mass spectrometry. J. Environ. Manage. 2018, 217, 629–645. 10.1016/j.jenvman.2018.04.006. [DOI] [PubMed] [Google Scholar]
- Verhoven S. M.; Groszek J. J.; Fissell W. H.; Seegmiller A.; Colby J.; Patel P.; Verstraete A.; Shotwell M. Therapeutic drug monitoring of piperacillin and tazobactam by RP-HPLC of residual blood specimens. Clin. Chim. Acta 2018, 482, 60–64. 10.1016/j.cca.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wang J.; Wu Q.; Li L.; Wang Y.; Yang H. Determination of kanamycin by high performance liquid chromatography. Molecules 2019, 24, 1902. 10.3390/molecules24101902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.-K.; Jung W.; Lee H.-J. Application of a solid-phase fluorescence immunoassay to determine amoxicillin residues in fish tissue. Acta Vet. Hung. 2010, 58, 83–89. 10.1556/avet.58.2010.1.9. [DOI] [PubMed] [Google Scholar]
- Reybroeck W.; Ooghe S.; De Brabander H. F.; Daeseleire E.. Validation of the Charm MRL-3 for fast screening of β-lactam antibiotics in raw milk. J. AOAC Int. 2011, 94, 373–−382., https://www.ncbi.nlm.nih.gov/pubmed/21563670. [PubMed] [Google Scholar]
- Beltrán M. C.; Romero T.; Althaus R. L.; Molina M. P. Evaluation of the Charm maximum residue limit β-lactam and tetracycline test for the detection of antibiotics in ewe and goat milk. J. Dairy Sci. 2013, 96, 2737–2745. 10.3168/jds.2012-6044. [DOI] [PubMed] [Google Scholar]
- Chan P.-H.; Liu H.-B.; Chen Y. W.; Chan K.-C.; Tsang C.-W.; Leung Y.-C.; Wong K.-Y. Rational design of a novel fluorescent biosensor for β-lactam antibiotics from a class A β-lactamase. J. Am. Chem. Soc. 2004, 126, 4074–4075. 10.1021/ja038409m. [DOI] [PubMed] [Google Scholar]
- Reder-Christ K.; Bendas G. Biosensor applications in the field of antibiotic research – a review of recent developments. Sensors 2011, 11, 9450–9466. 10.3390/s111009450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzuelo F.; Gamella M.; Campuzano S.; Martínez-Ruiz P.; Esteban-Torres M.; de las Rivas B.; Reviejo A. J.; Muñoz R.; Pingarrón J. M. Integrated amperometric affinity biosensors using Co2+-tetradenate nitrilotriacetic acid modified disposable carbon electrodes: application to the determination of β-lactam antibiotics. Anal. Chem. 2013, 85, 3246–3254. 10.1021/ac303604b. [DOI] [PubMed] [Google Scholar]
- Tsang M.-W.; So P.-K.; Liu S.-Y.; Tsang C.-W.; Chan P.-H.; Wong K.-Y.; Leung Y.-C. Catalytically impaired fluorescent class C β-lactamase enables rapid and sensitive cephalosporin detection by stabilizing fluorescence signals: implications for biosensor design. Biotechnol. J. 2015, 10, 126–135. 10.1002/biot.201400140. [DOI] [PubMed] [Google Scholar]
- Blair J. M. A.; Webber M. A.; Baylay A. J.; Ogbolu D. O.; Piddock L. J. V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Chan P.-H.; So P.-K.; Ma D.-L.; Zhao Y.; Lai T.-S.; Chung W.-H.; Chan K.-C.; Yiu K.-F.; Chan H.-W.; Siu F.-M.; Tsang C.-W.; Leung Y.-C.; Wong K.-Y. Fluorophore-labeled β-lactamase as a biosensor for β-lactam antibiotics: a study of the biosensing process. J. Am. Chem. Soc. 2008, 130, 6351–6361. 10.1021/ja076111g. [DOI] [PubMed] [Google Scholar]
- Wong W.-T.; Au H.-W.; Yap H.-K.; Leung Y.-C.; Wong K.-Y.; Zhao Y. Structural studies of the mechanism for biosensing antibiotics in a fluorescein-labeled β-lactamase. BMC Struct. Biol. 2011, 11, 15. 10.1186/1472-6807-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadway R. J. The amino acid sequence of penicillinase from Bacillus licheniformis. Biochem. J. 1969, 115, 12P–13P. 10.1042/bj1150012pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madgwick P. J.; Waley S. G. β-lactamase I from Bacillus cereus. Structure and site-directed mutagenesis. Biochem. J. 1987, 248, 657–662. 10.1042/bj2480657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076 10.1128/aac.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke C. L.; Hinchliffe P.; Bragginton E. C.; Colenso C. K.; Hirvonen V. H. A.; Takebayashi Y.; Spencer J. β-lactamases and β-lactamase inhibitors in the 21st century. J. Mol. Biol. 2019, 431, 3472–3500. 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen M.; Wong K. Y.; Leung Y. C.; Wong W. T.; Chan P. H.; Andresen-Vasquez M.; Alegria L.; Silva C.; Tapia P.; Downey P.; Soto D. Method based on the β-lactamase PenPC fluorescent labeled for β-lactam antibiotic quantification in human plasma. BioMed Res. Int. 2016, 2016, 4307987. 10.1155/2016/4307987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen M.; Araos J.; Wong K. Y.; Leung Y. C.; So L. Y.; Wong W. T.; Cabrera S.; Silva C.; Alegria L.; Bruhn A.; Soto D. Evaluation of meropenem pharmacokinetics in an experimental acute respiratory distress syndrome (ARDS) model during extracorporeal membrane oxygenation (ECMO) by using a PenP β-lactamase biosensor. Sensors 2018, 18, 1424. 10.3390/s18051424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. G.; Shanker S.; Prasad B. V. V.; Palzkill T. Structural and biochemical evidence that a TEM-1 β-lactamase N170G active site mutant acts via substrate-assisted catalysis. J. Biol. Chem. 2009, 284, 33703–33712. 10.1074/jbc.m109.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzke L. E.; Chen C. C. H.; Banerjee S.; Li Z.; Wäsch S.; Kapadia G.; Moult J.; Herzberg O. Elimination of the hydrolytic water molecule in a class A β-lactamase mutant: crystal structure and kinetics. Biochemistry 1996, 35, 16475–16482. 10.1021/bi962242a. [DOI] [PubMed] [Google Scholar]
- Escobar W. A.; Tan A. K.; Fink A. L. Site-directed mutagenesis of β-lactamase leading to accumulation of a catalytic intermediate. Biochemistry 1991, 30, 10783–10787. 10.1021/bi00108a025. [DOI] [PubMed] [Google Scholar]
- Vanhove M.; Houba S.; Lamotte-Brasseur J.; Frère J. M. Probing the determinants of protein stability: comparison of class A β-lactamase. Biochem. J. 1995, 308, 859–864. 10.1042/bj3080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.