Abstract
Metabolism and gene expression, which are two fundamental biological processes that are essential to all living organisms, reciprocally regulate each other to maintain homeostasis and regulate cell growth, survival and differentiation. Metabolism feeds into the regulation of gene expression via metabolic enzymes and metabolites, which can modulate chromatin directly or indirectly — through regulation of the activity of chromatin trans-acting proteins, including histone-modifying enzymes, chromatin-remodelling complexes and transcription regulators. Deregulation of these metabolic activities has been implicated in human diseases, prominently including cancer.
Genomic DNA in eukaryotic cells is packaged into chromatin by the formation of nucleosomes. Each nucleosome core particle consists of a histone octamer wrapped by ~146 base pairs of DNA1. A histone octamer is composed of two copies each of the core histones H2A, H2B, H3 and H4. Histone amino-terminal tails, which protrude from the nucleosome, and the globular histone cores are subjected to a wide array of covalent modifications2, including acetylation, methylation, phosphorylation, sumoylation, ubiquitylation, succinylation, glutarylation, propionylation, butyrylation, crotonylation, 2-hydroxyisobutyrylation, β-hydroxybutyrylation, malonylation, formylation, citrullination, hydroxylation, O-GlcNAcylation and ADP ribosylation, which engage more than 600 post-translational modification sites of eukaryotic histones3–5. In addition to histones, DNA is also subject to modifications, with four different DNA modifications described in mammals: methylation of the 5-carbon on cytosine residues, which yields 5-methylcytosine (5mC) in CpG dinucleotides; 5mC is oxidized to 5-hydroxymethylcytosine (5hmC), which is further oxidized to 5-formylcytosine (5fC) and eventually to 5-carboxylcytosine (5caC)6,7. These different chromatin modifications regulate chromatin structure, which in turn modulates the accessibility of DNA and histones for binding proteins and subsequently contributes to the regulation of all DNA-based processes, including gene expression, DNA replication and DNA damage response5,8–10. Aberrant regulation of DNA modifications and histone post-translational modifications has been linked to a number of diseases, including cancer as well as developmental and neurological disorders10–13.
Regulation of cell metabolism to support all instrumental cellular activities is essential for maintenance of cell homeostasis, growth, proliferation, migration, differentiation and apoptosis. Cell metabolism can be regulated at the gene expression level in response to extracellular and intracellular signalling to cope with the metabolic needs of cells and to adapt to the changing environment. In turn, cell metabolism — through the activity of metabolic enzymes and metabolites — can, directly or indirectly, impact chromatin modifications (FIG. 1) and thereby influence chromatin functions. Thus, metabolism and DNA-based processes are closely integrated to regulate physiological responses to extracellular stimuli, and deregulation of these interconnected pathways can have profound consequences, leading to disease development.
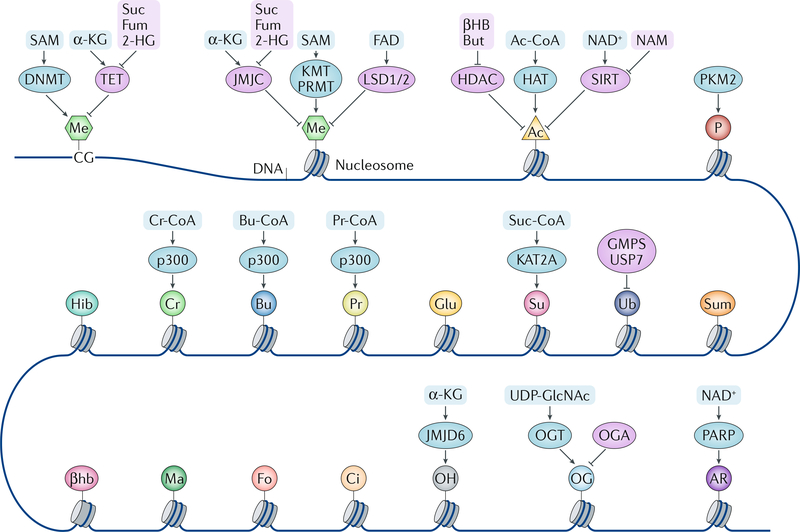
Fig. 1 |. Chromatin modulation by metabolites.
DNA and histones are modified by different writers and erasers, and the enzymatic activities of these modifiers are regulated by metabolites and metabolic enzymes. Some metabolic enzymes can act as direct modifiers (writers) of the histone code as well. α-KG, α-ketoglutarate; βHB, β-hydroxybutyrate; βhb, β-hydroxybutyrylation; 2-HG, 2-hydroxyglutarate; Ac, acetylation; Ac-CoA, acetyl-CoA; AR, ADP ribosylation; Bu, butyrylation; Bu-CoA, butyryl-CoA; But, butyrate; Ci, citrullination; Cr, crotonylation; Cr-CoA, crotonyl-CoA; DNMT, DNA methyltransferase; Fo, formylation; Fum, fumarate; Glu, glutarylation; GMPS, GMP synthase; HAT, histone acetyltransferase; HDAC, histone deacetylase; Hib, 2-hydroxyisobutyrylation; JMJC, Jumonji C domain-containing demethylase; JMJD6, Jumonji domain-containing 6; KMT, lysine methyltransferase; LSD, lysine-specific histone demethylase; Ma, malonylation; Me, methylation; NAM, nicotinamide; OG, O-GlcNAcylation; OGA, O-GlcNAcase; OGT, O-GlcNAc transferase; OH, hydroxylation; P, phosphorylation; PARP, poly(ADP) ribose polymerase; PKM2, pyruvate kinase M2 isoform; Pr, propionylation; Pr-CoA, propionyl-CoA; PRMT, peptidyl-arginine methyltransferase; SAM, S-adenosylmethionine; SIRT, sirtuin; Su, succinylation; Suc, succinate; Suc-CoA, succinyl-CoA; Sum, sumoylation; TET, ten-eleven translocation (DNA demethylase); Ub, ubiquitylation; UDP-GlcNAc, uridine diphosphate N-acetylglucosamine; USP7, ubiquitin-specific processing protease 7.
In this Review, we discuss the current understanding of the molecular mechanisms underlying metabolic enzyme-mediated and metabolite-mediated modulation of chromatin. We focus here on the regulation of gene expression by metabolism during normal cellular function and in disease, especially cancer, and discuss examples of direct as well as indirect — through modulation of chromatin-remodelling proteins — roles of metabolic enzymes and metabolites in chromatin regulation. In particular, we highlight recent evidence reported by our groups and others indicating that metabolic enzymes can be recruited to the nucleus to locally generate key metabolites involved in chromatin modifications. This crosstalk between metabolism and epigenetic regulation of chromatin has important roles in both physiological and pathological processes, and understanding it will open new possibilities for targeted therapeutic interventions for a variety of human diseases.
Metabolites in chromatin modification
Metabolites serve as important cofactors and regulators of various enzymes, including those implicated in chromatin modifications. Accordingly, covalent modifications of DNA and histones, including methylation, hydroxylation, acetylation, crotonylation, β-hydroxybutyrylation, 2-hydroxyisobutyrylation, O-GlcNAcylation and poly(ADP) ribosylation (PARylation), have been shown to be influenced by metabolic processes (FIG. 1), with subsequent effects on gene expression (TABLE 1).
Table 1 |.
Regulation of gene expression by metabolites and metabolic enzymes
| Metabolite or enzyme | Regulation of gene expression | Mechanism | Links to disease |
|---|---|---|---|
| Metabolites | |||
| SAM | Histone methylation-dependent genes↑↓ | Methyl donor for methyltransferases | NA |
| FAD | Histone methylation-dependent genes↑↓ | Positive regulator of LSD1 and LSD2 | NA |
| α-KG | • HIF1-targeted genes↓ • Histone methylation-dependent genes↑↓ |
Cofactor for α-KG-utilizing dioxygenases | NA |
| Succinate | • HIF1-targeted genes↓ • Histone methylation-dependent genes↑↓ |
Inhibition of α-KG-utilizing dioxygenases | NA |
| Fumarate | • HIF1-targeted genes↑ • ATF2-targeted genes↑ • NRF2-targeted genes↑ • Histone methylation-dependent genes↑↓ |
• Inhibition of α-KG-utilizing dioxygenases • Activation of NRF2 |
NA |
| 2-HG | • HIF1-targeted genes↑ • Histone methylation-dependent genes↑↓ |
Inhibition of α-KG-utilizing dioxygenases | NA |
| Acetyl-CoA | • HIF2-targeted genes↑ • TFEB-targeted genes↑ • Memory-related neuronal genes↑ • Glucose metabolism-related genes↑ • Cell cycle progression-related genes↑ • AhR-targeted genes↑ • Other histone acetylation-dependent genes↑ |
Acetyl donor for acetyltransferases | NA |
| NAD+ | • SIRT-regulated and histone acetylation-dependent genes↓ • Genes regulated by PARylation of histones and transcription regulators (CLOCK, NELFA, NELFE and KDM5B)↑↓ |
Activation of histone deacetylase (SIRT) and PARP | NA |
| NAM, β-hydroxybutyrate and butyrate | Histone acetylation-dependent genes↑ | Inhibition of histone deacetylase | NA |
| Succinyl-CoA | Regulation of more than 7,000 genes↑↓ | Histone succinylation | NA |
| Metabolic enzymes | |||
| PKM2 | • CCND1↑ • MYC↑ • STAT3-targeted genes (MEK5)↑ • HIF1-targeted genes↑ • AhR-targeted genes↑ • OCT4-targeted genes↓ |
• Phosphorylation of H3T11 • Activation of β-catenin, STAT3, HIF1 and AhR • Suppression of OCT4 |
Tumour development |
| ACSS2 | • HIF2-targeted genes↑ • TFEB-targeted genes↑ • Memory-related neuronal genes↑ |
Histone acetylation | • Anaemia • Tumour development • Memory disorder |
| ACLY | Glucose metabolism-related genes↑ | Histone acetylation | Tumour development |
| PDC | • Cell cycle progression-related genes↑ • AhR-targeted genes↑ |
Histone acetylation | ND |
| FH | • HIF1-targeted genes↑a • ATF2-targeted genes↑a • NRF2-targeted genes↑a • Histone methylation-dependent genes↑↓a |
• Inhibition of α-KG-utilizing dioxygenases • Activation of NRF2 |
Tumour development associated with FH loss of function |
| SDH, IDH1 and IDH2 mutants | • HIF1-targeted genes↑a • Histone methylation-dependent genes↑↓a |
Inhibition of α-KG-utilizing dioxygenases | • Tumour development associated with loss of function of the SDHgenes • Tumour development associated with IDH1 and/or IDH2 mutation |
| FBP1 | HIF1-targeted genes↓ | Binding and inhibiting HIF1α and HIF2α | Tumour development associated with FBP1 genomic loci loss |
| PFKFB4 | ATF4-targeted genes↑ | Stabilizing the recruitment of SRC3 and ATF4 to target gene promoters | Tumour growth and metastasis |
| GAPDH | H2B gene↑ | Recruitment to the H2B promoter | ND |
| α-KGDH | Regulation of more than 7,000 genes↑↓ | Histone succinylation | ND |
| IMPDH | Histone genes and E2F genes↓ | Binding to single-stranded CT-rich DNA elements | ND |
| GMPS | • PRC1 and PRC2-regulated homeotic genes↓ • Ecdysone-inducible genes↓ |
Deubiquitylation of p53 and histone H2B | ND |
| MATII | • HMOX1 gene↓ • Histone methylation-regulated genes↑↓ |
Histone methylation | ND |
α-KG, α-ketoglutarate; α-KGDH, α-KG dehydrogenase; ACLY, ATP citrate synthase; ACSS2, acetyl-CoA synthetase short-chain family member 2; AhR, aryl hydrocarbon receptor; ATF, activating transcription factor; CLOCK, circadian locomotor output cycles protein kaput; FBP1, fructose-1,6-bisphosphatase 1; FH, fumarase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GMPS, GMP synthase; H3T11, histone H3 threonine 11; HIF, hypoxia-inducible factor; IDH, isocitrate dehydrogenase; IMPDH, inosine 5′-monophosphate dehydrogenase; KDM5B, lysine-specific demethylase 5B; LSD, lysine-specific histone demethylase; MATII, methionine adenosyltransferase II; NA, not applicable; NAD, nicotinamide adenine dinucleotide; NAM, nicotinamide; ND, not determined; NELF, negative elongation factor; NRF2, nuclear factor erythroid 2-related factor 2; OCT4, octamer-binding protein 4; PDC, pyruvate dehydrogenase complex; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4; PKM2, pyruvate kinase M2 isoform; PRC, Polycomb repressive complex; SAM, S-adenosylmethionine; SDH, succinate dehydrogenase; SIRT, sirtuin; SRC3, steroid receptor co-activator protein 3; STAT3, signal transducer and activator of transcription 3; TFEB, transcription factor EB.
Effects observed in mutants (FH and SDH loss of function and IDH1 and/or IDH2 mutants).
Histone and DNA methylation
A key chromatin modification that is strongly interconnected with metabolism is methylation (FIG. 2). A number of methyltransferases including DNA methyltransferases (DNMTs), lysine methyltransferases (KMTs) and peptidyl-arginine methyltransferases (PRMTs) are involved in epigenetic gene regulation14. These enzymes deposit methyl groups on DNA and histones (FIG. 2a), leading to complex changes in chromatin accessibility, transcription factor binding and gene expression15. Methylation is regulated by S-adenosylmethionine (SAM) abundance, whereby SAM serves as a universal methyl donor and is synthesized from methionine and ATP by methionine adenosyltransferases (MATs)16. This reaction produces another metabolite, S-adenosylhomocysteine (SAH), which is a potent inhibitor of all methyltransferases. Thus, the intracellular SAM:SAH ratio, which is tightly regulated by metabolism of methionine, threonine and serine, dynamically regulates methyltransferase activity17–19 (FiG. 2a). Accordingly, consumption of food that is rich in methyl-donating nutrients (SAM, folic acid and vitamin B) induces DNA and histone methylation and influences gene expression20. Intriguingly, SAM metabolism provides a functional link between histone and phospholipid methylation21. Methylation of phosphoethanolamine (PE) generates phosphatidylcholine (PC), the most abundant phospholipid for constructing cellular membranes. PE methylation consumes SAM and releases SAH, which is further metabolized for synthesis of cysteine and glutathione to protect against oxidative stress. Deficiencies of PE methylation lead to elevated intracellular SAM and increased methylation of histones. Thus, PE functions as a methyl sink that maintains intracellular homeostasis by counteracting increased SAM21. Overall, membrane lipid production, glutathione metabolism and histone methylation are linked through coordinated regulation of histone and phospholipid methylation (FIG. 2a).
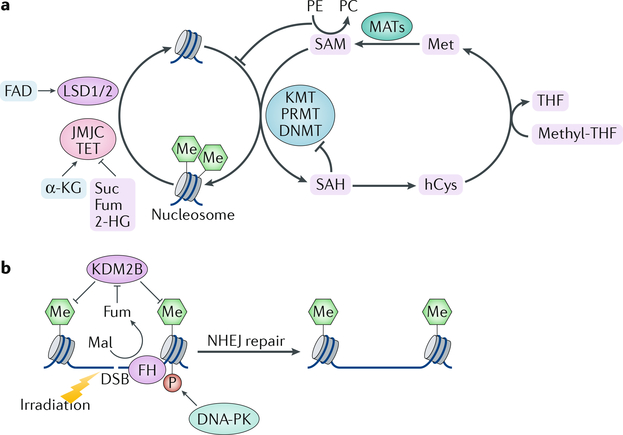
Fig. 2 |. Regulation of chromatin methylation by metabolic enzymes and metabolites and their roles in DNA repair.
a | Histone and DNA methylation are regulated by methyltransferases (lysine methyltransferases (KMTs), peptidyl-arginine methyltransferases (PRMTs) or DNA methyltransferases (DNMTs)) and demethylases (lysine-specific demethylase 1A (LSD1) and LSD2, Jumonji C (JMJC) family proteins or ten-eleven translocation (TET) proteins) in a metabolite-dependent manner. Histone and DNA methylation are inhibited when S-adenosylmethionine (SAM) levels are low. Production of phosphatidylcholine (PC) from phosphoethanolamine (PE) consumes SAM, thereby providing a link between chromatin methylation and lipid metabolism. b | Histone methylation allows recruitment of DNA-dependent protein kinase (DNA-PK) to DNA breaks, thereby fuelling DNA repair by nonhomologous end joining (NHEJ). Local generation of fumarate (Fum) at the DNA damage regions by fumarase (FH) promotes DNA repair through inhibition of histone H3 demethylation. In a feedforward mechanism, DNA-PK phosphorylates fumarase and promotes its binding to DNA lesions. Succinate (Suc), Fum and 2-hydroxyglutarate (2-HG) are competitive inhibitors for the α-ketoglutarate (α-KG)-dependent demethylases. DSB, double-strand break; hCys, homocysteine; KDM2B, lysine-specific demethylase 2B; Mal, malate; MAT, methionine adenosyltransferase; Me, methylation; Met, methionine; SAH, S-adenosylhomocysteine; THF, tetrahydrofolate.
Histone and DNA methylation can be removed by demethylases that use different metabolites as cofactors (FIG. 2a). Lysine-specific histone demethylase 1A (LSD1; also known as KDM1A) and LSD2 (also known as KDM1B) catalyse an amine oxidation reaction dependent on FAD22,23, whereas the Jumonji C (JMJC) family members and the ten-eleven translocation (TET) methylcytosine hydroxylases use a dioxygenation reaction that requires Fe2+, O2 and α-ketoglutarate (α-KG)24. High levels of α-KG maintained by glucose and glutamine catabolism promote demethylation of trimethylated histone H3 lysine 27 (H3K27me3), H3K9me3, H4K20me3 and TET-dependent DNA demethylation and have been shown to contribute to the regulation of pluripotency-associated gene expression25,26. α-KG-dependent dioxygenases are inhibited by metabolites structurally related to α-KG, including succinate, fumarate and 2-hydroxyglutarate (2-HG), the latter of which is an oncometabolite that is primarily produced in cancer cells bearing gain-of-function mutations in the isocitrate dehydrogenase genes, IDH1 and IDH2 (REFS27,28). In addition, high succinate and fumarate levels induced by tumour-associated loss-of-function mutations in succinate dehydrogenase (which is encoded by SDH genes) and fumarase (also known as fumarate hydratase; which is encoded by FH) inhibit TET family and JMJC family proteins, respectively, and contribute to the hypermethylation phenotype and aggressiveness of SDH-deficient and FH-deficient tumours19,29 (FIG. 2a) (see also the Conclusions section).
Thus, histone methylation and DNA methylation are reversibly regulated by dynamic modulation of methyltransferase and demethylase activities, and these modulations are governed by a variety of metabolites. Production of these metabolites is subject to regulation by cell signalling, and the producing enzymes can be subject to mutagenesis, thereby determining biological outcomes.
Histone hydroxylation
As described above, histone demethylation by JMJC dioxygenases results in histone hydroxylation, which is typically transient, as hydroxymethyl groups are immediately processed to formaldehyde. However, it has been revealed that Jumonji domain-containing protein 6 (JMJD6), which belongs to the JMJC superfamily, is capable of catalysing stable lysyl hydroxylation as well as arginyl demethylation on diverse protein substrates30, and it has been shown to bind histone proteins and hydroxylate multiple lysyl residues of histone H3 and H4 tails in vitro31. Comparative amino acid component analysis of purified histones from wild-type and Jmjd6 knockout mice revealed differences in monohydroxylation of multiple lysine residues of the histones H3, H4, H2A and H2B. JMJD6 overexpression increases the amount of 5-hydroxylysine in histones in human embryonic kidney 293 cells, and high levels of JMJD6 correlate with high levels of 5-hydroxylysine in histones of murine testis31. 5-Hydroxylation of lysine residues can inhibit subsequent acetylation and methylation at the same residues, implying a role of histone hydroxylation in the structural alteration of chromatin and the epigenetic regulation of gene expression31. Like other JMJC proteins, JMJD6 can potentially be regulated by α-KG24 (FIG. 1).
Histone acetylation
Histone acetylation, which is one of the most intensely studied post-translational modifications of histones, is a dynamic and reversible process primarily regulated by the activity of a pair of enzyme families, histone acetyltransferases (HATs) and histone deacetylases (HDACs), which acetylate and deacetylate histones, respectively. Histone acetylation-dependent gene expression is sensitive to the availability of metabolites that serve as substrates or regulators of HATs and HDACs. Class I and IIa HDACs can be directly inhibited by butyrate and β-hydroxybutyrate (βHB), which is produced by fatty acid hydrolysis32. Nuclear enzymes that deacetylate histones, including sirtuin 1 (SIRT1) and SIRT2, consume and require oxidized NAD (NAD+) for their activity, being activated by high NAD+ levels and inhibited by nicotinamide (NAM), a precursor of NAD+ (REF.33). Thus, metabolites can exert their regulation on histone acetylation via modulation of HDAC activity (FIG. 1).
HATs acetylate conserved lysine residues on histones by transferring an acetyl group from acetyl-CoA. Acetyl-CoA is produced from acetate, citrate and pyruvate by acetyl-CoA synthetase short-chain family member (ACSS), ATP citrate synthase (ACLY) and the pyruvate dehydrogenase complex (PDC), respectively. It can also be generated from fatty acid β-oxidation and metabolism of amino acids and ketone bodies34. Metabolic regulation of acetyl-CoA production is instrumental for HAT activity35. Acetyl-CoA abundance can impact global histone acetylation levels. In Saccharomyces cerevisiae, glucose, galactose and glycolysis end products, such as ethanol, acetate and lactate, substantially increase intracellular acetyl-CoA levels. High acetyl-CoA levels promote the activity of KAT2A-containing SAGA (SPT–ADA–GCN5 acetyltransferase) complex to catalyse histone acetylation, which is important for key cellular processes, such as cell growth35. Therefore, the metabolic production of acetyl-CoA can regulate histone acetylation via modulation of HAT activity (FIG. 1). Accordingly, acetyl-CoA metabolism is tightly regulated both spatially and temporally to elicit responses of transcription machinery to nutrient availability34.
Histone acylation
In addition to acetylation, histone lysine residues can be post-translationally modified by other acyl groups, resulting in crotonylation, propionylation, butyrylation, glutarylation, malonylation, succinylation, β-hydroxybutyrylation and 2-hydroxyisobutyrylation of histones3,36–38. The overall class of these modifications is named histone acylation, and it is implicated in diverse physiological functions, such as spermatogenesis, tissue injury and metabolic stress and is tightly regulated by metabolic processes39.
Crotonylation has been shown to depend on histone acetyltransferase p300, which is one of the main HATs but in addition can also act as a histone crotonyltransferase to induce transcription to a level exceeding that regulated by p300-mediated histone acetylation40 (FIG. 1). Levels of histone crotonylation are regulated by the cellular concentration of crotonyl-CoA, which can be altered through genetic and environmental perturbations, such as availability of extracellular sodium crotonate, and the different acyl-CoA groups compete for the acyltransferase activity of p300 (REFS39,40). It was shown that reduction of nucleocytosolic acetyl-CoA by ACLY or PDC depletion decreases H3K18 acetylation and increases H3K18 crotonylation40. By contrast, depletion of ACSS2, which in addition to acetyl-CoA synthesis also catalyses crotonyl-CoA production from crotonate, decreases H3K18 crotonylation at gene promoters and attenuates gene expression40.
Propionylation of histone lysines was detected in mammalian cells and is regarded as a mark of active chromatin41. In the presence of propionyl-CoA, p300 and the acetyltransferase cAMP-responsive element-binding protein (CBP) catalyse histone lysine propionylation in vitro36,42. Intriguingly, p300-mediated H3 propionylation at K23 can be removed by SIRT2 in the presence of NAD+ in vitro42, suggesting that SIRT2 is involved in histone depropionylation.
Similar to histone propionylation, histone butyrylation was shown to be catalysed by p300 and CBP using butyryl-CoA37. Histone H3 butyrylation at K14 and histone H4 butyrylation at K5 and K18 have been linked to active gene expression43,44.
Lysine β-hydroxybutyrylation (Kβhb) is another form of histone acylation driven by ketogenesis under low nutrient conditions41. Ketone bodies are generated in the liver from fatty acids during starvation, prolonged intense exercise or under carbohydrate restrictive diets. In addition, serum concentrations of ketone bodies can be dramatically increased in untreated individuals with diabetes45–47. A number of Kβhb sites have been detected on histones at levels comparable to those of other known histone acyl marks41. Kβhb is dramatically induced in response to elevated βHB levels in livers from mice subjected to prolonged fasting or streptozotocin-induced diabetic ketoacidosis, and H3K9-βhb is associated with increased expression of starvation pathway-related genes41.
Lysine 2-hydroxyisobutyrate is a structural isomer of lysine βHB. Although little is known about metabolic regulation of hydroxyisobutyrate-CoA, mass spectrometry analysis of human and mouse histones identified 63 lysine 2-hydroxyisobutyrylation (Khib) sites, and histone Khib shows distinct genomic distributions from histone acetylation and histone crotonylation during male germ cell differentiation. In addition, H4K8hib is associated with active gene transcription in meiotic and post-meiotic cells, suggesting an important role for H4K8hib in the regulation of gene expression37.
Histone O-GlcNAcylation
O-GlcNAcylation is a noncanonical glycosylation that involves the attachment of single O-linked N-acetylglucosamine (O-GlcNAc) moieties to serine and threonine residues of cytoplasmic, nuclear and mitochondrial proteins48. O-GlcNAcylation is the product of nutrient flux through the hexosamine biosynthetic pathway, which integrates glucose, amino acid, fatty acid and nucleotide metabolism to generate uridine diphosphate GlcNAc (UDP-GlcNAc), the donor substrate for O-GlcNAcylation. A single pair of enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), which add and remove O-GlcNAc groups to and from proteins, respectively, controls the dynamic cycling of this protein modification in a nutrient-responsive and stress-responsive manner48 (FIG. 1). The O-GlcNAcylation of the histone core is extensive and interplays with other post-translational modifications of histones, such as phosphorylation, methylation, acetylation and ubiquitylation, thereby regulating gene expression49. Besides histones, various epigenetic regulators can be O-GlcNAcylated. For example, methylcytosine dioxygenase TET1, TET2 and TET3 interact with OGT and are extensively O-GlcNAcylated48,49. TET proteins, in turn, recruit OGT to chromatin, where OGT and O-GlcNAcylated TET regulate the conversion of 5mC into 5hmC. In addition, subunits of Polycomb repressive complex 1 (PRC1) and PRC2, which are required for maintenance of repression of homeotic genes during embryonic development, cell proliferation and differentiation, are regulated by O-GlcNAcylation that targets PRC1 to specific sets of genes49,50.
Histone poly(ADP) ribosylation
PARylation is a posttranslational modification by which poly(ADP) ribose, or PAR, which is derived from NAD+, is covalently attached to proteins by a large family of PAR polymerase (PARP) enzymes that includes PARP1, PARP2 and PARP3 (FIG. 1). Diverse physiological situations — such as high-fat diet, oxidative stress, DNA damage and ageing — are associated with increased PARP activity, which regulates a number of biological processes, including chromatin reorganization, transcription regulation, DNA repair, apoptosis and mitosis51. PARPs are dependent on NAD+ for their catalytic activity. As such, the dynamics of NAD+ synthesis and consumption as well as its subcellular localization are important for PARP function. NAD+ levels can be modulated by NAD+ ‘consumers’, such as PARPs themselves, sirtuins and CD38, which hydrolyse NAD+ (REF.52).
The histones H1 and H2B are the most extensively PARylated histones in vivo and are the preferred targets of PARP1 in vitro. Histone PARylation factor 1 (HPF1) was shown to be a co-regulator of PARP1-dependent histone PARylation by restricting PARP1 automodification, thereby promoting histone PARylation53. PARylation of polynucleosomes by purified PARP1, which mimics the effects of depletion of linker histone (H1) from higher-order chromatin, results in polynucleosome decondensation and histone release from destabilized nucleosomes to further expose DNA, thereby providing greater access of various DNA-acting protein machineries to their targets51,54. Phosphorylation of the catalytic domain of PARP1 by cyclin-dependent kinase 2 (CDK2), which depends on the hormone progestin, increases PARP1 activity to promote the displacement of histone H1, thereby regulating progestin-responsive genes and cell cycle progression of breast cancer cells55.
In addition to regulating histone PARylation, PARP1 catalyses PARylation of transcription factors, such as circadian locomotor output cycles protein kaput (CLOCK) for regulation of circadian rhythms, and interacts with nuclear receptors and transcription factors, such as peroxisome proliferator-activated receptor-γ (PPARγ), oestrogen receptor (ER) and forkhead box protein O1 (FOXO1), to regulate gene expression56,57. PARP1 mediates PARylation of negative elongation factor A (NELFA) and NELFE, which are both components of a protein complex that regulates promoter-proximal pausing by RNA polymerase II (Pol II), for productive transcription elongation58. PARP1 can also indirectly alter chromatin architecture by modulating the activity of histone-modifying enzymes and chromatin remodellers, such as lysine-specific demethylase 5B (KDM5B), chromatin-remodelling complex ATPase chain ISWI and chromodomain-helicase-DNA-binding protein 1-like (ALC1)52. PARylation of KDM5B inhibits its binding to chromatin and its histone demethylase activity, leading to an increase in H3K4me3 and increased gene expression59.
Metabolic enzymes in the nucleus
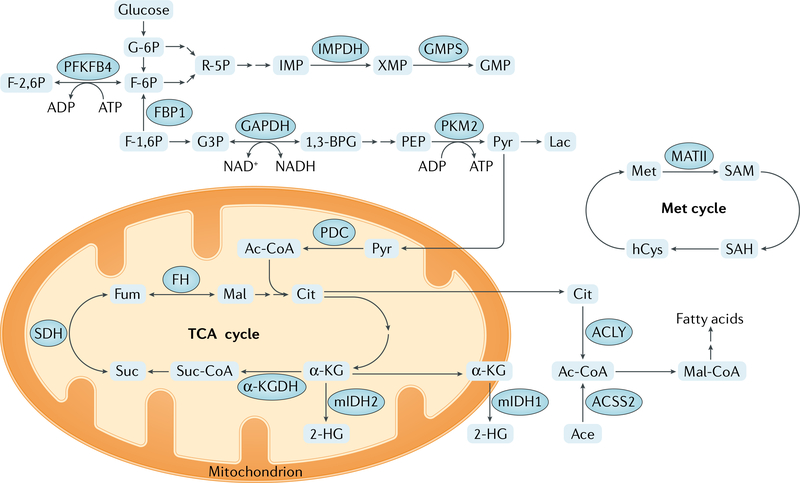
It has recently been appreciated that various metabolic enzymes, which typically localize to the cytoplasm or mitochondria, also can be found in the nucleus. These enzymes include glycolytic enzymes such as pyruvate kinase M2 isoform (PKM2), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4), fructose-1,6-bisphosphatase 1 (FBP1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH); tricarboxylic acid cycle (TCA) (also known as citric acid cycle or Krebs cycle) enzymes such as α-ketoglutarate dehydrogenase (α-KGDH) and fumarase; nucleotide synthesis enzymes such as inosine 5′-monophosphate (IMP) dehydrogenase (IMPDH) and GMP synthase (GMPS); and other metabolic enzymes, prominently including MATII (also known as MAT2A) (FIG. 3). In the nucleus, these enzymes can supply metabolites to regulate the activity of chromatin-modifying enzymes or transcription regulators to regulate chromatin structure and gene expression. They can also have other roles in regulating chromatin and gene expression that are independent of the production of metabolites, including direct modifications of histones and transcription regulators (TABLE 1).
Fig. 3 |. Core metabolic functions of metabolic enzymes that also function in epigenetic modifications.
Main metabolic pathways regulated by metabolic enzymes involved in epigenetic modifications are shown. α-KG, α-ketoglutarate; α-KGDH,α-KG dehydrogenase; 1,3-BPG, 1,3-bisphosphoglycerate; 2-HG, 2-hydroxyglutarate; Ac-CoA, acetyl-CoA; Ace, acetate; ACLY, ATP citrate synthase; ACSS2, acetyl-CoA synthetase short-chain family member 2; Cit, citrate; F-1,6 P, fructose 1,6-bisphosphate; F-2,6 P, fructose 2,6-bisphosphate; F-6P, fructose 6-phosphate; FH, fumarase; Fum, fumarate; G3P, glyceraldehyde 3-phosphate; G-6P, glucose 6-phosphate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GMPS, GMP synthase; hCys, homocysteine; IMP, inosine 5′-monophosphate; IMPDH, IMP dehydrogenase; Lac, lactate; Mal, malate; Mal-CoA, malonyl-CoA; MATII, methionine adenosyltransferase II; Met, methionine; mIDH1, isocitrate dehydrogenase [NADP], cytoplasmic mutant; mIDH2, isocitrate dehydrogenase [NADP], mitochondrial mutant; PDC, pyruvate dehydrogenase complex; PEP, 2-phosphoenolpyruvate; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4; PKM2, pyruvate kinase M2 isoform; Pyr, pyruvate; R-5P, ribose 5-phosphate; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SDH, succinate dehydrogenase; Suc, succinate; Suc-CoA, succinyl-CoA; TCA, tricarboxylic acid; XMP, xanthosine monophosphate.
Pyruvate kinase
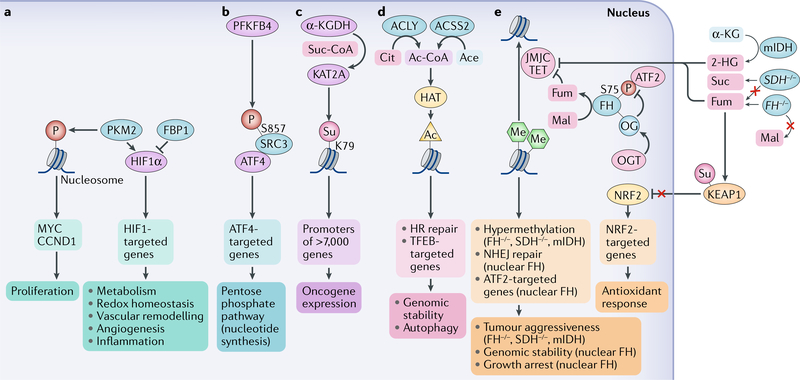
Pyruvate kinase (PKM) regulates the final rate-limiting step of glycolysis by catalysing the transfer of a phosphate group from phosphoenolpyruvate to adenosine diphosphate to produce pyruvate and adenosine triphosphate60. Alternative splicing of PKM pre-mRNA by heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) and HNRNPA2 and polypyrimidine-tract binding (PTB) splicing factors results in the generation of exon 10-included PKM2 or exon 9-included PKM isozyme 1 (PKM1) (REF.61). PKM2 expression, which is upregulated by receptor tyrosine kinase activation, is increased in 16 cancer types, with an isoform switch from PKM1 to PKM2 in glioblastomas62–64.
PKM2 is a cytosolic enzyme. However, in response to growth factor stimulation, PKM2 translocates into the nucleus and promotes cell proliferation (FIG. 4). Upon activation of epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR), which are induced by their ligands or by expression of the constitutively activated EGFR variant III (EGFRvIII) mutant (which is commonly found in many types of cancer)64,65, activated ERK1 and/or ERK2 binds to the region that is encoded by exon 10 of PKM2 via a docking groove in ERK. This interaction results in ERK-mediated phosphorylation of S37 of PKM2 but not of PKM1. Phosphorylated PKM2 then recruits peptidylprolyl cis-trans isomerase NIMA-interacting 1 (PIN1) to induce its own isomerization, leading to conversion of PKM2 from a tetramer into a monomer, exposure of its nuclear localization sequence (NLS) for recruitment of importin α5 and subsequent nuclear translocation of PKM2 (REFS66,67). Phosphorylation-independent modification of PKM2 was also reported to support nuclear translocation of monomeric PKM2, whereby p300-mediated acetylation of PKM2 at K433 prevents PKM2 from binding to FBP1 and from forming tetramers68,69. In addition, Jumonji domain-containing protein 5 (JMJD5), expression of which is upregulated by hypoxia, interacts directly with the intersubunit interface region of PKM2, thereby hindering PKM2 tetramerization and promoting its nuclear translocation70 (FIG. 4).
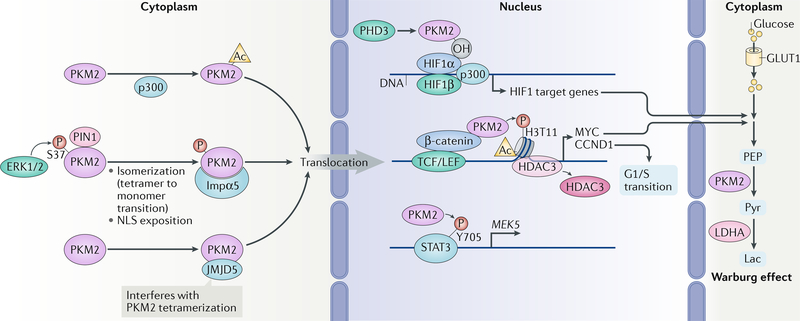
Fig. 4 |. The roles of nuclear PKM2 in gene expression.
The nuclear translocation of pyruvate kinase M2 isoform (PKM2) is regulated by several mechanisms. ERK1 and ERK2 phosphorylate PKM2, which promotes its isomerization (transition from tetramer to monomer) mediated by peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (PIN1), resulting in the exposition of nuclear localization signal (NLS) of PKM2 and its interaction with importin α5 (Impα5). Nuclear translocation of PKM2 is also promoted by acetylation (Ac) mediated by histone acetyltransferase p300 and by the interaction of PKM2 with Jumonji domain-containing protein 5 (JMJD5). In the nucleus, PKM2 interacts with β-catenin and binds to β-catenin-regulated promoters, where it phosphorylates histone H3 at T11 and subsequently removes histone deacetylase 3 (HDAC3) to promote histone H3 acetylation and expression of genes regulating the Warburg effect and cell cycle progression. PKM2 also directly increases the transcriptional activity of signal transducer and activator of transcription 3 (STAT3) by phosphorylation (P) of STAT3. PKM2 hydroxylated by prolyl hydroxylase 3 (PHD3) binds to hypoxia-inducible factor 1α (HIF1α) to promote recruitment of HIF1α and p300 to the promoters of HIF1α-regulated genes for expression. CCND1, G1/S-specific cyclin D1; GLUT1, glucose transporter type 1, erythrocyte/brain (also known as SLC2A1); Lac, lactate; LDHA, l-lactate dehydrogenase A chain; LEF, lymphoid enhancer factor; MYC, MYC proto-oncogene protein; OH, hydroxylation; PEP, 2-phosphoenolpyruvate; Pyr, pyruvate; TCF, T cell factor.
In the nucleus, K433 of PKM2 binds to β-catenin, which has been phosphorylated at Y333 by the protooncogene tyrosine-protein kinase SRC. PKM2-β-catenin complex is then recruited to the CCND1 and MYC promoter regions, where PKM2 binds directly to histone H3 and functions as a protein kinase to phosphorylate histone H3 at T11. This phosphorylation promotes dissociation of HDAC3 from the CCND1 and MYC promoter regions, thereby supporting acetylation of histone H3 at K9 in these regions and expression of cyclin D1 (CCND1) and MYC proto-oncogene protein. CCND1 then promotes G1/S phase transition, whereas MYC increases glycolytic gene expression to induce the Warburg effect, which is reflected by increased glucose uptake and lactate production even in the presence of ample oxygen (FIG. 4). Overall, nuclear PKM2 supports cell cycle progression and tumour cell proliferation and has been implicated in brain tumorigenesis66,71,72. Although a study failed to identify protein kinase activity of PKM2 from crude whole cell lysate or nuclear extracts of PKM2-deficient mouse fibroblasts on commercially available calf thymus histones73, a later report further validated the protein kinase activity of the yeast homologue of PKM2 (pyruvate kinase (Pyk1)) and demonstrated that Pyk1, in a complex with histone lysine N-methyltransferase, H3 lysine 4-specific (Setl), SAM synthetases, serine metabolic enzymes and an acetyl-CoA synthetase, phosphorylates histone H3 at T11 and upregulates H3K4me3 levels74,75. These findings further highlight the role of PKM2 as a histone code writer and epigenetic regulator of gene expression.
In addition to directly modifying histone H3 via phosphorylation, PKM2 acts as a transcription coactivator by directly phosphorylating signal transducer and activator of transcription 3 (STAT3) at Y705 to activate transcription of MEK5 (also known as MAP2K5)76. The role of PKM2 in gene regulation is further demonstrated by its binding to hypoxia-inducible factor 1α (HIF1α) when hydroxylated on P403 and P408. This binding promotes transactivation of HIF1α target genes by enhancing HIF1α binding and p300 recruitment to hypoxia response elements and increases glucose uptake and lactate production in cancer cells77 (FIG. 4).
Nuclear PKM2 has also been shown to act as a coactivator of aryl hydrocarbon receptor (AhR) and to interact with the pluripotency factor octamer-binding protein 4 (OCT4; also known as POU5F1) to inhibit OCT4-dependent gene expression, thereby inducing the differentiation of glioma spheroids78–80. Outside of the nucleus, monomeric PKM2 functions as a cytosolic binding protein for thyroid hormone, although the effect of this interaction on thyroid hormone-regulated gene expression remains to be defined78–80.
Overall, these studies elucidated an instrumental role for nuclear PKM2 in regulation of gene expression. This role of PKM2 is especially evident in cancer cells, suggesting the potential of targeting nuclear translocation of PKM2 as a therapeutic approach in human cancers.
6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4
PFKFB4 is a member of the family of bifunctional 6-phosphofructo 2-kinase:fructose-2,6-biphosphatase enzymes, which catalyse both the synthesis and the degradation of fructose-2,6-bisphosphate using independent catalytic domains. PFKFB4 uses its kinase activity to synthesize fructose-2,6-bisphosphate from fructose-6-phosphate and ATP and its phosphatase activity to hydrolyse fructose-2,6-bisphosphate into fructose-6-phosphate and inorganic phosphate81. A kinome-wide RNAi-based screening determined that PFKFB4 is required for the activation of steroid receptor co-activator protein 3 (SRC3; also known as NCOA3), which co-regulates ER. SRC3 is a transcription co-activator that contains several nuclear receptor interacting domains and intrinsic HAT activity. Of note, PFKFB4 functions as a protein kinase and phosphorylates SRC3 at S857. This phosphorylation increases the interaction between SRC3 and the transcription factor ATF4 at gene promoters to increase gene expression of metabolic enzymes, such as transketolase, AMP deaminase 1 (AMPD1) and xanthine dehydrogenase/oxidase (XDH), thereby driving glucose flux towards the pentose phosphate pathway and purine synthesis. Expression of the SRC3-S857A mutant blunted breast tumour growth and lung metastasis in mice. In addition, PFKFB4 and phosphorylated SRC3 levels are increased in ER-positive tumours, and their expression correlates with poor survival of patients with the basal subtype of breast cancer81. These findings demonstrate that PFKFB4, as another example of a glycolytic enzyme other than PKM2, acts as a protein kinase to regulate gene expression, and thereby tumour growth, by direct phosphorylation of transcription co-activator SRC3.
α-Ketoglutarate dehydrogenase complex
Histone lysine succinylation is a newly identified histone acylation that occurs frequently82,83. However, the mechanism underlying histone lysine succinylation and its functional consequences are unknown. A recent report demonstrated that histone succinylation is regulated by the α-KGDH complex, which is composed of 2-oxoglutarate dehydrogenase, mitochondrial (OGDH), dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex (DLST) and dihydrolipoamide dehydrogenase (DLD) and is known to catalyse the conversion of α-KG into succinyl-CoA in the mitochondria84. In glioblastoma cells, a fraction of α-KGDH is distributed in the nucleus in a manner that requires the presence of the NLS in DLST. In the nucleus, α-KGDH interacts with the histone acetyltransferase KAT2A at gene promoters and locally supplies succinyl-CoA, which is then bound by KAT2A with high affinity. The crystal structure of the catalytic domain of KAT2A in a complex with succinyl-CoA demonstrates that succinyl-CoA binds to a deep cleft of KAT2A with the succinyl moiety pointing towards the end of the flexible loop 3, which adopts different structural conformations in succinyl-CoA-bound versus acetyl-CoA-bound forms. Y645 in this loop forms hydrogen bonds with succinyl-CoA and has an important role in the selective binding of KAT2A to succinyl-CoA over acetyl-CoA. The high binding affinity of succinyl-CoA for KAT2A and the high local concentrations of succinyl-CoA generated by the KAT2A-associated α-KGDH complex facilitate histone succinylation on K79 in the promoter regions of more than 7,000 genes despite low concentration of succinyl-CoA in the nucleus. Inhibition of nuclear entrance of the α-KGDH complex or expression of the KAT2A mutant with low binding affinity for succinyl-CoA (KAT2AY645A) reduces gene expression and inhibits tumour cell proliferation and glioma growth in mice84. These findings revealed that local generation of succinyl-CoA by the nuclear α-KGDH complex coupled with the succinyltransferase activity of KAT2A (FIG. 1) has an instrumental role in histone succinylation, which has a large impact on gene expression, particularly in the context of cancer, fuelling tumour cell proliferation and tumour development.
Fumarase
Fumarase catalyses the reversible hydration and dehydration of fumarate to malate in the TCA cycle to facilitate a transition step in the production of energy in the form of reduced NAD (NADH). In the cytosol, fumarase metabolizes fumarate, which is a by-product of the urea cycle and amino acid catabolism85. Fumarase translocates from the cytosol into the nucleus upon ionizing radiation-induced DNA damage86. In the nucleus, DNA-dependent protein kinase (DNA-PK) phosphorylates fumarase and promotes its binding to histone H2A.Z. This leads to the enrichment of fumarase at irradiation-induced double-strand break regions and subsequent, localized fumarate production. The locally accumulated fumarate inhibits α-KG-dependent lysine-specific demethylase 2B (KDM2B) histone demethylase activity and increases dimethylation of histone H3K36 and accumulation of the DNA-PK complex at DNA damage regions for subsequent nonhomologous end joining (NHEJ) DNA repair and cell survival85 (FIG. 2b). Fumarase deficiency promotes tumorigenesis partially owing to impairment of DNA repair and accumulation of mutations86. Biallelic inactivation of the FH gene, resulting in accumulation of fumarate, causes hereditary leiomyomatosis and renal cell carcinoma. The accumulated fumarate also upregulates expression of genes involved in antioxidant response by inducing succinylation of Kelch-like ECH-associated protein 1 (KEAP1). This succinylation abrogates the ability of KEAP1 to repress NRF2 (nuclear factor erythroid 2-related factor 2)), which activates the NRF2-mediated antioxidant response pathway87,88. These studies underscore the instrumental role of fumarase in promoting repair of irradiation-induced DNA damage and in regulating antioxidant response, which are processes often deregulated during tumorigenesis.
Chromatin-associated fumarase also directly participates in gene transcription. Under glucose deprivation, fumarase is phosphorylated by AMP-activated protein kinase (AMPK) at S75. The phosphorylated fumarase binds to the transcription factor ATF2 and translocates to ATF2-regulated gene promoter regions. There, fumarase-catalysed fumarate inhibits lysine-specific demethylase 2A (KDM2A), thereby promoting histone H3K36me2 and expression of genes mediating cell growth arrest89. By contrast, under glucose-sufficient conditions, S75 of fumarase is O-GlcNAcylated by OGT, leading to suppression of ATF2-regulated gene expression. Levels of O-GlcNAcylation of fumarase are higher in pancreatic cancer cells than in healthy human pancreatic duct epithelial cells and, to a large extent, these tumour cells exhibit resistance to glucose deprivation-induced increase in fumarase phosphorylation. Consistently, the levels of fumarase S75 phosphorylation are inversely correlated with OGT expression levels and with poor prognosis in patients with pancreatic cancer89. These findings uncovered an instrumental mechanism underlying transcription regulation by fumarase and a link between dysregulated OGT activity and the growth advantage of cancer cells exposed to glucose deficiency.
Fructose-1,6-bisphosphatase 1
FBP1 catalyses the hydrolysis of fructose 1,6-bisphosphate to fructose 6-phosphate in the presence of divalent cations, acting as a rate-limiting enzyme in gluconeogenesis90. Pan-metabolomic profiling and metabolic gene set analysis revealed that FBP1 depletion resulting from loss of the FBP1 loci on chromosome 9q22 occurred uniformly in 600 cases of clear cell renal cell carcinoma (ccRCC), the most common form of kidney cancer, but did not occur in normal kidney tissue, and that low FBP1 expression correlated strongly with advanced tumour stage and poor prognosis90. As a tumour suppressor, FBP1 inhibits glycolytic flux depending on its catalytic activity, thereby limiting energy supply and cell growth. Notably, in ccRCC cells with von Hippel-Lindau mutations, which occur in more than 90% of ccRCC, FBP1 is also present in the nucleus and inhibits the nuclear function of HIF1α and HIF2α (also known as EPAS1) via direct interaction of HIFs with the HIF-inhibitory domain of FBP1. This inhibition results in the further inhibition of glycolysis, the pentose phosphate pathway and of subsequent cell proliferation. These effects occur in a manner independent of FBP1 catalytic activity90. Thus, FBP1 exhibits dual tumour-suppressive functions in ccRCC through both enzymatic and non-enzymatic activities, with the latter depending on nuclear localization of FBP1.
Glyceraldehyde-3-phosphate dehydrogenase
The expression of histones H1, H2A, H2B, H3 and H4 is regulated at both the transcriptional and the posttranscriptional level, and occurs predominantly in an S phase-dependent and DNA replication-dependent fashion. GAPDH, which converts glyceraldehyde 3-phosphate into 1,3-bisphosphoglycerate in the glycolytic pathway, interacts directly with octamer-binding protein 1 (OCT1; also known as POU2F1) as well as protein NPAT, which is a substrate of G1/S-specific cyclin E-CDK2 complex and is broadly involved in histone gene transcription91. GAPDH is selectively recruited to the H2B promoter in S phase and is essential for S phase-specific H2B gene (HIST1H2B) transcription in vivo and in vitro. This activity of GAPDH is stimulated by NAD+ but inhibited by NADH91. Although the exact role of GAPDH in transactivation of this gene remains to be elucidated, this study linked HIST1H2B transcription upregulation to cell cycle regulators and GAPDH, thereby indicating that nuclear GAPDH has a role in coupling histone gene expression and DNA replication.
Inosine 5′-monophosphate dehydrogenase
In de novo synthesis of purine nucleotides, inosine 5′-monophosphate (IMP) is the branch-point metabolite for the synthesis of either guanine or adenine nucleotides. In the guanine nucleotide pathway, two enzymes are involved in converting IMP into GMP: IMPDH and GMPS (see below). IMPDH catalyses the NAD+-dependent oxidation of IMP to xanthosine monophosphate (XMP). GMPS then catalyses the amination of XMP to GMP The IMPDH-catalysed reaction is the first committed and rate-limiting step towards de novo guanine nucleotide biosynthesis and is crucial for maintaining the cellular guanine deoxynucleotide and ribonucleotide pools needed for DNA and RNA synthesis and cell proliferation. IMPDH is highly expressed in human cancers92. Although IMPDH is largely cytoplasmic, during the G2 phase of the cell cycle or upon replicative or oxidative stress, IMPDH accumulates in the nucleus, where it functions as a transcription factor and binds to single-stranded CT-rich DNA elements independently of its catalytic activity. IMPDH binds to and represses expression of histone genes and E2f, the master driver of the G1/S transition, thereby inhibiting cell proliferation93. Thus IMPDH, as a nucleotide biosynthetic enzyme, promotes cell proliferation. However, under stress conditions, nuclear IMPDH acts as a transcription factor to regulate gene expression for repression of cell proliferation93. Two substitution mutations in the cystathionine-β-synthase (CBS) subdomain of IMPDH1, which are associated with human autosomal dominant retinitis pigmentosa, impair nucleic acid binding but not enzymatic activity93, suggesting that the inability of mutant IMPDH to bind to DNA contributes to human disease.
Guanosine 5-monophosphate synthase
Apart from metabolic function in converting XMP into GMP, GMPS also has a non-metabolic role, whereby it forms a complex with nuclear ubiquitin-specific processing protease 7 (USP7). The GMPS-USP7 complex acts as a transcription co-repressor, which is required to catalyse deubiquitylation of the human tumour suppressor p53 and histone H2B and to epigenetically silence homeotic genes by PRC1 and PRC294,95. In addition, in flies, GMPS-USP7 binds to the ecdysone receptor, localizes at ecdysone-regulated loci and acts as a selective transcription co-repressor of ecdysone-inducible genes96. These results revealed cooperation between a metabolic enzyme and a ubiquitin protease in regulation of hormone receptor-mediated developmental gene expression.
Methionine adenosyltransferase
MAT, also known as SAM synthetase, produces SAM with methionine and ATP As a methyl donor, SAM enables methyltransferase-catalysed methylation of cytosines in DNA and of arginine and lysine residues in various proteins, including histones.
In yeast, MAT activity is incorporated in the serine-responsive SAM-containing metabolic enzyme (SESAME) complex, which also contains Pyk1, serine metabolic enzymes and an acetyl-CoA synthetase. The Set1 H3K4 methyltransferase complex interacts with SESAME and requires SAM synthesized from SESAME for H3K4 trimethylation and subsequent Pyk1-mediated H3T11 phosphorylation74.
Three distinct forms of MAT (MATI, MATII and MATIII) have been identified in mammals. MATI and MATIII are the gene products of MAT1A, while MATII is encoded by the gene MAT2A. MATIIα is the catalytic subunit of MATII and is inhibited by association with MATIIβ regulatory subunit97. MATIIα localizes in the nucleus and interacts with members of chromatin-modifying and chromatin-remodelling complexes, including the Polycomb group proteins, NuRD complex, SWi/SNF complex and PARP, as well as with the oncogenic transcription factor MAFK. MATIIα is recruited to the MAFK recognition element at the HMOX1 gene, which encodes haem oxygenase 1, an essential enzyme in haem catabolism. The catalytic activity of MATIIα as well as its interacting factors, including MATIIβ, BRG1-associated factor 53A (BAF53A; also known as ACTL6A), chromodomain-helicase-DNA-binding protein 4 (CHD4) and PARP1, is required for HMOX1 repression and positive regulation of haem levels, but the exact mechanisms for this repression are not known. Of note, methyltransferase activity was detected in the nuclear MATIIα complex, suggesting that MATIIα-associated histone methyltransferases are involved in histone methylation to regulate gene expression. Indeed, upon MATIIα depletion, H3K4me2 and H3K9me2 levels are decreased at the HMOX1 locus, with consequent derepression of HMOX1 (REF.97). Thus, MATII serves as a transcription co-repressor of MAFK, whereby MATIIα interacts with chromatin regulators, such as histone methyltransferases and supplies SAM for subsequent histone methylation.
Acetyl-CoA synthesizing enzymes
Recent studies demonstrate that enzymes involved in acetyl-CoA synthesis, such as ACSS2 (REFS98,99), ACLY100,101 and PDC102, translocate to the nucleus to regulate gene expression. This forms a sharp contrast to prevailing dogma, which places these enzymes in the mitochondria or cytoplasm, where they conduct their conventional metabolic functions in the TCA cycle and fatty acid and lipid synthesis.
In S. cerevisiae, Acs2p, the ACSS2 orthologue, is located in the nucleus of cells grown in rich media103. Acs2p deficiency causes global histone deacetylation, transcription inhibition and growth defects in HAT mutant background. Rapid histone deacetylation upon Acs2p inactivation suggests that nuclear Acs2p-mediated acetyl-CoA synthesis is rate limiting for histone acetylation103.
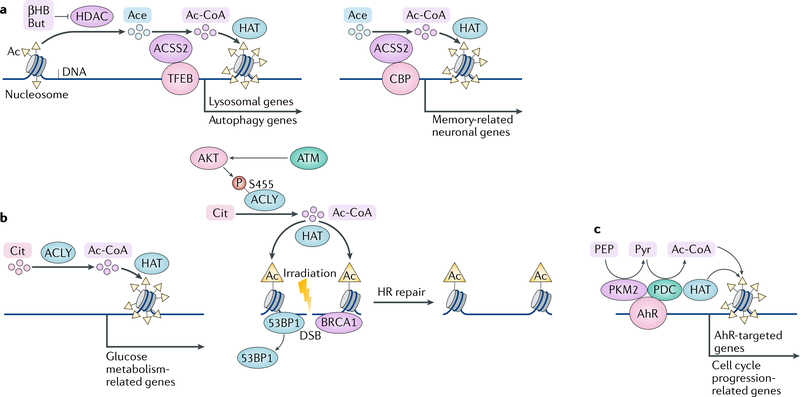
Metabolic stress, such as a limited supply of glucose and acetate, is common in rapidly growing tumours and induces nuclear translocation of cytosolic ACSS2; this process is mediated by phosphorylation of ACSS2 S659 by AMPK, exposure of the NLS of ACSS2 and subsequent ACSS2 binding to importin α5 (REF.98). In the nucleus, ACSS2 interacts with transcription factor EB (TFEB) — a master regulator of lysosomal and autophagy genes. These two proteins bind, in a mutually dependent manner, to the promoters of TFEB target genes, where ACSS2 converts acetate generated by HDAC-mediated histone deacetylation into acetyl-CoA that is recycled by HATs to locally acetylate histone H3 at TFEB-responsive promoters98,104. These findings suggest that ACSS2 utilizes acetate that is generated by HDAC-mediated global histone deacetylation to locally produce acetyl-CoA to support local histone acetylation, thereby promoting the expression of lysosomal and autophagy genes and subsequent lysosomal biogenesis and autophagy (FIG. 5a, left panel). Increased autophagy and lysosomal degradation may provide tumour cells with alternative sources of nutrients for tumour cell survival and have been shown to promote brain tumorigenesis in mice. Accordingly, analysis of human astrocytoma and glioblastoma samples revealed a direct correlation between ACSS2 S659 phosphorylation and tumour aggressiveness98,105, supporting a critical role of nuclear ACSS2 in the induction of a specific group of genes to counteract nutritional stress and support tumour growth.
Fig. 5 |. Acetylation of histones regulated by metabolic enzymes and metabolites.
a | Histone acetylation (Ac) is mediated by histone acetyltransferases (HATs) and reversed by histone deacetylases (HDACs). Both enzyme groups are metabolically regulated. HDACs are inhibited by β-hydroxybutyrate (βHB) and butyrate (But). Acetyl-CoA (Ac-CoA) synthetase short-chain family member 2 (ACSS2) fuels the activity of HATs by producing Ac-CoA from acetate (Ace) locally. By forming complexes with transcription factor EB (TFEB) (left panel) or cAMP-responsive element-binding protein (CBP) (right panel), ACSS2 regulates the expression of lysosomal and autophagy genes and memory-related neuronal genes, respectively. b | Nuclear ATP citrate (Cit) synthase (ACLY) produces nuclear Ac-CoA to regulate expression of glucose metabolism-related genes (left panel) or DNA repair by homologous recombination (HR) (right panel). In the context of DNA repair, ACLY is regulated by phosphorylation (P) mediated by protein kinase B (AKT), which acts downstream of DNA double-strand break (DSB)-activated ataxia-telangiectasia mutated (ATM) kinase. c | Pyruvate dehydrogenase complex (PDC) together with pyruvate kinase M2 isoform (PKM2) and HAT coordinately acetylates histones and activates expression of genes regulated by aryl hydrocarbon receptor (AhR) as well as genes involved in cell cycle progression. 53BP1, TP53-binding protein; BRCA1, breast cancer type 1 susceptibility protein; PEP, 2-phosphoenolpyruvate; Pyr, pyruvate.
Targeted gene expression regulated by nuclear ACSS2 is also observed in neuronal plasticity regulation in mice99. In differentiating neurons and in the hippocampus, nuclear ACSS2 together with the acetyltransferase CBP is recruited to the promoter regions of memory-related neuronal genes, where it supports histone acetylation, gene expression and hippocampus-mediated long-term memory consolidation. ACSS2 depletion specifically in the dorsal hippocampus dramatically reduces mouse performance in a novel object recognition task99, suggesting that the loss of ACSS2-mediated histone acetylation in hippocampal neurons results in impaired memory consolidation (FIG. 5a, right panel). Consequently, this pathway might be a promising novel therapeutic target in various neuropsychiatric disorders including age-dependent memory decline106, Alzheimer disease107–109 and substance use disorders110–114.
In addition to their role in histone acetylation, nuclear acetyl-CoA synthesis enzymes can promote acetylation of transcription factors. It was shown that ACSS2 increases HIF2α acetylation, resulting in HIF2α-CBP complex formation and recruitment of this complex to the erythropoietin gene (EPO) enhancer115. Notably, supplementation of acetate, which is a substrate of ACSS2, increases EPO expression, inducing higher haematocrit levels in an ACSS2-dependent manner115. Thus, acetate supplementation provides a potential approach to treat acute and chronic anaemic conditions. In addition, ACSS2-HIF2α-CBP signalling is also active and required for tumour cell proliferation during hypoxia and in the presence of low glucose116.
Overall, in response to various cellular signalling cues, nuclear ACSS2 can form distinct complexes with transcription regulators. ACSS2 is then able to support acetylation of its interacting partners as well as histones. The composition and modifications of such ACSS2-containing protein complexes specify their recruitment to the promoter regions of different sets of genes to coordinately and locally support histone acetylation and to activate specific gene expression programmes.
Under abundant nutrient and oxygen conditions, ACLY is a major source of acetyl-CoA. ACLY has been detected in the nucleus, although it lacks a canonical NLS, and the mechanism underlying its nuclear localization remains poorly understood100. ACLY-dependent production of acetyl-CoA contributes to increased histone acetylation and to selective regulation of genes involved in glucose metabolism during cellular response to growth factor stimulation and during adipocyte differentiation101 (FIG. 5b, left panel).
ACLY also has a role in the DNA damage response. Upon exposure to ionizing radiation, ACLY accumulates in nuclei in the S and G2 cell cycle phases (when repair through homologous recombination (HR) is favoured)100. AKT kinase, which is activated by ataxia-telangiectasia mutated (ATM) kinase in response to DNA double-strand breaks, phosphorylates ACLY at S455 and increases its catalytic activity. Activated ACLY increases DNA damage-induced acetylation of H4K16 at nucleosomes flanking sites of DNA double-strand breaks, which promotes the exclusion of TP53-binding protein (53BP1) and the recruitment of breast cancer type 1 susceptibility protein (BRCA1), thereby promoting HR100 (FIG. 5b, right panel).
PDC is a mitochondria-localized multi-enzyme complex. However, PDC has been found to enter the nucleus in response to stimulation with growth factors, oncogenic signalling and mitochondrial stress. Nuclear PDC has also been detected at different stages of embryogenesis102,117,118. PDC was suggested to translocate from the mitochondria to the nucleus in complex with heat shock protein 70 (HSP70)102. Nuclear PDC promotes acetylation of H3K9 and H3K18, phosphorylation of retinoblastoma protein and expression of transcription factor E2F, cyclin A and CDK2 (REF.102), which are important for cell cycle progression. In addition, nuclear PDC has been found to form a complex with p300 and PKM2 (REF.80). This complex is specifically recruited to the enhancer regions of AhR-targeted genes in an AhR-dependent manner and acetylates H3K9 to drive gene expression80. These results suggest cooperation between nuclear metabolic enzymes, whereby PKM2 produces local pyruvate to enable PDC-mediated local acetyl-CoA production and histone acetylation by p300 (REF.80) (FIG. 5c).
In sum, acetyl-CoA produced by distinct, nuclear pools of ACSS2, ACLY and PDC103,104, not only regulates global histone acetylation and gene expression but also modulates local histone acetylation and the expression of specific genes that facilitate an appropriate cellular response to availability of nutrients, different extracellular cues or stresses. This nuclear production of acetyl-CoA is regulated in a manner dependent on extracellular and intracellular stimuli, post-translational modifications of acetyl-CoA synthesis enzymes and dynamic regulation of the nuclear localization and chromatin recruitment of these enzymes in complex with distinct transcription co-regulators.
Conclusions and perspective
Closely integrated and mutual regulation between cell metabolism and gene expression is crucial for fundamental cellular activities. Immediate alteration of the enzymatic activity and the subcellular localization of metabolic enzymes occurs in response to availability of nutrients, growth factors and cytokines as well as physiological and pathological stimuli. These changes not only respond to cellular metabolic needs by direct regulation of metabolism but also regulate cellular homeostasis, growth, proliferation, survival and many other functions by reprogramming gene expression. Metabolic products of these enzymes, such as acyl-CoA, SAM, fumarate, succinate, 2-HG, NAD+, NAM and ketone bodies, either act directly as substrates for DNA or histone modifications or modulate the activity of DNA-modifying and histone-modifying enzymes. In addition to their originally defined metabolic roles, metabolic enzymes can perform non-metabolic functions, such as acting as protein kinases to directly phosphorylate histones or gene transcription machinery components, including transcription factors and transcription co-activators. In addition, nucleus-localized metabolic enzymes can form complexes with transcription factors and histone writers, readers and erasers.
Given the nature of the rapid, precise, dynamic and integrated regulation conveyed by metabolic enzymes and their metabolites, we expect more metabolic enzymes that have functions in the nucleus to be discovered. Importantly, the finding that the nuclear functions of metabolic enzymes and metabolites result in disease-related chromatin modulation and changes in gene expression will shed light on the mechanisms underlying disease development and will make these enzymes promising targets for new therapeutic interventions. As discussed throughout the Review, accumulating evidence indicates that aberrant mutual regulation of metabolism and gene expression promotes tumour development (FIG. 6). For instance, activation of overexpressed or mutated receptor tyrosine kinases results in nuclear translocation of PKM2 and nuclear PKM2-mediated gene expression via post-translational modifications of histones and transcription factors66,71,72. The nuclear α-KGDH–KAT2A complex acts as a histone H3 succinyltransferase to increase expression of many oncogenic genes and to promote tumorigensis84. Similarly, local acetyl-CoA produced by nuclear ACSS2 and ACLY regulates histone acetylation to promote tumour development and HR repair of irradiation-induced DNA damage, respectively98. There is also a strong link between tumorigenesis and the changes in gene expression regulated by fumarase activity85,86,88,89. High fumarate levels locally produced by nuclear fumarase promote JMJC-dependent NHEJ repair of irradiation-induced DNA damage and genomic stability. Nuclear fumarase also regulates genes involved in cell growth arrest downstream of ATF2 by locally interfering with histone demethylation at ATF2 promoters. O-GlcNAcylation of chromatin-associated fumarase, which is prominent in tumour cells, decreases the ability of fumarase to interact with ATF2, thereby counteracting the growth arrest. In some cancers, excess of cellular fumarate caused by tumour-associated loss of function of the FH gene and consequent perturbation of the TCA cycle drives the NRF2-mediated expression of antioxidant genes by promoting succinylation and inactivation of KEAP1. Furthermore, high fumarate levels caused by FH mutation, high succinate levels resulting from loss-of-function mutations in SDH and the production of 2-HG by mutated IDH1 and IDH2 inhibit α-KG-dependent dioxygenases to promote chromatin hypermethylation, which regulates gene expression in tumour cells19. Disruption of the nuclear entry and functions of these metabolic enzymes in the context of cancer is thus expected to provide unique approaches to counteract tumour development119. Structural elucidation of the binding and post-translational modifications of the nuclear protein and DNA substrates of nuclear metabolic enzymes will facilitate the identification of specific interventions that target their nuclear rather than cytosolic or mitochondrial activity, thereby providing selective treatment of human diseases without interfering with the primary role of these enzymes in metabolic reactions.
Fig. 6 |. Association of metabolites and metabolic enzymes with cancer development.
a | Nuclear pyruvate kinase M2 isoform (PKM2) promotes gene expression through phosphorylation (P) of histones or regulation of transcription factors, such as hypoxia-inducible factor 1α (HIF1α). HIF1α activity can also be upregulated by fructose-1,6-bisphosphatase 1 (FBP1) deficiency. b | 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) promotes gene expression through phosphorylation of steroid receptor co-activator protein 3 (SRC3) at S857. This phosphorylation increases the interaction between SRC3 and activating transcription factor 4 (ATF4), leading to gene expression. c | In the nucleus, α-ketoglutarate dehydrogenase (α-KGDH) interacts with histone acetyltransferase KAT2A and locally supplies succinyl-CoA (Suc-CoA). This high local concentration of Suc-CoA fuels succinylation (Su) of K79 in histones in the promoter regions of more than 7,000 genes, promoting oncogene expression. d | ATP citrate (Cit) synthase (ACLY) and nuclear acetyl-CoA (Ac-CoA) synthetase short-chain family member 2 (ACSS2) generate local Ac-CoA, which promotes histone acetylation (Ac) by histone acetyltransferases (HATs) and regulates homologous recombination (HR)-dependent DNA repair and the expression of lysosome and autophagy genes under the control of transcription factor EB (TFEB). e | α-KG dioxygenases, including the ten-eleven translocation (TET) and Jumonji C (JMJC) domain-containing demethylases are inhibited by high levels of succinate (Suc) induced by loss-of-function mutations in genes encoding succinate dehydrogenase (SDH−/−), by high levels of fumarate (Fum), which accumulate in the absence of fumarase (FH) activity in the tricarboxylic acid (TCA) cycle owing to loss-of-function mutations in the fumarase gene (FH−/−) as well as by aberrant production of 2-hydroxyglutarate (2-HG) induced by mutation of isocitrate dehydrogenases: cytoplasmic (mIDH1) and mitochondrial (mIDH2). This inhibition contributes to the hypermethylation phenotype and aggressiveness of tumours with SDH, FH or IDH1/2 mutations. High levels of Fum also increase the transcriptional activity of nuclear factor erythroid 2-related factor 2 (NRF2) and subsequent antioxidant response by promoting succinylation (Su) and, in turn, inhibition of Kelch-like ECH-associated protein 1 (KEAP1).Wild-type fumarase can be targeted to the nucleus, where it contributes to local production of Fum from malate (Mal) and inhibition of demethylases, which locally increases histone methylation (Me) to support nonhomologous end joining (NHEJ) DNA repair. In addition, wild-type fumarase as a tumour suppressor can be negatively regulated by O-GlcNAcylation (OG) mediated by O-GlcNAc transferase (OGT), which interferes with fumarase phosphorylation and the interaction of fumarase with ATF2, leading to suppression of ATF2-regulated cell growth arrest genes. Ace, acetate; CCND1, G1/S-specific cyclin D1.
Cofactors.
Non-protein chemical compounds or metallic ions that are required for the activity of an enzyme.
β-Oxidation.
The catabolic process by which fatty acid molecules are broken down in the cytosol in prokaryotes and in the mitochondria in eukaryotes to generate acetyl-CoA.
Ketone bodies.
Any of three related compounds (acetone, acetoacetic acid and β-hydroxybutyric acid) produced during the metabolism of fats.
Streptozotocin.
A naturally occurring alkylating anti-neoplastic agent that is particularly toxic to the insulin-producing β-cells of the pancreas in mammals.
Homeotic genes.
A group of genes that regulates the development of anatomical structures in various organisms.
Linker histone.
Member of a family of histones that bind to the nucleosomal core particle around the DNA entry and exit sites and serve as key components of chromatin. Also known as H1 histone.
β-Catenin.
A protein that regulates cell–cell adhesion and gene transcription by translocating to the nucleus and associating with T cell factor (TCF) and lymphoid enhancer factor (LEF) transcription factors; it is encoded by the CTNNB1 gene.
Warburg effect.
The elevated glucose uptake and lactate production observed in many cancer cell lines regardless of oxygen availability.
Aryl hydrocarbon receptor.
(AhR). A ligand-activated transcription factor involved in the regulation of biological responses to planar aromatic (aryl) hydrocarbons.
Pentose phosphate pathway.
A glycolysis-parallel metabolic pathway that generates NADPH, pentoses and ribose 5-phosphate for nucleotide synthesis.
DNA-dependent protein kinase.
(DNA-PK). A nuclear serine/ threonine-protein kinase that is activated upon DNA damage.
Histone H2A.Z.
An evolutionarily conserved histone variant involved in transcription regulation and genome stability.
Nonhomologous end joining.
(NHEJ). A pathway that repairs double-strand DNA breaks by direct ligation without the need for a homologous template.
Hereditary leiomyomatosis.
An autosomal dominant condition in which susceptible individuals are at risk of the development of cutaneous leiomyomas, early onset multiple uterine leiomyomas and an aggressive form of type 2 papillary renal cell cancer.
Gluconeogenesis.
A metabolic pathway that results in the generation of glucose from non-carbohydrate carbon substrates such as lactate, glycerol and glucogenic amino acids.
von Hippel–Lindau.
An inherited disorder characterized by the formation of tumours and fluid-filled sacs (cysts) in many different parts of the body.
Retinitis pigmentosa.
A genetic disorder of the eyes that causes loss of vision.
Ecdysone.
A steroid hormone secreted by the prothoracic gland that, in its active form, stimulates metamorphosis and regulates moulting in insects.
NuRD complex.
The nucleosome remodelling and deacetylase complex.
SWI/SNF complex.
An evolutionarily conserved multisubunit chromatinremodelling complex that uses the energy of ATP hydrolysis to mobilize nucleosomes and remodel chromatin.
Haem.
The component of haemoglobin (and other haemoproteins) responsible for binding oxygen.
Haematocrit.
The ratio of the volume of red blood cells to the total volume of blood.
Homologous recombination.
(HR). A type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA.
Ataxia-telangiectasia mutated (ATM) kinase.
A serine/threonine-protein kinase that is recruited and activated by DNA double-strand breaks.
TP53-binding protein.
(53BP1). A protein involved in DNA repair, which is encoded by the TP53BP1 gene.
Breast cancer type 1 susceptibility protein.
(BRCA1). A tumour suppressor protein involved in DNA repair.
Retinoblastoma protein.
A tumour suppressor protein that inhibits cell cycle progression and is dysfunctional in several major cancers.
Acknowledgements
This work was supported by National Institute of Neurological Disorders and Stroke grant R01 NS089754 (to Z.L.), NCI grants 2R01 CA109035 and R01CA204996 (to Z.L.) and the US National Institutes of Health (NIH) National Cancer Institute (NCI) under award number P30CA016672, 2P50 CA127001 (Brain Cancer SPORE). Z.L. is a Ruby E. Rutherford Distinguished Professor.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Molecular Cell Biology thanks Y. Zhao and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Tan M et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).This study describes a large number of novel histone marks, including lysine crotonylation and tyrosine hydroxylation as novel histone modifications.
- 4.Huang H, Sabari BR, Garcia BA, Allis CD & Zhao Y Snapshot: histone modifications. Cell 159, 458-458.e1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tessarz P & Kouzarides T Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol 15, 703–708 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB & Jones PA A decade of exploring the cancer epigenome — biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H & Zhang Y Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Li H, Patel DJ & Allis CD Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol 8, 983–994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi P, Allis CD & Wang GG Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson MA & Kouzarides T Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ronan JL, Wu W & Crabtree GR From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet 14, 347–359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis PW et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng X Structural and functional coordination of DNA and histone methylation. Cold Spring Harb. Perspect. Biol 6, a018747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger SL The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Grillo MA & Colombatto S S-adenosylmethionine and its products. Amino Acids 34, 187–193 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Mentch SJ et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell. Metab 22, 861–873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiraki N et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell. Metab 19, 780–794 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Janke R, Dodson AE & Rine J Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol 31, 473–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolinoy DC, Huang D & Jirtle RL Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl Acad. Sci. USA 104, 13056–13061 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye C, Sutter BM, Wang Y, Kuang Z & Tu B P A metabolic function for phospholipid and histone methylation. Mol. Cell 66, 180–193.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzger E et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature 464, 792–796 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Forneris F, Binda C, Vanoni MA, Mattevi A & Battaglioli E Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett 579, 2203–2207 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Shi YG & Tsukada Y The discovery of histone demethylases. Cold Spring Harb. Perspect. Biol 5, a017947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey BW, Finley LW, Cross JR, Allis CD & Thompson CB Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang IY et al. Psat1-Dependent Fluctuations in alpha-Ketoglutarate Affect the Timing of ESC Differentiation. Cell. Metab 24, 494–501 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Losman JA & Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salminen A, Kauppinen A, Hiltunen M & Kaarniranta K Krebs cycle intermediates regulate DNA and histone methylation: epigenetic impact on the aging process. Ageing Res. Rev 16, 45–65 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Sciacovelli M et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greer EL & Shi Y Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet 13, 343–357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unoki M et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem 288, 6053–6062 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimazu T et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339, 211–214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canto C, Menzies KJ & Auwerx J NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivanand S, Viney I & Wellen KE Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends Biochem. Sci 43, 61–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai L, Sutter BM, Li B & Tu BP Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 (2011).This study describes a link between lipid metabolism and histone methylation.
- 36.Chen Y et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteom 6, 812–819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai L et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nature Chem. Biol 10, 365–370 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Xie Z et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom 11, 100–107 (2012).References 36–38 identify new types of histone modification.
- 39.Sabari BR, Zhang D, Allis CD & Zhao Y Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol 18, 90–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabari BR et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Z et al. Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation. Mol. Cell 62, 194–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B et al. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem 284, 32288–32295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goudarzi A et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell 62, 169–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kebede AF et al. Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol 24, 1048–1056 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Cahill GF Jr. Fuel metabolism in starvation. Annu. Rev. Nutr 26, 1–22 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Laffel L Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab. Res. Rev 15, 412–426 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Robinson AM & Williamson DH Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol. Rev 60, 143–187 (1980). [DOI] [PubMed] [Google Scholar]
- 48.Yang X & Qian K Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol 18, 452–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardiville S & Hart GW Nutrient regulation of gene expression by O-GlcNAcylation of chromatin. Curr. Opin. Chem. Biol 33, 88–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gambetta MC & Muller J A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma 124, 429–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraus WL & Lis JT PARP goes transcription. Cell 113, 677–683 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Gupte R, Liu Z & Kraus WL PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibbs-Seymour I, Fontana P, Rack JGM & Ahel I HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell 62, 432–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C & Mandel P Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA 79, 3423–3427 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright RH et al. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 26, 1972–1983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibson BA & Kraus WL New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews. Mol. Cell Biol 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Bai P & Canto C The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell. Metab 16, 290–295 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Gibson BA et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353, 45–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krishnakumar R & Kraus WL PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang W & Lu Z Pyruvate kinase M2 at a glance. J. Cell Sci 128, 1655–1660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David CJ, Chen M, Assanah M, Canoll P & Manley JL HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desai S et al. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 5, 8202–8210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bluemlein K et al. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget 2, 393–400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W et al. EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell 48, 771–784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuan CT, Wikstrand CJ & Bigner DD EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr. Relat. Cancer 8, 83–96 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Yang W et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol 14, 1295–1304 (2012).This study describes an instrumental mechanism of the Warburg effect regulated by nuclear PKM2.
- 67.Lu Z & Hunter T Prolyl isomerase Pin1 in cancer. Cell Res. 24, 1033–1049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spoden GA et al. The SUMO-E3 ligase PIAS3 targets pyruvate kinase M2. J. Cell. Biochem 107, 293–302 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Lv L et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol. Cell 52, 340–352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang HJ et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1alpha-mediated glucose metabolism. Proc. Natl Acad. Sci. USA 111, 279–284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 (2012).This study discovers that a metabolic enzyme can directly phosphorylate histone proteins.
- 72.Yang W et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 480, 118–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosios AM, Fiske BP, Gui DY & Vander Heiden MG Lack of evidence for PKM2 protein kinase activity. Mol. Cell 59, 850–857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S et al. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell 60, 408–421 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Yu Q et al. Regulation of SESAME-mediated H3T11 phosphorylation by glycolytic enzymes and metabolites. PLoS ONE 12, e0175576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao X, Wang H, Yang JJ, Liu X & Liu ZR Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 (2012).This study discovers that PKM2 phosphorylates the STAT3 transcription factor.
- 77.Luo W et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kato H, Fukuda T, Parkison C, McPhie P & Cheng SY Cytosolic thyroid hormone-binding protein is a monomer of pyruvate kinase. Proc. Natl Acad. Sci. USA 86, 7861–7865 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morfouace M et al. Control of glioma cell death and differentiation by PKM2-Oct4 interaction. Cell Death Dis. 5, e1036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matsuda S et al. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Res. 44, 636–647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dasgupta S et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature 556, 249–254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weinert BT et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Zhang Z et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol 7, 58–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).This study discovers that KAT2A, which was already known as a HAT, is also a histone H3 succinyltransferase.
- 85.Jiang Y et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol 17, 1158–1168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yogev O et al. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 8, e1000328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ooi A et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20, 511–523 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Adam J et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang T et al. O-GlcNAcylation of fumarase maintains tumour growth under glucose deficiency. Nat. Cell Biol 19, 833–843 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Li B et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature 513, 251–255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng L, Roeder RG & Luo Y S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 (2003). [DOI] [PubMed] [Google Scholar]
- 92.Hedstrom L IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev 109, 2903–2928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kozhevnikova EN et al. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol. Cell 47, 133–139 (2012). [DOI] [PubMed] [Google Scholar]
- 94.Frappier L & Verrijzer CP Gene expression control by protein deubiquitinases. Curr. Opin. Genet. Dev 21, 207–213 (2011). [DOI] [PubMed] [Google Scholar]
- 95.van der Knaap JA et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695–707 (2005). [DOI] [PubMed] [Google Scholar]
- 96.van der Knaap JA, Kozhevnikova E, Langenberg K, Moshkin YM & Verrijzer CP Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Mol. Cell. Biol 30, 736–744 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katoh Y et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell 41, 554–566 (2011). [DOI] [PubMed] [Google Scholar]
- 98.Li X et al. Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol. Cell 66, 684–697.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mews P et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017).References 98 and 99 report that nuclear ACSS2 regulates histone acetylation in specific sets of gene promoter regions under physiological and pathological conditions.
- 100.Sivanand S et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell 67, 252–265.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wellen KE et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sutendra G et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Takahashi H, McCaffery JM, Irizarry RA & Boeke JD Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 (2006).References 100–103 reveal important roles of nuclear acetyl-CoA synthesizing enzymes in histone acetylation.
- 104.Bulusu V et al. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 18, 647–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li X, Qian X & Lu Z Local histone acetylation by ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Autophagy 13, 1790–1791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peleg S et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 (2010). [DOI] [PubMed] [Google Scholar]
- 107.Bennett DA et al. Epigenomics of Alzheimer’s disease. Transl Res. 165, 200–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fischer A Targeting histone-modifications in Alzheimer’s disease. What is the evidence that this is a promising therapeutic avenue? Neuropharmacology 80, 95–102 (2014). [DOI] [PubMed] [Google Scholar]
- 109.Lu X et al. Histone acetyltransferase p300 mediates histone acetylation of PS1 and BACE1 in a cellular model of Alzheimer’s disease. PLoS ONE 9, e103067 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Egervari G, Ciccocioppo R, Jentsch JD & Hurd YL Shaping vulnerability to addiction - the contribution of behavior, neural circuits and molecular mechanisms. Neurosci. Biobehav. Rev 85, 117–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Egervari G et al. A functional 3’UTR polymorphism (rs2235749) of prodynorphin alters microRNA-365 binding in ventral striatonigral neurons to influence novelty seeking and positive reward traits. Neuropsychopharmacology 41, 2512–2520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Egervari G et al. Striatal H3K27 acetylation linked to glutamatergic gene dysregulation in human heroin abusers holds promise as therapeutic target. Biol Psychiatry 81, 585–594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koo JW et al. Epigenetic basis of opiate suppression of Bdnf gene expression in the ventral tegmental area. Nat. Neurosci 18, 415–422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Robison AJ & Nestler EJ Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci 12, 623–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu M et al. An acetate switch regulates stress erythropoiesis. Nat. Med 20, 1018–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen R et al. The acetate/ACSS2 switch regulates HIF-2 stress signaling in the tumor cell microenvironment. PLoS ONE 10, e0116515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nagaraj R et al. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 168, 210–223.e1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shi WY, Yang X, Huang B, Shen WH & Liu L NOK mediates glycolysis and nuclear PDC associated histone acetylation. Front. Biosci 22, 1792–1804 (2017). [DOI] [PubMed] [Google Scholar]
- 119.Lu Z & Hunter T Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci 43, 301–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]