Abstract
N-linked glycoprotein is a highly interesting class of proteins for clinical and biological research. Over the last decade, large-scale profiling of N-linked glycoproteins and glycosylation sites from biological and clinical samples has been achieved through mass spectrometry-based glycoproteomic approaches. In this paper, we reviewed the human glycoproteomic profiles that have been reported in more than 80 individual studies, and mainly focused on the N-glycoproteins and glycosylation sites identified through their deglycosylated forms of glycosite-containing peptides. According to our analyses, more than 30,000 glycosite-containing peptides and 7,000 human glycoproteins have been identified from five different body fluids, twelve human tissues (or related cell lines), and four special cell types. As the glycoproteomic data is still missing for many organs and tissues, a systematical glycoproteomic analysis of various human tissues and body fluids using a uniform platform is still needed for an integrated map of human N-glycoproteomes.
Keywords: Glycosylation, glycoprotein, glycosylation site, glycoproteome, mass spectrometry, human body fluids, human tissues, human cell lines
Introduction
Glycosylation is one of the most prevalent post-translational modifications of proteins. It not only plays major roles in folding, transport and localization of proteins [1], but also regulates various biological processes such as cell growth, viral replication and immune defense [2–4]. Aberrant glycosylation is usually associated with the pathological progression of many diseases [5]. N-linked glycoproteins are widely distributed, ranging from surface of various types of cells to different human body fluids such as serum, cerebrospinal fluid and urine [5, 6]. Transmembrane or cell surface glycoproteins are easily accessible to therapeutic drugs, antibodies, and ligands while glycoproteins secreted in body fluids are thought to provide a detailed window into the state of health of an individual. These features make glycoproteins a highly interesting class of proteins for clinical and biological research.
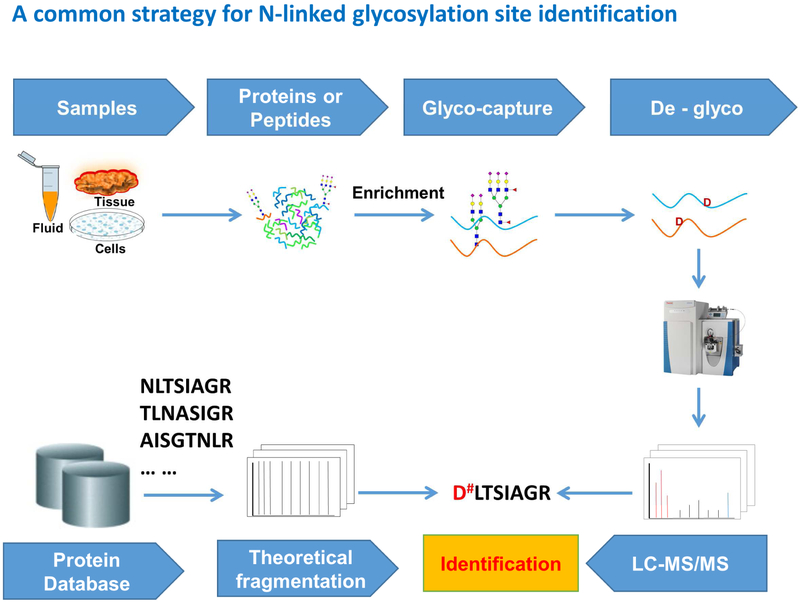
In the last decade, mass spectrometry (MS) has been widely used for large-scale glycoproteomic analysis. Typically, glycoproteins or glycopeptides are first enriched from complex samples using different enrichment methods, such as lectin affinity [7], hydrazide chemistry [8] and hydrophilic interaction liquid chromatography (HILIC) [9]; glycans are then cleaved from the glycoproteins or glycopeptides by enzymatic or chemical methods prior to MS analysis of deglycosylated peptides (Figure 1). As for N-linked glycosylation, the deamidation which occurs at the formerly N-glycosylation sites during the PNGase F treatment can serve as a mass tag (+0.98 Da, or +3 Da when performed in H218O) for N-glycosylation site identification using mass spectrometry [7, 8]. This simple but effective strategy has been widely applied into the large-scale analysis of N-linked glycoproteomes in various samples from human as well as other species.
Figure 1.
The most commonly used workflow for N-linked glycoprotein and glycosylation site identification. The glycopeptides or glycoproteins from complex samples are enriched, N-linked glycans are then removed from the peptide portion using PNGase F digestion. Deglycosylated peptides are analyzed by LC-MS/MS analysis, and the peptide portions of the glycopeptides are identified by database searching. The deamidation (+0.98 Da) occurs at the formerly N-glycosylation sites (after PNGase F) can be used as a mass tag for N-glycosylation site identification using mass spectrometry.
Using this strategy, thousands of N-linked glycoproteins have been identified from various human-derived samples through identifying their glycosite-containing peptides using mass spectrometry [12]. Recently, we have collected more than 30,000 human glycosite-containing peptides, representing >10,000 N-glycosites from >7,000 N-glycoproteins, from over 100 studies regarding to human glycosite-containing peptide analysis published since 2003. These human-derived samples include various body fluids, tissues and cell lines, which can be classified into more than 20 different sample type groups. The entire human glycoproteome database as well as sub-databases associated with individual body fluids or tissues can be found in the N-GlycositeAtlas website at nglycositeatlas.biomarkercenter.org.
In this paper, we summarized the progress of MS-based identification of human N-linked glycoproteins, focusing mainly on the identification of N-glycoproteins and glycosylation sites based on the deglycosylated forms of the glycosite-containing peptides. The glycoproteomic data were classified into three major groups based on their sample sources, including human body fluids, tissues (and related cell lines), and some special cell types such as blood platelets, B-cells, T-cells, as well as spermatozoa. We also discussed the glycosylation at some atypical N-glycosylation sites identified previously by mass spectrometry. Despite the development of novel methods for intact glycopeptide analysis in recent years, these data are beyond the scope of this review.
Glycoproteins in body liquids
Body fluid is one of the main resources for the identification of disease-related biomarkers. Now that N-linked glycoproteins account for a large portion of the protein content in the body fluids, identifying the glycoprotein components in these body fluids is essential for their clinical utility. Till now, N-glycoproteomic data identified by MS has been reported in serum/plasma, urine, saliva, cerebrospinal fluid (CSF) and milk samples (Table 1).
Table 1.
The numbers of unique N-linked glycoproteins and glycosylation sites identified from various human-derived samples including body fluids, tissues and related cell lines, as well as some special cell types.
| Category | Sample Source | Glycosites | Glycoproteins |
|---|---|---|---|
| Body Fluid | Serum or plasma | ~2,600 | ~1,500 |
| Urine | ~4,300 | ~2,600 | |
| Saliva | ~300 | ~200 | |
| Cerebrospinal fluid | ~1700 | ~700 | |
| Human milk | ~200 | ~100 | |
| Respiratory System | Lung | ~900 | ~600 |
| Digestive System | Liver | ~5,800 | ~2,500 |
| Pancreas | ~1,900 | ~1,200 | |
| Colon | ~2,100 | ~1,100 | |
| Reproductive System | Prostate | ~6,200 | ~3,300 |
| Ovary | ~5,700 | ~2,600 | |
| Cervix | ~1,300 | ~600 | |
| Urinary System | Kidney | ~1,100 | ~600 |
| Bladder | ~300 | ~200 | |
| Other Tissues | Breast | ~3,500 | ~1,700 |
| Thyroid | ~500 | ~300 | |
| Bone marrow | ~300 | ~200 | |
| Special Cell Types | Blood platelet | ~700 | ~400 |
| T cells | ~900 | ~600 | |
| B cells | ~2,200 | ~1,100 | |
| Spermatozoa | ~600 | ~300 |
The data were obtained from the N-glycositeAtlas website (glycositeatlas.biomarkercenter.org).
The numbers for glycosites and glycoproteins in tissues includes both tissue samples and related cell lines.
A. Serum or plasma
Serum and plasma glycoproteomic data can be obtained from dozens of papers, with one of the main reasons being that serum is an easily accessible sample and therefore was widely used for method development. For example, during the hydrazide chemistry method development for glycopeptide enrichment in 2003, Zhang et al. identified more than 300 deglycosylated peptides from 97 serum N-glycoproteins [8]. In 2004, plasma was again used as a test sample to evaluate the performance of the HILIC method for glycospeptide enrichment [9]. Using the HILIC method, the authors identified 62 N-glycosylation sites in 37 glycoproteins from plasma. In 2005, Liu et al. developed an approach mainly for plasma glycoprotein analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry [13]. In this study, the high abundance proteins were first subtracted from human blood plasma via immunoaffinity, then the remained glycoproteins were enriched by hydrazide chemistry method. After trypsin digestion and PNGase F treatment, the deglycosylated peptides were undertaken strong cation exchange (SCX) fractionation followed by LC−MS/MS analysis. Using this approach, they identified 639 N-glycosylation sites from 303 non-redundant glycoproteins.
In addition, N-glycoproteins from human plasma/serum have also been used for the development of many other methods, such as the glycoprotein/glycopeptide enrichment methods using acetone [14], hydrazide tips [15], multi-dimensional lectin affinity chromatography [16], and size exclusion chromatography [17]; methods for core-fucosylated glycoprotein/glycopeptide enrichment and identification [18–21]; methods for sialylated glycoprotein/glycopeptide enrichment using titanium dioxide(TiO2) chromatography [22] and strong cation exchange [23]; material synthesis and preparation such as magnetic microspheres [24] and nanoparticles [25] for glycoprotein/glycopeptide enrichment; methods for glycoprotein quantification including 18O stable isotope labeling [26] and SWATH-MS [27], et al. The numbers of the serum/plasma glycoproteins and glycosylation sites identified in these studies were among tens to hundreds. Human serum (or plasma) has also been widely used for biomarker discovery. Till now, mass spectrometry-based N-glycoprotein and glycosite analysis has been performed in the serum of HIV elite suppressors [28], patients with Parkinson’s disease [29], lung cancer [30, 31], liver cancer [32], as well as esophagus disease [33].
Altogether, a total of more than 3,000 glycosylation sites from ~1,800 glycoproteins have been identified from human serum and plasma. Part of these glycosite-containing peptides have been included in the UniPep [35] and PeptideAtlas [36] databases. Even though, more efforts may still be needed to maximize the identification coverage of serum/plasma glycoproteins and glycosites by using the combination of different strategies, such as different enzymatic digestion [37] and enrichment methods. This is particularly important for some low abundance glycoproteins. In addition, serum contains many high abundance glycoproteins, which might inhibit the identification of low abundance glycoproteins in serum. Removal of these high abundance proteins before mass spectrometry-based proteomic or sub-proteomic analyses would increase the coverage of the identified glycoproteins [38].
B. Urine
Urine is another potentially attractive fluid for disease biomarker discovery and diagnostic tests [39–43]. It is especially useful for the detection of lesions that are proximate to the urinary system, such as the kidney and prostate. There have been several publications focusing on the glycosylation site analysis of urinary glycoproteins in the last decade. In 2012, Yeh et al. identified 85 N-glycosites in 53 glycoproteins from urine samples using their novel magnetic bead-based zwitterionic HILIC (ZIC-HILIC) material [44]. One year later, Zhou et al. identified 865 N-glycosites from three urine samples using the GlycoFilter method [45]. In another paper, our group reported a total of 2,923 unique glycosite-containing peptides identified from the urine samples of prostate cancer patients with different Gleason scores using solid-phase extraction of N-linked glycosite-containing peptides (SPEG) and 2D-LC-MS/MS analysis [46]. Importantly, by comparing the identified glycoproteins among urine, tissues and serum from the same patients’ samples, we revealed that the majority of aggressive prostate cancer associated glycoproteins were more readily detected in patient’s urine than serum samples, suggesting the great potential of urinary glycoproteins for biomarker discovery.
C. Saliva
Whole saliva is a slightly cloudy colorless liquid which is mainly comprised of the secretions of the parotid, submandibular, sublingual and minor salivary glands, and it exhibits multiple host defense functions in the maintenance of oral health [47]. It has been reported that the protein content of saliva has a significant overlap with proteins from plasma, suggesting that saliva is also very promising in disease-related biomarker discovery as well as diagnostic tests [48, 49].
At least six mass spectrometry-based salivary glycoproteomic datasets have been published. In 2006, Ramachandran et al. enriched N-glycopeptides from the whole saliva using hydrazide chemistry method and analyzed the released formerly N-glycosylated peptides by LC−MS/MS [50]. With this approach, they first reported 84 deglycosylated peptides from 45 unique salivary N-glycoproteins. They subsequently extended the salivary glycoprotein catalogue using a modified hydrazide chemistry method and identified a total of 156 deglycosylated peptides, representing 77 unique N-glycoproteins in salivary fluid [51]. In 2007, by isolating the sialylated glycopeptides from whole salivary using the titanium dioxide chromatography, Larsen et al. identified 97 glycosylation sites from salivary glycoproteins [22], most of which were supposed to be sialic acid-containing glycopeptides (the sialiome) based on the principle of the enrichment method. Although titanium dioxide chromatography was initially developed to enrich the phosphorylated peptides, it was used for sialylated glycopeptide enrichment in this study. In 2014, by using a novel hexapeptide library method, Bandhakavi et al. further increased the number of identified salivary glycoproteins to 192 [52]. In the same year, with the combination of two complementary glycopeptide isolation methods, hydrazide chemistry and HILIC, our group identified a total of 156 non-redundant deglycosylated peptides (representing 164 unique N-glycosylation sites) and 85 N-linked glycoproteins in human whole saliva from six different gender and age groups [53].
D. Cerebrospinal fluid (CSF) and human milk
Several papers have been published on the MS-based analysis of formerly N-glycosylated peptides and glycosites from human CSF and milk. From CSF, a total of >1,600 glycosites from ~700 glycoproteins were identified in health and disease-related samples using multiple strategies, including gel separation, reversed phase-anion exchange and hydrazide-based glycopeptide capture [54–57]. In human milk, 240 N-glycosylation sites from 114 glycoproteins were identified in human colostrum and mature milk using HILIC enrichment, deglycosylation via PNGase F treatment and MS analysis [58–60]. These data might provide insight into the potential applications of some N-glycoproteins in infant formulae at different stages of development.
As preferred specimens for biomarker discovery, the analysis of human body fluids is very technically challenging because they contain a large number of proteins that could be modified in a variety of forms. Besides, detection of different diseases may require different body fluids but glycoproteomes of many other body fluids remain unknown, which include tear fluid, bile, bronchoalveolar lavage fluid, synovial fluid, nipple aspirate fluid, amniotic fluid, and so on [61, 62]. Despite these challenges, glycoproteomics of body fluid is still one of the most important and promising ways for the study of human diseases and disease-related biomarker discovery. For the biological studies and potentially clinical utilities, comprehensive and deeper glycoproteomic analysis from body fluids is still needed.
Glycoproteins in human tissues and related cell lines
Resolving the molecular details of (glyco)proteome variation in different tissues and organs of the human body is critical for the understanding of human biology and disease [63]. Glycoproteomic analysis of disease-related tissues and cells has provided valuable information to identify promising targets for their diagnosis, prognosis and therapy. In this part, we summarized recent glycoproteomic studies in 12 different tissues (and related cell lines), involving some closely related diseases (Table 1).
A. Respiratory system
Lung.
Various glycoproteomic technologies have been used for the biomarker discovery of lung cancers. By using lectin affinity chromatography, Hirao et al. identified 1092 AAL-bound glycoproteins and 948 HHL/ConA-bound glycoproteins in non-small cell lung carcinoma (NSCLC) in 2014. Unfortunately, the results lack the glycosite information of the identified glycoproteins [64]. Recently, by coupling lectin on magnetic nanoprobes, Waniwan et al. performed site-specific glycosylation analysis in drug-sensitive and resistant NSCLC cell lines, in which 2290 and 2767 non-redundant glycopeptides were confidently identified (Byonic score ≥100) in EGFR-TKI-sensitive PC9 and resistant PC9-IR cells, respectively [65]. In addition, glycoproteomic analyses have also been performed on tissues from different subtypes of NSCLC tissues, fetal lung fibroblasts and NSCLC cell lines with KRAS and EGFR mutations [66–68]. During the process, a few glycoproteins have been approved in Japan for clinical examinations of lung cancer, including SLX, CYFRA 21.1, SCC (squamous cell antigen), NCAM, Pro-gastrin-releasing peptide (ProGRP), NSE and carcinoembryonic antigen (CEA) [69].
B. Digestive system
Liver.
Besides the well-known marker AFP and AFP-L3, many biomarkers for HCC monitoring and diagnosis remain under research to improve the sensitivity and specificity of their use, which include several proteins with fucosylation, such as haptoglobin, kininogen and alpha-1-antitrypsin [70]. So far, many glycoproteins and glycopeptides have been determined to be altered in samples from HCC patients. For further information, the glycoproteomic studies of HCC and all methods involved have been well-summarized in a recent review by Zhu et al., 2018 [70].
Pancreas.
Currently, CA19–9 is the only clinical biomarker for management of pancreatic cancer, but it still suffers from the low sensitivity and specificity [71]. There have been several glycoproteomic analyses conducted in normal and disease-related pancreas samples. In 2011, Danzer et al. identified 956 unique N-linked glycoproteins including 611 N-linked glycoproteins in mouse MIN6 β-cells and 545 N-linked glycoproteins in human pancreatic islets [72]. Thereafter, to seek sialoglycoproteins associated with pancreatic cancer, Tian et al. analyzed the sialylated glycoproteins in metabolically oligosaccharide engineered pancreatic cells and identified 75 sialylated glycopeptides from 55 glycoproteins by lectin affinity chromatography combined with mass spectrometry [73]. More information can be obtained from the review by Pan et al. [71].
Colon.
As one of the most common malignant tumors of the gastrointestinal cancer, many attempts have been made to search for biomarkers for colorectal cancer (CRC) using glycoproteomic approaches [74, 75]. In 2009, Zhang et al. enriched the glycopeptides from human CRC tissues using a boronic acid functionalized core-satellite composite nanoparticles, in which they identified 194 unique glycosylation sites in 155 glycoproteins [76]. In 2011 Nagano et al. identified 219 glycosylated proteins by the monomeric avidin labeling and glycoprotein capturing in HCT-116 cells, and 312 N-glycosylated proteins in xenograft samples [77]. In 2014, Nicastri et al. used solid-phase extraction and 18O stable isotope labeling method and quantitatively identified 1459 glycopeptides and 770 glycoproteins, among which 54 glycoproteins were found to be up-regulated in colorectal cancer samples [78]. So far, there is still no effective early diagnostic markers for CRC and profiling of glycoproteome in CRC tissues/cells would provide new insights for the discovery of new biomarkers.
C. Reproductive system
Prostate.
Being one of the most important, death-related diseases in the male population, a number of glycoproteomic studies have been reported in prostate cancer. In 2012, a work by Whitmore et al. using hydrazide-based chemistry revealed 200 glycopeptides from 104 glycoproteins in 22Rv1 prostate cancer cells cultured in vitro [79]. Later, Liu et al. comparatively analyzed glycoproteomes of normal prostate, non-aggressive and metastatic prostate cancer tumor tissues and identified 5422 N-glycosites from 2460 glycoproteins using SWATH mass spectrometry, which essentially covers ~50% of the annotated human N-glycoproteome [80]. In 2015, we performed a quantitative glycoproteomic analysis on LNCaP and PC3 prostate cancer cell lines using solid-phase extraction and iTRAQ-labeled peptides followed by LC-MS/MS analysis. A total of 1810 unique N-glycopeptides from 653 identified N-glycoproteins were identified, among which 176 glycoproteins were observed to be differentially expressed between the two cell lines [81]. Due to their close relations, the glycosylation in bladder and prostate proteome is often analyzed together. In 2009, Goo et al. analyzed secreted glycoproteomes from both human prostate and bladder stromal cells, in which 116 prostate cell secreted glycoproteins and 84 bladder cell secreted glycoproteins were identified with Protein Prophet probability scores ≥ 0.9 using glycopeptide-capture method followed by mass spectrometry [82]. Biomarkers for prostate cancer have recently been reviewed by Intasqui et al., in which a list of clinical useful biomarkers/glycol-related biomarkers were also summarized [83].
Ovary.
Till now, the glycoprotein Cancer Antigen 125 (CA125) is the most frequently used biomarker for ovarian cancer detection and different glycoforms of CA125 have been investigated to improve its sensitivity and specificity [84, 85]. In 2011, our team analyzed three cases of normal ovary and ovarian cancer tissues and identified 368 N-glycosylation sites in 286 glycoproteins using a combined method of solid-phase extraction and iTRAQ labeled glycoproteins [86]. In 2013, we developed an approach for quantitative N-glycoproteomic analysis based on genomic N-glycosite prediction, which improved over three times the quantity of N-deglycopeptide assignments from ovarian cancer cell lines compared to the traditional method [87]. Later, we also developed a chemoenzymatic method called solid-phase extraction of N-linked glycans and glycosite-containing peptides (NGAG) for the comprehensive characterization of glycoproteins and identified 2,044 unique glycosite-containing peptides with an N-X-S/T motif from OVCAR-3 ovarian cancer cells [88].
Cervix.
The cervix is the lower part of the uterus in the human female reproductive system [89]. Infection with the human papillomavirus (HPV) can cause changes in the epithelium lined on the cervical canal, which can lead to cervix cancer [90]. In 2009, McDonald et al. identified 240 glycoproteins from the HeLa cells derived from cervical cancer cells, in which they combined the approaches of periodate/hydrazide chemistry and the M. amurensis lectin affinity chromatography [91]. In 2015, Weng et al. profiled the N-glycosylation sites of Hela cell line with combination of commonly applied protocol and a new method (N-terminal succinylation assisted enzymatic deglycosylation). In total, 1230 glycopeptides from 511 glycoproteins were mapped from HeLa cell lysate, which achieved deep coverage of glycosylation at N-terminal Asparagine of the glycopeptides [92].
D. Urinary system
Kidney.
The kidney is a main organ of the human body for urine generation. The human embryonic kidney 293T cell line (HEK 293T) has been used in the development of several glycoproteomic methods. In 2011, Chen et al. developed a novel two-step protease digestion (Lys-C and trypsin) and glycopeptide capture approach and applied it for the analysis of HEK 293T cells, where a total of 359 glycosites from 143 N-glycoproteins were identified [93]. In 2014, HEK 293T cell line was again analyzed by an integrated hydrophilic interaction chromatography solid-phase interaction (HILIC-SPE) and mass spectrometric strategy, and 811 N-glycosites from 567 proteins were identified [94]. The next year, cell surface N-glycoproteins from HEK293T were analyzed by integrating metabolic labeling, copper-free click chemistry, and MS-based proteomics methods and lead to the identification of 144 unique N-glycopeptides from 110 glycoproteins containing 152 N-glycosylation sites [95]. Unfortunately, no data is available yet for the glycoproteome of human renal tissues.
E. Other tissues
Breast.
Breast-related cancer has a high incidence rate in female and a large number of studies have been conducted on the diagnosis, occurrence, and treatment of breast cancer. In 2012, Yen et al. identified 486 glycoproteins from 14 breast cancer cell lines using hydrazide magnetic beads, in which a panel of differentially expressed glycoproteins were determined to allow the classification of the breast cancer [96]. One year later, Boersema et al. identified and quantified 1398 N-glycosylation sites from the supernatant of 11 cell lines that represent different stages of breast cancer development using the N-glyco-FASP technology coupled with super-SILAC accurate quantification [97]. In addition, glycoproteomic analysis was also performed in triple-negative and luminal breast tumors, in which a panel of glycoproteins were determined to be differentially expressed [98, 99].
Thyroid.
The glycoproteomic analysis of thyroid tissues and cell lines is still very limited. In 2009, glycoproteome profiles of cell surface and secreted proteins were reported using a serial of thyroid cancer cell lines, including papillary thyroid cancer (TPC-1), follicular thyroid cancer (FTC-133), Hürthle cell carcinoma (XTC-1), and anaplastic thyroid cancer (ARO and DRO-1), which represented the range of thyroid cancers of follicular cell origin [100]. An average of 150 glycoproteins were identified in each cell line but with distinct glycoproteomic patterns, of which more than 57% were known as cell surface or secreted glycoproteins. Based on the results obtained, a set of glycoprotein biomarker candidates, such as CD44, galectin 3 and metalloproteinase inhibitor 1, have been proposed for thyroid cancer diagnosis.
Bone marrow.
To understand the chemoresistance in the chemotherapy of leukemia, Zhang et al. investigated the differences between the glycoproteomes of adriamycin-sensitive cell line K562S and the adriamycin-resistant leukemia cell line K562A using the cell-surface capturing method and quantitative proteomics [101]. A total of 134 glycopeptides from 94 glycoproteins in K562S cells and 244 glycopeptides from 180 glycoproteins in K562A cells were identified with FDR<1% at the peptide level. Using two quantitative methods (isotopic dimethyl labeling and SWATH), 15 glycoproteins were found to display a consistent significant change trend between these cell lines. These 15 proteins included classical multidrug resistance-related glycoproteins such as ABCB1 as well as three glycoproteins (CTSD, FKBP10, and SLC2A1) that were shown to be novel participants in the chemoresistance of leukemia cancer cells.
With the rapid improvements of mass spectrometry and glycoproteomic technologies, more and more MS data related to the glycosites and glycoforms from different tissues will be generated. Therefore, how to take advantage of these data and solve problems in diseases and fundamental biology will be the next challenge.
Glycoproteins in special cell types
There are some special cell types in human that researchers are particularly interested in. Hereby, glycoproteomic studies in special cell types are summarized, including human blood platelets, T cells, B cells and spermatozoa (Table 1).
As a critical component of human blood, more than 1000 N-glycosylation sites have been identified from blood platelets by MS. In 2006, Lewandrowski et al. reported the first deglycopeptide dataset from human blood platelets by using both lectin and hydrazide chemistry enrichment methods, PNGase F treatment and mass spectrometry [102]. Using this approach, they identified 70 different glycosylation sites from 41 different platelet glycoproteins. One year later, they published the second paper reporting 148 N-linked glycosylation sites on 79 glycoproteins from the platelet membrane [103]. In 2008, the same group reported 125 glycosylation sites on 66 proteins from platelets by glycopeptides enrichment using electrostatic repulsion hydrophilic interaction chromatography (ERLIC) [104]. In total, more than 250 glycosylation sites annotated for platelets were identified using three different enrichment strategies (four different enrichment methods) [104]. In 2017, our group published another MS dataset related to platelet glycoproteins. By using iTRAQ labeling and SPEG (hydrazide chemistry) enrichment, we identified 799 unique N-linked glycosylation sites in platelets [105], which further extended the depth of the glycoprotein coverage of human platelets. Up to now, more than one thousand N-glycoproteins have been identified from human platelets by MS.
Besides blood platelets, glycoproteomic analysis on T cells, B cells and spermatozoa have been reported as well. From T cell line ACH-2, Yang et al. identified 563 unique glycopeptides from 247 unique glycoproteins using hydrazide chemistry enrichment method [106]. In 2014, Deeb et al. identified 2383 unique glycosylation sites on 1321 glycoproteins from the cell surface of diffuse large B-cell lymphoma subtypes by glyco-FASP method and MS analysis [107]. Using glyco-FASP method coupled with LC-MS/MS, Wang et al. identified 554 N-glycosylation sites and 297 N-glycoproteins from human spermatozoa [108].
Atypical N-glycosylation sites
In addition to the canonical N-linked glycosylation motif asparagine-X-serine/threonine (N-XS/T, X is any amino acid except proline) [5, 109], many sites with atypical sequons have been identified by MS as N-linked glycosylation sites. These reported atypical glycosites include NX-C [110], N-X-V [111, 112] and N-X-G [112]. Except for the N-X-C motif, which has been confirmed in several known glycoproteins [110], all other atypical motifs were only identified based on the deamidation of asparagine (N) residues in the peptides after PNGase F treatment (with/without 18O labeling) using mass spectrometry-based glycoproteomics [111, 112]. However, the atypical sites identified based on deamidation of N are potentially false positives as it could occur naturally or be induced during sample preparation [10, 11].
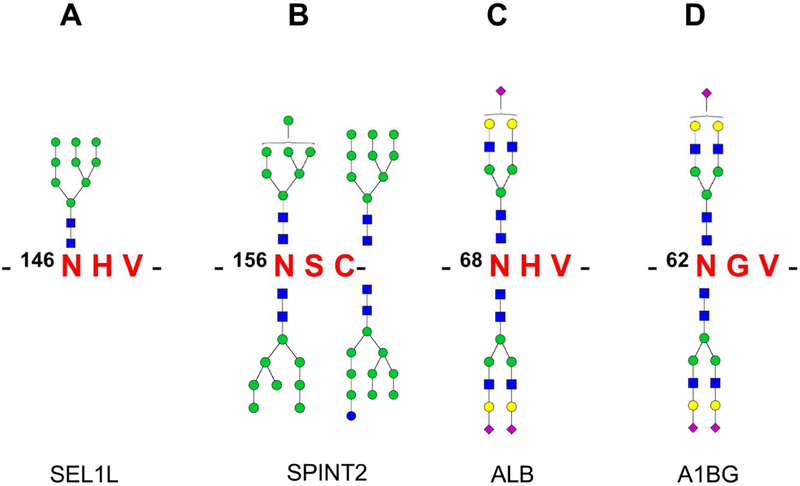
Recently, by using the NGAG method coupled with high resolution mass spectrometry, we identified two atypical N-glycosites with 146N#HV and 156N#SC motifs on Protein sel-1 homolog 1 (SEL1L) and Kunitz-type protease inhibitor 2 (SPINT2) from OVCAR-3 cell line, respectively [113]. These two atypical glycosites were further confirmed via the direct identification of their intact N-glycopeptide forms (with glycan still attached at the glycosylation site). The results showed that the glycosite 146N#HV was modified by the glycan Man9 (Figure 2A) while the glycosite 156N#SC was modified by four different oligo-mannose glycans (HexNAc2Hex7 -HexNAc2Hex10, Figure 2B). Using the same strategy, another two atypical N-glycosites with the N-X-V motif, which included 68N#EV on Albumin (ALB) and 62N#GV on Alpha-1B-glycoprotein (A1BG), were identified and verified from human serum [34]. Both glycosites were modified by complex glycans N4H5S1 and N4H5S2 (Figure 2C and 2D).
Figure 2.
Atypical N-glycosites identified from human glycoproteins. Four glycosites identified from (A) Protein sel-1 homolog 1 (SEL1L), (B) Kunitz-type protease inhibitor 2 (SPINT2), (C) Albumin (ALB), and (D) Alpha-1B-glycoprotein (A1BG).
Compared to previously reported atypical N-glycosites that were identified based on the deamidation of asparagine residues after PNGase F treatment [111, 112], these two studies further validated the identified atypical motif glycosites by directly identifying their intact glycopeptides. Since the deamidation of N can sometimes cause false-positives, direct identification of the intact glycopeptides with glycan attached is particularly important for confirmation of these atypical N-glycosylation sites detected by mass spectrometry-based glycoproteomics.
Future perspectives
Owing to the huge improvement of mass spectrometry technology, liquid chromatography separation, analysis software and glycoprotein/glycosite-containing peptide isolation methods in recent years, many mass spectrometry methods and strategies have been developed for large-scale profiling of N-linked glycoproteins and glycosylation sites. Prior to MS analysis, glycopeptide enrichment is usually performed to increase the concentration of the glycopeptides. Then glycans are normally removed from the glycosite-containing peptides to eliminate the micro-heterogeneity of the glycosylation. By using this strategy, large-scale glycosylation analysis has been achieved by employing proteomic approaches, and the glycan attached sites can be identified through the deamidation (with +0.98 Da shift) at the formerly glycosylation sites [7, 8]. The majority of human N-linked glycoproteins and glycosylation sites known nowadays were identified though this strategy. This strategy, however, would cause false identification when the mass accuracy of the detected peptide is not high enough. The high resolution and accuracy of mass spectrometers developed in recent years have greatly increased the identification confidence of the glycosites and glycoproteins, and increased the numbers of identified glycosite-containing peptides, but the false positive rates of identified glycosylation sites using this strategies are still much higher than we expected, as deamidation of asparagine could be induced during sample preparation, and even occur naturally in human body, as reported in recent studies [10, 11]. The naturally occurred deamidation can be influenced by several factors, such as pH (in particular pH conditions for PNGase F driven deglycosylation), temperature, ionic strength, and others [10]. Therefore, appropriate controls and extreme care have to be taken in both sample preparation and data interpretation to prevent false-positive identifications [11].
Based on the data summarized above, although several thousand glycoproteins could be identified from one single body fluid or tissue, there are still many tissues (such as heart, stomach, brain, and kidney) in which no mass spectrometry-based glycosite data has been generated yet. In addition, the identification numbers of glycoproteins and glycosylation sites are largely determined by the approaches of sample preparation, mass spectrometry analysis as well as data analysis. Therefore, a systematically glycoproteomic analysis of various human body fluids and tissues using a uniform platform may still be needed. This uniform platform may need to combine several different strategies, such as using different enzymatic digestion [37], enrichment methods, as well as intact glycopeptide analysis, to maximize the identification coverage of human glycoproteins and glycosites. Such work will help to create a high quality of human glycoproteome database to facilitate the future studies of glycoprotein structures and functions.
Along with glycosylation sites, characterization of the glycans attached is also essential for the understanding the biological roles of a glycoprotein. Although the overall glycoform diversities can be revealed by analyzing the released glycans or de-glycosylated peptides, the “true” glycan heterogeneity still need to be uncovered at the intact glycopeptides or glycoproteins level. In recent years, direct analysis of intact glycopeptide/glycoproteins was enabled with the development of mass spectrometry technologies and softwares [88, 125–129]. Detailed information is available from reviews by Cao et al., 2016 and Yang et al., 2017 [130, 131]. Intact glycopeptide analysis can provide additional site-specific glycosylation information, which allows us to know which glycans are attached to which glycosylation sites [88]. These technologies will indisputably push glycoproteomic researches to a higher stage.
HIGHLIGHTS.
Large-scale profiling of N-linked glycoproteins and glycosylation sites from various human–derived samples has been achieved by mass spectrometry.
Glycoproteomic profiles from five body fluids, 12 human tissues, and four special cell types were summarized.
Mass spectrometry has identified more than 30,000 glycosite-containing peptides from >7,000 human N-glycoproteins.
N-glycosylation at the atypical sites with N-X-C and N-X-V sequons has been confirmed in several human glycoproteins.
A systematical glycoproteomic analysis of all kinds of body fluids and human tissues using a uniform platform would be necessary for an integrated map of human N-glycoproteomes.
ACKNOWLEDGEMENTS
This work was supported by National Natural Science Foundation of China (Grant No. 91853123, 21705127, 81773180 and 81800655), and Natural Science Foundation of Shaanxi Province (Grant No. 2018JM7086074). Dr. Hui Zhang was supported by the National Institutes of Health, National Cancer Institute, the Early Detection Research Network (EDRN, U01CA152813), the Clinical Proteomic Tumor Analysis Consortium (CPTAC, U24CA210985), National Heart Lung and Blood Institute, Programs of Excellence in Glycosciences (PEG, P01HL107153), and the National Institute of Allergy and Infectious Diseases (R21AI122382).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
The authors declare that they have no competing interests.
REFERENCES
- [1].Hirabayashi J, Arata Y, Kasai K, Proteomics 1 (2001) 295. [DOI] [PubMed] [Google Scholar]
- [2].Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA, Science 291 (2001) 2370. [DOI] [PubMed] [Google Scholar]
- [3].Haltiwanger RS, Current Opinion in Structural Biology 12 (2002) 593. [DOI] [PubMed] [Google Scholar]
- [4].Haltiwanger RS, Lowe JB, Annual Review of Biochemistry 73 (2004) 491. [DOI] [PubMed] [Google Scholar]
- [5].Ohtsubo K, Marth JD, Cell 126 (2006) 855. [DOI] [PubMed] [Google Scholar]
- [6].Zhang H, Chan DW, Cancer Epidemiology Biomarkers & Prevention 16 (2007) 1915. [DOI] [PubMed] [Google Scholar]
- [7].Madera M, Mechref Y, Novotny MV, Analytical Chemistry 77 (2005) 4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang H, Li XJ, Martin DB, Aebersold R, Nature Biotechnology 21 (2003) 660. [DOI] [PubMed] [Google Scholar]
- [9].Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P, Journal of Proteome Research 3 (2004) 556. [DOI] [PubMed] [Google Scholar]
- [10].Palmisano G, Melo-Braga MN, Engholm-Keller K, Parker BL, Larsen MR, Journal of Proteome Research 11 (2012) 1949. [DOI] [PubMed] [Google Scholar]
- [11].Hao P, Ren Y, Alpert AJ, Sze SK, Molecular & Cellular Proteomics 10 (2011) O111. 009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tian Y, Zhang H, Proteomics 13 (2013) 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu T, Qian WJ, Gritsenko MA, Camp DG, Monroe ME, Moore RJ, Smith RD, Journal of Proteome Research 4 (2005) 2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Takakura D, Harazono A, Hashii N, Kawasaki N, Journal of Proteomics 101 (2014) 17. [DOI] [PubMed] [Google Scholar]
- [15].Chen J, Shah P, Zhang H, Analytical chemistry 85 (2013) 10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Qiu RQ, Regnier FE, Analytical Chemistry 77 (2005) 2802. [DOI] [PubMed] [Google Scholar]
- [17].Alvarez-Manilla G, Atwood J, Guo Y, Warren NL, Orlando R, Pierce M, Journal of Proteome Research 5 (2006) 701. [DOI] [PubMed] [Google Scholar]
- [18].Ma C, Zhang Q, Qu JY, Zhao XY, Li X, Liu YP, Wang PG, Journal of Proteomics 114 (2015) 61. [DOI] [PubMed] [Google Scholar]
- [19].Tan ZJ, Yin HD, Nie S, Lin ZX, Zhu JH, Ruffin MT, Anderson MA, Simone DM, Lubman DM, Journal of Proteome Research 14 (2015) 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jia W, Lu Z, Fu Y, Wang HP, Wang LH, Chi H, Yuan ZF, Zheng ZB, Song LN, Han HH, Liang YM, Wang JL, Cai Y, Zhang YK, Deng YL, Ying WT, He SM, Qian XH, Molecular & Cellular Proteomics 8 (2009) 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hagglund P, Matthiesen R, Elortza F, Hojrup P, Roepstorff P, Jensen ON, Bunkenborg J, Journal of Proteome Research 6 (2007) 3021. [DOI] [PubMed] [Google Scholar]
- [22].Larsen MR, Jensen SS, Jakobsen LA, Heegaard NHH, Molecular & Cellular Proteomics 6 (2007) 1778. [DOI] [PubMed] [Google Scholar]
- [23].Lewandrowski U, Zahedi RP, Moebius J, Walter U, Sickmann A, Molecular & Cellular Proteomics 6 (2007) 1933. [DOI] [PubMed] [Google Scholar]
- [24].Wang M, Zhang X, Deng C, Proteomics 15 (2015) 2158. [DOI] [PubMed] [Google Scholar]
- [25].Wang Y, Liu M, Xie L, Fang C, Xiong H, Lu H, Analytical Chemistry 86 (2014) 2057. [DOI] [PubMed] [Google Scholar]
- [26].Liu Z, Cao L, He Y, Qiao L, Xu C, Lu H, Yang P, Journal of Proteome Research 9 (2010) 227. [DOI] [PubMed] [Google Scholar]
- [27].Liu Y, Huettenhain R, Surinova S, Gillet LCJ, Mouritsen J, Brunner R, Navarro P, Aebersold R, Proteomics 13 (2013) 1247. [DOI] [PubMed] [Google Scholar]
- [28].Yang W, Laeyendecker O, Wendel SK, Zhang B, Sun S, Zhou J-Y, Ao M, Moore RD, Jackson JB, Zhang H, Theranostics 4 (2014) 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pan C, Zhou Y, Dator R, Ginghina C, Zhao Y, Movius J, Peskind E, Zabetian CP, Quinn J, Galasko D, Stewart T, Shi M, Zhang J, Journal of Proteome Research 13 (2014) 4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zeng X, Hood BL, Sun M, Conrads TP, Day RS, Weissfeld JL, Siegfried JM, Bigbee WL, Journal of Proteome Research 9 (2010) 6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim JY, Kim SK, Kang D, Moon MH, Analytical Chemistry 84 (2012) 5343. [DOI] [PubMed] [Google Scholar]
- [32].Lee HJ, Na K, Choi E-Y, Kim KS, Kim H, Paik Y-K, Journal of Proteome Research 9 (2010) 308. [DOI] [PubMed] [Google Scholar]
- [33].Song E, Zhu R, Hammond ZT, Mechref Y, Journal of Proteome Research 13 (2014) 4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sun S, Hu Y, Jia L, Eshghi ST, Liu Y, Shah P, Zhang H, Analytical Chemistry 90 (2018) 6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang H, Loriaux P, Eng J, Campbell D, Keller A, Moss P, Bonneau R, Zhang N, Zhou Y, Wollscheid B, Cooke K, Yi EC, Lee H, Peskind ER, Zhang J, Smith RD, Aebersold R, Genome Biology 7 (2006) R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmström J, Ossola R, Molecular & Cellular Proteomics 10 (2011) M110. 006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H, J Proteome Res 8 (2009) 651. [DOI] [PubMed] [Google Scholar]
- [38].Liu T, Qian WJ, Gritsenko MA, Camp DG, Monroe ME, Moore RJ, Smith RD, Journal of proteome research 4 (2005) 2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pang JX, Ginanni N, Dongre AR, Hefta SA, Opiteck GJ, Journal of Proteome Research 1 (2002) 161. [DOI] [PubMed] [Google Scholar]
- [40].Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield X, Widmark A, British journal of cancer 100 (2009) 1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ploussard G, de la Taille A, Nature Reviews Urology 7 (2010) 101. [DOI] [PubMed] [Google Scholar]
- [42].Haj-Ahmad TA, Abdalla MA, Haj-Ahmad Y, Journal of Cancer 5 (2014) 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Struck-Lewicka W, Kordalewska M, Bujak R, Yumba Mpanga A, Markuszewski M, Jacyna J, Matuszewski M, Kaliszan R, Markuszewski MJ, J Pharm Biomed Anal 111 (2015) 351. [DOI] [PubMed] [Google Scholar]
- [44].Yeh CH, Chen SH, Li DT, Lin HP, Huang HJ, Chang CI, Shih WL, Chern CL, Shi FK, Hsu JL, Journal of Chromatography A 1224 (2012) 70. [DOI] [PubMed] [Google Scholar]
- [45].Zhou H, Froehlich JW, Briscoe AC, Lee RS, Molecular & Cellular Proteomics 12 (2013) 2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jia X, Chen J, Sun S, Yang W, Yang S, Shah P, Hoti N, Veltri B, Zhang H, Proteomics 16 (2016) 2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Edgar WM, British Dental Journal 172 (1992) 305. [DOI] [PubMed] [Google Scholar]
- [48].Malamud D, Dental clinics of North America 55 (2011) 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Loo JA, Yan W, Ramachandran P, Wong DT, Journal of Dental Research 89 (2010) 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA, Journal of Proteome Research 5 (2006) 1493. [DOI] [PubMed] [Google Scholar]
- [51].Ramachandran P, Boontheung P, Pang E, Yan W, Wong D, Loo J, Clinical Proteomics 4 (2008) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bandhakavi S, Van Riper SK, Tawfik PN, Stone MD, Haddad T, Rhodus NL, Carlis JV, Griffin TJ, Journal of Proteome Research 10 (2010) 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sun S, Zhao F, Wang Q, Zhong Y, Cai T, Wu P, Yang F, Li Z, Clinical Proteomics 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Guldbrandsen A, Vethe H, Farag Y, Oveland E, Garberg H, Berle M, Myhr KM, Opsahl JA, Barsnes H, Berven FS, Molecular & Cellular Proteomics 13 (2014) 3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Goyallon A, Cholet S, Chapelle M, Junot C, Fenaille F, Rapid Communications in Mass Spectrometry 29 (2015) 461. [DOI] [PubMed] [Google Scholar]
- [56].Kroksveen AC, Guldbrandsen A, Vaudel M, Lereim RR, Barsnes H, Myhr KM, Torkildsen O, Berven FS, J Proteome Res 16 (2017) 179. [DOI] [PubMed] [Google Scholar]
- [57].Wang JX, Cunningham R, Zetterberg H, Asthana S, Carlsson C, Okonkwo O, Li LJ, Proteomics Clinical Applications 10 (2016) 1225. [DOI] [PubMed] [Google Scholar]
- [58].Picariello G, Ferranti P, Mamone G, Roepstorff P, Addeo F, Proteomics 8 (2008) 3833. [DOI] [PubMed] [Google Scholar]
- [59].Cao X, Yang M, Yang N, Liang X, Tao D, Liu B, Wu J, Yue X, Food Chem 276 (2019) 266. [DOI] [PubMed] [Google Scholar]
- [60].Cao X, Song D, Yang M, Yang N, Ye Q, Tao D, Liu B, Wu R, Yue X, J Agric Food Chem 65 (2017) 10360. [DOI] [PubMed] [Google Scholar]
- [61].Ahn SM, Simpson RJ, Proteomics Clin Appl 1 (2007) 1004. [DOI] [PubMed] [Google Scholar]
- [62].Hu S, Loo JA, Wong DT, Proteomics 6 (2006) 6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Science 347 (2015) 1260419. [DOI] [PubMed] [Google Scholar]
- [64].Hirao Y, Matsuzaki H, Iwaki J, Kuno A, Kaji H, Ohkura T, Togayachi A, Abe M, Nomura M, Noguchi M, Ikehara Y, Narimatsu H, J Proteome Res 13 (2014) 4705. [DOI] [PubMed] [Google Scholar]
- [65].Waniwan JT, Chen YJ, Capangpangan R, Weng SH, Chen YJ, Journal of Proteome Research 17 (2018) 3761. [DOI] [PubMed] [Google Scholar]
- [66].Clark DJ, Mei Y, Sun S, Zhang H, Yang AJ, Mao L, Theranostics 6 (2016) 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yang S, Chen L, Chan DW, Li QK, Zhang H, Clin Proteomics 14 (2017) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Takakura D, Tada M, Kawasaki N, Proteomics 16 (2016) 47. [DOI] [PubMed] [Google Scholar]
- [69].Togayachi A, Iwaki J, Kaji H, Matsuzaki H, Kuno A, Hirao Y, Nornura M, Noguchi M, Ikehara Y, Narimatsu H, Journal of Proteome Research 16 (2017) 4495. [DOI] [PubMed] [Google Scholar]
- [70].Zhu J, Warner E, Parikh ND, Lubman DM, Mass Spectrom Rev (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pan S, Brentnall TA, Chen R, World Journal of Gastroenterology 22 (2016) 9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Danzer C, Eckhardt K, Schmidt A, Fankhauser N, Ribrioux S, Wollscheid B, Muller L, Schiess R, Zullig R, Lehmann R, Spinas G, Aebersold R, Krek W, J Proteome Res 11 (2012) 1598. [DOI] [PubMed] [Google Scholar]
- [73].Tian Y, Almaraz RT, Choi CH, Li QK, Saeui C, Li D, Shah P, Bhattacharya R, Yarema KJ, Zhang H, Clin Proteomics 12 (2015) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lee PY, Chin SF, Low TY, Jamal R, J Proteomics 187 (2018) 93. [DOI] [PubMed] [Google Scholar]
- [75].Sethi MK, Hancock WS, Fanayan S, Acc Chem Res 49 (2016) 2099. [DOI] [PubMed] [Google Scholar]
- [76].Zhang L, Xu Y, Yao H, Xie L, Yao J, Lu H, Yang P, Chemistry - A European Journal 15 (2010) 10158. [DOI] [PubMed] [Google Scholar]
- [77].Nagano K, Shinkawa T, Kato K, Inomata N, Yabuki N, Haramura M, Journal of Proteomics 74 (2011) 1985. [DOI] [PubMed] [Google Scholar]
- [78].Nicastri A, Gaspari M, Sacco R, Elia L, Gabriele C, Romano R, Rizzuto A, Cuda G, Journal of Proteome Research 13 (2014) 4932. [DOI] [PubMed] [Google Scholar]
- [79].Whitmore TE, Peterson A, Holzman T, Eastham A, Amon L, McIntosh M, Ozinsky A, Nelson PS, Martin DB, J Proteome Res 11 (2012) 2653. [DOI] [PubMed] [Google Scholar]
- [80].Liu Y, Chen J, Sethi A, Li QK, Chen L, Collins B, Gillet LC, Wollscheid B, Zhang H, Aebersold R, Mol Cell Proteomics 13 (2014) 1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Shah P, Wang X, Yang W, Eshghi ST, Sun S, Hoti N, Chen L, Yang S, Pasay J, Rubin A, Zhang H, Molecular & Cellular Proteomics 14 (2015) 2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Goo YA, Lilu AY, Ryu S, Shaffer SA, Malmstrom L, Page L, Nguyen LT, Doneanu CE, Goodlett DR, Prostate 69 (2009) 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Intasqui P, Bertolla RP, Sadi MV, Expert Rev Proteomics 15 (2018) 65. [DOI] [PubMed] [Google Scholar]
- [84].Elzek MA, Rodland KD, Cancer Metastasis Rev 34 (2015) 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Swiatly A, Plewa S, Matysiak J, Kokot ZJ, J Ovarian Res 11 (2018) 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tian Y, Yao Z, Roden R, Zhang H, Proteomics 11 (2011) 4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sun S, Zhang B, Aiyetan P, Zhou JY, Shah P, Yang W, Levine DA, Zhang Z, Chan DW, Zhang H, Journal of Proteome Research 12 (2015) 5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sun S, Shah P, Eshghi ST, Yang W, Trikannad N, Yang S, Chen L, Aiyetan P, Hoti N, Zhang Z, Chan DW, Zhang H, Nature Biotechnology 34 (2016) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Goldenberg RL, Culhane JF, Iams JD, Romero R, Lancet 371 (2008) 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tran NP, Hung CF, Roden RBS, Wu TC, Recent results in cancer research 193 (2014) 149. [DOI] [PubMed] [Google Scholar]
- [91].McDonald CA, Yang JY, Marathe V, Yen TY, Macher BA, Mol Cell Proteomics 8 (2009) 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Weng Y, Sui Z, Jiang H, Shan Y, Chen L, Zhang S, Zhang L, Zhang Y, Sci Rep 5 (2015) 9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chen Y, Cao J, Yan G, Lu H, Yang P, Talanta 85 (2011) 70. [DOI] [PubMed] [Google Scholar]
- [94].Chen R, Seebun D, Ye M, Zou H, Figeys D, J Proteomics 103 (2014) 194. [DOI] [PubMed] [Google Scholar]
- [95].Smeekens JM, Chen W, Wu R, Journal of the American Society for Mass Spectrometry 26 (2015) 604. [DOI] [PubMed] [Google Scholar]
- [96].Yen TY, Macher BA, McDonald CA, Alleyne-Chin C, Timpe LC, J Proteome Res 11 (2012) 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Boersema PJ, Geiger T, Wisniewski JR, Mann M, Mol Cell Proteomics 12 (2013) 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chen X, Wu J, Huang H, Ding Q, Liu X, Chen L, Zha X, Liang M, He J, Zhu Q, Wang S, Xia T, Cell Physiol Biochem 38 (2016) 110. [DOI] [PubMed] [Google Scholar]
- [99].Hill JJ, Tremblay TL, Fauteux F, Li J, Wang E, Aguilar-Mahecha A, Basik M, O’Connor-McCourt M, J Proteome Res 14 (2015) 1376. [DOI] [PubMed] [Google Scholar]
- [100].Arcinas A, Yen TY, Kebebew E, Macher BA, Journal of Proteome Research 8 (2009) 3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sun Z, Dong J, Zhang S, Hu Z, Cheng K, Li K, Xu B, Ye M, Nie Y, Fan D, Zou H, Journal of Proteome Research 13 (2014) 1593. [DOI] [PubMed] [Google Scholar]
- [102].Lewandrowski U, Moebius J, Walter U, Sickmann A, A Glycoproteomic Approach 5 (2006) 226. [DOI] [PubMed] [Google Scholar]
- [103].Lewandrowski U, Zahedi RP, Moebius J, Walter U, Sickmann A, Molecular & Cellular Proteomics 6 (2007) 1933. [DOI] [PubMed] [Google Scholar]
- [104].Lewandrowski U, Lohrig K, Zahedi RP, Wolters D, Sickmann A, Clinical Proteomics 4 (2008) 25. [Google Scholar]
- [105].Shah P, Yang W, Sun S, Pasay J, Faraday N, Zhang H, Proteomics 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yang W, Zhou JY, Chen L, Ao M, Sun S, Aiyetan P, Simmons A, Zhang H, Jackson JB, Clinical Proteomics 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Deeb SJ, Cox J, Schmidt-Supprian M, Mann M, Molecular & Cellular Proteomics 13 (2014) 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang GG, Wu YB, Zhou T, Guo YS, Zheng B, Wang J, Bi Y, Liu FJ, Zhou ZM, Guo XJ, Sha JH, Journal of Proteome Research 12 (2013) 5750. [DOI] [PubMed] [Google Scholar]
- [109].Ajit V, Richard C, Jeffrey E, Hudson F, Gerald H, Jamey M, Cold Spring Harbor Laboratory Press, New York: (2009). [Google Scholar]
- [110].Bause E, Legler G, Biochemical Journal 195 (1981) 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Baycin-Hizal D, Tian Y, Akan I, Jacobson E, Clark D, Wu A, Jampol R, Palter K, Betenbaugh M, Zhang H, Analytical Chemistry 83 (2011) 5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zielinska DF, Gnad F, Wi M Mann, Cell 141 (2010) 897. [DOI] [PubMed] [Google Scholar]
- [113].Sun S, Zhang H, Analytical Chemistry 87 (2015) 11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Shah M, Telang S, Raval G, Shah P, Patel PS, Cancer 113 (2008) 336. [DOI] [PubMed] [Google Scholar]
- [115].Holst S, Deuss AJ, van Pelt GW, van Vliet SJ, Garcia-Vallejo JJ, Koeleman CA, Deelder AM, Mesker WE, Tollenaar RA, Rombouts Y, Wuhrer M, Mol Cell Proteomics 15 (2016) 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhou J, Yang W, Hu Y, Hoti N, Liu Y, Shah P, Sun S, Clark D, Thomas S, Zhang H, Anal Chem 89 (2017) 7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Jia L, Zhang J, Ma T, Guo Y, Yu Y, Cui J, Front Oncol 8 (2018) 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Miyoshi E, Moriwaki K, Nakagawa T, J Biochem 143 (2008) 725. [DOI] [PubMed] [Google Scholar]
- [119].Miyoshi E, Moriwaki K, Terao N, Tan CC, Terao M, Nakagawa T, Matsumoto H, Shinzaki S, Kamada Y, Biomolecules 2 (2012) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Vajaria BN, Patel KR, Begum R, Patel PS, Pathol Oncol Res 22 (2016) 443. [DOI] [PubMed] [Google Scholar]
- [121].Park JJ, Lee M, Gut Liver 7 (2013) 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Liu YC, Yen HY, Chen CY, Chen CH, Cheng PF, Juan YH, Chen CH, Khoo KH, Yu CJ, Yang PC, Hsu TL, Wong CH, Proc Natl Acad Sci U S A 108 (2011) 11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH, Nucleic Acids Res 42 (2014) D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Hayes CA, Karlsson NG, Struwe WB, Lisacek F, Rudd PM, Packer NH, Campbell MP, Bioinformatics 27 (2011) 1343. [DOI] [PubMed] [Google Scholar]
- [125].Bern M, Kil YJ, Becker C, Current Protocols in Bioinformatics (2012) 13.20. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kolarich D, Jensen PH, Altmann F, Packer NH, Nature protocols 7 (2012) 1285. [DOI] [PubMed] [Google Scholar]
- [127].Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ, Journal of Proteome Research 12 (2013) 5791. [DOI] [PubMed] [Google Scholar]
- [128].Yang W, Shah P, Toghi Eshghi S, Yang S, Sun S, Ao M, Rubin A, Jackson JB, Zhang H, Analytical chemistry 86 (2014) 6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wu SW, Pu TH, Viner R, Khoo KH, Analytical Chemistry 86 (2014) 5478. [DOI] [PubMed] [Google Scholar]
- [130].Cao L, Qu Y, Zhang Z, Wang Z, Prytkova I, Wu S, Expert Rev Proteomics 13 (2016) 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yang Y, Franc V, Heck AJR, Trends Biotechnol 35 (2017) 598. [DOI] [PubMed] [Google Scholar]