Abstract
ATP-binding cassette (ABC) transporters use the energy of ATP hydrolysis to transport a large diversity of molecules actively across biological membranes. A combination of biochemical, biophysical, and structural studies has established the maltose transporter MalFGK2 as one of the best characterized proteins of the ABC family. MalF and MalG are the transmembrane domains, and two MalKs form a homodimer of nucleotide-binding domains. A periplasmic maltose-binding protein (MalE) delivers maltose and other maltodextrins to the transporter, and triggers its ATPase activity. Substrate import occurs in a unidirectional manner by ATP-driven conformational changes in MalK2 that allow alternating access of the substrate-binding site in MalF to each side of the membrane. In this review, we present an integrated molecular mechanism of the transport process considering all currently available information. Furthermore, we summarize remaining inconsistencies and outline possible future routes to decipher the full mechanistic details of transport by MalEFGK2 complex and that of related importer systems.
Keywords: ABC transporter, Substrate-binding protein, Importer, EPR spectroscopy, Single molecule fluorescence, smFRET
The ATP-binding cassette (ABC) proteins constitute one the largest protein superfamilies present in all kingdoms of life. These proteins utilize the energy from ATP hydrolysis to perform diverse functions such as nutrient uptake, antibiotic and drug resistance, antigen presentation, cell-volume regulation and many others [1]. Most ABC proteins are membrane transporters, functioning as either exporters or importers, while others are soluble proteins that are involved in various cellular processes [2] such as ribosome quality control and maintenance [3].
ABC transporters minimally contain two transmembrane domains (TMDs) that form a substrate translocation pathway, and two cytoplasmic nucleotide-binding domains (NBDs) that energize the system via ATP hydrolysis [1]. These four core domains can exist in multiple arrangements either as a single polypeptide or as separate subunits [4], [5]. ABC transporters are usually specific for a particular substrate class, resulting in little sequence conservation among the TMDs, whereas much greater sequence conservation exists among the NBDs that carry out the ATPase function [5]. Importers are largely found in bacteria where they mediate the uptake of a variety of molecules, ions, amino acids, peptides, sugars, vitamins, co-factors and siderophores [1]. Based on functional and structural distinctions, importers can be divided into three major classes [6].
Most importers have an extracellular substrate-binding protein or domain (both abbreviated SBPs here) [7], [8]. In these binding protein-dependent transport systems, the SBPs not only deliver substrates to the transporter, they also control and trigger conformational changes required for transport [1]. In Gram-negative bacteria, SBPs are usually soluble proteins located in the periplasmic space, whereas in Gram-positive bacteria, they are lipoproteins tethered to the membrane or they are fused to the TMDs [8]. The binding protein-dependent transporters can be divided into two classes termed type I and type II importers [6], [9]. The TMDs of type I importers, exemplified by the structures of the molybdate/tungstate, maltose and methionine transporters, contain a minimal core of five TM helices and dimerize to form the translocation pathway [5]. Type II importers, first recognized as a distinct sub-class with the structure determination of the Escherichia coli vitamin B12 transporter BtuCD [10], typically contain larger TMDs with a distinct architecture and coupling mechanism, and in general transport larger substrates (metal chelates, heme or vitamins) [4].
More recently, a third class of ABC importers, the type III importers or energy-coupling factor (ECF) transporters, was identified which do not make use of soluble extracellular SBPs [11]. ECF-type transporters have a small membrane-integrated subunit (S-component) that acts as a high-affinity binder for the transported ligand. The precise transport mechanism of ECF-type transporters has yet to be clarified [11].
This review will discuss properties and the transport mechanism of a prototype type I ABC importer: the maltose transporter from E. coli.
1. The maltose ABC transporter: a brief history
Over several decades a wealth of knowledge has been accumulated on the E. coli maltose/maltodextrin transport system MalEFGK2. Much of this derives from genetic and biochemical studies that were conducted long before any structural and biophysical information of ABC proteins was available (see for reviews [12], [13], [14]). The transporter initially attracted attention as a model for positively regulated regulons and because of the enigmatic relationship between maltose metabolism and susceptibility to λ phage infection. The λ phage receptor was later identified as an outer membrane porin (LamB, also called maltoporin) responsible for the uptake of maltose and maltodextrins into the periplasm [15], [16], [17]. Extensive studies of bacterial transport systems mediating the uptake of nutrients were performed in the 1970s (see for review [18]). In the next decade, the gene sequences of the components of the maltose import system were determined [19], [20], [21], [22].
The periplasmic SBP, MalE [23], was shown to be essential for maltose uptake in vivo, since a MalE knock-out strain was unable to grow on maltose at external concentration of 25 mM [24]. MalE was the first component of the maltose import system to be extensively characterized and crystallized [25], [26] (for details see review [1]). In addition to maltose, MalE binds several maltodextrins with affinities in the micromolar range. After induction by maltose, MalE copy numbers in the periplasm suggested concentrations as high as 1 mM [27], [28].
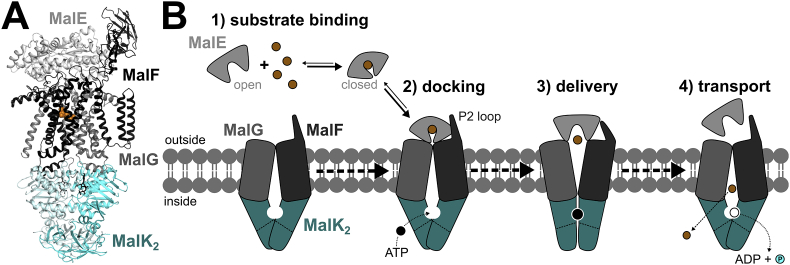
Studies on the histidine and maltose transporters independently demonstrated that ATP hydrolysis was the energy source for uphill transport by ABC transport systems [29], [30]. Overexpression, purification, and reconstitution of the maltose transporter in proteoliposomes showed that ATPase activity and maltose transport were strongly dependent on MalE and substrate [31], [32]. The subunit stoichiometry of the transporter was determined after immunoprecipitation of the complex from detergent-solubilized E. coli membranes [32]. In this multi-subunit complex, MalF and MalG, the two TMDs, were shown to be present in single copies, and MalK, the NBD, was present as a homodimer (Fig. 1A) [33].
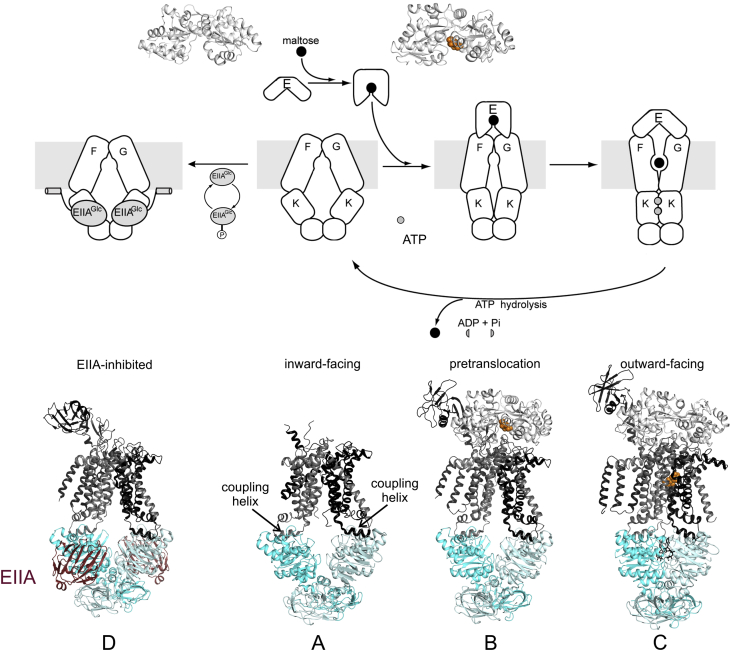
Fig. 1.
Maltose transport by MalEFGK2. A, Crystal structure of MalEFGK2 in complex with ATP and maltose. MalE is docked onto MalFG in the outward ATP-occluded state, and a maltose molecule is colored in orange spheres (PDB: 2R6G); B, Schematic of the import mechanism of maltose. The four steps depicted are discussed in detail in this review. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Based on the wealth of biochemical, biophysical and structural data that we will detail below, different and even partially opposing models for transport have been proposed. To develop a conclusive mechanism of maltose/maltodextrin uptake by MalEFGK2, we dissect the uphill transport process involving ATP binding, hydrolysis and alternating access in four essential sequential steps: 1) substrate recognition by binding to MalE, 2) docking of MalE to the TMDs, 3) substrate delivery to the TMDs, and 4) substrate import (see Fig. 1B).
2. Step 1: Substrate selection and binding by MalE
2.1. LamB protein facilitates maltose and maltodextrins diffusion through the E. coli outer membrane
Maltose and maltodextrins enter the periplasm via the outer membrane protein LamB [17]. LamB is a trimeric protein, and each monomer is a β-sheet made of 18 strands folded in a toroidal motif (β-barrel). Four long uncoiled loops (L1, L3 and L6 together with L2 from an adjacent LamB monomer) inwardly form a diameter constriction delineating a channel mainly occupied by aromatic and polar residues [16]. The latter constitute “polar tracks” that can dynamically establish hydrogen bonds with the sugars, thereby promoting their correct orientation with respect to the axis of the barrel [34]. The pattern made of aromatic amino acids are arranged to form a “greasy side” in the channel and facilitate their sliding along [16]. Maltose and maltodextrins up to seven glucose moieties are specifically transported by LamB and not by generic porins [35]. Its role is crucial in facilitating the diffusion of maltodextrins and especially the ones with higher exclusion limit, ensuring that the transport rate is not limited by their passive diffusion through the outer membrane in maltose/maltodextrin fully induced strain [12], [36].
2.2. Sugar binding to MalE
In E. coli, MalE is the primary receptor for maltose/maltodextrins and freely diffuses in the periplasm in 20–40 times excess of the MalFGK2 transporter [28]. MalE also functions as the chemoreceptor for maltose during chemotaxis by interacting with the methyl-accepting chemotaxis protein, Tar [12], [37].
Several studies have examined the specificity of sugar binding to MalE. Schwartz et al. [36] reported an average apparent dissociation constant (Kd) of 3.5 μM for maltose at 4 °C and 2.2 μM at room temperature using equilibrium dialysis, values that are similar to those obtained with spectrofluorometry [38], [39]. A summary of Kd values for MalE substrates as determined by fluorescence quenching by Ferenci et al., as well as using equilibrium and rapid-kinetic techniques by Quiocho et al., are shown in Table 1. For other related sugars such as glucose and lactose there is not yet convincing evidence that they can bind to MalE [40].
Table 1.
Comparison of dissociation constants (Kd values) of different MalE substrates. Determination was done via fluorescence quenching [38] or via equilibrium and rapid-kinetics (stopped flow spectrometer for fluorescence measurements) [39].
| Substrate | MalE protein Kd [μM] |
MalE protein Kd [μM] |
|---|---|---|
| Ferenci et al. 1986 | Quiocho et al. 1997 | |
| Maltose | 1.0 | 3.5 |
| Maltotriose | – | 0.16 |
| Maltotetraose | 1.6 | 2.3 |
| Maltopentaose | – | 5.0 |
| Maltohexaose | 2.8 | 3.4 |
| Maltoheptaose | – | 1.6 |
| a-Cyclodextrin (cyclic maltohexaose) | – | 4.0 |
| b-Cyclodextrin (cyclic maltoheptaose) | – | 1.8 |
2.3. Structure of MalE and conformational changes
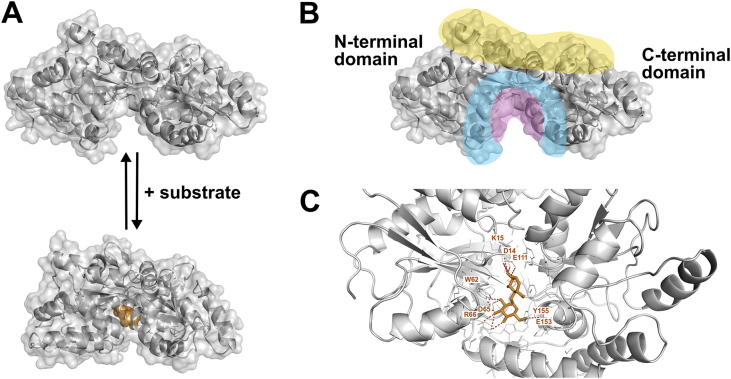
MalE consists of two globular lobes (the N-terminal and the C-terminal domains) that form a substrate-binding pocket at their interface (Fig. 2). Structural and biochemical studies of MalE allowed to distinguish different regions within the protein, which are relevant for substrate binding and resulting conformational changes (Fig. 2A). In addition to a high-affinity substrate-binding pocket (Fig. 2B, purple), these regions include the “lip region”, broadly defined as the area outside and around the binding pocket (Fig. 2B, blue), and the “balancing interface”, an area on the opposite side of the substrate-binding pocket including the hinge (Fig. 2B, yellow).
Fig. 2.
MalE structure and conformational states.A, Crystal structures of MalE, without substrate bound (PDB: 1OMP) and with maltose bound (PDB: 1ANF). The “closure” around the ligand within the binding pocket led to the term “venus flytrap mechanism”. B, Regions often mentioned in the literature are indicated in different colors: hinge region or balancing interface (yellow), binding pocket (red) and lip region (blue). As there is no consensus about the concrete residues assembling each region, the cartoon was kept simple and is just an indication including residues named in different publications. C, MalE binding pocket with maltose (shown in orange) bound. Red dashed lines indicate hydrogen bonds to subsite B (D14, K15, E111) or both, subsite B and C (E153, Y155). Subsite A and D are not involved and do not have relevance for maltose binding in E. coli MalE (PDB: 1ANF). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Early structural and biophysical studies suggested that substrate binding to MalE is coupled to conformational changes [36]. Substrate binding induces bending of the central beta-sheet hinge, thereby trapping the substrate at the interface of the two lobes (Fig. 2A). The resemblance of the process to the Venus flytrap plant capturing its prey has led it to be known as the Venus flytrap mechanism. The interaction between maltose/maltodextrins and MalE occurs through residues in one loop and three helical segments located in the binding site (Fig. 2C). A comparison of MalE and other maltose-binding proteins that bind monosaccharides, disaccharides, or trisaccharides revealed the presence of four subsites that bind individual glucose ring units. Subsite A forms hydrogen bonds with the first glucose ring in maltose and maltodextrins. It is occluded in MalE by residues D14 and K15, which contribute, with E111, to subsite B. E153 and Y155 form contacts with the second glucose ring and are found in both subsite B and C. Subsite D can interact with larger substrates, e.g. maltotriose, but is not used for maltose [41]. Additional hydrogen bonds between the ligand and residues outside of the different subsites are shown in the crystal structure (Fig. 2C).
Due to the variety of ligands bound by MalE and the difficulty in assessing all the substrate-bound conformational states by crystallography, the relationship between MalE conformational state and the competency of that state to initiate transport via MalFGK2 has been difficult to determine. Early studies using small-angle X-ray scattering (SAXS) confirmed the existence in solution of the closed and open conformations seen in crystal structures, where the former ligand-bound conformation was stabilized by sugar–protein interactions and the latter by interdomain contacts [42]. These findings were refined using wide-angle X-ray scattering (WAXS), with the focus on the hinge region, showing a 35° bending of the hinge in parallel to a 8° rotation of the two domains between open and close state. The latter is slightly larger than expected from crystallography experiments [43]. In parallel, more ligands and their effects on the degree of closure were tested by intrinsic fluorescent spectroscopy and electron paramagnetic resonance (EPR), suggesting that sugars that bind to MalE without causing the closure of its two lobes are not transported into the cytoplasm [44], [45]. These findings and the improved knowledge regarding MalE conformations soon made it a model protein to test experimental methods for investigating conformational dynamics. Especially for Nuclear Magnetic Resonance (NMR) Spectroscopy, MalE was a useful tool to refine the method’s settings, e.g. for fast backbone dynamics in ps to ns time scales, which are similar for the apo and holo states. But, as described above, it again also revealed significant differences in domain orientation (about 10° more closed) between solution and crystalline states of the β-cyclodextrin bound MalE [46]. In addition to the 1H, 15N, 13Cα, and 13Cβ chemical shifts, 129Xe and stereo-array isotope labeling (SAIL) were exploited as useful probes to detect conformational changes of MalE and similar proteins [47], [48]. In the NMR studies, a small population of partially closed apo MalE (approx. 5%) was identified. This intermediate conformation was found to be dynamic in ns to μs time scale rendering this conformation impossible to be crystallized [49]. Molecular Dynamics (MD) studies indicated a closure rate of 30–50 ns. In presence of a ligand the occurrence of the closed conformation of MalE is clearly preferred, whereas, in the absence of ligand, the open conformation is largely predominant [50]. Accelerated MD studies later confirmed the data of a preexisting dynamical exchange, yet showing that the stability of a semi-open conformation is lower than of the open state. This is revealing the importance of hydrophobic residues in the hinge region to stabilize this state without a ligand, while the fully closed state can never be reached without a suitable ligand [51].
2.4. Role of MalE in transport selectivity
MalE’s ability to bind sugars is not the sole determinant of whether the sugar is transported. Linear maltooligosaccarides and maltodextrins are transported if linked through two to seven α-1, 4 glycosidic bonds. Longer maltooligosaccharides or maltodextrin analogs with modified glucosyl residues at the reducing end can bind to MalE, but are not transported by MalFGK2.
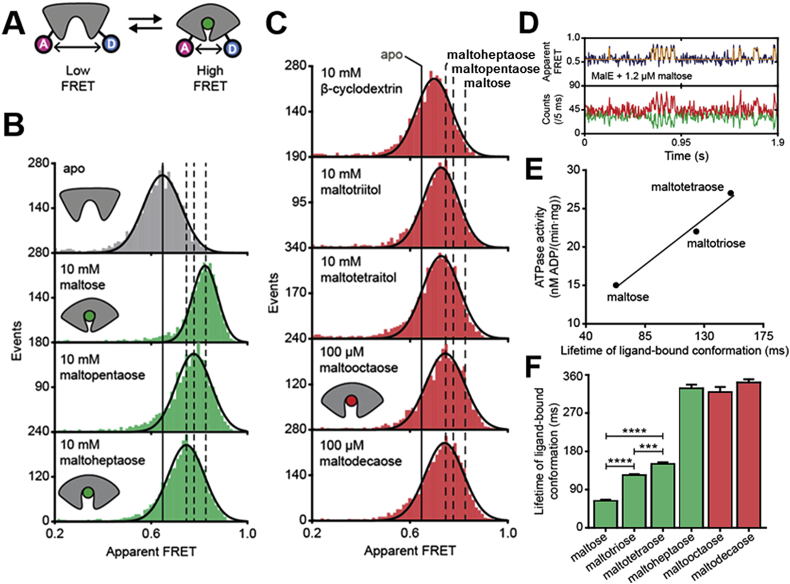
Recently, single-molecule Förster Resonance Energy Transfer (smFRET, Fig. 3A) has been used to characterize the different conformational states of SBPs (including MalE) and their equilibrium [52], [53], [54]. Comparison of absolute distance changes from FRET and crystallography data suggested that smFRET allows the quantification of the degree of closure of SBPs upon substrate binding. De Boer et al. [52] showed that both transported (Fig. 3B) and non-transported substrates (Fig. 3C) induce conformational changes in MalE, but there was no clear relationship between the degree of closure and MalFGK2 ATPase activity.
Fig. 3.
smFRET to study the closure of MalE in the presence of various sugars. A, smFRET assay for characterization of MalE closure by using fluorophore proximity for B, Transported (green) and C, non-transported (red) ligands. D, Real-time monitoring of ligand-driven conformational changes in isolated MalE by smFRET with example of maltose. E, Positive correlation between ligand release times derived from data similar to that of D from isolated MalE in comparison to ATPase activity of MalEFGK2. F, Ligand-release times of various transported (maltose, maltotriose, maltotetraose, maltoheptaose) and non-transported ligands (maltooctaose, maltodecaose). All panels are reprinted from de Boer et al. [52]. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In contrast to other SBPs [52], intrinsic conformational dynamics for wild type MalE were difficult to observe in the absence of ligand, and only the open conformation was visible [52], [54]. However, the use of mutations in the hinge region could exacerbate the ability of MalE to sample both the open and closed conformations in the absence of sugars. For instance, a MalE double mutant (A96W/I329W) showed a 90% population of closed conformation in the absence of sugars [54]. De Boer et al., showed that the degree of closing observed in the absence of ligand was indeed identical to that of the ligand-bound conformation for both the mutant and wild type [52]. Interestingly, the MalFGK2 ATPase activity seen with both wild type MalE and A96W/I329W MalE in the absence of maltose were similar, but only wild type MalE resulted in a large stimulation of activity upon addition of maltose [55]. This suggests that the closed form of unliganded A96W/I329W MalE can only stimulate MalFGK2 ATPase activity to a small extent. This can be interpreted such that the allosteric trigger for ATP hydrolysis is the presence of both MalE and a transported ligand. The fact that MalE showed a single (low-FRET, thus open) apo state and that the transition into the closed-liganded conformation was concentration dependent suggested that ligands are bound by an induced-fit mechanism [52], [54], unless fast conformational fluctuations occur on a sub-100 μs timescale.
Further details on the selectivity of the transport process will be discussed in sections 3 and 4: the interaction of MalE with MalFG (i.e., docking, Fig. 1, step 2) might be hindered for longer substrates [38], [56] and some sugars may not be accommodated in the TMDs.
3. Step 2: Interaction of MalE with MalFGK2
Once MalE is bound to a substrate molecule (Fig. 1, step 1), an interaction between MalE and the TMDs is required to initiate transport via alternating access of the ligand-binding site within the TMDs. How MalE docks to the TMDs and which factors govern efficient transport are key mechanistic questions (Fig. 1, step 2). Biochemical evidence, such as cysteine cross-linking experiments [57], showed that unliganded and liganded MalE can interact with the transporter (see also review [14]). Also, mathematical analysis of transport kinetics suggests models that are only consistent with interactions with both unliganded and liganded MalE [58], [59]. Increasing the concentration of MalE inhibited transport in vesicles when maltose concentration was kept constant at a sub-stoichiometric level, indicating that unliganded MalE competes with liganded MalE for interaction with MalFGK2 [60]. In addition, these models predict that Km for transport approaches the Kd of MalE toward maltose when the amount of binding protein increases [58], [60], [61].
3.1. Structural elements for MalFGK2 interaction with MalE
In a binding protein-dependent molybdate transporter, a surface with charged residues facilitating SBP-TMD interactions has been identified and may serve as a docking interface [62]. Yet, MalFGK2 has additional domains to facilitate the recruitment of MalE. MalF has four periplasmic loops, of which the P2 loop (residues 91 to 276) has high relevance for MalE interactions. Isothermal titration calorimetry experiments demonstrated a direct interaction between the isolated P2 loop and both unliganded and liganded MalE (Kd of 22 μM and 7 μM, respectively) [63]. The P2 loop folds into an Ig-like domain that extends about 3 nm into the periplasmic space [33]. In addition, MalG has three periplasmic loops [64], of which P1 and P3 are important for MalE docking and substrate delivery in different manner.
Cross-linking experiments suggest that MalF P2 loop and MalG P1 loop interact with the N-lobe of MalE throughout the catalytic cycle [57], [65]. Proteolysis and fluorescent labeling experiments showed that the accessibility of these loops were higher in the presence of ATP, i.e. when MalK2 was in a closed dimerized state [66], suggesting local conformational changes in these loops during the catalytic cycle.
A recent comparison of MalEFGK2 with a homologous maltose transporter from Bdellovibrium bacteriovorus further supports the conclusions drawn on the role of the P2 loop in the MalE-transporter interaction. In the B. bacteriovorus transporter, the SBP homolog to MalE (388 amino acids with 26% sequence identity to E. coli MalE) is fused to the MalF protein, suggesting possible differences in its transport mechanism as compared to MalEFGK2. To clarify the implications of this arrangement, the isolated B. bacteriovorus-MalE domain was expressed and purified [67]. Although the fusion to MalF sets the ratio of MalE to transporter in B. bacteriovorus to precisely 1:1, these experiments showed that the ATPase activity of the transporter was further stimulated by adding an excess of soluble MalE from either B. bacteriovorus or E. coli. For the E. coli transporter, however, only increased concentrations of its cognate SBP increased ATPase activity. The failure of the B. bacteriovorus-MalE to stimulate the ATPase activity of the E. coli transporter was suggested to be due to an inability of the protein to interact with the P2 loop of MalF. The important contact residues in the E. coli transporter (E72, D82, E278 and N278) [67] are not conserved in the B. bacteriovorus protein.
After an initial interaction of the P2 loop with MalE N-lobe, several pieces of evidence support that MalE docks when its C-lobe engages with the transporter [68], [69], [70]. It is also possible that such docking promotes a tighter interaction of the N-lobe with MalFG.
The periplasmic P3 loop of MalG (residues 236 to 262), connecting TM5 and TM6 helices, inserts into the substrate-binding site of MalE during the transport cycle. This ensures the transfer of maltose from the MalE binding pocket into the binding site in the TMDs. For this reason, the P3 loop was also called “scoop loop” [33].
Taken together, these results suggest that productive docking and substrate release to the TMD requires not only the P2 loop of MalF and P3 loop of MalG as essential elements, but also structural rearrangements at the SBP-MalFG interface (notably involving MalE C-lobe, see also section 4 for details).
3.2. Ligand-bound MalE is required for MalFGK2 function
Biochemical studies established early on that MalFGK2 alone shows very low ATPase and transport activities, even in the presence of maltose [31], [71]. However, both were strongly stimulated in the presence of maltose and MalE in lipid-reconstituted systems. It is important to emphasize that liganded MalE stimulates the ATPase activity ∼7–10 times stronger than unliganded MalE [68], [71], [72]. The difference in stimulation between liganded and unliganded MalE might even have been underestimated, since a fraction of MalE tends to retain ligands even after several purification steps. A denaturation/renaturation step for MalE is thus required to obtain a fully ligand-free preparation of MalE [55]. Transport assays and kinetic models suggested that both unliganded and liganded forms of MalE interact with the transporter although the liganded form may bind with higher affinity [60]. Because the liganded form of MalE stimulates the ATPase activity of the transporter much more efficiently, ATPase and transport activities are likely initiated by liganded MalE docking onto the transporter or by binding of sugar to the unliganded MalE that is already bound to the P2 loop. In our view, both scenarios can happen, depending on the relative concentrations of MalE and maltose in the periplasm.
This interpretation, i.e. closed MalE initiates transport, is also supported by recent single-molecule studies that show a correlation between ligand-induced conformational changes, ATPase activity, and transport (Fig. 3D, E) [52]. As mentioned in section 2, both transported and non-transported ligands trigger the formation of closed MalE conformations, albeit with different degrees of closure (Fig. 3B, C). The differences in the degree of MalE closure could provide a selectivity filter for transport by conformational control, i.e., only some conformations induce ATPase activity and transport (Fig. 3B, green ligands vs. Fig. 3C, red ligands). Interestingly, the degree of closure is not the sole determinant of substrate selectivity because longer maltodextrins with >7 glucosyl units (e.g. maltooctaose) induced a maltose-like closing of MalE but are not transported. For such ligands, the critical step in selection might occur later during substrate delivery to the TMDs.
The existence of a maltose-binding site within the TMDs of the transporter was strongly suggested with the early isolation of binding-protein independent MalFGK2-mutants containing a pair of mutations that mapped to either malF or malG genes [73]. The strains carrying these mutants transported maltose with a much higher Km (∼2 mM) than wild type (∼1 μM), suggesting that the TMDs contained a low affinity maltose-binding site [74]. In addition, these mutant MalFGK2 complexes when reconstituted in liposomes displayed ATP hydrolysis activity in the absence of MalE [71]. Structural studies with MalEFGK2 complex later showed that MalF has a binding site for maltose and maltodextrins [33]. Additionally, MalG was shown to interact with the reducing end of the large malto-oligosaccharide maltoheptaose when it protrudes from MalE [56]. Thus, MalF and MalG provide a third level at which substrate selection takes place, in addition to the ones provided by LamB and MalE.
From bulk biochemical studies, it is known that a relatively high concentration of liganded MalE is required to achieve half-maximal transport activity (15–100 μM) [28], [55], [75]. This suggests that MalE and the transporter initially interact with low affinity [76]. The opening of MalE and the associated changes in the transporter generate a tight interaction that promotes catalysis by stabilizing the transition state for ATP hydrolysis [72]. Consistent with this tight interaction, the apparent Km for stimulation of MalFGK2 ATPase activity by unliganded (i.e. open) MalE was below 1 μM [55]. Opening of MalE in the transition state triggers the release of maltose from the binding protein directly into the transporter [76] (Fig. 1). Single-molecule studies suggest that substrate release times from SBPs can be a rate-limiting step for transport [52], [53], [77]. It should however be noted, that this conclusion was derived from experiments on isolated SBPs.
4. Step 3 & 4: Substrate delivery and import mechanism
After substrate binding and MalE docking, a pivotal mechanism ensures unidirectional maltose import by concerted conformational changes and ATP hydrolysis. In this section, we will review what is known about substrate delivery to the TMDs and transmembrane transport.
4.1. Structural characterization of the ATPase subunits
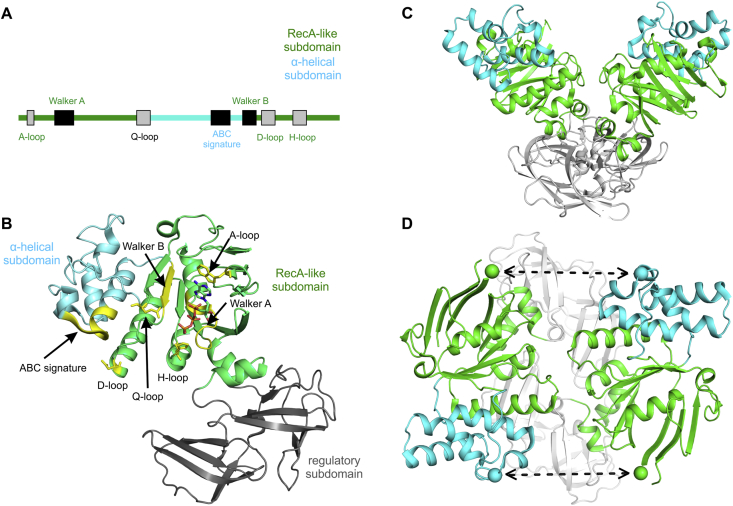
Crystal structures of the full transporter in different conformational states, as well as those of the isolated MalK2 domains, form the basis for understanding the alternating access mechanism employed by MalEFGK2 to transport maltose and other sugars. The MalK subunit consists of a NBD fold and a C-terminal regulatory domain, RD (Fig. 4) [78]. The NBD is comprised of a RecA-like subdomain that is commonly found in ATPases and a α-helical subdomain, which is unique to ABC proteins. The RecA-like subdomain contains several conserved sequence motifs required for ATP hydrolysis: the Walker A motif with the consensus sequence GX4GKT/S, where position X can be occupied by any amino acid; the Walker B motif, which has four hydrophobic residues followed by an aspartate; and several loop motifs named after their conserved residues (A, D, Q, H) (Fig. 4). The helical subdomain contains the LSGGQ ABC signature motif that identifies the ABC proteins [79], [80].
Fig. 4.
MalK structure with conserved motifs. A, Conserved motifs in the NBD of MalK. B, ATP-bound MalK structure (PDB: 1Q12) showing the subdomains and conserved motifs (in yellow) [78]. C, Open MalK dimer viewed from the side (PDB: 1Q1E). D, Open MalK dimer viewed from the top. The green and cyan spheres represent the V16 and R129 residues, respectively, that were used to monitor distance changes during closure of the dimer interface via EPR spectroscopy [99]. Closure of the MalK dimer is indicated by the dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Crystal structures of the isolated MalK dimers, in which the monomers adopt a head-to-tail arrangement, were obtained in distinct conformations and nucleotide-occupancy states [78]. The interactions between RD domains stabilize the MalK dimer even in the absence of the TMDs. In the absence of nucleotides, the MalK dimer was crystallized in two conformations: one with well-separated NBDs (termed open MalK state, Fig. 4C), and one with an intermediate degree of opening (termed semi-open MalK state). In the presence of ATP, MalK2 forms a closed dimer in which two ATP molecules are sandwiched (“occluded”) between the Walker A and LSGGQ motifs contributed by each monomer. In the presence of ADP, the dimer was found to be semi-open [81]. The overall structure of the MalK dimer led to it being compared to a pair of tweezers: the RDs being the point at which the NBD arms are joined (Fig. 4C). Sampling of the three conformational states requires a rotation of the entire NBD relative to the RD (Fig. 4D). This interdomain movement is accompanied by a second rigid-body rotation of the RecA-like and helical subdomains relative to each other. During closure, this second rotation results in the proper orientation of the LSGGQ motif at the closed dimer interface for catalytic activity. Closing of the MalK dimer in the presence of ATP and its reopening subsequent to ATP hydrolysis (or ADP/Pi release) suggested a mechanism by which conformational changes in the NBDs could potentially power substrate translocation in the TMDs by conformational coupling [78].
4.2. An alternating access mechanism for maltose transport
The import of maltose proceeds by a classical alternating access mechanism in which the TMDs switch between inward- and outward-facing conformations, alternately exposing the substrate-binding site to opposite sides of the membrane (Fig. 5). As detailed above, the conformational changes in the TMD are driven by ATP binding and hydrolysis events. Because a number of recent reviews have been dedicated to the detailed structural aspects of this transition [82], [83], we will here only summarize these structural observations.
Fig. 5.
Structures of the maltose transporter in various states of the transport cycle. Crystal structures of MalE are shown in its open (maltose free; PDB: 1OMP) and closed (maltose bound; PDB: 1ANF) conformations. The maltose transporter was crystallized in several conformational states: Inward-facing MalFGK2 in its resting state (PDB: 3FH6, see A), MalEFGK2 in a pretranslocation state (PDB: 3PV0 or 3PUZ, see B), MalEFGK2 outward-facing state (PDB: 2R6G, see C) and MalFGK2 in inward-facing state inhibited by its EIIAGlc regulatory protein (PDB: 4JBW, see D). A scheme of maltose delivery by MalE to MalFGK2 is displayed based on the crystal structures.
The inward-facing state of the intact transporter was crystallized in the absence of nucleotides and MalE, and was interpreted as being the resting state of the system [84]. The NBDs of the intact transporter were well separated, and similar to the open conformation of the MalK dimer (Fig. 4C). In this state, the TMDs form an inward-facing cavity that exposes a MalF transmembrane maltose-binding site to the cytoplasm (Fig. 5, see A). MalF and MalG form a gate that shields the inward-facing cavity from the periplasm. In addition, the maltose transporter has a unique extended periplasmic domain, the MalF P2 loop, connecting the third and fourth TM helices of MalF, as already discussed in section 3.1. In subsequent transport states, the P2 loop is involved in mediating interactions between the TMDs and MalE. Most of the flexible P2 loop was not resolved in the resting-state crystal structure, but the loop in its entirety was resolved in other structures, for instance in the inward-facing structure where the transporter was stabilized by the EIIAGlc protein involved in catabolic repression [85] (Fig. 5, see D). As mentioned in section 3.1, the affinity of the isolated P2 loop for MalE has been estimated to be 7 and 22 μM in the presence and absence of maltose, respectively [63]. Considering the high concentration of MalE in the periplasm under maltose induction (∼1 mM) [27], [28], the population of transporters is likely to be saturated with MalE under these conditions. Consistent with this assertion, EPR spectroscopy experiments suggested that MalE is bound to the P2 loop throughout the entire transport cycle [57], [63], [65], [86]. However, this does not mean that MalE is fully docked or engaged with MalFGK2 throughout the cycle.
The nature of NBD/TMD interface turned out to be remarkably similar to early predictions based on the biochemical experiments of Dassa and collaborators [21]. Dassa and Hofnung identified a conserved sequence (EAA---G---------I-LP, the EAA loop) in the TMDs of binding protein-dependent ABC importers [21]. This region was predicted to form two amphipathic α-helices connected by a loop containing an invariant glycine, and was hypothesized to be an interaction site between the NBDs and TMDs [87]. Paired mutations in the EAA loops of MalF and MalG resulted in a loss of in vivo maltose transport activity and disengagement of MalK from the TMDs [88]. The effect of these mutations was suppressed by mutations in the helical subdomain of MalK that restore assembly of MalK onto MalF and MalG mutant proteins. The helical subdomains of the NBDs of ABC transporters are the most variable region of these proteins [89], [90]. This is consistent with their role in coupling of NBDs to the TMDs, as the latter are poorly conserved among ABC transporters. Cross-linking studies have also suggested a direct interaction between the EAA loops and the helical subdomains in MalK [91]. Because cross-linking patterns between these two regions were modulated by ATP-Mg2+, Dassa and collaborators proposed that the helical subdomain was involved in the coupling between ATP binding/hydrolysis and transport [91], [92]. Years later, these conclusions were verified by crystal structures of the transporter that showed the coupling helices and the EAA motifs docked into hydrophobic clefts formed between the RecA-like and helical subdomains of each MalK subunit [33]. Indeed, the structure revealed that the EAA motifs were present within MalF and MalG coupling helices, which dock into hydrophobic clefts at the interface between the RecA-like and helical subdomains of each MalK subunit. A salt bridge between the conserved glutamate in the EAA motifs and a conserved arginine in MalK stabilized this coupling interface (Fig. 5).
The maltose transporter was also crystallized in an outward-facing state [33] (Fig. 5, see C). This was achieved by co-crystallizing a catalytic mutant of the transporter, MalFGK2 (E159Q), in the presence of ATP, maltose and MalE. The E159Q mutation was introduced to prevent ATP hydrolysis and to stabilize a tight ATP-bound NBD dimer as seen in other ABC transporters [93], [94]. The addition of maltose-bound MalE was predicted to stabilize the outward-facing conformation [72], [95]. The structure of the outward-facing conformation strongly supported the formerly proposed concerted model of transport [72]. Indeed, upon vanadate-induced trapping, MalE was found tightly associated to the MalFGK2 transporter and in a low affinity state for maltose [72]. Based on these observations, the tightly bound MalE was proposed to be in an open state, while MalK2 was simultaneously suggested to be in a closed conformation. Similar to the closed isolated MalK2 structure, the MalK dimer in the outward-facing structure displayed two ATP molecules at the interface between the Walker A and LSGGQ motifs. A large upper cavity formed by MalE, MalF, and MalG extends halfway across the predicted membrane bilayer. This cavity can readily accommodate maltoheptaose, the longest maltodextrin that can be transported. At the bottom of the cavity, maltose forms hydrogen bonds and van der Waals interactions with nine residues of MalF while its reducing-end participates in stacking interactions with two MalF residues [33], [83]. Six of these residues were identified in mutational studies where they severely impair maltose transport [96], [97]. Remarkably, no interaction between maltose and MalG was observed. The existence of a single transmembrane maltose-binding site suggested that only one maltose is imported per transport cycle. The large outward-facing cavity was shielded from the periplasmic side by MalE thus preventing release of the substrate back into the periplasm. The MalG P3 or “scoop loop” was found to be inserted into the substrate-binding site of MalE, and it was suggested that it ensures transfer of maltose from MalE to the maltose-binding site in MalF. This speculation was later confirmed by EPR spectroscopy and mutagenesis. Deletion of the P3 loop abolished transport and maltose was found retained in the binding pocket of MalE. The mutant, however, still displayed ∼50% of the ATPase activity compared with the wild type [98]. A similar phenotype of uncoupled ATPase activity was observed with mutation of a MalF residue that is essential for binding of maltose in the transmembrane maltose-binding site.
The structure of another distinct intermediate referred to as the pretranslocation state was solved in the presence of maltose-bound MalE and in the absence of nucleotides [70] (Fig. 5, see B). Liganded MalE was in a closed conformation, docked to MalFG, with the P2 loop making extensive contacts with the N-terminal lobe of MalE. The inward-facing cavity formed by the TMDs was narrower, and the MalK dimer was partially closed. In this semi-open MalK dimer, several catalytic residues necessary for ATP hydrolysis are brought in contact with the neighboring NBD. Indeed, the conserved D-loop of one MalK subunit hydrogen bonds with both the Walker motif S38 and the H-loop motif H192 from the other NBD. Given that both S38 and H192 also interact with the γ-phosphate of ATP in the closed MalK dimer primed for ATP hydrolysis, binding of two ATP molecules was predicted to disrupt the pretranslocation intermediate [70].
Based on these structures, and supported by other biochemical/biophysical techniques described below, the maltose transporter likely cycles between inward- and outward-facing conformations, alternately exposing the transmembrane substrate-binding site to the two sides of the membrane. The concerted closure of the MalK dimer and opening of MalE release maltose to the transmembrane cavity (Fig. 5, see C). ATP hydrolysis and ADP/Pi release are expected to open the MalK dimer, and in so doing convert the transporter to an inward-facing conformation, permitting release of maltose into the cytoplasm. It is important to stress that the transitions between the distinct conformational states discussed above have not yet been observed directly, however, the model that has thus far emerged remains the most plausible explanation of all the available structural data.
4.3. Conformational dynamics and substrate delivery
Because crystal structures of the maltose transporter provide molecular details of snapshots during the transport cycle, biochemical and biophysical studies are still required to understand the conformational dynamics of ATP hydrolysis and maltose translocation. These studies would provide information on intermediate states not captured by crystallography. For instance, there is no crystal structure of ATP-bound MalFGK2 and the effect of ATP binding to the transporter had to be investigated by other methods. In addition, knowledge of the extent and frequency of conformational motions are critical for understanding the transport mechanism and its energetics. As are the order in which the conformational states arise during the transport cycle and their connection to specific biochemical events such as ATP binding and hydrolysis. Finally, the dynamics of a specific membrane protein in detergent or lipids can be different, as it was observed with the maltose transporter [68].
Site-directed spin labeling and EPR spectroscopy have been employed to study conformational changes at strategic positions in MalEFGK2. Inter-residue distance measurements were used to track the opening and closing events of MalK2 during the catalytic cycle of the full transporter (Fig. 4D) [99]. In this context, MalK2 was observed in three different conformations: open, closed and semi-open. In the absence of ligand, MalK2 remained in an open conformation, as seen in the resting state (inward-facing) structure of MalFGK2 [84]. In contrast to what was observed using crystallography with the isolated MalK dimer [78], binding of ATP alone did not induce MalK closure in the intact transporter; however, the addition of both ATP and maltose-bound MalE stabilized a closed MalK dimer. These results strongly support the idea that maltose-bound MalE stimulates the ATPase activity of the transporter by promoting MalK closure [99]. This was later corroborated by the EPR work of Bordignon’s team [86] and more recently by molecular dynamic simulations [100]. Similarly, using spin-labeled MalE [45], other studies showed that ATP binding leads to the opening of MalE [68], [95]. These results are consistent with a concerted mechanism [72], in which closing of the MalK dimer is concomitant with MalE opening and vice-versa. Interestingly, the conformational states of MalK correlate with the distinct conformations of the P2 loop [86], in agreement with the notion of concerted motions of the intracellular and periplasmic components of the transporter [69].
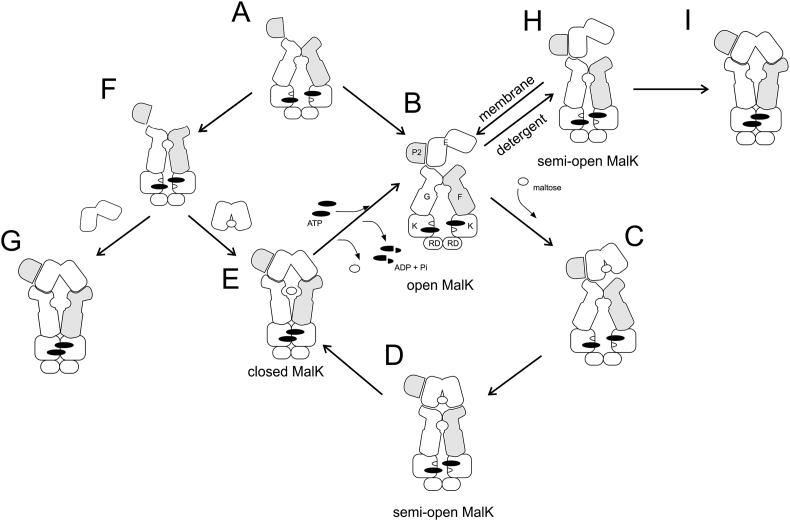
To further investigate the progression from the inward-facing state to the outward-facing state, Orelle et al. attempted to trap and analyze an intermediate between these two states [101]. To this end the authors employed a MalE variant (G69C/S337C) that can be locked in a closed conformation with a disulfide bond [102], and investigated the impact of this mutant on the conformation of MalK2. The cross-link prevents the two globular lobes of MalE from opening and maintains the protein in a conformation mimicking the maltose-bound wild type [70]. With wild type MalE that is able to open, the addition of ATP drives the transporter to the outward-facing state. The hope was that if MalE cannot open, an intermediate state would be revealed. The transporter in detergent or nanodiscs [103] was spin-labeled to measure either MalK2 closure or the relative movement between the helical and RecA-like subdomains [101]. The movement of these two subdomains was thus far only examined in isolated NBDs [78] and its relevance during the transport cycle was never examined by distance measurements. The addition of AMP-PNP-Mg2+ to the mutant complex maintained MalK2 in a semi-open conformation (Fig. 6, state D). In contrast, as expected, the addition of AMP-PNP-Mg2+ to the wild type complex triggered MalK2 closure. These observations indicate that closed MalE participates in the stabilization of the semi-open MalK2 [68], [101]. Furthermore, this study showed that complete closure of MalK2 (from semi-open to closed) was accompanied by a relative inward rotation of the helical subdomain towards the RecA-like subdomain (or reciprocally, as later proposed [104]). This conformational change is coupled to the reorientation of the TMDs and thus the opening of MalE, events that promote transfer of maltose to the transporter. These data are summarized in Fig. 6 (see path ABCDE). Importantly, it should be kept in mind that, in vivo, the amount of MalE and its saturation by maltose in the periplasm depend on complex regulation pathways based notably on catabolic repression (see section 5). If most of MalE population is liganded with maltose, the preferential path would be ACDE (Fig. 6).
Fig. 6.
Integrated mechanism of maltose transport based on structural and biochemical studies. Cartoons A, D, E are directly derived from crystal structures (A, PDB: 3FH6; D, 3PUZ; E, 3RLF or 2R6G). EPR studies from Davidson and Bordignon’s groups suggested that the main conformational route occurs through ABCDEB, or ACDEB if maltose is highly available [68], [69], [86], [99], [101]. Step C is probably transient but is depicted for clarity. In detergent, the ATPase activity of MalEFGK2 was insensitive to the presence of maltose, and EPR studies suggested that this was due to the increased ability of MalEFGK2 in detergent to transition through H in the absence of maltose [68]. Note that conformations I and G are also observed by EPR and cross-linking studies [69]. A different model was proposed by the group of Duong, in which ATP alone induces the outward-facing conformation [105]. However, this model was later refined to a route through F, in which MalK2 is asymmetric and semi-open in the presence of ATP [107]. The interaction of liganded MalE then stabilizes this conformation (similar as in D) to promote the conversion toward E, a route that is consistent with EPR data, especially if F is a small or transient population.
Duong and co-workers suggested a mechanistically distinct model. Here, ATP alone triggers formation of the outward-facing conformation onto which unliganded open MalE docks (Fig. 6, cartoon G). Maltose then binds to MalE to initiate transport [105]. The critical evidence supporting this interpretation comes from cross-linking and fluorescence studies. The inconsistencies of such a model with the EPR results presented above and past biochemical/structural work has been discussed elsewhere [68]. Moreover, it is probable that the aforementioned cysteine cross-linking experiments trapped transient states of the transporter that were not visible by EPR. Based on recent work in collaboration with Rouiller’s team, the Duong group has refined their model and it is now more compatible with the model suggested by the crystal structures and EPR data (Fig. 6, path AFE). They employed negative stain single-particle electron microscopy to probe the conformations of MalEFGK2 in nanodiscs [106] in the presence of ligands [107]. According to the authors, binding of ATP may drive closure of MalK2 but this conformation is unstable and reverts to an asymmetrical semi-closed state that was, to the best of our knowledge, not reported before. The additional presence of MalE stabilizes the closed conformation of MalK2.
4.4. Mechanism of ATPase stimulation by substrate-bound MalE
In the absence of MalE, the TMDs impose structural constraints that prevent MalK2 closure [84], and reciprocally, nucleotide-free MalK promotes an inward-facing conformation of the TMDs [108]. The low basal ATPase activity of the complex is thus a consequence of the low likelihood of reaching the closed state where ATP hydrolysis can occur [109]. When the transporter is in membranes, only the substrate-bound form of MalE efficiently stimulates ATPase activity [68], [71], [106]. However, this is not the case when the transporter is in detergent, where MalE-stimulated ATPase activity was independent of maltose [68].
These observations are consistent with recent smFRET studies that show that the ATPase activity of the transporter was dependent on the propensity of substrate-bound MalE to adopt one of several closed conformations [52]. Only closed MalE conformations (Fig. 3B, C) seem to induce transport and substantial ATPase activity. Interestingly, it was found that the ATPase activity of the transporter increased with slower ligand release times as determined for freely diffusing MalE (Fig. 3D, E) with maltose, maltotriose, and maltotetraose as substrates. According to previous studies, however, faster ligand release should result in higher ATPase activity and transport [53], [77]. One possible explanation for this discrepancy is that the longer maltodextrins make additional interactions with the TMDs, subtly altering the conformation of the complex and thus promoting ATP hydrolysis. Maltoheptaose and maltooctaose induce similar conformational changes in MalE and were released with similar rates (Fig. 3F), but maltooctaose is not transported, suggesting substrate selectivity is to some degree determined by the TMD [52].
Based on the findings described above, maltose-bound MalE stimulates the ATPase activity of MalFGK2 by two mechanisms. First, its interaction with the transporter facilitates closure of the ATP-bound MalK dimer [33], [86], [99], [110]. It does so by stabilizing the semi-open state of the MalK dimer when the transporter is in a lipid environment, and it should be noted that maltose is not required for this stabilization when the transporter is in detergent [68] (Fig. 6, states H and I). This stabilizing property is a key mechanistic element for the coupling of maltose transport to ATP hydrolysis. The crystal structure of the pretranslocation state gives structural insights into this transition. Docking of both lobes of maltose-bound MalE facilitates partial rotations of both the TMDs and the MalK dimer, bringing critical catalytic residues to the NBD dimer interface [70]. Importantly, it should be noted that crystals of the pretranslocation intermediate state of MalEFGK2 were obtained under two conditions: either with liganded MalE alone (PDB 3PV0) or with AMP-PNP-Mg2+ in conjunction with a liganded MalE locked in a closed conformation (PDB 3PUZ). However, the pretranslocation state was not observed by EPR under the conditions where only liganded MalE was present [69], [99]. The pretranslocation state with a semi-open MalK dimer was only seen in EPR when AMP-PNP-Mg2+ was bound and MalE was prevented from opening [101] (Fig. 6, state D). Consistent with the EPR observations, Shilton proposed, by analogy with enzymes, that conformational changes in the maltose transporter are likely promoted by the stabilization of higher-energy intermediate conformations [111]. It is speculated that the pretranslocation state of MalEFGK2 would have a relatively high energy as compared to the inward and outward-facing conformations, thereby presenting an energetic barrier that limits uncoupled ATP hydrolysis. According to this hypothesis, liganded MalE is unlikely to interact with the inward-facing resting state conformation (and in doing so promoting the formation of outward-facing conformation). Indeed, if maltose-bound MalE was to bind preferentially to the resting state of MalFGK2, it would decrease ATPase activity by stabilizing a low-energy conformation that is not able to hydrolyze ATP. Shilton therefore hypothesized that binding to a higher-energy occluded conformation, intermediate between open-in and open-out, offers an energetically reasonable pathway for liganded MalE to promote conformational changes in the system [111]. Thus, maltose-bound MalE and ATP might synergistically initiate the catalytic cycle by binding to, and lowering the energy of, the occluded conformation, facilitating the transition to the outward-facing conformation. When the NBDs of the pretranslocation state are closer together [70], [101], ATP can complete the progression to the outward-facing state in which MalE opens to deliver maltose. In this configuration, open MalE also participates in the stimulation of ATPase activity by stabilizing the transition state for ATP hydrolysis [69], [72], [95], [112]. In agreement with this idea, several studies suggested that the maltose transporter binding-protein independent (BPI) mutants may shift the equilibrium from the inward-facing state to the outward-facing state [66], [110], [113], [114], [115], [116], thereby explaining how these mutants hydrolyze ATP in the absence of MalE [71], [73], [117]. Accordingly, most of the primary BPI mutations were found at the interface between MalF and MalG and were predicted to destabilize the inward-facing conformation [84].
Therefore, the stimulation of ATPase activity found in productive maltose transport is governed by direct interactions of maltose-bound MalE with the transporter; however, maltose-free MalE is capable of stimulating low levels of ATPase activity suggesting a conformational plasticity of the transporter [69] and that open MalE might interact with a small population of transporters in the outward-facing conformation [55]. Maltose-bound MalE is much more efficient in stimulating the ATPase activity of MalFGK2 because the substrate directly affects the conformation of MalE and thus its capacity to interact productively with the transporter [70].
Pre-steady state studies of the maltose transporter suggested that basal ATPase activity of MalFGK2 is very low because the cleavage of ATP is rate limiting. The binding of unliganded MalE to the transporter increases ATP hydrolysis and Pi release becomes rate limiting. Liganded MalE, however, stimulates release of Pi [118].
In summary, substrate-bound MalE stabilizes the semi-open configuration of MalK2, whereas open MalE stabilizes the closed state of MalK2. Thus, both MalE conformations participate in the stimulation of MalFGK2 ATPase activity.
4.5. Mechanism of ATP hydrolysis
The mechanism of ATP hydrolysis by the maltose transporter was elucidated by Oldham and Chen [119]. During progression to the transition state, the geometry of the γ-phosphate changes from tetrahedral to trigonal bipyramidal and several compounds can be used to trap the various stages of the hydrolysis reaction. Beryllium fluoride (BeF3) forms a complex with ADP with tetrahedral geometry, thereby mimicking the ground state. Orthovanadate (Vi) and aluminum tetrafluoride (AlF4), however, form trigonal bipyramidal and octahedral complexes, respectively, in the transition state. Structures of the maltose transporter were obtained with AMP-PNP and all three compounds [119]. The glutamate adjacent to the Walker B motif, E159, was found either bonded to a properly positioned attacking water molecule or the equivalent Vi apical oxygen, consistent with a general base mechanism. In contrast, whereas both the histidine of the H-loop and the serine of the signature sequence interacted with the nucleotide, neither was bonded to the attacking water molecule, precluding a substrate-assisted or serine-assisted mechanism. The position of the signature motif (LSGGQ) was similar to the “arginine finger” found in other RecA-like ATPases. Interestingly, in contrast to what is generally observed with isolated NBDs, the Q-loop, which joins the helical and RecA-like subdomains (Fig. 4), was well stabilized and interacted with both Mg2+ and the γ-phosphate in the structures [119]. The presence of the TMDs thus ensured a proper orientation of the full active site. Therefore, the low ATPase activities measured for isolated NBDs may result from a poorly stabilized Q-loop motif and the improper positioning of the helical subdomain carrying the LSSGQ motif.
The overall structures of the maltose transporter using mimics of both the ground (ADP-BeF3, AMP-PNP) and transition states (ADP-VO4, ADP-ALF4) were similar, suggesting that no major structural rearrangements occur during formation of the transition state. The transition state may only be a chemical intermediate with no special role in substrate translocation. Thus, reversion of the transporter to its inward-facing state likely follows release of inorganic phosphate and/or ADP molecules.
5. Transcriptional and functional regulation of maltose/maltodextrin transport
Complex pathways regulate the utilization of carbon sources depending on their availability in the external cellular (growth) medium. Bacteria prefer to use primary sources, such as glucose and glucose 6-P, rather than secondary sources like maltose or maltodextrins. This preferential “uptake-dependent” usage of catabolites by bacteria, where some are more rapidly metabolized than others, was recognized early on [120], [121]. Thus, under conditions of primary nutrients deficiency, the uptake of maltose/maltodextrins in E. coli results from the synergistic action of LamB and MalEFGK2 system, which are both controlled at the transcriptional level [122], [123]. More precisely, the expression level and activity of both LamB and MalEFGK2 need to be adjusted according to (i) the abundance of maltodextrins in the medium and/or (ii) the levels of primary metabolizable carbon sources, also known as phosphotransferase system (PTS) substrates [124]. As explained below, the integration of positive and negative control factors not only fine-tunes the transcription of both LamB and MalEFGK2, but also modulates the functional activity of the maltose transporter.
5.1. Catabolic repression
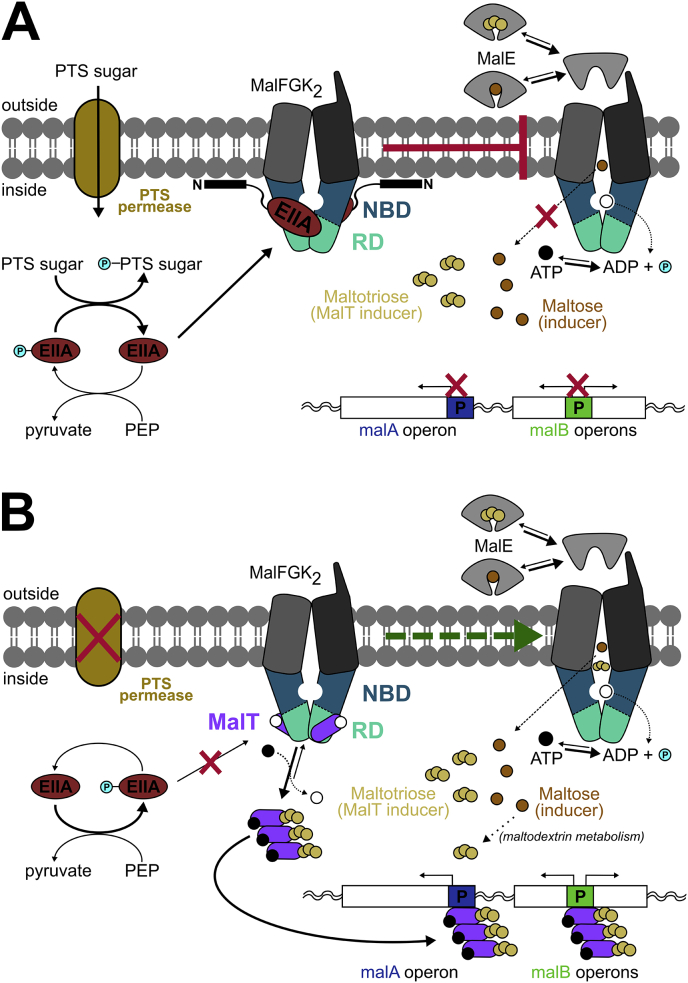
The PTS consists of a series of chain-phosphorylation reactions, which produces phosphorylated sugars that are available as cellular energy sources. Uptake and metabolism of maltose and maltodextrins (the so-called non-PTS sugars) are regulated by phosphorylation of a key PTS-intermediate, the glucose-specific enzyme EIIAglc (also known as IIIglc). EIIAglc is unphosphorylated when glucose is in excess and inhibits maltose/maltodextrins import by a direct 1:1 interaction with each subunit of the MalK homodimer [85] (Fig. 5, see D). Mechanistically, this interaction arrests the MalFGK2 complex in the inward-facing conformation and thus prohibits transport of maltodextrins. Maltose and maltodextrins such as maltotriose induce the machinery required for their uptake and metabolism by promoting directly, or indirectly through the maltodextrins metabolism, the transcription of both the malA and malB chromosomal regions that contain the inducible MalEFGK2 operon, and genes encoding LamB and maltose/maltodextrins metabolic enzymes [12], [125], [126]. For this reason, the EIIAglc-triggered prohibition of maltodextrins uptake is also known as “inducer exclusion” mechanism or catabolite repression [124], [127] (Fig. 7A). Dean et al. [122] studied inducer exclusion of the MalEFGK2 transporter reconstituted in EIIAglc-filled liposomes by measuring 14C-maltose uptake, and estimated the inhibitor constant Ki for EIIAglc of ∼40 μM, a value in accordance with intracellular concentration of the enzyme [128].
Fig. 7.
Mechanism for the control of maltose uptake. A, Schematic model of the inducer exclusion mechanism. PTS permeases facilitate PTS sugar uptake into the cell. EIIAglc is dephosphorylated, interacts with NBD and RD from different monomers of MalK, and is anchored to the inner leaflet of the membrane. The maltose transporter MalFGK2 is thus trapped in an inactive conformation and maltodextrins uptake is inhibited. Consequently, maltodextrins (e.g. maltose, maltotriose) cannot further induce the maltose system by promoting the expression of malA and malB genes. B, Representation of the MalT positive regulation. In the absence of PTS sugars, the equilibrium of EIIAglc is shifted towards its phosphorylated state (EIIAglc-P) which cannot interact with MalK; therefore, transport can occur. Under these conditions, maltose induces the expression of malA and malB regions through its channeling into a metabolic pathway mediated by MalT-inducible enzymes. MalT is released from MalK and can be activated by (cognate) maltotriose imported or produced from the maltodextrin metabolism. MalT multimerization (upon its activation) allows its binding to the malA and malB promoters thereby inducing gene transcription. Of note, the divergent operon that belongs to the malA region and contains the malT gene is not shown, since it does not belong to the maltose regulon. Moreover, MalT-inducible operons located in other loci than malA and malB are also not shown for simplicity.
Genetic and structural approaches such as site-directed mutagenesis, protein docking prediction, and X-ray crystallography, revealed the β-sheet sandwich topology, and 3D structure of the glucose-specific enzyme EIIAglc [129], [130]. Other studies have revealed the three main regions of EIIAglc that are involved in binding to MalK [131]. One interaction site is essential and specific for MalK binding, whereas the other two regions partially overlap with the EIIAglc binding surfaces for other PTS-sensitive targets (e.g. lactose permease, glycerol kinase) and PTS phosphocarriers (Hpr protein or EIIBglc). Transport activity and MalE-stimulation were recovered with EIIA mutants containing substitutions in these critical regions [131]. More recently, by a combination of in silico approaches, e.g., docking predictions, and site-directed cross-linking experiments (using single cysteine mutants of MalK and EIIAglc), the mode of interaction between MalK and EIIA was confirmed [129].
Chen et al. [85] determined the structure of the inward-facing MalFGK2 bound to EIIAglc. From the superimposition of the EIIAglc structures in its free and transporter-bound state, it became evident that EIIAglc does not undergo significant conformational changes upon binding to MalK. It was also hypothesized that phosphorylation of the H90 residue of EIIA greatly reduces its affinity for MalK. It was reasoned that the introduction of a negatively charged phosphate group on H90 would abolish its interactions with Q122 of MalK by sterical and charge-induced disruptions of the surrounding hydrophobic interface. Notably, this structure showed that EIIA interacts with both the NBD (55% of its buried surface) and the RD (45%) of MalFGK2, as was also proposed from previous site-directed mutagenesis studies [122], [129], [131]. Interestingly, these interactions involve the NBD and the RD from opposite monomers of MalK, thus hindering the NBD interdomain-rotation with respect to the RDs-pivot, that normally facilitates the transition from the resting (inward-facing) to the active (outward-facing) state of the maltose transporter (Fig. 5; section 4). This model is in accordance with EPR studies that employed double-spin-labeled versions of MalK (V17C and E128C variants) to analyze their inter-distance distributions in MalFGK2 in nanodisc in the presence of maltose-bound MalE [132]. These results confirmed the hypothesis that EIIAglc reduces transport activity by inhibiting ATP hydrolysis, without affecting the binding affinity of ATP for MalFGK2. Comparable affinities for ATP were indeed reported in the presence and absence of EIIAglc using the MalFGK2 system reconstituted in nanodiscs [129].
In addition to direct physical contact between EIIAglc and MalK, an interaction between EIIAglc and the surrounding membrane, in particular membrane enriched in phosphatidylglycerol (PG), also contributes to the stability of the complex [129], [133]. This membrane interaction is mediated by the first 18 N-terminal amino acids of EIIAglc, which form a α-helix upon proximity to the negatively charged headgroups of PG (and other phospholipids); the interaction occurs mainly thanks to the presence of lysine residues in this region of the protein. This interaction has been observed in structural analyses [85] and in co-sedimentation and detergent versus proteoliposomes/nanodiscs functional assays that employed N-truncated versions of EIIAglc [85], [129], [132]. Taken together, these studies suggest that the N-terminal amphipathic α-helix plays an essential role in inducer exclusion of the MalEFGK2 transporter, essentially by stabilizing the interaction with MalK.
5.2. Positive regulation by MalT
In addition to the negative control exerted indirectly on the maltose regulon by EIIAglc, there is also direct positive regulation, unlike in other non-PTS expression systems, e.g. the arabinose operon [123]. In enteric bacteria (e.g. E. coli, Salmonella spp., Vibrio spp.) this positive control is exerted by the transcription factor MalT [134], [135]. MalT is composed of an NBD, two linker regions (one winged-helix and one flexible arm), a domain for sensing maltotriose (the MalT activator), and a DNA-binding domain [136]. It binds to promoter “initiator” regions (MalT boxes) of the malA and malB operons to effect transcription of their genes [12], [123]. The equilibrium between a resting ADP- and an active ATP-bound state of MalT is an essential feature of its activation: once MalT is ATP-bound it multimerizes and undergoes cooperative interaction with its DNA target sequences [137]. The multimerization of MalT is induced by the presence of maltotriose in the cytoplasm [125], [137] (Fig. 7B). Thus, maltotriose represents the only direct inducer of the maltose system by interacting with MalT and thereby promoting its activation. The presence of maltotriose in the cell originates from several sources, as it can be: (i) directly imported via MalFGK2; (ii) produced as intermediate of maltose/maltodextrins metabolism [12], [125]; (iii) generated as a product of glycogen catabolism [138]. Recently, it was proposed that MalK sequesters MalT when the maltose transporter is in its resting state (Fig. 7B). This interaction between MalT and MalK would prevent MalT from binding maltotriose thereby limiting the expression of the maltose regulons [136]. The MalK–MalT interaction has been corroborated in mutagenesis studies, which have identified the residues that form the contact interface between MalK and MalT [139], [140]. Although biochemical information on MalT and its DNA binding to the MalT boxes are available, various mechanistic and structural details are still to be elucidated. Current understanding of how maltose regulon functions has allowed its exploitation for the selective cytoplasmic internalization of synthetic maltotriose-conjugated molecules, an example of how the maltose uptake system can be used as a self-powered molecular machine for the selective delivery of drugs and other molecules into cells [141].
6. Discussion and outlook
In our review, we have presented an integrated mechanism for the maltose transporter based on a combination of biochemical, structural and biophysical studies. A critical question is whether we have come to full and satisfying understanding of the transport mechanism. There is a saying in engineering that one cannot understand what one cannot build. If we manipulate the maltose import system by mutagenesis, varying the experimental environment, or using promiscuous ligands, can we predict the response of the transporter? We are probably far from that goal. For instance, manipulating the affinity of MalE or MalF for maltodextrins, engineering new substrate specificities, and altering the dynamics of MalE or MalK2 closure/opening may be investigated to assess their impact on transport kinetics and properties. We consider the following aspects crucial for further mechanistic insights and advancement of the transport model:
-
(i)
Are one or two ATP molecules hydrolyzed per transport cycle? If two ATP molecules are required for MalK2 closure, a key question is whether they are hydrolyzed simultaneously or sequentially. A molecular dynamics study of MalEFGK2 suggested that opening of both ATP-binding sites in the MalK2 dimer is necessary for the transporter to return to the inward-facing state [104]. However, the study does not exclude the possibility that on longer timescales hydrolysis of a single ATP molecule could drive the opening of both sites, as seen with isolated MalK2 [142]. It also still unclear at which catalytic step maltose is translocated into the cell, although it is likely to occur during release of the ATP hydrolysis products (Pi and/or ADP). In the presence of maltose, EPR studies suggest that the ADP-bound post-hydrolysis conformation is inward-facing with a semi-open MalK2 [69], [99]. Thus, the post-hydrolysis conformation might resemble the pretranslocation state, with a semi-open MalK dimer coupled to inward-facing TMDs. In this configuration, dissociation of ADP and/or MalE is likely to occur thereby promoting a full opening of MalK2.
-
(ii)
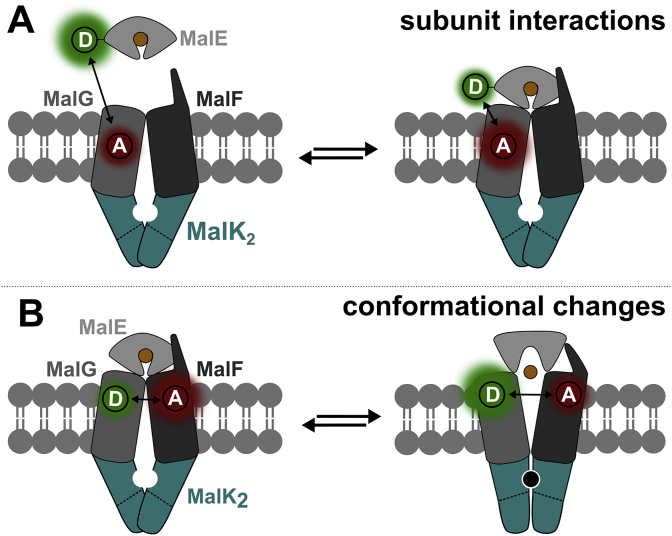
What are the relevant conformational dynamics required for substrate translocation? In our view, this question would be best addressed using single-molecule methodology as used for the soluble MalE (Fig. 3). Such studies would focus on the ligand-dependent transitions to determine transition rates between different conformational states. This would provide opportunities for assessing the dominant states of the transporter and discovering minor sub-populations or transient intermediates that occur during the transport cycle. For instance, it would be important to determine whether MalE induces conformational changes in MalFGK2 or stabilizes some high-energy and transient states of the transporter. Inter- and intramolecular approaches (Fig. 8) would allow a direct visualization of subunit interactions, for instance, docking (Fig. 8A) of MalE to MalFG, conformational changes in TMDs (Fig. 8B) and NBDs, or the interaction between EIIA and MalK.
Fig. 8.
Proposed future smFRET studies of MalEFGK2. A, Visualization of one of the many interactions that occur between subunits (complex formation) using smFRET. Depicted assay would monitor interactions between MalE and MalF or MalG to understand further the docking process. B, Study of intra- or interdomain conformational changes via FRET. Figure adapted from Mächtel et al. [166].
In an early unpublished study from the Davidson lab, the NBDs of nanodisc-reconstituted maltose transporter were labeled with Cy3/Cy5 to monitor effects of nucleotide addition using TIRF microscopy [143]. This work established a solid basis for the overexpression of MalFGK2 and for the flexible fluorophore labeling needed for smFRET. It also provided the first exciting insights into the conformational heterogeneity of the NBDs under various conditions (apo, ATP-bound, vanadate, maltose-bound MalE, unliganded MalE etc...). It remains, however, challenging to observe conformational dynamics in real time and to interpret the observed dynamics in light of the transport cycle. TMD movements or MalE-TMD interactions, as illustrated in Fig. 8, have to the best of our knowledge, not thus far been reported. Recently, smFRET was used to study isolated MalK2 to further elucidate the details of its ATPase catalytic function [144]. In accordance with existing crystal structures of isolated MalK2, three different conformational states were observed with TIRF-smFRET. The population distributions showed sensitivity to addition of ATP, nucleotide analogs and catalytic mutations.
As illustrated by the few examples reported thus far, smFRET studies (and this may be true of other biophysical approaches) remain challenging for membrane transporters. One key problem is achieving efficient fluorescence labeling of a functional transporter. And this may explain the rather limited number of published smFRET studies on ABC transporters labeled in the NBDs or TMDs [145], [146], [147], [148]. In the future, successful characterization of conformational dynamics may depend on alternative assays based on approaches that make quality sample preparation more readily achievable, such as the single label methods, protein induced fluorescence enhancement [149], [150] and quenching [151].
Molecular dynamics (MD) simulations are also useful for structural and dynamic investigations at the atomic level and may beneficially complement experimental approaches. They can in principle provide detailed information on conformational dynamics, energetics, populations, and allosteric effects during the transport cycle, although substantial technical limitations exist [152].
-
(iii)
How are substrate binding, energy utilization and translocation coordinated? A more comprehensive model of transport would require direct observation of the coordination of transport events, i.e., how substrate and ATP binding, as well as how ATP hydrolysis are coupled and transferred into conformational changes that drive substrate transport. To answer these questions, not only structural movements of the transporter have to be monitored, but also its biochemical activity (ATPase activity and transport) would need to be visualized on the single-molecule level. The first, and to our knowledge only study to report single-molecule visualization of conformational dynamics and transporter activity, albeit not simultaneously, was reported for the vitamin B12 importer BtuCD-F [153]. Another recent example investigated the functional dynamics of a eukaryotic primary active transporter, the Arabidopsis thaliana H+-adenosine triphosphatase (ATPase) isoform 2 (AHA2) [154]. In this study, the authors used single-molecule fluorescence microscopy to monitor single AHA2 containing proteoliposomes that had been tethered to a passivated glass slide to allow high throughput imaging. Changes in the H+ concentration were measured with a lipid-conjugated pH-sensitive fluorophore [154]. Their results, evaluated in the light of a physical non-equilibrium model of vesicle acidification, revealed that transport was stochastically interrupted by long-lived inactive or leaky states. Such approaches, more widely applied to ABC transporters, represent in our view, the most promising route to a detailed elucidation of the biochemistry of membrane transport.
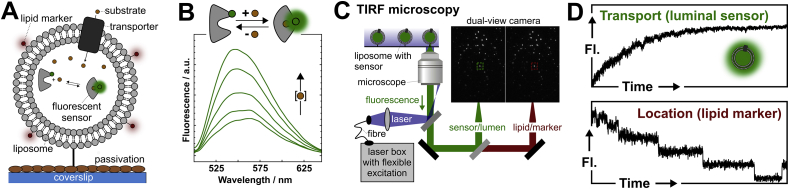
In future work, the Cordes lab envisions to monitor the biochemical activity of transporters and in particular that of the maltose import system using single liposome approaches (Fig. 9) similar to published work from Stamou and co-workers [154]. The central idea of this will be the visualization of the biochemical activity of one transporter (and different mechanistic aspects of it) in a Poisson-dilution regime in a single liposome, which is visualized by sensitive fluorescence video microscopy. In these single liposome assays, ‘fluorescent substrate sensors’ (Fig. 9A, B) are deposited in the lumen at high concentrations. The fluorescence will be recorded using TIRF microscopes in combination with (multi-color) detection (Fig. 9C). In this way fluorescent lipid markers such as DiD (Fig. 9A) can be used for verification of the liposome location and simultaneous analysis of luminal sensor dye(s). Key to success of such an assay are the fluorescent substrate sensors. The sensors should change their photophysical parameters whenever the substrate, ATP, or co-substrate concentration is altered inside the liposome due to transport. In analogy to published studies [154], chemical fluorescent sensors for transporter substrates could be employed in the lumen of the liposome or via lipid anchoring.
Fig. 9.
Proposed future single transporter recordings using periplasmic substrate binding protein-based fluorescent biosensors. A, Schematic of the proposed experimental set-up for single-transporter recordings. Single-transporters are reconstituted into proteoliposomes at low protein-to-lipid ratios to have one (or none) transporter per liposome, providing positive and negative control experiments. The position of the liposome can be detected via a fluorescent marker. Liposomes are filled with a large quantity of fluorescent sensors such as SBPs that detect changes in the concentration of the substrate, here via increase in their signal. B, Assay design for use of MalE as maltose-sensor using an environmentally-sensitive fluorophore (blue: IANBD). Fluorescence spectra at increasing maltose concentration from 0 (low fluorescence) to 1 mM maltose (high fluorescence). C, Scheme of the TIRF-microscope with laser box and dual-view allowing simultaneous recording of two emission channels, exemplified by data from a lipid marker DiD and luminal dsDNA-fluorescein. D, Example data of fluorescent markers in lumen and the liposome leaflet as schematics for expected signals upon single-transporter recordings. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In particular, SBP-based biosensors represent a great tool for versatile sensing of various ligands in transport assays. SBPs have been exploited as biosensors before [155] and in these efforts substrate induced conformational changes were converted into fluorescence [156], [157], electrical [158] or magnetic signals [159]. Preliminary data of our lab show the basics of this idea by conversion of MalE (Fig. 9B) into a fluorescent turn-on sensor for maltose using covalent labeling of the protein with IANBD-fluorophore. Alternative other fluorescent sensor principles could use any photophysical effects (quenching, energy transfer [52] etc.) that converts the conformational motion of the large array of binding proteins [7], [8] into conformational changes.
-
(iv)
How general is the mechanism of MalEFGK2 and what does it tell us about the mechanism of other type I importers? The large body of work on the maltose transporter has provided many valuable insights into the molecular mechanisms at play in ABC proteins. Work on the transporter has contributed to a general understanding of the mechanism of ATP hydrolysis (e.g. [99], [109], [119]) and substrate import, from genetic [12], biochemical (e.g. [71], [72], [88]) and structural viewpoints [33], [78], [82]. Various aspects of the transport cycle have been shown to be similar to those of other type I transporters [160], [161], [162]. For instance, similar conformational changes have been observed in the histidine permease, HisQMP2 [160], [161]. This is remarkable given that HisQMP2, like most type I transporters, differs in several important respects from the maltose transporter, for instance, in lacking a large P2 loop, the scoop loop and the C-terminal regulatory domains of the NBDs. However, large differences exist between the mechanism of MalEFGK2 and that of type II transporters [163], [164]. An elegant comparative study of two molybdate/tungstate importers (one from type I and the other from type II) highlighted their mechanistic differences despite having the same substrate specificity [162]. The latter system forms a high-affinity, slow-dissociating complex with its SBP that was destabilized by nucleotide and substrate binding (similar to BtuCDF [164]). The former displayed a low-affinity, transient complex that is stabilized by ligands (similar to MalEFGK2). However, given the mechanistic diversity in ABC transporters, the study of many more ABC systems, such as the type I l-glutamine transporter, GlnPQ, that has different covalently linked SBDs [77], [165], will illuminate similarities and differences in their mechanism.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was financed by an ANR grant (No. ANR-17-CE11-0045-01 to C.O.), an ERC Starting Grant (No. 638536 – SM-IMPORT; to T.C.), Deutsche Forschungsgemeinschaft (GRK2062 project C03 to T.C.; a GRK2062 visiting scholarship to C.O.; SFB863 project A13 to T.C.). T.C. was further supported by the Center of Nanoscience Munich (CeNS), LMUexcellent and the DFG excellence cluster Center for integrated protein science Munich (CiPSM).
Contributor Information
Thorben Cordes, Email: cordes@bio.lmu.de.
Cédric Orelle, Email: cedric.orelle@ibcp.fr.
References
- 1.Davidson A.L., Dassa E., Orelle C., Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [table of contents] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dassa E. Natural history of ABC systems: not only transporters. Essays Biochem. 2011;50:19–42. doi: 10.1042/bse0500019. [DOI] [PubMed] [Google Scholar]
- 3.Gouridis G., Hetzert B., Kiosze-Becker K., de Boer M., Heinemann H., Nurenberg-Goloub E. ABCE1 controls ribosome recycling by an asymmetric dynamic conformational equilibrium. Cell Rep. 2019;28:723–734 e6. doi: 10.1016/j.celrep.2019.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locher K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 5.ter Beek J., Guskov A., Slotboom D.J. Structural diversity of ABC transporters. J Gen Physiol. 2014;143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice A.J., Park A., Pinkett H.W. Diversity in ABC transporters: type I, II and III importers. Crit Rev Biochem Mol Biol. 2014;49:426–437. doi: 10.3109/10409238.2014.953626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berntsson R.P., Smits S.H., Schmitt L., Slotboom D.J., Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Scheepers G.H., Lycklama A.N.J.A., Poolman B. An updated structural classification of substrate-binding proteins. FEBS Lett. 2016;590:4393–4401. doi: 10.1002/1873-3468.12445. [DOI] [PubMed] [Google Scholar]
- 9.Lewinson O., Livnat-Levanon N. Mechanism of action of ABC importers: conservation, divergence, and physiological adaptations. J Mol Biol. 2017;429:606–619. doi: 10.1016/j.jmb.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Locher K.P. Review. Structure and mechanism of ATP-binding cassette transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:239–245. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rempel S., Stanek W.K., Slotboom D.J. Energy-coupling factor-type ATP-binding cassette transporters. Annu Rev Biochem. 2019;88:551–576. doi: 10.1146/annurev-biochem-013118-111705. [DOI] [PubMed] [Google Scholar]
- 12.Boos W., Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordignon E., Grote M., Schneider E. The maltose ATP-binding cassette transporter in the 21st century--towards a structural dynamic perspective on its mode of action. Mol Microbiol. 2010;77:1354–1366. doi: 10.1111/j.1365-2958.2010.07319.x. [DOI] [PubMed] [Google Scholar]
- 14.Shilton B.H. The dynamics of the MBP-MalFGK(2) interaction: a prototype for binding protein dependent ABC-transporter systems. Biochim Biophys Acta. 2008;1778:1772–1780. doi: 10.1016/j.bbamem.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Luckey M., Nikaido H. Specificity of diffusion channels produced by lambda phage receptor protein of Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schirmer T., Keller T.A., Wang Y.F., Rosenbusch J.P. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 17.Szmelcman S., Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975;124:112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins C.F. ABC transporters: physiology, structure and mechanism--an overview. Res Microbiol. 2001;152:205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 19.Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- 20.Froshauer S., Beckwith J. The nucleotide sequence of the gene for malF protein, an inner membrane component of the maltose transport system of Escherichia coli. Repeated DNA sequences are found in the malE-malF intercistronic region. J Biol Chem. 1984;259:10896–10903. [PubMed] [Google Scholar]
- 21.Dassa E., Hofnung M. Sequence of gene malG in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. Embo J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilson E., Nikaido H., Hofnung M. Sequence of the malK gene in E.coli K12. Nucleic Acids Res. 1982;10:7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellermann O., Szmelcman S. Active transport of maltose in Escherichia coli K12. Involvement of a "periplasmic" maltose binding protein. Eur J Biochem. 1974;47:139–149. doi: 10.1111/j.1432-1033.1974.tb03677.x. [DOI] [PubMed] [Google Scholar]
- 24.Shuman H.A. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J Biol Chem. 1982;257:5455–5461. [PubMed] [Google Scholar]
- 25.Sharff A.J., Rodseth L.E., Spurlino J.C., Quiocho F.A. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 26.Spurlino J.C., Lu G.Y., Quiocho F.A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 27.Dietzel I., Kolb V., Boos W. Pole cap formation in Escherichia coli following induction of the maltose-binding protein. Arch Microbiol. 1978;118:207–218. doi: 10.1007/BF00415731. [DOI] [PubMed] [Google Scholar]
- 28.Manson M.D., Boos W., Bassford P.J., Jr., Rasmussen B.A. Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J Biol Chem. 1985;260:9727–9733. [PubMed] [Google Scholar]
- 29.Dean D.A., Davidson A.L., Nikaido H. Maltose transport in membrane vesicles of Escherichia coli is linked to ATP hydrolysis. Proc Natl Acad Sci U S A. 1989;86:9134–9138. doi: 10.1073/pnas.86.23.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishop L., Agbayani R., Jr., Ambudkar S.V., Maloney P.C., Ames G.F. Reconstitution of a bacterial periplasmic permease in proteoliposomes and demonstration of ATP hydrolysis concomitant with transport. Proc Natl Acad Sci U S A. 1989;86:6953–6957. doi: 10.1073/pnas.86.18.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson A.L., Nikaido H. Overproduction, solubilization, and reconstitution of the maltose transport system from Escherichia coli. J Biol Chem. 1990;265:4254–4260. [PubMed] [Google Scholar]
- 32.Davidson A.L., Nikaido H. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 33.Oldham M.L., Khare D., Quiocho F.A., Davidson A.L., Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 34.Dumas F., Koebnik R., Winterhalter M., Van Gelder P. Sugar transport through maltoporin of Escherichia coli. Role of polar tracks. J Biol Chem. 2000;275:19747–19751. doi: 10.1074/jbc.M000268200. [DOI] [PubMed] [Google Scholar]
- 35.Wandersman C., Schwartz M., Ferenci T. Escherichia coli mutants impaired in maltodextrin transport. J Bacteriol. 1979;140:1–13. doi: 10.1128/jb.140.1.1-13.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz M., Kellermann O., Szmelcman S., Hazelbauer G.L. Further studies on the binding of maltose to the maltose-binding protein of Escherichia coli. Eur J Biochem. 1976;71:167–170. doi: 10.1111/j.1432-1033.1976.tb11102.x. [DOI] [PubMed] [Google Scholar]
- 37.Koiwai O., Hayashi H. Studies on bacterial chemotaxis. IV. Interaction of maltose receptor with a membrane-bound chemosensing component. J Biochem. 1979;86:27–34. [PubMed] [Google Scholar]
- 38.Ferenci T., Muir M., Lee K.-S., Maris D. Substrate specificity of the Escherichia coli maltodextrin transport system and its component proteins. Biochim Biophys Acta (BBA) – Biomembranes. 1986;860:44–50. doi: 10.1016/0005-2736(86)90496-7. [DOI] [PubMed] [Google Scholar]
- 39.Quiocho F.A., Spurlino J.C., Rodseth L.E. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 40.Ferenci T. The recognition of maltodextrins by Escherichia coli. Eur J Biochem. 1980;108:631–636. doi: 10.1111/j.1432-1033.1980.tb04758.x. [DOI] [PubMed] [Google Scholar]
- 41.Cuneo M.J., Changela A., Beese L.S., Hellinga H.W. Structural adaptations that modulate monosaccharide, disaccharide, and trisaccharide specificities in periplasmic maltose-binding proteins. J Mol Biol. 2009;389:157–166. doi: 10.1016/j.jmb.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Shilton B.H., Flocco M.M., Nilsson M., Mowbray S.L. Conformational changes of three periplasmic receptors for bacterial chemotaxis and transport: the maltose-, glucose/galactose- and ribose-binding proteins. J Mol Biol. 1996;264:350–363. doi: 10.1006/jmbi.1996.0645. [DOI] [PubMed] [Google Scholar]
- 43.Fischetti R.F., Rodi D.J., Gore D.B., Makowski L. Wide-angle X-ray solution scattering as a probe of ligand-induced conformational changes in proteins. Chem Biol. 2004;11:1431–1443. doi: 10.1016/j.chembiol.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Hall J.A., Gehring K., Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Correlation with the structure of ligands and the structure of binding protein. J Biol Chem. 1997;272:17605–17609. doi: 10.1074/jbc.272.28.17605. [DOI] [PubMed] [Google Scholar]
- 45.Hall J.A., Thorgeirsson T.E., Liu J., Shin Y.K., Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Electron paramagnetic resonance study of ligand-induced global conformational changes by site-directed spin labeling. J Biol Chem. 1997;272:17610–17614. doi: 10.1074/jbc.272.28.17610. [DOI] [PubMed] [Google Scholar]
- 46.Evenas J., Tugarinov V., Skrynnikov N.R., Goto N.K., Muhandiram R., Kay L.E. Ligand-induced structural changes to maltodextrin-binding protein as studied by solution NMR spectroscopy. J Mol Biol. 2001;309:961–974. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]
- 47.Kainosho M., Torizawa T., Iwashita Y., Terauchi T., Mei Ono A., Guntert P. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 48.Rubin S.M., Spence M.M., Dimitrov I.E., Ruiz E.J., Pines A., Wemmer D.E. Detection of a conformational change in maltose binding protein by (129)Xe NMR spectroscopy. J Am Chem Soc. 2001;123:8616–8617. doi: 10.1021/ja0110325. [DOI] [PubMed] [Google Scholar]
- 49.Tang C., Schwieters C.D., Clore G.M. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 50.Stockner T., Vogel H.J., Tieleman D.P. A salt-bridge motif involved in ligand binding and large-scale domain motions of the maltose-binding protein. Biophys J. 2005;89:3362–3371. doi: 10.1529/biophysj.105.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bucher D., Grant B.J., Markwick P.R., McCammon J.A. Accessing a hidden conformation of the maltose binding protein using accelerated molecular dynamics. PLoS Comput Biol. 2011;7:e1002034. doi: 10.1371/journal.pcbi.1002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Boer M., Gouridis G., Vietrov R., Begg S.L., Schuurman-Wolters G.K., Husada F. Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. Elife. 2019;8 doi: 10.7554/eLife.44652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Husada F., Gouridis G., Vietrov R., Schuurman-Wolters G.K., Ploetz E., de Boer M. Watching conformational dynamics of ABC transporters with single-molecule tools. Biochem Soc Trans. 2015;43:1041–1047. doi: 10.1042/BST20150140. [DOI] [PubMed] [Google Scholar]
- 54.Kim E., Lee S., Jeon A., Choi J.M., Lee H.S., Hohng S. A single-molecule dissection of ligand binding to a protein with intrinsic dynamics. Nat Chem Biol. 2013;9:313–318. doi: 10.1038/nchembio.1213. [DOI] [PubMed] [Google Scholar]
- 55.Gould A.D., Telmer P.G., Shilton B.H. Stimulation of the maltose transporter ATPase by unliganded maltose binding protein. Biochemistry. 2009;48:8051–8061. doi: 10.1021/bi9007066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oldham M.L., Chen S., Chen J. Structural basis for substrate specificity in the Escherichia coli maltose transport system. Proc Natl Acad Sci. 2013;110:18132–18137. doi: 10.1073/pnas.1311407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daus M.L., Grote M., Schneider E. The MalF P2 loop of the ATP-binding cassette transporter MalFGK2 from Escherichia coli and Salmonella enterica serovar typhimurium interacts with maltose binding protein (MalE) throughout the catalytic cycle. J Bacteriol. 2009;191:754–761. doi: 10.1128/JB.01439-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohl E., Shuman H.A., Boos W. Mathematical treatment of the kinetics of binding protein dependent transport systems reveals that both the substrate loaded and unloaded binding proteins interact with the membrane components. J Theor Biol. 1995;172:83–94. doi: 10.1006/jtbi.1995.0006. [DOI] [PubMed] [Google Scholar]
- 59.Shilton B.H., Mowbray S.L. Simple models for the analysis of binding protein-dependent transport systems. Protein Sci. 1995;4:1346–1355. doi: 10.1002/pro.5560040710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merino G., Boos W., Shuman H.A., Bohl E. The inhibition of maltose transport by the unliganded form of the maltose-binding protein of Escherichia coli: experimental findings and mathematical treatment. J Theor Biol. 1995;177:171–179. doi: 10.1006/jtbi.1995.0236. [DOI] [PubMed] [Google Scholar]
- 61.Bohl E., Boos W. Quantitative analysis of binding protein-mediated ABC transport systems. J Theor Biol. 1997;186:65–74. doi: 10.1006/jtbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 62.Hollenstein K., Frei D.C., Locher K.P. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 63.Jacso T., Grote M., Daus M.L., Schmieder P., Keller S., Schneider E. Periplasmic loop P2 of the MalF subunit of the maltose ATP binding cassette transporter is sufficient to bind the maltose binding protein MalE. Biochemistry. 2009;48:2216–2225. doi: 10.1021/bi801376m. [DOI] [PubMed] [Google Scholar]
- 64.Dassa E., Muir S. Membrane topology of MalG, an inner membrane protein from the maltose transport system of Escherichia coli. Mol Microbiol. 1993;7:29–38. doi: 10.1111/j.1365-2958.1993.tb01094.x. [DOI] [PubMed] [Google Scholar]
- 65.Daus M.L., Berendt S., Wuttge S., Schneider E. Maltose binding protein (MalE) interacts with periplasmic loops P2 and P1 respectively of the MalFG subunits of the maltose ATP binding cassette transporter (MalFGK(2)) from Escherichia coli/Salmonella during the transport cycle. Mol Microbiol. 2007;66:1107–1122. doi: 10.1111/j.1365-2958.2007.05982.x. [DOI] [PubMed] [Google Scholar]
- 66.Daus M.L., Landmesser H., Schlosser A., Muller P., Herrmann A., Schneider E. ATP induces conformational changes of periplasmic loop regions of the maltose ATP-binding cassette transporter. J Biol Chem. 2006;281:3856–3865. doi: 10.1074/jbc.M511953200. [DOI] [PubMed] [Google Scholar]
- 67.Licht A., Bommer M., Werther T., Neumann K., Hobe C., Schneider E. Structural and functional characterization of a maltose/maltodextrin ABC transporter comprising a single solute binding domain (MalE) fused to the transmembrane subunit MalF. Res Microbiol. 2019;170:1–12. doi: 10.1016/j.resmic.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Alvarez F.J., Orelle C., Huang Y., Bajaj R., Everly R.M., Klug C.S. Full engagement of liganded maltose-binding protein stabilizes a semi-open ATP-binding cassette dimer in the maltose transporter. Mol Microbiol. 2015;98:878–894. doi: 10.1111/mmi.13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bohm S., Licht A., Wuttge S., Schneider E., Bordignon E. Conformational plasticity of the type I maltose ABC importer. Proc Natl Acad Sci U S A. 2013;110:5492–5497. doi: 10.1073/pnas.1217745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oldham M.L., Chen J. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science. 2011;332:1202–1205. doi: 10.1126/science.1200767. [DOI] [PubMed] [Google Scholar]
- 71.Davidson A.L., Shuman H.A., Nikaido H. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci U S A. 1992;89:2360–2364. doi: 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J., Sharma S., Quiocho F.A., Davidson A.L. Trapping the transition state of an ATP-binding cassette transporter: evidence for a concerted mechanism of maltose transport. Proc Natl Acad Sci U S A. 2001;98:1525–1530. doi: 10.1073/pnas.041542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Treptow N.A., Shuman H.A. Genetic evidence for substrate and periplasmic-binding-protein recognition by the MalF and MalG proteins, cytoplasmic membrane components of the Escherichia coli maltose transport system. J Bacteriol. 1985;163:654–660. doi: 10.1128/jb.163.2.654-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Panagiotidis C.H., Shuman H.A. Maltose transport in Escherichia coli: mutations that uncouple ATP hydrolysis from transport. Methods Enzymol. 1998;292:30–39. doi: 10.1016/s0076-6879(98)92005-5. [DOI] [PubMed] [Google Scholar]
- 75.Dean D.A., Hor L.I., Shuman H.A., Nikaido H. Interaction between maltose-binding protein and the membrane-associated maltose transporter complex in Escherichia coli. Mol Microbiol. 1992;6:2033–2040. doi: 10.1111/j.1365-2958.1992.tb01376.x. [DOI] [PubMed] [Google Scholar]
- 76.Davidson A.L. Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J Bacteriol. 2002;184:1225–1233. doi: 10.1128/JB.184.5.1225-1233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gouridis G., Schuurman-Wolters G.K., Ploetz E., Husada F., Vietrov R., de Boer M. Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat Struct Mol Biol. 2015;22:57–64. doi: 10.1038/nsmb.2929. [DOI] [PubMed] [Google Scholar]
- 78.Chen J., Lu G., Lin J., Davidson A.L., Quiocho F.A. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 79.Jones P.M., O’Mara M.L., George A.M. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem Sci. 2009;34:520–531. doi: 10.1016/j.tibs.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 80.Orelle C., Mathieu K., Jault J.M. Multidrug ABC transporters in bacteria. Res Microbiol. 2019;170:358–365. doi: 10.1016/j.resmic.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 81.Lu G., Westbrooks J.M., Davidson A.L., Chen J. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc Natl Acad Sci U S A. 2005;102:17969–17974. doi: 10.1073/pnas.0506039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J. Molecular mechanism of the Escherichia coli maltose transporter. Curr Opin Struct Biol. 2013;23:492–498. doi: 10.1016/j.sbi.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orelle C., Oldham M.L., Davidson A.L. The maltose ABC transporter: where structure meets function. In: Krämer R., Ziegler C., editors. Membrane transport mechanism: 3D structure and beyond. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 181–205. [Google Scholar]
- 84.Khare D., Oldham M.L., Orelle C., Davidson A.L., Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell. 2009;33:528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen S., Oldham M.L., Davidson A.L., Chen J., Lafayette W., Lafayette W. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature. 2013;499:364–368. doi: 10.1038/nature12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grote M., Polyhach Y., Jeschke G., Steinhoff H.J., Schneider E., Bordignon E. Transmembrane signaling in the maltose ABC transporter MalFGK2-E: periplasmic MalF-P2 loop communicates substrate availability to the ATP-bound MalK dimer. J Biol Chem. 2009;284:17521–17526. doi: 10.1074/jbc.M109.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saurin W., Koster W., Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 88.Mourez M., Hofnung M., Dassa E. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. Embo J. 1997;16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bouige P., Laurent D., Piloyan L., Dassa E. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr Protein Pept Sci. 2002;3:541–559. doi: 10.2174/1389203023380486. [DOI] [PubMed] [Google Scholar]
- 90.Dassa E., Bouige P. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 91.Hunke S., Mourez M., Jehanno M., Dassa E., Schneider E. ATP modulates subunit-subunit interactions in an ATP-binding cassette transporter (MalFGK2) determined by site-directed chemical cross-linking. J Biol Chem. 2000;275:15526–15534. doi: 10.1074/jbc.275.20.15526. [DOI] [PubMed] [Google Scholar]
- 92.Mourez M., Jehanno M., Schneider E., Dassa E. In vitro interaction between components of the inner membrane complex of the maltose ABC transporter of Escherichia coli: modulation by ATP. Mol Microbiol. 1998;30:353–363. doi: 10.1046/j.1365-2958.1998.01070.x. [DOI] [PubMed] [Google Scholar]
- 93.Moody J.E., Millen L., Binns D., Hunt J.F., Thomas P.J. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Orelle C., Dalmas O., Gros P., Di Pietro A., Jault J.M. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J Biol Chem. 2003;278:47002–47008. doi: 10.1074/jbc.M308268200. [DOI] [PubMed] [Google Scholar]
- 95.Austermuhle M.I., Hall J.A., Klug C.S., Davidson A.L. Maltose-binding protein is open in the catalytic transition state for ATP hydrolysis during maltose transport. J Biol Chem. 2004;279:28243–28250. doi: 10.1074/jbc.M403508200. [DOI] [PubMed] [Google Scholar]
- 96.Ehrle R., Pick C., Ulrich R., Hofmann E., Ehrmann M. Characterization of transmembrane domains 6, 7, and 8 of MalF by mutational analysis. J Bacteriol. 1996;178:2255–2262. doi: 10.1128/jb.178.8.2255-2262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Steinke A., Grau S., Davidson A., Hofmann E., Ehrmann M. Characterization of transmembrane segments 3, 4, and 5 of MalF by mutational analysis. J Bacteriol. 2001;183:375–381. doi: 10.1128/JB.183.1.375-381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cui J., Qasim S., Davidson A.L. Uncoupling substrate transport from ATP hydrolysis in the Escherichia coli maltose transporter. J Biol Chem. 2010;285:39986–39993. doi: 10.1074/jbc.M110.147819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Orelle C., Ayvaz T., Everly R.M., Klug C.S., Davidson A.L. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc Natl Acad Sci U S A. 2008;105:12837–12842. doi: 10.1073/pnas.0803799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsu W.L., Furuta T., Sakurai M. The mechanism of nucleotide-binding domain dimerization in the intact maltose transporter as studied by all-atom molecular dynamics simulations. Proteins. 2018;86:237–247. doi: 10.1002/prot.25433. [DOI] [PubMed] [Google Scholar]
- 101.Orelle C., Alvarez F.J., Oldham M.L., Orelle A., Wiley T.E., Chen J. Dynamics of alpha-helical subdomain rotation in the intact maltose ATP-binding cassette transporter. Proc Natl Acad Sci U S A. 2010;107:20293–20298. doi: 10.1073/pnas.1006544107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y., Mannering D.E., Davidson A.L., Yao N., Manson M.D. Maltose-binding protein containing an interdomain disulfide bridge confers a dominant-negative phenotype for transport and chemotaxis. J Biol Chem. 1996;271:17881–17889. doi: 10.1074/jbc.271.30.17881. [DOI] [PubMed] [Google Scholar]
- 103.Alvarez F.J., Orelle C., Davidson A.L. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J Am Chem Soc. 2010;132:9513–9515. doi: 10.1021/ja104047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wen P.C., Tajkhorshid E. Conformational coupling of the nucleotide-binding and the transmembrane domains in ABC transporters. Biophys J. 2011;101:680–690. doi: 10.1016/j.bpj.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bao H., Duong F. ATP alone triggers the outward facing conformation of the maltose ATP-binding cassette transporter. J Biol Chem. 2013;288:3439–3448. doi: 10.1074/jbc.M112.431932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bao H., Dalal K., Wang V., Rouiller I., Duong F. The maltose ABC transporter: action of membrane lipids on the transporter stability, coupling and ATPase activity. Biochim Biophys Acta. 2013;1828:1723–1730. doi: 10.1016/j.bbamem.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 107.Fabre L., Bao H., Innes J., Duong F., Rouiller I. Negative stain single-particle EM of the maltose transporter in nanodiscs reveals asymmetric closure of MalK2 and catalytic roles of ATP, MalE, and maltose. J Biol Chem. 2017;292:5457–5464. doi: 10.1074/jbc.M116.757898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bao H., Duong F. Nucleotide-free MalK drives the transition of the maltose transporter to the inward-facing conformation. J Biol Chem. 2014;289:9844–9851. doi: 10.1074/jbc.M113.545525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fetsch E.E., Davidson A.L. Vanadate-catalyzed photocleavage of the signature motif of an ATP-binding cassette (ABC) transporter. Proc Natl Acad Sci U S A. 2002;99:9685–9690. doi: 10.1073/pnas.152204499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mannering D.E., Sharma S., Davidson A.L. Demonstration of conformational changes associated with activation of the maltose transport complex. J Biol Chem. 2001;276:12362–12368. doi: 10.1074/jbc.M011686200. [DOI] [PubMed] [Google Scholar]
- 111.Shilton B.H. Active transporters as enzymes: an energetic framework applied to major facilitator superfamily and ABC importer systems. Biochem J. 2015;467:193–199. doi: 10.1042/BJ20140675. [DOI] [PubMed] [Google Scholar]
- 112.Gould A.D., Shilton B.H. Studies of the maltose transport system reveal a mechanism for coupling ATP hydrolysis to substrate translocation without direct recognition of substrate. J Biol Chem. 2010;285:11290–11296. doi: 10.1074/jbc.M109.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Daus M.L., Grote M., Muller P., Doebber M., Herrmann A., Steinhoff H.J. ATP-driven MalK dimer closure and reopening and conformational changes of the “EAA” motifs are crucial for function of the maltose ATP-binding cassette transporter (MalFGK2) J Biol Chem. 2007;282:22387–22396. doi: 10.1074/jbc.M701979200. [DOI] [PubMed] [Google Scholar]
- 114.Hall J.A., Ganesan A.K., Chen J., Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Functional significance in active transport. J Biol Chem. 1997;272:17615–17622. doi: 10.1074/jbc.272.28.17615. [DOI] [PubMed] [Google Scholar]
- 115.Bajaj R., Park M.I., Stauffacher C.V., Davidson A.L. Conformational dynamics in the binding-protein-independent mutant of the Escherichia coli maltose transporter, MalG511, and its interaction with maltose binding protein. Biochemistry. 2018;57:3003–3015. doi: 10.1021/acs.biochem.8b00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carlson M.L., Bao H., Duong F. formation of a chloride-conducting state in the maltose ATP-binding cassette (ABC) transporter. J Biol Chem. 2016;291:12119–12125. doi: 10.1074/jbc.M115.711622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Covitz K.M., Panagiotidis C.H., Hor L.I., Reyes M., Treptow N.A., Shuman H.A. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. Embo J. 1994;13:1752–1759. doi: 10.1002/j.1460-2075.1994.tb06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bao H., Dalal K., Cytrynbaum E., Duong F. Sequential action of MalE and maltose allows coupling ATP hydrolysis to translocation in the MalFGK2 transporter. J Biol Chem. 2015;290:25452–25460. doi: 10.1074/jbc.M115.671826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oldham M.L., Chen J. Snapshots of the maltose transporter during ATP hydrolysis. Proc Natl Acad Sci U S A. 2011;108:15152–15156. doi: 10.1073/pnas.1108858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3:371–394. [Google Scholar]
- 121.Schaechter M. A brief history of bacterial growth physiology. Front Microbiol. 2015;6:1–5. doi: 10.3389/fmicb.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dean D.A., Reizer J., Nikaido H., Saier M.H., Jr. Regulation of the maltose transport system of Escherichia coli by the glucose-specific enzyme III of the phosphoenolpyruvate-sugar phosphotransferase system. Characterization of inducer exclusion-resistant mutants and reconstitution of inducer exclusion in proteoliposomes. J Biol Chem. 1990;265:21005–21010. [PubMed] [Google Scholar]
- 123.Débarbouillé Dominant constitutive mutations in maIT , the positive regulator gene of the maltose regulon in Escherichia coli. The J Mol Biol. 1978;124:359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- 124.Saier M.H., Jr., Crasnier M. Inducer exclusion and the regulation of sugar transport. Res Microbiol. 1996;147:482–489. doi: 10.1016/s0923-2508(96)90150-3. [DOI] [PubMed] [Google Scholar]
- 125.Raibaud O., Richet E. Maltotriose is the inducer of the maltose regulon of Escherichia coli. J Bacteriol. 1987;169:3059–3061. doi: 10.1128/jb.169.7.3059-3061.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiss S.C., Skerra A., Schiefner A. Structural basis for the interconversion of maltodextrins by MalQ, the amylomaltase of Escherichia coli. J Biol Chem. 2015;290:21352–21364. doi: 10.1074/jbc.M115.667337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wanner B.L., Kodaira R., Neidhardt F.C. Regulation of lac operon expression : reappraisal of the theory of catabolite repression. J Bacteriol. 1978;136:947–954. doi: 10.1128/jb.136.3.947-954.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scholte B.J., Schuitema A.R., Postma P.W. Isolation of IIGIc of the phosphoenolpyruvate-dependent glucose phosphotransferase system of Salmonella typhimurium. J Bacteriol. 1981;148:257–264. doi: 10.1128/jb.148.1.257-264.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bao H., Duong F. Phosphatidylglycerol directs binding and inhibitory action of EIIAGlc protein on the maltose transporter. J Biol Chem. 2013;288:23666–23674. doi: 10.1074/jbc.M113.489567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Feese M.D., Comolli L., Meadow N.D., Roseman S., James Remington S. Structural studies of the Escherichia coli signal transducing protein IIA(Glc): implications for target recognition. Biochemistry. 1997;36:16087–16096. doi: 10.1021/bi971999e. [DOI] [PubMed] [Google Scholar]
- 131.Blüschke B., Volkmer-engert R., Schneider E. Topography of the surface of the signal-transducing protein EIIA Glc that interacts with the MalK subunits of the maltose ATP-binding cassette transporter (MalFGK 2) of Salmonella typhimurium. J Biol Chem. 2006;281:12833–12840. doi: 10.1074/jbc.M512646200. [DOI] [PubMed] [Google Scholar]
- 132.Wuttge S., Licht A., Timachi M.H., Bordignon E., Schneider E. Mode of interaction of the signal-transducing protein EIIAGlc with the maltose ABC transporter in the process of inducer exclusion. Biochemistry. 2016;55:5442–5452. doi: 10.1021/acs.biochem.6b00721. [DOI] [PubMed] [Google Scholar]
- 133.Wang G., Peterkofsky A., Clore G.M. A novel membrane anchor function for the N-terminal amphipathic sequence of the signal-transducing protein IIAGlucose of the Escherichia coli phosphotransferase system. J Biol Chem. 2000;275:39811–39814. doi: 10.1074/jbc.C000709200. [DOI] [PubMed] [Google Scholar]
- 134.Hofnung M. Divergent operons and the genetic structure of the maltose B region in Escherichia coli K12. Genetics. 1974;76:169–184. doi: 10.1093/genetics/76.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Richet E., Raibaud O. Purification and properties of the MalT protein, the transcription activator of the Escherichia coli maltose regulon. J Biol Chem. 1987;262:12647–12653. [PubMed] [Google Scholar]
- 136.Richet E., Davidson A.L., Joly N. The ABC transporter MalFGK(2) sequesters the MalT transcription factor at the membrane in the absence of cognate substrate. Mol Microbiol. 2012;85:632–647. doi: 10.1111/j.1365-2958.2012.08137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marquenet E., Richet E. How integration of positive and negative regulatory signals by a STAND signaling protein depends on ATP hydrolysis. Mol Cell. 2007;28:187–199. doi: 10.1016/j.molcel.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 138.Dippel R., Boos W. The maltodextrin system of Escherichia coli: metabolism and transport. J Bacteriol. 2005;187:8322–8331. doi: 10.1128/JB.187.24.8322-8331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bohm A., Diez J., Diederichs K., Welte W., Boos W. Structural model of MalK, the ABC subunit of the maltose transporter of Escherichia coli: implications for mal gene regulation, inducer exclusion, and subunit assembly. J Biol Chem. 2002;277:3708–3717. doi: 10.1074/jbc.M107905200. [DOI] [PubMed] [Google Scholar]
- 140.Kuhnau S., Reyes M., Sievertsen A., Shuman H.A., Boos W. The activities of the Escherichia coli MalK protein in maltose transport, regulation, and inducer exclusion can be separated by mutations. J Bacteriol. 1991;173:2180–2186. doi: 10.1128/jb.173.7.2180-2186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dumont E., Vergalli J., Pajovic J., Bhamidimarri S.P., Morante K., Wang J. Mechanistic aspects of maltotriose-conjugate translocation to the Gram-negative bacteria cytoplasm. Life Sci Alliance. 2019;2:e201800242. doi: 10.26508/lsa.201800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wen P.C., Tajkhorshid E. Dimer opening of the nucleotide binding domains of ABC transporters after ATP hydrolysis. Biophys J. 2008;95:5100–5110. doi: 10.1529/biophysj.108.139444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang S. Purdue University; 2014. Application of Single-Molecule Fluorescence Resonance Energy Transfer to Maltose Transporter. [Google Scholar]
- 144.Wu S., Liu J., Wang W. Dissecting the conformational dynamics-modulated enzyme catalysis with single-molecule FRET. J Phys Chem B. 2018;122:6179–6187. doi: 10.1021/acs.jpcb.8b02374. [DOI] [PubMed] [Google Scholar]
- 145.Husada F., Bountra K., Tassis K., de Boer M., Romano M., Rebuffat S. Conformational dynamics of the ABC transporter McjD seen by single-molecule FRET. EMBO J. 2018;37 doi: 10.15252/embj.2018100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Verhalen B., Ernst S., Borsch M., Wilkens S. Dynamic ligand-induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J Biol Chem. 2012;287:1112–1127. doi: 10.1074/jbc.M111.301192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang M., Livnat Levanon N., Acar B., Aykac Fas B., Masrati G., Rose J. Single-molecule probing of the conformational homogeneity of the ABC transporter BtuCD. Nat Chem Biol. 2018;14:715–722. doi: 10.1038/s41589-018-0088-2. [DOI] [PubMed] [Google Scholar]
- 148.Liu Y., Liu Y., He L., Zhao Y., Zhang X.C. Single-molecule fluorescence studies on the conformational change of the ABC transporter MsbA. Biophys Rep. 2018;4:153–165. [Google Scholar]
- 149.Lerner E., Ploetz E., Hohlbein J., Cordes T., Weiss S. A quantitative theoretical framework for protein-induced fluorescence enhancement-forster-type resonance energy transfer (PIFE-FRET) J Phys Chem B. 2016;120:6401–6410. doi: 10.1021/acs.jpcb.6b03692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ploetz E., Lerner E., Husada F., Roelfs M., Chung S., Hohlbein J. Forster resonance energy transfer and protein-induced fluorescence enhancement as synergetic multi-scale molecular rulers. Sci Rep. 2016;6:33257. doi: 10.1038/srep33257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rashid F., Raducanu V.S., Zaher M.S., Tehseen M., Habuchi S., Hamdan S.M. Initial state of DNA-Dye complex sets the stage for protein induced fluorescence modulation. Nat Commun. 2019;10:2104. doi: 10.1038/s41467-019-10137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Szollosi D., Rose-Sperling D., Hellmich U.A., Stockner T. Comparison of mechanistic transport cycle models of ABC exporters. Biochim Biophys Acta Biomembr. 2018;1860:818–832. doi: 10.1016/j.bbamem.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goudsmits J.M.H., Slotboom D.J., van Oijen A.M. Single-molecule visualization of conformational changes and substrate transport in the vitamin B12 ABC importer BtuCD-F. Nat Commun. 2017;8:1652. doi: 10.1038/s41467-017-01815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Veshaguri S., Christensen S.M., Kemmer G.C., Ghale G., Moller M.P., Lohr C. Direct observation of proton pumping by a eukaryotic P-type ATPase. Science. 2016;351:1469–1473. doi: 10.1126/science.aad6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hirshberg M., Henrick K., Haire L.L., Vasisht N., Brune M., Corrie J.E. Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate. Biochemistry. 1998;37:10381–10385. doi: 10.1021/bi980428z. [DOI] [PubMed] [Google Scholar]
- 156.Brune M., Hunter J.L., Corrie J.E., Webb M.R. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 157.Wolf A., Shaw E.W., Oh B.H., De Bondt H., Joshi A.K., Ames G.F. Structure/function analysis of the periplasmic histidine-binding protein. Mutations decreasing ligand binding alter the properties of the conformational change and of the closed form. J Biol Chem. 1995;270:16097–16106. doi: 10.1074/jbc.270.27.16097. [DOI] [PubMed] [Google Scholar]
- 158.Benson D.E., Conrad D.W., de Lorimier R.M., Trammell S.A., Hellinga H.W. Design of bioelectronic interfaces by exploiting hinge-bending motions in proteins. Science. 2001;293:1641–1644. doi: 10.1126/science.1062461. [DOI] [PubMed] [Google Scholar]
- 159.Luck L.A., Johnson C. Fluorescence and 19F NMR evidence that phenylalanine, 3-L-fluorophenylalanine and 4-L-fluorophenylalanine bind to the L-leucine specific receptor of Escherichia coli. Protein Sci. 2000;9:2573–2576. doi: 10.1110/ps.9.12.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Heuveling J., Frochaux V., Ziomkowska J., Wawrzinek R., Wessig P., Herrmann A. Conformational changes of the bacterial type I ATP-binding cassette importer HisQMP2 at distinct steps of the catalytic cycle. Biochim Biophys Acta. 2014;1838:106–116. doi: 10.1016/j.bbamem.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 161.Sippach M., Weidlich D., Klose D., Abe C., Klare J., Schneider E. Conformational changes of the histidine ATP-binding cassette transporter studied by double electron-electron resonance spectroscopy. Biochim Biophys Acta. 2014;1838:1760–1768. doi: 10.1016/j.bbamem.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 162.Vigonsky E., Ovcharenko E., Lewinson O. Two molybdate/tungstate ABC transporters that interact very differently with their substrate binding proteins. Proc Natl Acad Sci U S A. 2013;110:5440–5445. doi: 10.1073/pnas.1213598110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Korkhov V.M., Mireku S.A., Veprintsev D.B., Locher K.P. Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F. Nat Struct Mol Biol. 2014;21:1097–1099. doi: 10.1038/nsmb.2918. [DOI] [PubMed] [Google Scholar]
- 164.Lewinson O., Lee A.T., Locher K.P., Rees D.C. A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation. Nat Struct Mol Biol. 2010;17:332–338. doi: 10.1038/nsmb.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lycklama A.N.J.A., Vietrov R., Schuurman-Wolters G.K., Poolman B. Energy coupling efficiency in the type I ABC transporter GlnPQ. J Mol Biol. 2018;430:853–866. doi: 10.1016/j.jmb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 166.Mächtel R., Gebhardt C., Cordes T. Konformationsbewegungen von aktiven Membrantransportern. BIOspektrum. 2018;24:495–497. [Google Scholar]