Abstract
Aim
To investigate the mRNA expression and clinical significance of structural maintenance of chromosomes protein 4 (SMC4) in breast cancer.
Methods
A total of 23 paired samples were sequenced, and data from the Cancer Genome Atlas were analyzed.
Results
SMC4 mRNA level was significantly upregulated in breast cancer tissues (P < 0.001). Patients with high mRNA expression of SMC4 had significantly poor survival (P = 0.012). Subgroup analyses show that in nontriple negative breast cancer (non-TNBC) patients, the high SMC4 mRNA expression, older age (>65), negative progesterone receptor, and advanced stages (III-IV) were independent risk factors (HR = 3.293, 95% CI 1.257-8.625, P = 0.015). In patients with TNBC, high mRNA expression of SMC4 correlated with better survival rate (P < 0.046).
Conclusion
SMC4 mRNA level is a good prognostic biomarker for patients with breast cancer.
1. Introduction
Breast cancer is the most prevalent cancer among women globally. Recent data shows that about 2,080,000 new cases of breast cancer are diagnosed yearly, while more than 626,000 patients are dying of this disease each year [1]. The evolution of molecular subtype classification has incentivized the design of more precise and effective therapies for patients. Currently, it has been reported that early surgical treatment of breast cancer leads to good prognostic outcomes. However, about 10% of patients experience metastasis before death within 5 years after surgery [2–4]. Therefore, identification of new biomarkers is needed to facilitate the development of more effective therapeutic strategies.
Structural maintenance of chromosomes protein 4 (SMC4) is encoded by SMC4, which located in 3q25.33 (https://www.ncbi.nlm.nih.gov/gene/10051). SMC4 is a member of the SMC family. This ATPase family maintains the stability of chromosomal structure and participates in mitosis of eukaryotic cells [5, 6]. It has been shown that condensin, a heterodimer composed of SMC4 and SMC2, is involved in chromatin condensation and gene regulation [5, 7]. Other researchers have found a relationship between SMC4 and tumors. Zhou et al. discovered that SMC4 is highly impressed in human primary hepatocellular carcinoma [8]. High expression of SMC4 is an independent predictor of poor survival in patients with colorectal cancer [9, 10]. Recently, Jiang et al. found that the high expression of SMC4 is related to the aggressiveness of glioma [11]. However, the mechanisms have not been resolved.
In patients with breast cancer, SMC4 is thought to be one of the 28 genes related to paclitaxel resistance [12]. It has also been suggested that SMC4 contributes to the risk of distant metastasis of lymph node-negative primary breast cancer [13]. Nevertheless, the succinct role of SMC4 in breast cancer and the relationship between SMC4 expression and clinical outcomes remain unknown.
In this study, we analyzed the expression of SMC4 in breast cancer tissues and adjacent noncancerous tissue. We further explored the clinical value of SMC4 expression.
2. Materials and Methods
2.1. Patient Specimen
A total of 23 breast cancer tissues and 23 paired adjacent noncancerous tissues were obtained from patients in the First Affiliated Hospital of Wenzhou Medical University. The tissues were harvested during surgery and immediately snap-frozen in liquid nitrogen. Total RNA was then extracted from the tissue samples using TRIzol agent (Life Technologies, California, USA). The complementary DNA (cDNA) libraries were prepared for single-end sequencing using Ion Total RNA-Seq Kit v2.0 (Life Technologies). The cDNA libraries were then processed for the Proton (Life Technologies) sequencing process. Data acquisition and patient enrollment were approved by the clinical ethics committee of the First Affiliated Hospital of Wenzhou Medical University.
2.2. Retrieval of Publicly Available Data
Public data of SMC4 gene expression and clinicopathological characteristics for patients with breast cancer were obtained from the Cancer Genome Atlas (TCGA) database. The expression of human epidermal growth factor receptor 2 (HER-2) determined by immunohistochemical assay was further adjusted to the fluorescence of in situ hybridization assays. A total of 992 female patients enrolled from 2001 to 2013. Among them, 113 patients had paired cancerous and normal sample data.
2.3. Statistical Analysis
Paired continuous variables were analyzed by paired t-test. Categorical variables were expressed as number of cases (percentages) and analyzed using the χ2 test. Survival analyses were performed by Kaplan-Meier survival analysis and multivariable Cox regression analysis, along with hazard rate (HR) and 95% confidence interval (95% CI). SPSS version 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism version 7.0 (GraphPad Software Inc., La Jolla, CA, USA) were used for data analyses. A two-tailed P < 0.05 was considered significantly different.
3. Results
3.1. SMC4 Is Upregulated in Breast Cancerous Tissue
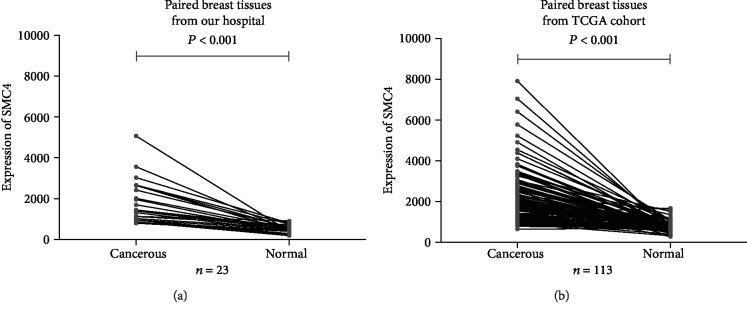
Results showed that SMC4 mRNA expression was higher in all the 23 breast cancerous tissues than in all the 23 paired adjacent noncancerous tissues (Figure 1(a)). In most of the breast cancerous tissues (18/23), the expression was 2-fold higher compared to the paired nonbreast cancer tissues. The log2 fold change was about 1.75. In the TCGA data, SMC4 mRNA expression was similarly higher in cancerous tissues than in paired noncancerous tissues (P < 0.001) (Figure 1(b)).
Figure 1.
The mRNA expression levels of SMC4 upgraded in cancerous tissues, both in the 23 paired human specimens (a) (P < 0.001) and in the TCGA dataset (b) (P < 0.001).
3.2. Relationship between SMC4 mRNA Expression and Clinicopathological Characteristics
In TCGA cohort, 992 female patients were enrolled from 2001 to 2013. As shown in Table 1, there were no significant differences in age, menopause, lymph node metastasis, distant metastasis, and HER-2 state, between the high SMC4 mRNA expression group and the low SMC4 mRNA expression group (all P values > 0.05). However, there were significant differences between the two groups when adjusted to tumor categories, ER state or PR state (all P values < 0.05).
Table 1.
The relationship between SMC4 expression and clinicopathological characteristics in female breast cancer patients from the TCGA cohort enrolled from 2001 to 2013 (n = 992).
| Clinicopathological characteristic | Low SMC4 (%) (n = 614) | High SMC4 (%) (n = 378) | χ 2 | P |
|---|---|---|---|---|
| Age | 2.465 | 0.117 | ||
| ≤65 | 428 (60.4%) | 281 (39.6%) | ||
| >65 | 186 (65.7%) | 97 (34.3%) | ||
| Menopause | 2.165 | 0.141 | ||
| Premenopause | 146 (57.5%) | 108 (42.5%) | ||
| Postmenopause | 387 (62.8%) | 229 (37.2%) | ||
| Tumor categories | 8.993 | 0.003 | ||
| T1 | 181 (69.6%) | 79 (30.4%) | ||
| T2-T4 | 432 (59.1%) | 299 (40.9%) | ||
| Lymph node metastasis | 1.359 | 0.244 | ||
| None | 289 (59.8%) | 194 (40.2%) | ||
| Present | 166 (31.6%) | 227 (18.2%) | ||
| Distant metastasis | 0.077 | 0.781 | ||
| No | 489 (59.1%) | 339 (40.9%) | ||
| Yes | 10 (62.5%) | 6 (37.5%) | ||
| Clinical stage | 1.954 | 0.162 | ||
| I-II | 449 (60.7%) | 291 (39.3%) | ||
| III-IV | 159 (65.7%) | 83 (34.3%) | ||
| Estrogen receptor | 71.256 | <0.001 | ||
| Negative | 80 (37.4%) | 134 (62.6%) | ||
| Positive | 508 (69.2%) | 226 (30.8%) | ||
| Progesterone receptor | 58.413 | <0.001 | ||
| Negative | 141 (44.9%) | 173 (55.1%) | ||
| Positive | 445 (70.5%) | 186 (29.5%) | ||
| HER-2 | 0.036 | 0.849 | ||
| Negative | 457 (62.1%) | 279 (37.9%) | ||
| Positive | 111 (61.3%) | 70 (38.7%) | ||
| TNBC | 72.197 | <0.001 | ||
| Yes | 50 (32.1%) | 106 (67.9%) | ||
| No | 517 (68.3%) | 240 (31.7%) |
HER-2 = human epidermal growth factor receptor 2; TNBC = triple negative breast cancer; SMC4 = structural maintenance of chromosomes protein 4 mRNA.
3.3. Survival Analyses
As shown in Figure 2, the 5-year overall survival (OS) rate of the low SMC4 group was 89.71%, whereas the OS rate of the high SMC4 group was 77.00%, indicating a significantly lower OS rate in the high SMC4 group than in the low SMC4 group (P = 0.012).
Figure 2.
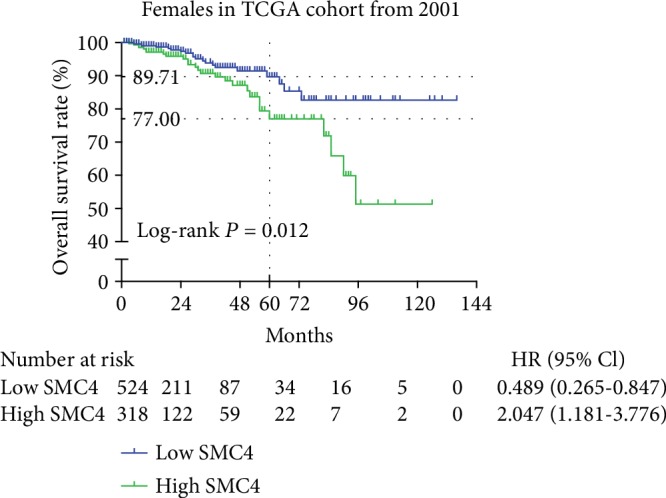
Kaplan-Meier analysis indicated that breast cancer patients with higher mRNA expression of SMC4 had worse survival in TCGA dataset (P = 0.012).
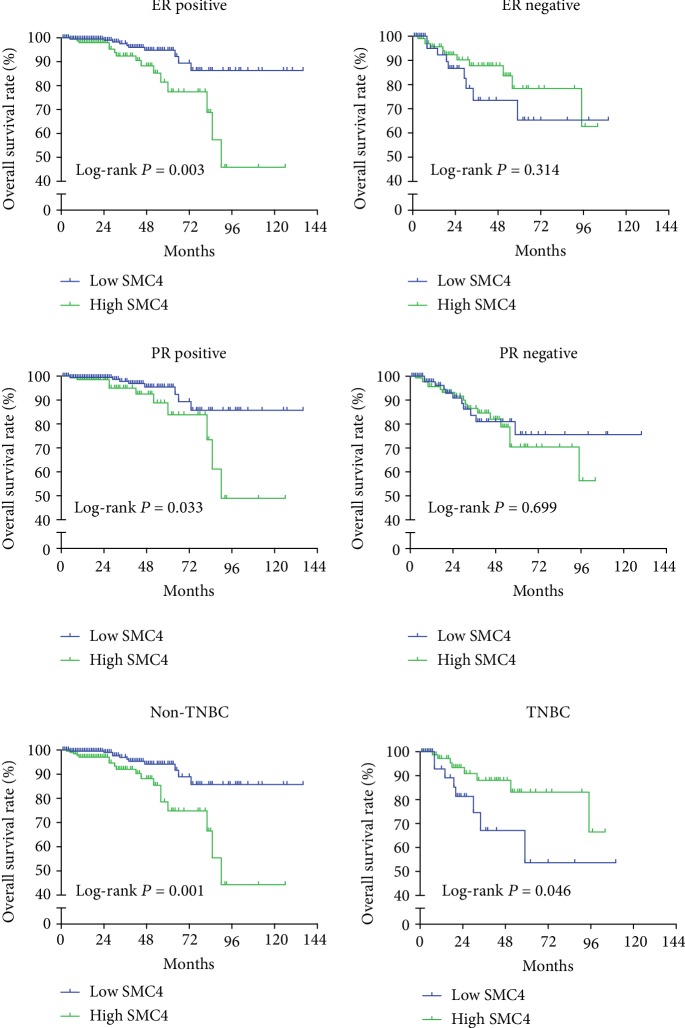
Further subgroup analyses revealed that the OS rates were markedly lower in high SMC4 expression groups in patients with ER positive or PR positive (P = 0.003 and P = 0.033, respectively) (Figure 3). Nevertheless, there were no significant differences about OS rates in patients with ER negative or PR negative (all P > 0.05) (Figure 3). In patients with nontriple negative breast cancer (non-TNBC), the high SMC4 expression group had poor prognosis (P = 0.001) (Figure 3). However, among the patients with TNBC, the high SMC4 expression group had better outcomes (P = 0.046) (Figure 3).
Figure 3.
Subgroup analyses indicated that in patients with ER positive or PR positive or nontriple negative breast cancer (non-TNBC), higher mRNA expression of SMC4 was associated with lower survival rate (P = 0.003, P = 0.033, and P = 0.001, respectively). In patients with TNBC, higher mRNA expression of SMC4 correlated with higher survival rate (P = 0.046). There was no significant difference in ER/PR-negative patients between the higher SMC4 mRNA expression group and the lower SMC4 mRNA expression group.
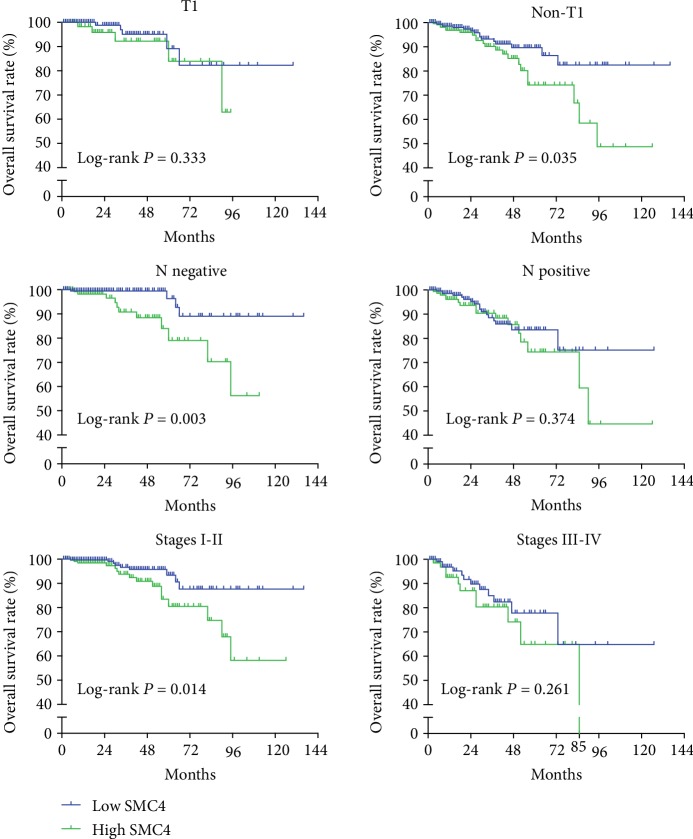
Figure 4 shows that there was no significant difference between the high SMC4 mRNA expression group and the low SMC4 mRNA expression group in terms of tumor category 1 (T1), lymph node (N) metastases, or stages III-IV (all P > 0.05). However, other subgroups according to non-T1, lymph node negative, or stages I-II showed positive results that high SMC4 expression groups had significantly lower OS rates (P = 0.035, P = 0.003, and P = 0.014, respectively).
Figure 4.
Subgroup analyses indicated that in breast cancer patients with larger tumors (>T1), without lymph node metastasis, or stages I-II, higher mRNA expression of SMC4 was associated with low survival rate (P = 0.035, P = 0.003, and P = 0.014, respectively). In patients with T1, lymph node metastasis, or stages III-IV, there was no significant difference in T1, lymph node metastasis, or stages III-IV, between the higher SMC4 mRNA expression group and the lower SMC4 mRNA expression group.
As shown in Table 2, when adjusted to non-TNBC, age, stage, PR state, and the SMC4 mRNA expression were found to be significant independent risk factors of prognosis (hazard ratio (HR) = 7.372, 95% confidence interval (CI) 2.902-18.727, P < 0.001; HR = 4.370, 95% CI 1.745-10.949, P = 0.002; HR = 4.296, 95% CI 1.628-11.336, P = 0.003; and HR = 3.293, 95% CI 1.257-8.625, P = 0.015, respectively).
Table 2.
Multivariate Cox regression analysis for risk factors associated with OS in female non-TNBC patients from the TCGA cohort from 2001 to 2013 (n = 992).
| Characteristics | HR | 95% CI | P |
|---|---|---|---|
| Age >65 vs. ≤65 | 7.372 | 2.902-18.727 | <0.001 |
| Stages III-IV vs. stages I-II | 4.370 | 1.745-10.949 | 0.002 |
| PR negative vs. PR positive | 4.296 | 1.628-11.336 | 0.003 |
| High SMC4 vs. low SMC4 | 3.293 | 1.257-8.625 | 0.015 |
TNBC = triple negative breast cancer; HR = hazard ratio; CI = confidence intervals; PR = progesterone receptor; SMC4 = structural maintenance of chromosomes protein 4. Wald forward with SMC4 expression and all the characteristics.
4. Discussion
Breast cancer is the most prevalent malignance in women. Since 1989, it was reported that the mortality rate due to this cancer has been decreasing over the years. However, this trend has slowed down [14]. Therefore, new strategies are needed.
SMC4 had been found highly expressed in several cancers, such as glioma, colorectal carcinoma, and hepatocellular carcinoma [8, 9, 11]. 3q25, the locus of SMC4 belonging to, had been proved to be involved in high level of recurrent DNA amplifications in breast cancer cell lines [15, 16]. Here, we show that the mRNA expression of SMC4 was upregulated in invasive breast cancer cells. Upregulated SMC4 mRNA level could improve the sensitivity of Cdk1 to drive chromatin compaction at mitotic entry [17] and increase the aggressiveness, proliferation, and dedifferentiation of cancer cells [11, 18]. This may explain why larger tumors have higher SMC4 mRNA expression in our study. Moreover, the high expression of SMC4 may increase double-stranded DNA breaks by enhancing the action of topoisomerase II [19] and cause mutations and mismatches resulting in unique chromosomal rearrangements in breast epithelial cells [20]. And it has been reported that overexpression of SMC4 activates JAK2/Stat3 and TGFβ/Smad pathway and promotes aggressiveness of cancer cells [8, 11].
Previous studies reported that in ER-positive breast cancer, PLK1 increased ER transcriptional activity and ER expression and then induced the invasion [21, 22]. PLK1 could be upregulated by the high expression of SMC4 [23]. That may be the reason why high SMC4 expression levels were associated with worse survival in ER/PR-positive patients in the present study. And in HER-2-positive breast cancer, PLK1-siRNA suppresses cancer growth and metastasis [24]. In the present study, there was a trend that HER-2-positive patients with high mRNA expression of SMC4 may suffer lower survival rate (Supplementary Fig. ()). These findings indicate that overexpression of SMC4 may lead to cancer progression and poor prognosis through PLK1 in non-TNBC. Together, our results suggest that SMC4 has the potential to be an independent prognostic predictor and therapeutic target in non-TNBC.
However, in patients with TNBC, overexpression of SMC4 mRNA correlated with better survival. It may be related to SMC4 modulating the sensitivity of breast cancer cells to paclitaxel treatment [12]. Moreover, SMC4 overexpression triggers the formation of double-stranded DNA breaks and unique chromosomal rearrangements, leading to impaired DNA mismatch repair [19, 20]. While DNA mismatch repair plays a central role in the development of drug resistance in TNBC cancer cells, the high expression of SMC4 may lead to long-term effect of chemotherapy.
Additionally, we found that in T2-3N0 or ER/PR-positive patients, higher mRNA expression level of SMC4 was associated with worse survival rates. This implies that the mRNA expression of SMC4 would be a useful tool to identify patients who need more aggressive therapy and those with low relapse risk.
In conclusion, we found that mRNA expression of SMC4 was upregulated in invasive breast cancer cells. Furthermore, patients with high mRNA level of SMC4 suffered different survival with TNBC and non-TNBC. And SMC4 could be a good biomarker for predicting the prognosis and potential for therapeutic target.
Acknowledgments
This work was supported by the Foundation of Wenzhou Municipal Science and Technology Bureau, China (Grant No. Y20170184).
Data Availability
The data used to support the findings of this study are included within the supplementary materials.
Conflicts of Interest
The authors declare that they have no competing interests.
Supplementary Materials
There was only a trend that HER-2-positive patients with high mRNA expression of SMC4 may suffer lower survival rate whether ER/PR positive or not (all P > 0.05).
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P. D., et al. Cancer statistics in China, 2015. CA: a Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis C. E., Fedewa S. A., Goding Sauer A., Kramer J. L., Smith R. A., Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA: a Cancer Journal for Clinicians. 2016;66(1):31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Angulo A. M., Morales-Vasquez F., Hortobagyi G. N. Overview of resistance to systemic therapy in patients with breast cancer. Advances in Experimental Medicine and Biology. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 5.Losada A., Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes & Development. 2005;19(11):1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 6.Nishiwaki T., Daigo Y., Kawasoe T., et al. Isolation and characterization of a human cDNA homologous to the Xenopus laevis XCAP-C gene belonging to the structural maintenance of chromosomes (SMC) family. Journal of Human Genetics. 1999;44(3):197–202. doi: 10.1007/s100380050142. [DOI] [PubMed] [Google Scholar]
- 7.Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes & Development. 2012;26(15):1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou B., Chen H., Wei D., et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research. 2014;33(1):p. 55. doi: 10.1186/1756-9966-33-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng X. D., Song Q., Li C. W., et al. Structural maintenance of chromosomes 4 is a predictor of survival and a novel therapeutic target in colorectal cancer. Asian Pacific Journal of Cancer Prevention. 2014;15(21):9459–9465. doi: 10.7314/apjcp.2014.15.21.9459. [DOI] [PubMed] [Google Scholar]
- 10.Jinushi T., Shibayama Y., Kinoshita I., et al. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Medicine. 2014;3(6):1544–1552. doi: 10.1002/cam4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L., Zhou J., Zhong D., et al. Overexpression of SMC4 activates TGFβ/Smad signaling and promotes aggressive phenotype in glioma cells. Oncogene. 2017;6(3):p. e301. doi: 10.1038/oncsis.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H., Jeung H. C., Jung J. J., Kim T. S., Rha S. Y., Chung H. C. Identification of genes associated with chemosensitivity to SAHA/taxane combination treatment in taxane-resistant breast cancer cells. Breast Cancer Research and Treatment. 2011;125(1):55–63. doi: 10.1007/s10549-010-0825-z. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Klijn J. G., Zhang Y., et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 14.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 15.Rosa-Rosa J. M., Pita G., Urioste M., et al. Genome-wide linkage scan reveals three putative breast-cancer-susceptibility loci. American Journal of Human Genetics. 2009;84(2):115–122. doi: 10.1016/j.ajhg.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forozan F., Mahlamäki E. H., Monni O., et al. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Research. 2000;60(16):4519–4525. [PubMed] [Google Scholar]
- 17.Robellet X., Thattikota Y., Wang F., et al. A high-sensitivity phospho-switch triggered by Cdk1 governs chromosome morphogenesis during cell division. Genes & Development. 2015;29(4):426–439. doi: 10.1101/gad.253294.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B., Yuan T., Liu M., et al. Overexpression of the structural maintenance of chromosome 4 protein is associated with tumor de-differentiation, advanced stage and vascular invasion of primary liver cancer. Oncology Reports. 2012;28(4):1263–1268. doi: 10.3892/or.2012.1929. [DOI] [PubMed] [Google Scholar]
- 19.Wang J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Reviews. Molecular Cell Biology. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 20.Kulawiec M., Safina A., Desouki M. M., et al. Tumorigenic transformation of human breast epithelial cells induced by mitochondrial DNA depletion. Cancer Biology & Therapy. 2008;7(11):1732–1743. doi: 10.4161/cbt.7.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang G. Y., Lee E. R., Kim J. H., et al. Downregulation of PLK-1 expression in kaempferol-induced apoptosis of MCF-7 cells. European Journal of Pharmacology. 2009;611(1-3):17–21. doi: 10.1016/j.ejphar.2009.03.068. [DOI] [PubMed] [Google Scholar]
- 22.Bhola N. E., Jansen V. M., Bafna S., et al. Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer. Cancer Research. 2015;75(2):405–414. doi: 10.1158/0008-5472.CAN-14-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., Kuang M., Li M., Feng L., Zhang K., Cheng S. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Scientific Reports. 2016;6(1) doi: 10.1038/srep34508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao Y. D., Sun T. M., Huang S. Y., et al. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Science Translational Medicine. 2012;4(130):p. 130ra48. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
There was only a trend that HER-2-positive patients with high mRNA expression of SMC4 may suffer lower survival rate whether ER/PR positive or not (all P > 0.05).
Data Availability Statement
The data used to support the findings of this study are included within the supplementary materials.