Highlights
-
•
Plant microbiomes have an unexplored potential to control root parasitic weeds.
-
•
Understanding the mechanisms by which microbes can control parasitic weeds is largely elusive.
-
•
Members of the root microbiome can interfere with host-parasite chemical communication.
-
•
Direct and indirect modes of action can work synergistically in microbe-mediated weed control.
Abstract
Microbiomes can significantly expand the genomic potential of plants, contributing to nutrient acquisition, plant growth promotion and tolerance to (a)biotic stresses. Among biotic stressors, root parasitic weeds (RPWs), mainly of the genera Orobanche, Phelipanche and Striga, are major yield-limiting factors of a wide range of staple crops, particularly in developing countries. Here, we provide a conceptual synthesis of putative mechanisms by which soil and plant microbiomes could be harnessed to control RPWs. These mechanisms are partitioned in direct and indirect modes of action and discussed in the context of past and present studies on microbe-mediated suppression of RPWs. Specific emphasis is given to the large but yet unexplored potential of root-associated microorganisms to interfere with the chemical signalling cascade between the host plant and the RPWs. We further provide concepts and ideas for future research directions and prospective designs of novel control strategies.
Current Opinion in Microbiology 2019, 49:26–33
This review comes from a themed issue on Environmental microbiology
Edited by Roeland Berendsen and Klaus Schlaeppi
For a complete overview see the Issue and the Editorial
Available online 23rd October 2019
https://doi.org/10.1016/j.mib.2019.09.006
1369-5274/© 2019 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
The economically most important root parasitic weeds (RPWs) belong to the family Orobanchaceae, encompassing the genera Orobanche,Striga and Phelipanche. These RPWs have a hidden but devastating effect on host plants as a large part of its life cycle occurs belowground. Once the parasite emerges aboveground, the adverse impact on crop productivity has already taken place. Striga species, also known as witchweeds, are widely distributed in Sub-Saharan Africa, India and Southeast Asia [1], affecting cereal crops such as maize, rice, millets, sorghum and the legume cowpea. Striga causes yield losses up to 80%, often resulting in field abandonment by local farmers. For Striga hermonthica it has been estimated that 50–300 million hectares of field soils in Africa are currently infested [2]. In addition to Striga spp., also the broomrapes Phelipanche and Orobanche are widely distributed and their hosts are not limited to cereals and legumes, but also comprise Solanaceae(e.g. tomato, tobacco), Asteraceae (e.g. sunflower), and Cucurbitaceae (e.g. watermelon). They substantially affect crop production in Western Africa, the Mediterranean area but also occur in Australia, America and Asia. For Orobanche crenata, legume crop losses of up to 100% have been reported in Morocco, Portugal, Spain and Syria [2].
Despite their wide geographic distribution and host range, the RPW’s life cycles and infection strategies have common traits. For obligate RPWs, seed germination relies on host-derived signals released by the roots, in particular the strigolactones. The primary eco-evolutionary role of these multi-functional phytohormones is to initiate, under low nutrient conditions, a symbiotic association with arbuscular mycorrhizal fungi (AMF) [3]. Hence, obligate RPWs hijack these signals for infection, repurposing this ancient beneficial signalling mechanism [4]. The germination signal is perceived by the RPWs via strigolactone receptors [5], but the downstream signalling is not yet fully resolved [6]. Following seed germination, an important second step in root infection by RPWs is haustoria formation. Also here the underlying chemistry has received considerable attention and various haustorium-inducing factors have been identified, including quinones (e.g. 2,6-dimethoxy-1,4-benzoquinone), phenolic compounds (e.g. syringic acid, vanillic acid, vanillin), and anthocyanins (e.g. peonidin, pelargonidin) [7,8]. Other key stages of the life cycle that are promising targets for control include the seed bank in soils and the production of new seeds [9]. Current control strategies include breeding for host resistance, cultural methods such as hand weeding and alternative cropping practices, and chemical control. Each of these strategies is not singularly effective and not always available to smallholder farmers [9]. Hence, a systems approach is needed to provide effective and sustainable control of RWPs.
In this opinion article, we provide a conceptual framework to explore the yet-untapped potential of soil and root-associated microbes to interfere with the chemical signalling cascade and to induce physiological and phenotypic changes in the host plant to suppress RPWs. We discuss direct and indirect modes of action in the ecological context of the tripartite interaction between host, parasite and microbiome. We argue that understanding the intricate eco-evolutionary, chemical and genetic mechanisms operating at the root-soil interface constitutes an essential step towards developing new integrated strategies to mitigate the adverse impacts of RPWs on crop production.
Microbe-mediated mechanisms of root parasitic weed control
Microbes can directly and indirectly interfere in the RPW’s life cycle, either by deterring the parasite or by triggering processes that impair infection of the host roots (Figure 1). Direct modes of action are those in which the microbe or microbiome interact directly with the parasite: these include (1) pathogenicity towards the RPW, (2) antagonism towards RPWs via secondary metabolites, and (3) interference with host-parasite signalling. We refer to indirect modes of action as those in which the microbe or microbiome affect the parasite through interactions with the host and/or local environment. These modes of action include (4) enhancement of nutrient acquisition by the host, in particular phosphorous (P) and nitrogen (N), (5) modulation of host root physiology, that is, alteration of exudation or root architecture, and (6) induced systemic resistance (ISR). Importantly, these different mechanisms are not mutually exclusive and likely work in sequence, simultaneously or even synergistically during the RPW life cycle (Figure 2).
Figure 1.
Microbe-mediated mechanisms for root parasitic weed (RPW) control. The conceptual figure depicts examples of direct modes of action that target the RPWs by hindering or disrupting the RPW’s life-cycle. Indirect modes of action comprise those in which microbes affect the soil nutrient pool bioavailable to the plant, affect plant physiology or induce local and systemic resistance against RPW infections.
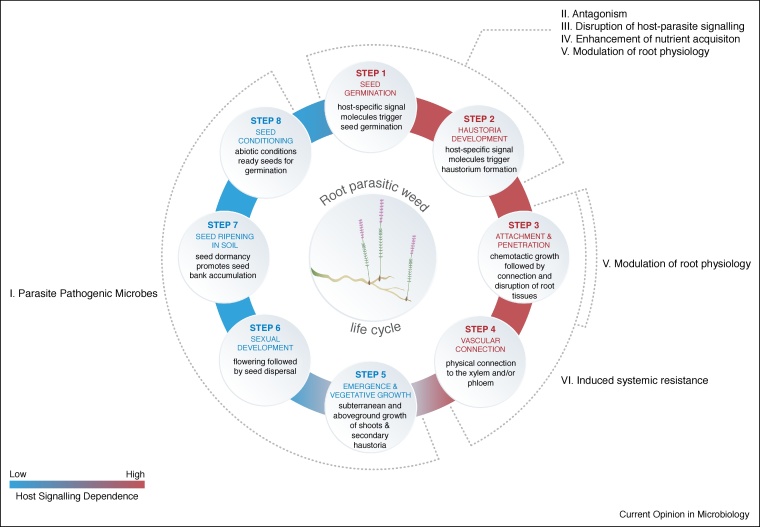
Figure 2.
Signalling and life cycle of root parasitic weeds.
(1) Host plant roots release signalling molecules (i.e. strigolactones) that induce the germination of root parasitic weed (RPW) seeds in the root-soil interface. (2) After germination, the parasite forms radicles and haustoria, the formation of which are induced by molecules known as haustorium-inducing factors. (3) The haustorium connects to and penetrates host roots reaching the vascular tissues. (4) RPWs establish a vascular connection with the xylem and/or xylem and phloem (this is dependent on the photosynthetic capability of the RPW species) in order to syphon water and photosynthates from the host plant. (5) Once a functional vascular connection is established, the RPW undergoes vegetative growth, followed by emergence from the soil; in some cases, secondary haustoria are formed allowing for additional connections with the host(s). (6) After weeks of vegetative growth, the RPWs flower and set seeds. (7) The newly formed RPW seeds are deposited in the soil, where they can remain dormant (i.e. RPW seed bank), (8) Before being able to respond to host signals, RPW seeds require a pre-conditioning stage that is provided by specific abiotic soil conditions, that is, moisture and temperature. Note that for facultative RPWs step 1 is not dependent on host-specific signal molecules, as it is for obligate RPWs, but can be triggered endogenously.
The general life cycle of a root parasitic weed (RPW). Schematic presentation of the different steps of an obligate RPW’s life cycle and its dependency on host signals. The warmth of the colours (blue to red) in the outer circle indicates how dependent the RPW is on signalling molecules from the host to serve as cues for its development and to complete its life cycle. Microbe-mediated mechanisms and their most preferred timing to control RPWs are indicated along the dotted line.
Direct modes of action
RPW pathogens
There are two important considerations with respect to the use of pathogens to control RPWs: (1) host specificity of the pathogen, and (2) stage of the RPW’s life cycle affected by the pathogen. One of the most studied RPW pathogens is the fungus Fusarium, with ca. 15 species tested against parasitic weeds from the genera Orobanche, Striga and Phelipanche [10]. Only Fusarium oxysporum f. sp. strigae was shown to be specific to S. hermonthica, with the exception of some solanaceous plants which also can be colonised by this fungus [11]. In a consortium consisting of three strains of F. oxysporum f. sp. strigae (called Foxy T14), the overproduction of tyrosine, leucine and/or methionine (due to metabolic imbalances and inhibitory feedbacks [12]) was significantly related to reduced emergence of Striga and consequently increased yields of maize [13••]. Also other fungal species including Alternaria, Aspergillus and Verticillium were reported as pathogens of Striga spp., with emphasis on S. hermonthica, resulting in a significant reduction of RPW emergence and biomass [10]. For O. crenata, the fungus Ulocladium atrum was shown to infect vegetative structures, such as shoots and tubercules, thus hindering RPW infection and development [14]. An excellent example of a pathogen acting at early stages of RPW development is the fungus F. oxysporum f. sp. orthoceras, which colonises seeds of Orobanche cumana, and act by dissolving the seed endosperm and metabolizing cytoplasmic compounds [15]. Next to fungi, several bacterial genera such as Bacillus, albeit not pathogenic sensu stricto, can cause seed decay of S. hermonthica by extracellular xylanases, pectinases, and amylases [16]. Interestingly, the implications of such findings can also be translated into the development of new control strategies that target the seed bank in highly infested and abandoned field sites. Despite some studies investigating the potential use of viruses to control weeds [17], their efficacy in controlling RPWs remains to be explored.
Antagonism via secondary metabolites and volatile organic compounds (VOCs)
Recent high-throughput screenings of chemical libraries have led to the discovery of several compounds that interfere with strigolactone signalling. These include compounds inducing the germination of S. hermonthica, such as sphynolactone-7 [18••], inhibiting a strigolactone receptor in S. hermonthica, such as soporidine [19] and simple β-lactones [20], or inhibiting receptors of strigolactones from a range of other plant species, such as derivatives of N-phenylanthranilic acid [21]. Soil and plant-associated microbes can make structurally similar compounds. For example, bacterial strains of the genera Streptomyces and Arthrobacter produce anthranilic acid derivatives. Also, β-lactone derivatives are produced by bacteria and fungi, such as hymeglusin by Fusarium, obafluorin by Pseudomonas fluorescens, lipstatin and belactosins by Streptomyces spp. [22]. Other fungal metabolite classes that hold potential to suppress RPWs include sesquiterpenoids, tricothecenes (e.g. HT-2 toxin, neosolaniol, nivalenol, roridin A and verrucarins A, B, M), in addition to amino acid overproduction as highlighted above [12]. Tricothecenes are broadly distributed across the fungal genera Fusarium and Myrothecium, which are well-known RPW antagonists. As strigolactones are sesquiterpene lactones, it would be interesting to investigate if the observed suppressive effect of tricothecene-producing fungal RPW antagonists can be explained, in part, by competition for binding sites of the strigolactone receptors. Plant-associated strains from a range of bacterial genera, such as Streptomyces, Azospirillum, Pseudomonas and Rhizobium, have been tested for activity against RPWs. In most of these studies, however, the underlying mechanisms and metabolites were not characterized in detail. Nevertheless, a small lipophilic compound [23] and a small peptide [24] of Azospirillum brasilense were implicated in germination arrest of S. hermonthica and P. aegyptiaca, respectively.
A separate class of microbial metabolites for RPW control are the volatile organic compounds (VOCs). VOCs are chemically diverse small molecules with low vapour pressure that can, from a distance, regulate plant growth and root development [25]. The best example of a microbial VOC that can trigger suicidal germination of RPW seeds is ethylene [26]. Ethylene was successfully used as a soil fumigant to eradicate Striga asiatica in North and South Carolina [27], but this technology is not easily applicable in developing countries due to high costs and non-target effects on soil (micro-)biology. Alternatively, there is a high number of microbes able to produce ethylene. For example, ethylene produced in vitro by Pseudomonas syringae pv. glycinea [26] and Klebsiella sp. [28] induced seed germination of several Striga species, including S. aspera, S. hermonthica and S. gesnerioides. In addition, the sulphurous microbial VOC dimethyldisulfide produced by various bacterial genera such as Burkholderia [29] was implicated in P. aegyptiaca control [10]. Collectively these studies exemplify that soil and root-associated microbiomes hold a yet-untapped metabolic repertoire to (1) induce RPW seed germination in the absence of its host [30], referred to as suicidal germination, (2) suppress RPW seed germination, or (3) hinder the development of radicles and/or haustoria [12].
Disruption of host-parasite signalling
Since seed germination and haustoria formation are crucial steps in the infection process of RPWs, it is interesting to explore the capability of soil and root-associated microbes to interfere with or disrupt this chemical signalling cascade. For example, after growing bacterial epiphytes from sorghum seeds in sorghum root exudates, the induction of S. hermonthica germination by the root exudate decreased almost completely and a reduced number of Striga attachments to the host root was observed. These findings were, to some extent, related to changes in the composition of phenolic compounds in the exudates [31]. In another example, when fungal strains (i.e. F. oxysporum, Fusarium solani, Botrytis cinerea, Trichoderma harzianum) were grown in liquid culture, the germination stimulants strigol, 5-deoxystrigol, 4-deoxyorobanchol, and the synthetic analogue GR24 were significantly degraded [32]. A myriad of signalling molecules (e.g. sterols, isothyacyanates, organic acids) that can induce RPW seed germination and haustorium formation are released in the root-soil interface. Because of antimicrobial properties [33], several of these signalling molecules may also indirectly affect RPWs via changes in the composition and activity of plant-associated microbial communities or via affecting the association with AMF. Although microbe-mediated chemical modifications or degradation of signals seem to work effectively in in vitro assays, the efficacy in planta as well as the impact on the mutualistic interactions between the plant and symbionts, such as AMF, are still underexplored areas of research in microbe-mediated RPW control.
Indirect modes of action
Enhancement of host nutrient acquisition
Exudation of strigolactones is induced by phosphorous (P) and, to some extent, by nitrogen (N) starvation [34], resulting in a ‘nutrient-dependent strigolactone negative feedback’. In other words, when a host plant is nutrient starved, it will start recruiting AMF via increased strigolactone exudation, which are then hijacked by RPWs as a signal of host presence. In line with this, exudates of P-starved tomatoes induced higher Phelipanche ramosa germination [35], however, when plants were colonised by AMF the biosynthesis of strigolactones was halted [36]. Moreover, some AMF were shown to increase root nodulation [37], which can improve both P and N uptake. This finding is particularly relevant for leguminous host plants of Striga gesneroides and O. crenata. Chemical fertilization (P and N) can negatively affect S. hermonthica germination, attachment and emergence [38]. Therefore, microbe-mediated provision of the host with labile sources of P and N is a potential mechanism that, indirectly, hampers the signalling between host and RPWs. Since AMF also depend on strigolactones to initiate symbiosis, working towards P provision via AMF association may not be a viable option as they might be outcompeted by RPWs. However, different strigolactone exudation profiles were observed for maize cultivars resistant and susceptible to S. hermonthica, dominated by sorgomol and 5-deoxystrigol, respectively. These exudates differentially affected seed germination of S. hermonthica and only minimally influenced AMF colonization [39•]. These findings point towards a need to better understand the specificity of distinct strigolactones on AMF symbiosis and RPW infections [40]. Apart from the well-known benefits of AMF, various other fungal and bacterial genera are effective P-solubilizers, through the production of organic acids such as citric, lactic and oxalic acids [41]. These include the fungi Fusarium, Trichoderma, and Myrothecium, and a wide range of bacteria such as Pseudomonas, Streptomyces, Burkholderia¸ and Rhizobium — all of which have been linked to suppression of various RPWs. For these other fungi, however, the link between P-solubilisation and reduced RPW infection has not yet been established.
Modulation of root physiology
Root-associated microbes can modulate root physiology and exudation both quantitatively and qualitatively [42,43]. For instance, upon AMF (Glomus intraradices) colonization of tomato, the level of strigolactones in exudates (i.e. solanacol, didehydro-orobanchol) was significantly reduced, resulting in lower seed germination of P. ramosa [36]. Whether this effect is indirectly caused by phosphate nutrition or directly via AMF colonization was not resolved in this study. Microbes may also modulate other root exudates with allelopathic properties that influence RPWs. An example is the sesquiterpene inuloxin C from the medicinal composite plant Inulaviscosa (syn. Dittrichia viscosa Greuter), which was shown to hinder seed germination of P. ramosa and several Orobanche species, even in the presence of strigolactones [44]. Other examples are the rye-cyanatines from cereals, which had an adverse effect on broomrape germination and development [45], and 6-chloroacetyl-2-benzoxazolinone, a derivative of 2-benzoxazalinone described as inhibitor of germination and radicle development of O. crenata [46]. Moreover, root exudation can also be influenced by aboveground pathogens and herbivores leading to changes in the composition and activity of root-associated microbes [47, 48, 49]. In addition to changes in root exudation, microorganisms can also induce changes in root architecture [50,51], and possibly root tissue distribution and chemical depositions (e.g. callose, suberin and phenolic compounds) that can act as physical barriers to RPW infections [52]. For example, the AMF Gigaspora margarita was shown to induce lateral root formation in Lotus japonicus via exudates and volatiles emitted from germinating spores [51]. Such shifts in root architecture can potentially lead to variation in RPW infection sites. For instance, it was shown that O. cumana had a preference for infecting younger thinner roots of sunflower, likely due to increased lignification of older root tissues [53]. It is noteworthy, however, that in these experiments it is challenging to disentangle microbe-induced effects on root chemistry from plant responses to RPWs and/or to the local environment.
Induced systemic resistance
Several root-associated microorganisms can induce systemic resistance in plants against root and leaf pathogens [54]. Induced resistance responses are accompanied by substantial transcriptional changes in plant defence pathways, in particular salicylic acid and jasmonic acid, as well as changes in physiology and cell wall chemistry [54]. Recent studies have shown that salicylic acid and to some extent jasmonic acid signalling pathways can also be important for defence against parasitic plants [reviewed in Ref. 55]. When inoculated onto pea roots challenged with O. crenata, Rhizobium leguminosarum led to the induction of several defence-related enzymes and metabolites such as polyphenoloxidase, H2O2, lipoxygenase and the phytoalexin pisatin [56]. Similarly, Streptomyces enissocaesilis triggered polyphenoloxidase in sunflower, the host of O. cumana [57]. Although microbes can induce defence responses in multiple plant species that are hosts for RPWs, the underlying signal-transduction pathways and their conclusive role in suppression of RPWs have, to our knowledge, not yet been resolved.
Outstanding questions and concluding remarks
Despite the mounting examples of soil and root-associated microbes influencing the life cycle of RPWs, there is still a scarcity of information on the underlying mechanisms by which these microbes operate. Moreover, many other outstanding questions remain to be answered. For example, what is the frequency of RPW-pathogenic and antagonistic microorganisms in the plant root microbiome? And, what is the impact of RPW infection on the host microbiome composition and antagonistic activity? In this context, there are a few intriguing recent studies. For example, it was shown that Orobanche and Phelipanche infections led to a significant decrease of microbial cell densities in the rhizosphere of parasitized plants [58]. Furthermore, two studies found that upon infection of tomato plants by Phelipanche aegyptiaca [59••] and of Nitraria tangutorum by Cynomorium songaricum [60], the endophytic microbiome (bacteria [59••] and fungi [60]) became more similar between the parasite and host plants. Hence, one may speculate that RPWs can, to some extent, modulate the host microbiome systemically and likely at the infection sites for their own benefit. Because of the physical connection of RPWs with their host through their vascular systems, this also enables the exchange of antagonistic microbes (e.g. endophytes) and compounds from the host to the RPW.
To date, most studies on microbe-RPW interactions focus on single microbes. However, the use of single members of the plant microbiome has proven to be an inconsistent strategy, particularly in field settings. Hence, designing functional synthetic microbial communities (SynComs) [61•,62] may be the way forward to more consistently suppress RPWs. To this end, the design should involve microbes with complementary modes of action (Figure 1) that act together or synergistically, and preferably at different stages of the parasite’s life cycle. In line with that, Oyserman et al. [63] recently introduced the concept of microbiome-associated phenotypes (MAPs), where modular microbiomes are engineered in concert with the host genotype to increase the efficacy of the desired trait. This reinforces the need to understand how each ‘module’ (or trait) behaves across different conditions, that is, the ecological context of trait function. Moreover, a microbial-mediated strategy for RPWs control should also take into account other commonly used agricultural practices (such as the use of organic amendments [64]), for instance by promoting the selective enrichment of microbes/SynComs with RPW suppressive functions. Current agricultural management practices used to control RPWs (e.g. crop rotation, trap/catch cropping) do not take into account the untapped importance of the microbiome. Considering the largely unexplored potential of microbiomes indigenous to the geographic regions where RPWs cause major crop losses, these microbiome-based strategies hold promise for developing and integrating novel and sustainable strategies for RPW control.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Bill and Melinda Gates Foundation, Seattle, WA via grant OPP1082853: ‘RSM Systems Biology for sorghum’. This is publication number 6808 of the NIOO-KNAW.
Contributor Information
Raul Masteling, Email: r.masteling@nioo.knaw.nl.
Jos M Raaijmakers, Email: j.raaijmakers@nioo.knaw.nl.
References
- 1.Spallek T., Mutuku M., Shirasu K. The genus Striga : a witch profile. Mol Plant Pathol. 2013;14:861–869. doi: 10.1111/mpp.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vurro M., Boari A., Thiombiano B., Bouwmeester H. Strigolactones and parasitic plants. In: Koltai H., Prandi C., editors. Strigolactones - Biology and Applications. Springer International Publishing; 2019. pp. 89–120. [Google Scholar]
- 3.Akiyama K., Matsuzaki K.I., Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 4.Westwood J.H., Yoder J.I., Timko M.P., dePamphilis C.W. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Toh S., Holbrook-Smith D., Stogios P.J., Onopriyenko O., Lumba S., Tsuchiya Y., Savchenko A., McCourt P. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science. 2015;350:203–207. doi: 10.1126/science.aac9476. [DOI] [PubMed] [Google Scholar]
- 6.Lumba S., Subha A., McCourt P. Found in translation: applying lessons from model systems to strigolactone signaling in parasitic plants. Trends Biochem Sci. 2017;42:556–565. doi: 10.1016/j.tibs.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Chang M., Lynn D.G. The haustorium and the chemistry of host recognition in parasitic angiosperms. J Chem Ecol. 1986;12:561–579. doi: 10.1007/BF01020572. [DOI] [PubMed] [Google Scholar]
- 8.Cui S., Wada S., Tobimatsu Y., Takeda Y., Saucet S.B., Takano T., Umezawa T., Shirasu K., Yoshida S. Host lignin composition affects haustorium induction in the parasitic plants Phtheirospermum japonicum and Striga hermonthica. New Phytol. 2018;218:710–723. doi: 10.1111/nph.15033. [DOI] [PubMed] [Google Scholar]
- 9.Ejeta G. Breeding for Striga resistance in sorghum: exploitation of an intricate host–parasite biology. Crop Sci. 2007;47(Suppl. 3):S216–S227. [Google Scholar]
- 10.Joel D.M., Gressel J., Musselman L.J. Springer; Berlin Heidelberg: 2013. Parasitic Orobanchaceae. [Google Scholar]
- 11.Zarafi A.B., Elzein A., Abdulkadir D.I., Beed F., Akinola O.M. Host range studies of Fusarium oxysporum f. sp. strigae meant for the biological control of Striga hermonthica on maize and sorghum. Arch Phytopathol Plant Prot. 2015;48:1–9. [Google Scholar]
- 12.Vurro M., Boari A., Evidente A., Andolfi A., Zermane N. Natural metabolites for parasitic weed management. Pest Manag Sci. 2009;65:566–571. doi: 10.1002/ps.1742. [DOI] [PubMed] [Google Scholar]
- 13••.Nzioki H.S., Oyosi F., Morris C.E., Kaya E., Pilgeram A.L., Baker C.S., Sands D.C. Striga biocontrol on a toothpick: a readily deployable and inexpensive method for smallholder farmers. Front Plant Sci. 2016;7:1–8. doi: 10.3389/fpls.2016.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a consortia (Foxy T14) of amino acid overproducing Fusarium oxysporum f. sp. strigae resulted in a reduction of Striga hermonthica infection, and increased maize yields across seasons in smallholder farms.
- 14.Linke K., Scheibel C., Saxena M.C., Sauerborn J. Fungi occurring on Orobanche spp. and their preliminary evaluation for Orobanche control. Trop Pest Manag. 1992;38:127–130. [Google Scholar]
- 15.Thomas H., Heller A., Sauerborn J., Müller-Stöver D. Fusarium oxysporum f. sp. orthoceras, a potential mycoherbicide, parasitizes seeds of Orobanche cumana (Sunflower Broomrape): a cytological study. Ann Bot. 1999;83:453–458. [Google Scholar]
- 16.Neondo J.O., Alakonya A.E., Kasili R.W. Screening for potential Striga hermonthica fungal and bacterial biocontrol agents from suppressive soils in Western Kenya. BioControl. 2017;62:705–717. [Google Scholar]
- 17.Harding D.P., Raizada M.N. Controlling weeds with fungi, bacteria and viruses: a review. Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Uraguchi D., Kuwata K., Hijikata Y., Yamaguchi R., Imaizumi H., Sathiyanarayanan A.M., Rakers C., Mori N., Akiyama K., Irle S. A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science. 2018;362:1301–1305. doi: 10.1126/science.aau5445. [DOI] [PubMed] [Google Scholar]; Authors describe a molecule able to specifically induce germination of Striga hermonthica in vitro and in situ with no adverse effects to either host or AMF.
- 19.Holbrook-Smith D., Toh S., Tsuchiya Y., McCourt P. Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat Chem Biol. 2016;12:724–729. doi: 10.1038/nchembio.2129. [DOI] [PubMed] [Google Scholar]
- 20.Xiang H., Yao R., Quan T., Wang F., Chen L., Du X., Zhang W., Deng H., Xie D., Luo T. Simple β-lactones are potent irreversible antagonists for strigolactone receptors. Cell Res. 2017;27:1525–1528. doi: 10.1038/cr.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamiaux C., Drummond R.S.M., Luo Z., Lee H.W., Sharma P., Janssen B.J., Perry N.B., Denny W.A., Snowden K.C. Inhibition of strigolactone receptors by N-phenylanthranilic acid derivatives: Structural and functional insights. J Biol Chem. 2018;293:6530–6543. doi: 10.1074/jbc.RA117.001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson S.L., Christenson J.K., Wackett L.P. Biosynthesis and chemical diversity of β-lactone natural products. Nat Prod Rep. 2019;36:458–475. doi: 10.1039/c8np00052b. [DOI] [PubMed] [Google Scholar]
- 23.Miché L., Bouillant M.L., Rohr R., Sallé G., Bally R. Physiological and cytological studies on the inhibition of Striga seed germination by the plant growth-promoting bacterium Azospirillum brasilense. Eur J Plant Pathol. 2000;106:347–351. [Google Scholar]
- 24.Dadon T., Nun N.B., Mayer A.M. A factor from Azospirillum brasilense inhibits germination and radicle growth of Orobanche aegyptiaca. Isr J Plant Sci. 2004;52:83–86. [Google Scholar]
- 25.Tyc O., Song C., Dickschat J.S., Vos M., Garbeva P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017;25:280–292. doi: 10.1016/j.tim.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Berner D.K., Schaad N.W., Völksch B. Use of ethylene-producing bacteria for stimulation of Striga spp. seed germination. Biol Control. 1999;15:274–282. [Google Scholar]
- 27.Tasker A.V., Westwood J.H. The U.S. Witchweed eradication effort turns 50: a retrospective and look-ahead on parasitic weed management. Weed Sci. 2012;60:267–268. [Google Scholar]
- 28.Hassan M.M., Abdelgani M.E., Babiker A.E., Osm M.G. Effect of Klebsiella spp. and different ethylene inhibitors on Striga hermonthica Benth. (Del.) seeds germination. Asian J Agric Sci. 2010;2(3):94–98. [Google Scholar]
- 29.Carrión V.J., Cordovez V., Tyc O., Etalo D.W., de Bruijn I., de Jager V.C.L., Medema M.H., Eberl L., Raaijmakers J.M. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 2018;12:2307–2321. doi: 10.1038/s41396-018-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwanenburg B., Mwakaboko A.S., Kannan C. Suicidal germination for parasitic weed control. Pest Manag Sci. 2016;72:2016–2025. doi: 10.1002/ps.4222. [DOI] [PubMed] [Google Scholar]
- 31.Ali H.A., Elamin H.B., Dirar H.A. Biological control of Striga hermonthica Del. Bendth : screening for bacteria scavenging Strigol. Am J Biochem. 2013;3:89–92. [Google Scholar]
- 32.Boari A., Ciasca B., Pineda-Martos R., Lattanzio V.M., Yoneyama K., Vurro M. Parasitic weed management by using strigolactone-degrading fungi. Pest Manag Sci. 2016;72:2043–2047. doi: 10.1002/ps.4226. [DOI] [PubMed] [Google Scholar]
- 33.Aires A., Mota V.R., Saavedra M.J., Monteiro A.A., Simões M., Rosa E.A.S., Bennett R.N. Initial in vitro evaluations of the antibacterial activities of glucosinolate enzymatic hydrolysis products against plant pathogenic bacteria. J Appl Microbiol. 2009;106:2096–2105. doi: 10.1111/j.1365-2672.2009.04181.x. [DOI] [PubMed] [Google Scholar]
- 34.Yoneyama K.K., Xie X., Kim H., II, Kisugi T., Nomura T., Sekimoto H., Yokota T., Yoneyama K.K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta. 2012;235:1197–1207. doi: 10.1007/s00425-011-1568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Ráez J.A., Charnikhova T., Gómez-Roldán V., Matusova R., Kohlen W., De Vos R., Verstappen F., Puech-Pages V., Bécard G., Mulder P. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 2008;178:863–874. doi: 10.1111/j.1469-8137.2008.02406.x. [DOI] [PubMed] [Google Scholar]
- 36.López-Ráez J.A., Charnikhova T., Fernández I., Bouwmeester H., Pozo M.J. Arbuscular mycorrhizal symbiosis decreases strigolactone production in tomato. J Plant Physiol. 2011;168:294–297. doi: 10.1016/j.jplph.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 37.De Boer W., Folman L.B., Summerbell R.C., Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Jamil M., Kanampiu F.K., Karaya H., Charnikhova T., Bouwmeester H.J. Striga hermonthica parasitism in maize in response to N and P fertilisers. Field Crop Res. 2012;134:1–10. [Google Scholar]
- 39•.Yoneyama K., Arakawa R., Ishimoto K., Kim H., II, Kisugi T., Xie X., Nomura T., Kanampiu F., Yokota T., Ezawa T. Difference in Striga-susceptibility is reflected in strigolactone secretion profile, but not in compatibility and host preference in arbuscular mycorrhizal symbiosis in two maize cultivars. New Phytol. 2015;206:983–989. doi: 10.1111/nph.13375. [DOI] [PubMed] [Google Scholar]; Two maize cultivars, one resistant and one susceptible to Striga hermonthica had different strigolactone exudation profiles, significantly affecting Striga germination but only marginally AMF colonization.
- 40.Cardoso C., Ruyter-Spira C., Bouwmeester H.J. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 2011;180:414–420. doi: 10.1016/j.plantsci.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Alori E.T., Glick B.R., Babalola O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X., Chaparro J.M., Reardon K.F., Zhang R., Shen Q., Vivanco J.M. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany. 2014;92:267–275. [Google Scholar]
- 43.Etalo D.W., Jeon J.-S., Raaijmakers J.M. Modulation of plant chemistry by beneficial root microbiota. Nat Prod Rep. 2018;35:398–409. doi: 10.1039/c7np00057j. [DOI] [PubMed] [Google Scholar]
- 44.Cimmino A., Fernández-Aparicio M., Andolfi A., Basso S., Rubiales D., Evidente A. Effect of fungal and plant metabolites on broomrapes (Orobanche and Phelipanche spp.) seed germination and radicle growth. J Agric Food Chem. 2014;62:10485–10492. doi: 10.1021/jf504609w. [DOI] [PubMed] [Google Scholar]
- 45.Cimmino A., Fernández-Aparicio M., Avolio F., Yoneyama K., Rubiales D., Evidente A. Ryecyanatines A and B and ryecarbonitrilines A and B, substituted cyanatophenol, cyanatobenzo[1,3]dioxole, and benzo[1,3]dioxolecarbonitriles from rye (Secale cereale L.) root exudates: Novel metabolites with allelopathic activity on Orobanche seed germination. Phytochemistry. 2015;109:57–65. doi: 10.1016/j.phytochem.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 46.Fernández-Aparicio M., Cimmino A., Evidente A., Rubiales D. Inhibition of Orobanche crenata seed germination and radicle growth by allelochemicals identified in cereals. J Agric Food Chem. 2013;61:9797–9803. doi: 10.1021/jf403738p. [DOI] [PubMed] [Google Scholar]
- 47.Rudrappa T., Czymmek K.J., Pare P.W., Bais H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim B., Song G.C., Ryu C.-M. Root exudation by aphid leaf infestation recruits root-associated Paenibacillus spp. to lead plant insect susceptibility. J Microbiol Biotechnol. 2016;26:549–557. doi: 10.4014/jmb.1511.11058. [DOI] [PubMed] [Google Scholar]
- 49.Yi H.-S., Yang J.W., Ghim S.-Y., Ryu C.-M. A cry for help from leaf to root. Plant Signal Behav. 2011;6:1192–1194. doi: 10.4161/psb.6.8.15780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng X., Etalo D.W., van de Mortel J.E., Dekkers E., Nguyen L., Medema M.H., Raaijmakers J.M. Genome-wide analysis of bacterial determinants of plant growth promotion and induced systemic resistance by Pseudomonas fluorescens. Environ Microbiol. 2017;19:4638–4656. doi: 10.1111/1462-2920.13927. [DOI] [PubMed] [Google Scholar]
- 51.Guang Sun X., Bonfante P., Tang M. Effect of volatiles versus exudates released by germinating spores of Gigaspora margarita on lateral root formation. Plant Physiol Biochem. 2015;97:1–10. doi: 10.1016/j.plaphy.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida S., Shirasu K. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol. 2009;183:180–189. doi: 10.1111/j.1469-8137.2009.02840.x. [DOI] [PubMed] [Google Scholar]
- 53.Musselman L.J. The biology of Striga, Orobanche, and other root-parasitic weeds. Annu Rev Phytopathol. 1980;18:463–489. [Google Scholar]
- 54.Pieterse C.M.J., Zamioudis C., Berendsen R.L., Weller D.M., Van Wees S.C.M., Bakker P.A.H.M. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 55.Yoder J.I., Scholes J.D. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr Opin Plant Biol. 2010;13:478–484. doi: 10.1016/j.pbi.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Mabrouk Y., Simier P., Delavault P., Delgrange S., Sifi B., Zourgui L., Belhadj O. Molecular and biochemical mechanisms of defence induced in pea by Rhizobium leguminosarum against Orobanche crenata. Weed Res. 2007;47:452–460. [Google Scholar]
- 57.Chen J., Xue Q.H., McErlean C.S.P., Zhi J.H., Ma Y.Q., Jia X.T., Zhang M., Ye X.X. Biocontrol potential of the antagonistic microorganism Streptomyces enissocaesilis against Orobanche cumana. BioControl. 2016;61:781–791. [Google Scholar]
- 58.Hristeva T., Dekalska T., Denev I. Structural and functional biodiversity of microbial communities in the rhizosphere of plants infected with broomrapes (Orobanchaceae) Biotechnol Biotechnol Equip. 2013;27:4082–4086. [Google Scholar]
- 59••.Iasur Kruh L., Lahav T., Abu-Nassar J., Achdari G., Salami R., Freilich S., Aly R. Host-parasite-bacteria triangle: the microbiome of the parasitic weed Phelipanche aegyptiaca and tomato - Solanum lycopersicum (Mill.) as a host. Front Plant Sci. 2017;8:1–9. doi: 10.3389/fpls.2017.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]; The microbiomes of the host plants and parasite become more similar after infection and hold functional potential to suppress the parasitic weed Phelipanche aegyptiaca.
- 60.Cui J.-L., Vijayakumar V., Zhang G. Partitioning of fungal endophyte assemblages in root-parasitic plant Cynomorium songaricum and its host Nitraria tangutorum. Front Microbiol. 2018;9:1–11. doi: 10.3389/fmicb.2018.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Vorholt J.A., Vogel C., Carlström C.I., Müller D.B. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22:142–155. doi: 10.1016/j.chom.2017.07.004. [DOI] [PubMed] [Google Scholar]; Comprehensive review on the factors to consider for the design of synthetic microbial communities for predictable plant phenotypes.
- 62.Vannier N., Agler M., Hacquard S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oyserman B.O., Medema M.H., Raaijmakers J.M. Road MAPs to engineer host microbiomes. Curr Opin Microbiol. 2018;43:46–54. doi: 10.1016/j.mib.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 64.Bonanomi G., Lorito M., Vinale F., Woo S.L. Organic amendments, beneficial microbes, and soil microbiota: toward a unified framework for disease suppression. Annu Rev Phytopathol. 2018;56:1–20. doi: 10.1146/annurev-phyto-080615-100046. [DOI] [PubMed] [Google Scholar]