Abstract
Blastocystis sp. is a protozoan parasite, commonly found in the gastrointestinal tracts of animals and humans globally. The parasitic species has wide genetic diversity. Currently the mammalian and avian isolates of the parasite are grouped into 17 well known subtypes (STs), of which ten (ST1-ST9, ST12) are reported in humans. To assess the genetic diversity of Blastocystis sp. in wildlife, a total of 200 fresh fecal samples were collected from 32 mammalian wildlife species in Bangladesh National Zoo. Blastocystis sp. was screened and subtyped by PCR amplification and sequencing of the small subunit ribosomal RNA (SSU rRNA) gene. The minimum prevalence of Blastocystis sp. infection was 15.5% (31/200) in zoo animals. Eight out of 32 wildlife animal species (25.0%) were infected with Blastocystis sp. Among them, the occurrence of Blastocystis sp. was higher in non-human primates (NHPs) (31.8%) than that in herbivores (4.9%) and carnivores (0). Nucleotide sequence analysis of the SSU rRNA gene revealed seven different Blastocystis sp. subtypes, such as ST1, ST2, ST3, ST10, ST11, ST13 and ST14 in the wild animals. ST3 was the dominant subtype (41.9%, 13/31) being detected in NHPs. Of the 31 Blastocystis sp. isolates from the wild animals, 24 (77.4%) isolates belonged to the most common subtypes (ST1 to ST3) found in humans. This is the first molecular study of Blastocystis sp. in wild animals in Bangladesh. This study highlights the remarkable genetic diversity in Blastocystis sp. isolates from zoo animals and provides the first molecular evidence from spotted deer, gayal and grey langur. Due to circulation of large percentage of potentially zoonotic subtypes in the wild animals, there is a higher risk of zoonotic transmission of Blastocystis sp. in the zoo keepers and visitors.
Keywords: Blastocystis, Subtypes, Wild animals, Zoonotic potential, Bangladesh
Graphical abstract

Highlights
-
•
First report of Blastocystis sp. infection in wild animals in Bangladesh.
-
•
Minimum prevalence of Blastocystis sp. infection was 15.5% in wildlife.
-
•
ST3 was the dominant subtype in wildlife in Bangladesh.
-
•
Majority of the Blastocystis sp. isolates belonged to the most common human subtypes.
-
•
Non-human primates with high prevalence and zoonotic significance.
1. Introduction
Blastocystis sp. is one of the most commonly found eukaryotic protists in the intestinal tract of humans and animals worldwide (Stensvold and Clark, 2016; Javanmard et al., 2018). The parasite transmits through fecal-oral route, for example, through contaminated water (Leelayoova et al., 2008). The prevalence of Blastocystis sp. in humans varies from 0.5% to 30% in industrialized countries, and from 30% to 76% in developing countries (Audebert et al., 2016). Such a high prevalence of Blastocystis sp. obviously invokes the question of the impact of this parasite on human health. In fact, the common occurrence of the parasite in asymptomatic carriage doubts about its role in human health and disease (Cian et al., 2017). However, recent genomic data combined with in vitro and in vivo studies allowed the identification of potential virulence factors and revealed the damaging effects of the parasite on the intestinal barrier, leading to credible models of pathogenesis (Skotarczak, 2018). Furthermore, the colonization of Blastocystis sp. is observed to be associated with increased diversity of human gut bacterial microbiota, indicating the need of high overall microbial diversity for the parasite to become established in the human colon (Audebert et al., 2016; Stensvold and Clark, 2016; Cian et al., 2017). Recent reports also suggest that this parasite should be related with non-specific gastrointestinal symptoms like diarrhea, abdominal pain, and vomiting, and is suspected to be linked to irritable bowel syndrome (IBS) and urticaria (Poirier et al., 2012; Lepczynska et al., 2016). As in many parasitic infections, immunocompromized individuals such as AIDS and cancer patients, malnourished persons, children and the elderly are found to be more susceptible to Blastocystis sp. infection (Lepczynska et al., 2017). The parasite is also known to infect a wide range of non-human hosts, including wildlife and zoo animals (Cian et al., 2017).
Detection methods of Blastocystis sp. mainly include direct smear examination by light microscopic or xenic in vitro culture. However, the methods seem to have largely underestimated Blastocystis sp. in the context of enteric parasite diagnosis because of the fact of occurrence of different parasitic forms (especially the hardly recognizable cystic form), deterioration caused by environmental conditions or drug treatment and the fact that the parasite can be confused with other enteric microorganisms (Tan, 2008; Wawrzyniak et al., 2013). Moreover, culturing this parasite is time consuming and can bias subsequent genotyping due to the different ability of isolates to grow in selective medium (Roberts et al., 2011). Therefore, to overcome these limitations, currently molecular assays are used that are commonly based on the PCR amplification of the small subunit ribosomal RNA (SSU rRNA) gene (Scicluna et al., 2006). The genetic heterogeneity of the SSU rRNA gene revealed seventeen subtypes (ST1-ST17) of Blastocystis sp. in various hosts (Stensvold and Clark, 2016). Among the subtypes, ST1 to ST9 and ST12 were observed in humans, however 95% of the human infections were related to just four of these subtypes (ST1 to ST4) (Alfellani et al., 2013a; Andersen and Stensvold, 2016; Stensvold; Clark, 2016). The four most common subtypes in humans have also been identified in other hosts, the most frequent of which were other primates, but they have also been found in numerous hoofed mammals, rodents and birds (Skotarczak, 2018). On the contrary, the rarer subtypes in humans (ST5-ST8) were also more commonly discovered in other hosts: ST5 in hoofed animals, ST6 and ST7 in birds, and ST8 in non-human primates (NHPs) (Stensvold and Clark, 2016), suggesting their zoonotic derivation. Furthermore, the frequent observation of some subtypes in persons having close contact with animals, such as ST5 in piggery workers (Wang et al., 2014), and ST3 and ST8 in zookeepers (Parkar et al., 2010; Alfellani et al., 2013b), indicates the presence of a potential transmission cycle between humans and animals.
Until now, only two reports of Blastocystis sp. infection in humans are found in Bangladesh. The first study subtyped 26 isolates collected from symptomatic and asymptomatic patients and identified two subtypes ST1 (7.69%) and ST3 (92.31%) (Yoshikawa et al., 2004). The second study was based on in vitro culture and direct microscopy that recorded the prevalence of Blastocystis sp. as 14% in 5,679 stool samples of slum-dwelling infants, with 14.76% prevalence in asymptomatic samples and 6.99% in symptomatic samples (Barua et al., 2015).
Bangladesh National Zoo (situated in Dhaka) is a pleasant recreation center for people of all ages and corners in this country. The zoo contains many kinds of wildlife and hosts about four million visitors every year. Wildlife species have been shown to be an important source of emerging zoonotic diseases, and most emerging and reemerging pathogens originate from the animal reservoirs (Miller et al., 2013). However, there is no organized report on the zoonotic diseases, including Blastocystis sp. infection in the animals of Bangladesh National Zoo. Therefore, the aim of the present study was to assess the genetic diversity of Blastocystis sp. in the wild animals of Bangladesh National Zoo.
2. Materials and methods
2.1. Ethics statement
Ethical clearance was obtained from the Institutional Committee on Animal Care and Use in Research (ICACUR), Bangabandhu Sheikh Mujibur Rahman Agricultural University (BSMRAU), Bangladesh. Appropriate permission was obtained from the Ministry of Livestock and Fisheries and Department of Livestock Services (DLS), Bangladesh, and Curator of Bangladesh National Zoo before the collection of fecal samples from the animals.
2.2. Sample collection
Bangladesh National Zoo contains approximately 2,150 individuals of 191 animal species in a land area of 75 ha (186 acres) (https://bnzoo.org/). The animals investigated in this study were predominantly born in this zoo. The animals are kept separately according to the species. The animals are housed in large spaces with reconstruction of natural habitats suitable for each species. The animals live in accommodations provided with shelter places. Some of the animals live individually in single cages while others live in groups.
A total of 200 fresh fecal samples were collected from 32 captive mammalian animal species under carnivores, herbivores and NHPs during September to October 2018 (supplementary materials: Table 1S). In case of group housing, fresh fecal deposits were collected in the early morning, since the floor of animal cages was cleaned every evening. To avoid repeated sampling, only the fresh fecal deposits were selected based on differentiation by careful visual and physical observations (amount, color, consistency etc). The number of collected samples were in proportion (10%–100%) to the number of animals raised in group housing. For animals that were kept in the pens during the day, fecal samples were collected from individual boxes where they spent the night. The samples were collected with the help of respective animal attendants to minimize unnecessary fear due to strangers (collectors) in the houses.
Table 1.
Occurrence and subtype distributions of Blastocystis sp. in captive wild animals in Bangladesh National Zoo.
| Animal type | Common name | Scientific name | Sample number | Positive number (%) | Isolate (subtype)/Subtype (no.) |
|---|---|---|---|---|---|
| Herbivore | Waterbuck | Kobus ellipsiprymnus | 7 | 1 (14.3%) | ZH-46 (ST10) |
| Herbivore | Barking deer | Muntiacus muntjak | 6 | 0 | - |
| Herbivore | Spotted deer | Axis axis | 30 | 1 (3.3%) | ZH-77 (ST14) |
| Herbivore | Camel | Camelus dromedarious | 2 | 0 | - |
| Herbivore | Rhinoceros | Rhinoceros unicornis | 1 | 0 | - |
| Herbivore | Impala | Aepyceros melampus | 2 | 0 | - |
| Herbivore | Donkey | Equus asinus | 8 | 0 | - |
| Herbivore | Hippopotamus | Hippopotamus amphibius | 2 | 0 | - |
| Herbivore | Elephant | Elephas maximus | 3 | 1 (33.3%) | ZH-15 (ST11) |
| Herbivore | Horse | Equus ferus caballus | 5 | 0 | - |
| Herbivore | Wild beast | Connochaetes taurinus | 2 | 0 | - |
| Herbivore | Common eland | Taurotragus opyx | 2 | 0 | - |
| Herbivore | Zebra | Equus quagga | 3 | 0 | - |
| Herbivore | Greater kudu | Tragelaphus strepsiceros | 1 | 0 | - |
| Herbivore | Giraffe | Giraffe camelopardalis | 4 | 0 | - |
| Herbivore | Gayal | Bos frontalis | 4 | 1 (25.0%) | ZH-57 (ST14) |
| Carnivore | Fishing cat | Prionailurus viverrinus | 5 | 0 | - |
| Carnivore | Asian black bear | Ursus thibetanus | 4 | 0 | - |
| Carnivore | Asiatic lion | Panthera leo leo | 2 | 0 | - |
| Carnivore | Asiatic lion | Panthera leo | 4 | 0 | - |
| Carnivore | Royal bengal tiger | Panthera tigris tigris | 1 | 0 | - |
| Carnivore | Dog | Canis lupus | 2 | 0 | - |
| Carnivore | Spotted hyena | Crocuta crocuta | 1 | 0 | - |
| Carnivore | Strioed hyena | Hyena hyena | 2 | 0 | - |
| Carnivore | Jackal | Canis mesomelas | 12 | 0 | - |
| NHPs | Rhesus macaque | Macaca mulatta | 62 | 20 (32.3%) | ZP-122 (ST1); ZP-126 (ST1); ZP-135 (ST3); ZP-137 (ST3); ZP-149 (ST3); ZP-150 (ST3); ZP-151 (ST3); ZP-152 (ST2); ZP-156 (ST2); ZP-157 (ST2); ZP-159 (ST3); ZP-161 (ST3); ZP-164 (ST3); ZP-165 (ST3); ZP-168 (ST3); ZP-169 (ST3); ZP-178 (ST1); ZP-180 (ST1); ZP-181 (ST1); ZP-182 (ST3) |
| NHPs | Vervet monkey | Chlorocebus pygerythrus | 7 | 3 (42.9%) | ZP-183 (ST3); ZP-184 (ST13); ZP-185 (ST2) |
| NHPs | Langur | Trachypithecus johnii | 5 | 3 (60.0%) | ZP-186 (ST1); ZP-187 (ST13); ZP-188 (ST13) |
| NHPs | Pangolins | Manis paleojavanica | 2 | 0 | - |
| NHPs | Hamadryas baboon | Papio hamadryas | 5 | 0 | - |
| NHPs | Grey langur | Semnopithecus schistaceus | 2 | 1 (50.0%) | ZP-199 (ST1) |
| NHPs | Northern pig-tailed macaque | Macaca leonina | 2 | 0 | |
| Total | 200 | 31 (15.5%) | ST1 (7); ST2 (4); ST3 (13); ST10 (1); ST11 (1); ST13 (3); ST14 (2) | ||
Each sample of about 10 gm was collected in a clean plastic zipper bag marked with relative information and shipped in cool condition to the Laboratory of Veterinary Medicine, Bangabandhu Sheikh Mujibur Rahman Agricultural University. The fecal samples were then stored in 2.5% (w/v) potassium dichromate solution at 4 °C until DNA extraction. During sample collection, no obvious clinical symptoms were observed, except in five rhesus macaques, which were hospitalized due to some clinical and surgical problems other than diarrhea.
2.3. DNA extraction and PCR amplification
Using the centrifugation at 10,000 x g for 2 min, each fecal sample (~200 mg) was washed at least three times with sterilized distilled water to remove the potassium dichromate which would decrease the efficiency of DNA extraction. Total DNA was extracted with the E.Z.N.A.R® Stool DNA Kit (Omega Biotek Inc., Norcross, Georgia, USA), according to the manufacturer's instructions. The extracted DNA was stored at −20 °C until PCR amplification.
Blastocystis sp. was screened and subtyped with the PCR amplification of an approximately 600 bp region of the SSU rRNA gene using the forward primer RD5: ATCTGGTTGATCCTGCCAGT, and the reverse primer BhRDr: GAGCTTTTTAACTGCAACAACG (Scicluna et al., 2006). After electrophoresis in a 1.5% agarose gel stained with ethidium bromide, the PCR products were visualized on a UV transilluminator. The PCR amplification of each sample was performed at least for three times.
2.4. Nucleotide sequencing and phylogenetic analysis
The positive PCR amplicons were sequenced on an ABI PRISM™ 3730 XL DNA Analyzer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The sequence accuracy was confirmed with bidirectional sequencing, and the sequences obtained were aligned and analyzed with MUSCLE embedded in the MEGA 7 program (http://www.megasoftware.net/). The consensus sequences were then compared to similar sequences in GenBank database using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/). The subtypes of Blastocystis sp. isolates were identified by an online platform: Blastocystis locus/sequence definitions database (https://pubmlst.org/bigsdb?db=pubmlst_blastocystis_seqdef). Finally, phylogenetic analysis was used to determine the exact position and identity of the Blastocystis sp. isolates with novel sequences.
The phylogenetic analysis was performed using maximum likelihood method implemented in the MEGA 7 program. Twenty representative sequences from this study and 42 reference sequences from GenBank database were included in the phylogenetic analysis. The sequences were aligned with MUSCLE in MEGA 7 program. The alignment was trimmed using the trimAI v1.2 software (http://trimal.cgenomics.org/downloads). All positions containing gaps were eliminated and the phylogenetic inference was restricted to 120 sites that could be unambiguously aligned. A bootstrap method was used to assess the robustness of the clusters using 1,000 replicates. Tree construction was performed using the Karotomorpha sp. and Protoopalina intestinalis as the outgroups.
2.5. Nucleotide sequence accession numbers
The representative nucleotide sequences obtained in this study were submitted to the National Center for Biotechnology Information (NCBI) and deposited in the GenBank database under the accession numbers: MN338073 to MN338089.
3. Results
3.1. Occurrence of Blastocystis sp. in wild animals
The PCR and sequence based screening of 200 fecal samples collected from 32 wild animal species in Bangladesh National Zoo revealed that eight of the animal species (25.0%, 8/32) were infected with Blastocystis sp. The waterbuck, spotted deer, elephant, gayal, rhesus monkey, vervet monkey, langur and grey langur were infected with Blastocystis sp. with the infection rates of 14.3% (1/7), 3.3% (1/30), 33.3% (1/3), 25.0% (1/4), 32.3% (20/62), 42.9% (3/7), 60.0% (3/5) and 50.0% (1/2), respectively. Overall, NHPs had higher rate of Blastocystis infection (31.8%, 27/85) compared to herbivorous (4.9%, 4/82) and carnivorous animals (0/33). Altogether, the occurrence of Blastocystis sp. was 15.5% (31/200) in wildlife in Bangladesh National Zoo (Table 1).
3.2. Subtype characterization of Blastocystis sp.
Nucleotide sequence analysis of the SSU rRNA gene showed high genetic variability among the Blastocystis sp. isolates from zoo animals. A total of seven known subtypes, such as ST1 (n = 7), ST2 (n = 4), ST3 (n = 13), ST10 (n = 1), ST11 (n = 1), ST13 (n = 3) and ST14 (n = 2), were identified out of 31 Blastocystis sp. isolates. Among the subtypes, ST3 was the dominant one being detected in 13 NHP isolates out of 31wild animal isolates (41.9%) (Table 1). Of the 31 isolates, 24 (77.4%) isolates belonged to the most common human associated subtypes, ST1 to ST3.
The homology analysis of the SSU rRNA gene revealed that among the seven ST1 isolates obtained from NHPs, four and three isolates produced the same sequences MN338073 and MN338074, respectively, with the former and later identical to those from a Macaque (MK357786) and Kangaroo (MK930347) in China, respectively. Three representative sequences such as MN338075 (n = 2), MN338076 (n = 1) and MN338077 (n = 1) obtained from four ST2 isolates of NHPs were identical to that from a human in Japan (AB070987), monkey in Philippines (EU445491) and human in Italy (MF184970), respectively. The highest sequence diversity was observed in ST3 isolates that yielded seven representative sequences in thirteen NHP isolates. Of the seven representative sequences, MN338078 (n = 7), MN338080 (n = 1), MN338081 (n = 1), MN338083 (n = 1) and MN338084 (n = 1) had 100% similarity with that from a patas monkey in China (MK930350), Philippine long-tailed macaques in Philippines (KY929119 and KY929120), human in China (MK934333) and cattle in Malaysia (MG831425), respectively. However, the remaining two sequences MN338079 (n = 1) and MN338082 (n = 1) had the largest similarity (99.83% and 99.67%) to that from a red monkey (MK357784) and human in China (MK934333), with one and two nucleotide substitutions, respectively. The only ST10 sequence (MN338085) obtained from a waterbuck had 99.18% homology with that from a deer in China (MK357785), with five nucleotide substitutions being observed. The single sequence (MN338089) of ST11 isolate found in an elephant showed a homology of 96.39% to the sequence of ST1 isolated from a monkey in Philippines (EU445488), with 22 single nucleotide polymorphisms (SNPs). Meanwhile, three NHP-derived identical ST13 sequences (MN338086) had 98.51% similarity with that from a mousedeer in UK (KC148209). In the case of two ST14 isolates, two representative sequences MN338087 and MN338088 were obtained from a gayal and spotted dear, respectively and the same sequences had been described in a sheep in UAE (MH807191) and cattle in China (MF974619), respectively.
3.3. Phylogeny of Blastocystis sp. isolates
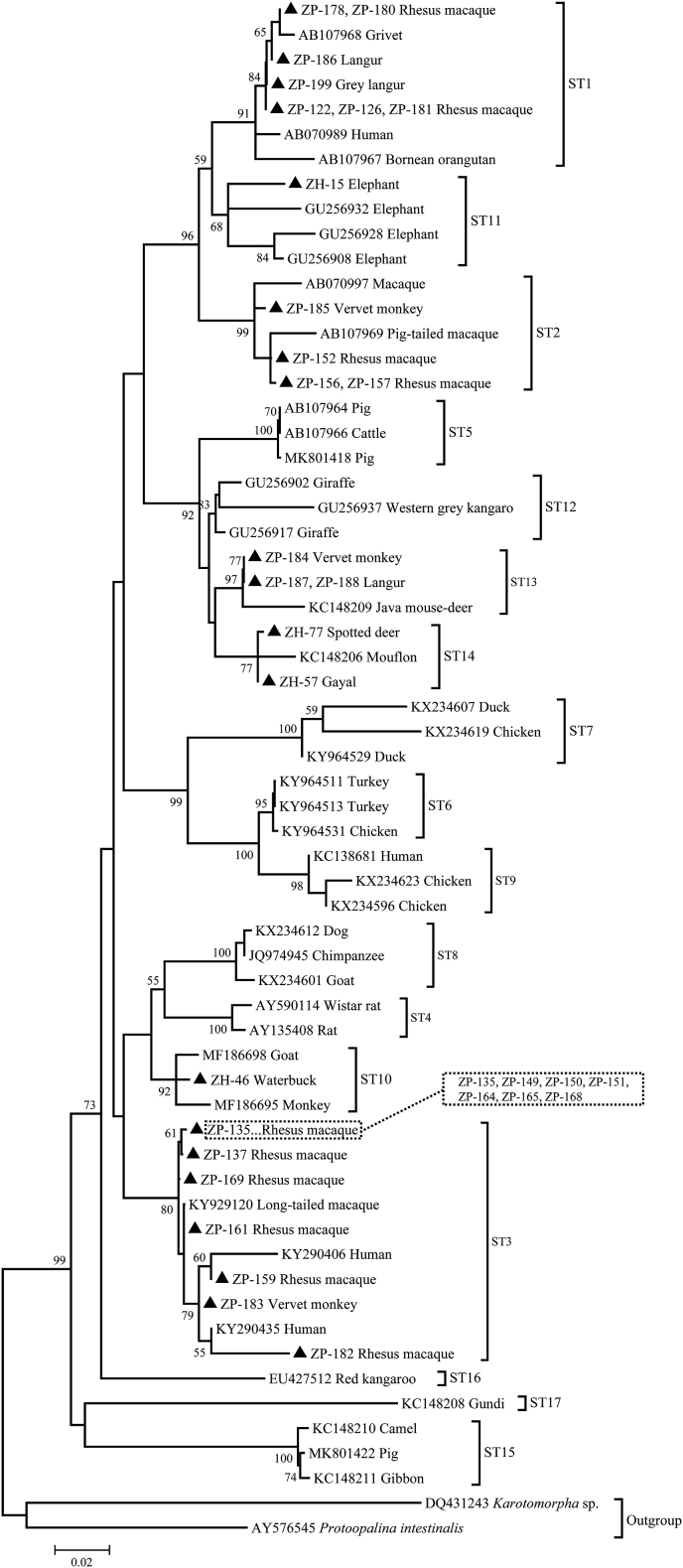
The phylogenetic analysis using 20 representative sequences and 42 reference sequences, including two outgroups clearly demonstrated that the sequences of six well-known subtypes (ST1, ST2, ST3, ST10, ST13 and ST14) obtained from this study were clustered with their reference subtypes (Fig. 1). However, the sequence of elephant-derived isolate ZH-15, which had a homology of 96.39% to the sequence of ST1, was clustered with reference sequences of ST11 obtained from elephants, thus suggesting its identity as ST11. Based on the constructed tree, the isolates included in the same subtype clustered with each other with good bootstrap support, and therefore the seven subtypes were seen as seven independent monophyletic groups.
Fig. 1.
Phylogenetic tree of the Blastocystis sp. isolates and reference SSU rRNA gene sequences from GenBank based on maximum likelihood analysis. The tree was rooted on Karotomorpha sp. and Protoopalina intestinalis. Bootstrap values > 50% from 1,000 replicates are shown on the nodes. Reference sequences from GenBank have accession number and host designation. The isolates of seven subtypes, with their host designations, are indicated by triangle shape.
4. Discussion
In the last few decades, many researchers have performed numerous studies on different aspects of Blastocystis sp., however there are still controversy on classification, pathogenicity, genotyping and role of this organism (Clark et al., 2013; Stensvold and Clark, 2016). Molecular based detection methods have led to a recognition that the organism is much more common than previously thought, at least in some geographic regions and some groups of individuals (Stensvold and Clark, 2016). Although molecular studies have generated a large amount of data on subtypes of Blastocystis sp. isolated from various non-human hosts in recent years, there is still lack of run-through information on the prevalence as well as genetic diversity of Blastocystis sp. worldwide (Noël et al., 2005; Roberts et al., 2013; Stensvold et al., 2009; Yoshikawa et al., 2016).
In this study, the minimum prevalence of Blastocystis sp. was 15.5% (31/200) in mammalian wildlife in Bangladesh National Zoo. This result is comparable with the previous results observed in various animals worldwide. Almost similar infection rate of Blastocystis sp. was recorded in domestic animals (buffaloes, cattle, goats and pigs) (15.4%) in Nepal (Lee et al., 2012). However, the higher infection rates were reported in wild animals (40.2%) in Qinling Mountains, China (Zhao et al., 2017), wild animals in two zoos (32.2%, 99/307) in France (Cian et al., 2017), wild chimpanzees (21.9%, 25/114) in southeast Cameroon (Drakulovski et al., 2014), rhesus monkeys (100%, 10/10) in Nepal (Yoshikawa et al., 2009), wild animals (34.4%, 115/334) in Brazil (Valença-Barbosa et al., 2019), wild boars (25.0%, 3/12) in western Iran (Solaymani-Mohammadi et al., 2004), dairy cattle in Japan (54.1%, 72/133), Lebanon (63.4%, 161/254) (Masuda et al., 2018; Greige et al., 2019), yaks (27.07%, 278/1027) in China (Ren et al., 2019), diverse animals (20.18%, 23/114, with 22.7% in cattle, 63.6% in sheep, 33.3% in rabbits, 37.5% in rodents and 21.2% in rodents) in UAE (AbuOdeh et al., 2019), domestic and companion animals (45.79%, 98/214) in Thailand (Udonsom et al., 2018), street dogs (23.8%, 19/80) in India (Wang et al., 2013). Conversely, the lower infection rates were also reported in cattle (9.5%, 14/147), pigs (8.8%, 6/68), sheep (5.5%, 6/109) in China (Wang et al., 2018). These differences in the infection rates might be associated with the species, immune status and husbandry practices of animals, as well as detection methods, sample size and sampling location and time (Wang et al., 2018).
Of the 32 wild animal species included in this study, eight species (25.0%) were infected with Blastocystis sp., including waterbuck (14.3%), spotted deer (3.3%), elephant (33.3%), gayal (25.0%), rhesus monkey (32.3%), vervet monkey (42.9%), langur and grey langur (50.0%). Among them, the spotted deer, gayal and grey langur were reported with the parasitic infection for the first time in this study. The prevalence of Blastocystis sp. in waterbuck, elephant, rhesus monkey, vervet monkey and langur was lower compared with that (50.0%, 55.0%, 96.6%, 51.4% and 83.3%, respectively) for the same species in China and Australia (Roberts et al., 2013; Zhao et al., 2017). Regarding animal type, the highest rate of Blastocystis sp. infection was observed in NHPs (31.8%) followed by herbivores (4.9%). Similar to our study, higher rate of Blastocystis sp. infection was reported in NHPs compared to herbivorous animals (Zhao et al., 2017; Deng et al., 2019), indicating the common occurrence of this parasite in primates. However, compared to our result, the previous studies recorded much higher infection rate of the parasite in herbivorous animals (Betts et al., 2018; Udonsom et al., 2018; Deng et al., 2019; AbuOdeh et al., 2019; Greige et al., 2019). Surprisingly, Blastocystis sp. was detected in none of the carnivores of our study. Similarly, the parasite was also not reported in carnivorous animals in few previous studies (Abe et al., 2002; Lim et al., 2008; Parkar et al., 2010; Alfellani et al., 2013a; Zhao et al., 2017). However, some studies documented various infection rates of the parasite in carnivores: 2.83% in China (Deng et al., 2019), 2.94% in England (Betts et al., 2018), 10.68% in USA (Ruaux and Stang, 2014), 23.8% in India (Wang et al., 2013) and 69.35% in Australia (Duda et al., 1998).
In the present study, seven Blastocystis sp. subtypes, such as ST1, ST2, ST3, ST10, ST11, ST13 and ST14 were detected in 31 sequencing positive samples from captive mammalian wildlife. Of them, subtype ST3 was the predominant one being identified in 41.9% (13/31) of the positive samples that merely detected in NHPs. Similarly, this was the most commonly detected subtype in humans in various studies (Skotarczak, 2018). The only report on Blastocystis sp. subtypes in Bangladesh also showed the dominance of ST3 (92.3%) in humans (Yoshikawa et al., 2004), indicating the wide distribution and zoonotic significance of this subtype in this area. Furthermore, ST3 was frequently reported in captive wild animals, nonhuman primates, cattle, goats, and rodents in various geographical areas (Cian et al., 2017; Song et al., 2017; Zhao et al., 2017; Wang et al., 2018; Valença-Barbosa et al., 2019). Although, ST1 was detected at the second highest frequency (22.6%, 7/31) in this study, it was also only observed in the fecal samples of NHPs. This subtype was previously reported in humans of Bangladesh, suggesting its zoonotic potential in this country (Yoshikawa et al., 2004). The preceding studies conducted in various locations also reported the ST1 in the captive wild animals, NHPs, sheep, goats, pigs, water voles, marsupials and birds (Cian et al., 2017; Song et al., 2017; Zhao et al., 2017; Betts et al., 2018; Li et al., 2018; Wang et al., 2018; Valença-Barbosa et al., 2019). The subtype ST2 obtained from the three fecal samples of rhesus monkeys and one fecal sample of vervet monkey was previously reported alone in the NHPs among the wild animals studied (Valença-Barbosa et al., 2019). It is worth noting that subtypes ST1 and ST2 are among the most common subtypes in humans, which have been described as having a low host specificity and probable zoonotic implication (Skotarczak, 2018).
The subtype ST13 found in three NHP samples was formerly delineated in golden snub-nosed monkeys in China, mouse deer in UK and Libya and Kangaroo in Australia (Parkar et al., 2010; Alfellani et al., 2013a; Zhao et al., 2017). Meanwhile, the subtype ST14 that obtained from one spotted deer and one gayal was previously reported in captive wild animals, cattle, sheep, goats, deer, kangaroo and rodents (Cian et al., 2017; Song et al., 2017; Zhao et al., 2017; Betts et al., 2018; Li et al., 2018; Wang et al., 2018).
In this study, ST10 and ST11 were found at the lowest frequency; former detected in a waterbuck sample and later in an elephant sample. However, ST10 was reported as the predominant subtype in the wild animal species and yaks in China, fallow deer in Mauritius, cattle and camels in Libya, cattle and sheep in UK and cattle in Denmark (Stensvold et al., 2009; Alfellani et al., 2013a; Zhao et al., 2017; Ren et al., 2019). On the other hand, the host specific subtype ST11 was only previously detected in Asian elephants in Australia (Parkar et al., 2010). Our observation of ST11 in elephant alone further confirms the host specificity of this subtype.
Of the seven known subtypes identified in this study, ST1 to ST3 that constituted 77.4% (24/31) of Blastocystis sp. isolates, all of which from NHPs, belonged to the most common human subtypes (Alfellani et al., 2013a; Andersen and Stensvold, 2016). By combining our data with previous data, and comparing the summarized subtype distribution between animals and humans, it appears that NHPs and livestock may serve as reservoirs for human infection (Skotarczak, 2018). The zoonotic transmission of the Blastocystis sp. subtypes was also evidenced by the previous studies that detected same subtypes in both humans and their contacting animals (Yoshikawa et al., 2009; Parkar et al., 2010; Lee and Bak, 2011; Lee et al., 2012; Wang et al., 2014). The earlier report of higher incidence of Blastocystis sp. infection in the zoo keepers having close contact with animal enclosures indicates the possibility of zoonotic transmission via fecal-oral route (Parkar et al., 2010). Furthermore, the finding of ST2 in monkeys and children in a study in Nepal (Yoshikawa et al., 2009) clarifies the zoonotic transmission cycle of Blastocystis sp. in the zoo facility.
In this study, 24 sequences of 31 Blastocystis sp. isolates were previously reported in various animals and humans. The known sequences belong to subtypes ST1 (n = 7), ST2 (n = 4), ST3 (n = 11) and ST14 (n = 2). The remaining seven new sequences are designated as subtypes ST3 (n = 2), ST10 (n = 1), ST11 (n = 1) and ST13 (n = 3). The analysis of ST3 and ST10 new sequences revealed one to five polymorphic sites, with genetic diversity less than 1% for either of them. The new sequences of ST13 were differed from the known one by about 1.5%. Meanwhile, the new sequence of the elephant isolate had a genetic diversity of 3.61% with 22 SNPs compared to a monkey-derived sequence of ST1 (EU445488). In phylogenetic analysis, the sequence was clustered with known elephant-derived sequences of subtype ST11. Despite having the considerable genetic diversity, the sequence should have been propounded as ST11 because of the fact that it was grouped with ST11 with good bootstrap support in phylogenetic tree. It is worth noting here that because the sequence is derived from the same host as the rest of ST11 sequences and because the SSU rDNA sequence is not complete, it is not clear at this time if this is indeed a new subtype or if it is a divergent sequence of ST11. It is well suggested that new subtype assignments should be based on complete or essentially complete SSU-rDNA sequences (Clark et al., 2013). Previous studies reported that genetic diversity within some subtypes (e.g., ST3) may amount to about 3% (Clark et al., 2013). Thus, Clark and associates proposed that a nearly complete sequence of the suspected new subtype must be compared to the nearly complete sequences of all other subtypes and if the new sequence differs by 4% or more, it can be considered as new subtype with confidence (Clark et al., 2013). Therefore, in spite of having new sequences, no new Blastocystis sp. subtype was surely identified in the mammalian wildlife in Bangladesh National Zoo.
In conclusion, this is the first report of Blastocystis sp. infection and subtype distribution in wildlife in Bangladesh. The data demonstrate that Blastocystis sp. could be maintained and transmitted between wildlife, with the attendant and visitor risk of outbreaks originating in zoo facilities. Since the wildlife health is intrinsically important to human health, the present results will provide fundamental materials for the protection of wild animals, evaluation of potential zoonotic transmission and eventually preservation of human heath from Blastocystis sp. infection.
Funding
This study was supported by Doctoral Startup Foundation of Henan University of Chinese Medicine (00104311-2019-31) and the Special Allocation from Ministry of Science and Technology, Bangladesh.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank the Director General of DLS, Bangladesh, and curator, zoo officers and supporting staff of Bangladesh National Zoo for their full cooperation in data and sample collection. We are grateful to Dr. Abu Nasar Md. Aminoor Rahman, Professor, Wildlife Reproduction & Conservation Laboratory, Department of Gynecology, Obstetrics & Reproductive Health, BSMRAU, Bangladesh for providing the photo for graphical abstract.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.11.003.
Contributor Information
Md. Robiul Karim, Email: vet_robiul@bsmrau.edu.bd.
Longxian Zhang, Email: zhanglx8999@henau.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abe N., Nagoshi M., Takami K., Sawano Y., Yoshikawa H. A survey of Blastocystis sp. in livestock, pets, and zoo animals in Japan. Vet. Parasitol. 2002;106:203–212. doi: 10.1016/s0304-4017(02)00050-x. [DOI] [PubMed] [Google Scholar]
- AbuOdeh R., Ezzedine S., Madkour M., Stensvold C.R., Samie A., Nasrallah G., AlAbsi E., ElBakri A. Molecular subtyping of Blastocystis from diverse animals in the United Arab Emirates. Protist. 2019;170:125679. doi: 10.1016/j.protis.2019.125679. [DOI] [PubMed] [Google Scholar]
- Alfellani M.A., Jacob A.S., Perea N.O., Krecek R.C., Taner-Mulla D., Verweij J.J., Levecke B., Tannich E., Clark C.G., Stensvold C.R. Diversity and distribution of Blastocystis sp. subtypes in nonhuman primates. Parasitology. 2013;140:966–971. doi: 10.1017/S0031182013000255. [DOI] [PubMed] [Google Scholar]
- Alfellani M.A., Taner-Mulla D., Jacob A.S., Imeede C.A., Yoshikawa H., Stensvold C.R., Clark C.G. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. 2013;164:497–509. doi: 10.1016/j.protis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Andersen L.O., Stensvold C.R. Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J. Clin. Microbiol. 2016;54:524–528. doi: 10.1128/JCM.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert C., Even G., Cian A., Blastocystis Investigation Group, Loywick A., Merlin S., Viscogliosi E., Chabé M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep. 2016;6:25255. doi: 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua P., Khanum H., Haque R., Najib F., Kabir M. Establishment of Blastocystis hominis in-vitro culture using fecal samples from infants in slum area of Mirpur, Dhaka, Bangladesh. Acta Med. Int. 2015;2:40–47. [Google Scholar]
- Betts E.L., Gentekaki E., Thomasz A., Breakell V., Carpenter A.I., Tsaousis A.D. Genetic diversity of Blastocystis in non-primate animals. Parasitology. 2018;145:1228–1234. doi: 10.1017/S0031182017002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian A., El Safadi D., Osman M., Moriniere R., Gantois N., Benamrouz-Vanneste S., Delgado-Viscogliosi P., Guyot K., Li L.L., Monchy S., Noël C., Poirier P., Nourrisson C., Wawrzyniak I., Delbac F., Bosc S., Chabé M., Petit T., Certad G., Viscogliosi E. Molecular epidemiology of Blastocystis sp. in various animal groups from two French zoos and evaluation of potential zoonotic risk. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C.G., van der Giezen M., Alfellani M.A., Stensvold C.R. Recent developments in Blastocystis research. Adv. Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- Deng L., Chai Y., Zhou Z., Liu H., Zhong Z., Hu Y., Fu H., Yue C., Peng G. Epidemiology of Blastocystis sp. infection in China: a systematic review. Parasite. 2019;26:41. doi: 10.1051/parasite/2019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakulovski P., Bertout S., Locatelli S., Butel C., Pion S., Mpoudi-Ngole E., Delaporte E., Peeters M., Mallié M. Assessment of gastrointestinal parasites in wild chimpanzees (Pan troglodytes troglodytes) in southeast Cameroon. Parasitol. Res. 2014;113:2541–2550. doi: 10.1007/s00436-014-3904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda A., Stenzel D.J., Boreham P.F. Detection of Blastocystis sp. in domestic dogs and cats. Vet. Parasitol. 1998;76:9–17. doi: 10.1016/s0304-4017(97)00224-0. [DOI] [PubMed] [Google Scholar]
- Greige S., El Safadi D., Khaled S., Gantois N., Baydoun M., Chemaly M., Benamrouz-Vanneste S., Chabé M., Osman M., Certad G., Hamze M., Viscogliosi E. First report on the prevalence and subtype distribution of Blastocystis sp. in dairy cattle in Lebanon and assessment of zoonotic transmission. Acta Trop. 2019;194:23–29. doi: 10.1016/j.actatropica.2019.02.013. [DOI] [PubMed] [Google Scholar]
- Javanmard E., Niyyati M., Ghasemi E., Mirjalali H., Asadzadeh Aghdaei H., Zali M.R. Impacts of human development index and climate conditions on prevalence of Blastocystis: a systematic review and meta-analysis. Acta Trop. 2018;185:193–203. doi: 10.1016/j.actatropica.2018.05.014. [DOI] [PubMed] [Google Scholar]
- Lee B.J., Bak Y.T. Irritable bowel syndrome, gut microbiota and probiotics. J. Neuro Gastroenterol. Motil. 2011;17:252–266. doi: 10.5056/jnm.2011.17.3.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.L., Tan T.C., Tan P.C., Nanthiney D.R., Biraj M.K., Surendra K.M., Suresh K.G. Predominance of Blastocystis sp. subtype 4 in rural communities, Nepal. Parasitol. Res. 2012;110:1553–1562. doi: 10.1007/s00436-011-2665-0. [DOI] [PubMed] [Google Scholar]
- Leelayoova S., Siripattanapipong S., Thathaisong U., Naaglor T., Taamasri P., Piyaraj P., Mungthin M. Drinking water: a possible source of Blastocystis spp. subtype 1 infection in schoolchildren of a rural community in central Thailand. Am. J. Trop. Med. Hyg. 2008;79:401–406. [PubMed] [Google Scholar]
- Lepczynska M., Bialkowska J., Dzika E., Piskorz-Ogórek K., Korycinska J. Blastocystis: how do specific diets and human gut microbiota affect its development and pathogenicity? Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1–10. doi: 10.1007/s10096-017-2965-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepczynska M., Chen W.C., Dzika E. Mysterious chronic urticarial caused by Blastocystis spp.? Int. J. Dermatol. 2016;55:259–266. doi: 10.1111/ijd.13064. [DOI] [PubMed] [Google Scholar]
- Li W.C., Wang K., Gu Y. Occurrence of Blastocystis sp. and pentatrichomonas hominis in sheep and goats in China. Parasites Vectors. 2018;11:93. doi: 10.1186/s13071-018-2671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.A., Ngui R., Shukri J., Rohela M., Mat Naim H.R. Intestinal parasites in various animals at a zoo in Malaysia. Vet. Parasitol. 2008;157:154–159. doi: 10.1016/j.vetpar.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Masuda A., Sumiyoshi T., Ohtaki T., Matsumoto J. Prevalence and molecular subtyping of Blastocystis from dairy cattle in Kanagawa, Japan. Parasitol. Int. 2018;67:702–705. doi: 10.1016/j.parint.2018.07.005. [DOI] [PubMed] [Google Scholar]
- Miller R.S., Farnsworth M.L., Malmberg J.L. Diseases at the livestock-wildlife interface: status, challenges, and opportunities in the United States. Prev. Vet. Med. 2013;110:119–132. doi: 10.1016/j.prevetmed.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël C., Dufernez F., Gerbod D., Edgcomb V.P., Delgado-Viscogliosi P., Ho L.C., Singh M., Wintjens R., Sogin M.L., Capron M., Pierce R., Zenner L., Viscogliosi E. Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J. Clin. Microbiol. 2005;43:348–355. doi: 10.1128/JCM.43.1.348-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkar U., Traub R.J., Vitali S., Elliot A., Levecke B., Robertson I., Geurden T., Steele J., Drake B., Thompson R.C. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet. Parasitol. 2010;169:8–17. doi: 10.1016/j.vetpar.2009.12.032. [DOI] [PubMed] [Google Scholar]
- Poirier P., Wawrzyniak I., Vivares C.P., Delbac F., El Alaoui H. New insights into Blastocystis spp.: a potential link with irritable bowel syndrome. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Song J.K., Yang F., Zou M., Wang P.X., Wang D., Zhang H.J., Zhao G.H., Lin Q. First genotyping of Blastocystis in yaks from Qinghai province, northwestern China. Parasites Vectors. 2019;12:171. doi: 10.1186/s13071-019-3436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T., Barratt J., Harkness J., Ellis J., Stark D. Comparison of microscopy, culture, and conventional polymerase chain reaction for detection of Blastocystis sp. in clinical stool samples. Am. J. Trop. Med. Hyg. 2011;84:308–312. doi: 10.4269/ajtmh.2011.10-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T., Stark D., Harkness J., Ellis J. Subtype distribution of Blastocystis isolates from a variety of animals from new South Wales, Australia. Vet. Parasitol. 2013;196:85–89. doi: 10.1016/j.vetpar.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Ruaux C.G., Stang B.V. Prevalence of Blastocystis in shelter-resident and client-owned companion animals in the US Pacific Northwest. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scicluna S.M., Tawari B., Clark C.G. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Skotarczak B. Genetic diversity and pathogenicity of Blastocystis. Ann. Agric. Environ. Med. 2018;25:411–416. doi: 10.26444/aaem/81315. [DOI] [PubMed] [Google Scholar]
- Solaymani-Mohammadi S., Rezaian M., Hooshyar H., Mowlavi G.R., Babaei Z., Anwar M.A. Intestinal protozoa in wild boars (Sus scrofa) in western Iran. J. Wildl. Dis. 2004;40:801–803. doi: 10.7589/0090-3558-40.4.801. [DOI] [PubMed] [Google Scholar]
- Song J.K., Yin Y.L., Yuan Y.J., Tang H., Ren G.J., Zhang H.J., Li Z.X., Zhang Y.M., Zhao G.H. First genotyping of Blastocystis sp. in dairy, meat, and cashmere goats in northwestern China. Acta Trop. 2017;176:277–282. doi: 10.1016/j.actatropica.2017.08.028. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Alfellani M.A., Nørskov-Lauritsen S., Prip K., Victory E.L., Maddox C., Nielsen H.V., Clark C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009;39:473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Clark C.G. Current status of Blastocystis: a personal view. Parasitol. Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Tan K.S. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008;21:639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udonsom R., Prasertbun R., Mahittikorn A., Mori H., Changbunjong T., Komalamisra C., Pintong A.R., Sukthana Y., Popruk S. Blastocystis infection and subtype distribution in humans, cattle, goats, and pigs in central and western Thailand. Infect. Genet. Evol. 2018;65:107–111. doi: 10.1016/j.meegid.2018.07.007. [DOI] [PubMed] [Google Scholar]
- Valença-Barbosa C., do Bomfim T.C.B., Teixeira B.R., Gentile R., Neto S.F.D.C., Magalhães B.S.N., Balthazar D.A., da Silva F.A., Biot R., d'Avila Levy C.M., Santos H.L.C. Molecular epidemiology of Blastocystis isolated from animals in the state of Rio de Janeiro, Brazil. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Gong B., Yang F., Zhang W., Zheng Y., Liu A. Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China's Heilongjiang Province. Infect. Genet. Evol. 2018;57:171–176. doi: 10.1016/j.meegid.2017.11.026. [DOI] [PubMed] [Google Scholar]
- Wang W., Cuttell L., Bielefeldt-Ohmann H., Inpankaew T., Owen H., Traub R.J. Diversity of Blastocystis subtypes in dogs in different geographical settings. Parasites Vectors. 2013;6:215. doi: 10.1186/1756-3305-6-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Owen H., Traub R.J., Cuttell L., Inpankaew T., Bielefeldt-Ohmann H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet. Parasitol. 2014;203:264–269. doi: 10.1016/j.vetpar.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak I., Poirier P., Viscogliosi E., Dionigia M., Texier C., Delbac F., Alaoui H.E. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Ther. Adv. Infect. Dis. 2013;1:167–178. doi: 10.1177/2049936113504754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H., Koyama Y., Tsuchiya E., Takami K. Blastocystis phylogeny among various isolates from humans to insects. Parasitol. Int. 2016;65:750–759. doi: 10.1016/j.parint.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Wu Z., Kimata I., Iseki M., Ali I.K., Hossain M.B., Zaman V., Haque R., Takahashi Y. Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol. Res. 2004;92:22–29. doi: 10.1007/s00436-003-0995-2. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Wu Z., Pandey K., Pandey B.D., Sherchand J.B., Yanagi T., Kanbara H. Molecular characterization of Blastocystis isolates from children and rhesus monkeys in Kathmandu, Nepal. Vet. Parasitol. 2009;160:295–300. doi: 10.1016/j.vetpar.2008.11.029. [DOI] [PubMed] [Google Scholar]
- Zhao G.H., Hu X.F., Liu T.L., Hu R.S., Yu Z.Q., Yang W.B., Wu Y.L., Yu S.K., Song J.K. Molecular characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol. Res. 2017;116:2327–2333. doi: 10.1007/s00436-017-5506-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.