Abstract
Members of the Fanconi anemia (FA) protein family are involved in multiple cellular processes including response to DNA damage and oxidative stress. Here we show that a major FA protein, Fancd2, plays a role in mitochondrial biosynthesis through regulation of mitochondrial translation. Fancd2 interacts with Atad3 and Tufm, which are among the most frequently identified components of the mitochondrial nucleoid complex essential for mitochondrion biosynthesis. Deletion of Fancd2 in mouse hematopoietic stem and progenitor cells (HSPCs) leads to increase in mitochondrial number, and enzyme activity of mitochondrion-encoded respiratory complexes. Fancd2 deficiency increases mitochondrial protein synthesis and induces mitonuclear protein imbalance. Furthermore, Fancd2-deficient HSPCs show increased mitochondrial respiration and mitochondrial reactive oxygen species. By using a cell-free assay with mitochondria isolated from WT and Fancd2-KO HSPCs, we demonstrate that the increased mitochondrial protein synthesis observed in Fancd2-KO HSPCs was directly linked to augmented mitochondrial translation. Finally, Fancd2-deficient HSPCs are selectively sensitive to mitochondrial translation inhibition and depend on augmented mitochondrial translation for survival and proliferation. Collectively, these results suggest that Fancd2 restricts mitochondrial activity through regulation of mitochondrial translation, and that augmented mitochondrial translation and mitochondrial respiration may contribute to HSC defect and bone marrow failure in FA.
Keywords: Fanconi anemia, Hematopoietic stem and progenitor cells, Mitochondrial translation, Proliferation, Survival
1. Introduction
Fanconi anemia (FA) is a genetic disorder associated with congenital developmental defects, bone marrow (BM) failure and predisposition to cancers (Bagby, 2003; Tischkowitz and Hodgson, 2003; Kennedy and D’Andrea, 2000; Green and Kupfer, 2009). FA is genetically heterogeneous, with at least 22 complementation groups (FANCA-FANCW) identified thus far (Dong et al., 2015; Bogliolo et al., 2013; Sawyer et al., 2015; Knies et al., 2017). The biological function of these FA proteins has been subjected to intensive investigation. Studies have shown that eight of the FA proteins (FANCA, B, C, E, F, G, L, and M) form the FA core complex that functions as an ubiquitin ligase. In response to DNA damage or DNA replication stress, the FA core complex monoubiquitinates the D-I complex formed by two downstream FA proteins, FANCD2 and FANCI, which then recruit other downstream FA proteins and additional DNA repair factors, to nuclear loci containing damaged DNA and consequently influence important cellular processes such as DNA replication, cell-cycle control, and DNA damage response and repair (Kottemann and Smogorzewska, 2013; Deans and West, 2011; Kim and D’Andrea, 2012).
One of the important clinical features of FA is hematological. FA commonly progresses from BM failure to a pre-leukemic myelodysplastic syndrome (MDS) stage and finally evolves to acute myeloid leukemia (AML). These hematological manifestations of FA are believed to be resulted from defects in hematopoietic stem cells (HSCs). Indeed, studies in patients and knockout mice have shown that FA deficiency leads to severe defects in both quantity (frequencies and absolute numbers) and quality (such as the ability to reconstitute hematopoiesis) of the HSCs (Haneline et al., 1999; Kelly et al., 2007; Pulliam et al., 2008; Haneline et al., 2003; Du et al., 2013; Du et al., 2015). In addition, FA HSCs have high risk of clonal evolution (Haneline et al., 2003; Li et al., 2007). This latter HSC phenotype is correlated with the very high incidence of MDS and AML that is observed in FA patients (Auerbach and Allen, 1991; Kutler et al., 2003). Finally, allogeneic HSC transplantation can cure the progressive marrow aplasia in FA patients (Mehta et al., 2010; Smith & Wagner, 2012), also invoking a HSC–specific deficiency phenotype.
Emerging evidence has revealed that resting quiescent HSCs possess a distinct metabolic profile with a preference for anaerobic glycolysis rather than mitochondrial oxidative phosphorylation (OXPHOS) (Simsek et al., 2010; Takubo et al., 2013; Kohli and Passegué, 2014). Indeed, HSCs with low mitochondrial activity have been shown to correlate with their functionality (Simsek et al., 2010). Moreover, altered metabolic energetics has been demonstrated in HSCs at different stages of their life cycle and in certain blood disorders (Suda et al., 2011; Warr et al., 2011; Baumann, 2013). Thus, new insights into the metabolic differences between normal and diseased HSCs may prove valuable for developing better therapeutic strategies for hematologic diseases like BM failure and leukemia. In this report, we show that mouse hematopoietic stem and progenitor cells (HSPCs) deficient for Fancd2 show increase in mitochondrial protein synthesis and depend on augmented mitochondrial translation for survival and proliferation. Our results suggest that Fancd2 plays a role in regulation of mitochondrial translation.
2. Results
2.1. Fancd2 interacts with mitochondrial proteins Atad3 and Tufm
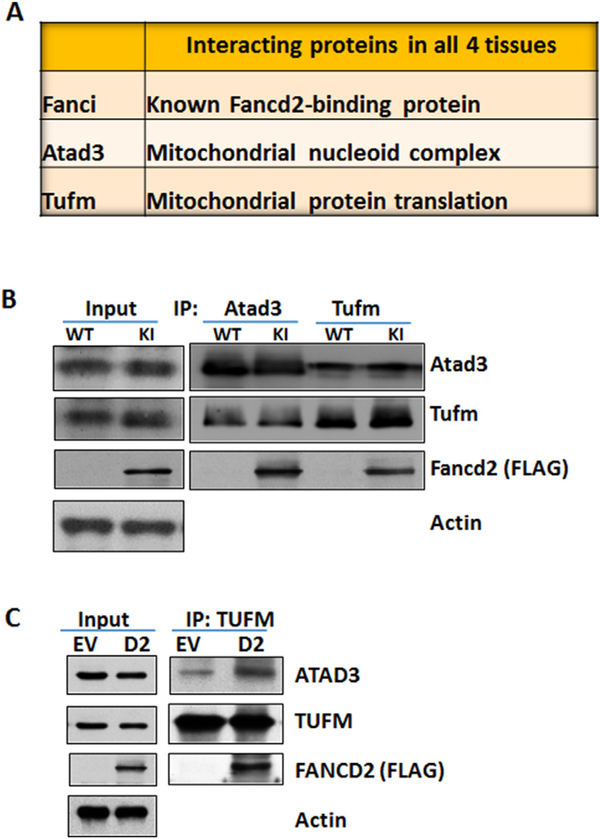
We recently generated a Fancd2 knock-in mouse model, in which a duel tandem (3XFLAG and HA) tag was inserted at the C-terminus of the endogenous Fancd2 locus (Zhang et al., 2017). We showed that the tagged Fancd2 protein expressed in the Fancd2-KI mice retained the ability to induce monoubiquitination and form DNA damage foci in nuclei in response to DNA damage, and could be efficiently pulled down by FLAG and HA antibodies using 2-step immunoprecipitation. By using this Fancd2-KI mouse model and proteomic approach, we have shown that many of the Fancd2-interacting proteins were mitochondrion-specific. We have also shown that Fancd2 was localized in the mitochondrion and associated with the nucleoid complex components Atad3, Tufm (mitochondrial Tu translation elongation factor) and Tfam, all of which are required for mitochondrial biosynthesis (Li et al., 2014; He et al., 2012; Valente et al., 2007; Smeitink et al., 2006; Antonicka et al., 2006; Ryan and Hoogenraad, 2007). Moreover, the Atad3-Tufm-Tfam complex was disrupted in Fancd2-KO mice and the mice deficient for the FA core component Fanca. Significantly, we found three proteins, Fanci, Atad3, and Tufm that interacted with Fancd2 in all four tissues (ES cells, E11.5 embryos, testes and spleen; Fig. 1A). We note that we did not pursue Fanci further, because it is the well-known Fancd2-binding partner (Kottemann and Smogorzewska, 2013; Deans and West, 2011; Kim and D’Andrea, 2012). We turn our attention to the two mitochondrial Fancd2-interactors, Atad3 and Tufm. Reciprocal immunoprecipitation showed that the endogenous Atad3 and Tufm associate with Fancd2 (Fig. 1B). We also addressed whether the loss of Fancd2 affects these interactors. Due to the lack of workable antibody for mouse Fancd2, we chose a FA-D2 patient-derived lymphoblast cell line that had been constituted with a functional FANCD2 (with a 3 × FLAG tag) gene (Timmers et al., 2001). FANCD2 deficiency does not affect the stability of either ATAD3 or TUFM; however, loss of FANCD2 reduces the ATAD3-TUFM complex (Fig. 1C). These results suggest that Fancd2 may play a role in the formation or stability of mitochondrial ATAD3-TUFM complex.
Fig. 1.
Fancd2 interacts with mitochondrial proteins Atad3 and Tufm. (A) Fancd2-interacting proteins in all four tested tissues. Fancd2-containing complexes from ES cells, E11.5 embryos, testes and spleen of Fancd2-KI mice were purified by two-step (anti-FLAG then anti-HA), and the identities of Fancd2-associated proteins were determined by liquid chromatography and tandem mass spectrometry (LC–MS/MS). Note that three proteins, Fanci, Atad3 and Tufm interacted with Fancd2 in all four tissues. (B) Reciprocal immunoprecipitation of FLAG-tagged Fancd2 with endogenous Atad3 and Tufm proteins. Protein extracts of spleen mono-nucleated cells from Fancd2-KI mice were subjected to immuno-precipitation (IP) with anti-Atad3 and anti-Tufm antibodies, and then immunoblotting with antibodies against Atad3 and Tufm and Fancd2. (C) Loss of Fancd2 disrupts the Atad3-Tufm complex. The human FA-D2 lymphoblast lysates infected with empty vector (EV) or 3 × FLAG-FANCD2 (D2) lentivirus were immunoprecipitated with TUFM antibody and blotted with the indicated antibodies.
2.2. Increased mitochondrial number and protein synthesis in Fancd2-KO HSPCs
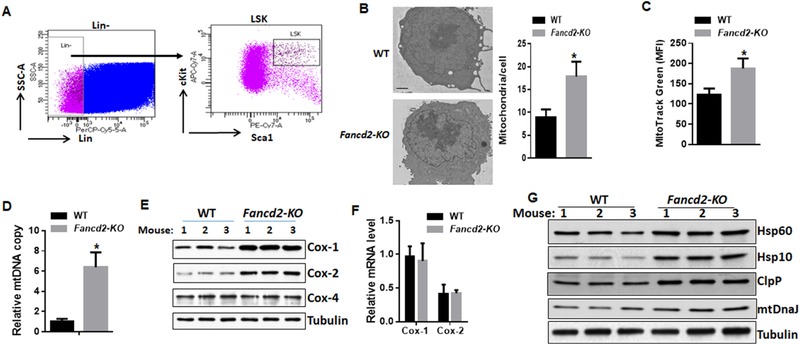
The observation that Fancd2 interacts with the Atad3-Tufm mitochondrial nucleoid complex prompted us to address whether loss of Fancd2 altered mitochondrial properties in HSPCs. Indeed, we found a marked increase in mitochondrial number in Fancd2-KO LSK (Lin−Sca1+c-kit+; Fig. 2A) cells compared to WT cells (Fig. 2B). Consistently, we found a significant increase in mitochondrial mass in Fancd2-KO LSK cells, as determined by MitoTracker Green staining (Fig. 2C), and mitochondrial DNA quantification (Fig. 2D). Since Atad3 and Tufm are required for the mitochondrial coupled-transcription and translation process (Li et al., 2014; He et al., 2012; Valente et al., 2007; Smeitink et al., 2006; Antonicka et al., 2006; Ryan and Hoogenraad, 2007), we asked whether loss of Fancd2 altered expression of proteins whose translation is known to be encoded by nuclear or mitochondrial DNA (mtDNA). We chose three subunits of respiratory complex IV as surrogates: mtDNA-encoded Cox-1, Cox-2 and nuclear-encoded Cox-4 (Skrtić et al., 2011). We observed much higher levels of mtDNA-encoded Cox-1 and Cox-2 proteins relative to constant amounts of nuclear DNA-encoded Cox-4 in Fancd2-KO BM HSPCs (Fig. 2E). However, this increase in protein levels was not accompanied by an increase in mitochondrial transcription (Fig. 2F). These results suggest that Fancd2 deficiency might induce mitonuclear protein imbalance, a stoichiometric imbalance between nuclear and mitochondrially encoded proteins (Houtkooper et al., 2013; Jovaisaite et al., 2014). To test this notion, we analyzed several mitochondrial stress response proteins, including HSP60, HSP10, ClpP and mtDnaJ, in BM Lin- cells from WT and Fancd2-KO mice. Indeed, all these mitochondrial stress response proteins were elevated in Fancd2-KO HSPCs (Fig. 2G). Thus, deletion of Fancd2 induces mitonuclear protein imbalance in murine HSPCs. Taken together, these results suggest that Fancd2 may be a negative regulator of mitochondrial translation.
Fig. 2.
Loss of Fancd2 increases mitochondrial number and protein synthesis. (A) Gating strategy for sorting HSPCs (Lin−Sca1+c-kit+; LSK). (B) Increased mitochondrial number in Fancd2-KO HSPCs. Shown are representative transmission electron microscopy images (left; scale bar 1 μm) and quantification (right). Results are presented as mean ± SD of two independent experiments (n=4 mice per group). (C-D) Increased mitochondrial mass in Fancd2-KO LSK cells, as determined by MitoTracker Green staining (C) and mitochondrial DNA quantification (D). The relative mtDNA copy number was calculated as a ratio of mtDNA/nuclear DNA (n=6 mice per group). (E) Increased mitochondrial protein synthesis in Fancd2-KO HSPCs. Protein lysates of Lin− BM cells were analyzed for Cox-1, Cox-2, Cox-4, and tubulin. (F) Fancd2 loss does not affect mitochondrial transcription. mRNA levels of Cox-1 and Cox-2 were determined in BM LSK cells by qRT-PCR and normalized relative to mitochondrial 16S rRNA (n=6). (G) Increased mitochondrial stress response proteins in Fancd2-KO HSPCs. Protein lysates of Lin− BM cells were analyzed for HSP60, HSP10, ClpP, mtDnaJ and tubulin. ∗p < .05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Increased enzyme activity of respiratory complexes I and IV in Fancd2-KO HSPCs
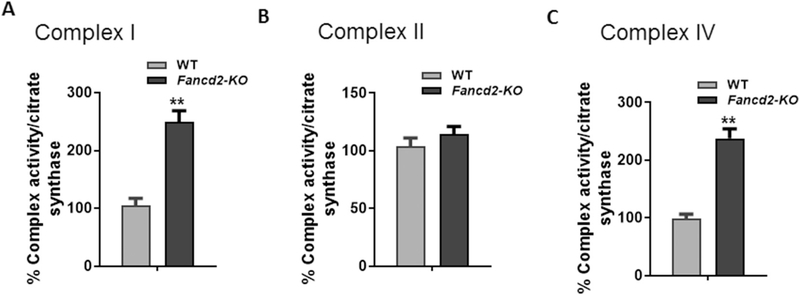
Because loss of Fancd2-KO HSCPs showed elevated mitochondrial respiratory complex proteins, we next asked whether Fancd2 deficiency would affect the enzymatic activity of mitochondrial respiratory complexes. We chose three respiratory complexes I, II and IV for analysis because respiratory complexes I and IV contain proteins encoded by the mitochondrion genome; in contrast, respiratory complex II does not contain mitochondrially encoded subunits in its substructure (Ott and Herrmann, 2010). We observed significantly increase in the enzyme activity of respiratory complexes I (Fig. 3A) and IV (Fig. 3C) but not in that of the complex II in Fancd2-KO HSPCs (Fig. 3B). Thus, loss of Fancd2 specifically affects the enzyme activity of respiratory complexes containing subunits encoded by the mitochondrion genome.
Fig. 3.
Increased enzyme activity of mitochondrial respiratory complexes in Fancd2-KO HSPCs. (A-C) Effect of Fancd2 deficiency on the enzyme activities of mitochondrial respiratory chain complex I (A), II (B), and IV (C). BM LSK cells from WT and Fancd-KO mice were sorted by FACS, and the enzyme activity of complexes I, II, and IV was measure as described in Materials and Methods, and is expressed in relative to citrate synthase activity. Values shown are average of three independent experiments. ∗∗p < .01.
2.4. Loss of Fancd2 increases OXPHOS and mtROS
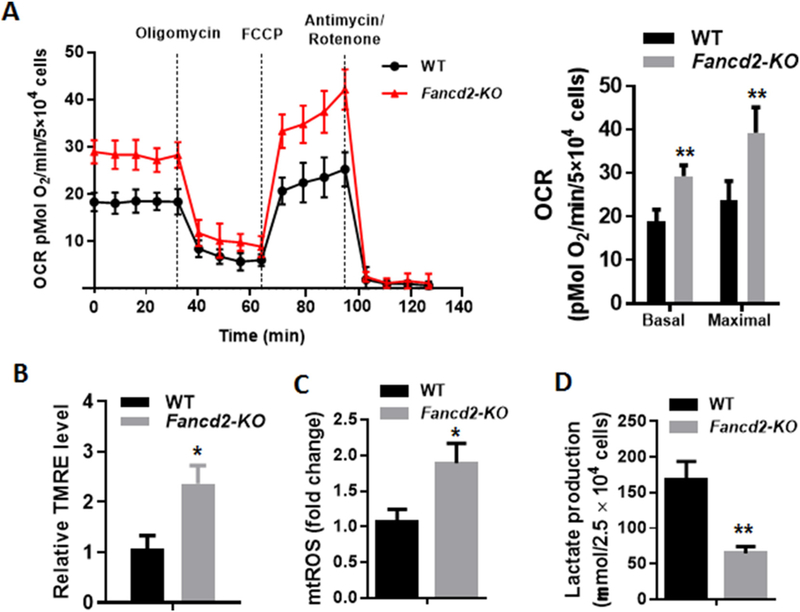
To determine whether the increase in mitochondrial protein synthesis and the enzymatic activity of respiratory complexes observed in Fancd2-KO HSPCs correlated with increased OXPHOS, we measured mitochondrial respiration and found significant increase in both basal and maximal oxygen consumption rate (OCR) in Fancd2-KO LSK cells compared to WT cells (Fig. 4A). In addition, we found that Fancd2-KO LSK cells had much higher mitochondrial membrane potential than their WT controls (Fig. 4B). Fancd2-KO LSK cells also showed significant increase in mitochondrial ROS (mtROS) (Fig. 4C), and marked reduction in lactate production (indicative of decreased glycolysis; Fig. 4D). We also measured OCR and mtROS in LSK cells isolated from Fanca-KO and Fancc-KO mice, and found that consistent with Fancd2-KO LSK cells, loss of Fanca or Fancc led to significant increase in both mitochondrial respiration and mtROS (Supplemental Fig. 1). These data suggest that the FA proteins may negatively regulate mitochondrial OXPHOS in HSPCs.
Fig. 4.
Loss of Fancd2 increases OXPHOS and mtROS. (A) Increased mitochondrial respiration in Fancd2-KO HSPCs. Oxygen consumption rates (OCR) were measured in BM LSK cells using the Seahorse XF96 analyzer. A representative experiment of three is shown. Basal OCR and Maximal OCR are shown on the right panel. (B) Increased mitochondrial membrane potential, measured using Tetramethylrhodamine ethyl ester perchlorate (TMRE), in Fancd2-KO LSK cells. Results are presented as mean ± SD of three independent experiments. (C) Increased mitochondrial ROS, measured using mitoSOX, in Fancd2-KO LSK cells. Results are presented as mean ± SD of three independent experiments. (D) Decreased lactate production, measured using the Biovision Lactate Assay kit, in Fancd2-KO LSK cells. Results are presented as mean ± SD of three independent experiments. ∗p < .05, ∗∗p < .01.
2.5. Fancd2-KO HSPCs are selectively sensitive to mitochondrial translation inhibition
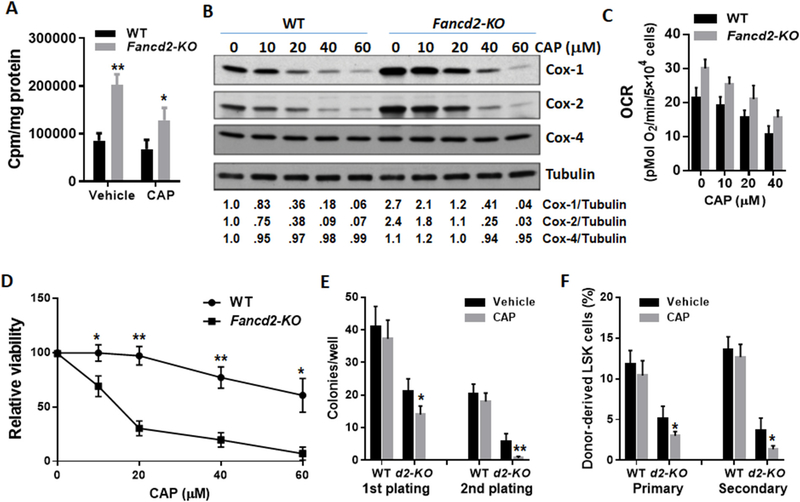
To determine if the increased mitochondrial protein synthesis observed in Fancd2-KO HSPCs was directly linked to dysregulation of mitochondrial translation, we performed a cell-free assay to analyze mitochondrial translation with mitochondria isolated from WT and Fancd2-KO BM Lin− cells (enriched for HSPCs). We found a significant increase in mitochondrial translation in Fancd2-KO HSPCs compared to WT cells (Fig. 5A). Consistently, treatment with low dose of the mitochondrion-specific translation inhibitor Chloramphenicol (CAP; Nagiec et al., 2005; McKee et al., 2006) confirmed the effect of Fancd2 deficiency on mitochondrial translation (Fig. 5A). To assess the functional consequence of the augmented mitochondrial translation, we treated WT and Fancd2-KO BM Lin− cells with the mitochondrial translation inhibitor, and found that incubation of the cells with CAP at 20 μM for 36 h was able to reduce the levels of mtDNA-encoded Cox-1 and Cox-2 subunits in Fancd2-KO HSPCs to approximately the levels in untreated WT cells but did not affect the level of nuclear DNA-encoded Cox-4 (Fig. 5B). Furthermore, 20 μM CAP reduced the basal OCR in Fancd2-KO LSK cells to the similar level seen in untreated WT cells (Fig. 5C). We next compared the effect of the mitochondrial translation inhibitor on the survival of WT and Fancd2-KO LSK cells. We found that Fancd2-KO LSK cells were extremely sensitive to CAP even at low doses (10 and 20 μM), which had no effect on the viability of WT cells (Fig. 5D). It is noteworthy that this result does not exclude the possibility that the reduction in the basal OCR in Fancd2-KO LSK cells by CAP treatment (Fig. 5C) might be due to a decrease in cell viability. To determine the effect of mitochondrial translation inhibition on the proliferation of Fancd2-KO HSPCs, we performed serial plating and BM transplantation experiments using the cells treated with CAP at 20 μM for 36 h. To our surprise, CAP did not improve the function of Fancd2-KO HSPCs; instead, it significantly decreased colony generation of Fancd2-KO HSPCs in the first plating and almost completely eliminated their colony-generating capacity in the second plating (Fig. 5E). Similar results were observed in serial BM transplantation experiments, in which CAP inhibited both short-term and long-term repopulating ability of Fancd2-KO HSCs (Fig. 5F). These results indicate that Fancd2-KO HSPCs depend on augmented mitochondrial translation for survival and proliferation.
Fig. 5.
Fancd2-KO HSPCs depend on augmented mitochondrial translation and OXPHOS for proliferation and survival. (A) Augmented mitochondrial translation in Fancd2-KO HSPCs. Mitochondrial isolated from BM Lin− cells were incubated with protein synthesizing medium in the presence of vehicle (DMSO) or the mitochondrial translation inhibitor Chloramphenicol (CAP; 10 μM) and [3H]-leucine. Incorporation of [3H]-leucine was measured after 60 min. (B) Effect of increasing concentrations of the mitochondrial translation inhibitor Chloramphenicol (CAP; 36 h) on protein levels of Cox-1, Cox-2, Cox-4, and tubulin in WT and Fancd2-KO Lin- cells. The relative levels of Cox-1, Cox-2 or Cox-4 to tubulin are indicated below the blot. (C) Inhibition of mitochondrial respiration by CAP. Levels of basal OCR were measured in in WT and Fancd2-KO LSK cells treated with increasing concentrations of CAP for 36 h. (D) Effect of mitochondrial translation inhibition on HSPC survival. WT and Fancd2-KO LSK cells were treated with increasing concentrations of CAP for 36 h, and the proportion of viable cells was measured by Annexin-PI flow cytometry to calculate the yield of viable cells shown as percent viable vehicle-treated cells in the same experiment. (E) CAP decreases colony generation of Fancd2-KO HSPCs. WT and Fancd2-KO LSK cells were treated with 20 mM CAP for 36 h and subjected to serial plating. Data represent mean ± SD of 3 independent experiments. (F) CAP impairs the self-renewal ability of Fancd2-KO HSCs. WT and Fancd2-KO LSK cells were treated with 20 mM CAP for 36 h and used for Serial BM transplantation. Donor-derived LSK cells in the primary and secondary recipients were analyzed by Flow cytometry at 16 weeks post-transplant. Results are mean ± SD of three independent experiments (n = 6–9 per group for each BMT). *p < .05, **p < .01.
3. Discussion
The role of the FA pathway in HSC energy metabolism is largely unknown. We recently found that FA HSCs are more dependent on OXPHOS relative to glycolysis in their resting state for energy metabolism (Du et al., 2016). However, the mechanistic underpinning of the altered bioenergetics program in FA HSCs has not been defined. Using the innovative Fancd2-KI mouse model, we establish biochemical interaction between Fancd2 and the Atad3-Tufm complex essential for mitochondrial translation (29; Fig. 1). In addition, loss of Fancd2 leads to a significant increase in mitochondrial number, protein synthesis, enzyme activity of mitochondrion-encoded respiratory complexes, and consequently OXPHOS and mtROS in HSPCs (Figs. 2, 3, 4). Surprisingly, Fancd2-deficient HSPCs are hypersensitive to mitochondrial translation inhibition and appear to depend on augmented mitochondrial translation for survival and proliferation. These novel findings indicate a functional link between Fancd2 and mitochondrial translation in HSPCs, and suggest that Fancd2 could play a role in restricting mitochondrial activity through repressing mitochondrial translation and OXPHOS in HSC maintenance.
We previously showed that a significant portion of Fancd2 is localized in the mitochondrion (Zhang et al., 2017). We envision that interaction between the Fancd2 and the mitochondrial Atad3-Tufm Nucleoid complex plays a role in the maintenance of mitochondrial function. Indeed, loss of Fancd2 leads to a marked increase in mitochondrial protein synthesis, OXPHOS and ROS in HSPCs. Elevated ROS and consequently oxidative stress is now recognized as one of important phenotypic hallmarks in FA (Park et al., 2004; Saadatzadeh et al., 2004; Du et al., 2008). Since mitochondria represent the main source of both cellular energetic metabolism and ROS production (85–90%) (Balaban et al., 2005), and since the role of mitochondria in the onset of oxidative stress is well established (Balaban et al., 2005; Addabbo et al., 2009; Lambert and Brand, 2009), the link between FA and mitochondrial dysfunction has long been suspected. Indeed, over the last decade, mitochondrial dysfunction and oxidant hypersensitivity has been documented in many studies using FA mouse models and primary and immortalized cell cultures as well as ex vivo materials from FA patients (Pagano et al., 2013; Usai et al., 2015; Ravera et al., 2013; Kumari et al., 2014; Ponte et al., 2012; Mukhopadhyay et al., 2006; Bogliolo et al., 2002; Zanier et al., 2004; Rousset et al., 2002; Pagano et al., 2014). However, these studies were performed on heterogeneous cell populations and it remains to be seen if similar phenotypes of mitochondrial dysfunction exist in FA HSCs. In this context, our results provide molecular and cellular evidence for the function of Fancd2 in the regulation of mitochondrial metabolism and the role of this regulatory mechanism in HSC maintenance.
By using a cell-free assay with mitochondria isolated from WT and Fancd2-KO HSPCs, we demonstrate that the increased mitochondrial protein synthesis observed in Fancd2-KO HSPCs was directly linked to augmented mitochondrial translation. Because Fancd2-deficient HSCs show augmented mitochondrial translation, we postulated that normalization of mitochondrial translation by specific inhibitor CAP could improve the function of Fancd2-deficient HSCs. Surprisingly, restoring mitochondrial protein synthesis of Fancd2-KO HSPCs to WT levels by CAP at 20 μM, which also reduced OXPHOS to WT levels, not only failed to improve but worsened the defect of Fancd2-KO HSCs in short-term and long-term hematopoietic repopulation, as assessed by serial BM transplantation (Fig. 5). These results indicate that the augmented mitochondrial translation renders Fancd2-KO HSPCs sensitive to mitochondrial translation inhibition, and suggest that Fancd2-KO HSPCs may have depended on augmented mitochondrial translation/OXPHOS for proliferation and survival.
HSC defect is currently considered the driver of FA BM failure and leukemogenesis (Bagby, 2003; Tischkowitz and Hodgson, 2003; Kennedy and D’Andrea, 2000; Green and Kupfer, 2009); whereas the role of FA mitochondrial dysfunction in FA disease progression has long been suspected but remained undefined. Our unbiased proteomics and functional studies identified interplay between the FA pathway and mitochondrial function in mouse HSC maintenance. Whether the augmented mitochondrial translation/OXPHOS observed in Fancd2-KO HSPCs has functional implication for the FA disease remains further investigation. Nevertheless, our results argue a potential contribution of mitochondrial dysfunction to the pathophysiology of the FA disease. Furthermore, currently, the mechanism by which FA disease progresses through BMF-MDS-AML has been focused on the defects in DNA damage repair and genomic maintenance. Thus, our findings shed new light on the role of FA protein FANCD2 in mitochondrial function and suggest that targeting mitochondrial dysfunction may be therapeutically valuable for the disease.
4. Materials and methods
4.1. Mice
Fancd2KI/KI mice were generated in our laboratory (Zhang et al., 2017). Fancd2+/− mice were provided by Dr. Markus Grompe (Oregon Health & Sciences University; Houghtaling et al., 2003). We used 8–16 week-old age-matched mice in all experiments. Mice were maintained on C57BL/6J background in the animal barrier facility at Cincinnati Children’s Hospital Medical Center. Animals were kept in accordance with the protocol approved by the CCHMC Institutional Animal Care and Use Committees.
4.2. Isolation of bone marrow cells and flow cytometry analysis
The femora and tibiae were harvested from the mice immediately after their sacrifice with CO2. Bone marrow (BM) cells were flushed from bones into Iscove’s modified Dulbecco’s medium (IMDM; Invitrogen) containing 10% FCS, using a 21-guage needle and syringe. Low-density BM mononuclear cells (LDBMMNCs) were separated by Ficoll Hypaque density gradient (Sigma-Aldrich, St. Louis, MO) and washed with IMDM medium.
For flow analysis and cell sorting, BM cells from mice of the indicated genotype were stained with the following antibodies (all from BioLegend, San Diego, CA): Ter119 (#79748), CD45R/B220 (#79752), CD3e (#79751), Gr1 (#79750), CD11b (#79749), Ly-6A/E (Sca1, #108114), CD117 (c-Kit, 105826). Flow cytometric analyses were done using BD LSRII flow cytometer (BD Bioscience). For cell sorting, lineage negative cells were enriched using lineage depletion reagents (StemCell Technologies) according to the manufacturer’s instruction. The Linnegative and LSK populations were acquired by using the FACSAria II sorter (BD Biosciences).
4.3. In vitro cell culture and treatment
Briefly, BM Lin− or LSK cells were maintained in StemSpan medium supplemented with 50 ng/ml murine rTpo (Preprotech, Rocky Hill, NJ), 50 ng/ml murine rSCF (Preprotech, Rocky Hill, NJ) and 1% BSA at 37 °C in normoxia (21% O2, 5% CO2). Cells with the indicated genotype were treated with increasing doses of the mitochondrial translation inhibitor Chloramphenicol (CAP; 0–60 μM) for 36 h followed by survival and Oxygen Consumption Rate (OCR) assays.
4.4. Cell viability assay
Cell death was measured by Annexin V fluoroscein isothiocyanate (FITC) and Propidium Iodide (PI) (Biovision Research Products, Mountain View, CA) staining using flow cytometry according to the manufacturer’s instructions.
4.5. Colony forming unit assay
For the in vitro colony forming unit (CFU) assay, 1000 sorted LSK cells were seeded in MethoCult GF M3434 (STEMCELL Technologies) according to the manufacturer’s recommendations. Colonies were visualized and counted at day 7. The experiment was performed in triplicate for each sample.
4.6. Serial bone marrow transplantation (BMT)
1000–2000 LSK cells (CD45.2+), along with 200,000 c-Kit-depleted protector cells, were transplanted into lethally irradiated BoyJ (CD45.1+) mice. The recipients were subjected to Flow cytometric analysis for donor-derived LSK cells 16 weeks after BMT. For secondary BMT, one million BM cells from the primary recipient mice were transplanted into sublethally irradiated secondary CD45.1+ recipient mice. Four months later, the recipients were subjected to Flow cytometric analysis for donor-derived LSK cells.
4.7. Oxygen consumption rate
Measurement of oxygen consumption was performed using a Seahorse XF96 analyzer (Seahorse Bioscience, North Billerica, MA, USA). BM LSK cells were seeded in XF96 plates. Cells were equilibrated to the un-buffered medium for 45 min at 37 °C in a CO2- free incubator before being transferred to the XF96 analyzer. We measured the basal Oxygen Consumption Rate (OCR), and then sequentially injected Oligomycin (ATP synthase inhibitor; 0.5 μM) and the electron transport chain accelerator ionophore FCCP (Trifluorocarbonylcyanide Phenylhydrazone; 0.5 μM), which measured the Maximal OCR. Finally, the reaction was stopped by adding the electron transport chain inhibitors Rotenone and Antimycin A (1 μM each).
4.8. Mitochondrial mass measurements
We assessed mitochondrial mass using two assays: (Bagby, 2003) mitochondrial DNA (mtDNA) copy number - genomic DNA was extracted from primary cells using the DNeasy Blood and Tissue kit (Qiagen MD, USA). The relative mtDNA copy number was determined by a real-time polymerase chain reaction (qPCR), and compared relative to nuclear DNA. The primer sequences were forward primer (Cox-2-F), 5′-AATTGCTCTCCCCTCTCTACG-3′; reverse primer (Cox-2-R), 5′-GTAGCTTCA GTATCATTGGTGC-3′, forward primer (ApoB-F), 5′-CGTGGGCTCCAG CATTCTAAC-3′; reverse primer (ApoB-R), 5′-TCACCAGTCATTTCTGCCTTTG-3′. (Tischkowitz and Hodgson, 2003) Mito Tracker Green (GTG) - cells were stained with 50 nM of Mitotracker Green FM (Invitrogen, Carlsbad, CA) in PBS buffer at 37 °C for 30 min, and samples were analyzed by flow cytometry.
4.9. Mitochondrial membrane potential, mitochondrial ROS and lactate measurements
For mitochondrial membrane potential, BM LSK cells were incubated with 50 nM Tetramethylrhodamine ethyl ester perchlorate (TMRE; Invitrogen, Carlsbad, CA) for 20 min at 37 °C, and fluorescence of TMRE was determined flow cytometrically. For mitochondrial ROS, BM LSK cells were stained with 5 μM MitoSOX (Invitrogen, Carlsbad, CA). Lactate levels were measured using the Lactate kit according to manufacturer’s instructions (Biovision, Milpitas, CA).
4.10. Mitochondrial enzymatic assays
The activity of the mitochondrial respiratory chain complex I (NADH dehydrogenase) was measured by monitoring rotenone-sensitive 2,6-dichloroindophenol reduction by electrons accepted from decylubiquinol reduced after oxidation of NADH by complex I (Janssen et al., 2007). The activity of complex II (succinate dehydrogenase) was determined by monitoring malonate-sensitive reduction of 2,6-dichloroindophenol when coupled to complex II-catalyzed reduction of decylubiquinol (Jung et al., 2000). The activity of complex IV (cytochrome c oxidase) was measured by KCN-sensitive oxidation of ferrocytochrome c (Trounce et al., 1996). The activity of citrate synthase was analyzed by tracking the reduction of 5,5′-dithiobis-2-nitrobenzoic acid at 412 nm in the presence of acetyl-CoA and oxaloacetate. The enzyme activity of complexes I, II, and IV was normalized to citrate synthase activity.
4.11. Mitochondrial translation assay
Mitochondria were isolated from BM Lin− cells using the Mitochondria Isolation kit (Miltenyi Biotech Inc) following manufacturer instructions. Cell-free mitochondrial translation was carried out using a modified protocol described previously (McKee et al., 2006). Briefly, mitochondria were incubated at 3 mg protein/ml in 30 μL protein synthesizing medium. The mitochondrial mix was incubated at 30 °C in the presence of vehicle (DMSO) or the mitochondrial translation inhibitor Chloramphenicol (CAP; 10 μM) for 5 min, and 4 μL (4 μCi) [3H]-leucine was then added to the mix and incubation continued for 60 min. The incorporation of [3H]-leucine into mitochondrial protein was determined by counting the [3H]-leucine-labeled proteins in a Beckman LS6000SC liquid scintillaton counter.
4.12. Western blotting and immunoprecipitation
4.12.1. Preparation of cell extracts, immunoblotting and immunoprecipitation
To prepare protein lysates, cells were washed with ice-cold PBS, and resuspended in ice-cold lysis buffer containing 50 mM Tris-HCL (pH 7.4), 0.1% NP40, and 1 M NaCl supplemented with protease and phosphatase inhibitors [10 μg/mL aprotinin, 25 μg/mL leupeptin, 10 μg/mL pepstatin A, 2 mM phenylmethylsulfonyl fluoride, 0.1 M NaP2O4, 25 mM NaF, and 2 mM sodium orthovandate] for 30 min on ice. Cell debris was removed from the lysates by centrifuging them at 14,000 rpm for 30 min. Protein concentration was quantified by using Bio-Rad reagent and resolved on SDS-PAGE and transferred onto nitrocellulose membranes. Immunoblots were then incubated with primary antibodies specific for COX-1 (Abcam), COX-2 (Santa Cruz), COX-4 (Santa Cruz), β-actin (Sigma-Aldrich), FLAG M2 (sigma), ATAD3A (Abcam), TUFM (ThermoFisher), HSP60 (Invitrogene), HSP10 (Sigma-Aldrich), ClpP (Abcam), or mtDnaJ (Abcam). Quantification of Western blot was conducted with ImageJ software (NIH) by measuring the density of each single band.
For immunoprecipitation, protein A/G agarose beads (Santa Cruz Biotechnologies, Dallas, Texas) precleaned cell lysates were incubated with ATAD3A (Abcam) or TUFM (ThermoFisher) primary antibody by gentle rocking overnight at 4 °C followed by incubation with protein A/G agarose beads for additional 1 h at 4 °C. Pellets were then washed with 500 μl of lysis buffer and resuspended with 20 μl of 4× SDS sample buffer. Immunoblots were then incubated with antibodies specific for ATAD3A (Abcam) or TUFM (ThermoFisher), or FLAG M2 (sigma) antibodies for 12 to 16 h at 4 degree. Signals were revealed after incubation with anti-mouse or anti-rabbit secondary antibodies.
4.13. Statistical analysis
Student’s t-test was performed using GraphPad Prism v6 (GrapPad software). Comparaison of > 2 groups was analyzed by one-way Anova test. Values of p < .05 were considered statistically significant. Results are presented as mean ± SD. * indicates p < .05; ** = p < .01; *** = p < .001.
Supplementary Material
Acknowledgements
We thank Dr. Markus Grompe (Oregon Health & Sciences University) for the Fancd2+/−mice, Dr. Taosheng Huang (Cincinnati Children’s Hospital Medical Center) for mitochondrion study, and the Comprehensive Mouse and Cancer Core of the Cincinnati Children’s Research Foundation (Cincinnati Children’s Hospital Medical Center) for bone marrow transplantation service. This investigation was partially supported by NIH grants R01 HL076712 and R01 HD089932. Q.P. was supported by a Leukemia and Lymphoma Scholar award.
Footnotes
Declaration of Competing Interest
The authors declare no competing interests.
Ethics approval and consent to participate
All animal procedures were approved by the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center prior to study initiation (IACUC protocol # 2013-0159).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2019.101550.
References
- Addabbo F, Montagnani M, Goligorsky MS, 2009. Mitochondria and reactive oxygen species. Hypertension 53, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H, Sasarman F, Kennaway NG, Shoubridge EA, 2006. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum. Mol. Genet. 15, 1835–1846. [DOI] [PubMed] [Google Scholar]
- Auerbach AD, Allen RG, 1991. Leukemia and preleukemia in Fanconi anemia patients. A review of the literature and report of the International Fanconi Anemia Registry. Cancer Genet. Cytogenet. 51, 1–12. [DOI] [PubMed] [Google Scholar]
- Bagby GC, 2003. Genetic basis of Fanconi anemia. Curr. Opin. Hematol. 10, 68–76. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T, 2005. February 25. Mitochondria, oxidants, and aging. Cell 120 (4), 483–495. [DOI] [PubMed] [Google Scholar]
- Baumann K, 2013. Stem cells: a metabolic switch. Nat Rev Mol Cell Biol 14 (2), 64–65. [DOI] [PubMed] [Google Scholar]
- Bogliolo M, Borghini S, Abbondandolo A, Degan P, 2002. January. Alternative metabolic pathways for energy supply and resistance to apoptosis in Fanconi anaemia. Mutagenesis. 17 (1), 25–30. [DOI] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, et al. , 2013. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92 (5), 800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC, 2011. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 11 (7), 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nebert DW, Bruford EA, Thompson DC, Joenje H, Vasiliou V, 2015. November 24. Update of the human and mouse Fanconi anemia genes. Hum Genomics. 9 (1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Adam Z, Rani R, Zhang X, Pang Q, 2008. Oxidative stress in Fanconi anemia hematopoiesis and disease progression. Antioxidants & Redox Signaling 10 (11) (1909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Amarachintha S, Sipple J, Schick J, Pang Q, 2013. Inflammation-mediated Notch signaling skews Fanconi Anemia hematopoietic stem cell differentiation. J. Immunol. 191 (5), 2806–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Amarachintha S, Erden O, Wilson A, Meetei AR, Andreassen PR, Namekawa SH, Pang Q, 2015. Fancb deficiency impairs hematopoietic stem cell function. Sci Rep. 5, 18127 December 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Amarachintha S, Wilson AF, Pang Q, 2016. SCO2 mediates oxidative stress-induced glycolysis to OXPHOS switch in hematopoietic stem cells. Stem Cells 34 (4), 960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AM, Kupfer GM, 2009. April. Fanconi anemia. Hematol. Oncol. Clin. North Am. 23 (2), 193–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneline LS, Gobbett TA, Ramani R, Carreau M, Buchwald M, Yoder MC, Clapp DW, 1999. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood 94 (1), 1–8. [PubMed] [Google Scholar]
- Haneline LS, Li X, Ciccone SL, Li Y, Chen S, Srour EF, Yang FC, Broxmeyer HE, Clapp DW, 2003. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc−/− hematopoietic stem cells and decreases the risk of clonal evolution. Blood 101 (4), 1299–1307. [DOI] [PubMed] [Google Scholar]
- He J, et al. , 2012. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic acids research 40, 6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M, 2003. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17, 2021–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, 2013. May 23. Auwerx. JMitonuclear protein imbalance as a conserved longevity mechanism. Nature 497 (7450), 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen AJ, Trijbels FJ, Sengers RC, Smeitink JA, van den Heuvel LP, Wintjes LT, Stoltenborg-Hogenkamp BJ, Rodenburg RJ, 2007. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin. Chem. 53, 729–734. [DOI] [PubMed] [Google Scholar]
- Jovaisaite V, Mouchiroud L, Auwerx J, 2014. January 1. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 217 (Pt 1), 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Higgins CM, Xu Z, 2000. Measuring the quantity and activity of mitochondrial electron transport chain complexes in tissues of central nervous system using blue native polyacrylamide gel electrophoresis. Anal. Biochem. 286, 214–223. [DOI] [PubMed] [Google Scholar]
- Kelly PF, Radtke S, von Kalle C, Balcik B, Bohn K, Mueller R, Schuesler T, Haren M, Reeves L, Cancelas JA, Leemhuis T, Harris R, Auerbach AD, Smith FO, Davies SM, Williams DA, 2007. Stem cell collection and gene transfer in Fanconi anemia. Mol. Ther. 15 (1), 211–219. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, D’Andrea AD, 2000. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19, 2925–2940. [DOI] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD, 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26 (13), 1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies K, Inano S, Ramírez MJ, Ishiai M, Surrallés J, Takata M, Schindler D, 2017. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Invest. 127 (8), 3013–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli L, Passegué E, 2014. Surviving change: the metabolic journey of hematopoietic stem cells. Trends Cell Biol 24 (8), 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A, 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493 (7432), 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari U, Ya Jun W, Huat Bay B, Lyakhovich A, 2014. January 9. Evidence of mitochondrial dysfunction and impaired ROS detoxifying machinery in Fanconi anemia cells. Oncogene. 33 (2), 165–172. [DOI] [PubMed] [Google Scholar]
- Kutler DI, Singh B, Satagopan J, et al. , 2003. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101, 1249–1256. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD, 2009. Reactive oxygen species production by mitochondria. Methods Mol. Biol. 554, 165–181. [DOI] [PubMed] [Google Scholar]
- Li J, Sejas DP, Zhang X, et al. , 2007. TNF-α induces leukemic clonal evolution ex vivo in Fanconi anemia group C stem cells. J. Clin. Invest. 117, 3283–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, et al. , 2014. ATAD3 is a limiting factor in mitochondrial biogenesis and adipogenesis of white adipocyte-like 3T3-L1 cells. Mol. Cell. Biol. 37, e01170–13. [DOI] [PubMed] [Google Scholar]
- McKee EE, Ferguson M, Bentley AT, Marks TA, 2006. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob. Agents Chemother. 50, 2042–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Locatelli F, Stary J, Smith FO, 2010. Bone marrow transplantation for inherited bone marrow failure syndromes. Pediatr. Clin. N. Am. 57 (1), 147–170. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay SS, Leung KS, Hicks MJ, Hastings PJ, Youssoufian H, Plon SE, 2006. October 23. Defective mitochondrial peroxiredoxin-3 results in sensitivity to oxidative stress in Fanconi anemia. J Cell Biol. 175 (2), 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec EE, Wu L, Swaney SM, Chosay JG, Ross DE, Brieland JK, Leach KL, 2005. Oxazolidinones inhibit cellular proliferation via inhibition of mitochondrial protein synthesis. Antimicrob. Agents Chemother. 49, 3896–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Herrmann JM, 2010. June. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim. Biophys. Acta 1803 (6), 767–775. [DOI] [PubMed] [Google Scholar]
- Pagano G, Talamanca AA, Castello G, d’Ischia M, Pallardó FV, Petrović S, Porto B, Tiano L, Zatterale A, 2013. August. Bone marrow cell transcripts from Fanconi anaemia patients reveal in vivo alterations in mitochondrial, redox and DNA repair pathways. Eur. J. Haematol. 91 (2), 141–151. [DOI] [PubMed] [Google Scholar]
- Pagano G, Shyamsunder P, Verma RS, Lyakhovich A, 2014. April 21. Damaged mitochondria in Fanconi anemia - an isolated event or a general phenomenon? Oncoscience 1 (4), 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Ciccone SL, Beck BD, Hwang B, Freie B, Clapp DW, Lee SH, 2004. Oxidative stress/damage induces multimerization and interaction of Fanconi anemia proteins. J. Biol. Chem. 279, 30053–30059. [DOI] [PubMed] [Google Scholar]
- Ponte F, Sousa R, Fernandes AP, Gonçalves C, Barbot J, Carvalho F, Porto B, 2012. May 16. Improvement of genetic stability in lymphocytes from Fanconi anemia patients through the combined effect of α-lipoic acid and N-acetylcysteine. Orphanet J Rare Dis 7 (28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam AC, Hobson MJ, Ciccone SL, Li Y, Chen S, Srour EF, Yang FC, Broxmeyer HE, Clapp DW, 2008. AMD3100 synergizes with G-CSF to mobilize repopulating stem cells in Fanconi anemia knockout mice. Exp. Hematol. 36 (9), 1084–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravera S, Vaccaro D, Cuccarolo P, Columbaro M, Capanni C, Bartolucci M, Panfoli I, Morelli A, Dufour C, Cappelli E, Degan P, 2013. October. Mitochondrial respiratory chain Complex I defects in Fanconi anemia complementation group A. Biochimie. 95 (10), 1828–1837. [DOI] [PubMed] [Google Scholar]
- Rousset S, Nocentini S, Rouillard D, Baroche C, Moustacchi E, 2002. Mitochondrial alterations in Fanconi anemia fibroblasts following ultraviolet A or psoralen photoactivation. Photochem. Photobiol. 75, 159–166. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ, 2007. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76, 701–722. [DOI] [PubMed] [Google Scholar]
- Saadatzadeh MR, Bijangi-Vishehsaraei K, Hong P, Bergmann H, Haneline LS, 2004. April 16. Oxidant hypersensitivity of Fanconi anemia type C-deficient cells is dependent on a redox-regulated apoptotic pathway. J. Biol. Chem. 279 (16), 16805–16812. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kähkönen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian Genomics, et al. , 2015. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 5 (2), 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T, et al. , 2010. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7 (3), 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtić M, Sriskanthadevan S, Jhas B, Gebbia M, et al. , 2011. November 15. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20 (5), 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink JA, et al. , 2006. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. American journal of human genetics 79, 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, Wagner JE, 2012. Current clinical management of Fanconi anemia. Expert Rev Hematol 5 (5), 513–522. [DOI] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL, 2011. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310. [DOI] [PubMed] [Google Scholar]
- Takubo K, Kocabas F, Zheng J, et al. , 2013. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12 (1), 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, et al. , 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7, 241–248. [DOI] [PubMed] [Google Scholar]
- Tischkowitz MD, Hodgson SV, 2003. Fanconi anaemia. J. Med. Genet. 40, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounce IA, Kim YL, Jun AS, Wallace DC, 1996. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 264, 484–509. [DOI] [PubMed] [Google Scholar]
- Usai C, Ravera S, Cuccarolo P, Panfoli I, Dufour C, Cappelli E, Degan P, 2015. January 28. Dysregulated Ca2+ homeostasis in Fanconi anemia cells. Sci Rep. 5, 8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, et al. , 2007. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. American journal of human genetics 80, 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Pietras EM, Passegué E, 2011. Nov-Dec. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med. 3 (6), 681–701. [DOI] [PubMed] [Google Scholar]
- Zanier R, Briot D, Dugas du Villard JA, Sarasin A, Rosselli F, 2004. Fanconi anemia C gene product regulates expression of genes involved in differentiation and inflammation. Oncogene 23, 5004–5013. [DOI] [PubMed] [Google Scholar]
- Zhang T, Du W, Wilson AF, Namekawa SH, Andreassen PR, Meetei AR, Pang Q, 2017. April 5. Fancd2 in vivo interaction network reveals a non-canonical role in mitochondrial function. Sci. Rep. 7, 45626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.