Summary
Most genetic variants that contribute to disease1 are challenging to correct efficiently and without excess byproducts2–5. Here we describe prime editing, a versatile and precise genome editing method that directly writes new genetic information into a specified DNA site using a catalytically impaired Cas9 fused to an engineered reverse transcriptase, programmed with a prime editing guide RNA (pegRNA) that both specifies the target site and encodes the desired edit. We performed >175 edits in human cells including targeted insertions, deletions, and all 12 types of point mutations without requiring double-strand breaks or donor DNA templates. We applied prime editing in human cells to correct efficiently and with few byproducts the primary genetic causes of sickle cell disease (requiring a transversion in HBB) and Tay-Sachs disease (requiring a deletion in HEXA), to install a protective transversion in PRNP, and to precisely insert various tags and epitopes into target loci. Four human cell lines and primary post-mitotic mouse cortical neurons support prime editing with varying efficiencies. Prime editing shows higher or similar efficiency and fewer byproducts than homology-directed repair, complementary strengths and weaknesses compared to base editing, and much lower off-target editing than Cas9 nuclease at known Cas9 off-target sites. Prime editing substantially expands the scope and capabilities of genome editing, and in principle can correct up to 89% of known genetic variants associated with human diseases.
The ability to make virtually any targeted change in the genome of any living cell or organism is a longstanding aspiration of the life sciences. Despite rapid advances in genome editing technologies, the majority of the >75,000 known human genetic variants associated with diseases1 remain difficult to correct or install in most therapeutically relevant cell types (Fig. 1a). Programmable nucleases such as CRISPR-Cas9 make double-strand DNA breaks (DSBs) that can disrupt genes by inducing mixtures of insertions and deletions (indels) at target sites2–4. DSBs, however, are associated with undesired outcomes including complex mixtures of products, translocations5, and p53 activation6,7. Moreover, the vast majority of pathogenic alleles arise from specific insertions, deletions, or base substitutions that require more precise editing technologies to correct (Fig. 1a, Supplementary Discussion). Homology-directed repair (HDR) stimulated by DSBs8 has been widely used to install precise DNA changes. HDR, however, relies on exogenous donor DNA repair templates, typically generates an excess of indels from end-joining repair of DSBs, and is inefficient in most therapeutically relevant cell types (T cells and some types of stem cells being important exceptions)9,10. While enhancing the efficiency and precision of DSB-mediated editing remains the focus of promising efforts11–15, these challenges motivate the exploration of alternative precision genome editing strategies.
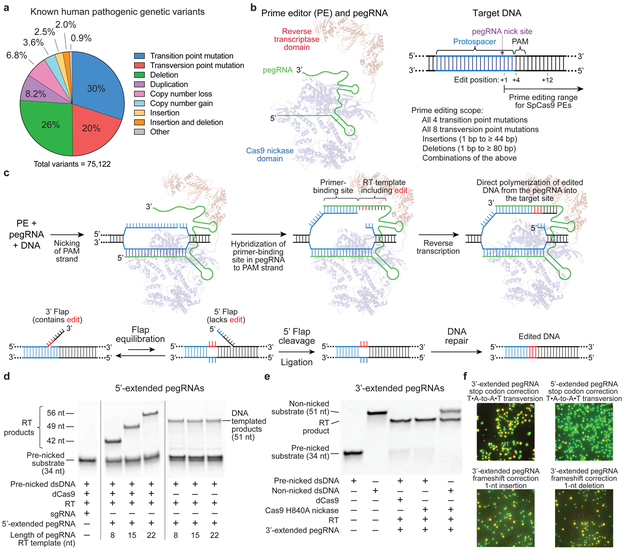
Figure 1. Overview of prime editing and feasibility studies in vitro and in yeast cells.
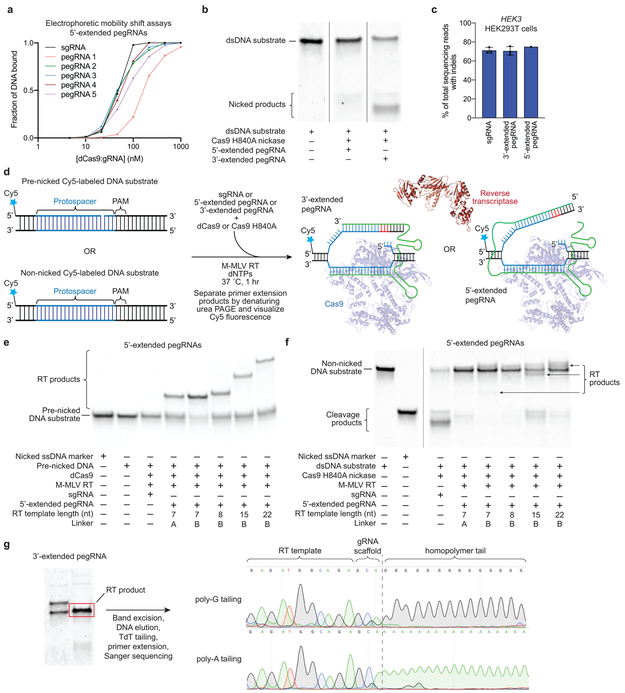
(a) The 75,122 known pathogenic human genetic variants in ClinVar (accessed July, 2019), classified by type. (b) A prime editing complex consists of a prime editor (PE) protein containing an RNA-guided DNA-nicking domain, such as Cas9 nickase, fused to a reverse transcriptase domain and complexed with a prime editing guide RNA (pegRNA). The PE:pegRNA complex enables a variety of precise DNA edits at a wide range of positions. (c) The PE:pegRNA complex binds the target DNA and nicks the PAM-containing strand. The resulting 3’ end hybridizes to the primer-binding site, then primes reverse transcription of new DNA containing the desired edit using the RT template of the pegRNA. Equilibration between the edited 3’ flap and the unedited 5’ flap, cellular 5’ flap cleavage and ligation, and DNA repair results in stably edited DNA. (d) In vitro primer extension assays with 5’-extended pegRNAs, pre-nicked dsDNA substrates containing 5’-Cy5 labeled PAM strands, dCas9, and a commercial M-MLV RT variant (RT, Superscript III). dCas9 was complexed with pegRNAs, then added to DNA substrates along with the indicated components. After 1 hour, reactions were analyzed by denaturing PAGE, visualizing Cy5 fluorescence. (e) Primer extension assays performed as in (d) using 3’-extended pegRNAs pre-complexed with dCas9 or Cas9 H840A nickase, and pre-nicked or non-nicked dsDNA substrates. (f) Yeast colonies transformed with GFP–mCherry fusion reporter plasmids edited in vitro with pegRNAs, Cas9 nickase, and RT. Plasmids containing nonsense or frameshift mutations between GFP and mCherry were edited with pegRNAs that restore mCherry translation via transversion, 1-bp insertion, or 1-bp deletion. GFP and mCherry double-positive cells (yellow) reflect successful editing. Images in (d-f) are representative of n=2 independent replicates. For gel source data, see Supplementary Figure 1.
Base editing can efficiently install the four transition mutations (C→T, G→A, A→G, and T→C) without requiring DSBs in many cell types and organisms, including mammals16–19, but cannot currently perform the eight transversion mutations (C→A, C→G, G→C, G→T, A→C, A→T, T→A, and T→G), such as the T•A-to-A•T mutation needed to directly correct the most common cause of sickle cell disease (HBB E6V). In addition, no DSB-free method has been reported to perform targeted deletions, such as the removal of the 4-base duplication that causes Tay-Sachs disease (HEXA 1278+TATC), or targeted insertions, such as the 3-base insertion required to directly correct the most common cause of cystic fibrosis (CFTR ΔF508). Targeted transversions, insertions, and deletions thus are difficult to install or correct efficiently and without excess byproducts in most cell types, even though they collectively account for most known pathogenic alleles (Fig. 1a).
Here we describe the development of prime editing, a “search-and-replace” genome editing technology that mediates targeted insertions, deletions, all 12 possible base-to-base conversions, and combinations thereof in human cells without requiring DSBs or donor DNA templates. Prime editors (PEs), initially exemplified by PE1, use a reverse transcriptase (RT) fused to an RNA-programmable nickase and a prime editing guide RNA (pegRNA) to directly copy genetic information from an extension on the pegRNA into the target genomic locus. PE2 uses an engineered RT to increase editing efficiencies, while PE3 nicks the non-edited strand to induce its replacement and further increase editing efficiency, typically to 20-50% with 1-10% indel formation in human HEK293T cells. Prime editing offers much lower off-target activity than Cas9 at known Cas9 off-target loci, far fewer byproducts and higher or similar efficiency compared to Cas9-initiated HDR, and complementary strengths and weaknesses compared to base editors. By enabling precise targeted insertions, deletions, and all 12 possible classes of point mutations without requiring DSBs or donor DNA templates, prime editing has the potential to advance the study and correction of the vast majority of pathogenic alleles.
Results
Prime editing strategy
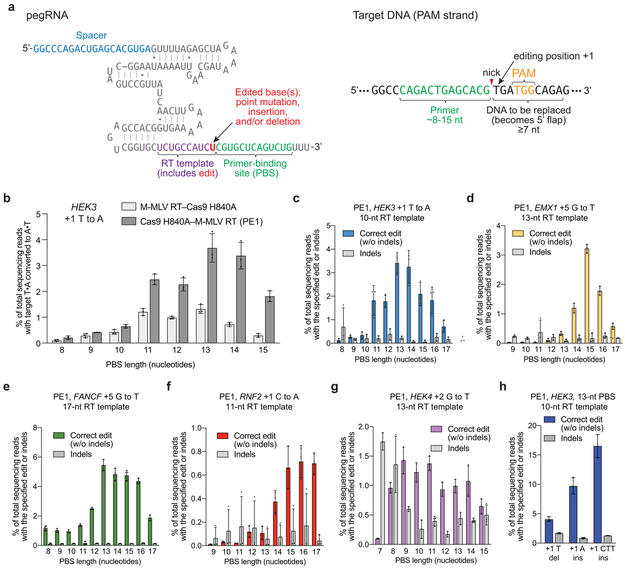
Cas9 targets DNA using a guide RNA containing a spacer sequence that hybridizes to the target DNA site2–4,20,21. We envisioned engineering guide RNAs that both specify the DNA target and contain new genetic information that replaces target DNA nucleotides. To transfer information from these engineered guide RNAs to target DNA, we proposed that genomic DNA, nicked at the target site to expose a 3’-hydroxyl group, could be used to prime the reverse transcription of an edit-encoding extension on the engineered guide RNA (hereafter referred to as the prime editing guide RNA, or pegRNA) directly into the target site (Fig. 1b,c, Supplementary Discussion).
These initial steps result in a branched intermediate with two redundant single-stranded DNA flaps: a 5’ flap that contains the unedited DNA sequence, and a 3’ flap that contains the edited sequence copied from the pegRNA (Fig. 1c). While hybridization of the perfectly complementary 5’ flap to the unedited strand is likely to be thermodynamically favored, 5’ flaps are the preferred substrate for structure-specific endonucleases such as FEN122, which excises 5’ flaps generated during lagging-strand DNA synthesis and long-patch base excision repair. Alternatively, the redundant unedited DNA may be removed by 5’ exonucleases such as EXO123. We reasoned that preferential 5’ flap excision and 3’ flap ligation could drive the incorporation of the edited DNA strand, creating heteroduplex DNA containing one edited strand and one unedited strand (Fig. 1c). DNA repair to resolve the heteroduplex by copying the information in the edited strand to the complementary strand would permanently install the edit (Fig. 1c). Based on a similar strategy we developed to favorably resolve heteroduplex DNA during base editing16–18, we hypothesized that nicking the non-edited DNA strand might bias DNA repair to preferentially replace the non-edited strand.
Validation in vitro and in yeast
First, we tested if the 3’ end of the PAM-containing DNA strand cleaved by Cas9’s RuvC nuclease domain is sufficiently accessible to prime reverse transcription. We designed pegRNAs by adding to sgRNAs a primer binding site (PBS) that allows the 3’ end of the nicked DNA strand to hybridize to the pegRNA, and a RT template containing the desired edit (Fig. 1c). We constructed candidate pegRNAs by extending sgRNAs on either end with a PBS sequence (5-6 nucleotides, nt) and an RT template (7-22 nt), and confirmed that 5’-extended pegRNAs support Cas9 binding to target DNA in vitro, and that both 5’-extended and 3’-extended pegRNAs support Cas9-mediated DNA nicking in vitro and DNA cleavage in mammalian cells (Extended Data Fig. 1a–c). Next, we tested the compatibility of these candidate pegRNAs with reverse transcription using pre-nicked 5’-Cy5-labeled dsDNA substrates, catalytically dead Cas9 (dCas9), and a commercial Moloney murine leukemia virus (M-MLV) RT variant (Extended Data Fig. 1d). When all components were present, the labeled DNA strand was efficiently converted into longer DNA products with gel mobilities consistent with reverse transcription along the RT template (Fig. 1d, Extended Data Fig. 1d–e). Omission of dCas9 led to nick translation products from RT-mediated DNA polymerization on the DNA template, with no pegRNA information transfer. No DNA polymerization products were observed when the pegRNA was replaced by a conventional sgRNA (Fig. 1d). These results demonstrate that nicked DNA exposed by dCas9 is competent to prime reverse transcription from a pegRNA.
Next, we tested non-nicked dsDNA substrates with a Cas9 H840A nickase that nicks the PAM-containing strand2. In these reactions, 5’-extended pegRNAs generated reverse transcription products inefficiently (Extended Data Fig. 1f), but 3’-extended pegRNAs enabled efficient Cas9 nicking and reverse transcription (Fig. 1e). The use of 3’-extended pegRNAs generated only a single apparent product, despite the theoretical possibility that reverse transcription could terminate anywhere within the pegRNA. DNA sequencing of reactions with Cas9 nickase, RT, and 3’-extended pegRNAs revealed that the complete RT template sequence was reverse transcribed into the DNA substrate (Extended Data Fig. 1g). These experiments establish that 3’-extended pegRNAs can direct Cas9 nickase and template reverse transcription in vitro.
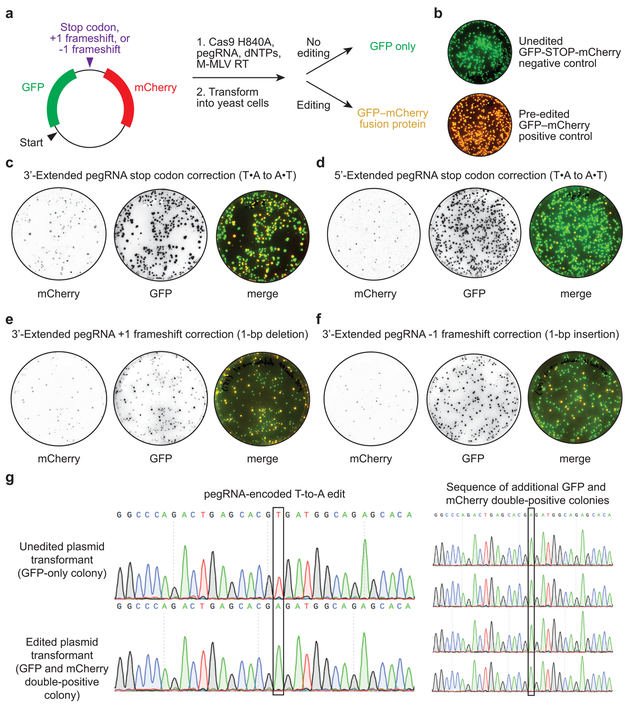
To evaluate eukaryotic cell DNA repair outcomes of 3’ flaps produced by pegRNA-programmed reverse transcription in vitro, we performed in vitro prime editing on reporter plasmids, then transformed the reaction products into yeast cells (Extended Data Fig. 2). We constructed reporter plasmids encoding EGFP and mCherry separated by a linker containing an in-frame stop codon, +1 frameshift, or −1 frameshift. When plasmids were edited in vitro with Cas9 nickase, RT, and 3’-extended pegRNAs encoding a transversion that corrects the premature stop codon, 37% of yeast transformants expressed both GFP and mCherry (Fig. 1f, Extended Data Fig. 2). Editing reactions with 5’-extended pegRNAs yielded fewer GFP and mCherry double-positive colonies (9%). Productive editing was also observed using 3’-extended pegRNAs that insert a single nucleotide (15%) or delete a single nucleotide (29%) to correct frameshift mutations (Fig. 1f, Extended Data Fig. 2). These results demonstrate that DNA repair in eukaryotic cells can resolve 3’ DNA flaps from prime editing to incorporate precise transversions, insertions, and deletions.
Prime editor 1 (PE1)
Encouraged by these observations, we sought to develop a prime editing system with a minimum number of components capable of editing genomic DNA in mammalian cells. We transfected HEK293T cells with one plasmid encoding a fusion of wild-type M-MLV RT through a flexible linker to either terminus of Cas9 H840A nickase, and a second plasmid encoding a pegRNA (Extended Data Fig. 3a). Initial attempts led to no detectable editing.
Extension of the PBS in the pegRNA to 8-15 bases, however, led to detectable installation of a transversion at the HEK293 site 3 (hereafter referred to as HEK3) target site, with higher efficiencies when the RT was fused to the C-terminus of Cas9 nickase compared to N-terminal RT–Cas9 nickase fusions (Extended Data Fig. 3b). These results suggest that wild-type M-MLV RT fused to Cas9 requires longer PBS sequences for genome editing in human cells compared to what is required in vitro using the commercial variant of M-MLV RT supplied in trans. We designated this M-MLV RT fused to the C-terminus of Cas9 H840A nickase as PE1.
We tested the ability of PE1 to introduce transversion point mutations at four additional genomic sites specified by the pegRNA (Fig. 2a). Editing efficiency at these sites was dependent on PBS length, with maximal editing efficiencies reaching 0.7-5.5% (Fig. 2a). Indels from PE1 were minimal, averaging 0.2±0.1% for the five sites under conditions that maximized each site’s editing efficiency (Extended Data Fig. 3a–f). PE1 also mediated targeted insertions and deletions with 4-17% efficiency at the HEK3 locus (Fig. 2a). These findings establish the ability of PE1 to directly install targeted transversions, insertions, and deletions without requiring DSBs or DNA templates.
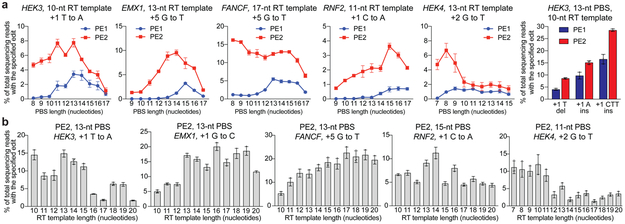
Figure 2. Prime editing of genomic DNA in human cells by PE1 and PE2.
(a) Use of an engineered M-MLV reverse transcriptase (D200N, L603W, T306K, W313F, T330p) in PE2 substantially improves prime editing efficiencies at five genomic sites in HEK293T cells, and small insertion and small deletion edits at HEK3. (b) PE2 editing efficiencies with varying RT template lengths at five genomic sites in HEK293T cells. Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Prime editor 2 (PE2)
We hypothesized that engineering the RT in PE1 might improve the efficiency of DNA synthesis during prime editing. M-MLV RT mutations that increase thermostability24,25, processivity24, and DNA:RNA substrate affinity26, and that inactivate RNaseH activity27 have been reported. We constructed 19 PE1 variants containing a variety of RT mutations to evaluate their editing efficiency in human cells.
First, we investigated M-MLV RT variants that support reverse transcription at elevated temperatures24. Introduction of D200N+L603W+T330P into M-MLV RT, hereafter referred to as M3, led to a 6.8-fold average increase in transversion and insertion editing efficiency across five genomic loci in HEK293T cells compared to PE1 (Extended Data Fig. 4).
We tested additional RT mutations that were previously shown to enhance binding to template:PBS complex, enzyme processivity, and thermostability26. Among the 14 additional mutants analyzed, adding T306K and W313F to M3 improved editing efficiency an additional 1.3-fold to 3.0-fold for six transversion or insertion edits across five genomic sites (Extended Data Fig. 4). This pentamutant RT incorporated into PE1 (Cas9 H840A–M-MLV RT D200N+L603W+T330P+T306K+W313F) is hereafter referred to as PE2.
PE2 installs single-nucleotide transversion, insertion, and deletion mutations with substantially higher efficiency than PE1, and is compatible with shorter PBS sequences, consistent with enhanced engagement of transient genomic DNA:PBS complexes (Fig. 2a). On average, PE2 led to a 1.6- to 5.1-fold improvement in prime editing point mutation efficiency over PE1. PE2 also performed targeted insertions and deletions more efficiently than PE1 (Fig. 2a, Extended Data Fig. 4d).
Optimization of pegRNAs
We systematically probed the relationship between pegRNA structure and PE2 editing efficiency. Priming regions with lower G/C content generally required longer PBS sequences, consistent with the energetic requirements of hybridization of the nicked DNA strand to the pegRNA PBS (Fig. 2a). No PBS length or G/C content level was strictly predictive of editing efficiency, suggesting that other factors such as DNA primer or RT template secondary structure also influence editing activity. We recommend starting with a PBS length of ~13 nt, and testing different PBS lengths if the priming region deviates from ~40-60% G/C.
Next, we systematically evaluated pegRNAs with RT templates 10-20 nt at five genomic target sites using PE2 (Fig. 2b), and with RT templates up to 31 nt at three genomic sites (Extended Data Fig. 5a–c). As with PBS length, RT template length also could be varied to maximize prime editing efficiency, although many RT template lengths ≥10 nt perform comparably. Since some target sites preferred longer RT templates (>15 nt) (FANCF, EMX1), while other loci preferred shorter RT templates (HEK3, HEK4 (HEK293 site 4)) (Fig. 2b), we recommend starting with ~10-16 nt and testing shorter and longer RT templates during pegRNA optimization.
Importantly, RT templates that place a C adjacent to the 3’ hairpin of the sgRNA scaffold generally resulted in lower editing efficiency (Extended Data Fig. 5a–c). We speculate that a C as the first nucleotide of the 3’ extension can disrupt guide RNA structure by pairing with G81, which normally forms a pi stack with Y1356 in Cas9 and a non-canonical base pair with sgRNA A6828. Since many RT template lengths support prime editing, we recommend designing pegRNAs such that the first base of the 3’ extension is not C.
Prime editor 3 systems (PE3, PE3b)
Resolution of heteroduplex DNA from PE2 containing one edited and one non-edited strand determines long-term editing outcomes. Previously, to optimize base editing we used Cas9 nickase to nick the non-edited strand, directing DNA repair to that strand using the edited strand as a template16–18. To apply this strategy to enhance prime editing, we tested nicking the non-edited strand using the Cas9 H840A nickase already present in PE2 and a simple sgRNA (Fig. 3a). Since the edited DNA strand is also nicked to initiate prime editing, we tested a variety of non-edited strand nick locations to minimize DSBs that lead to indels.
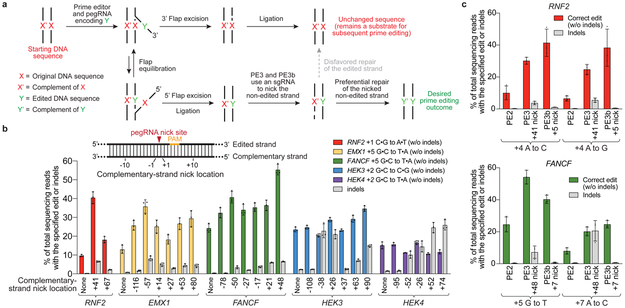
Figure 3. PE3 and PE3b systems nick the non-edited strand to increase prime editing efficiency.
(a) Overview of prime editing by PE3. After initial synthesis of the edited strand, 5’ flap excision leaves behind a DNA heteroduplex containing one edited strand and one non-edited strand. Mismatch repair resolves the heteroduplex to give either edited or non-edited products. Nicking the non-edited strand favors repair of that strand, resulting in preferential generation of duplex DNA containing the desired edit. (b) The effect of complementary strand nicking on prime editing efficiency and indel formation. “None” refers to PE2 controls, which do not nick the complementary strand. (c) Comparison of editing efficiencies with PE2, PE3, and PE3b (edit-specific complementary strand nick). Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
We first tested this strategy, designated PE3, at five genomic sites in HEK293T cells using sgRNAs that induce nicks 14-116 nt away from the site of the pegRNA-induced nick. In four of the five sites tested, nicking the non-edited strand increased editing efficiency by 1.5- to 4.2-fold compared to PE2, to as high as 55% (Fig. 3b). While the optimal nicking position varied depending on the genomic site (Supplementary Discussion), nicks positioned 3’ of the edit ~40-90 bp from the pegRNA-induced nick generally increased editing efficiency (averaging 41%) without excess indel formation (6.8% average indels for the sgRNA resulting in the highest editing efficiency) (Fig. 3b). We recommend starting with non-edited strand nicks ~50 bp from the pegRNA-mediated nick, and testing alternative nick locations if indel frequencies exceed acceptable levels.
Nicking the non-edited strand only after edited strand flap resolution should minimize the presence of concurrent nicks, minimizing DSB and indel formation. To achieve this goal, we designed sgRNAs with spacers that match the edited strand, but not the original allele. Using this “PE3b” strategy, mismatches between the spacer and the unedited allele should disfavor sgRNA nicking until after editing of the PAM strand takes place. PE3b resulted in 13-fold lower average indels (0.74%) compared to PE3, without any evident decrease in editing efficiency (Fig. 3c). When the edit lies within a second protospacer, we recommend the PE3b approach.
Together, these findings establish that PE3 systems improve editing efficiencies ~3-fold compared with PE2, albeit with a higher range of indels than PE2. When it is possible to nick the non-edited strand with an sgRNA that requires editing before nicking, the PE3b system offers PE3-like editing levels while greatly reducing indel formation.
To demonstrate the targeting scope and versatility of prime editing with PE3, we performed all 24 possible single-nucleotide substitutions across the +1 to +8 positions (counting the first base 3’ of the pegRNA-induced nick as position +1) of the HEK3 target site using PE3 and pegRNAs with 10-nt RT templates (Fig. 4a). These 24 edits collectively cover all 12 possible transition and transversion mutations, and proceeded with editing efficiencies (containing no indels) averaging 33±7.9%, with 7.5±1.8% average indels.
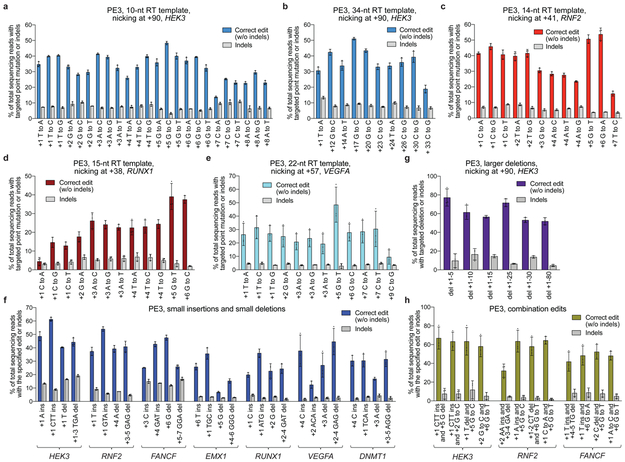
Figure 4. Targeted insertions, deletions, and all 12 types of point mutations with PE3 at seven endogenous genomic loci in HEK293T cells.
(a) All 12 types of single-nucleotide edits from position +1 to +8 of the HEK3 site using a 10-nt RT template, counting the first nucleotide following the pegRNA-induced nick as position +1. (b) Long-range PE3 edits at HEK3 using a 34-nt RT template. (c-e) PE3-mediated transition and transversion edits at the specified positions for (c) RNF2, (d) RUNX1, and (e) VEGFA. (f) Targeted 1- and 3-bp insertions, and 1- and 3-bp deletions with PE3 at seven endogenous genomic loci. (g) Targeted precise deletions of 5-80 bp at HEK3. (h) Combination edits at three endogenous genomic loci. Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Importantly, long-distance RT templates can also give rise to efficient prime editing. Using PE3 with a 34-nt RT template, we installed point mutations at positions +12, +14, +17, +20, +23, +24, +26, +30, and +33 in the HEK3 locus with 36±8.7% average efficiency and 8.6±2.0% indels (Fig. 4b). Other RT templates ≥30 nt at three other genomic sites also support prime editing (Extended Data Fig. 5a–c). Since an NGG PAM on either DNA strand occurs on average every ~8 bp, far less than edit-to-PAM distances that support efficient prime editing, prime editing is not substantially constrained by the availability of a nearby PAM sequence, in contrast to other precision editing methods11,15,16. Given the presumed relationship between RNA secondary structure and prime editing efficiency, when designing pegRNAs for long-range edits we recommend testing RT templates of various lengths and, if necessary, sequence compositions (e.g., using synonymous codons).
To further test the scope and limitations of PE3 for introducing point mutations, we tested 72 additional edits covering all possible types of point mutations across six additional genomic target sites (Fig. 4c–e, Extended Data Fig. 5d–f). Editing efficiency averaged 25±14%, while indel formation averaged 8.3±7.5%. Since the pegRNA RT template includes the PAM sequence, prime editing can induce PAM sequence changes. In these cases, we observed higher editing efficiency (averaging 39±9.7%) and lower indel generation (averaging 5.0±2.9%) (Fig. 4, mutations at +5 or +6), potentially due to the inability of Cas9 nickase to re-bind and nick the edited strand prior to the repair of the complementary strand. We recommend editing the PAM, in addition to other desired changes, whenever possible.
Next, we performed 28 targeted small insertions and small deletions at seven genomic sites using PE3 (Fig. 4f). Targeted 1-bp and 3-bp insertions proceeded with an average efficiency of 32±9.8% and 39±16%, respectively. Targeted 1-bp and 3-bp deletions were also efficient, averaging 29±14% and 32±11% editing, respectively. Indel generation (beyond the target insertion or deletion) averaged 6.8±5.4%. Since insertions and deletions between positions +1 and +6 alter PAM location or structure, we speculate that insertions or deletions at these positions are more efficient by preventing re-engagement of the edited strand.
We also tested PE3 for its ability to mediate larger precise deletions of 5 bp to 80 bp at the HEK3 site (Fig. 4g). We observed very high editing efficiencies (52-78%) for precise 5-, 10-, 15-, 25-, and 80-bp deletions, with indels averaging 11±4.8%. Finally, we tested the ability of PE3 to mediate 12 combinations of insertions, deletions, and/or point mutations across three genomic sites. These combination edits were also very efficient, averaging 55% editing with 6.4% indels (Fig. 4h). Together, the 156 distinct edits in Fig. 4 and Extended Data Fig. 5d–f establish the versatility, precision, and targeting flexibility of PE3 systems.
Prime editing compared with base editing
Cytidine base editors (CBEs) and adenine base editors (ABEs) can install transition mutations efficiently and with few indels16–18. The application of base editing can be limited by unwanted bystander edits from the presence of multiple cytidine or adenine bases within the base editing activity window16–18,29, or by the absence of a PAM positioned ~15±2 nt from the target nucleotide16,30. We anticipated that prime editing could complement base editing when bystander edits are unacceptable, or when the target site lacks a suitably positioned PAM.
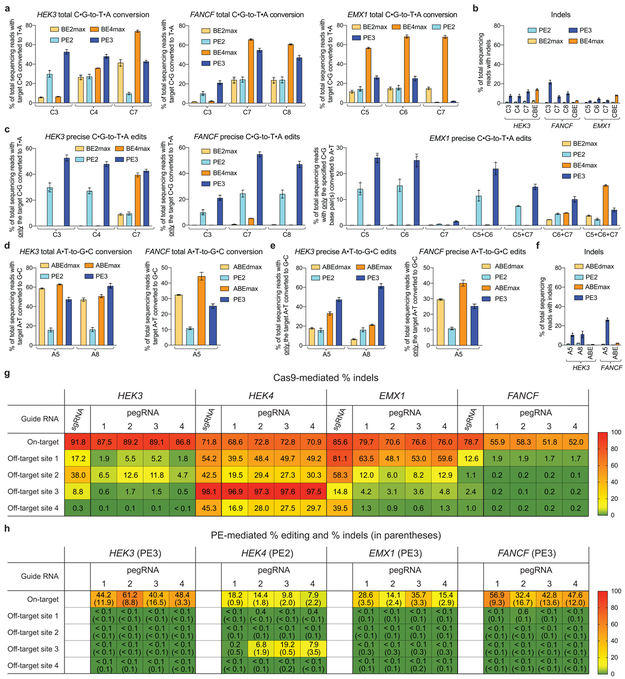
We compared PEs and CBEs at three genomic loci that contain multiple target cytosines in the canonical base editing window (protospacer positions 4-8, counting the PAM as positions 21-23) using current-generation CBEs31 without or with nickase activity (BE2max and BE4max, respectively), or using analogous PE2 and PE3 prime editing systems. Among the nine total cytosines within the base editing windows of the three sites, BE4max yielded 2.2-fold higher average total C•G-to-T•A conversion than PE3 for bases in the center of the base editing window (protospacer positions 5-7, Extended Data Fig. 6a). However, PE3 outperformed BE4max by 2.7-fold at cytosines positioned outside the center of the base editing window. Overall, indel frequencies for PE2 were very low (averaging 0.86±0.47%), and for PE3 were similar to or modestly higher than that of BE4max (PE3: 2.5-21%; BE4max: 2.5-14%) (Extended Data Fig. 6b).
For installation of precise edits (with no bystander editing), the efficiency of prime editing greatly exceeded that of base editing at the above sites, which like most genomic DNA sites contain multiple cytosines within the base editing window. BE4max generated few products containing only the single target base pair conversion with no bystander edits. In contrast, prime editing at this site could be used to selectively install a C•G-to-T•A edit at any position or combination of positions (Extended Data Fig. 6c).
We also compared nicking and non-nicking adenine base editors (ABEs) with PE3 and PE2, with similar results (Extended Data Fig. 6d–f, Supplementary Discussion). Collectively, these results indicate that base editing and prime editing offer complementary strengths and weaknesses for making targeted transition mutations. When a single target nucleotide is present within the base editing window, or when bystander edits are acceptable, current base editors are typically more efficient and generate fewer indels than prime editors. When multiple cytosines or adenines are present and bystander edits are undesirable, or when PAMs that position target nucleotides for base editing are not available, prime editors offer substantial advantages.
Off-target prime editing
Prime editing requires target DNA:pegRNA spacer complementary for the Cas9 domain to bind, target DNA:pegRNA PBS complementarity to initiate pegRNA-templated reverse transcription, and target DNA:RT product complementarity for flap resolution. To test if these three distinct DNA hybridization steps reduce off-target prime editing compared to editing methods that only require target:guide RNA complementarity, we treated HEK293T cells with PE3 or PE2 and 16 total pegRNAs that target four genomic loci, each of which have at least four well-characterized Cas9 off-target sites32,33. We also treated cells with Cas9 nuclease and the same 16 pegRNAs, or with Cas9 and four sgRNAs targeting the same four protospacers (Supplementary Table 1).
Consistent with previous studies32, Cas9 and sgRNAs targeting HEK3, HEK4, EMX1, and FANCF modified the top four known Cas9 off-target loci for each sgRNA with an average frequency of 16±16%, 60±26%, 48±28%, and 4.3±5.6%, respectively (Extended Data Fig. 6g). Cas9 with pegRNAs modified on-target sites with similar efficiency as Cas9+sgRNAs, while Cas9+pegRNAs modified off-target sites at 4.4-fold lower average efficiency than Cas9+sgRNAs.
Strikingly, PE3 or PE2 with the same 16 pegRNAs containing these four target spacers resulted in detectable off-target editing at only 3/16 off-target sites, with only 1/16 showing off-target editing efficiency ≥1% (Extended Data Fig. 6h). Average off-target prime editing for pegRNAs targeting HEK3, HEK4, EMX1, and FANCF at the top four known Cas9 off-target sites for each protospacer was <0.1%, <2.2±5.2%, <0.1%, and <0.13±0.11%, respectively (Extended Data Fig. 6h). Notably, at the HEK4 off-target 3 site that Cas9+pegRNA1 edits with 97% efficiency, PE2+pegRNA1 results in only 0.2% off-target editing despite sharing the same pegRNA, demonstrating how the two additional hybridization events required for prime editing can greatly reduce off-target modification. Taken together, these results suggest that prime editing induces much lower off-target editing than Cas9 at known Cas9 off-target sites.
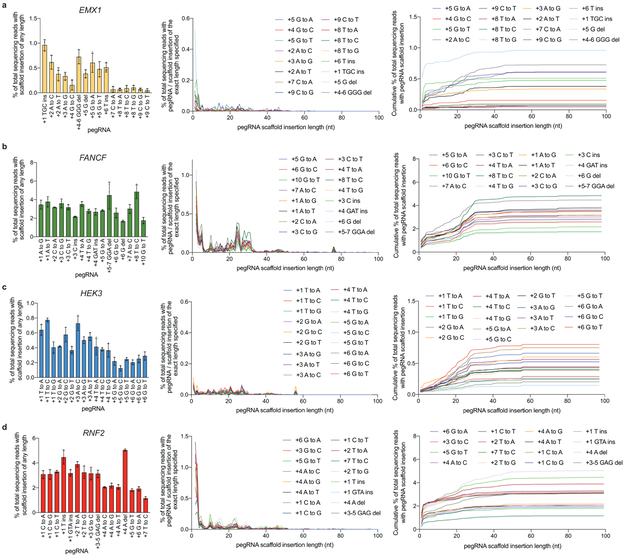
Reverse transcription of 3’-extended pegRNAs in principle can proceed into the guide RNA scaffold, resulting in scaffold sequence insertion that contributes to indels at the target locus. We analyzed 66 PE3 editing experiments at four loci in HEK293T cells and observed 1.7±1.5% average total insertion of any number of pegRNA scaffold nucleotides (Extended Data Fig. 7). We speculate that inaccessibility of the guide RNA scaffold to reverse transcription due to Cas9 domain binding, and cellular excision of the mismatched 3’ end of 3’ flaps that extend into the pegRNA scaffold, minimize products that incorporate pegRNA scaffold nucleotides.
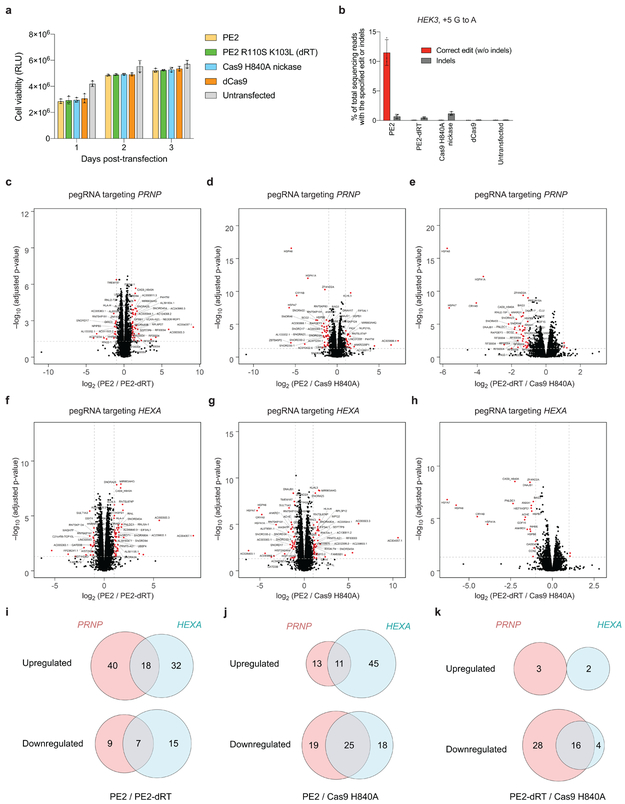
The presence of endogenous human reverse transcriptases from retroelements34 and telomerase suggests RT activity is not inherently toxic to human cells. Indeed, we observed no differences in HEK293T cell viability expressing dCas9, Cas9 H840A nickase, PE2, or PE2 with R110S+K103L mutations (PE2-dRT) that inactivate RT and abolish prime editing35 (Extended Data Fig. 8a,b). To evaluate cellular transcriptome changes from prime editing, we performed RNA-seq on HEK293T cells expressing PE2, PE2-dRT, or Cas9 H840A nickase together with a PRNP-targeting or HEXA-targeting pegRNA (Extended Data Fig. 8c–k), and observed that active PE2 minimally perturbs the transcriptome relative to Cas9 nickase or a control lacking active RT (Supplementary Discussion).
Prime editing pathogenic mutations
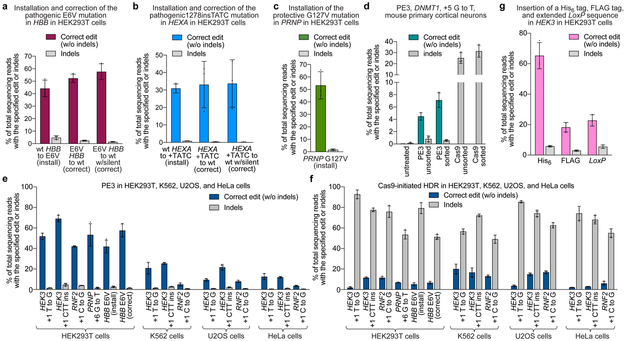
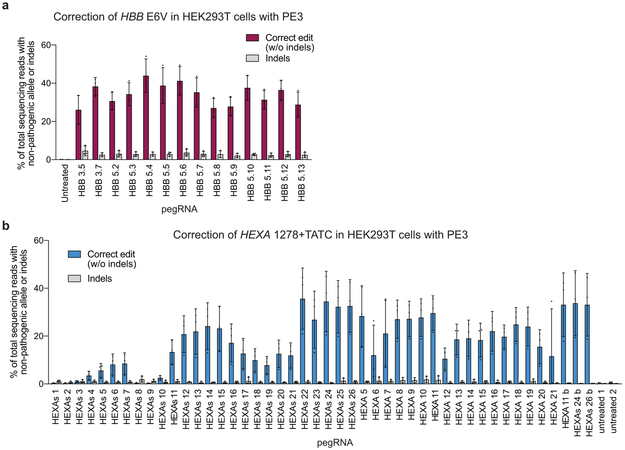
We tested the ability of PE3 to directly install or correct in human cells transversion, insertion, and deletion mutations that cause genetic diseases. Sickle cell disease is caused by a A•T-to-T•A transversion mutation in HBB, resulting in an E6V mutation in beta-globin (Supplementary Discussion). We used PE3 to install the HBB E6V mutation in HEK293T cells with 44% efficiency and 4.8% indels (Fig. 5a) and isolated from a single prime editing experiment six HEK293T cell lines that are homozygous (triploid) for the HBB E6V allele (Supplementary Note 1). To correct the HBB E6V allele to wild-type HBB, we treated homozygous HBB E6V HEK293T cells with PE3 and a pegRNA programmed to directly revert the HBB E6V mutation to wild-type HBB. All 14 tested pegRNAs mediated efficient correction of HBB E6V to wild-type HBB (26-52% efficiency), and indel levels averaging 2.8±0.70% (Extended Data Fig. 9a). Introduction of a PAM-modifying silent mutation improved editing efficiency and product purity to 58% correction with 1.4% indels (Fig. 5a).
Figure 5. Prime editing of pathogenic mutations, prime editing in primary mouse cortical neurons, and comparison of prime editing and HDR in four human cell lines.
(a) Installation (via T•A-to-A•T transversion) and correction (via A•T-to-T•A transversion) of the pathogenic E6V mutation in HBB in HEK293T cells. Correction either to wild-type HBB, or to HBB containing a PAM-disrupting silent mutation, is shown. (b) Installation (via 4-bp insertion) and correction (via 4-bp deletion) of the pathogenic HEXA 1278+TATC allele in HEK293T cells. Correction either to wild-type HEXA, or to HEXA containing a PAM-disrupting silent mutation, is shown. (c) Installation of the protective G127V variant in PRNP in HEK293T cells via G•C-to-T•A transversion. (d) Installation of a G•C-to-T•A transversion in DNMT1 of mouse primary cortical neurons using a split-intein PE3 lentivirus system (see Methods). Sorted values reflect editing or indels from GFP-positive nuclei, while unsorted values are from all nuclei. (e) PE3 editing and indels or (f) Cas9-initiated HDR editing and indels at endogenous genomic loci in HEK293T, K562, U2OS, and HeLa cells. (g) Targeted insertion of a His6 tag (18 bp), FLAG epitope tag (24 bp), or extended LoxP site (44 bp) in HEK293T cells by PE3. Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting, except where specified in (e). Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
The most common mutation that causes Tay-Sachs disease is a 4-bp insertion in HEXA (HEXA 1278+TATC). We used PE3 to install this 4-bp insertion into HEXA with 31% efficiency and 0.8% indels (Fig. 5b), and isolated two HEK293T cell lines that are homozygous for HEXA 1278+TATC (Supplementary Note 1). We used these cells to test 43 pegRNAs and three nicking sgRNAs with PE3 or PE3b systems for correction of the pathogenic insertion in HEXA (Extended Data Fig. 9b). Nineteen of the 43 pegRNAs tested resulted in ≥20% editing. Correction to wild-type HEXA with the best pegRNA proceeded with 33% efficiency with 0.32% indels using PE3b (Fig. 5b and Extended Data Fig. 9b).
Finally, we used PE3 to install a protective G•C-to-T•A transversion (G127V) into PRNP in HEK293T cells, introducing a G127V mutant allele that confers resistance to prion disease in humans36 and mice37 (Supplementary Discussion). We evaluated four pegRNAs and three nicking sgRNAs. The most effective pegRNA with PE3 resulted in 53% installation of G127V, with 1.7% indels (Fig. 5c). Taken together, these results establish the ability of prime editing in human cells to install or correct transversion, insertion, or deletion mutations that cause or confer resistance to disease efficiently, and with few byproducts.
Other cell lines and primary neurons
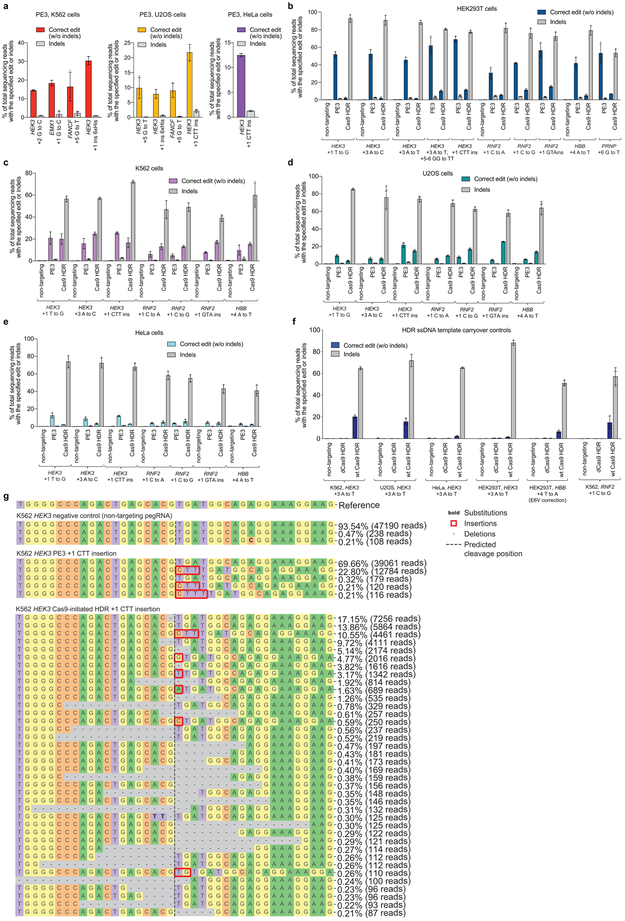
Next, we tested prime editing at endogenous sites in three additional human cell lines (Extended Data Fig. 10a, Supplementary Discussion). In K562 cells, PE3 achieved three transversion edits and a 6xHis tag insertion with 15-30% editing efficiency and 0.85-2.2% indels (Extended Data Fig. 10a). In U2OS cells, we installed transversion mutations, as well as a 3-bp insertion and 6xHis tag insertion, with 7.9-22% editing efficiency and 0.13-2.2% indels (Extended Data Fig. 10a). Finally, in HeLa cells we performed a 3-bp insertion with 12% average efficiency and 1.3% indels (Extended Data Fig. 10a). Collectively, these data indicate that cell lines beyond HEK293T support prime editing, although editing efficiencies vary by cell type and are generally less efficient than in HEK293T cells. Editing:indel ratios remained favorable in all tested human cell lines.
To determine if prime editing is possible in post-mitotic, terminally differentiated primary cells, we transduced primary cortical neurons from E18.5 mice with a PE3 lentiviral delivery system in which PE2 protein components are expressed from the neuron-specific synapsin promoter38 along with a GFP marker (Methods). Nuclei were isolated two weeks after transduction and sequenced directly, or sorted for GFP expression before sequencing. We observed 7.1% average prime editing of DNMT1 with 0.58% average indels in sorted cortical neuron nuclei (Fig. 5d). Cas9 nuclease in the same lentivirus system resulted in 31% average indels among sorted nuclei (Fig. 5d). These data indicate that post-mitotic, terminally differentiated primary cells can support prime editing.
Prime editing compared with HDR
Finally, we compared the performance of PE3 with that of optimized Cas9-initiated HDR11,14 in mitotic cell lines that support HDR14. We treated HEK293T, HeLa, K562 and U2OS cells with Cas9 nuclease, an sgRNA, and a ssDNA donor template designed to install a variety of transversion and insertion edits (Fig. 5e,f, Extended Data Fig. 10). Cas9-initiated HDR in all cases successfully installed the desired edit, but with far higher levels of indel byproducts than with PE3, as expected given that Cas9 induces DSBs. In HEK293T cells, the ratio of editing:indels for HBB E6V installation, HBB E6V correction, and PRNP G127V installation on average was 270-fold higher for PE3 than for Cas9-initiated HDR.
Comparisons between PE3 and HDR in human cell lines other than HEK293T showed similar results, although with lower PE3 editing efficiencies (Fig. 5e,f, Supplementary Discussion). Collectively, these data indicate that HDR typically results in similar or lower editing efficiencies with far higher indels than PE3 in four cell lines (Extended Data Fig. 10).
Discussion and future directions
The ability to insert arbitrary DNA sequences with single-nucleotide precision is an especially enabling prime editing capability. For example, we used PE3 in HEK293T cells to precisely insert into HEK3 a His6 tag (18 bp, 65% efficiency), a FLAG epitope tag (24 bp, 18% efficiency), and an extended Cre recombinase LoxP site (44 bp, 23% efficiency) with 3.0-5.9% indels (Fig. 5g). We anticipate many biotechnological and therapeutic applications will be enabled by the ability to efficiently and precisely insert new DNA sequences into target sites in living cells.
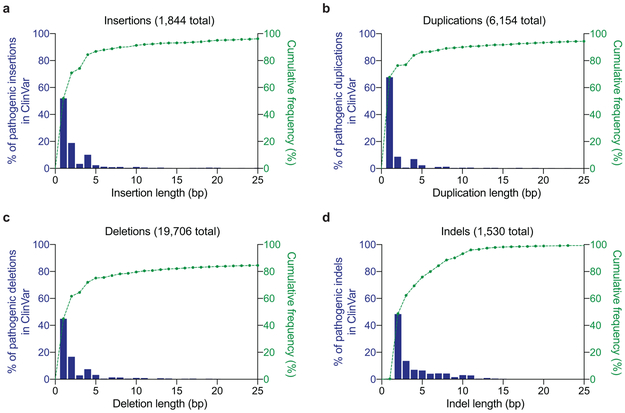
Collectively the prime editing experiments described in this study performed 19 insertions up to 44 bp, 23 deletions up to 80 bp, 119 point mutations including 83 transversions, and 18 combination edits at 12 endogenous loci in the human and mouse genomes at locations ranging from 3 bp upstream to 29 bp downstream of a PAM without making explicit DSBs. These results establish prime editing as a remarkably versatile genome editing method. Because 85-99% of insertions, deletions, indels, and duplications in ClinVar are ≤30 bp in length (Extended Data Fig. 11), in principle prime editing can correct up to ~89% of the 75,122 pathogenic human genetic variants in ClinVar (Fig. 1a).
Prime editing offers many possible choices of pegRNA-induced nick locations, sgRNA-induced second nick locations, PBS lengths, RT template lengths, and which strand to edit first. This flexibility, which contrasts with more limited options typically available for other precision editing methods11,15,16, allows editing efficiency, product purity, DNA specificity, and other parameters to be optimized to suit a given application (Extended Data Fig. 9).
Much additional research is needed to further understand and improve prime editing in a broad range of cell types and organisms, to assess off-target prime editing in a genome-wide manner, and to further characterize the extent to which prime editors might affect cells. Interfacing prime editing with additional in vitro and in vivo delivery strategies is critical to explore the potential of prime editing to enable applications including the study and treatment of genetic diseases. By enabling precise targeted transitions, transversions, insertions, and deletions in the genomes of mammalian cells without requiring DSBs, donor DNA templates, or HDR, however, prime editing provides a new “search-and-replace” capability that substantially expands the scope of genome editing.
Online Content
Methods, along with any additional Extended Data display items, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Methods
General methods.
DNA amplification was conducted by PCR using Phusion U Green Multiplex PCR Master Mix (ThermoFisher Scientific) or Q5 Hot Start High-Fidelity 2x Master Mix (New England BioLabs) unless otherwise noted. DNA oligonucleotides, including Cy5-labeled DNA oligonucleotides, dCas9 protein, and Cas9 H840A protein were obtained from Integrated DNA Technologies. Yeast reporter plasmids were derived from previously described plasmids39 and cloned by the Gibson assembly method. All mammalian editor plasmids used in this work were assembled using the USER cloning method as previously described40. Plasmids expressing sgRNAs were constructed by ligation of annealed oligonucleotides into BsmBI-digested acceptor vector (Addgene plasmid #65777). Plasmids expressing pegRNAs were constructed by Gibson assembly or Golden Gate assembly using a custom acceptor plasmid (see Supplementary Note 3). Sequences of sgRNA and pegRNA constructs used in this work are listed in Supplementary Tables 2 and 3. All vectors for mammalian cell experiments were purified using Plasmid Plus Midiprep kits (Qiagen) or PureYield plasmid miniprep kits (Promega), which include endotoxin removal steps. All experiments using live animals were approved by the Broad Institute Institutional and Animal Care and Use Committees. Wild-type C57BL/6 mice were obtained from Charles River (#027).
In vitro biochemical assays.
pegRNAs and sgRNAs were transcribed in vitro using the HiScribe T7 in vitro transcription kit (New England Biolabs) from PCR-amplified templates containing a T7 promoter sequence. RNA was purified by denaturing urea PAGE and quality-confirmed by an analytical gel prior to use. 5’-Cy5-labeled DNA duplex substrates were annealed using two oligonucleotides (Cy5-AVA024 and AVA025; 1:1.1 ratio) for the non-nicked substrate or three oligonucleotides (Cy5-AVA023, AVA025 and AVA026; 1:1.1:1.1) for the pre-nicked substrate by heating to 95 °C for 3 minutes followed by slowly cooling to room temperature (Supplementary Table 2). Cas9 cleavage and reverse transcription reactions were carried out in 1× cleavage buffer41 supplemented with dNTPs (20 mM HEPES-K, pH 7.5; 100 mM KCl; 5% glycerol; 0.2 mM EDTA, pH 8.0; 3 mM MgCl2; 0.5 mM dNTP mix; 5 mM DTT). dCas9 or Cas9 H840A (5 μM final) and the sgRNA or pegRNA (5 μM final) were pre-incubated at room temperature in a 5 μL reaction mixture for 10 minutes prior to the addition of duplex DNA substrate (400 nM final), followed by the addition of Superscript III reverse transcriptase (ThermoFisher Scientific), an undisclosed M-MLV RT variant, when applicable. Reactions were carried out at 37 °C for 1 hour, then diluted to a volume of 10 μL with water, treated with 0.2 μL of proteinase K solution (20 mg/mL, ThermoFisher Scientific), and incubated at room temperature for 30 minutes. Following heat inactivation at 95 °C for 10 minutes, reaction products were combined with 2x formamide gel loading buffer (90% formamide; 10% glycerol; 0.01% bromophenol blue), denatured at 95 °C for 5 minutes, and separated by denaturing urea-PAGE gel (15% TBE-urea, 55 °C, 200V). DNA products were visualized by Cy5 fluorescence signal using a Typhoon FLA 7000 biomolecular imager.
Electrophoretic mobility shift assays were carried out in 1× binding buffer (1× cleavage buffer + 10 μg/mL heparin) using pre-incubated dCas9:sgRNA or dCas9:pegRNA complexes (concentration range between 5 nM and 1 μM final) and Cy5-labeled duplex DNA (Cy5-AVA024 and AVA025; 20 nM final). After 15 minutes of incubation at 37 °C, the samples were analyzed by native PAGE gel (10% TBE) and imaged for Cy5 fluorescence.
For DNA sequencing of reverse transcription products, fluorescent bands were excised and purified from urea-PAGE gels, then 3’ tailed with terminal transferase (TdT; New England Biolabs) in the presence of dGTP or dATP according to the manufacturer’s protocol. Tailed DNA products were diluted 10-fold with binding buffer (40% saturated aqueous guanidinium chloride + 60% isopropanol) and purified by QIAquick spin column (Qiagen), then used as templates for primer extension by Klenow fragment (New England Biolabs) using primer AVA134 (A-tailed products) or AVA135 (G-tailed products) (Supplementary Table 2). Extensions were amplified by PCR for 10 cycles using primers AVA110 and AVA122, then sequenced with AVA037 using the Sanger method (Supplementary Table 2).
Yeast fluorescent reporter assays.
Dual fluorescent reporter plasmids containing an in-frame stop codon, a +1 frameshift, or a −1 frameshift were subjected to 5’-extended pegRNA or 3’-extended pegRNA prime editing reactions in vitro as described above. Following incubation at 37 °C for 1 hour, the reactions were diluted with water and plasmid DNA was precipitated with 0.3 M sodium acetate and 70% ethanol. Resuspended DNA was transformed into S. cerevisiae by electroporation as previously described42 and plated on synthetic complete media without leucine (SC(glucose), L-). GFP and mCherry fluorescence signals were visualized from colonies with the Typhoon FLA 7000 biomolecular imager.
General mammalian cell culture conditions.
HEK293T (ATCC CRL-3216), U2OS (ATTC HTB-96), K562 (CCL-243), and HeLa (CCL-2) cells were purchased from ATCC and cultured and passaged in Dulbecco’s Modified Eagle’s Medium (DMEM) plus GlutaMAX (ThermoFisher Scientific), McCoy’s 5A Medium (Gibco), RPMI Medium 1640 plus GlutaMAX (Gibco), or Eagle’s Minimal Essential Medium (EMEM, ATCC), respectively, each supplemented with 10% (v/v) fetal bovine serum (Gibco, qualified) and 1× Penicillin Streptomycin (Corning). All cell types were incubated, maintained, and cultured at 37 °C with 5% CO2. Cell lines were authenticated by their respective suppliers and tested negative for mycoplasma.
HEK293T tissue culture transfection protocol and genomic DNA preparation.
HEK293T cells were seeded on 48-well poly-D-lysine coated plates (Corning). 16-24 h post-seeding, cells were transfected at approximately 60% confluency with 1 μL of Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocols and 750 ng of PE plasmid, 250 ng of pegRNA plasmid, and 83 ng of sgRNA plasmid (for PE3 and PE3b). Unless otherwise stated, cells were cultured 3 days following transfection, after which the media was removed, the cells were washed with 1× PBS solution (Thermo Fisher Scientific), and genomic DNA was extracted by the addition of 150 μL of freshly prepared lysis buffer (10 mM Tris-HCl, pH 7.5; 0.05% SDS; 25 μg/mL Proteinase K (ThermoFisher Scientific)) directly into each well of the tissue culture plate. The genomic DNA mixture was incubated at 37 °C for 1-2 hrs, followed by an 80 °C enzyme inactivation step for 30 min. Primers used for mammalian cell genomic DNA amplification are listed in Supplementary Table 4. For HDR experiments in HEK293T cells, 231 ng of nuclease-expression plasmid, 69 ng of sgRNA expression plasmid, 50 ng (1.51 pmol) 100-nt ssDNA donor template (PAGE-purified; Integrated DNA Technologies) was lipofected using 1.4 μL Lipofectamine 2000 (ThermoFisher) per well. Genomic DNA from all HDR experiments was purified using the Agencourt DNAdvance Kit (Beckman Coulter), according to the manufacturer’s protocol.
High-throughput DNA sequencing of genomic DNA samples.
Genomic sites of interest were amplified from genomic DNA samples and sequenced on an Illumina MiSeq as previously described with the following modifications17,18. Briefly, amplification primers containing Illumina forward and reverse adapters (Supplementary Table 4) were used for a first round of PCR (PCR 1) amplifying the genomic region of interest. 25-μL PCR 1 reactions were performed with 0.5 μM of each forward and reverse primer, 1 μL of genomic DNA extract and 12.5 μL of Phusion U Green Multiplex PCR Master Mix. PCR reactions were carried out as follows: 98 °C for 2 min, then 30 cycles of [98 °C for 10 s, 61 °C for 20 s, and 72 °C for 30 s], followed by a final 72 °C extension for 2 min. Unique Illumina barcoding primer pairs were added to each sample in a secondary PCR reaction (PCR 2). Specifically, 25 μL of a given PCR 2 reaction contained 0.5 μM of each unique forward and reverse Illumina barcoding primer pair, 1 μL of unpurified PCR 1 reaction mixture, and 12.5 μL of Phusion U Green Multiplex PCR 2x Master Mix. The barcoding PCR 2 reactions were carried out as follows: 98 °C for 2 min, then 12 cycles of [98 °C for 10 s, 61 °C for 20 s, and 72 °C for 30 s], followed by a final 72 °C extension for 2 min. PCR products were evaluated analytically by electrophoresis in a 1.5% agarose gel. PCR 2 products (pooled by common amplicons) were purified by electrophoresis with a 1.5% agarose gel using a QIAquick Gel Extraction Kit (Qiagen), eluting with 40 μL of water. DNA concentration was measured by fluorometric quantification (Qubit, ThermoFisher Scientific) or qPCR (KAPA Library Quantification Kit-Illumina, KAPA Biosystems) and sequenced on an Illumina MiSeq instrument according to the manufacturer’s protocols.
Sequencing reads were demultiplexed using MiSeq Reporter (Illumina). Alignment of amplicon sequences to a reference sequence was performed using CRISPResso243. For all prime editing yield quantification, prime editing efficiency was calculated as: % of [# of reads with the desired edit that do not contain indels] ÷ [# of total reads]. For quantification of point mutation editing, CRISPResso2 was run in standard mode with “discard_indel_reads” on. Prime editing for installation of point mutations was then explicitly calculated as: [frequency of specified point mutation in non-discarded reads] × [# of non-discarded reads] ÷ [total reads]. For insertion or deletion edits, CRISPResso2 was run in HDR mode using the desired allele as the expected allele (e flag), and with “discard_indel_reads” on. Editing yield was calculated as: [# of HDR aligned reads] ÷ [total reads]. For all experiments, indel yields were calculated as: [# of indel-containing reads] ÷ [total reads].
Nucleofection of U2OS, K562, and HeLa cells.
Nucleofection was performed in all experiments using K562, HeLa, and U2OS cells. For PE conditions in these cell types, 800ng prime editor-expression plasmid, 200ng PEgRNA-expression plasmid, and 83ng nicking plasmid was nucleofected in a final volume of 20uL in a 16-well nucleocuvette strip (Lonza). For HDR conditions in these three cell types, 350 ng nuclease-expression plasmid, 150 ng sgRNA-expression plasmid and 200 pmol (6.6 μg) 100-nt ssDNA donor template (PAGE-purified; Integrated DNA Technologies) was nucleofected in a final volume of 20 μL per sample in a 16-well Nucleocuvette strip (Lonza). K562 cells were nucleofected using the SF Cell Line 4D-Nucleofector X Kit (Lonza) with 5 × 105 cells per sample (program FF-120), according to the manufacturer’s protocol. U2OS cells were nucleofected using the SE Cell Line 4D-Nucleofector X Kit (Lonza) with 3—4 × 105 cells per sample (program DN-100), according to the manufacturer’s protocol. HeLa cells were nucleofected using the SE Cell Line 4D-Nucleofector X Kit (Lonza) with 2 × 105 cells per sample (program CN-114), according to the manufacturer’s protocol. Cells were harvested 72 hours after nucleofection for genomic DNA extraction.
Genomic DNA extraction for HDR experiments.
Genomic DNA from all HDR comparison experiments in HEK293T, HEK293T HBB E6V, K562, U2OS, and HeLa cells was purified using the Agencourt DNAdvance Kit (Beckman Coulter), according to the manufacturer’s protocol.
Comparison between PE2, PE3, BE2, BE4max, ABEdmax, and ABEmax.
HEK293T cells were seeded on 48-well poly-D-lysine coated plates (Corning). After 16-24 h, cells were transfected at approximately 60% confluency. For base editing with CBE or ABE constructs, cells were transfected with 750 ng of base editor plasmid, 250 ng of sgRNA expression plasmid, and 1 μL of Lipofectamine 2000 (Thermo Fisher Scientific). PE transfections were performed as described above. Genomic DNA extraction for PE and BE was performed as described above.
Determination of PE3 activity at known Cas9 off-target sites.
To evaluate PE3 off-target editing activity at known Cas9 off-target sites, genomic DNA extracted from HEK293T cells 3 days after transfection with PE3 was used as template for PCR amplification of 16 previously reported Cas9 off-target genomic sites32,33 (the top four off-target sites each for the HEK3, EMX1, FANCF, and HEK4 spacers; primer sequences are listed in Supplementary Table 4). These genomic DNA samples were identical to those used for quantifying on-target PE3 editing activities shown in Fig. 4 or Extended Data Fig. 5d–e; pegRNA and nicking sgRNA sequences are listed in Supplementary Table 3. Following PCR amplification of off-target sites, amplicons were sequenced on the Illumina MiSeq platform as described above (see High-throughput DNA sequencing of genomic DNA samples). For determining Cas9 nuclease, Cas9 H840A nickase, dCas9, and PE2-dRT on-target and off-target editing activity, HEK293T cells were transfected with 750 ng of editor plasmid (Cas9 nuclease, Cas9 H840A nickase, dCas9, or PE2-dRT), 250 ng of pegRNA or sgRNA plasmid, and 1 μL of Lipofectamine 2000. Genomic DNA was isolated from cells 3 days after transfection as described above. On-target and off-target genomic loci were amplified by PCR using primer sequences in Supplementary Table 4 and sequenced on an Illumina MiSeq.
HTS data analysis was performed using CRISPResso243. The editing efficiencies of Cas9 nuclease, Cas9 H840A nickase, and dCas9 were quantified as the percent of total sequencing reads containing indels. For quantification of PE3 and PE3-dRT off-targets, aligned sequencing reads were examined for point mutations, insertions, or deletions that were consistent with the anticipated product of pegRNA reverse transcription initiated at the Cas9 nick site. Single nucleotide variations occurring at <0.1% overall frequency among total reads within a sample were excluded from analysis. For reads containing single nucleotide variations that both occurred at frequencies ≥ 0.1% and were partially consistent with the pegRNA-encoded edit, t-tests (unpaired, one-tailed, α = 0.5) were used to determine if the variants occurred at significantly higher levels compared to samples treated with pegRNAs that contained the same spacer but encoded different edits. To avoid differences in sequencing errors, comparisons were made between samples that were sequenced simultaneously within the same MiSeq run. Variants that did not meet the criteria of p-value > 0.05 were excluded. Off-target PE3 editing activity was then calculated as the percentage of total sequencing reads that met the above criteria.
Generation of a HEK293T cell line containing the HBB E6V mutation using Cas9-initiated HDR.
HEK293T cells were seeded in a 48-well plate and transfected at approximately 60% confluency with 1.5 μL of Lipofectamine 2000, 300 ng of Cas9 D10A nickase plasmid, 100 ng of sgRNA plasmid, and 200 ng of 100-mer ssDNA donor template (Supplementary Table 5). Three days after transfection, media was exchanged for fresh media. Four days after transfection, cells were dissociated using 30 μL of TrypLE solution and suspended in 1.5 mL of media. Single cells were isolated into individual wells of two 96-well plates by fluorescence-activated cell sorting (FACS) (Beckman-Coulter Astrios). See Supplementary Note 1 for representative FACS sorting examples. Cells were expanded for 14 days prior to genomic DNA sequencing as described above. Of the isolated clonal populations, none was found to be homozygous for the HBB E6V mutation, so a second round of editing by lipofection, sorting, and outgrowth was repeated in a partially edited cell line to yield a cell line homozygous for the E6V allele.
Generation of a HEK293T cell line containing the HBB E6V mutation using PE3.
2.5 × 104 HEK293T cells were seeded on 48-well poly-D-lysine coated plates (Corning). 16-24 h post-seeding, cells were transfected at approximately 70% confluency with 1 μL of Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocols and 750 ng of PE2-P2A-GFP plasmid, 250 ng of pegRNA plasmid, and 83 ng of sgRNA plasmid. After 3 days, cells were washed with 1× PBS (Gibco) and dissociated using TrypLE Express (Gibco). Cells were then diluted with DMEM plus GlutaMax (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS (Gibco) and passed through a 35-μm cell strainer (Corning) prior to sorting. Flow cytometry was carried out on a LE-MA900 cell sorter (Sony). Cells were treated with 3 nM DAPI (BioLegend) 15 minutes prior to sorting. After gating for doublet exclusion, single DAPI-negative cells with GFP fluorescence above that of a GFP-negative control cell population were sorted into 96-well flat-bottom cell culture plates (Corning) filled with pre-chilled DMEM with GlutaMax supplemented with 10% FBS. See Supplementary Note 1 for representative FACS sorting examples and allele tables. Cells were cultured for 10 days prior to genomic DNA extraction and characterization by HTS, as described above. A total of six clonal cell lines were identified that are homozygous for the E6V mutation in HBB.
Generation of a HEK293T cell line containing the HEXA 1278+TATC insertion using PE3.
HEK293T cells containing the HEXA 1278+TATC allele were generated following the protocol described above for creation of the HBB E6V cell line; pegRNA and sgRNA sequences are listed in Supplementary Table 3 under the Figure 5 subheading. After transfection and sorting, cells were cultured for 10 days prior to genomic DNA extraction and characterization by HTS, as described above. Two heterozygous cell lines were isolated that contained 50% HEXA 1278+TATC alleles, and two homozygous cell lines containing 100% HEXA 1278+TATC alleles were recovered.
Cell viability assays.
HEK293T cells were seeded in 48-well plates and transfected at approximately 70% confluency with 750 ng of editor plasmid (PE3, PE3 R110S K103L, Cas9 H840A nickase, or dCas9), 250 ng of HEK3-targeting pegRNA plasmid, and 1 μL of Lipofectamine 2000, as described above. Cell viability was measured every 24 hours post-transfection for 3 days using the CellTiter-Glo 2.0 assay (Promega) according to the manufacturer’s protocol. Luminescence was measured in 96-well flat-bottomed polystyrene microplates (Corning) using a M1000 Pro microplate reader (Tecan) with a 1-second integration time.
Lentivirus production.
Lentivirus was produced as previously described44. T-75 flasks of rapidly dividing HEK293T cells (ATCC; Manassas, VA, USA) were transfected with lentivirus production helper plasmids pVSV-G and psPAX2 in combination with modified lentiCRISPRv2 genomes carrying intein-split PE2 editor using FuGENE HD (Promega, Madison, WI, USA) according to the manufacturer's protocol. Four split-intein editor constructs were designed: 1) a viral genome encoding a U6-pegRNA expression cassette and the N-terminal portion (1-573) of Cas9 H840A nickase fused to the Npu N-intein, a self-cleaving P2A peptide, and GFP-KASH; 2) a viral genome encoding the Npu C-intein fused to the C-terminal remainder of PE2; 3) a viral genome encoding the Npu C-intein fused to the C-terminal remainder of Cas9 for the Cas9 control; and 4) a nicking sgRNA for DNMT1 (derived from Addgene plasmid #52963). The split-intein45 mediates trans splicing to join the two halves of PE2 or Cas9, while the P2A GFP-KASH enables co-translational production of a nuclear membrane-localized GFP. After 48 h, supernatant was collected, centrifuged at 500 g for 5 minutes to remove cellular debris, and filtered using a 0.45 μm filter. Filtered supernatant was concentrated using the PEG-it Virus Precipitation Solution (System Biosciences, Palo Alto, CA, USA) according to the manufacturer's directions. The resulting pellet was resuspended in Opti-MEM (Thermo Fisher Scientific, Waltham, MA, USA) using 1% of the original media volume. Resuspended pellet was flash-frozen and stored at −80°C until use.
Mouse primary cortical neuron dissection and culture.
E18.5 dissociated cortical cultures were harvested from timed-pregnant C57BL/6 mice (Charles River). Embryos were harvested from pregnant mice after euthanasia by CO2 followed by decapitation. Cortical caps were dissected in ice-cold Hibernate-E supplemented with penicillin/streptomycin (Life Technologies). Following a rinse with ice-cold Hibernate-E, tissue was digested at 37 °C for 8 minutes in papain/DNase (Worthington/Sigma). Tissue was triturated in NBActiv4 (BrainBits) supplemented with DNase. Cells were counted and plated in 24-well plates at 100,000 cells per well. Half of the media was changed twice per week.
Prime editing in primary neurons and nuclei isolation.
At DIV 1, 15 μL of lentivirus was added at 10:10:1 ratio of N-terminal:C-terminal:nicking sgRNA. At DIV 14, neuronal nuclei were isolated using the EZ-PREP buffer (Sigma D8938) following the manufacturer’s protocol. All steps were performed on ice or at 4 °C. Media was removed from dissociated cultures, and cultures were washed with ice-cold PBS. PBS was aspirated and replaced with 200 μL EZ-PREP solution. Following a 5-minute incubation on ice, EZ-PREP was pipetted across the surface of the well to dislodge remaining cells. The sample was centrifuged at 500 g for 5 minutes, and the supernatant removed. Samples were washed with 200 μL EZ-PREP and centrifuged again at 500 g for 5 minutes. Samples were resuspended with gentle pipetting in 200 μL ice-cold Nuclei Suspension Buffer (NSB) consisting of 100 μg/mL BSA and 3.33 μM Vybrant DyeCycle Ruby (Thermo Fisher) in 1×PBS, then centrifuged at 500 g for 5 minutes. The supernatant was removed and nuclei were resuspended in 100 μL NSB and sorted into 100 μL Agencourt DNAdvance lysis buffer using a MoFlo Astrios (Beckman Coulter) at the Broad Institute flow cytometry facility. Genomic DNA was purified according to the manufacturer’s Agencourt DNAdvance instructions.
RNA-sequencing and data analysis.
HEK293T cells were co-transfected with PRNP-targeting or HEXA-targeting pegRNAs and PE2, PE2-dRT, or Cas9 H840A nickase. 72 h following transfection, total RNA was harvested from cells using TRIzol reagent (Thermo Fisher) and purified with RNeasy Mini kit (Qiagen) including on-column DNaseI treatment. Ribosomes were depleted from total RNA using the rRNA removal protocol of the TruSeq Stranded Total RNA library prep kit (Illumina) and subsequently washed with RNAClean XP beads (Beckman Coulter). Sequencing libraries were prepared using ribo-depleted RNA on a SMARTer PrepX Apollo NGS library prep system (Takara) following the manufacturer’s protocol. Resulting libraries were visualized on a 2200 TapeStation (Agilent Technologies), normalized using a Qubit dsDNA HS assay (Thermo Fisher), and sequenced on a NextSeq 550 using high output v2 flow cell (Illumina) as 75-bp paired-end reads. Fastq files were generated with bcl2fastq2 version 2.20 and trimmed using TrimGalore version 0.6.2 (https://github.com/FelixKrueger/TrimGalore) to remove low-quality bases, unpaired sequences, and adaptor sequences. Trimmed reads were aligned to a Homo sapiens genome assembly GRCh38 with a custom Cas9 H840A gene entry using RSEM version 1.3.146. The limma-voom47 package was used to normalize gene expression levels and perform differential expression analysis with batch effect correction. Differentially expressed genes were called with FDR-corrected p-value < 0.05 and fold-change > 2 cutoffs, and results were visualized in R.
ClinVar analysis.
The ClinVar variant summary was downloaded from NCBI (accessed July 15, 2019), and the information contained therein was used for all downstream analysis. The list of all reported variants was filtered by allele ID in order to remove duplicates and by clinical significance in order to restrict the analysis to pathogenic variants. The list of pathogenic variants was filtered sequentially by variant type in order to calculate the fraction of pathogenic variants that are insertions, deletions, etc. Single nucleotide variants (SNVs) were separated into two categories (transitions and transversions) based on the reported reference and alternate alleles. SNVs that did not report reference or alternate alleles were excluded from the analysis.
The lengths of reported insertions, deletions, and duplications were calculated using reference/alternate alleles, variant start/stop positions, or appropriate identifying information in the variant name. Variants that did not report any of the above information were excluded from the analysis. The lengths of reported indels (single variants that include both insertions and deletions relative to the reference genome) were calculated by determining the number of mismatches or gaps in the best pairwise alignment between the reference and alternate alleles. Frequency distributions of variant lengths were calculated using GraphPad Prism 8.
Data availability.
High-throughput sequencing data have been deposited to the NCBI Sequence Read Archive database PRJNA565979. Plasmids encoding PE1, PE2/PE3, and pegRNA expression vectors have been deposited to Addgene for distribution.
Code availability.
The script used to quantify pegRNA scaffold insertion is provided as Supplementary Note 4.
Extended Data
Extended Data Figure 1. In vitro prime editing validation studies with fluorescently labeled DNA substrates.
(a) Electrophoretic mobility shift assays with dCas9, 5’-extended pegRNAs and 5’-Cy5-labeled DNA substrates. pegRNAs 1 through 5 contain a 15-nt linker sequence (linker A for pegRNA 1, linker B for pegRNAs 2 through 5) between the spacer and the PBS, a 5-nt PBS sequence, and RT templates of 7 nt (pegRNAs 1 and 2), 8 nt (pegRNA 3), 15 nt (pegRNA 4), and 22 nt (pegRNA 5). pegRNAs are those used in (e) and (f); full sequences are listed in Supplementary Table 2. (b) In vitro nicking assays of Cas9 H840A using 5’-extended and 3’-extended pegRNAs. Data in (a-b) are representative of n=2 independent replicates. (c) Cas9-mediated indel formation in HEK293T cells at HEK3 using 5’-extended and 3’-extended pegRNAs. Values and error bars reflect mean±s.d. of n=3 independent biological replicates. (d) Overview of prime editing in vitro biochemical assays. 5’-Cy5-labeled pre-nicked and non-nicked dsDNA substrates were tested. sgRNAs, 5’-extended pegRNAs, or 3’-extended pegRNAs were pre-complexed with dCas9 or Cas9 H840A nickase, then combined with dsDNA substrate, Superscript III M-MLV RT, and dNTPs. Reactions were allowed to proceed at 37 °C for 1 hour prior to separation by denaturing urea PAGE and visualization by Cy5 fluorescence. (e) Primer extension reactions using 5’-extended pegRNAs, pre-nicked DNA substrates, and dCas9 lead to significant conversion to RT products. (f) Primer extension reactions using 5’-extended pegRNAs as in (b), with non-nicked DNA substrate and Cas9 H840A nickase. Product yields are greatly reduced by comparison to pre-nicked substrate. (g) An in vitro primer extension reaction using a 3’-pegRNA generates a single apparent product by denaturing urea PAGE. The RT product band was excised, eluted from the gel, then subjected to homopolymer tailing with terminal transferase (TdT) using either dGTP or dATP. Tailed products were extended by poly-T or poly-C primers, and the resulting DNA was sequenced. Sanger traces indicate that three nucleotides derived from the pegRNA scaffold were reverse transcribed (added as the final 3’ nucleotides to the DNA product). Note that in mammalian cell prime editing experiments, pegRNA scaffold insertion is much rarer than in vitro (Extended Data Fig. 6), potentially due to the inability of the tethered reverse transcriptase to access the Cas9-bound guide RNA scaffold, and/or cellular excision of mismatched 3’ ends of 3’ flaps containing pegRNA scaffold sequences. Data in (e-g) are representative of n=2 independent replicates. For gel source data, see Supplementary Figure 1.
Extended Data Figure 2. Cellular repair in yeast of 3’ DNA flaps from in vitro prime editing reactions.
(a) Dual fluorescent protein reporter plasmids contain GFP and mCherry open reading frames separated by a target site encoding an in-frame stop codon, a +1 frameshift, or a −1 frameshift. Prime editing reactions were carried out in vitro with Cas9 H840A nickase, pegRNA, dNTPs, and M-MLV reverse transcriptase, then transformed into yeast. Colonies that contain unedited plasmids produce GFP but not mCherry. Yeast colonies containing edited plasmids produce both GFP and mCherry as a fusion protein. (b) Overlay of GFP and mCherry fluorescence for yeast colonies transformed with reporter plasmids containing a stop codon between GFP and mCherry (unedited negative control, top), or containing no stop codon or frameshift between GFP and mCherry (pre-edited positive control, bottom). (c-f) Visualization of mCherry and GFP fluorescence from yeast colonies transformed with in vitro prime editing reaction products. (c) Stop codon correction via T•A-to-A•T transversion using a 3’-extended pegRNA or (d) a 5’-extended pegRNA. (e) +1 frameshift correction via a 1-bp deletion using a 3’-extended pegRNA. (f) −1 frameshift correction via a 1-bp insertion using a 3’-extended pegRNA. (g) Sanger DNA sequencing traces from plasmids isolated from GFP-only colonies in (b) and GFP and mCherry double-positive colonies in (c). Data in (b-g) are representative of n=2 independent replicates.
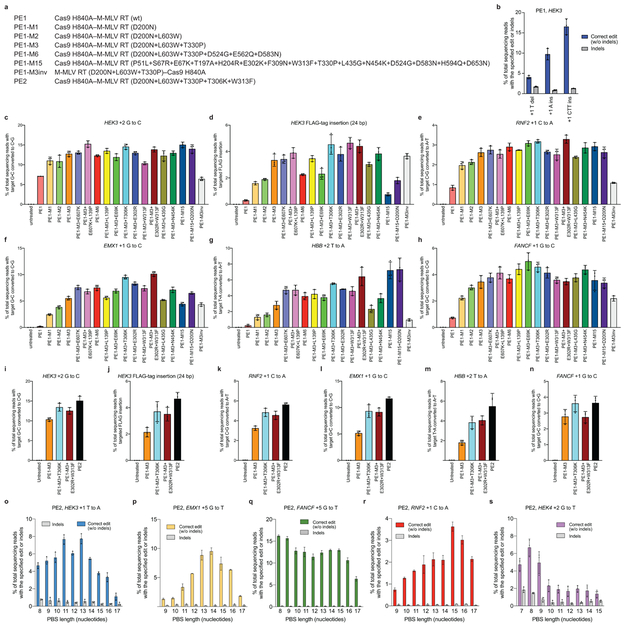
Extended Data Figure 3. Prime editing of genomic DNA in human cells by PE1.
(a) pegRNAs contain a spacer sequence, an sgRNA scaffold, and a 3’ extension containing a reverse transcription (RT) template (purple), which contains the edited base(s) (red), and a primer-binding site (PBS, green). The primer-binding site hybridizes to the nicked target DNA strand. The RT template is homologous to the DNA sequence downstream of the nick, with the exception of the encoded edited base(s). (b) Installation of a T•A-to-A•T transversion at the HEK3 site in HEK293T cells using Cas9 H840A nickase fused to wild-type M-MLV reverse transcriptase (PE1) and pegRNAs with varying PBS lengths. (c) T•A-to-A•T transversion editing efficiency and indel generation by PE1 at the +1 position of HEK3 using pegRNAs containing 10-nt RT templates and a PBS sequences ranging from 8-17 nt. (d) G•C-to-T•A transversion editing efficiency and indel generation by PE1 at the +5 position of EMX1 using pegRNAs containing 13-nt Rt templates and a PBS sequences ranging from 9-17 nt. (e) G•C-to-T•A transversion editing efficiency and indel generation by PE1 at the +5 position of FANCF using pegRNAs containing 17-nt RT templates and a pBs sequences ranging from 8-17 nt. (f) C•G-to-A•T transversion editing efficiency and indel generation by PE1 at the +1 position of RNF2 using pegRNAs containing 11 -nt RT templates and a PBS sequences ranging from 9-17 nt. (g) G•C-to-T•A transversion editing efficiency and indel generation by PE1 at the +2 position of HEK4 using pegRNAs containing 13-nt RT templates and a PBS sequences ranging from 7-15 nt. (h) PE1-mediated +1 T deletion, +1 A insertion, and +1 CTT insertion at the HEK3 site using a 13-nt PBS and 10-nt RT template. Sequences of pegRNAs are those used in Fig. 2a (see Supplementary Table 3). Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Extended Data Figure 4. Evaluation of M-MLV RT variants for prime editing.
(a) Abbreviations for prime editor variants used in this figure. (b) Targeted insertion and deletion edits with PE1 at the HEK3 locus. (c-h) Comparison of 18 prime editor constructs containing M-MLV RT variants for their ability to install (c) a +2 G•C-to-C•G transversion edit at HEK3, (d) a 24-bp FLAG insertion at the +1 position of HEK3, (e) a +1 C•G-to-A•T transversion edit at RNF2, (f) a +1 G•C-to-C•G transversion edit at EMX1, (g) a +2 T•A-to-A•T transversion edit at HBB, and (h) a +1 G•C-to-C•G transversion edit at FANCF. (i-n) Comparison of four prime editor constructs containing M-MLV variants for their ability to install the edits shown in (c-h) in a second round of independent experiments. (o-s) PE2 editing efficiency at five genomic loci with varying PBS lengths. (o) +1 T•A-to-A•T at HEK3. (p) +5 G•C-to-T•A at EMX1. (q) +5 G•C-to-T•A at FANCF. (r) +1 C•G-to-A•T at RNF2. (s) +2 G•C-to-T•A at HEK4. Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
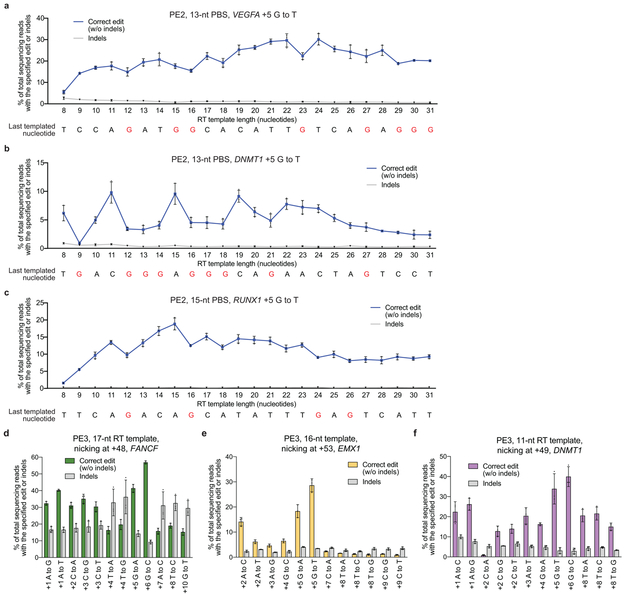
Extended Data Figure 5. Design features of pegRNA PBS and RT template sequences, and additional editing examples with PE3.
(a) PE2-mediated +5 G•C-to-T•A transversion editing efficiency (blue line) at VEGFA in HEK293T cells as a function of RT template length. Indels (gray line) are plotted for comparison. The sequence below the graph shows the last nucleotide templated for synthesis by the pegRNA. G nucleotides (templated by a C in the pegRNA) are highlighted in red; RT templates that end in C should be avoided during pegRNA design to maximize prime editing efficiencies. (b) +5 G•C-to-T•A transversion editing and indels for DNMT1 as in (a). (c) +5 G•C-to-T•A transversion editing and indels for RUNX1 as in (a). PE3-mediated transition and transversion edits at the specified positions for (d) FANCF, (e) EMX1, and (f) DNMT1. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Extended Data Figure 6. Comparison of prime editing and base editing, and off-target editing by Cas9 and prime editors at known Cas9 off-target sites.
(a) C•G-to-T•A editing efficiency at the same target nucleotides for PE2, PE3, BE2max, and BE4max at endogenous HEK3, FANCF, and EMX1 sites in HEK293T cells. (b) Indel frequency from treatments in (a). (c) Editing efficiency of precise C•G-to-T•A edits (without bystander edits or indels) at HEK3, FANCF, and EMX1. (d) Total A•T-to-G•C editing efficiency for PE2, PE3, ABEdmax, and ABEmax at HEK3 and FANCF. (e) Precise A•T-to-G•C editing efficiency without bystander edits or indels at HEK3 and FANCF. (f) Indel frequency from treatments in (d). (g) Average triplicate Cas9 nuclease editing efficiencies (indel frequencies) in HEK293T cells at four endogenous on-target sites and their 16 known top off-target sites32,33. For each on-target site, Cas9 was paired with an sgRNA or with each of four pegRNAs that recognize the same protospacer. (h) Average triplicate on-target and off-target editing efficiencies and indel efficiencies (below in parentheses) in HEK293T cells for PE2 or PE3 paired with each pegRNA in (g). Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Off-target editing efficiencies in (h) reflect off-target locus modification consistent with prime editing. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Extended Data Figure 7. Incorporation of pegRNA scaffold sequence into target loci.
HTS data were analyzed for pegRNA scaffold sequence insertion as described in Supplementary Note 4. (a) Analysis for the EMX1 locus. Shown is the % of total sequencing reads containing one or more pegRNA scaffold sequence nucleotides within an insertion adjacent to the RT template (left); the percentage of total sequencing reads containing a pegRNA scaffold sequence insertion of the specified length (middle); and the cumulative total percentage of pegRNA insertion up to and including the length specified on the X axis. (b) As in (a) for FANCF. (c) As in (a) for HEK3. (d) As in (a) for RNF2. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Extended Data Figure 8. Effects of PE2, PE2-dRT, Cas9 H840A nickase, and dCas9 on cell viability and on transcriptome-wide RNA abundance.
HEK293T cells were transiently transfected with plasmids encoding PE2, PE2 R110S K103L, Cas9 H840A nickase, or dCas9, together with a HEK3-targeting pegRNA plasmid. Cell viability was measured for the bulk cellular population every 24 hours post-transfection for 3 days using the CellTiter-Glo 2.0 assay (Promega). (a) Viability, as measured by luminescence, at 1, 2, or 3 days post-transfection. Values and error bars reflect mean±s.e.m. of n=3 independent biological replicates, each performed in technical triplicate. (b) Percent editing and indels for PE2, PE2 R110S K103L, Cas9 H840A nickase, or dCas9, together with a HEK3-targeting pegRNA plasmid that encodes a +5 G to A edit. Editing efficiencies were measured on day 3 post-transfection from cells treated alongside of those used for assaying viability in (a). Values and error bars reflect mean±s.d. of n=3 independent biological replicates. (c-k) Analysis of cellular RNA, depleted for ribosomal RNA, isolated from HEK293T cells expressing PE2, PE2-dRT, or Cas9 H840A nickase and a PRNP-targeting or HEXA-targeting pegRNA. RNAs corresponding to 14,410 genes and 14,368 genes were detected in PRNP and HEXA samples, respectively. (c-h) Volcano plot displaying the −log10 FDR-adjusted p-value vs. log2-fold change in transcript abundance for each RNA, comparing (c) PE2 vs. pE2-dRT with PRNP-targeting pegRNA, (d) PE2 vs. Cas9 H840A with PRNP-targeting pegRNA, (e) PE2-dRT vs. Cas9 H840A with PRNP-targeting pegRNA, (f) PE2 vs. PE2-dRT with HeXa-targeting pegRNA, (g) PE2 vs. Cas9 H840A with HEXA-targeting pegRNA, (h) PE2-dRT vs. Cas9 H840A with HEXA-targeting pegRNA. Red dots indicate genes that show ≥2-fold change in relative abundance that are statistically significant (FDR-adjusted p < 0.05). (i-k) Venn diagrams of upregulated and downregulated transcripts (≥2-fold change) comparing PRNP and HEXA samples for (i) PE2 vs PE2-dRT, (j) PE2 vs. Cas9 H840A, and (k) PE2-dRT vs. Cas9 H840A. Values for each RNA-seq condition reflect the mean n=5 biological replicates. Differential expression was assessed using a two-sided t-test with empirical Bayesian variance estimation.
Extended Data Figure 9. PE3-mediated HBB E6V correction and HEXA 1278+TATC correction by various pegRNAs.
(a) Screen of 14 pegRNAs for correction of the HBB E6V allele in HEK293T cells with PE3. All pegRNAs evaluated convert the HBB E6V allele back to wild-type HBB without the introduction of any silent PAM mutation. (b) Screen of 41 pegRNAs for correction of the HEXA 1278+TATC allele in HEK293T cells with pE3 or PE3b. Those pegRNAs labeled HEXAs correct the pathogenic allele by a shifted 4-bp deletion that disrupts the PAM and leaves a silent mutation. Those pegRNAs labeled HEXA correct the pathogenic allele back to wild-type. Entries ending in “b” use an edit-specific nicking sgRNA in combination with the pegRNA (the PE3b system). Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Extended Data Figure 10. PE3 activity in human cell lines and comparison of PE3 and Cas9-initiated HDR.
(a) Prime editing in K562 (leukemic bone marrow), U2OS (osteosarcoma), and HeLa (cervical cancer) cells. Efficiency of generating the correct edit (without indels) and indel frequency for PE3 and Cas9-initiated HDR in (b) HEK293T cells, (c) K562 cells, (d) U2OS cells, and (e) HeLa cells. Each bracketed editing comparison installs identical edits with PE3 and Cas9-initiated HDR. Non-targeting controls are PE3 and a pegRNA that targets a non-target locus. (f) Control experiments with non-targeting pegRNA+PE3, and with dCas9+sgRNA, compared with wild-type Cas9 HDR experiments confirming that ssDNA donor HDR template, a common contaminant that artificially elevates apparent HDR efficiencies, does not contribute to the HDR measurements in (a-d). (g) Example HEK3 site allele tables from genomic DNA samples isolated from K562 cells after editing with PE3 or with Cas9-initiated HDR. Alleles were sequenced on an Illumina MiSeq and analyzed with CRISPResso243. The reference HEK3 sequence from this region is at the top. Allele tables are shown for a non-targeting pegRNA negative control, a +1 CTT insertion at HEK3 using PE3, and a +1 CTT insertion at HeK3 using Cas9-initiated HDR. Allele frequencies and corresponding Illumina sequencing read counts are shown for each allele. All alleles observed with frequency ≥0.20% are shown. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.
Extended Data Figure 11∣. Distribution by length of pathogenic insertions, duplications, deletions, and indels in the ClinVar database.
The ClinVar variant summary was downloaded from NCBI July 15, 2019. The lengths of reported insertions, deletions, and duplications were calculated using reference and alternate alleles, variant start and stop positions, or appropriate identifying information in the variant name. Variants that did not report any of the above information were excluded from the analysis. The lengths of reported indels (single variants that include both insertions and deletions relative to the reference genome) were calculated by determining the number of mismatches or gaps in the best pairwise alignment between the reference and alternate alleles. (a) Length distribution of insertions. (b) Length distribution of duplications. (c) Length distribution of deletions. (d) Length distribution of indels.
Supplementary Material
Acknowledgements
We thank J.M. Madison for neuron cell culture advice. This work was supported by the Merkin Institute of Transformative Technologies in Healthcare, U.S. NIH U01AI142756, RM1HG009490, R01EB022376, and R35GM118062, and HHMI. A.V.A. acknowledges a Jane Coffin Childs postdoctoral fellowship. P.B.R. and A.R. acknowledge NIH T32 GM095450. A.A.S. acknowledges NIH T32 GM007726. P.J.C. and A.R. acknowledge an NSF graduate fellowships. C.W. acknowledges a Damon Runyon Cancer Research Foundation fellowship (DRG-2343-18). G.A.N acknowledges a Helen Hay Whitney postdoctoral fellowship.
Footnotes
Supplementary Information is available in the online version of the paper.
The authors declare competing financial interests: authors through the Broad Institute have filed patent applications on prime editing. D.R.L. is a consultant and co-founder of Prime Medicine, Beam Therapeutics, Pairwise Plants, and Editas Medicine, companies that use genome editing.
References
- 1.Landrum MJ et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P et al. RNA-Guided Human Genome Engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haapaniemi E, Botla S, Persson J, Schmierer B & Taipale J CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med 24, 927–930 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Ihry RJ et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med 24, 939–946 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Rouet P, Smih F & Jasin M Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci 91, 6064–6068 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman JR, Taylor MRG & Boulton SJ Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Cox DBT, Platt RJ & Zhang F Therapeutic genome editing: prospects and challenges. Nat. Med 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paquet D et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Chu VT et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol 33, 543–548 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Maruyama T et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol 33, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rees HA, Yeh W-H & Liu DR Development of hRad51–Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat. Commun 10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen MW et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rees HA & Liu DR Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet 19, 770 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marraffini LA & Sontheimer EJ CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Kao H-I & Bambara RA Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem 73, 589–615 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Keijzers G, Bohr VA & Rasmussen LJ Human exonuclease 1 (EXO1) activity characterization and its function on flap structures. Biosci. Rep 35, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baranauskas A et al. Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Eng. Des. Sel 25, 657–668 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Gerard GF et al. The role of template-primer in protection of reverse transcriptase from thermal inactivation. Nucleic Acids Res. 30, 3118–3129 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arezi B & Hogrefe H Novel mutations in Moloney Murine Leukemia Virus reverse transcriptase increase thermostability through tighter binding to template-primer. Nucleic Acids Res. 37, 473–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotewicz ML, Sampson CM, D’Alessio JM & Gerard GF Isolation of cloned Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 16, 265–277 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimasu H et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 156, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thuronyi BW et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol (2019) doi: 10.1038/s41587-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol 35, 371–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koblan LW et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol (2018) doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinstiver BP et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannert N & Kurth R Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci 101, 14572–14579 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halvas EK, Svarovskaia ES & Pathak VK Role of Murine Leukemia Virus Reverse Transcriptase Deoxyribonucleoside Triphosphate-Binding Site in Retroviral Replication and In Vivo Fidelity. J. Virol 74, 10349–10358 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mead S et al. A Novel Protective Prion Protein Variant that Colocalizes with Kuru Exposure. N. Engl. J. Med 361, 2056–2065 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Asante EA et al. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature 522, 478–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kügler S, Kilic E & Bähr M Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 10, 337–347 (2003). [DOI] [PubMed] [Google Scholar]
Methods References
- 39.Anzalone AV, Lin AJ, Zairis S, Rabadan R & Cornish VW Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat. Methods 13, 453–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badran AH et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533, 58–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders C & Jinek M Chapter One - In Vitro Enzymology of Cas9. in Methods in Enzymology (eds. Doudna JA & Sontheimer EJ) vol. 546 1–20 (Academic Press, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirakitikulr N, Ostrov N, Peralta-Yahya P & Cornish VW PCRless library mutagenesis via oligonucleotide recombination in yeast. Protein Sci. Publ. Protein Soc 19, 2336–2346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy JM & Nicoll RA Membrane-associated guanylate kinase dynamics reveal regional and developmental specificity of synapse stability. J. Physiol 595, 1699–1709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zettler J, Schütz V & Mootz HD The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data have been deposited to the NCBI Sequence Read Archive database PRJNA565979. Plasmids encoding PE1, PE2/PE3, and pegRNA expression vectors have been deposited to Addgene for distribution.