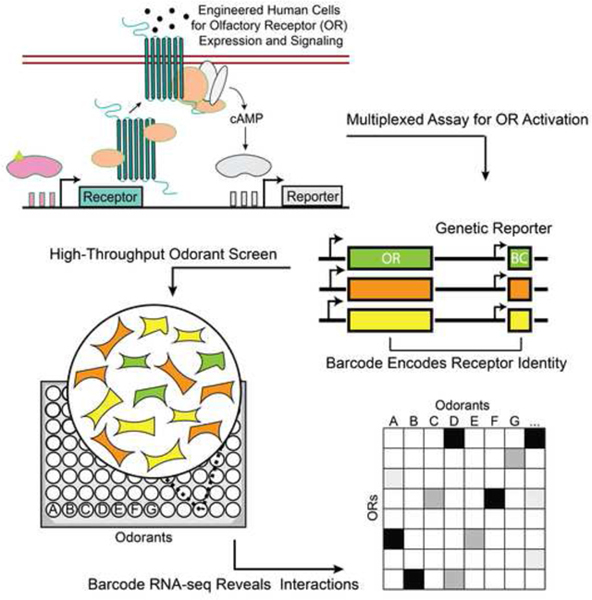
Summary:
G-protein coupled receptors (GPCRs) are central to how mammalian cells sense and respond to chemicals. Mammalian olfactory receptors (ORs), the largest family of GPCRs, mediate the sense of smell through activation by small molecules, though for most bonafide ligands have not been identified. Here, we introduce a platform to screen large chemical panels against multiplexed GPCR libraries using next-generation sequencing of barcoded genetic reporters in stably-engineered human cell lines. We mapped 39 mammalian olfactory receptors against 181 odorants and identified 79 interactions that have not been reported to our knowledge, including ligands for 15 previously orphaned receptors. This multiplexed receptor assay allows the cost-effective mapping of large chemical libraries to receptor repertoires at scale.
Keywords: Olfaction, GPCR, high throughput, NGS, multiplex, synthetic biology
Graphical abstract
Introduction
Interactions between small molecules and receptors underpin an organism’s ability to communicate with both its internal state and the environment. For many drugs and natural products, the ability to modulate many biological targets at once is crucial for their efficacy (Fang et al., 2017; Reddy and Zhang, 2013; Roth et al., 2004). Thus, to understand the effect of many small molecules, we need to comprehensively characterize their functional interactions with biological targets. This many-on-many problem is laborious to study one interaction at a time, and is especially salient in the mammalian sense of smell (Anighoro et al., 2014; Malnic et al., 1999).
Olfaction is mediated by a class of G protein-coupled receptors (GPCRs) known as olfactory receptors (ORs) (Buck and Axel, 1991). GPCRs are a central player in small molecule signaling and are currently targeted by 34% of US Food and Drug Administration (FDA) approved drugs (Hauser et al., 2017). ORs are a large family of class A GPCRs with approximately 396, 1130, and 1948 intact receptors in humans, mice, and elephants respectively (Niimura et al., 2014). Each OR can potentially interact with many odorants, and inversely, each odorant with many ORs. The majority of ORs have no known ligand (i.e., orphan) because de-ophanization is difficult. In particular, recapitulating mammalian GPCR function in vitro is challenging (Lu et al., 2003; Peterlin et al., 2014) and screening each receptor against each chemical individually becomes difficult and expensive to scale. In addition, no experimentally determined structures for any mammalian OR are available, hindering computational efforts to predict which odorants can activate each OR (Bushdid et al., 2018).
Most OR assays test chemicals against each receptor individually using transient mammalian cell line-based luciferase assays. Large-scale in vitro screens have been a useful tool for mapping chemicals to the receptors they activate (Mainland et al., 2014; Saito et al., 2009), as well as to better understand how genetic variation in ORs affect our ability to perceive odors (Mainland et al., 2014; Saito et al., 2009). Receptor-ligand pairs identified by these in vitro screens have also been shown to be activated in vivo by these same ligands. Various studies have examined in vivo murine olfactory sensory neuron activation by odorants, identified the OR expressed in these neurons, and verified the interaction in vitro (Jiang et al., 2015; Shirasu et al., 2014). Furthermore, allelic variation of ORs in humans corresponds with human odor perception and with in vitro data (Keller et al., 2007; Mainland et al., 2014; Menashe et al., 2007). However, given the size of mammalian OR repertoires and possible chemicals that might interact with them, screening these interactions one at a time is impractical.
Multiplexed assays, where the reporters can be measured in the same well, would increase the throughput. Multiplexed GPCR activity assays have previously been attempted (Botvinik and Rossner, 2012; Galinski et al., 2018). In these works, each cell expresses a single type of receptor and, upon activation, transcribes a short barcode sequence that identifies the particular receptor expressed in that cell. The enrichment of barcoded transcripts corresponding to each receptor’s activation are then measured by microarrays or next-generation sequencing. However, these assays are difficult to perform, especially in olfaction, for several reasons. First, ORs, like many GPCRs, are difficult to express in their non-native contexts and often require specialized accessory factors and signaling proteins to function heterologously (Zhuang and Matsunami, 2007). Second, transient transfection must be performed for tens to hundreds of individual cell lines each time an assay is performed. Thus, experimental protocols for such multiplexed screens are expensive, labor intensive, and often carried out in a low-throughput manner. Using stable lines would alleviate these burdens, but building stable and functional OR reporter lines is challenging and has only worked in one reported case for a single OR (Belloir et al., 2017; Cook et al., 2008).
Results
Here we report a new high-throughput screen to characterize small molecule libraries against mammalian OR libraries in multiplex (summarized in Fig. 1A). To do this, we developed both a stable cell line capable of functional OR expression (ScL21) and a multiplexed reporter for OR activity (Fig. S1A–S1E, S2A–S2D, Data S1). Activation of each OR induces G-protein signaling which leads to the expression of a genetic reporter transcript with a unique 15-nucleotide barcode sequence. The barcoded reporter and OR are cloned into the same vector, mapped to each other, and genomically integrated into a human cell line. Each barcode identifies the OR expressed in that cell; this enables OR activation to be measured by quantifying differential barcode expression with RNA-seq. This technology enables the simultaneous profiling of a single chemical’s activity against a library of receptors in a single well.
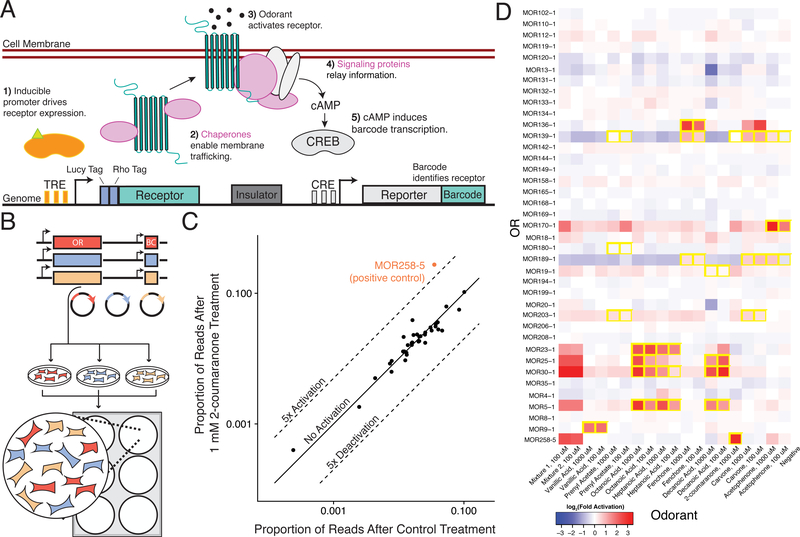
Figure 1. Overview of a Multiplexed Platform for Mammalian Olfactory Receptor Activation.
(A) Schematic of the synthetic circuit for stable OR expression and function in an engineered HEK293T cell line (ScL21). Heterologous accessory factors expressed include (pink): RTP1S, RTP2, Gαolf, and Ric8b. (B) Experimental workflow for OR library generation and pilot-scale screening. To perform assay, we cloned OR genes and barcodes into plasmids, engineered cell lines via individual transposition of plasmids, pooled cell lines and performed screen in 6-well plates. (C) The proportion of barcode reads for each OR after treatment with 1mM 2-coumaranone versus after Control treatment. Highlighted point is an OR known to be activated by 2-coumaranone. (D) Heatmap displaying 39 pooled receptors’ activity against 9 odorants and 2 mixtures (1: 2-coumaranone, Decanoic Acid, Acetophenone, 2: 2-coumaranone, Decanoic Acid). Interactions are colored by the log2-fold activation of the genetic reporter (see methods). Odorant interactions previously identified(Saito et al., 2009) are boxed in yellow.
To test the ability of the platform to simultaneously assay multiple receptors, we chose 42 phylogenetically divergent murine ORs with both known and unknown chemical specificities and created a library of OR-expressing cell lines (Figure S3A). To create the individual cell lines, we first cloned and mapped the ORs to their corresponding barcodes and transposed the plasmids individually into the genomes of ScL21 cells (Li et al., 2013). After selection we pooled the cell lines together, generating assay-ready libraries for repeated testing (Figure 1B). Of note, the receptor lines we pooled together were not isoclonal and likely contain a range of receptor/reporter copy number. Unlike a luciferase reporter assay, each well contains the entire OR library and a single chemical’s activity is measured against the entire library of ORs in a single well (Figures 1C, S3B–S3D). We seeded the cell library in 6-well culture dishes and assayed odorants known to activate specific ORs in our library. Analysis of the sequencing readout recapitulated previously identified odorant-receptor pairs (Saito et al., 2009), and chemical mixtures appropriately activated multiple ORs (Figure 1D). We found that the assay was robust to chemicals such as the adenylate cyclase stimulator, forskolin, which non-specifically induces barcode transcription independent of the OR each cell expresses (Figure S3E). This is likely because our library-based approach measures the relative activation of ORs to each other, normalizing any global effects.
Next, we adapted the platform for high-throughput screening in 96-well format. To decrease reagent cost and assay time, we optimized an in-lysate reverse transcription protocol and used dual indexing to uniquely link the barcode sequences to the correct well after samples were mixed for high throughput sequencing (see Methods). With these improvements, the assay is able to recapitulate dose-response curves for known odorant-receptor pairs (Figure S3E). We observed reproducible results between identically treated but biologically independent wells; all but 3 ORs passed quality filtering to obtain reliable estimates of activation (See Methods, Fig. S4A–S4F)
We subsequently screened 181 odorants with both known and unknown receptor specificity at three concentrations in triplicate against the 39-member OR cell library, or 81,012 wells if each combination had been tested individually including controls (Figure 2A,Tables S1 and S2). The odorants were chosen based on those available on hand and easily acquirable and therefore likely do not represent an unbiased sample of odorant space. Each 96-well plate in the assay contained independent positive control odorants and solvent (DMSO) for normalization (Figure S4E). We used a generalized linear model to determine OR-odorant interactions (see Methods) (McCarthy et al., 2012).
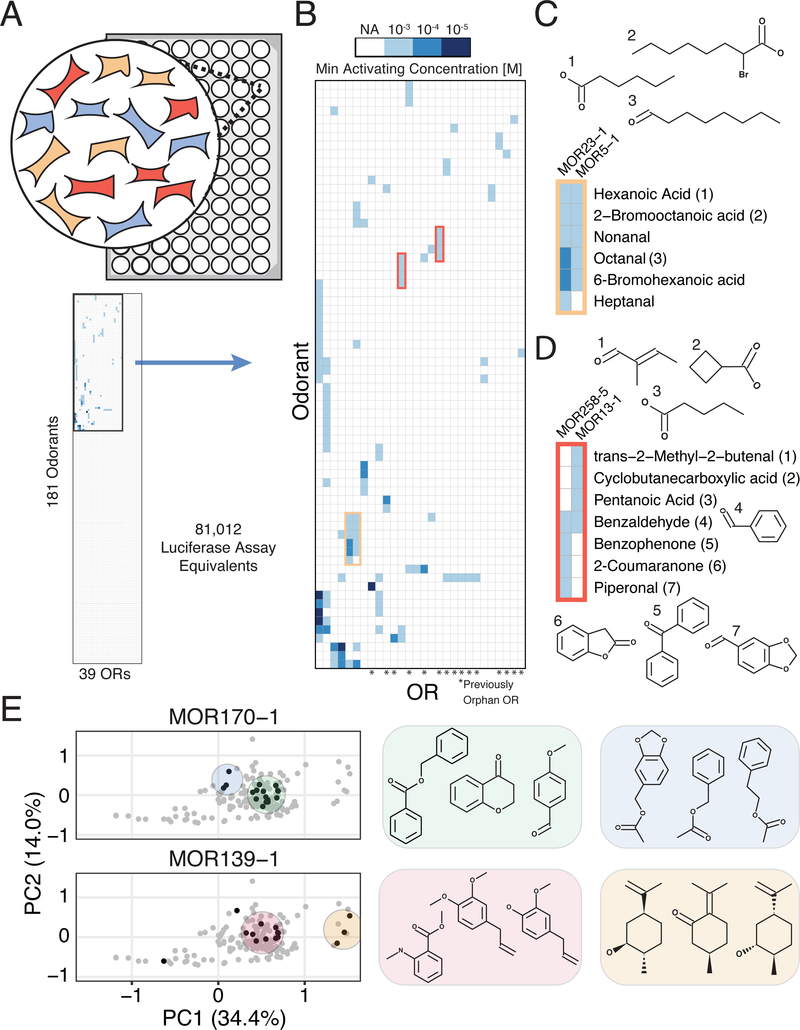
Figure 2. Large-Scale, Multiplexed Screening of Olfactory Receptor-Odorant Interactions.
(A) Overview of high-throughput screening platform. (Top) We optimized a protocol for and performed the screen in 96 well plates. We assayed the equivalent of 81,012 wells of a screen where interactions are tested individually, testing 181 odorants in three concentrations against 39 receptors. (Bottom) Graphic displaying the total interaction space traversed, highlighting the minor portion of the space where interactions are observed. (B) Heatmap of interactions from the screen clustered by odorant and receptor responses, and shaded by the minimum activating odorant concentration that triggered reporter activity. Only ORs and chemicals that registered at least one interaction are shown. (C) Chemical names and structures for odorants that activate MOR23–1 and MOR5–1. (D) Chemical names and structures for odorants that activate MOR258–5 and MOR13–1. (E) Chemical hits identified for MOR170–1 and MOR139–1 (black) mapped onto a PCA projection of the chemical space of our odorant panel (grey). Shaded areas highlight hits that cluster together in chemical space. See also Figures S5–S8.
As our goal was to identify as many interactions between olfactory receptors and chemicals as possible, we screened for a positive interaction at only three widely spaced odorant concentrations. Thus we did not fit a classic dose-response curve to such a small number of points. Instead, we use a semi-quantitative measure of the minimum concentration of odorant required to activate a particular receptor. As a semiquantitative readout, the minimum activating concentration (MAC) serves to indicate positive interactions and the approximate concentration ranges that should be used in further targeted assays. Additionally, the MAC metric attempts to reduce false positives by requiring that two different concentrations of a chemical or simply the highest concentration (1000 uM in our experiment) activates the receptor in order to qualify as activating. This metric is robust for monotonic dose-response curves but may fail for non-monotonic curves that decrease rapidly after saturation (Conolly and Lutz, 2004). Using this metric, we found 112 significant interactions (out of >7,000 combinations), of which 79 were previously unreported, and 24 that target 15 orphan receptors (Benjamini-Hochberg corrected FDR = 1%; Figures 2B, S5A, and Tables S3–S6) (Benjamini and Hochberg, 1995). Overall, 28 of 39 receptors were activated by at least one odorant, and 67 of 181 odorants activated at least one OR (Table S3).
To validate our assay, we compared results to a previous study and also analyzed individual interactions in a different experimental context. First, we chose 36 interactions (the number we could practically perform) with different fold inductions and p-values to retest individually in a previously developed transient OR activation system (Zhuang and Matsunami, 2008) (Figure S5B, S6). Of the 27 significant interactions at an FDR of 1%, 20 of them replicated in this orthogonal system, showing that the assay and MAC metric are mostly calling attention to real interactions. Notably, some of the seven interactions which did not replicate in this orthogonal system, appear to be true hits. For instance, our assay registered two hits for MOR19–1 with high chemical similarity (methyl salicylate and benzyl salicylate), suggesting they are likely not false positives (Figure S6). Additionally, three of nine interactions not passing the 1% FDR threshold showed activation in the orthogonal assay, indicating that a conservative FDR threshold likely generated some false negatives. A previous large-scale OR deorphanization study screened some of the same receptors and chemicals, and we found that 9/12 of their reported interactions with EC50 below 100μM were also detected in our platform, though we did not identify most of the previous low affinity interactions (Saito et al., 2009) (Figure S7A and S7B). Conversely, we also detected 14 positive interactions absent from the previous study. Finally, our assay replicated the vast majority of non-interacting odorant-OR pairs (493/507).
Using the data generated by this high throughput assay, we found that chemicals with similar features activate the same ORs, including those receptors we deorphanize in this study (Figure 2C). For example, the previously orphan MOR19–1 has clear affinity for the salicylate functional group, while MOR13–1 is activated by four chemicals with hydrogen bond accepting groups attached and--in three cases--to stiff non-rotatable scaffolds (Data S2). We also detect ORs with partial overlap in chemical specificity; MOR13–1 detects compounds with terminal carbonyls while MOR258–5 detects cyclic conjugated molecules (Figure 2D). Benzaldehyde, an intermediate size carbonyl, activates both ORs.
To more systematically understand how chemical similarity relates to receptor activation, we used a recently developed molecular autoencoder (Gómez-Bombarelli et al., 2018) to computationally map each tested chemical onto a ~292-dimensional continuous representation of chemical space and visualized the results with Principal Components Analysis (Figure S8). Chemicals for 11/17 multi-hit receptors cluster together across the first two principal components (Figure S9). For instance, of 13 aliphatic aldehydes or carboxylic acids with >5 carbons in our chemical panel, 10 activate MOR5–1 (Figure 2C, Data S2). This analysis also highlights the instances where ORs are sensitive to several distinct sets of chemicals (Figure 2E). For example, MOR139–1 is activated by compounds that belong to two distinct clusters: one with benzene rings and the other with cyclohexane rings, hinting at the selective features of these odorants. Similarly, MOR170–1 exhibits a broad activation pattern: this receptor responds to ~50% of all odorants in our panel that contains both a benzene ring, and either a carbonyl or ether group. Most of these odorants form a single cluster with the exception of the acetate compounds that form a separate cluster. Understanding the global chemical space that activates each OR establishes the groundwork for the prediction of novel odorant-OR interactions.
Discussion
In this study, we develop and validate an engineered, mammalian cell line and corresponding high throughput assay that enables the systematic interrogation of OR-chemical interactions in multiplex. This multiplexed screen decreases the cost and labor required per interaction and expands the number of possible interactions probed per experiment. For example, the large-scale screen conducted here was performed in 21 96-well plates and would have required ~800 plates if each interaction was screened individually. Aside from screening capacity, the assay is also resistant to compounds that non-specifically activate the reporter independently of the receptor because receptor activations are measured relative to each other. As the vast majority of ORs in humans and other animals remain orphans, a multiplexed screening platform is an enabling technology for ligand discovery and adds to our understanding odor perception.
However, this method still has several limitations. First, OR activity within our in vitro assay may not directly relate to the ORs ability to detect those chemicals in vivo. Recapitulating the endogenous expression levels of olfactory receptors and signaling pathways in vitro is difficult, and thus this assay is best-suited for identifying receptor-ligand interactions from the vast space of possible interactions. Indeed, previous interactions identified by in vitro assays have shown good correspondence with in vivo data (Jiang et al., 2015; Shirasu et al., 2014). Second, we found our assay was poor at recapitulating weak (EC50 > 0.3 mM) interactions found in transient assays (Fig. S6), likely because weak interactions require very high OR expression that are not achieved in our stable cell-lines. Lastly, as mentioned earlier, each OR must be expressed in sufficient numbers of cells in the mixed cell library to be accurately detected by the assay, and this becomes more critical as library sizes increases. We were unable to quantify three of receptors due to these issues, and this problem might be exasperated as we continue to scale the assay in the future.
Looking forward, we anticipate that this platform can be scaled and generalized to other applications. First, it could be used to test the 396-member human OR repertoire and comprehensively define OR response to any odorant of interest. Such scaling would require improving the number of cells assayed per well, further improving signal to noise, or using clonal instead of polyclonal cell lines. Second, such multiplexed assays could be extended to the ~400 non-olfactory GPCRs, many of which are clinically relevant (Hauser et al., 2017). This would require engineering of new multiplexed signaling reporters that these receptors use. Such genetic reporters exist for most other G protein subtypes (NFAT for Gq, SRE/CRE for Gi/o) and can be substituted into our system (Azimzadeh et al., 2017; Cheng et al., 2010). Finally, this technique can theoretically be applied to any receptor class with a genetic reporter, offering a scalable solution to generate large-scale datasets that will help guide both empirical and algorithmic efforts to better dissect the complex interactions between small molecules and biological targets (Colwell, 2018).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sriram Kosuri (sri@ucla.edu).
Experimental Model and Subject Details
Cell Culture
Human embryonic kidney cells 293T (HEK 293T) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin and streptomycin (Thermo Fisher Scientific). All cells were kept at 37°C in a humidified atmosphere containing 5% CO2 and were frozen in DMEM + 10% dimethyl sulfoxide (Sigma Aldrich). Trypsin-EDTA solution (Thermo Fisher Scientific) was used to detach cells from culture dishes.
Method Details
Summary of Reporter System Strategy
We first engineered a stable HEK293T-derived cell line, ScL21, capable of functionally expressing genomically-integrated ORs and responding to odorant stimuli by transcribing an RNA barcode. We found that multi-copy integration and inducible receptor expression are both essential for reporter activation, but individually neither of these features is sufficient to generate a response (Figure S1A and S1B).
To allow larger OR repertoires to be assayed, we added features known to improve OR function (Saito et al., 2004; Von Dannecker et al., 2005; Zhuang and Matsunami, 2007) (Figure S1C). We stably integrated a pool of 4 accessory factors at multi-copy under inducible expression into HEK293T-derived cell line stably expressing m2rtTA: Golf and Ric8b for signal transduction, and RTP1S and RTP2 to promote surface expression. To select a single line for further use (ScL21), we isolated clones and screened for robust activation of two ORs known to require accessory factors to function heterologously (Figure S1D and 1E). In summary, upon stable transposition of the OR/reporter construct into ScL21, doxycycline induces expression of the receptor and accessory factors (Figures 1A and S2A). Then, G protein signaling triggers genetic reporter expression and barcode transcription.
In constructing the multiplexed reporter, we incorporated multiple improvements to maximize signal-to-noise (Figures S2B–S2D). We fused two N-terminal protein trafficking tags to our receptors, the Rho and Lucy tags, previously shown to increase GPCR and OR surface expression (Krautwurst et al., 1998; Shepard et al., 2013). We partitioned sequence elements on the vector with CHS4 insulator sequences to reduce background reporter activation (Gaszner and Felsenfeld, 2006). Lastly, we added additional copies of the cAMP response element (CRE) to the genetic reporter in order to improve reporter signal. The OR expression and genetic reporter cassette along with these improvements were combined into a single transposable vector to speed cell line development (Data S1). We validated our system on three murine ORs with known ligands, and observed induction-and dose-dependent activation (Figure S2E). Notably, we observed that integrated OR constructs required higher odorant concentrations to achieve activation compared to transient transfection experiments, likely due to lower copy number (compare Figure S1C and S2E).
Odorant-Receptor Activation Luciferase Assay (Transient)
The Dual-Glo Luciferase Assay System (Promega) was used to measure OR-odorant responses as previously described(Saito et al., 2009). HEK293T (ATCC #11268) cells HEK293T-derived Hana3a cells(Zhuang et al., 2008) were plated in poly-D-lysine coated white 96-well plates (Corning) at a density of 7,333 cells per well in 100 ul DMEM+10% FBS (Thermo Fisher Scientific). 24 hours later, cells were transfected using lipofectamine 2000 (Thermo Fisher Scientific) with 5 ng/well of plasmids (pCI) encoding ORs and 10 ng/well of luciferase driven by a cyclic AMP response element (pCRE-Luc) or 10 ng/well of a plasmid encoding both the OR and the luciferase gene, and in both cases 5 ng/well of a plasmid (pRLSV40) encoding Renilla luciferase. Experiments conducted with accessory factors included 5 ng/well of plasmids (pCI) encoding RTP1S (Gene ID: 132112) and RTP2 (Gene ID: 344892). Inducibly expressed ORs were transfected with 1 ug/ml doxycycline (Sigma-Aldrich) added to the transfection media. 10–100 mM odorant stocks were established in DMSO or ethanol. 24 h after transfection, transfection medium was removed and replaced with 25 ul/well of the appropriate concentration of odorant diluted from the stocks into CD293 (Thermo Fisher Scientific). Four hours after odorant stimulation, the Dual-Glo Luciferase Assay kit was administered according to the manufacturer’s instructions. Luminescence was measured using the M1000 plate reader (Tecan). All luminescence values were normalized to Renilla luciferase activity to control for transfection efficiency in a given well. Data were analyzed with Microsoft Excel and R.
Odorant-Receptor Activation Luciferase Assay (Integrated)
HEK293T and HEK293T derived cells integrated with the combined receptor/reporter plasmids were plated at a density of 7333 cells/well in 100 uL DMEM+10in poly-D-lysine coated 96-well plates. 24 hours later, 1 ug/ml doxycycline was added to the well medium. Odorant stimulation, luciferase reagent addition, and luminescence measurements were carried out in the same manner as the transient assays. Constitutively expressed ORs were assayed in the same manner without doxycycline addition. Data were analyzed with Microsoft Excel and R.
Odor Stimulation and RNA Extraction for Pilot-Scale Multiplexed Odorant Screening
HEK293T and HEK293T-derived cells transposed with the combined receptor/reporter plasmid were plated at a density of 200k cells/well in a 6 well plate in 2 mL DMEM+10%FBS. 24 hours later, 1 ug/ml doxycycline was added to the well medium. 10–100 mM odorant stocks were diluted in DMSO or ethanol. 24 hours after doxycycline addition, odorants were diluted in OptiMEM and media was aspirated and replaced with 1 mL of the odorant-OptiMEM solution. 3 hours after odor stimulation, odor media was aspirated and 600 uL of buffer RLT (Qiagen) was added to each well. Cells were lysed with the Qiashredder Tissue and Cell Homogenizer (Qiagen),and RNA was purified using the RNEasy MiniPrep Kit (Qiagen) with the optional on-column DNAse step according to the manufacturer’s protocol.
Pilot Scale Library Preparation and RNA-seq
5 ug of total RNA per sample was reverse transcribed with Superscript IV (ThermoFisher) using a gene specific primer for the barcoded reporter gene (OL003). The reaction conditions are as follows: annealing: [65°C for 5 min, 0°C for 1 min] extension: [52°C for 60 min, 80°C for 10 min]. 10% of the cDNA library volumes were amplified for 5 cycles (OL004F and R) using HiFi Master Mix (Kapa Biosystems). The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 5 cycles of 98°C for 20 seconds, 59°C for 15 seconds, and 72°C for 10 seconds, followed by an extension of 72°C for 1 minute. The PCR products were purified using the DNA Clean & Concentrator kit (Zymo Research) into 10 uL and 1 uL of each sample was amplified (OL005F and R) using the SYBR FAST qPCR Master mix (Kapa Biosystems) with a CFX Connect Thermocycler (Biorad) to determine the number of PCR cycles necessary for library amplification. The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 40 cycles of 95°C for 3 seconds and 60°C for 20 seconds. After qPCR, 5 uL of the pre-amplified cDNA libraries were amplified a second time at the same cycling conditions as the first amplification with the same primers used for qPCR for 4 cycles greater than the previously determined Cq. The PCR products were then gel isolated from a 1% agarose gel with the Zymoclean Gel DNA Recovery Kit (Zymo Research). Library concentrations were quantified using a TapeStation 2200 (Agilent) and loaded at equimolar ratios onto a HiSeq 3000 with a 20% PhiX spike-in and sequenced with custom primers: Read 1 (OL003) and i7 Index (OL006).
Pilot Scale Data Analysis
To determine fold activation of each OR treated with each chemical, we first calculated the fraction of barcodes (composition) corresponding to each OR in the control treatment (DMSO). Then, we calculated the fold change in the composition of each OR in each a specific condition. As the barcode reads from activated ORs can dominate the composition of all reads and change the effective library size, we then normalized the activation of each OR by the median activation for each well. To be effective, this normalization assumes that fewer than half of the ORs are activated by an odorant.
OR Library Cloning
The backbone plasmid (all genetic elements except the OR and barcode) was created using isothermal assembly with the Gibson Assembly HiFi Mastermix (SGI-DNA). A short fragment was amplified with a primer containing 15 random nucleotides to create the barcode sequence (OL007F and R) using HiFi Master Mix. The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 35 cycles of 98°C for 20 seconds, 60°C for 15 seconds, and 72°C for 20 seconds, followed by an extension of 72°C for 1 minute. The amplicon and the backbone plasmid were digested with restriction enzymes MluI and AgeI (New England Biolabs) and ligated together with T4 DNA ligase (New England Biolabs). DH5α E.coli competent cells (New England Biolabs) were transformed directly into liquid culture with antibiotic to maintain the diversity of the barcode library.
OR genes were isolated from genomic DNA as described in a previous publication(Saito et al., 2009). OR genes were amplified individually with primers (OL008) adding homology to the barcoded backbone plasmid using HiFi Master Mix. The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 35 cycles of 98°C for 20 seconds, 61°C for 15 seconds, and 72°C for 30 seconds, followed by an extension of 72°C for 1 minute. The amplified ORs were purified with DNA Clean and Concentrator Kit (Zymo Research) and pooled together. The barcoded backbone plasmid was digested with NdeI and SbfI and the OR amplicon pool was cloned into it using isothermal assembly with the Gibson Assembly Hifi Mastermix. DH5α E.coli competent cells were transformed with the assembly and antibiotic resistant clones were picked and grown up in 96-well plates overnight. The plasmid DNA was prepped with the Zyppy-96 Plasmid Miniprep Kit (Zymo Research). Plasmids were Sanger sequenced (OL109–111) both to associate the barcode with the reporter gene and identify error-free ORs.
Single Copy Genomic Integration System Development
The H11 locus was edited using TALEN plasmids received from Addgene (#51554, #51555). HEK293T cells were seeded at a density of 75k cells in a 24-well plate. 24 hours after seeding cells were transfected with 50 ng LT plasmid, 50 ng RT plasmid, and 400 ng of the Linearized Landing Pad using Lipofectamine 2000. 2 days after transfection, cells were expanded to a 6-well plate and one day after expansion 500 ug/ml hygromycin B (Thermo FIsher Scientific) was added to the media. Cells were grown under selection for 10 days. After selection, cells were seeded in a 96-well plate at a density of 0.5 cells/well. Wells were examined for single colonies after 3 days and expanded to 24-well plates after 7 days. gDNA was purified using the Quick-gDNA Miniprep kit (Zymo Research) from the colonies and PCR was performed with Hifi Master Mix to ensure the landing pad was present at the correct locus. The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 35 cycles of 98°C for 20 seconds, 63°C for 15 seconds, and 72°C for 40 seconds, followed by an extension of 72°C for 2 minutes. To ensure a single landing pad was present per cell, HEK293T cell lines with both singly and doubly-integrated landing pads along with untransduced (WT) HEK293T cells were plated at 4 × 10^5 cells per 6-well. All landing pad cells were transfected the next day with 1.094 μg of both an attB-containing eGFP and mCherry donor plasmid and 0.3125 μg of the BxB1 expression vector or a pUC19 control. Two singly-integrated landing pad cell samples were also transfected with 2.1875 μg of either an attB-containing eGFP and mCherry donor plasmid with 0.3125 μg of the BxB1 expression vector. Cells were transfected at a 1:1.5 Duvenaud ratio with Lipofectamine 3000. 2 days later cells were passaged at 1:10 and were analyzed using flow cytometry 10 days later after 4 total passages. Samples were flown using the LSRII at the UCLA Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research Flow Cytometry Core. Cytometer settings were adjusted to the settings: FSC – 183 V, SSC – 227 V, PE-Texas Red – 336 V, Alexa Fluor 488 – 275 V.
Stable Single Copy OR Genomic Integration
HEK293T derived cells engineered to contain the Bxb1 Recombinase site at the H11 locus were seeded at a density of 350k cells in a 6-well plate (Corning). 24 hours after seeding cells were transfected with 2 ug Donor plasmid and 500 ng plasmid encoding the Bxb1 recombinase using Lipofectamine 3000 (Thermo Fisher Scientific). 3 days after transfection cells were expanded to a T-75 flask (Corning) and 8 ug/ml blasticidin (Thermo Fisher Scientific) was added one day after expansion. Cells were kept under selection 7–10 days and passaged twice 1:10 to ensure removal of transient plasmid DNA.
Stable Multi-Copy OR Genomic Integration
HEK293T cells and HEK293T-derived cells were seeded at a density of 350k cells/well in a 6-well plate in 2 mL DMEM+10% FBS. 24 hours after seeding, cells were transfected with plasmids encoding receptor/reporter transposon and the Super PiggyBac Transposase (Systems Bioscience) according to the manufacturer’s instructions(Li et al., 2013). 1 ug of transposon DNA and 200 ng of transposase DNA were transfected per well with Lipofectamine 3000 (Thermo Fisher Scientific). 3 days after transfection, cells were passaged 1:10 into a 6-well plate, and one day after passaging 8 ug/mL blasticidin were added to the cells. Cells were grown with selection for 7–10 days. The OR library was transposed individually and pooled together at equal cell numbers.
Accessory Factor Cell Line Generation
HEK293T cells stably expressing m2rtTA were transposed with plasmids encoding the accessory factor genes RTP1S, RTP2, Gαolf (Gene ID: 2774), and Ric8b (Gene ID: 237422) inducibly driven by the Tet-On promoter (see Supplementary Data 2). The 4 accessory factor plasmids were transfected at equimolar concentration according to the transposition protocol in the Stable Multi-Copy OR Genomic Integration section. Cells were selected with 2 ug/mL puromycin (Thermo Fisher). After selection, cells were seeded in a 96-well plate at a density of 0.5 cells/well. Wells were examined for single colonies after 3 days and expanded to 24-well plates after 7 days. Clones were screened for accessory factor expression by screening them for robust activation of MOR258–5/Olfr62 and OR7D4 with a transient luciferase assay (Supplementary Fig. 1). The clone with the highest fold activation for both receptors and no salient growth defects was established for the multiplexed screen.
Transposon Copy Number Verification
gDNA was purified from cells transposed with the OR reporter vector and from cells containing the single copy landing pad with the Quick-gDNA Miniprep kit. 50 ng of gDNA was amplified with primers annealing to the regions of the exogenous DNA from each sample using the SYBR FAST qPCR Master Mix (Kapa Biosystems) on a CFX Connect Thermocycler using the manufacturer’s protocol. The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 40 cycles of 95°C for 3 seconds and 60°C for 20 seconds. Cq values for the transposed ORs were normalized to the single copy landing pad to determine copy number.
Lentiviral Transduction-Stable m2rtTA Expression
The pM2rtTA lentivirus was a gift from the lab of Donald Kohnd(Das et al., 2016). Lentiviral vector was produced by transient transfection of 293T cells with lentiviral transfer plasmid, pCMVΔR8.91 and pCAGGS-VSV-G using Mirus TransIT-293. HEK293T cells were transduced to express the m2rtTA transcription factor (Tet-On) at 50% confluency and seeded one day prior to transduction. Clones were isolated by seeding cells in a 96-well plate at a density of 0.5 cells/well. Wells were examined for single colonies after 7 days and expanded to 24 well plates. Clones were assessed for m2rtTA expression by screening for robust activation of MOR42–3 (Gene ID: 257926) with a transient luciferase assay.
High-throughput Odorant Screening
The OR library cell line was thawed from a liquid nitrogen frozen stock into a T-225 flask (Corning) three days before seeding into a 96-well plate for screening. The library was seeded at 6,666 cells per well in 100 uL of DMEM+10% FBS. 24 hours later a working concentration of 1 ug/mL of doxycycline in DMEM+10% FBS was added to the wells. 24 hours after induction, the media was removed from each plate and replaced with 25 ul of odorant diluted in OptiMEM. Each odor was added at three different concentrations (10 uM, 100 uM, 1 mM) in triplicate with the same amount of final DMSO (1%). Each plate contained two control odorants at a three concentration (10 uM, 100 uM, 1 mM) in triplicate and three wells containing 1% DMSO dissolved in media. The library was incubated with odorants for three hours in a cell culture incubator with the lids removed. This was because lids of 96-well culture plates still allow air flow between wells and we thought that cross contamination of odorants might occur more readily if the odorants were trapped in the small volume of the plate.
After odor incubation, media was pipetted out of the plates and cells were lysed by adding 25 uL of ice-cold Cells-to-cDNA II Lysis Buffer (Thermo Fisher) and pipetting up and down to homogenize and lyse cells. The lysate was then heated to 75°C for 15 minutes and flash frozen with liquid nitrogen and kept at −80C until further processing. Then 0.5 uL DNase I (New England Biolabs) was added to lysate, and incubated at 37°C for 15 minutes. To anneal the RT primer, 5 ul of lysate from each well was combined with 2.5 uL of 10 mM dNTPs (New England Biosciences), 1 uL of 2 uM gene specific RT primer (OL003), and 1.5 uL of H2O. The reaction was heated to 65°C for 5 min and cooled back down to 0°C. After annealing, 1 uL of M-MuLV Reverse Transcriptase (Enzymatics), 1 uL of buffer, and 0.25 ul of RNase Inhibitor (Enzymatics) were added to each reaction. Reactions were incubated at 42°C for 60 min and the RT enzyme was heat inactivated at 85°C for 10 min.
For each batch, qPCR was performed on a few wells (OL005F and OL013) with SYBR FAST qPCR Mastermix to determine the number of cycles necessary for PCR based library preparation. The reaction and cycling conditions are optimized as follows: 95°C for 3 minutes, 40 cycles of 95°C for 3 seconds and 60°C for 20 seconds. After qPCR, 5 uL of each RT reaction was combined with 0.4 uL of 10 uM primers containing sequencing adaptors (OL005F and OL013), 10 uL of NEB-Next Q5 Mastermix (New England Biosciences) and 4.2 uL H2O, the PCR was carried out according to the manufacturer’s protocol. The forward primer contains the P7 adaptor sequence and an index identifying the well in the assay and the reverse primer contains the P5 adaptor sequence and an index identifying the plate in the assay. PCR products were pooled together by plate and purified with the DNA Clean and Concentrator Kit. Library concentrations were quantified using a TapeStation 2200 and a Qubit (Thermo Fisher). The libraries were sequenced with two index reads and a single end 75-bp read on a NextSeq 500 in high-output mode (Illumina).
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of Next-Generation Sequencing Data
Samples were demultiplexed index adapters unique for each well (5’ end) and unique for each plate (3’ end). The well barcodes followed the 7bp indexing scheme in (Illumina Sequencing Library Preparation for Highly Multiplexed Target Capture and Sequencing Matthias Meyer, Martin Kircher, Cold Spring Harb Protoc; 2010; doi:10.1101/pdb.prot5448). The plate indexing scheme followed the Illumina indexing scheme (see Table S4). After demultiplexing, 15bp barcode sequences were counted with only exact matches tabulated.
Count data was then analyzed using the differential expression package EdgeR(McCarthy et al., 2012). To filter out ORs with low representation, we empirically set a cutoff that an OR had to contain at least 0.5% of the reads from atleast 400 of the 1954 test samples. This filtered out 3 of 42 ORs which were underrepresented in the cell library (MOR172–1, MOR176–1 and MOR181–1).
Normalization factors were then determined using the EdgeR package function calcNormFactors which implements a trimmed mean of m-factors to correct for the compositional nature of the sequencing data. Then a negative binomial generalized loglinear model was fit to the read counts for each barcode. To do this, the glmFit function from EdgeR was used, and the dispersion set to the tagwise dispersion, since only 39 ORs were present but with many replicates. By fitting a generalized linear model to the count data to determine if odorants stimulated specific ORs, we were able to determine both the mean activation for each OR-odorant interaction and the p-value. We then corrected this p-value for multiple hypothesis testing using the p.adjust function with the Benjamini & Hochberg correction(Benjamini and Hochberg, 1995), yielding a False Discovery Rate (FDR). We set a cutoff of 1% to determine interacting odorant-OR pairs. For each interaction between an odorant and an OR, we further required that an OR-odorant interaction was above the cutoff in two different concentrations of odorant or in just the 1000 uM concentration. For each interaction passing these filters, we further determined Minimum Activating Concentration (MAC) by simply choosing the lowest concentration that was below the 1% FDR.
Molecular Autoencoder
We used an autoencoder as described in Gómez-Bombarelli et al. to visualize OR-chemical interactions in the context of chemical space(Gómez-Bombarelli et al., 2018). Following the authors’ advice, we used a reimplementation of autoencoder as the original implementation requires a defunct Python package (https://github.com/chembl/autoencoder_ipython). This model comes pre-trained to a validation accuracy of 0.99 on the entire ChEMBL 23 database with the exception of molecules whose SMILES are longer than 120 characters. We used this pretrained model to generate the latent representations of the 168 chemicals for which we could find SMILES representations and 250,000 randomly sampled chemicals from ChEMBL 23. We then used scikit-learn(Pedregosa et al., 2011) to perform principal component analysis to project the resulting matrix onto two dimensions.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| DH5α E.coli Competent Cells | New England Biolabs | C2987H |
| pM2rtTA lentivirus | Gift from Donald Kohn’s lab | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Doxycycline hyclate | Sigma-Aldrich | D9891 |
| Blasticidin S-HCl | Thermo Fisher Scientific | A1113902 |
| Puromycin Dihydrochloride | Thermo Fisher Scientific | A1113802 |
| Nonanedioic Acid | Sigma-Aldrich | 246379 |
| 2-coumaranone | Sigma-Aldrich | 124591 |
| Decanoic Acid | Sigma-Aldrich | C1875 |
| Acetophenone | Sigma-Aldrich | A10701 |
| Prenyl Acetate | Sigma-Aldrich | W420201 |
| Vanilic Acid | Sigma-Aldrich | H36001 |
| Forskolin | Sigma-Aldrich | F3917 |
| Critical Commercial Assays | ||
| Dual-Glo Luciferase Assay System | Promega | E2920 |
| White Poly-D-Lysine Coated 96-well Plates | VWR | 89092–710 |
| Lipofectamine 2000 | Thermo-Fisher Scientific | 11668027 |
| CD293 Medium | Thermo-Fisher Scientific | 11913019 |
| OptiMEM Medium | Thermo-Fisher Scientific | 31985062 |
| Qiashredder Tissue and Cell Homogenizer | Qiagen | 79654 |
| RNEasy MiniPrep Kit | Qiagen | 74104 |
| Superscript IV | Thermo-Fisher Scientific | 18090010 |
| HiFI MasterMix | KAPA Biosystems | KR0370 |
| ZymoClean Gel DNA Recovery Kit | Zymo Research | D4001T |
| Gibson Assembly HiFI Mastermix | SGI-DNA | Quote Needed |
| MluI Restriction Enzyme | New England Biolabs | R3198 |
| AgeI Restriction Enzyme | New England Biolabs | R3552 |
| T4 DNA ligase | New England Biolabs | M0202S |
| DNA Clean and Concentrator Kir | Zymo Research | D4013 |
| Zyppy-96 Plasmid Miniprep Kit | Zymo Research | D4041 |
| Lipofectamine 3000 | Thermo Fisher Scientific | L3000008 |
| Quick-gDNA Miniprep kit | Zymo Research | D4074 |
| SYBR FAST qPCR Master Mix | Kapa Biosystems | KR0389 |
| Cells-to-cDNA II Lysis Buffer | Thermo Fisher Scientific | AM8723 |
| DNase I | New England Biolabs | M0303S |
| dNTPs | New England Biolabs | N0447 |
| M-MuLV Reverse Transcriptase | Enzymatics | P7040 |
| Rnase Inhibitor | Enzymatics | Y9240 |
| NEB-Next Q5 Mastermix | New England Biolabs | M0544 |
| Experimental Models: Cell Lines | ||
| Human: 293T Cells | ATCC | CRL-3216 |
| Oligonucleotides | ||
| Full list of primers is presented in Table S4 | ||
| Recombinant DNA | ||
| pCRE-Luc | Stratagene | 219076 |
| pRL-SV40 | Promega | E2231 |
| pCI | Promega | E1731 |
| Super PiggyBac Transposase | Systems Bioscience | PB210PA-1 |
| Software and Algorithms | ||
| sklearn | Pedregosa et al., 2011 | http://scikit-learn.org |
| EdgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
Acknowledgements:
We thank the Kosuri Lab for helpful discussions, the UCLA Broad Stem Cell Research Center Sequencing Core, and the Technology Center for Genomics and Bioinformatics for providing next-generation sequencing. We thank Mengjue Ni, Jeong Hoon Ko, and Aaron Cooper for expert technical assistance. We thank Jason Kakoyiannis for providing perfumes as a gift.
Funding: National Science Foundation, Brain Initiative (1556207 to S.K and 1556207/151801 to H.M.), the Jane Coffin Child Memorial Foundation Postdoctoral Fellowship (R.J.), Ruth L. Kirschstein National Research Service Award (GM007185 to N.L.), the USPHS National Research Service Award (5T32GM008496 to E.J.), the NIH (DP2GM114829 to S.K. and DC014423 to H.M.) and UCLA.
Footnotes
Data and materials availability: Processed data and analysis scripts are available on https://github.com/KosuriLab/olfaction. Raw data is available with accession number PRJNA521800. Plasmids are available from Addgene and cell lines upon request.
Declaration of Interests:
E.J. and N.L. are employed by and hold equity and R.J., J.B. and S.K. consult for and hold equity in Octant Inc. to which patent rights based on this work have been licensed.
All other authors declare that they have no competing interests.
DATA AND SOFTWARE AVAILABILITY
All data and analysis scripts are available on https://github.com/KosuriLab/olfaction. Plasmids are available from Addgene and cell lines upon request.
References:
- Anighoro A, Bajorath J, and Rastelli G. (2014). Polypharmacology: challenges and opportunities in drug discovery. J. Med. Chem 57, 7874–7887. [DOI] [PubMed] [Google Scholar]
- Azimzadeh P, Olson JA Jr, and Balenga N. (2017). Reporter gene assays for investigating GPCR signaling. Methods Cell Biol. 142, 89–99. [DOI] [PubMed] [Google Scholar]
- Belloir C, Miller-Leseigneur M-L, Neiers F, Briand L, and Le Bon A-M (2017). Biophysical and functional characterization of the human olfactory receptor OR1A1 expressed in a mammalian inducible cell line. Protein Expr. Purif 129, 31–43. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol 57, 289–300. [Google Scholar]
- Botvinik A, and Rossner MJ (2012). Linking cellular signalling to gene expression using EXT-encoded reporter libraries. Methods Mol. Biol 786, 151–166. [DOI] [PubMed] [Google Scholar]
- Buck L, and Axel R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- Bushdid C, de March CA, Matsunami H, and Golebiowski J. (2018). Numerical Models and In Vitro Assays to Study Odorant Receptors. In Olfactory Receptors: Methods and Protocols, F.M. Simoes de Souza, and G. Antunes, eds. (New York, NY: Springer New York), pp. 77–93. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Garvin D, Paguio A, Stecha P, Wood K, and Fan F. (2010). Luciferase Reporter Assay System for Deciphering GPCR Pathways. Curr. Chem. Genomics 4, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell LJ (2018). Statistical and machine learning approaches to predicting proteinligand interactions. Curr. Opin. Struct. Biol 49, 123–128. [DOI] [PubMed] [Google Scholar]
- Conolly RB, and Lutz WK (2004). Nonmonotonic dose-response relationships: mechanistic basis, kinetic modeling, and implications for risk assessment. Toxicol. Sci 77, 151–157. [DOI] [PubMed] [Google Scholar]
- Cook BL, Ernberg KE, Chung H, and Zhang S. (2008). Study of a synthetic human olfactory receptor 17–4: expression and purification from an inducible mammalian cell line. PLoS One 3, e2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Tenenbaum L, and Berkhout B. (2016). Tet-On Systems For Doxycycline-inducible Gene Expression. Curr. Gene Ther 16, 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Liu C, Wang Q, Lin P, and Cheng F. (2017). In silico polypharmacology of natural products. Brief. Bioinform [DOI] [PubMed] [Google Scholar]
- Galinski S, Wichert SP, Rossner MJ, and Wehr MC (2018). Multiplexed profiling of GPCR activities by combining split TEV assays and EXT-based barcoded readouts. Sci. Rep 8, 8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, and Felsenfeld G. (2006). Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet 7, 703–713. [DOI] [PubMed] [Google Scholar]
- Gómez-Bombarelli R, Wei JN, Duvenaud D, Hernández-Lobato JM, Sánchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP, and Aspuru-Guzik A. (2018). Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent Sci 4, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, and Gloriam DE (2017). Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov 16, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, and Matsunami H. (2015). Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat. Neurosci 18, 1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, and Matsunami H. (2007). Genetic variation in a human odorant receptor alters odour perception. Nature 449, 468–472. [DOI] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, and Reed RR (1998). Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95, 917–926. [DOI] [PubMed] [Google Scholar]
- Li X, Burnight ER, Cooney AL, Malani N, Brady T, Sander JD, Staber J, Wheelan SJ, Joung JK, McCray PB, Jr, et al. (2013). piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U. S. A 110, E2279–E2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Echeverri F, and Moyer BD (2003). Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic 4, 416–433. [DOI] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WLL, Zhuang H, et al. (2014). The missense of smell: functional variability in the human odorant receptor repertoire. Nat. Neurosci 17, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, and Buck LB (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Abaffy T, Hasin Y, Goshen S, Yahalom V, Luetje CW, and Lancet D. (2007). Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 5, e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, and Touhara K. (2014). Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24, 1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- Peterlin Z, Firestein S, and Rogers ME (2014). The state of the art of odorant receptor heterologously : a report from the orphanage. J. Gen. Physiol 143, 527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, and Zhang S. (2013). Polypharmacology: drug discovery for the future. Expert Rev. Clin. Pharmacol 6, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Sheffler DJ, and Kroeze WK (2004). Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov 3, 353–359. [DOI] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, and Matsunami H. (2004). RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, and Mainland JD (2009). Odor coding by a Mammalian receptor repertoire. Sci. Signal 2, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard BD, Natarajan N, Protzko RJ, Acres OW, and Pluznick JL (2013). A cleavable N-terminal signal peptide promotes widespread olfactory receptor surface expression in HEK293T cells. PLoS One 8, e68758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu M, Yoshikawa K, Takai Y, Nakashima A, Takeuchi H, Sakano H, and Touhara K. (2014). Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron 81, 165–178. [DOI] [PubMed] [Google Scholar]
- Von Dannecker LEC, Mercadante AF, and Malnic B. (2005). Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactoryspecific G-protein Galphaolf. J. Neurosci 25, 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H, and Matsunami H. (2007). Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem 282, 15284–15293. [DOI] [PubMed] [Google Scholar]
- Zhuang H, and Matsunami H. (2008). Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc 3, 1402–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.