Abstract
Hosts respond to viral infection by expressing interferon-stimulated genes, of which IFITs are potent inhibitors of viral RNA translation. Johnson et al. (2018) solved the structure of the IFIT1-IFIT3 complex bound cap 0 RNA and explored their concerted antiviral activity.
A key aspect of cytosolic RNA-sensing machinery is the ability to discriminate self versus non-self RNA during viral infection. Many viruses often carry uncapped RNA (5′-triphosphorylated, 5′-ppp) or incompletely capped RNA, termed cap 0 (guanine N7-methylated but not ribose 2′-O-methylated), whereas higher eukaryotes contain fully methylated 5′-end-capped cellular mRNA, termed cap 1 or cap 2. The unique structure of the viral RNA 5′-cap allows cytosolic RNA sensors to preferentially detect foreign nucleic acids in order to restrict viral infection. Interferon-induced protein with tetratricopeptide repeats (IFIT) family members are one of the cytosolic RNA-sensing molecules that are rapidly induced during infection through interferon-dependent and -independent pathways. The human genome encodes five paralogues (IFIT1, IFIT2, IFIT3, and IFIT5, along with the uncharacterized IFIT1B), whereas mice possess six Ifit genes (Ifit1, Ifit2, and Ifit3, as well as the less-characterized Ifit1b, Ifit1c, and Ifit3b) (Daugherty et al., 2016). IFIT-family proteins share a common tetratricopeptide repeat (TPR) domain. This domain contains 34 amino acid repeats that fold into a helix-turn-helix structure and mediate protein-protein interactions. Mechanistically, IFIT-family proteins directly detect and sequester viral single-stranded RNA bearing 5′-ppp or cap 0, thereby out-competing the translation initiation complex for viral RNA translation or sequestering viral RNA from active replication (Daffis et al., 2010; Pichlmair et al., 2011). Interestingly, Pichlmair et al. (2011) have previously observed that the heteromeric complex formation of IFIT proteins is essential for an optimal and full IFIT-mediated antiviral effect. However, the detailed molecular basis and functional significance of IFIT complex formation has not been fully understood. In a study published in this issue of Immunity, Johnson et al. (2018) examined how IFIT1 cooperatively associates with IFIT3 to exert a potent antiviral activity against viral RNA lacking 2′-O methylation in their cap structure.
The authors demonstrated that IFIT1 specifically makes a tight complex with IFIT3, but not IFIT2 or IFIT5, by in vitro co-precipitation assays incorporating bacterially purified proteins. The high binding affinity in the IFIT1-IFIT3 complex was revealed by isothermal titration calorimetry to be in the low nanomolar range. The authors also found that the carboxyl-terminal domain (CTD) of IFIT3 is required for the tight interaction with IFIT1. Because the murine orthologue Ifit3 lacks the cognate CTD region, it does not bind Ifit1. This indicates that there are species-specific functions for the IFIT proteins. In the present study, Johnson et al. (2018) demonstrated the molecular basis of the heterotrimeric complex by resolving the X-ray co-crystal structure of IFIT1, the single stranded 5′-cap 0 RNA, and the CTD of IFIT3 at 2.55 Å resolution.
To define critical residues within the interface between IFIT1 and cap 0 RNA or the CTD of IFIT3, the authors performed hydrogen-deuterium exchange with mass spectrometry and computational analysis. IFIT1 preferentially captures viral RNA with cap 0 structure through a central RNA-binding channel that interacts with the triphosphate bridge via electrostatic interactions and a cap-binding pocket required for N7-methylated guanosine accommodation via hydrophobic interactions. The structure reported by Johnson et al. (2018) was compared with a previously described structure of the amino-terminal IFIT1 alone (Abbas et al., 2013). Johnson et al. (2018) clearly showed that the cap 0 RNA-bound IFIT1 undergoes several conformational rearrangements within subdomain II to compactly associate with the N7-methyl guanosine moiety of cap 0 RNA. The authors also confirmed that IFIT1 has a much higher binding affinity to cap 0 RNA than cap 1 or 5′ppp RNA, which is consistent with previous studies (Habjan et al., 2013).
The authors further examined the a helical TPR IFIT1 subdomains that contact the CTD of IFIT3 via hydrophobic interactions and hydrogen bonds. Critical residues within the interface between IFIT3 and IFIT1 were predicted and confirmed by mutagenesis studies. Most importantly, the IFIT3 interaction conferred specificity on IFIT1 for cap 0 RNA binding. Using quantitative filter-binding assays, the authors found that the IFIT1-IFIT3 complex has a higher binding affinity for cap 0 RNA than IFIT1 alone, whereas a weak association of IFIT1 with ppp-RNA is undetectable in the presence of IFIT3. To understand the structural effect of IFIT3 binding on IFIT1, the authors compared the trimeric structure with previously reported IFIT structures, including 5′-ppp RNA bound to IFIT5 (Abbas et al., 2013), cap 0 RNA-bound dimeric IFIT1, and monomeric mutant IFIT1 (L457E and L464E) (Abbas et al., 2017). There are obvious structural changes in the subdomain III of IFIT1 upon association with IFIT3, which contributes to the formation of a highly compact RNA-binding channel. On the basis of these findings, Johnson et al. (2018) propose that IFIT3 binding allosterically modulates the RNA-binding domains of IFIT1 and gives rise to specific recognition of cap 0 RNA.
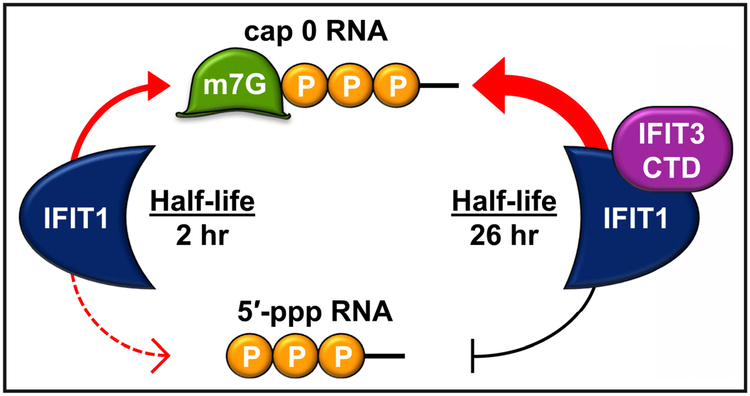
It has long been known that many viral families, including Flaviviridae, Coronaviridae, and Poxviridae, have evolved to evade host immune sensing by expressing a 2′-O-methyltransferase to modify their caps to have 2′-O-methylation (Daffis et al., 2010). Accordingly, mutant viruses, such as West Nile virus (WNV NS5 E218A) and Zika virus (ZIKV NS5 E218A), that lack 2′-O-methylation on their cap structure become susceptible to IFIT-mediated antiviral immune responses. Johnson et al. (2018) utilized these mutant viruses to study the functional significance of IFIT1-IFIT3 complex formation. IFIT1 or IFIT3 expression lowers infectivity of mutant WNV and ZIKV lacking 2′-O methylation of 5′-RNA caps, whereas wild-type viral infectivity was unaffected, indicating that IFIT1 and IFIT3 specifically target viruses bearing cap 0 RNA structure. Similar results were obtained with the alphavirus Venezuelan equine encephalitis virus (VEEV) TC 83 strain, which is susceptible to IFIT1 recognition (Hyde et al., 2014). Most notably, Johnson et al. (2018) found that in the presence of IFIT3 expression, IFIT1-mediated inhibition of infection with 2′-O-methylation-deficient WNV, ZIKV, or VEEV was enhanced. Accordingly, the antiviral effect of IFIT3 required IFIT1 expression, indicating that complex formation is necessary for viral restriction. Furthermore, IFIT1-binding-diminishing deletion or point mutations of the carboxyl-terminal tail of IFIT3 also failed to modulate IFIT1-mediated antiviral activity, supporting the biological relevance of the IFIT1-IFIT3 complex formation. The authors found that IFIT3 modulates IFIT1 by two mechanisms (Figure 1): IFIT3 binding not only increases the stability of IFIT1by extending its half-life in cells but also allosterically regulates the RNA-binding channel of IFIT1, leading to enhanced and preferential recognition of cap 0 RNA over 5′-ppp or cap 1 RNA.
Figure 1. The IFIT1-IFIT3 Complex Preferentially Binds to Cap 0 RNA.
On the left side of the panel, IFIT1 alone can bind to both cap 0 (guanine N7-methylated) and 5′-ppp RNA molecules, but the cap 0 binding has a higher affinity. On the right side is the complex of IFIT1 with the C-terminal domain (CTD) of IFIT3. This complex binds to cap 0 RNA with higher affinity than IFIT1 alone does and loses detectable binding to 5′-ppp RNA. The complex formation further increases IFIT1 stability.
Johnson et al. (2018) provide structural and mechanistic insight into how IFIT family proteins cooperativity enable the host to recognize specific viral RNA ligands to restrict infection. The authors point out that previous studies have demonstrated conflicting results regarding IFIT1 and IFIT3 antiviral activity in vitro and in vivo, and they propose possible explanations, including functional differences between human IFIT and murine Ifit as well as potential different experimental conditions, such as ectopic expression versus a doxycycline-inducible expressing system. Because numerous viruses have mechanisms to escape IFIT-mediated restriction and because the sequence homology between individual IFIT genes is low, it is possible that IFIT proteins might have multiple species-specific functions. A recent phylogenetic study revealed that mouse Ifit1 is actually an ortholog of human IFIT1B, not IFIT1 (Daugherty et al., 2016), and thus provided a possible explanation as to why Ifit3 binding activity is dispensable for Ifit1 function. However, resolving the remaining discrepancies in IFIT1-IFIT3-complex-mediated restriction of vesicular stomatitis virus containing cap 1 RNA requires further study. Besides the ability to sense viral RNA, IFIT proteins have been reported to achieve their antiviral activity through other mechanisms, such as binding to additional cellular factors (Hui et al., 2003; Liu et al., 2011). The work presented here raises the possibility that besides individual IFIT action, heteromeric IFIT complexes could add versatility and modularity to host antiviral activity. Indeed, the IFIT1-IFIT3 duo shows that when patrolling for viral RNA, two is better than one.
REFERENCES
- Abbas YM, Pichlmair A, Górna MW, Superti-Furga G, and Nagar B (2013). Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 494, 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas YM, Laudenbach BT, Martínez-Montero S, Cencic R, Habjan M, Pichlmair A, Damha MJ, Pelletier J, and Nagar B (2017). Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc. Natl. Acad. Sci. USA 114, E2106–E2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Schaller AM, Geballe AP, and Malik HS (2016). Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife 5, e14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil-Cruz C, Ziebuhr J, et al. (2013). Sequestration by IFIT1 impairs translation of 2’O-unmethylated capped RNA. PLoS Pathog. 9, e1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DJ, Bhasker CR, Merrick WC, and Sen GC (2003). Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem 278, 39477–39482. [DOI] [PubMed] [Google Scholar]
- Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C, Tonelli M, Paessler S, Takeda K, et al. (2014). A viral RNA structural element alters host recognition of nonself RNA. Science 343, 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B, VanBlargan LA, Xu W, White JP, Shan C, Shi P-Y, Zhang R, Adhikari J, Gross ML, Leung DW, et al. (2018). Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity 47, this issue, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Chen W, Wei B, Shan YF, and Wang C (2011). IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J. Immunol 187, 2559–2568. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Górna MW, Baumann CL, Burkard TR, Bürckstümmer T, Stefanovic A, Krieger S, Bennett KL, et al. (2011). IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol 12, 624–630. [DOI] [PubMed] [Google Scholar]