Abstract
Background:
The objectives of this study were to compare the interferon-induced protein 44-like (IFI44L) promoter methylation level between systemic lupus erythematosus (SLE) patients and healthy controls and to evaluate its diagnostic value in SLE.
Materials and Methods:
The IFI44L promoter methylation level was measured in 49 patients with SLE and 50 healthy controls. Quantitative analysis of promoter methylation IFI44L gene in genomic DNA samples extracted from peripheral blood mononuclear cells was examined in SLE patients and healthy controls. The level of DNA methylation was compared between SLE patients and healthy controls as well as within SLE patient groups based on the presence of renal involvement. Moreover, diagnostic values of IFI44L were calculated.
Results:
The IFI44L promoter methylation level in SLE patients was significantly lower than healthy controls (median, 43.8 vs. 57, respectively; P = 0.008). The level of IFI44L promoter methylation was not significantly different between SLE patients with renal involvement and SLE patients without renal involvement (84.6% vs. 92.7%, respectively; P = 0.774). The IFI44L promoter methylation level ≤94.3% was the best cutoff point with a sensitivity of 91.8% and a specificity of 38% to distinguish patients with SLE from healthy individuals.
Conclusion:
The level of IFI44L promoter methylation from whole peripheral blood in Iranian SLE patients was significantly lower than healthy controls. Furthermore, the DNA methylation level of IFI44L promoter was not associated with renal damage in patients with SLE.
Keywords: Autoimmune diseases, DNA methylation, interferon-induced protein 44-like, systemic lupus erythematosus
INTRODUCTION
Systemic lupus erythematosus (SLE), as a chronic, remitting and relapsing, multisystem autoimmune disease with a complex and multifactorial pathogenesis, including interactions between genetic susceptibility, epigenetics, hormones, and environmental factors, is much more prevalent in women.[1,2,3] The prevalence of SLE varied in different geographic regions, and it is estimated that 20–150 cases per 100,000 individuals in the world are affected by this disease.[1,4]
Autoantibodies play an important role in the pathogenesis of SLE. They primarily targeted against nuclear material of the cell, and the deposition of immune complexes induces inflammation in various tissues such as skin, kidney, brain, and other organ systems.[2] The presence of autoantibodies, as a main feature of this disease, is used as a common serological marker in SLE. Antinuclear antibodies (ANAs), anti-double-stranded DNA (dsDNA) antibodies, and anti-Smith (anti-Sm) antibodies are the main groups that are used as autoantibody screening markers.[5,6] These three autoantibodies are currently used as diagnostic markers of the disease, but each has significant limitations. Both anti-dsDNA antibody and Anti-Sm antibody have high specificity (94% and 99%, respectively) in differentiating large percentage of suspected people who are not affected. However, these markers are not very sensitive for SLE, which can lead to classification of many actual affected patients as healthy people.[7,8] On the other hand, one study reported that ANA has sensitivity of 100% to identified patients with SLE, but low specificity of 65% can lead to increased number of healthy people diagnosed as affected.[9] Hence, the search for new markers to aid in the diagnosis of SLE still continues.
In the past decade, epigenetic modifications such as DNA methylation, histone modification, and microRNA dysregulation became a new area of investigation into the pathogenesis of SLE.[10] In the recent years, more than 50 genetic loci associated with SLE have been identified.[11] Interferon-induced protein 44-like (IFI44L) gene is one of the important genes, and its hypomethylation was reported to be correlated with SLE susceptibility. Hypomethylated sites observed in the promoter region of IFI44L gene in SLE patients have been reported in recent studies.[12,13,14,15,16] In a more recent study, Zhao et al. assessed IFI44L promoter methylation as a blood biomarker for SLE. They examined the methylation status of two CpG sites located within the IFI44L promoter in DNA. They found significant hypomethylation of IFI44L promoter in both sites in SLE patients and suggested that it can be used as a diagnostic marker for SLE in Chinese population with high sensitivity and specificity.[17]
As mentioned above, studies on the diagnostic role of IFI44L in SLE patients as a new marker are limited. On the other hand, it has been shown that DNA methylation contributes to some of the phenotypic differences between individuals and ethnicities. Therefore, this study was carried out with the objective of assessing the level of IFI44L promoter methylation and its sensitivity and specificity as a diagnostic marker in a sample of Iranian patients with SLE.
MATERIALS AND METHODS
This was a case–control study. Between October 2017 and December 2018, a total of 49 unrelated SLE patients from rheumatology clinics and inpatient ward at Al-Zahra Hospital, Isfahan, Iran, were recruited consecutively. The selection of SLE patients was based on the American College of Rheumatology (ACR) criteria (1997) for SLE.[18] Patients with no presence of malignant tumors, no presence of infection in the past month, and no presence of other autoimmune diseases or oxidative stresses were eligible. As for the control group, fifty unrelated healthy individuals (from hospital personnel or students) were recruited from the same hospital. SLE-free controls, in addition to eligibility criteria for patients, were chosen if they did not have any kinship with patients and did not have history of lupus or other autoimmune diseases in their families. This study was performed with the approval of the Institutional Review Board of Isfahan University of Medical Sciences (IR.MUI.REC.1396.3.615), and all participants provided written informed consent.
Demographic and clinical data were obtained from all participants, including age, sex, body mass index (BMI, calculated as weight [kg] divided by height [m] squared), blood pressure, and the presence of diabetes mellitus (DM) or hyperlipidemia. White blood cell, hemoglobin, platelets, creatinine, fasting blood sugar, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglyceride (TG), and blood urea nitrogen were assessed using standard methods. Serum levels of C-reactive protein (CRP), rheumatoid factor (RF), complement component 3 (C3), and complement component 4 (C4) were determined using nephelometric assay. Both C3 and C4 were categorized into two groups as follows: C3 as <90 or ≥ 90 mg/dL and C4 as <10 or ≥10 mg/dL. Erythrocyte sedimentation rate (ESR) was determined according to the instructions described by the manufacturer. Anti-dsDNA antibodies were quantified using enzyme-linked immunoassay and were considered significant when titers ≥20 IU/mL. Patients with the following criteria, based on ACR 1997, were defined with renal damage: persistent proteinuria >0.5 g/day or >3+ by dipstick and/or cellular casts including red blood cells, hemoglobin, granular, tubular, or mixed.
Whole peripheral blood (5 cc) was collected in EDTA from cases and controls and stored at −20°C until usage.
Isolation of blood mononuclear cells from peripheral blood
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient of Ficoll. After withdrawal, blood was transferred into a 50 mL tube, diluted in PBS (1:1 dilution), carefully layered over a Ficoll medium, and spun at 833 × g. The tubes were carefully removed from the centrifuge without disturbing the layering. The PBMC layer was carefully removed from the tube and transferred into a new conical tube. The PBMCs were washed by adding enough PBS to make up 15 mL and spun at 425 × g for 10 min, the supernatant was discarded, and the cells were resuspended in the appropriate volume of PBS.
DNA was extracted from the PBMCs using the GeNet Bio DNA extraction Kit (Korea) according to the manufacturer's protocol. The DNA was then quantified and qualified using a NanoDrop spectrophotometer method (according to a ratio of absorbance of 260/280 nm), and its integrity was determined by electrophoresis on 1% agarose gel.
Methylation of genes in both healthy and diseased population was assessed by MethyQESD (methylation-quantification of endonuclease-resistant DNA) technique. Methylation-sensitive restriction enzyme Hin6I was combined with real-time polymerase chain reaction (RT-PCR); we specifically checked the level of methylation. Note that the methylated DNA was resistant to digestion with Hin6I. This approach was done by Keith Methyl II DNA Restriction Kit (EpiTect), based on Bettstetter Protocol (Bettstette2008). Two enzymatic digestion was performed for each sample. The first was methylation quantification digestion (MQD) with methylation-sensitive enzyme, (Hin6I). The restriction enzyme, which has no effect on GCGC methylated sequence, was used to cut nonmethylated GCGC regions. Then, the uncut methylated DNA was amplified by RT-PCR. Second, methylation-independent calibrator digestion (CalD), including Xba1 and Dra1 enzymes, serves as an internal control and calibrator. Their recognition sites would not be present within the amplicon and were used to digest total DNA. These enzymes increased the efficiency of RT-PCR in methylation-insensitive group and lead to amplification of interested region and our selected piece amplified without any changes, thus leading to identical conditions in both the groups. Each batch contained 5 μl DNA and was digested in a 20 μl reaction volume in 10 × buffer Tango at 37°C for 13 h in thermocycler instruments (Bio Rad T100). Digestion in MQD batch was done by 1.5 μl endonuclease Hin6I which digests only unmethylated CGCG recognition sites, while in CalD batch, digestion was done by 0.75 μl endonuclease XbaI and 0.75 μl endonuclease DraI. Following this, enzymes were deactivated by incubating at 70°C for 20 min, and the samples were stored at − 20°C. A positive control with up to 1 μg digested DNA was built by CpG methyltransferase (M. SssI) enzyme (Fermentas), according to manufacturer's instruction. Nonmethylated blood DNA from healthy controls was used as a negative control. Negative and positive controls were run in parallel with our sample. In order to evaluate the quality of digestion, agarose gel electrophoresis was used.
The following equation was used to calculate methylation percentage:
EΔCt × 100. ΔCt = Ct Calibrator − Ct methylation quantification, and E designated PCR efficiency.
The statistical analysis was performed using MedCalc Statistical Software version 10.2.0 (MedCalc Software bv, Ostend, Belgium; https://www.medcalc.org). The normality of distribution was checked by the Kolmogorov–Smirnov test. Ordinal variables were reported as number (%) and were analyzed using the Chi-square test or Fisher's exact test when appropriate. Continuous variables were reported as mean ± standard deviation and were analyzed using independent sample t-test. Because of abnormal distribution of IFI44L in both groups, Mann–Whitney U-test was used to compare this variable between cases and controls. Logistic regression analysis was made to evaluate whether the laboratory variables were independently associated with SLE. A receiver operating characteristic (ROC) curve analysis was used to evaluate the areas under the ROC curve (AUC), which established the best cutoff values for % DNA methylation to diagnose SLE. Sensitivity, specificity, positive predictive values (PPVs), and negative predictive values (NPVs) were then calculated. The results were considered statistically significant when P < 0.05.
RESULTS
Table 1 shows the characteristics of the 49 patients with SLE and 50 healthy controls. As shown, patients and controls did not differ on age, sex, BMI, and systolic and diastolic blood pressure (P > 0.05). Two (4.1%) patients had DM whereas controls did not have DM (P = 0.466). Four (8.2%) patients had hyperlipidemia where controls did not have hyperlipidemia (P = 0.115). Most of the studied patients presented oral ulcers (34 patients, 69.4%), skin symptoms (42 patients, 85.7%), and all patients had positive ANA (49 patients, 100%). Furthermore, 10 (10.2%) patients had neurological disorder (seizure). Serositis was observed in 11 (22.4%) patients. Test for lupus anticoagulant was positive in 5 (11.1%) patients. Among studied patients, 8.2% (4 patients) had high anti beta2 glycoprotein I and 8.2% (4 patients) had high anticardiolipin antibody.
Table 1.
Characteristics of patients with systemic lupus erythematosus and healthy controls
| SLE patients (n=49) | Healthy controls (n=50) | P | |
|---|---|---|---|
| Age (years) | 36.0±12.3 | 34.5±10.8 | 0.520 |
| Sex, male/female | 8 (16.3)/41 (83.7) | 9 (18)/41 (82) | 0.963 |
| BMI (kg/m2) | 26.3±7.1 | 24.9±6.8 | 0.319 |
| Age at onset of disease (years) | 28.9±12.8 | - | |
| Systolic blood pressure | 124.5±20.4 | 120.4±18.6 | 0.298 |
| Diastolic blood pressure | 80.7±11.6 | 79.8±9.9 | 0.678 |
| DM | 2 (4.1) | 0 | 0.466 |
| Hyperlipidemia | 4 (8.2) | 0 | 0.115 |
| Oral ulcers | 34 (69.4) | 0 | 0.0001 |
| Skin symptoms | 42 (85.7) | 0 | 0.0001 |
| Arthritis | 38 (77.5) | 0 | 0.0001 |
| Neurological disorder (seizure) | 10 (10.2) | 0 | 0.002 |
| Renal involvement | 17 (34.7) | - | - |
| Serositis | 11 (22.4) | 0 | 0.001 |
| Positive test for lupus anticoagulant | 5 (11.1) | 0 | 0.063 |
| High anti-beta-2 glycoprotein I | 4 (8.2) | 0 | 0.115 |
| High anticardiolipin antibody | 4 (8.2) | 0 | 0.115 |
| Positive ANA | 49 (100) | 0 | 0.0001 |
Data are mean±SD, or n (%). P values were calculated using independent sample t-test or Chi-square test. BMI=Body mass index; ANA=Antinuclear antibody; DM=Diabetes mellitus; SD=Standard deviation; SLE=Systemic lupus erythematosus
The laboratory characteristics of patients with SLE and healthy controls are presented in Table 2. ESR and CRP were significantly higher in the studied patients than healthy controls (P = 0.0001). Positive RF was observed in 3 (6.2%) patients. Hemoglobin and PLT were significantly lower in patients than healthy controls (P < 0.05). BUN in the studied patients was significantly more than healthy controls (P = 0.018). The level of creatinine in SLE patients was significantly higher than healthy controls (P = 0.046). White blood cell, FBS, HDL, LDL, and TG were not significantly different between patients and healthy controls (P > 0.05). The mean anti-dsDNA autoantibodies in the studied patients was 197.7 ± 208.8, whereas in healthy controls, it was 8.9 ± 4.4 (P = 0.0001). Forty-three (87.8%) patients presented seropositivity to anti-dsDNA autoantibodies where this factor in all healthy controls was negative (P = 0.0001). The mean C3 in patients was significantly lower than healthy controls (45.7 vs. 137.8, respectively; P = 0.0001). The level of C3 in all healthy controls was ≥90 mg/dL, but in patients, 83.7% had C3 lower than 90 mg/dL. The mean of C4 in patients was 10.4 ± 10.1, which was significantly lower than 16.3 ± 4.8 in healthy controls (P = 0.0001). The level of C4 in most of the patients (79.6%) was lower than 10 mg/dL, whereas all healthy controls had C4e ≥10 mg/dL. Furthermore, in logistic regression analysis, all laboratory variables were entered in the first model and then final model was run with associated variables in the first model including ESR, CRP, hemoglobin, BUN, and HDL. In the analysis, ESR, hemoglobin, and HDL levels were statistically significant and independently associated with SLE disease [Table 3].
Table 2.
Laboratory characteristics of patients with systemic lupus erythematosus and healthy controls
| SLE patients (n=49) | Healthy controls (n=50) | P | |
|---|---|---|---|
| ESR (mm/h) | 52.3±35.4 | 15.6±6.9 | 0.0001 |
| CRP (mg/l) | 15.4±17.4 | 4.2±2.6 | 0.0001 |
| RF, positive | 3 (6.2) | 0 | 0.119 |
| White blood cell (109/l) | 6.2±2.7 | 6.8±1.7 | 0.119 |
| Hemoglobin | 11.1±0.62 | 14.3±1.5 | 0.0001 |
| PLT (109/l) | 197.1±84.2 | 233.4±66.7 | 0.019 |
| BUN | 22.2±17.9 | 15.8±4.5 | 0.018 |
| Creatinine (mg/dL) | 1.39±1.81 | 0.87±0.21 | 0.046 |
| FBS | 89.7±20.4 | 98.1±42.5 | 0.214 |
| HDL | 52.4±13.5 | 47.5±12.0 | 0.059 |
| LDL | 100.7±24.1 | 106.2±38.8 | 0.335 |
| TG | 156.3±56.6 | 147.9±69.8 | 0.463 |
| Anti-dsDNA (IU/mL) | 197.7±208.8 | 8.9±4.4 | 0.0001 |
| Negative (<20) | 6 (12.2) | 49 (100) | 0.0001 |
| Positive (>20) | 43 (87.8) | 0 | |
| C3 (mg/dL) | 45.7±48.7 | 137.8±6.4 | 0.0001 |
| <90 | 41 (83.7) | 0 | - |
| ≥90 | 8 (16.3) | 50 (100) | |
| C4 (mg/dL) | 10.4±10.1 | 16.3±4.8 | 0.0001 |
| <10 | 39 (79.6) | 0 | - |
| ≥10 | 10 (20.4) | 50 (100) |
Data are mean±SD, or n (%). P values were calculated using independent sample t-test or Chi-square test. SLE=Systemic lupus erythematosus; SD=Standard deviation; ESR=Erythrocyte sedimentation rate; CRP=C-reactive protein; RF=Rheumatoid factor; BUN=Blood urea nitrogen; PLT=Platelet; HDL=High-density lipoprotein; LDL=Low-density lipoprotein; TG=Triglyceride; FBS=Fasting blood sugar; C3=Complement component 3; C4=Complement component 4; dsDNA=Double-stranded DNA
Table 3.
Results of the logistic regression analysis between systemic lupus erythematosus patients and healthy controls and laboratory variables
| OR | 95% CI | P | |
|---|---|---|---|
| ESR | 1.124 | 0.953–1.694 | 0.014 |
| CRP | 1.271 | 1.024–1.235 | 102 |
| Hemoglobin | 0.496 | 0.351–0.702 | 0.0001 |
| BUN | 1.087 | 0.957–1.234 | 0.201 |
| HDL | 1.076 | 1.018–1.138 | 0.010 |
OR=Odd ratio; CI=Confidence interval; ESR=Erythrocyte sedimentation rate; CRP=C-reactive protein; BUN=Blood urea nitrogen; HDL=High-density lipoprotein
Figure 1 shows the comparison of IFI44L promoter methylation level between case and control groups using Mann–Whitney U. The mean IFI44L promoter methylation level in the case group was 54.4% ± 50.9% (median: 43.8 and interquartile range [IQR]: 18.6–74.8) and in healthy controls was 89.8 ± 91.7 (median: 57 and IQR: 28.9–162.8); the difference between groups was statistically significant (P = 0.008). Furthermore, based on the results of ROC analyses presented in Figure 2, the AUC was 0.639 with 95% confidence interval (CI) between 0.536 and 0.733 (P = 0.012). The best optional cutoff point that statistically has high level of both sensitivity and specificity for IFI44L promoter methylation level to distinguish patients with SLE from healthy individuals was ≤94.3% with sensitivity of 91.8% (95% CI: 80.4–97/7) and specificity of 38% (95% CI: 24.7–52.8). The PPV was 59.2% (95% CI: 47.2–70.4) and NPV was 82.6% (95% CI: 61.2–94.9). The accuracy of IFI44L promoter methylation level to distinguish patients with systemic lupus was 61.7% (95% CI: 51.1%–71.5%).
Figure 1.
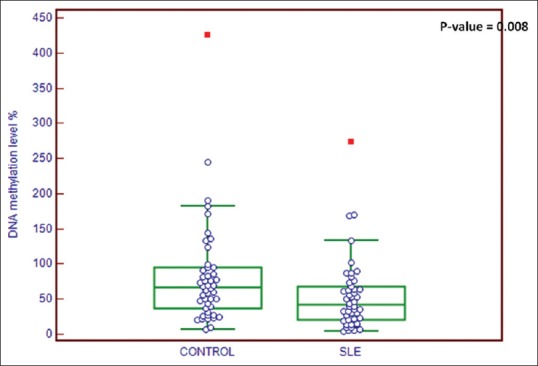
Comparison of interferon-induced protein 44-like promoter methylation level between patients with systemic lupus (median: 43.8 and IQR: 18.6–74.8) and healthy controls (median: 57 and IQR: 28.9–162.8). IQR = Interquartile range
Figure 2.
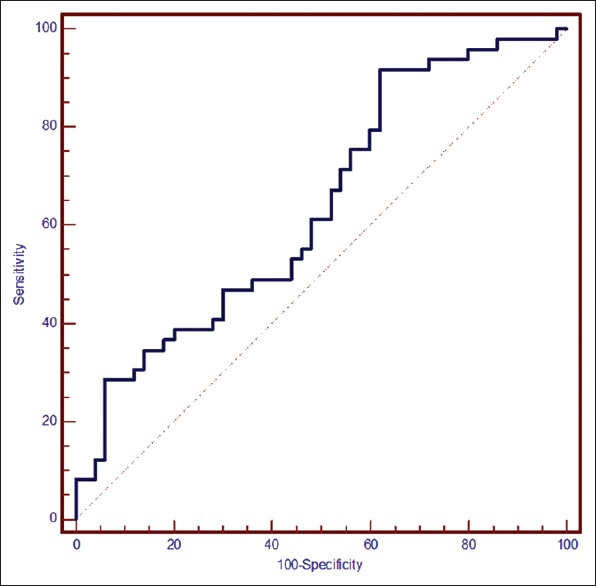
Receiver operating characteristic curves for % DNA methylation. Area under the receiver operating characteristic curve, 0.639 (95% CI: 0.536–0.733, P = 0.012), cutoff point ≤ 94.3, sensitivity of 91.8% (95% CI: 80.4–97.7), and specificity of 38% (95% CI: 24.7–52.8). CI = Confidence interval
Laboratory characteristics of patients with SLE based on renal involvement are presented in Table 4. ESR, CRP, and anti-dsDNA in patients with SLE with renal involvement were significantly more than patients without renal involvement (P < 0.001). Anti-dsDNA in all patients with renal involvement was positive. The level of C3 and C4 in patients with SLE with renal involvement was significantly lower than patients without renal involvement (P = 0.0001). All patients with renal involvement had C3 lower than 90 and C4 lower than 10 mg/dl. The mean of IFI44L promoter methylation level in SLE patients with renal involvement was not significantly lower than patients without renal involvement (84.6% vs. 92.7%, respectively; P = 0.774).
Table 4.
Laboratory characteristics of patients with systemic lupus erythematosus based on renal involvement
| With renal damage (n=17) | Without renal damage (n=32) | P | |
|---|---|---|---|
| ESR (mm/h) | 69.8±39.7 | 15.1±5.5 | 0.0001 |
| CRP (mg/l) | 21.9±19.3 | 2.5±1.4 | 0.001 |
| Negative (<6) | 0 | 32 (100) | 0.0001 |
| Positive (≥6) | 17 (100) | 0 | |
| C3 (mg/dL) | 28.8±29.0 | 127.9±36.8 | 0.0001 |
| <90 | 17 (100) | 24 (75) | 0.064 |
| ≥90 | 0 | 8 (25) | |
| C4 (mg/dL) | 7.8±8.8 | 21.7±8.9 | 0.0001 |
| <10 | 17 (100) | 22 (69) | 0.301 |
| ≥10 | 0 | 10 (31) | |
| Anti-dsDNA (IU/mL) | 285.8±253.4 | 12.6±4.5 | 0.0001 |
| Negative (<20) | 0 | 6 (19) | 0.203 |
| Positive (>20) | 17 (100) | 26 (81) | |
| ANA | 750.7±2724.0 | 776.3±2745.5 | 0.975 |
| DNA methylation (%) | 84.6±67.5 | 92.7±103.9 | 0.774 |
Data are mean±SD, or n (%). P values were calculated using independent sample t-test or Chi-square test. ESR=Erythrocyte sedimentation rate; CRP=C-reactive protein; SD=Standard deviation; ANA=Antinuclear antibody; C3=Complement component 3; C4=Complement component 4; dsDNA=Double-stranded DNA
DISCUSSION
The main objective of the present study was to compare the level of IFI44L promoter DNA methylation and its role in the diagnosis of SLE patients. Statistically significant difference in the DNA methylation level was observed between patients with SLE and healthy controls. However, our findings indicated that DNA methylation level has low sensitivity and specificity in separating SLE and healthy controls apart. An important finding was that DNA methylation level was not significantly different between SLE patients with renal involvement and SLE patients without renal involvement. Also, in regression analysis, ESR, hemoglobin, and HDL were significantly associated with SLE disease. It can be explained by the fact that most of the studied patients were hospitalized who had higher disease activity that can cause increasing ESR level and decreasing hemoglobin level; on the other hand, renal disease can decrease hemoglobin level in patients with SLE (17 patients in our study suffer from renal disease). The association between SLE and HDL and higher level of HDL in cases can be due to the fact that patients with SLE usually take antihyperlipidemic drugs as a medicine for the prevention of cardiovascular disease.
Luo et al.[19] induced IFI44L overexpression or its downregulation by siRNA in normal PBMCs and tested the key components of TBK1/IRF3 pathway as well as downstream cytokines to explore the function of IFI44L in SLE. They find that in dsDNA-stimulated human and mouse cells, TBK1 has been shown to move to cytoplasmic punctate structures, where it associates with STING to induce IRF3 activation. The IRFs are a class of master transcription factors that regulate pathogen-induced innate and acquired immune responses. IRF signaling pathway aberrations due to infection, genetic predisposition, or mutation lead to increased expression of type I interferon (IFN) genes, IFN-stimulated genes, and other pro-inflammatory cytokines/chemokines which are the basis for the development of autoimmune diseases and cancer. They reported that both mRNA and protein level of IFI44L were significantly elevated in PBMC from SLE patients as well as normal PBMCs stimulated by IFN-a and SLE serum. Moreover, the expression of IFI44L showed positive correlation with disease activity. They further validated that the overexpression of IFI44L was due to the hypomethylation of IFI44L promoter. IFI44L upregulation resulted in enhanced activation of TBK1/IRF3 pathway, including TBK1, IRF3, and STING, which are inducing some of the downstream pro-inflammatory cytokines such as tumor necrosis factor-alpha, CCL5, and IFN-beta. Therefore, IFI44L gene was considered a mechanistic link between IFN-inducible genes and the pathogenesis of SLE.
In concordance with previous studies, our results show that the level of Iranian patients' promoter methylation of IFI44L was significantly lower in SLE patients.[13,14,15,16,17] The difference in DNA methylation patterns by comparing monozygotic twins with and without SLE was first reported by Javierre et al. in 2010.[20] A genome-wide DNA methylation study by Coit et al. in 2013 showed a significant hypomethylation in IFN-regulated genes in naïve T cells from lupus patients, including IFI44L, compared to matched healthy controls.[13] The results of Coit et al. were confirmed in Absher et al.'s study.[14] The results of another study by Coit et al. observed more DNA hypomethylation in SLE patients than controls and reported IFI44L as one of the most hypomethylated genes.[15] More recently, Zhao et al. performed a genome-wide DNA methylation study in whole blood and reported significant hypomethylation of a CpG site within IFI44L promoter in SLE patients compared to healthy controls.[17]
In our study, the DNA methylation level of IFI44L promoter in SLE patients with renal involvement was lower than those patients without renal involvement, but was not statistically significant. This was inconsistent with those results reported in two recent studies. In Zhao et al.'s study, DNA methylation level was reported significantly lower in patients with SLE with renal involvement than in patients without renal involvement.[17] Furthermore, Coit et al. showed that DNA demethylation in IFN-regulated genes in SLE patients with renal disease was significantly lower than those patients without renal disease.[21] The difference between our results and the other two studies can be explained by the number of studied patients; in our study, only 17 patients had renal involvement that may be a factor in reduced statistical power to find significant differences between patients.
Validation of potential biomarkers and searching for additional markers for the diagnosis and prognosis of SLE is regarded as an urgent need due to its complex pathology and physiology. Previous studies focused on different genetic markers and various findings have been reported.[22,23] Recently, Zhao et al. examined the IFI44L promoter methylation as a noninvasive blood biomarker for SLE. They measured hypomethylation of two CpG sites within IFI44L promoter and reported that site 1 methylation with a sensitivity of 93.6% and a specificity of 96.8% at a cutoff DNA methylation level of 75.5% would be a decent novel biomarker to distinguish patients with SLE from healthy controls. Compared to Zhao et al.'s study, we found much lower sensitivity and specificity for IFI44L. The best cutoff point for IFI44L in our study was 94.3% with a sensitivity of 91.8% and a specificity of 38%. In contrast to Zhao et al.'s findings,[17] our study suggests that IFI44L could not be a good biomarker in Iranian patients with SLE. However, small sample size in our study in comparison to Zhao et al. might have led to low statistical power of our study. These data suggest that IFI44L methylation must be studied more thoroughly as a novel diagnostic test for SLE patients.
Our results should be interpreted in the context of its limitations. The main limitation of this study is small sample size that might not be enough and accordingly reduced statistical power to draw conclusions on some of the differences. Thus, further studies with larger sample sizes are required. In addition, patients with other autoimmune diseases such as rheumatoid arthritis or Sjogren's syndrome were not enrolled in our study; therefore, it is unknown whether the changes of DNA methylation are exclusively observed in SLE patients or not, and thus, further studies including patients with other autoimmune diseases are necessary to confirm the specificity of DNA methylation.
CONCLUSION
The level of IFI44L promoter methylation from whole peripheral blood in Iranian SLE patients was significantly lower than healthy controls. Furthermore, the DNA methylation level of IFI44L promoter was not associated with renal damage in patients with SLE. Moreover, in our study DNA methylation level of IFI44Lpromoter as a diagnostic biomarker of SLE exhibited low sensitivity and specificity, and we could not prove its reliability as diagnostic marker to distinguish patients with SLE from healthy people. Further studies are needed to establish the value of IFI44L promoter methylation as a diagnostic biomarker of SLE.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Gatto M, Zen M, Ghirardello A, Bettio S, Bassi N, Iaccarino L, et al. Emerging and critical issues in the pathogenesis of lupus. Autoimmun Rev. 2013;12:523–36. doi: 10.1016/j.autrev.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 3.Lockshin MD. Sex ratio and rheumatic disease: Excerpts from an institute of medicine report. Lupus. 2002;11:662–6. doi: 10.1191/0961203302lu274oa. [DOI] [PubMed] [Google Scholar]
- 4.Danchenko N, Satia JA, Anthony MS. Epidemiology of systemic lupus erythematosus: A comparison of worldwide disease burden. Lupus. 2006;15:308–18. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 5.Yu C, Gershwin ME, Chang C. Diagnostic criteria for systemic lupus erythematosus: A critical review. J Autoimmun. 2014;48-49:10–3. doi: 10.1016/j.jaut.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Egner W. The use of laboratory tests in the diagnosis of SLE. J Clin Pathol. 2000;53:424–32. doi: 10.1136/jcp.53.6.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bizzaro N, Villalta D, Giavarina D, Tozzoli R. Are anti-nucleosome antibodies a better diagnostic marker than anti-dsDNA antibodies for systemic lupus erythematosus? A systematic review and a study of metanalysis. Autoimmun Rev. 2012;12:97–106. doi: 10.1016/j.autrev.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Zieve GW, Khusial PR. The anti-Sm immune response in autoimmunity and cell biology. Autoimmun Rev. 2003;2:235–40. doi: 10.1016/s1568-9972(03)00018-1. [DOI] [PubMed] [Google Scholar]
- 9.Solomon DH, Kavanaugh AJ, Schur PH American College of Rheumatology Ad Hoc Committee on Immunologic Testing Guidelines. Evidence-based guidelines for the use of immunologic tests: Antinuclear antibody testing. Arthritis Rheum. 2002;47:434–44. doi: 10.1002/art.10561. [DOI] [PubMed] [Google Scholar]
- 10.Jeffries MA, Sawalha AH. Epigenetics in systemic lupus erythematosus: Leading the way for specific therapeutic agents. Int J Clin Rheumtol. 2011;6:423–39. doi: 10.2217/ijr.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–92. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+T-cell populations. PLoS Genet. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coit P, Yalavarthi S, Ognenovski M, Zhao W, Hasni S, Wren JD, et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J Autoimmun. 2015;58:59–66. doi: 10.1016/j.jaut.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363–72. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M, Zhou Y, Zhu B, Wan M, Jiang T, Tan Q, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis. 2016;75:1998–2006. doi: 10.1136/annrheumdis-2015-208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 19.Luo S, Zhao M, Lu Q. Upregulation of IFI44L induces inflammation by activating TBK1/IRF3 pathway in systemic lupus erythematosus. J Invest Dermatol. 2016;136:2. [Google Scholar]
- 20.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, et al. Renal involvement in lupus is characterized by unique DNA methylation changes in naïve CD4+T cells. J Autoimmun. 2015;61:29–35. doi: 10.1016/j.jaut.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shumnalieva R, Kachakova D, Shoumnalieva-Ivanova V, Miteva P, Kaneva R, Monov S. Whole peripheral blood miR-146a and miR-155 expression levels in systemic lupus erythematosus patients. Acta Reumatol Port. 2018;43:217–25. [PubMed] [Google Scholar]
- 23.Nambiar MP, Enyedy EJ, Fisher CU, Krishnan S, Warke VG, Gilliland WR, et al. Abnormal expression of various molecular forms and distribution of T cell receptor zeta chain in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:163–74. doi: 10.1002/1529-0131(200201)46:1<163::AID-ART10065>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]


