SUMMARY
Transcriptional regulation in eukaryotes occurs at promoter-proximal regions wherein transcriptionally engaged RNA Polymerase II (Pol II) pauses before proceeding towards productive elongation. The role of chromatin in pausing remains poorly understood. Here, we demonstrate that the histone deacetylase SIRT6 binds to Pol II and prevents the release of the Negative ELongation Factor NELF, thus stabilizing Pol II promoter-proximal pausing. Genetic depletion of SIRT6 or its chromatin deficiency upon glucose deprivation causes intragenic enrichment of acetylated histone H3 at lysines 9 (H3K9ac) and 56 (H3K56ac), activation of cyclin-dependent kinase 9 (CDK9) -which phosphorylates NELF and the carboxyl terminal domain of Pol II- and enrichment of the positive transcription elongation factors MYC, BRD4, PAF1 and the super elongation factors AFF4 and ELL2. These events lead to increased expression of genes involved in metabolism, protein synthesis, and embryonic development. Our results identified SIRT6 as a Pol II promoter-proximal pausing-dedicated histone deacetylase.
INTRODUCTION
Environmental adaptation is a fundamental trait of all living organisms. Individual cells rely on molecular sensors altering their activities in response to environmental dynamics, such as nutrient availability. The mammalian sirtuin family of proteins (SIRT1–7) are NAD+-dependent deacetylases capable of sensing changes in nutrient conditions to remodel cellular metabolism (Sebastian et al., 2012a; Choi and Mostoslavsky, 2014; Etchegaray and Mostoslavsky, 2016). SIRT6 is an histone H3K9ac and H3K56 deacetylase, affecting multiple gene networks involved in glucose metabolism, DNA repair, NF-κB signaling, tumorigenesis, early development and aging (Kawahara et al., 2009; Michishita et al., 2008; Michishita et al., 2009; Yang et al., 2009; Zhong et al., 2010b; Sebastian et al., 2012b; Kanfi et al., 2012; Toiber et al., 2013; Kugel and Mostoslavsky, 2014; Silberman et al., 2014; Etchegaray et al., 2015; Kugel et al., 2015; Kugel et al., 2016). The deacetylation activity of SIRT6 was also shown to protect telomeric regions from genomic instability (Michishita et al., 2008; Michishita et al., 2009), and to promote efficient repair of DNA double-strand breaks (Mao et al., 2011; Toiber et al., 2013). Additionally, SIRT6 can also deacetylate H3K18ac on pericentric chromatin as a potential mechanism to prevent DNA erosion upon cell division during cellular senescence (Tasselli et al., 2016). Our earlier work provided strong evidence for SIRT6 roles in repressing expression of glycolytic, ribosomal and developmental genes (Sebastian et al., 2012b; Zhong et al., 2010b; Etchegaray et al., 2015; Ferrer et al., 2018). Yet, the specific mechanisms underlying SIRT6-dependent transcriptional silencing remain unclear.
A key regulatory step during transcription occurs during promoter-proximal pausing of RNA polymerase II (Pol II), at approximately 30 to 60 nucleotides downstream of the transcription start site (TSS) (Rahl et al., 2010; Bartkowiak et al., 2010; Adelman and Lis, 2012). Pol II pausing is mainly regulated by two pausing-promoting factors, DSIF (DRB-sensitive inducing factor) and NELF (negative elongation factor), which is composed of four subunits (NELF-A, B, C/D, E) (Wada et al., 1998; Yamaguchi et al., 1999; Narita et al., 2003; Adelman and Lis, 2012). The release of paused Pol II into productive transcription elongation is facilitated by the recruitment of CDK9-containing P-TEFb (positive transcription elongation factor b), which phosphorylates the C-terminal domain (CTD) of Pol II at serine 2 (Ser2P) as well as the pausing factors resulting in the dissociation of NELF from chromatin and the conversion of DSIF into an elongation-stimulating factor (Marshall et al., 1996; Marshall and Price, 1995; Wei et al., 1998; Fujinaga et al., 2004; Adelman and Lis, 2012; Yamada et al., 2006).
Depending on genetic and cellular contexts, the recruitment of P-TEFb was proposed to be facilitated by c-MYC (MYC), BRD4, as well as different subunits from the Mediator complex including MED23, via interaction with MYC, MED26 and the Super Elongation Complex (SEC) (Rahl and Young, 2014; Liu et al., 2015; Li et al., 2018). BRD4 is a member of the bromodomain and extra-terminal domain (BET) family of transcription factors, which are recruited to acetylated chromatin to promote transcription elongation (Winter et al., 2017). However, the mechanisms downstream of BRD4 that govern release of Pol II pausing into productive elongation remain unclear. Notably, in vitro systems using reconstituted chromatinized templates demonstrated that the histone acetyltransferase p300 activity is required for efficient transcription, and served to elucidate the role of transcription factors involved in Pol II pausing/elongation (Kim et al., 2010; Pavri et al., 2006).
Acetylation of histones was also shown to stimulate productive transcriptional elongation in live cells (Stasevich et al., 2014). More recently, H3K9ac was shown to release Pol II pausing into transcription elongation by recruiting the SEC complex and thereby loss of H3K9ac caused an increase in Pol II pausing at specific genes in HeLa cells (Gates et al., 2017). These studies indicate that transcriptional pausing could be regulated at the level of chromatin deacetylation. However, the histone deacetylase(s) modulating the transition between transcriptional pausing and productive elongation remained unknown. Here, we show that the histone deacetylase SIRT6 interacts with Pol II and inhibits transcription elongation by decreasing intragenic levels of acetylated H3K9 and H3K56 to modulate recruitment of specific transcription factors. Our results identify SIRT6 as a dedicated chromatin deacetylase for modulating transcriptional pausing and elongation.
RESULTS
SIRT6-dependent histone deacetylation of H3K9ac and H3K56ac regulate transcriptional elongation
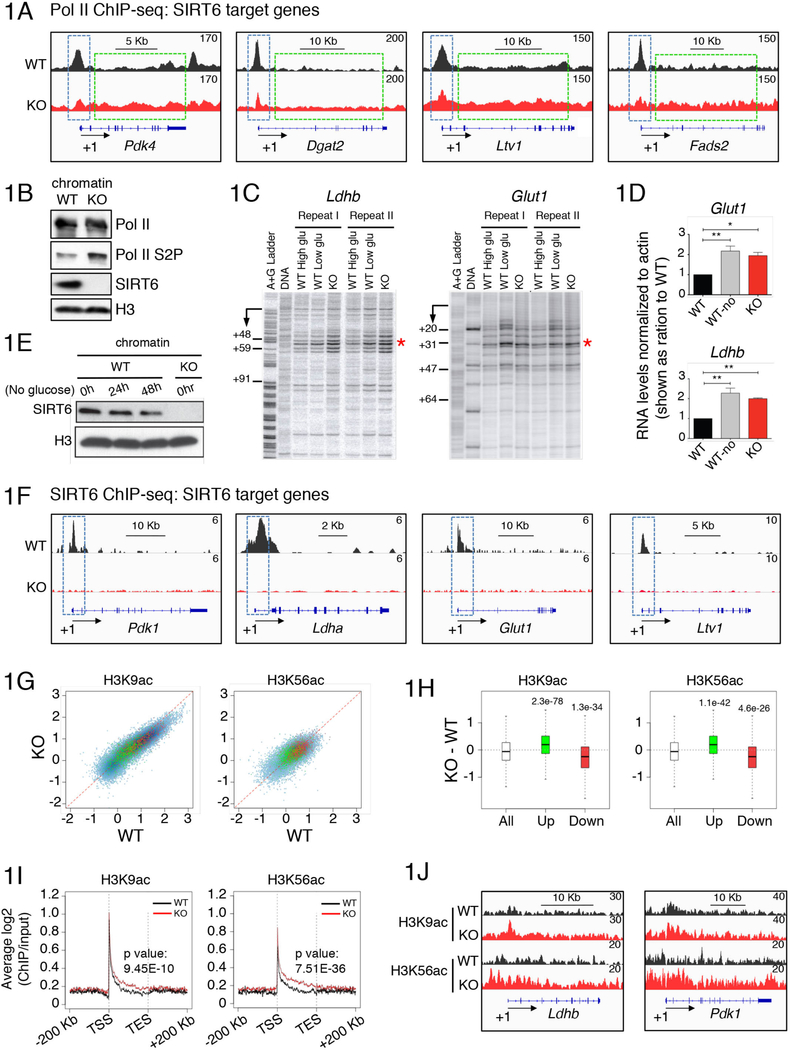
To investigate the role of SIRT6 in transcriptional regulation, we performed ChIP–seq analysis in wild-type (WT) and SIRT6 KO ESCs, using antibodies targeting RNA Polymerase II (Pol II) (Figure 1A). Notably, we found decreasing levels of Pol II at proximal-promoter sites (dotted blue line square) while increasing amounts within intragenic regions (dotted green line square) on multiple SIRT6 target genes, including Pdk4, Dgat2, Ltv1 and Fads2 in SIRT6 KO compared to WT ESCs. This differential pattern of gene-loaded Pol II between WT and SIRT6 KO suggested that SIRT6 could influence a transition from a promoter-proximal paused Pol II to an elongating state. The phosphorylation of Pol II within its carboxyl terminal domain at serine 2 (Pol II S2P) is a hallmark for productive transcription elongation (reviewed in (Jonkers and Lis, 2015). In this context, we found a global increase in the levels of Pol II S2P on chromatin from SIRT6 KO ESCs (Figure 1B).
Figure 1. SIRT6 regulates transcriptional pausing via deacetylation of H3K9ac and H3K56ac.
(A) Integrative Genomics Viewer (IGV) browser (Robinson et al., 2011; Thorvaldsdottir et al., 2013) images of read coverage across the genome. Images of tiled data files (TDFs) generated by the displaying the density tracks of reads aligned across the genome. The tracks show ChIP-seq maps of Pol II on SIRT6 target genes Pdk4, Dgat2, Ltv1 and Fads2 in WT (black) and SIRT6 KO (red) ESCs. (B) Western blots showing levels of Pol II and Pol II Ser2P in the chromatin fraction isolated from WT and SIRT6 KO ESCs. (C) Permanganate footprinting and (D) RTqPCR showing levels of Ldhb and Glut1 genes in WT ESCs grown in high glucose (black) or no glucose (grey), and SIRT6 KO ESCs (red). Samples were normalized to actin and shown as ratios over WT samples (*: p < 0.05, **: p< 0.01, n > 3. Error bar represents s.e.m). (E) Western blots showing levels of SIRT6 and H3 in chromatin fraction from WT ESCs grown in normal glucose (0hr) or under no glucose for 24hrs or 48hrs, and chromatin from SIRT6 KO ESCs grown in normal glucose conditions. (F) IGV browser images from SIRT6 ChIP-seq showing SIRT6 target genes Pdk1, Ldha and Glut1 and Ltv1 in WT (black) and in SIRT6 KO ESCs (red). (G) Scatter plot analysis showing significant increase (green) or decrease (red) in H3K9ac or H3K56ac on genes upregulated in SIRT6 KO versus WT ESCs (upper panels). (H) Graphical quantification of the data on (G). (I) Metagene analysis showing enrichment of H3K9ac or H3K56ac at intragenic regions in SIRT6 KO (red) compared to WT (black) ESCs. (J) IGV broswer images from H3K9ac and H3K56ac ChIP-seqs in SIRT6 KO (red) versus WT ESCs (black) on Ldhb and Pdk1 genes.
To further assess roles for SIRT6 in transcriptional regulation, we performed permanganate DNA footprinting, one of the most authoritative means to detect paused polymerase in vivo (Nechaev et al., 2010). We analyzed two representative SIRT6 target genes, lactose dehydrogenase b (Ldhb) and glucose transporter 1 (Glut1) (Figure 1C)( (Zhong et al., 2010a). We found Pol II pausing on both genes at promoter-proximal regions (between +48 and +59 on Ldhb and +31 on Glut1) in WT cells. Notably, SIRT6 KO ESCs exhibited a stronger signal within the same regions, showing increased trafficking of Pol II at promoter-proximal sites, indicative of increased transcriptional elongation (Samarakkody et al., 2015). Importantly, increased Pol II trafficking was also observed in WT ESCs grown under glucose deprivation (Figure 1C), demonstrating that SIRT6 deficiency mimics conditions of nutrient stress (Etchegaray and Mostoslavsky, 2016; Zhong et al., 2010b). Accordingly, increased mRNA levels for Ldhb and Glut1 were similar both in WT ESCs following glucose deprivation and SIRT6 KO ESCs (Figure 1D), in parallel to the global dissociation of SIRT6 from chromatin following glucose removal from the culturing media (Figure 1E). Using ChIP-seq analysis, we determined that SIRT6 localizes particularly at promoter-proximal regions of its target genes, including Pdk1, Ldha, Glut1 and Ltv1 (Figure 1F). Furthermore, by using ChIP-qPCR we also found that SIRT6 binding is lost in WT ESCs cultured under conditions of glucose starvation (Figure S1A). These results support a role for SIRT6 in regulating transcription elongation via Pol II promoter-proximal pausing.
As mentioned above, SIRT6 represses transcription by acting as a specific deacetylase for H3K9ac and H3K56ac (Zhong et al., 2010b; Sebastian et al., 2012b; Etchegaray et al., 2015; Kugel et al., 2016). Thus, we performed ChIP-seq experiments in WT and SIRT6 KO ESCs to identify the genomic regions targeted by SIRT6-dependent histone deacetylation impacting gene expression, as determined by RNA-seq analysis (Table S1). RNA sequencing identified 4938 genes upregulated and 4534 genes downregulated in SIRT6 KO cells. We focused on the upregulated genes, given the likelihood that downregulated genes cannot be explained as direct targets of a deleted histone deacetylase. From the nearly 5,000 genes upregulated in SIRT6 KO ESCs, about 1,200 genes have enriched levels of both H3K9ac and H3K56ac, and approximately 1,800 genes are enriched for either H3K9ac or H3K56ac (Figure S1B), indicating that most genes upregulated in the SIRT6 KO cells are direct targets of SIRT6. Furthermore, we observed a specific enrichment of these histone marks only on genes that are upregulated in SIRT6 KO ESCs, not the downregulated ones (Figures 1G and 1H; green dots compared to red dots, respectively).
Next, using metagene analysis, we found that the increase in H3K9ac and H3K56ac occurs mainly at promoter-proximal regions and gene bodies (Figures 1I, 1J and S1C). In addition to metabolism and pluripotency, the genes impacted by SIRT6 removal were also enriched for neural development ontologies (Table S2), which is consistent with the neural-skewed phenotype we observed upon differentiation of SIRT6 KO ESCs (Etchegaray et al., 2015). These results indicate that SIRT6 has a prominent role in modulating histone acetylation within intragenic regions encompassing promoter-proximal pausing and gene bodies. This ability of SIRT6 to regulate intragenic histone acetylation as a potential mean to modulate Pol II pausing/elongation is different from the known roles of Class I and Class II HDACs that work predominantly at TSS regions, affecting Pol II recruitment at promoter sites (Jones et al., 1998; Nan et al., 1998).
SIRT6 supports transcriptional promoter-proximal pausing
To identify the genes mostly affected by SIRT6-dependent Pol II pausing, we first mapped the binding of SIRT6 and Pol II in mouse ESCs in a genome-wide manner. Consistent with the distribution of histone marks, and in contrast to other HDACs (Ram et al., Cell 2011), SIRT6 localized at promoter-proximal pausing sites downstream of the TSS and closely following the positional pattern of Pol II throughout the genome (Figure 2A). Furthermore, when compared to WT, Pol II binding on SIRT6 KO cells exhibits a more diffuse pattern, suggesting increased intragenic binding (Figure 2A). When zoomed in at known SIRT6-targeted genes (Figure 2B–D), a prominent co-localization between Pol II and SIRT6 is observed near promoter-proximal sites. These results indicate that SIRT6 and Pol II are positioned together at the pausing site of these genes.
Figure 2. Global decrease of Pol II pausing in ESCs lacking SIRT6.
(A) Co-localization of SIRT6 and Pol II. Heat maps showing Pol II and SIRT6 ChIP-seq signal within 3kb genomic windows flanking the TSS (the TSS is denoted as an arrow) in WT and SIRT6 KO ESCs. The SIRT6 heat map on SIRT6 KO ESCs was included as a control. (B) IGV broswer images for of Pol II and SIRT6 ChIP-seq on the SIRT6 target genes Ldha and Ak4. (C) IGV broswer images from Pol II ChIP-seq showing increased levels on Pol II at intragenic regions (gene bodies) in SIRT6 KO (red) compared to WT ESCs (black) on SIRT6 target genes (Ldhb and Ak4). (D) IGV browser images from PRO-seq analysis showing increase levels on Pol II at gene bodies in SIRT6 KO (red) compared to WT ESCs (black) on SIRT6 target genes (Ldhb and Ak4). (E) Metagene profile, from Pol Ii ChIP-seq analysis, showing a decrase in Pol II levels near TSS in SIRT6 KO ESCs (red) compared to WT cells (black) and an overall increased intragenic Pol II levels in SIRT6 KO ESCs. The inset is a zoom-in of intragenic regions highlighting the higher levels of Pol II in SIRT6 KO cells (p<0.04). (F) Pausing Index, calculated from Pol II ChIP-seq analysis, is decreased in SIRT6 KO compared to WT (grey) ESCs. (G) Metagene profile, from PRO-seq analysis, showing an overall increased intragenic Pol II levels in SIRT6 KO (red) compared to WT (black) ESCs. The inset is a zoom-in of intragenic regions highlighting the higher levels of Pol II in SIRT6 KO cells (p<0.0001). (H) Pausing Index, calculated from PRO-seq analysis, is decreased in SIRT6 KO (red) compared to WT (black) ESCs ( p<0.0001). (I) Metagene profile, from PRO-seq analysis, showing an overall increased intragenic Pol II levels in glucose deprived WT ESCs (green) versus WT control cells (black) grwon under normal conditions. The inset is a zoom-in of intragenic regions highlighting the higher levels of Pol II in glucose deprived WT cells (p<0.0001). (J) Decreased Pausing Index, calculated from PRO-seq analysis, in glucose deprived WT ESCs (green) compared to WT control cells (black)(p<0.0001).
We next determined whether SIRT6 directly interact with Pol II. Indeed, endogenous SIRT6 co-immunoprecipitated with Pol II in WT ESCs, even in the presence of Ethidium Bromide, indicating that these proteins interact in a direct fashion (Figures S2C, S2D, S2E). We further validated this result by mass spectrometry analysis, and confirmed interaction of SIRT6 with one of the large subunits of Pol II, RPB2 (Figure S2F), as previously demonstrated by proteomic analysis (Miteva and Cristea, 2014). We next performed metagene analysis of Pol II ChIP-seq data from WT and SIRT6 KO ESCs and identified a significant increase in intragenic signal and a concomitant decrease of promoter-proximal signal for Pol II in SIRT6 KO cells (Figure 2E). Altogether, these results indicate a global role for SIRT6 in inhibiting Pol II transition from promoter-proximal regions to intragenic sites, a transition that is accompanied by active transcriptional elongation.
We next used the Pol II ChIP-seq analysis to calculate Pausing Index (PI) (Rahl et al., 2010) in WT and SIRT6 KO ESCs cells. Notably, SIRT6 KO cells exhibited a global decrease in PI (Figures 2F, S2G), with over two thousand genes shifted towards an increased productive elongation state. To establish whether these Pol II pausing dynamics correspond to transcriptional changes, we performed a comparative analysis between Pol II ChIP-seq and RNA-seq from WT and SIRT6 KO ESCs (Table S3). Nearly half of the upregulated genes (2,459 out of the 4,938 genes upregulated in SIRT6 KO ESCs, 49.7%) show greater than 2-fold decrease in PI compared to WT ESCs, providing evidence for their regulation by pausing/elongation (Figure S2G). Furthermore, these gene sets showing decreased PI include genes involved in metabolic processes and neural development, consistent with the role of SIRT6 as a key regulator of both metabolism (Kugel and Mostoslavsky, 2014) and early embryonic development (Etchegaray et al., 2015; Ferrer et al., 2018)(Figure S2A, S2B, Table S3).
To confirm these results using an orthogonal approach, we performed precision nuclear run-on sequencing (PRO-seq) analysis (Mahat et al., 2016). Pro-seq identifies nascent RNA in a way that defines mapping of RNA-Pol II with single base-pair resolution, allowing us to evaluate Pol II pausing and elongation in WT versus SIRT6 KO ESCs using an independent approach. Metagene analysis confirmed the decrease Pol II binding at TSS and concomitant increased in signal at gene bodies Fig. 2G, S2I, Table S4). Furthermore, from a total of 3659 upregulated genes (based on a minimum of 16 reads of Pol II binding, see Methods), and consistent with the Pol II ChIP seq data analysis described above, we found 2746 genes (75%) with significantly decreased PI (Figures 2H and S2H). Moreover, this molecular phenotype of decreased PI in SIRT6 KO ESCs was recapitulated in WT ESCs cultured under glucose deprivation (Figures 2I, 2J, S2I, Table S4). As control for this approach, we found no significant differences in the levels of intragenic Pol II on genes whose expression is unchanged between WT and SIRT6 KO (Figures, S2J, S2K). Importantly, between the two methodologies we used to determine PI (Pol II ChIP-seq and PRO-seq), we found an overlapping of 2022 genes (~ 80%) that exhibit lower PI in both assays (Table S5). Collectively, using two distinct approaches, we determined that a significant fraction of genes de-repressed in the absence of SIRT6 exhibit decreased transcriptional promoter-proximal pausing, resulting in more Pol II engaged in productive elongation.
SIRT6 supports Pol II pausing by preventing eviction of NELF-E from chromatin
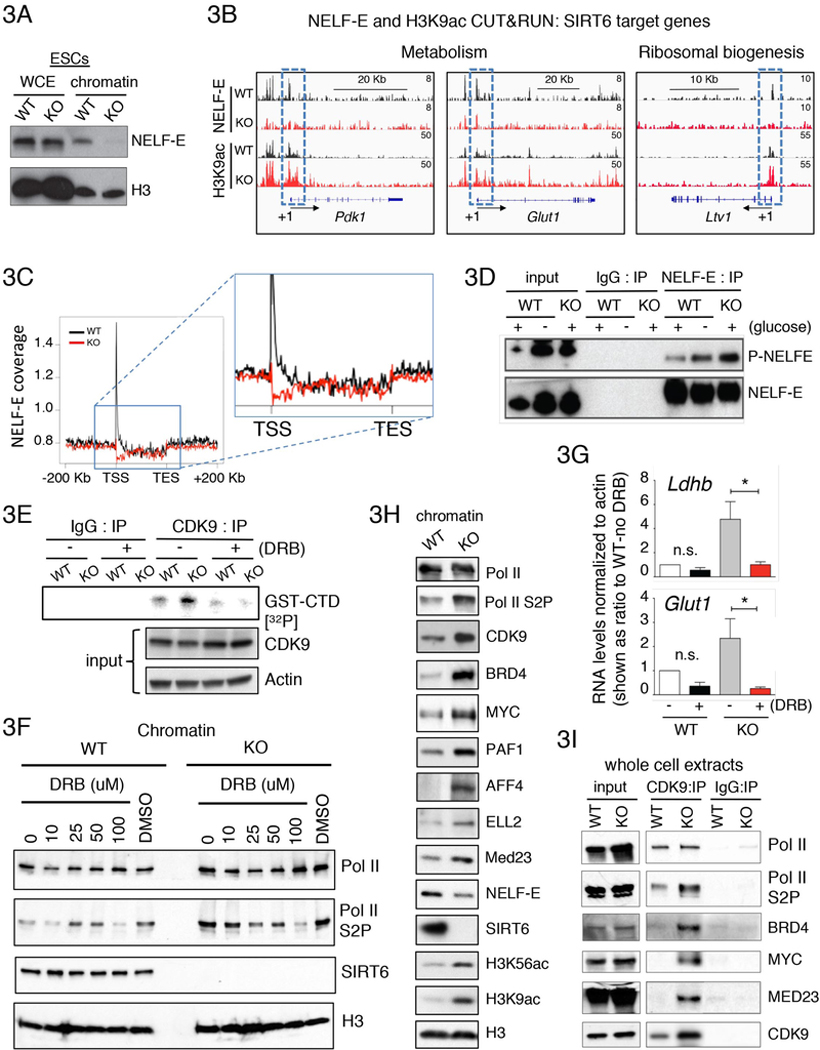
Promoter-proximal pausing requires the association of Pol II with the negative transcription elongation factors NELF and DSIF (Kwak and Lis, 2013). Subunits of these negative elongation factors, including NELF-E and SPT5, are phosphorylated by CDK9 (Fujinaga et al., 2004; Jonkers and Lis, 2015; Ping and Rana, 2001). Phosphorylated NELF-E dissociates from chromatin, while SPT5 phosphorylation converts DSIF into a positive elongation factor, leading to productive transcription elongation (Yamada et al., 2006; Gaertner and Zeitlinger, 2014). Given the above results, we next examined whether NELF was altered in SIRT6 KO cells. Strikingly, chromatin levels of NELF-E decreased in SIRT6 KO ESCs (Figure 3A). Similar results were also observed in mouse embryonic fibroblasts (MEFs) derived from SIRT6 KO mice (Figure S3A). Notably, this phenotype can be rescued by expressing a copy of WT SIRT6 (Figure S3A), indicating that the reduction in chromatin-bound NELF-E is dependent on SIRT6. Additionally, binding of NELF-E to chromatin was also gradually diminished upon glucose withdrawal in WT ESCs (Figure S3B).
Figure 3. SIRT6 is an integral component of the Pol II transcription pausing machinery.
(A) Western blot showing decreased levels of chromatin-bound NELF-E in SIRT6 KO compared to WT ESCs. (B) IGV broswer images from CUT&RUN assays for NELF-E and H3K9ac in WT (black) and SIRT6 KO ESCs (red). TSS regions are denoted as +1 and directionality of transcription by the arrow. Levels of NELF-E and H3K9ac near TSS are highlighted inside the blue dotted line square. (C) Metagene profile for NELF-E coverage in SIRT6 KO (red) versus WT ESCs shows a drastic decrease of NELF-E at promoter-proximal regions. (D) Phosphorylated NELF-E (P-NELF-E) levels increased in SIRT6 KO and glucose starved WT ESCs compared to WT controls. Western blots showing immunoprecipitation of NELF-E blotted with anti-Phospho-CDK9 substrate and anti-NELF-E antibodies in WT, WT no glucose and SIRT6 KO ESCs. (E) CDK9-dependent in vitro kinase assay. CDK9 was immunoprecipitated from either WT or SIRT6 KO ESCs and in vitro kinase assay was performed on beads conjugated with a synthetic GST-tagged carboxyl terminal domain of Pol II peptide (GST-CTD) in the presence of 32p-γATP, in the presence or absence of the CDK9 inhibitor DRB. A representative experiment is shown. (F) Western blot analysis showing decreasing chromatin levels of Pol II S2P by adding the CDK9 inhibitor DRB in a dose-dependent manner to WT and SIRT6 KO ESCs. (G) RTqPCR showing levels of Ldhb and Glut1 genes with or without DRB treatment in WT and SIRT6 KO ESCs. (H) Western blots showing chromatin levels for Pol II, Pol II Ser2P, CDK9, BRD4, MYC, PAF1, AFF4 and ELL2, MED23, NELF-E, SIRT6, H3K56ac, H3K9ac and total histone H3 in WT and SIRT6 KO ESCs. (I) Western blots showing increased co-immunoprecipitation of CDK9 with Pol II Ser2P, BRD4, MYC and MED23 in SIRT6 KO ESCs.
To specifically investigate NELF binding on SIRT6 target genes, we next performed both CUT&RUN (Skene et al., 2018) and ChIP-qPCR experiments. Indeed, we saw loss of NELF-E at the promoter-proximal regions of SIRT6 target genes in SIRT6 KO ESCs (Figure 3B; note the increased H3K9Ac in the same regions). From ~9,000 genes with lower levels of NELF-E in SIRT6 KO compared to WT ESCs, 1,040 genes have low PI (Figure S3C). Consistently, metagene analysis shows decrease levels of NELF-E in SIRT6 KO ESCs at promoter-proximal regions (Figure 3C; Table S6). We also observed a tight correlation between CUT&RUN and ChIP-seq for H3K9ac, thereby supporting the validity of the CUT&RUN approach (Figure S3D). Additionally, by ChIP-qPCR, we observed decreased levels of NELF-E near promoter-proximal regions, on Ldhb and Glut1 genes in SIRT6 KO ESCs as well as WT cells cultured under glucose deprivation (Figure S3E). These results indicate that chromatin-bound NELF depends on SIRT6 to maintain Pol II promoter-proximal pausing in ESCs.
Since CDK9-dependent phosphorylation is a pre-requisite for the release of NELF-E from chromatin (Fujinaga et al., 2004), we next examined the phosphorylation status of NELF-E in SIRT6 KO versus WT ESCs grown under normal or glucose-deprived conditions. Indeed, phosphorylation of NELF-E was increased in both SIRT6 KO ESCs and glucose-depleted WT ESCs, compared to WT ESCs grown under normal nutrient conditions (Figure 3D). These results, together with the increased Pol II phosphorylation we observe in the SIRT6 KO cells (Figure 1B), suggested that eviction or depletion of SIRT6 from its target genes triggers the activation and/or increase recruitment of CDK9. To evaluate this hypothesis, we used an in vitro kinase assay and found that CDK9 purified from SIRT6 KO ESCs exhibits increased kinase activity in comparison to CDK9 purified from WT ESCs (Figure 3E). This increased kinase activity is sensitive to the CDK9 inhibitor DRB (5,6-dichloro-1-B-D-ribofuranosyl-1H-benzimidazole). Accordingly, co-immunoprecipitation with CDK9 shows increased Pol II Ser2P levels in SIRT6 KO compared to WT ESCs (Figure S3F).
To further confirm that upregulation of SIRT6-targeted genes is CDK9-dependent, we treated ESCs with the CDK9 inhibitor DRB. Efficient inhibition of CDK9 was confirmed by measuring phosphorylation of Pol II (Ser2P) in a dose-dependent manner (Figure 3F). Treatment with DRB rescued the upregulated expression of Ldhb and Glut1 in SIRT6 KO cells (Figure 3G). These experiments indicate that SIRT6 sustains Pol II promoter-proximal pausing by preventing CDK9-dependent phosphorylation of NELF-E and Pol II (Ser2P). Importantly, in addition to the increased activity of CDK9, we observed higher levels of this kinase in the chromatin fraction of SIRT6 KO ESCs (Figure 3H).
Multiple transcription factors were shown to be involved in Pol II pausing release, including MYC (reviewed in Rahl and Young, 2014), the bromodomain protein BRD4 (Yang et al., 2005; Bisgrove et al., 2007); reviewed in Petesch and Lis, 2012), a subunit of the mediator complex, MED23, which has been associated with Pol II transcriptional pause release depending on BRD4 (Wang et al., 2005; Lu et al., 2016); AFF4 and ELL2, which are component of the multi-subunit Super Elongation Complex (SEC) (Lin et al., 2010; He et al., 2010; Lu et al., 2016), and the Pol II associated factor 1 (PAF1), which is a component of the PAF1 multi-subunit complex (PAF1C) (Van Oss et al., 2017). Strikingly, all of these factors exhibit increased chromatin levels in SIRT6 KO compared to WT ESCs (Figure 3H). Since P-TEFb, which contains CDK9, is recruited to promoter-proximal pausing sites by some of these transcription factors, such as MYC, BRD4 and MED23 (reviewed in Rahl and Young, 2014; Jonkers and Lis, 2015; Lu et al., 2016), we next assessed levels of these factors by co-immunoprecipitation with CDK9. Consistently, we find higher levels of BRD4, MYC and MED23 to co-immunoprecipitate with CDK9 in SIRT6 KO compared to WT ESCs (Figure 3I), further indicating that SIRT6 inhibits the formation of this multi-protein complex on chromatin.
Increased transcription elongation in SIRT6 KO ESCs is dependent on MYC, BRD4 and PAF1
MYC has been proposed as a key modulator of transcriptional elongation (Rahl et al., 2010). Following our observation of increased levels of MYC in the chromatin fraction of SIRT6 KO ESCs (Figure 3H), we investigated the role of MYC in the assembly of elongation factors and its effect on gene expression in SIRT6 KO ESCs. We first demonstrate that binding of MYC to chromatin is significantly increased in SIRT6 KO ESCs and in glucose-deprived WT ESCs paralleling the increase in H3K9ac and H3K56ac levels (Figure 4A). Furthermore, ChIP-qPCR experiments demonstrated increased binding of MYC to Ldhb and Glut1 genes in SIRT6 KO ESCs, particularly near promoter-proximal pausing sites (Figure 4B), supporting the known role of MYC as a transcription elongation promoting factor (Rahl et al., 2010). Consistently, MYC knockdown inhibited the expression of various SIRT6-targeted genes, including metabolic and ribosomal protein genes (Figure 4C). MYC deficiency also caused a reduction in CDK9 recruitment and the levels of Pol II (Ser2P), while total Pol II amounts remain unchanged in chromatin from WT and SIRT6 KO ESCs (Figures 4D, S4A). Consistent with the RT-qPCR results, the increase in protein levels of the SIRT6 targets LDHA and PDK1 was fully rescued upon MYC knockdown (Figure 4D, lower panel). Importantly, H3K9 acetylation was not affected by knocking down MYC in SIRT6 KO ESCs, confirming that recruitment of MYC occurs downstream of SIRT6 removal from chromatin and the concomitant increase in histone acetylation (Figure 4D). Together, these results suggest a positive feedback loop between CDK9 and MYC during transcriptional elongation, as previously proposed (Huang et al., 2014).
Figure 4. SIRT6 regulates transcription by modulating the levels of chromatin-bound MYC.
(A) Western blots showing chromatin bound MYC and levels H3K9ac and H3K56ac in WT, glucose deprived WT and SIRT6 KO ESCs. (B) MYC ChIP-qPCR experiments in Ldhb and Glut1 genes. Schematic representation for these genes are shown below. Samples were normalized to input and further normalized to IgG ChIP controls. (*: p < 0.05, **: p < 0.005, ***: p < 0.0005, n = 3. Error bar represents s.e.m). (C) RTqPCR analysis showing RNA levels for the metabolic genes Ldha, Ldhb, Glut1, Pdk1 and the ribosomal genes Rpl3, Rpl5, Rpl6 upon MYC knockdown (shMyc) or a control shRNA (shCtrl) in WT, glucose deprived WT and SIRT6 KO ESCs. (*: p < 0.05, n = 3. Error bar represents s.e.m). (D) Western blot analysis showing decreased Pol II S2P, BRD4 and CDK9 on chromatin of SIRT6 KO ESCs upon acute MYC knockdown using two distinct siRNA oligos. H3 is shown as loading control for chromatin fractions. Cytosolic fractions show the rescue of PDK1 and LDH levels upon accute Myc knockdown in SIRT6 KO ESCs. Actin is included as a loading control.
Given the enrichment of H3K9ac and H3K56ac intragenically (compare SIRT6 KO versus WT ESCs; Figures 1G–1J, S1C), we asked whether readers of these marks may be important in this phenotype. The bromodomain-containing factor BRD4 directly binds acetylated histones and can potentially influence the recruitment of MYC and the activation of CDK9, in turn facilitating transcriptional elongation (Kanno et al., 2014; Yang et al., 2005; Lu et al., 2015; Cheung et al., 2017). Additionally, inhibition of BRD4, using BET inhibitors such as JQ1, was shown to impair ESC self-renewal/pluripotency and to promote cell fate commitment to the neuroectodermal lineage (Di Micco et al., 2014), both phenotypes regulated by SIRT6 (Etchegaray et al., 2015; Ferrer et al., 2018).
We observed an increased recruitment of BRD4 in chromatin from SIRT6 depleted cells (ESCs and MEFs; Figure 3H and Figure S4B, respectively). Thus, we reasoned that BRD4 may play a role in the release of paused Pol II and tested whether deficiency of BRD4 could rescue gene expression and transcription elongation phenotypes in SIRT6 KO ESCs. While levels of Pol II remain unchanged, siRNA-mediated knockdown of BRD4 reduced Pol II (Ser2P) levels and decreased the expression of SIRT6 targets (Pdk1, Ldha and Ldhb) (Figures 5A–D, S4C), indicating a relevant role for BRD4 in SIRT6-dependent regulation of transcription elongation. Moreover, BRD4 knockdown lead to a decrease in chromatin MYC in the absence of SIRT6 (Figure 5D), thus supporting a model for BRD4-dependent recruitment of MYC that triggers the release of paused Pol II into transcription elongation. Notably, WT ESCs appear more resistant to BRD4 knockdown compared to SIRT6 KO ESCs, suggesting that highly acetylated chromatin, due to the absence of SIRT6, is more permissive of BRD4-mediated transcription (Figure 5D).
Figure 5. SIRT6 controls the assembly of transcription elongation and super elongation factors.
(A) Western blots from whole cell extracts show levels of PDK1 and LDHA rescued upon accute si RNA-mediated knockdown of BRD4 and/or PAF1 in SIRT6 KO ESCs. (B) RTqPCR analysis showing cDNA levels for Pdk1, Ldha and Ldhb in WT and SIRT6 KO ESCs following siRNA-mediated knockdown of BRD4 (siBrd4) and/or PAF1 (siPAF1). Samples were normalized to actin levels. Error bar represents s.e.m (*: p < 0.05, **: p< 0.01, ***: p< 0.001, n = 3). (C) Western blots showing decreased levels of Pol II Ser2P, in SIRT6 KO ESCs following accute siRNA-dependent BRD4 knockdown (siBRD4). (D) Western blots showing chromatin levels of BRD4, PAF1, MYC, AFF4, ELL2, SIRT6, and histone H3 in WT and SIRT6 KO ESCs upon acute siRNA-mediated knockdown of BRD4 and/or PAF1.
The PAF1 complex (PAF1C) has been implicated both as a positive (Yu et al., 2015) and negative regulator (Chen et al., 2015) of Pol II pausing release into productive transcription elongation. More recently, comparative three-dimensional studies of paused Pol II and elongating Pol II complexes clearly demonstrated a positive role for PAF1C in facilitating transcription elongation (Vos et al., 2018a; Vos et al., 2018b). Consistent with the positive role of PAF1C in transcription elongation, we found that upregulation of SIRT6-targets can be rescued by knocking down one of its subunits, PAF1 (Figures 5A and 5B). Furthermore, the recruitment of BRD4 and MYC along with the SEC components AFF4 and ELL2 was severely decreased on chromatin from SIRT6 KO cells upon PAF1 knockdown (Figure 5D). Importantly, knockdown of BRD4 also decreased binding of PAF1, MYC and the SEC factors (Figure 5D), indicating that both BRD4 and PAF1 may be reinforcing a positive feedback to sustain high levels of these factors on chromatin.
Next, to fully demonstrate a role for PAF1C in the context of SIRT6 regulation, we performed ChIP-seq analysis with an antibody targeting LEO1, a subunit of PAF1C (Van Oss et al., 2017). Consistently, in cells lacking SIRT6, we observed increased binding of LEO1 on multiple SIRT6-target genes, including metabolism, ribosomal biogenesis, neural development and pluripotency (Figure 6A and S4D; Table S7). The PAF1 complex has also been implicated in facilitating the recruitment of P-TEFb to facilitate phosphorylation of Pol II (Ser2P) (Lu et al., 2016). Indeed, we found that in the absence of SIRT6, the Pol II fraction interacting with different components of the PAF1 complex is highly enriched for Pol II (Ser2P) (Figure 6B). Moreover, higher levels of LEO1 co-immunoprecipitated with CDK9 in SIRT6 KO compared to WT ESCs (Figure S4E). Additionally, we found augmented levels of the SEC component AFF4 to co-immunoprecipitate with LEO1 and another subunit of the PAF1 complex, CDC73, in SIRT6 KO ESCs (Figure 6B). These results are consistent with previous studies showing that SEC factors can modulate P-TEFb recruitment via interaction with PAF1C (Lu et al., 2016).
Figure 6. The PAF1 complex is a positive regulator of transcription elongation in ESCs lacking SIRT6.
(A) Increased recruitment of the PAF1C subunit LEO1 in SIRT6 KO ESCs at specific genes. IGV broswer images of LEO1 ChIP-seq in SIRT6 KO (red) versus WT ESCs (black) on genes implicated in metabolism (Ldhb, Ak4, Car2) and neural development (Igfbp2, Crmp1, Zic5). (B) Western blot analysis showing increased co-imunoprecipitation of PAF1C components PAF1, LEO1 and CDC73 with Pol II, Pol II Ser2P and the super elongation factor AFF4 in chromatin fractions from SIRT6 KO compared to WT ESCs. (C) Enrichment of SEC complex at intragenic regions of SIRT6 target genes. ChIP-qPCR analysis of super elongation factors ELL2 and AFF4 at various genomic regions of Ldhb and Glut1 genes, in WT (black) versus SIRT6 KO (red) ESCs. Schematic representation of the Ldhb and Glut1 genes and the location of the regions targeted for qPCR is shown below.
Lastly, by employing ChIP-qPCR, we found an enrichment of the SEC factors ELL2 and AFF4 in SIRT6 KO ESCs specifically at intragenic regions within the SIRT6-target genes Ldhb and Glut1 (Figure 6C). All together, these results indicate that in the absence of SIRT6, both PAF1C and SEC complexes are recruited to chromatin in a coordinated fashion to facilitate transcription elongation of SIRT6-target genes.
SIRT6-dependent histone deacetylation aligns with H3K36me3 and H3K79me2 patterns on genes involved in metabolism and development
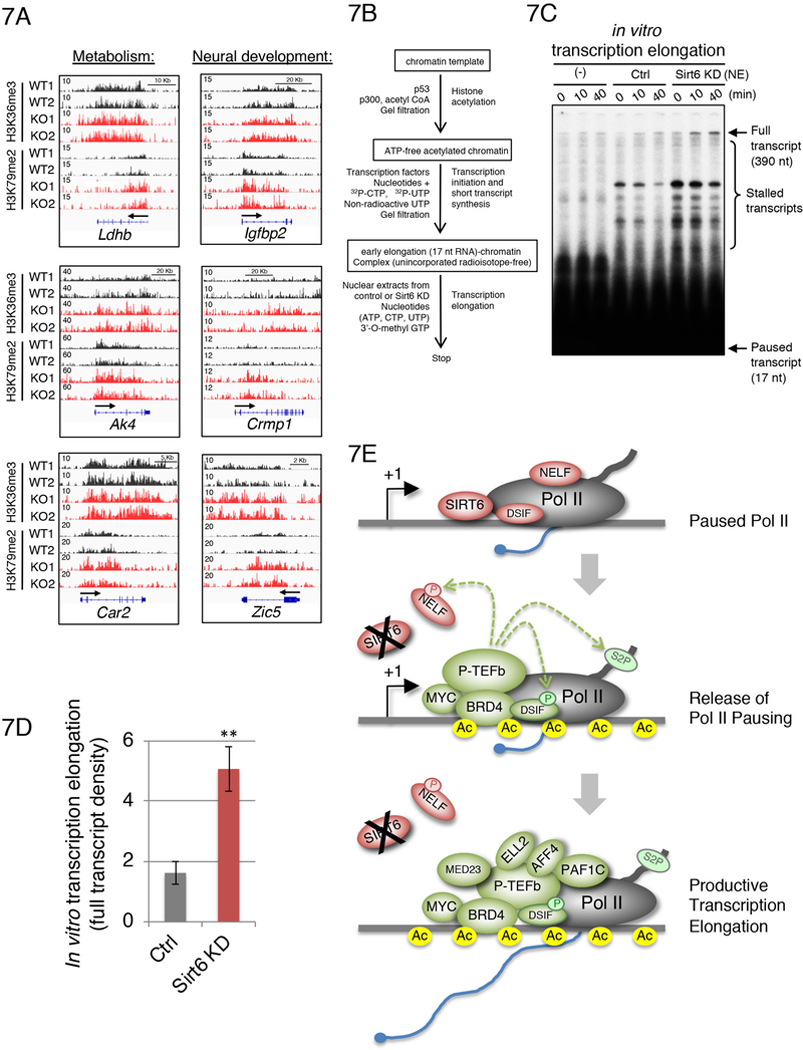
Methylation of H3 at lysine 36 (H3K36me3) and 79 (H3K79me2) has been previously associated with active transcriptional elongation (Guenther et al., 2007; Pokholok et al., 2005). We analyzed the genomic levels of these histone modifications in WT versus SIRT6 KO ESCs and found that many of the genes whose expression is upregulated in the absence of SIRT6 are enriched for H3K36me3 and/or H3K79me2 (Figure 7A; Table S2). Consistent with our previous studies (Sebastian et al., 2012b; Etchegaray et al., 2015), H3K36me3 and H3K79me2 are enriched at genes involved in metabolic pathways and neural development (Table S2). Although a modest number of genes are enriched for all these epigenetic marks (H3K9ac, H3K56ac, H3K36me3 and H3K79me2) (Figure S5A), a larger number of genes (~300 genes), whose expression is upregulated in SIRT6 KO ESCs, exhibit at least two of these epigenetic marks (Table S2). Plausibly, diverse combinations for these histone marks may be sufficient to trigger transcription elongation in the absence of SIRT6 (see Discussion below).
Figure 7. SIRT6-dependent chromatin dynamics regulates Pol II elongation.
(A) IGV broswer images showing the enrichment of H3K36me3 and H3K79me2 on metabolic (Ldhb, Ak4, Car2) and neural developmental (Igfbp2, Crmp1, Zic5) genes in WT (black) and SIRT6 KO (red) ESCs. (B) Scheme of the in vitro transcription elongation protocol using a chromatinized DNA template. (C) In vitro transcription elongation assay showing and augmentation of full elongation capacity in the absence of SIRT6. (D) Quantification of full elongation products from panel (C). (E) Model of SIRT6-dependent regulation of transcription elongation. Under normal nutrient conditions, SIRT6 forms a complex with Pol II at promoter-proximal regions where NELF is retained during transcriptional pausing. In this complex, SIRT6 maintains histone H3 in a deacetylated state on its targeted genes, thereby facilitating Pol II pausing. However, under poor nutrient conditions such as glucose starvation, SIRT6 dissociates from the chromatin, allowing acetylation of histone H3, which triggers the recruitment of BRD4, MYC. This facilitate the recruitment and activation of P-TEFb (containing CDK9), which causes the phosphorylation and chromatin eviction of NELF. Activated P-TEFb also phosphorylates the carboxyl terminal domain of Pol II at serine 2 (Ser2P), which triggers the release of transcriotional pausing. Subsequent recruitment of additional factors including PAF1C, MED23, along with super elongation factors AFF4 and ELL2 impel transcription into productive elongation mode.
SIRT6 depletion triggers increased transcriptional elongation in an in vitro assay
To further demonstrate that SIRT6 function as a regulator of Pol II pausing release, we used an in vitro transcription elongation assay containing a pre-assembled chromatinized DNA template reconstituted with purified transcription factors from HeLa cells, in the presence of p300/acetyl-CoA, as previously described (Kim et al., 2010). This in vitro elongation reaction was supplemented with nuclear extracts from SIRT6 knockdown versus control HeLa cells (Figure 7B). Strikingly, we observed a time-dependent increase of the fully elongated transcript (390 nucleotides) in the presence of the SIRT6-depleted nuclear extract, compared to control (Figures 7C and 7D). Comparable levels of the pre-initiated transcript (17 nucleotides) can be observed with a shorter exposure time (Figure S5B). This in vitro transcription elongation approach provides additional evidence that SIRT6 regulates transcription during the transition between pausing and productive elongation.
DISCUSSION
In this study we demonstrate that SIRT6 is a key regulator of Pol II promoter-proximal pausing. Under normal nutrient conditions, SIRT6 forms a complex with Pol II and maintains histone H3 in a deacetylated state to stabilize Pol II promoter-proximal pausing. Release of SIRT6 from chromatin results in augmented intragenic histone acetylation, thereby facilitating the recruitment of BRD4 and P-TEFb. Beyond increased recruitment of P-TEBb, we observed increased activity of the Cdk9 kinase. Notably, the inactive form of CDK9 within the P-TEFb complex, is maintained by its binding to HEXIM1 and the 7SK snRNP RNA (Guo and Price, 2013). However, co-IP experiments with CDK9 shows no difference in the levels of HEXIM1 interacting with CDK9 in WT and SIRT6 KO ESCs (Fig. S3G), suggesting that the increased activity of CDK9 observed in the KO cells is not dependent on HEXIM1 regulation, but rather may depend on the increased interaction with Pol II and the elongation promoting factors Myc, Brd4 and Med23 (Fig. 3I).
This in turn causes the phosphorylation and chromatin eviction of NELF-E, and the accumulation of Pol II (Ser2P), which facilitates the recruitment of elongation promoting factors including MYC, PAF1, MED23, and the SEC factors AFF4 and ELL2, together leading to the release of Pol II promoter-proximal pausing into productive transcription elongation (Figure 7E). Mechanistically, our results demonstrate that SIRT6-dependent histone deacetylation is critical for maintaining the chromatin landscape in a state that supports Pol II pausing at promoter-proximal regions and prevent productive elongation of target genes. A trigger to inactivate SIRT6 and promote elongation of specific gene sets may depend on cues originated from nutritional changes, developmental stages or proliferation signals.
Although years of research have demonstrated the requirement for histone acetylation in promoting transcription elongation in vivo and in vitro (Guermah et al., 2006; Kim et al., 2010; Pavri et al., 2006), the specific deacetylase(s) inhibiting transcription elongation at Pol II promoter-proximal pausing sites remained unknown. Our studies provide an in-depth mechanistic analysis of a histone deacetylase dedicated to controlling the release of paused Pol II into productive transcription elongation, a function that could confer cells with unique capabilities to fine-tune transcription in response to environmental cues. For instance, dynamic chromatin regulation of Pol II pausing can mediate faster and/or more synchronous modulation of gene expression in response to developmental and nutrient signals. Indeed, transcriptional synchronicity could ensure the orchestration of complex gene regulatory networks controlling mammalian embryogenesis, in accordance to earlier findings demonstrating fine-tuned orderly gene activation patterns in Drosophila embryos (Lagha et al., 2013; Boettiger and Levine, 2009). Overall, in the context of ESCs, we propose that this SIRT6-dependent chromatin structure/transcriptional mode may be context-specific. We consider this mechanism to have evolved in order to respond to cellular states and/or environmental cues, via the control of specific gene sets in a robust and synchronous manner. However, our data do not rule out the possibility that SIRT6 might also influence transcription initiation (we saw modest but consistent differences in histone acetylation and Pol II levels at TSS as well), a relevant question that will be addressed in future work. The involvement of other HDAC family members (class I and II) in transcriptional regulation has been shown to be predominantly at the level of gene promoters. For instance, the HDAC1-containing co-repressor complexes N-CoR and Sin3A limit the accessibility of transcription initiation factors to gene promoters (Nagy et al., 1997; Alland et al., 1997; Heinzel et al., 1997). The human NuRD co-repressor complex, containing both HDAC1 and HDAC2, represses transcription by limiting the chromatin access of basal transcription factors to promoter regions (Zhang et al., 1997; Tong et al., 1998; Xue et al., 1998).
The PAF1 complex has been proposed to function as an activator or repressor of transcription elongation depending on genetic/cellular contexts (Van Oss et al., 2017). PAF1 was also found at enhancer regions to restrain transcription on specific gene subsets (Chen et al., 2017). More recently, 3D studies demonstrated that PAF1C functions as a positive transcription elongation factor by displacing NELF from paused Pol II (Vos et al., 2018a; Vos et al., 2018b). Our findings support a role for PAF1C as an activator of transcription elongation, as originally determined in both mouse (Strikoudis et al., 2016) and human ESCs (Yu et al., 2015). Additionally, PAF1C is required for the maintenance of pluripotency in both mouse and human ESCs (Ding et al., 2009; Rigbolt et al., 2011; Ponnusamy et al., 2009). Finally, we demonstrated that levels of PAF1 in chromatin are increased in SIRT6 KO ESCs (Figure 5C), which we have previously showed to have an increased capacity to maintain expression of pluripotency genes (Etchegaray et al., 2015; Ferrer et al., 2018). Thereby, PAF1C might serve as part of a gatekeeper mechanism to uphold the pluripotency state in ESCs by promoting transcription elongation of pluripotency-related genes. In this context, we observed increased interaction of the PAF complex with members of the Super Elongation Complex (SEC)(Figure 6B) on chromatin, likely due to increase levels of these factors in the absence of SIRT6, further indicating enhanced elongation when SIRT6 is removed from chromatin. Of note, this function of PAF1C might differ upon differentiation, in other cell types or under conditions of disease.
The levels of H3K56ac were shown to correlate with transcription elongation in both yeast and Drosophila (Schneider et al., 2006). Thus, the role of H3K56ac as a positive modulator of transcriptional elongation seems to be evolutionary conserved. In addition, it was recently shown that H3K9ac is necessary for the release of paused Pol II by directly recruiting the super elongation complex (SEC) in HeLa cells (Gates et al., 2017). Removal of SIRT6 from chromatin increases acetylated levels of both H3K9 and H3K56, thereby explaining the enhanced recruitment of SEC components in SIRT6 KO ESCs. In addition, acetylation of histones serves as docking sites for the recruitment of BET bromodomain proteins, such as BRD4, which function as master transcription elongation factors (Winter et al., 2017). Importantly, we found that recruitment of MYC on chromatin is severely impaired upon BRD4 knockdown in the context of SIRT6 depletion, in line with a decreased expression of SIRT6 target genes (Figure 5A, 5B, 5D). This supports a model where BRD4 is responsible for recruiting MYC onto SIRT6-depleted chromatin to release Pol II pausing. BRD4 was shown to recruit the H3K36 methyltransferase NSD3 to increase gene expression (Rahman et al., 2011; Shen et al., 2015), while MYC interacts with DOT1L, the only known histone methyltransferase to catalyze methylation of H3K79, to induce transcription activation (Cho et al., 2015; Wong et al., 2017). Thus, the enhanced recruitment of BRD4 and MYC to the chromatin fraction that follows the increased levels of H3K9ac and/or H3K56ac upon SIRT6 depletion, may account for the enrichment of H3K36me3 and/or H3K79me2, thereby contributing to the up-regulated transcriptional elongation phenotype in SIRT6 KO ESCs. Given our previous studies defining SIRT6 as a co-repressor of MYC in development and cancer (Etchegaray et al., 2015; Kugel et al., 2015; Kugel et al., 2016; Sebastian et al., 2012b), our new results indicate that such modulation may depend on the ability of SIRT6 to fine-tune gene expression via transcriptional pausing/elongation to ensure proper cellular differentiation during embryonic development and to protect against malignant transformation.
STAR Methods
Contact for Reagents and Resources Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Raul Mostoslavsky (rmostoslavsky@mgh.harvard.edu).
Experimental Model and Subject details
Cell lines
SIRT6 KO and WT ESCs in 129 strain genetic background (Mostoslavsky et al., 2006) were maintained on γ-irradiated mouse embryonic fibroblasts (MEFs) in knockout DMEM medium (GIBCO) containing 15% ES-qualified FBS, 0.1 mM each of non-essential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 50 units ml−1 penicillin/streptomycin (Invitrogen) and supplemented with leukemia inhibiting factor (LIF). For all experiments described, cells were trypsinized and plated for 30 min on standard tissue culture dishes to remove feeder cells before floating ESCs were collected and re-plated on gelatin-coated dishes. For siRNA-mediated knockdowns of BRD4, MYC and/or PAF1 we transfected synthetic siRNA oligos using Lipofectamine 2000, according to the manufacturers protocol. The siRNA for BRD4 targets all mouse BRD4 isoforms (siGENOME SMARTpool from Dharmacon). Hence, BRD4 protein levels were almost undetectable by Western blot analysis. The expression of MYC and PAF1 was also knocked down using siGENOME SMARTpool siRNA oligos from Dharmacon. For MYC knockdown we used two siRNA systems, the oligo duplex (27-mers) from OriGene and siGENOME SMARTpool from Dharmacon. For glucose deprivation experiments, ESCs were cultured with glucose-free DMEM with 15% FBS 0.1 mM each of non-essential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 50 units ml−1 penicillin/streptomycin (Invitrogen) and supplemented with LIF for 2 days before harvesting cells for biochemical assays.
Method Details
Antibodies
The antibodies used for Western blot analysis are: anti-SIRT6 (Abcam, ab62739), anti-NELF-E (Santa Cruz, sc32912), anti-Pol II (Cell Signaling Tech., 2629), anti-Pol II S5P (Abcam, ab5131), anti-Pol II S2P (Millipore, 04–1571), anti-MYC (Abcam, ab32072), anti-BRD4 (Bethyl, 0301–985A50), anti-MED23 (BD Pharmingen, 550429), anti-PAF1(D9G9X) (Cell Signaling Tech., 12883S), anti-LEO1 (Bethyl, A300–175A), anti-CDK9 (Santa Cruz, sc-8338), anti-SPT5 (Bethyl., A300–868A), anti-PDK1 (Cell Signaling Tech., 3062S), anti-LDHA (Cell Signaling Tech., 2012S), anti-phospho-pan antibody (Invitrogen, 61–8300), anti-Tubulin (Abcam, ab6160), Rabbit plyclonal anti-AFF4 and anti-ELL2 antibodies (provided by Ali Shilatifard), anti-H3K9ac (Millipore, 07–352), anti-H3K56ac (Abcam, ab76307), anti-H3 (Abcam, ab1791) and anti-actin (Sigma-Aldrich, A2066). Protein-protein immunoprecipitations were performed as previously described (Lee et al., 2001). Antibodies targeting the super elongation factors AFF4 and ELL2 were provided by Ali Shilatifard.
The antibodies used for ChIP-RTqPCR, ChIP-seq and CUT&RUN analysis are: anti-MYC (Abcam, ab32072), anti-AFF4 and anti-ELL2 antibodies (provided by Ali Shilatifard), anti-Pol II (Cell Signaling Tech., 2629), anti-SIRT6 (Cell Signaling Tech., 12486S), anti-H3K9ac (Millipore 07–352), anti-H3K56ac (Abcam, ab76307), anti-H3K79me2 (Cell Signaling Tech., 5472), anti-H3K36me3 (Cell Signaling Tech., 4909), anti-LEO1 (Bethyl, A300–175A) and anti-NELF-E (Abcam, ab170104).
Immunoblotting
Immunoblots were developed with HRP conjugated goat anti-rabbit (Sigma-Aldrich, AQ132P), goat anti-mouse (Sigma-Aldrich, A3682) or goat anti-rat (Santa Cruz Biotech., sc-2006) secondary antibodies and ECL Plus (GE healthcare Lifescience, RPN2133).
Chromatin extraction and Western blot analysis
For chromatin extractions, cell pellets were resuspended in lysis buffer (10mM HEPES pH 7.4, 10 mM KCl, 0.05% NP-40) supplemented with a protease inhibitor cocktail (Complete EDTA-free, Roche Applied Science), 5μM TSA, 5mM sodium butyrate, 1mM DTT, 1mM PMSF, 50mM NaF, 0.2mM sodium orthovanadate and phosphatase inhibitors (Phosphatase Inhibitor Cocktail Sets I and II, Calbiochem) and incubated on ice for 20 min. The lysate was then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was removed (cytosolic fraction) and the pellet (nuclei) was acid-extracted using 0.2N HCl and incubated on ice for 20 min. The lysate was then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant (contains acid soluble proteins) was neutralized using 1M Tris-HCl pH 8. Protein concentration was measured by Biorad Protein Assay. Western blot analyses were performed as previously described (Zhong et al., 2010b). Briefly, 10 ug of protein was loaded on a 10–20% gradient polyacrylamide gel with SDS (Biorad) and electroblotted onto polyvinylidene difluoride membranes (PVDF) (Millipore). Membranes were blocked in TBS with 5% non-fat milk and 0.1% Tween and probed with antibodies. Bound proteins were detected with horseradish-peroxidase-conjugated secondary antibodies (Vector Biolaboratories) and SuperSignal West Pico Luminol/Enhancer Solution (Thermo Scientific).
CDK9 Kinase Assay
Cells were collected in cold PBS and lysed in cold EBC buffer (50 mM Tris-HCl pH 8.0, NaCl 120 mM, 0.5% NP-40, 5 mM DTT). After pre-clearing with protein-A beads for 2 hours at 4°C, the lysates were incubated with either 3 ug CDK9 antibody (Santa Cruz sc-8338x) or equal amount of IgG antibody control (Sigma-Aldrich, 12–370) under constant rotation at 4°C overnight. The beads were then washed 3 times with EBC + 0.05% SDS, then once with TKB/Mg buffer (50 mM HEPES pH 7.5, 10 mM MgCl2, 5 mM DTT). The beads were resuspend in 25 ul of TKB/Mg buffer plus 2.5 mM MnCl2, 5 uM ATP, 200ng GST-CTD (Abcam ab81888), and 5 uCi Y−32P-ATP and incubated at room temperature for 40 minutes with or without 50 uM of the CDK9 inhibitor DRB (5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside) (Sigma-Aldrich, D1916). The reactions were stopped by adding 18 ul of 4x SDS loading buffer and boil the sample 10 minutes at 95°C. Samples were loaded in a polyacrylamide gel, which was then fixed with 40% methanol + 10% acetic acid for 30 min. Gels were washed three times for 5 min with Milli-Q H2O and stained with BioRad Coomassie (161–0786) for 1 hour at room temperature. Additional washes with ddH2O were done to clear the background. Finally, the gel was dried under a vacuum system, exposed and developed.
Protein-protein Immunoprecipitations
Protein-protein immunoprecipitations were performed as previously described (Lee et al., 2001). Briefly, whole cell lysates or chromatin fractions were subjected to immunoprecipitation using magnetic Dynabeads Protein-A (Invitrogen, 10006D) or Protein-G (Invitrogen, 10007D) immunoprecipitation kits. Prior immunoprecipitation, magnetic beads were conjugated with specific primary antibodies (see list of antibodies above). Samples were incubated at 4°C with constant rotation overnight in IP buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 5 mM NaF, 1 mM DTT, 0.05% Triton X-100, 5% glycerol, 0.25 mM PMSF plus protease inhibitors cocktail tablets [Sigma-Aldrich, 4693116001]). Magnetic dynabeads containing immunoprecipitated samples were then washed twice with IP buffer and 3 times with washing buffer supplied in the Dynabead kit.
RNA extraction and quantitative PCR with reverse transcription
Total RNA was extracted with the TriPure Isolation Reagent (Roche) as described by the manufacturer. For cDNA synthesis, 1 mg of total RNA was retro-transcribed by using the QuantiTect Reverse Transcription Kit (Qiagen). Real-time PCR was performed using the SYBR green master mix (Roche), following the manufacturer’s instructions, with the exception that the final volume was 12.5 ml of SYBR green reaction mix. Real-time monitoring of PCR amplification was performed using the LightCycler 480 detection system (Roche). Data were expressed as relative mRNA levels normalized to the β-actin expression level in each sample. The primer sequences can be obtained on request. For RNA-Seq, total RNA purified with TriPure was cleaned up with RNeasy kits from Qiagene.
In vitro transcription elongation assay
The in vitro transcription elongation assay was performed and analyzed as described earlier (Kim et al., 2010). Briefly, after the second gel filtration, approximately 35 ng plasmid DNA-containing chromatin template was mixed with nuclear extract containing total 5 μg protein which were prepared from control or SIRT6 knockdown HeLa cells. The extracts were adjusted to TX-buffer condition prior to the addition. The reactions were 50ul in this step and incubated for 30 min at 30°C. The reactions were then added with nucleotides and RNasin in total 5 ul, and also adjusted to TX-buffer condition, which bring the final volume to 55ul, to allow the transcription elongation. At the indicated time point, the reactions were stopped and transcription products were resolved by acrylamide gel electrophoresis and visualized by X-ray film.
Permanganate footprinting
Ligation Mediated PCR (LM-PCR) based permanganate footprinting was done as described (Samarakkody et al., 2015). In brief, cells were washed with 1xPBS and treated with 20mM KMn04 for 60s on ice. Reactions were quenched with Stop buffer (1% SDS, 10mM Tris HCl pH 8.0, 20mM EDTA, 0.5 M 2-mercaptoethanol) and, after treatment with RNAse cocktail (Ambion), Proteinase K and extraction with phenol-chloroform, DNA was ethanol-precipitated and resuspended in water at approximately 100 ng/μl. Up to 5 μg of DNA was treated with 10% (v/v) piperidine in the final volume 100 μl, for 10 minutes at 90°C, after which DNA was extracted with chloroform twice, ethanol precipitated in the presence of 10 μg GlycoBlue carrier, and resuspended in water at approximately 50 ng/μl. Previously purified genomic DNA (naked DNA) was treated with permanganate and piperidine as above to establish background DNA reactivity. Template for the A+G sequencing ladder was generated by treating purified DNA with formic acid (Maxam and Gilbert, 1977) followed by piperidine treatment as above. Approximately 200 ng of piperidine-treated DNA was taken for each Ligation-mediated (LM) PCR reaction. LM PCR began with primer extension with Primer A with Phusion DNA polymerase followed by the addition of universal double-stranded linker (Samarakkody et al., 2015) and ligation with T4 DNA ligase. After ethanol precipitation, the ligated template was used for 22 cycles of PCR with gene-specific Primer B and Linker-specific primers, after which 5’−32P-labeled primer was added and PCR continued for another 2 cycles. After phenol extraction and ethanol precipitation, DNA was resuspended in 5 μl of 7M urea-1xTBE loading buffer and resolved on denaturing polyacrylamide sequencing gel followed by radioautography. Footprints were quantified using ImageJ by subtracting the signal from naked DNA from reactivity of cells within the pausing region. Primer sequences can be provided upon request.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (Gomes et al., 2006). Briefly, ESCs were crosslinked with 1% formaldehyde/PBS for 15 min at room temperature. Crosslinking was quenched by addition of 0.125 M glycine. ESCs were washes twice with ice-cold PBS, and then collected in RIPA buffer as described previously. Samples were then sonicated to generate DNA fragments of approximately 0.5 kb. Approximately, 1 μg of pre-cleared protein extract was used for immunoprecipitation with the following specific antibodies: anti-Pol II (Cell Signaling Tech., 2629), anti-SIRT6 (Cell Signaling Tech., 12486S), anti-NELF-E (Abcam), anti-MYC (Abcam, ab32072), anti- LEO1 (Bethyl, A300–175A). Antibodies targeting the super elongation factors AFF4 and ELL2 were provided by Ali Shilatifard. Immunoprecipitation was performed for approximately 12 hours at 4 °C using protein A/G plus agarose beads (Santa Cruz, sc2003). Bead-containing samples were then washed as previously described (Gomes et al., 2006). Immunocomplexes were eluted by incubation at 65 °C for 10 min in the presence of 1% SDS, and crosslinking was reversed by 6 h incubation at 65 °C in the presence of 200 mM NaCl. DNA was purified by the QIAquick spin kit (QIAGEN) and further assessed by qPCR using the LightCycler 480 system from Roche. Data were normalized to input and expressed relative to the nonspecific IgG or IgM ChIP controls. Primer sequences can be obtained on request.
ChIP-seq
Chromatin samples were prepared as previously described (Mishra et al., 2018). In brief, ESCs grown on 15 cm dish were crosslinked with 1% formaldehyde for 13 min at 37°C. Crosslinking was quenched with 0.125M glycine for 5 min at 37°C. Collected cell pellets from each 15 cm dish were resuspended in 2.5 ml lysis buffer supplemented with protease inhibitors (5 mM PIPES pH8, 85 mM KCL, 0.5% NP40) and incubated 5 min at 4°C. Chromatin was obtained by centrifugation at 800 rpm for 5 min at 4°C, where nuclear fraction from approximately 10 to 12 million cells were resuspended in 300 μl of nuclear lysis buffer (50 mM Tris-HCL pH8, 10 mM EDTA pH8, 0.2% SDS). Chromatin was then sonicated at 70% amplitude 15 sec ON and 45 sec OFF for 45 min. About 5 μl of sonicated chromatin was reverse-crosslinked with 1 μl of proteinase K in elution buffer (50 mM NaHCO3, 140 mM NaCl, 1% SDS) at 65°C overnight. After RNase treatment, DNA was isolated by phenol chloroform extraction and analyzed on 1% agarose gel electrophoresis. Only samples with smear below 300 bp were used for ChIP-seq analysis. Approximately 10 to 30 μg of pre-cleared chromatin was used per each immunoprecipitation with magnetic beads conjugated with ~2 μg of antibody in IP buffer (16.7 mM Tris HCL pH8, 1.2 mM EDTA pH8, 167 mM NaCl, 0.01% SDS, 1.1% Triton x-100) at 4°C overnight. The immunoprecipitated DNA was washed twice with IP buffer, once with TSE buffer (20 mM Tris HCL pH8, 2 mM EDTA pH8, 500 mM NaCl, 1% Triton x-100, 0.1% SDS), once with LiCl buffer ( 100 mM Tris HCL pH8, 500 mM LiCl, 1% deoxycholic acid, 1% NP40) and twice with TE-buffer (10 mM Tris-HCL pH8, 1 mM EDTA pH8) before elution by incubation in elution buffer (50 mM NaHCO3, 140 mM NaCl, 1% SDS) supplemented with 10 μg of proteinase K at 55°C for 1 hour. The samples were removed from the magnetic beads and reverse-crosslinked at 65°C for 4 hrs. Immunoprecipitated DNA was purified using PCR purification columns from QIAGEN. Construction of the DNA libraries for DIP sequencing were performed with Illumina reagents and sequencer equipment.
Chromatin immunoprecipitations were performed with the following antibodies: from Cell Signaling Technology we used anti-Pol II(Rpb1 CTD, 4H8) (Cat# 2629), anti-SIRT6(D8D12) (Cat# 12486S), anti-H3K79me2(D15E8) (Cat# 5472), anti-H3K36me3(D5A7) (Cat# 4909); Anti-H3K9ac (Millipore 07–352), anti-H3K56ac (abcam ab76307), and anti-LEO1 (Bethyl A300–175A).
ChIP-seq Heatmap Creation
Reads from RNA Pol II, SIRT6, and HDACI ChIP-seq for both WT and SIRT6 KO ESCs were aligned to mouse genome mm9 using bwa and duplicate reads were marked with Picard tools (http://picard.sourceforge.net). The heatmaps of Figure 2A were produced by calculating the mean coverage values of 100 b windows in the +/−3kb region centered on the TSS for all genes using the ChIP-seq data for SIRT6, Pol II, and HDACI in ESC cells. The heatmaps represent the mean enrichment by bin after correcting for the direction of the DNA strand. For the heatmap after filtering out genes with insufficient signal, coverage values for the genes z-score normalized and then ordered by hierarchical clustering using Euclidean distance and Ward method of the R heatmap.2 function. The list includes genes that passed the cutoffs for at least one of Pol II, SIRT6, and HDAC, and only the top 400 genes for each sample are displayed. To assess the enrichment of ChIP-seq signal across gene body regions (Fig. 2E), we calculated the ratio of ChIP to input sequencing tag number in the region of the gene body excluding the immediate 3 Kb proximity of TSS. These enrichment values for the gene set of interest were compared between Sirt6 KO and wild type using t-test.
CUT&RUN
These experiments were performed as described by Skene and Henikoff (PMID: 28079019) with modifications. Briefly, mouse ESCs (~1 million) were fixed with 1% paraformaldehyde (PFA) and quenched with 125 mM glycine. After centrifugation at ~400xg, cell pellets were washed three times with PBS and twice with HEPES buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5% BSA, 0.1% Tween 20). Activated concanavalin A-coated magnetic beads (Bangs Laboratories) were added to capture the cells, which were incubated at 4°C overnight with antibodies targeting H3K27me3 (cell Signaling Cat# 9733), H3K9ac (Millipore Cat# 07–352), NELF-E (Abcam Cat# ab170104), IgG control (Antibody-online Cat# ABIN101961). IgG and H3K27me3 antibodies served as negative and positive controls, respectively. Immunoprecipitated samples were supplemented with protein A-MNase (700 ng/ml) and incubated for 1 hour at 4°C and placed on ice. Protein A-MNase was activated by incubation with 2 mM CaCl2 on ice for 30 min. This reaction was terminated with stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.1% Tween 20, 50 ug/ml RNaseA, 50 ug/ml glycogene, 0.05 pg/ml heterologous yeast spike-in DNA). CUT&RUN fragments were released by incubation at 37°C for 10 min followed by centrifugation. Immunoprecipitated DNA was purified by QIAquick Gel extraction kit (QIAGEN Cat# 28706). Library preparations were done based on Skene and Henikoff method (PMID: 28079019) followed by Tapestation analysis to assess the quality of DNA libraries.
PRO-seq library preparation and data analysis
Wild-type mouse ESCs of (WT), or SIRT6-KO (KO) genotypes, or WT cells that were glucose-starved (No Glucose) for 48hr. were permeabilized at 4°C or on ice unless otherwise specified. Briefly, cells were washed once in ice-cold 1x PBS and resuspended in Buffer W (10 mM Tris-HCl pH 8.0, 10% glycerol, 250 mM sucrose, 10 mM KCl, 5 mM MgCl2, 0.5 mM DTT, protease inhibitors cocktail (Roche), and 4 u/mL RNase inhibitor [SUPERaseIN, Ambion]) at the cell density of 2 × 107 cells/mL. 9x volume of Buffer P (10 mM Tris-HCl pH 8.0, 10% glycerol, 250 mM sucrose, 10 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.1% Igepal, protease inhibitors cocktail (Roche), 4 u/mL RNase inhibitor [SUPERaseIN, Ambion]) was then immediately added. Cells were gently resuspended and incubated for up to 2 min on ice. Cells were then recovered by centrifugation (800 × g for 4 min) and washed in Buffer F (50 mM Tris-HCl pH 8.0, 40% glycerol, 5 mM MgCl2, 0.5 mM DTT, 4 u/mL RNase inhibitor [SUPERaseIN, Ambion]). Washed permeabilized cells were finally resuspended in Buffer F at a density of 1×106 cells/30 μL and immediately frozen in liquid nitrogen. Permeabilized cells were stored in −80°C until usage.
PRO-seq run-on reactions were carried out as follows: 1 × 106 permeabilized cells spiked with 5 × 104 permeabilized Drosophila S2 cells were added to the same volume of 2x Nuclear Run-On reaction mixture (10 mM Tris-HCl pH 8.0, 300 mM KCl, 1% Sarkosyl, 5 mM MgCl2, 1 mM DTT, 200 μM biotin-11-A/C/G/UTP (Perkin-Elmer), 0.8 u/μL SUPERaseIN inhibitor [Ambion]) and incubated for 5 min at 30°C. Nascent RNA was extracted using a Total RNA Purification Kit following the manufacturer’s instructions (Norgen Biotek Corp.). Extracted nascent RNA was fragmented by base hydrolysis in 0.25 N NaOH on ice for 10 min and neutralized by adding 1x volume of 1 M Tris-HCl pH 6.8. Fragmented nascent RNA was bound to 30 μL of Streptavidin M-280 magnetic beads (Thermo Fisher Scientific) in Binding Buffer (300 mM NaCl, 10 mM Tris-HCl pH 7.4, 0.1% Triton X-100). The beads were washed twice in High salt buffer (2 M NaCl, 50 mM Tris- HCl pH 7.4, 0.5% Triton X-100), twice in Binding buffer, and twice in Low salt buffer (5 mM Tris-HCl pH 7.4, 0.1% Triton X-100). Bound RNA was extracted from the beads using Trizol (Invitrogen) followed by ethanol precipitation.
For the first ligation reaction, fragmented nascent RNA was dissolved in H2O and incubated with 10 pmol of reverse 3’ RNA adaptor (5’p-rNrNrNrNrNrNrGrArUrCrGrUrCrGrGrArCrUrGrUrArGrArArCrUrCrUrGrArArC-/3’InvdT/) and T4 RNA ligase I (NEB) under manufacturer’s conditions for 2 h at 20°C. Ligated RNA was enriched with biotin-labeled products by another round of Streptavidin bead binding and washing (two washes each of High, Binding and Low salt buffers and one wash of 1x Thermo Pol Buffer (NEB)). To decap 5’ ends, the RNA products were treated with RNA 5’ Pyrophosphohydrolase (RppH, NEB) at 37°C for 30 min followed by one wash of High, Low and T4 PNK Buffer. To repair 5’ ends, the RNA products were treated with Polynucleotide Kinase (PNK, NEB) at 37°C for 30 min.
5’ repaired RNA was ligated to reverse 5’ RNA adaptor (5’-rCrCrUrUrGrGrCrArCrCrCrGrArGrArArUrUrCrCrA-3’) with T4 RNA ligase I (NEB) under manufacturer’s conditions for 2 h at 20°C. Adaptor ligated nascent RNA was enriched with biotin-labeled products by another round of Streptavidin bead binding and washing (two washes each of High, Binding and Low salt buffers and one wash of 1x SuperScript IV Buffer [Thermo Fisher Scientific]), and reverse transcribed using 25 pmol RT primer (5’- AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA-3’) for TRU-seq barcodes (RP1 primer, Illumina). A portion of the RT product was removed and used for trial amplifications to determine the optimal number of PCR cycles. For the final amplification, 12.5 pmol of RPI-index primers (for TRU-seq barcodes, Illumina) was added to the RT product with Phusion polymerase (NEB) under standard PCR conditions. Excess RT primer served as one primer of the pair used for the PCR. The product was amplified 12~14 cycles and beads size selected (ProNex Purification System, Promega) before being sequenced in NextSeq 500 machines in a mid-output 150-bp cycle run.
PRO-seq libraries from 3 independent biological replicates (WT and KO) or 2 biological replicates (NG) were generated. Paired-end reads were trimmed to 40 nt, for adapter sequence and low quality 3’ ends using cutadapt 1.14, discarding those containing reads shorter than 20 nt (-m 20 -q 10), and removing a single nucleotide from the 3’ end of all trimmed reads to allow successful alignment with Bowtie 1.2.2 (Langmead, Trapnell et al, Genome Biol. 10, R25, 2009). Remaining pairs were paired-end aligned to the dm3 genome index to determine spike-normalization ratios based on uniquely mapped reads. Mappable pairs were excluded from further analysis, and unmapped pairs were aligned to the mm9 genome assembly. Identical parameters were utilized in each alignment described above: up to 2 mismatches, maximum fragment length of 1000 nt, and uniquely mappable, and unmappable pairs routed to separate output files (-m1, -v2, -X1000, --un). Pairs mapping uniquely to mm9, representing biotin-labeled RNA 3’ ends, were separated, and strand-specific counts of the 3’ mapping positions determined at single nucleotide resolution, genome-wide, and expressed in bedGraph format with “plus” and “minus” strand labels swapped for each 3’ bedGraph, to correct for the “forward/reverse” nature of Illumina paired-end sequencing (Mahat et. al., 2016). Counts of pairs mapping uniquely to spike-in RNAs (drosophila genome) were determined for each sample. Uniquely mappable reads were determined, and a normalization factor calculated. In this case, the spike in reads exhibited no systematic differences across samples, thus normalization based on sequencing depth was used for each bedGraph. PRO-seq size factors used were: for WT 1.70255924, 3.35361899, 1.04422773; KO: 2.19052323, 3.46076838, 1.00000000; and NG: 3.22211594, 3.16824971. To account for the different number of replicates used in No Glucose condition (N=2) the WT and KO data sets (each N=3) were downsampled by 2/3 to facilitate direct comparisons. Combined bedGraphs were generated by summing counts per nucleotide of all replicates for each condition; and UCSC Genome Browser tracks displaying mean read coverage were generated from the combined replicates per condition, normalized as described above.
| Sample | Total reads | Uniquely mapped reads (Percentage of total) |
Agreement between replicates (Spearman’s rho) |
|---|---|---|---|
| WT (Wild-type) | 331,008,473 | 36.68% | >0.97 |
| KO (SIRT6-KO) | 340,699,369 | 33.10% | >0.97 |
| NG (No-Glucose) | 250,932,240 | 42.53% | 0.99 |
Pausing Indices and composite analyses of PRO-seq signals
TSSs were designated as bound by Pol II if there were a total of 16 reads in WT bedGraphs (without normalization) in the region ±150 bp from the annotated TSS. 12,941 of 17,032 TSSs (76.0%) were classified as Pol II-bound using this cutoff, of which 3,659 are upregulated genes in RNA-seq upon SIRT6-KO. Composite metagene distributions were generated by summing sequencing reads at each indicated position with respect to the TSS and dividing by the number of TSSs included within each group. These were plotted across a range of distances. Zoomed-in PRO-seq profiles (e.g. Figures 2G–2I) were smoothed by calculating a moving average across adjacent 5 bins.
Quantification and Statistical Analysis
Gene expression, transcription factor binding and in vitro transcription analysis
Standard 2-sample t-test assuming equal variance was used to calculate gene expression levels, transcription factor binding and elongation products, from RT-qPCR, ChIP-qPCR and in vitro transcription elongation experiments, respectively. The “n” values represent a minimum of 3 biological replicas, and the error bars correspond to s.e.m. calculated using analysis of variance (ANOVA).
ChIP-seq analysis
Sequencing reads were aligned against the mm9 reference genome using BWA (Li and Durbin, 2010). Alignments were filtered for uniquely mapped reads and duplicates were removed. Input-normalized coverage tracks were generated using SPP; enrichment peaks were called using SPP as broad regions of enrichment with FDR cutoff of 0.01 (Kharchenko et al., 2008). Regions of differential tag enrichment were determined based on tag counts in all identified peak regions using Diffbind (Ross-Innes et al., 2012) with FDR cutoff 0.1. These regions of differential enrichment between SIRT6 KO and WT ESCs were then analyzed for the proximity (within 20 Kb) to the differentially expressed genes detected in RNA-seq analysis. Reads from H3K9Ac, H3K56Ac, H3K36me3, H3K79me2, Pol II, and LEO1 ChIP-seq for WT and SIRT6 KO ESCs, and H3 control for mouse ESCs were aligned to mouse genome mm9 using bwa and duplicate reads were marked with Picard tools (http://picard.sourceforge.net). Peaks were called using MACS2 version 2.1 with the False Discovery Rates (FDR) q=0.01 with the ‘—broad’ flag. A subset of called peaks were checked visually with the IGV browser. The Bioconductor package DiffBind was used to find differentially bound ChIP-seq peaks for all the histone marks and factors using H3 ChIP-seq as an input control. Differential binding statistics in DiffBind were calculated with DESeq2 with an FDR threshold of 0.1. Differentially bound peaks with FDR<0.1 were assigned to genes if they fell within +/− 20kb of the gene body using Bioconductor ‘TxDb.Mmusculus.UCSC.mm9.knownGene’ annotation. Gene Ontology (GO) categories were assigned to genes using the functional annotation table method in DAVID functional annotation tool version 6.8 (david.ncifcrf.gov). p values were obtained using standard 2-sample t-test assuming equal variance. Pearson correlations between replicates were calculated by covering over 10Kb windows across the genome for all the different ChIP-seqs (see Figure S6).
Pausing Index calculated from Pol II ChIP-seq analysis
Reads from RNA Pol II ChIP-seq for WT and SIRT6 KO ESCs were aligned to mouse genome mm9 using bwa and duplicate reads were marked with Picard tools (http://picard.sourceforge.net). Pausing Indexes (PI) were calculated as the Pol II ChIP-seq density ratios between the promoter and the gene body regions in genes with significant Pol II signal in the promoter region (FDR<1e-3 from SISSRs) generally following the methods used in (Nechaev et al., 2010) and (Henriques and Adelman, 2013). The promoter was defined as the TSS +/− 150bp while the gene body was defined as +250bp to +2250bp (or the gene end if the gene is less than 2250bp). Figure 2E shows the fraction of RNA Pol II bound genes with a PI greater than or equal to the value of the PI value on the x-axis for WT and SIRT6 KO. Figure 2E shows average pausing for two replicates each of WT and SIRT6 KO ESC ChIP-seq data for RNA Pol II for genes that were up regulated in SIRT6 KO RNA-Seq samples with DESeq2 adjusted p-value<0.1 (4938 genes). There were 2459 unique genes that had greater than 2-fold decrease in pausing index for SIRT6 KO of the 4938 genes that were up regulated in SIRT6 KO ESCs (Tables S1, S3).
RNA-Seq data analysis
Read counts were calculated per gene, in a strand-specific manner, based upon alignments to the UCSC mouse mm9 transcriptome using tophat2 version 2.0.10 and htseq version 0.6.1, and finally differentially expressed genes were found using the Bioconductor package DESeq2. Differentially expressed genes were defined using an adjusted p-value threshold of <0.1. Of 23,366 genes, 4938 were identified as differentially expressed upon SIRT6-KO in mESC cells. p-values were obtained using standard 2-sample t-test assuming equal variance.
CUT&RUN data analysis
CUT&RUN sequenced reads were aligned to mouse mm9 and yeast S288C reference genome using BWA. Mouse alignments were filtered for uniquely mapped reads and duplicates were removed, then mouse profiles were normalized to the number of aligned heterologous yeast spike-in genomic DNA reads. Metagene profiles were generated using deeptools [PMID: 27079975].
Pausing Index calculated from PRO-seq analysis
Genomic statistical tests: Statistical significance for comparisons of promoter (±150 nt from TSS) and gene body (+250 to +1250 nt from TSS) between WT and KO or NG conditions was assessed by Mann-Whitney test. Statistical details and error bars are defined in each figure legend. Spearman correlations were calculated by using a +/− 250bp window around promoters (N=17032) of annotated genes for each of the replicates in WT, WT No Glucose or SIRT6-KO conditions (see Figure S7).
Data and Software availability
GEO Accession numbers
All the raw datasets for the different sequencing experiments have been deposited in NCBI under the following GEO accession numbers: GSE130689, GSE130690, GSE130691, and GSE130692.
Supplementary Material
RNA-seq analysis showing up-regulated genes in SIRT6 KO compared to WT ES cells, related to Figures 1-2.
First page: Chip-Seq analysis showing enrichment of H3K9ac and H3K56ac in SIRT6 KO ESCs, related to Figure 1. Second page: Comparative Analysis for the enrichment of H3K9Ac, H3K56Ac, H3K36me3 and H3K79me2 on up-regulated genes in SIRT6 KO ESCs, as determined by ChIP-seq, related to Figure 7.
List of genes with lower Pausing Index in SIRT6 KO ES cells based on Pol II ChIP-seq analysis, related to Figure 2.
List of genes with lower Pausing Index in SIRT6 KO ES cells and WT No Glucose cells (compared to WT cells) based on Pro-seq analysis, related to Figure 2.
Lower Pausing Index in SIRT6 KO compared to WT ESCs based on both Pol II ChIP-seq and PRO-seq analyses, related to Figure 2.
NELF-E binding in WT and SIRT6 KO ES cells, as determined by CUT&RUN, related to Figure 3.
Genes targeted for LEO1 in SIRT6 KO ES cells, as determined by ChIP-seq, related to Figure 6.
ACKNOWLEDGEMENTS
We would like to thank all the members of the Mostoslavsky lab for helpful discussions. R.M. is the Laurel Schwartz Endowed Chair in Oncology and supported by the Kristin and Bob Higgins Massachusetts General Hospital Research Scholar Award. Antibodies against the SEC complex were kindly provided by Ali Shilatifard. Protein A-MNase and heterologous yeast spike-in DNA for CUT&RUN experiments were kindly provided by Steven Henikoff. This work is supported by NIH grants (R01GM128448 and R01CA175727) to R.M, (R01GM097360) to J.R.W, R01CA129325 and R01DK071900 to R.G.R and the National Science Foundation Graduate Research Fellowship Grant DGE1745303 (to E.F.A.). We would also like to thank the HMS Nascent Transcriptomics Core for support of T.H and K.A.
Footnotes
Declaration of Interests: Dr. Mostoslavsky has a financial interest in Galilei Biosciences Inc., a company developing activators of the mammalian SIRT6 protein. Dr. Mostoslavsky’s interests were reviewed and are managed by MGH and Partners HealthCare in accordance with their conflict of interest policies.
REFERENCES
- Adelman K, and Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13, 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alland L, Muhle R, Hou H Jr., Potes J, Chin L, Schreiber-Agus N, and DePinho RA (1997). Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49–55. [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, and Greenleaf AL (2010). CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24, 2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, and Verdin E (2007). Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A 104, 13690–13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, and Levine M (2009). Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, Shiekhattar R, and Shilatifard A (2015). PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 162, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Xie P, Collings CK, Cao K, Aoi Y, Marshall SA, Rendleman EJ, Ugarenko M, Ozark PA, Zhang A, et al. (2017). PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 357, 1294–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KL, Zhang F, Jaganathan A, Sharma R, Zhang Q, Konuma T, Shen T, Lee JY, Ren C, Chen CH, et al. (2017). Distinct Roles of Brd2 and Brd4 in Potentiating the Transcriptional Program for Th17 Cell Differentiation. Mol Cell 65, 1068–1080 e1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ, Lee CH, Oh SH, Song YS, Kim HS, et al. (2015). DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun 6, 7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JE, and Mostoslavsky R (2014). Sirtuins, metabolism, and DNA repair. Curr Opin Genet Dev 26, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fontanals-Cirera B, Low V, Ntziachristos P, Yuen SK, Lovell CD, Dolgalev I, Yonekubo Y, Zhang G, Rusinova E, et al. (2014). Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep 9, 234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. (2009). A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4, 403–415. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A, et al. (2015). The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, and Mostoslavsky R (2016). Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell 62, 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer CM, Alders M, Postma AV, Park S, Klein MA, Cetinbas M, Pajkrt E, Glas A, van Koningsbruggen S, Christoffels VM, et al. (2018). An inactivating mutation in the histone deacetylase SIRT6 causes human perinatal lethality. Genes Dev 32, 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, and Peterlin BM (2004). Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B, and Zeitlinger J (2014). RNA polymerase II pausing during development. Development 141, 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, Sagum CA, Jung SY, Qin J, Tsai MJ, et al. (2017). Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem 292, 14456–14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, and Espinosa JM (2006). Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, and Young RA (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, and Roeder RG (2006). Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125, 275–286. [DOI] [PubMed] [Google Scholar]
- Guo J, and Price DH (2013). RNA polymerase II transcription elongation control. Chem Rev 113, 8583–8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, and Zhou Q (2010). HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38, 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brand G, Ngo SD, Davie JR, et al. (1997). A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Henriques T, and Adelman K (2013). Catching the waves: following the leading edge of elongating RNA polymerase II. Mol Cell 50, 159–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ, Kitzing T, Zhu N, de Stanchina E, Sawyers CL, et al. (2014). CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev 28, 1800–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, and Wolffe AP (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Jonkers I, and Lis JT (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, and Cohen HY (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, et al. (2014). BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 21, 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TLA, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KCL, Boxer LD, Chang HY, et al. (2009). SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappaB-Dependent Gene Expression and Organismal Life Span. Cell 136, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, and Park PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, and Roeder RG (2010). The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel S, Feldman JL, Klein MA, Silberman DM, Sebastian C, Mermel C, Dobersch S, Clark AR, Getz G, Denu JM, et al. (2015). Identification of and Molecular Basis for SIRT6 Loss-of-Function Point Mutations in Cancer. Cell Rep 13, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel S, and Mostoslavsky R (2014). Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci 39, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al. (2016). SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell 165, 1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, and Lis JT (2013). Control of transcriptional elongation. Annu Rev Genet 47, 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. (2013). Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, and Reppert SM (2001). Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867. [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu M, Chen LF, and Chen R (2018). P-TEFb: Finding its ways to release promoter-proximally paused RNA polymerase II. Transcription 9, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, and Shilatifard A (2010). AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kraus WL, and Bai X (2015). Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem Sci 40, 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xue Y, Yu GK, Arias C, Lin J, Fong S, Faure M, Weisburd B, Ji X, Mercier A, et al. (2015). Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife 4, e06535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhu X, Li Y, Liu M, Yu B, Wang Y, Rao M, Yang H, Zhou K, Wang Y, et al. (2016). Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res 44, 6853–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, and Lis JT (2016). Base-pair-resolution genome wide mapping of active RNA polymerases using precision nuclear run-on (Pro-seq). Nat. Protocols 11, 1455–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, and Gorbunova V (2011). SIRT6 Promotes DNA Repair Under Stress by Activating PARP1. Science 332, 1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, and Price DH (1996). Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271, 27176–27183. [DOI] [PubMed] [Google Scholar]
- Marshall NF, and Price DH (1995). Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270, 12335–12338. [DOI] [PubMed] [Google Scholar]
- Maxam AM, and Gilbert W (1977). A new method for sequencing DNA. Proc Natl Acad Sci U S A 74, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, and Chua KF (2009). Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Van Rechem C, Pal S, Clarke TL, Chakraborty D, Mahan SD, Black JC, Murphy SE, Lawrence MS, Daniels DL, et al. (2018). Cross-talk between Lysine-Modifying Enzymes Controls Site-Specific DNA Amplifications. Cell 174, 803–817 e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miteva YV and Christea IM (2014). A proteomic perspective of Sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol. Cell Proteomics 13, 168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. (2006). Genomic Instability and Aging-like Phenotype in the Absence of Mammalian SIRT6. Cell 124, 315–329. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, and Evans RM (1997). Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, and Bird A (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al. (2003). Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol 23, 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, and Adelman K (2010). Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, and Reinberg D (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, and Lis JT (2012). Overcoming the nucleosome barrier during transcript elongation. Trends Genet 28, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping YH, and Rana TM (2001). DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 276, 12951–12958. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527. [DOI] [PubMed] [Google Scholar]
- Ponnusamy MP, Deb S, Dey P, Chakraborty S, Rachagani S, Senapati S, and Batra SK (2009). RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells 27, 3001–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, and Young RA (2010). c-Myc regulates transcriptional pause release. Cell 141, 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, and Young RA (2014). MYC and transcription elongation. Cold Spring Harb Perspect Med 4, a020990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, and Howley PM (2011). The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31, 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, and Blagoev B (2011). System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4, rs3. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakkody A, Abbas A, Scheidegger A, Warns J, Nnoli O, Jokinen B, Zarns K, Kubat B, Dhasarathy A, and Nechaev S (2015). RNA polymerase II pausing can be retained or acquired during activation of genes involved in the epithelial to mesenchymal transition. Nucleic Acids Res 43, 3938–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, and Nomura M (2006). RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci U S A 103, 12707–12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Satterstrom FK, Haigis MC, and Mostoslavsky R (2012a). From sirtuin biology to human diseases: an update. J Biol Chem 287, 42444–42452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. (2012b). The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, and Vakoc CR (2015). NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol Cell 60, 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberman DM, Ross K, Sande PH, Kubota S, Ramaswamy S, Apte RS, and Mostoslavsky R (2014). SIRT6 is required for normal retinal function. PLoS One 9, e98831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, and Henikoff S (2018). Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nature protocols 13, 1006–1019. [DOI] [PubMed] [Google Scholar]
- Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, et al. (2014). Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275. [DOI] [PubMed] [Google Scholar]
- Strikoudis A, Lazaris C, Trimarchi T, Galvao Neto AL, Yang Y, Ntziachristos P, Rothbart S, Buckley S, Dolgalev I, Stadtfeld M, et al. (2016). Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat Cell Biol 18, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, and Chua KF (2016). SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol 23, 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. (2003). PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. (2013). SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51, 454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, and Schreiber SL (1998). Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921. [DOI] [PubMed] [Google Scholar]
- Van Oss SB, Cucinotta CE, and Arndt KM (2017). Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation. Trends Biochem Sci 42, 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, and Cramer P (2018a). Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612. [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, and Cramer P (2018b). Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. (1998). DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, and Berk AJ (2005). Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell 17, 683–694. [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, and Jones KA (1998). A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462. [DOI] [PubMed] [Google Scholar]
- Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, et al. (2017). BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell 67, 5–18 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Tee AEL, Milazzo G, Bell JL, Poulos RC, Atmadibrata B, Sun Y, Jing D, Ho N, Ling D, et al. (2017). The Histone Methyltransferase DOT1L Promotes Neuroblastoma by Regulating Gene Transcription. Cancer Res 77, 2522–2533. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, and Wang W (1998). NURD, a novel complex with both ATP-dependent chromatin-remodelling and histone deacetylase activities. Mol. Cell 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, and Handa H (2006). P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21, 227–237. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, and Handa H (1999). NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51. [DOI] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, and Lombard DB (2009). The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, and Zhou Q (2005). Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19, 535–545. [DOI] [PubMed] [Google Scholar]
- Yu M, Yang W, Ni T, Tang Z, Nakadai T, Zhu J, and Roeder RG (2015). RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350, 1383–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, and Reinberg D (1997). Histone deacetylase and SAP18, a novel polypedtide, are components of a human Sin3 complex. Cell 89, 357–364. [DOI] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. (2010a). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Urso AD, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. (2010b). The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1alpha. Cell 140, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-seq analysis showing up-regulated genes in SIRT6 KO compared to WT ES cells, related to Figures 1-2.
First page: Chip-Seq analysis showing enrichment of H3K9ac and H3K56ac in SIRT6 KO ESCs, related to Figure 1. Second page: Comparative Analysis for the enrichment of H3K9Ac, H3K56Ac, H3K36me3 and H3K79me2 on up-regulated genes in SIRT6 KO ESCs, as determined by ChIP-seq, related to Figure 7.
List of genes with lower Pausing Index in SIRT6 KO ES cells based on Pol II ChIP-seq analysis, related to Figure 2.
List of genes with lower Pausing Index in SIRT6 KO ES cells and WT No Glucose cells (compared to WT cells) based on Pro-seq analysis, related to Figure 2.
Lower Pausing Index in SIRT6 KO compared to WT ESCs based on both Pol II ChIP-seq and PRO-seq analyses, related to Figure 2.
NELF-E binding in WT and SIRT6 KO ES cells, as determined by CUT&RUN, related to Figure 3.
Genes targeted for LEO1 in SIRT6 KO ES cells, as determined by ChIP-seq, related to Figure 6.
Data Availability Statement
GEO Accession numbers
All the raw datasets for the different sequencing experiments have been deposited in NCBI under the following GEO accession numbers: GSE130689, GSE130690, GSE130691, and GSE130692.