Abstract
The apolipoprotein L1 (APOL1) risk variants G1 and G2 are associated with high rates of kidney disease in African Americans in genetic studies. However, our understanding of APOL1 biology has lagged far behind. Here we report that engineering G1 and G2 mutations on unnatural haplotype backgrounds instead of on the specific G1 and G2 haplotype backgrounds that occur in nature profoundly alters APOL1-mediated cytotoxicity in experimental systems. Thus, in addition to helping resolve some important controversies in the APOL1 field, our demonstration of the critical influence of haplotype background may apply more generally to the study of other genetic variants that cause or predispose to human disease.
Keywords: chronic kidney disease, FSGS
Introduction
Genetic variants in the ApolipoproteinL1 (APOL1) gene are associated with high rates of kidney disease in African Americans.1, 2 Two APOL1 risk alleles greatly increase risk of FSGS, H-ESRD, HIV nephropathy, and several glomerular phenotypes with features of collapsing nephropathy.1, 3, 4 Most but not all data to date support a model in which APOL1 kidney risk variants exhibit toxic, gain-of-function properties despite their generally recessive mode of inheritance.3 However, some investigators have observed no increase in risk variant-induced toxicity relative to wild-type APOL1, leaving open the possibility that loss-of-function mechanisms explain kidney injury.5 Even among proponents of gain-of-function models, inconsistent results regarding APOL1 trafficking, binding partners, intracellular location, and mechanisms driving cell death are difficult to reconcile, impeding our understanding of disease mechanism.
Many reports have demonstrated experimental differences between the behavior of wild-type APOL1 (also referred to as G0, or non-risk), and those of risk variants G1 and G2.6–16 We have previously observed that human variation includes APOL1 proteins containing multiple coding variants organized into distinct haplotypes, including at least 8 relatively common non-risk haplotypes with different coding sequences (Supplementary Tables 1 and 2). In other words, there is no canonical “wild-type” APOL1 but rather many non-risk haplotypes that differ from each other by at least one and sometimes several amino acids.17 Both risk variants G1 (S342G and I384M, two amino acid substitutions that nearly always occur together) and G2 (del388-9, a two amino acid deletion) are inherited on one specific human haplotype background. One non-risk APOL1 sequence, which we refer to as the Reference APOL1 haplotype, is the nucleotide and protein sequences found in most databases and encoded by most commercially available APOL1 cDNA plasmids. Studying G1 or G2 behavior by generating the G1 or G2 mutations on the Reference haplotype background creates APOL1 sequences that, to our knowledge, have not yet been observed in human genomes. We hypothesized that use of many different non-risk APOL1 amino acid sequences as “wild-type” APOL1, together with study of G1 and G2 variants on a variety of haplotype backgrounds (including ones that do not occur naturally), may have contributed to the range of APOL1 experimental results reported to date by different laboratories.
To that end, we compared the experimental consequences of overexpressing multiple different non-risk APOL1 haplotypes. We further examined the properties of G1 and G2 risk variants on different haplotype backgrounds, comparing the natural human haplotypes with artificial haplotypes rarely if ever observed in nature but sometimes inadvertently studied in previous publications. Our results demonstrate a strong influence of haplotype background on APOL1 function for both wild-type and risk variant forms of APOL1.
Results
We previously showed that transient expression in HEK293 cells of the Reference APOL1 haplotype was much less toxic than G1 or G2 on their native haplotype background.11 Reference (Ref) APOL1 is actually a minor APOL1 haplotype rarely found in Africans, but present in ~ 20-25% of non-African chromosomes, likely entering the human genome from Neandertal after the out-of-Africa expansion.17 We found that transient expression of Reference APOL1, found chiefly in people from regions without trypanosomal disease, caused less toxicity than did the most common APOL1 haplotype worldwide (labelled G0 in our figures; Fig. 1A and 1B). We therefore generated plasmids encoding the common APOL1 wild-type/non-risk variants (Supplemental Table 1). These non-risk APOL1 wild-type alleles differed widely in the toxicity they induced when transiently expressed in cells (Fig. 1B), though none were as cytotoxic as G1 or G2.
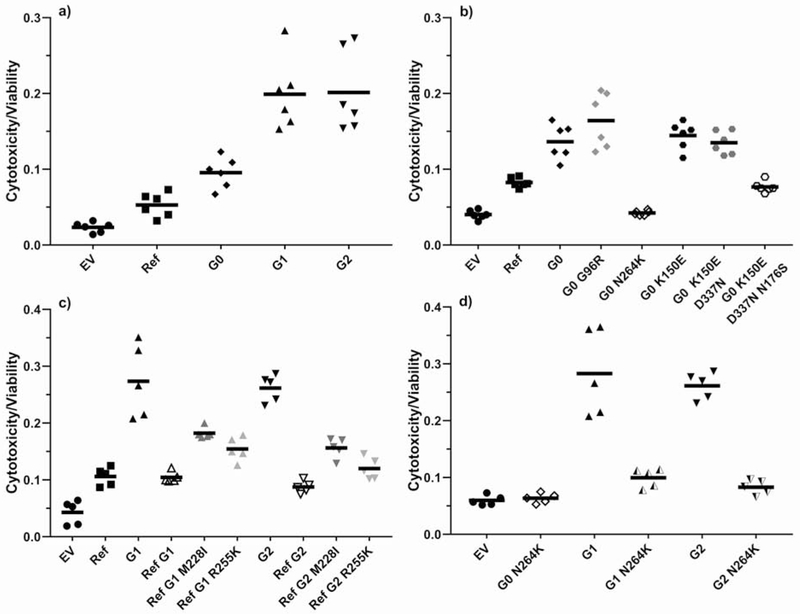
Figure 1.
APOLl-induced cytotoxicity in HEK293 cells transiently transfected using Lipofectamine 2000. The cytotoxicity-to-viability ratio was measured after 28 h using the MultiTox-Fluor Multiplex Cytotoxicity Assay.
a) Risk variants G1 and G2 demonstrate higher cytotoxicity than non-risk variants Ref and G0. P<0.001 for risk variants G1 and G2 vs non-risk variants Ref and G0.
b) Common APOL1 non-risk haplotypes demonstrate variable cytotoxicity but do not reach the toxicity levels of risk variants G1 and G2. P<0.001 for G0 vs Ref, G0 N264K, and G0 K150E D337N N176S.
c) G1 and G2 mutations on a Ref background show no difference in cytotoxicity compared to Ref APOL1 (M228/R255). Mutating either amino acid 228 (M to I) or 255 (R to K) in these artificial Ref G1 and Ref G2 variants back to the natural haplotype background residue restores about half the toxicity difference between the artificial G1 and G2 constructs on the Ref background versus their natural backgrounds. P<0.01 for G1 vs Ref G1, Ref G1 M228I, and Ref G1 R225K. P<0.001 for G2 vs. Ref G2, Ref G2 M228I, and Ref G2 R225K.
d) A N264K mutation on a G1 or G2 background completely abrogates risk variant toxicity. P<0.001 for G1 vs G1 N264K and for G2 vs. G2 N264K.
Since G1 and G2 occur naturally on one specific haplotype background, we compared the toxicity of natural G1 and G2 haplotypes with artificial haplotypes created by mutagenic insertion of G1 and G2 amino acid variants onto other non-risk APOL1 haplotypes. We found that G1 and G2 toxicity was dramatically reduced when expressed on either the common Reference haplotype background or the haplotype defined by N264K previously associated with loss-of-function for trypanolysis (Figs. 1C, 1D, S1).18 The Reference APOL1 haplotype includes both I228M and K255R polymorphisms not found in any other common APOL1 haplotypes. Each of the two polymorphisms independently explained about half of the reduction in toxicity of G1 and G2 on the Reference background (Fig. 1C). These results demonstrate that amino acid changes quite distant from the G1 (S342G and I384M) or G2 (del388-9) markedly affect the propensity of G1 and G2 to injure cells.
We also generated cell lines that stably express these different APOL1 haplotypes under the control of a tetracycline-inducible promoter to validate our findings in the absence of lipid transfection reagents or plasmids (bacterial DNA). We observed that the cytotoxicity difference between G0 and the risk variants G1 and G2 was larger in the stable inducible system than in transient transfections, but Reference and G0 were indistinguishable (Fig. 2A). Again we found that G1 and G2 were much less toxic on artificial haplotype backgrounds than when expressed on their native, naturally-occurring haplotypes (Fig. 2B, 2C, S2–4). We found consistent differences of much smaller magnitude when expressing G1 or G2 on a background containing E150 (natural) vs K150 (artificial) (Supplementary Figure S5). We confirmed these findings using a second type of cell death assay (MTT) (Fig. 2D, 2E).
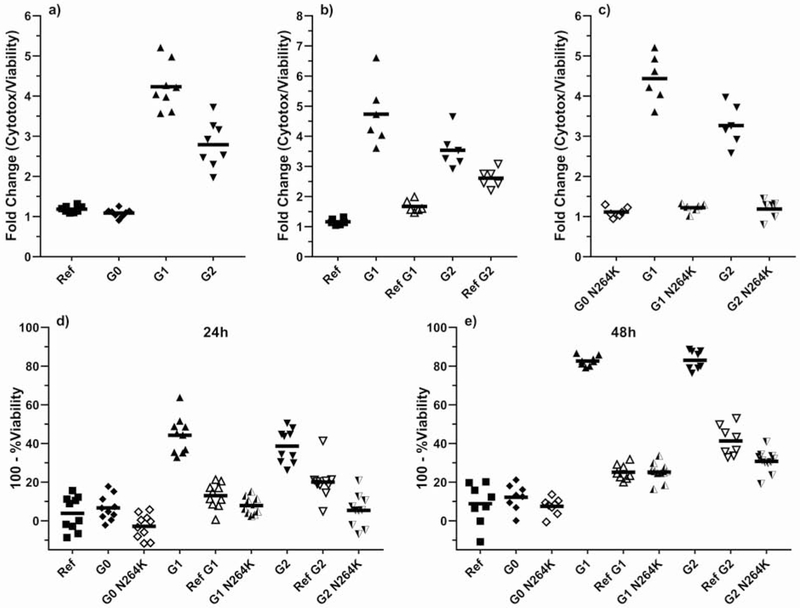
Figure 2.
Toxicity in inducible T-Rex 293 cells stably expressing APOL1 of different haplotypes. APOL1 expression was induced by addition of tetracycline. Cytotoxicity was measured using either the cytotoxicity-to-viability ratio (a-c) or by MTT assay (d-e). Each cell line’s data are normalized to the corresponding no-tetracycline control. Similar APOL1 expression levels among cell lines are demonstrated in Supplementary Figures S2 (mRNA) and S3 (protein); Supplementary Figure S3 demonstrates that APOL1 transgene expression does not activate endogenous APOL1 expression.
a) Risk variants G1 and G2 demonstrate higher cytotoxicity than non-risk variants Ref and G0. P<0.001 for risk variants G1 and G2 vs. non-risk variants Ref and G0.
b) G1 and G2 mutations on a Ref haplotype background demonstrate reduced cytotoxicity compared to naturally-occurring G1 and G2. P<0.001 for G1 vs. Ref G1. P<0.01 for G2 vs. Ref G2.
c) G1 or G2 mutations on the N264K haplotype background shows no difference in cytotoxicity as compared to G0 N264K. P<0.001 for G1 vs G1 N264K and for G2 vs. G2 N264K.
d) After 24 hours of tetracycline induction, cells expressing G1 or G2 exhibit the lowest viability, with near-complete rescue by engineering G1 and G2 into Ref or G0 N264K backgrounds. P<0.001 for G1 vs Ref G1 and G1 N264K. P<0.001 for G2 vs. Ref G2 and G2 N264K.
e) After 48 hours of tetracycline induction, cells expressing G1 and G2 exhibit more toxicity than G1 and G2 on the Ref or G0 N264K haplotype backgrounds. P<0.001 for G1 vs. Ref G1 and G1 N264K. P<0.001 for G2 vs. Ref G2 and G2 N264K.
Discussion
The disease-relevant biology of APOL1 remains elusive nearly a decade after its discovery as an important kidney disease risk factor in African Americans, at least in part because of many conflicting published experimental results. Some of these discrepancies may reflect investigators working with different APOL1 constructs, including sequences that probably do not occur in nature (summarized in Supplementary Table 3). In most reports the full APOL1 sequences used in experiments are not provided, further complicating data interpretation. Our in vitro findings cannot yet be extrapolated directly to human disease but are highly relevant for understanding the function of APOL1.
We explored natural human variation in the APOL1 gene and protein with respect to their propensity to cause cytotoxicity, a key phenotype in the gain-of-function model. We found that haplotype background profoundly influences the effects of transient and stable APOL1 overexpression. One APOL1 haplotype, which by chance became the Reference APOL1 sequence, was likely inherited by humans from Neandertal via introgression long after modern humans migrated out of Africa.17 Though modern humans have on average ~2-3% Neandertal DNA, Reference APOL1 haplotype has increased in frequency to >20% in much of Europe and Asia, suggesting positive selection in environments without trypanosomes. That some non-risk APOL1 haplotypes may confer toxicity in humans is consistent with loss of the APOL1 gene from genomes of several primate species. A second APOL1 haplotype (tagged by N264K) in our experiments that was minimally toxic and mitigated toxicities of G1 and G2 was recently reported by Cuypers et al.18 to lack activity against certain trypanosomes, again supporting a possible link between APOL1 cytotoxicity and trypanolytic potency.
Our experiments with natural haplotypes and haplotype switches emphasize the large effects on G1- and G2-mediated cytotoxicity of other amino acid residues, even those quite distant from G1 or G2 in the linear sequence. These results suggest a more complicated view of risk variant toxicity that will require structural studies of the entire APOL1 protein, rather than of the C-terminal region alone, to understand functional differences among G0, G1, and G2.
Our work demonstrates that so-called “wild-type” APOL1 comprises a set of different haplotypes with clearly demonstrable biological differences in human kidney cells, and that G1 and G2 are best viewed as haplotypes rather than as specific mutations. We further show that insertion of G1 or G2 mutation into the Reference wild-type APOL1 haplotype background generates risk variant APOL1 polypeptides with properties markedly different from those of naturally-occurring APOL1 risk variants. While we think it is possible that APOL1 risk variants may also have some loss-of-function properties, our data support the gain-of-function hypothesis for APOL1 risk variants and may explain why some reports do not observe enhanced risk variant toxicity. We propose that conducting experiments with the APOL1 haplotypes found in nature, and accurately reporting haplotypes chosen for experimentation, could minimize differences in experimental results among groups and accelerate progress towards a more complete understanding of APOL1 biology and pathobiology. Our findings regarding the effect of APOL1 haplotype background may have important implications for the study of many other disease-causing genes as the catalog of human variation (both individual variants and haplotype structure) continues to grow.
Short Methods
Cytotoxicity Assays
Cytotoxicity/Viability ratios were measured using the MultiTox-Fluor Multiplex Cytotoxicity Assay (Promega) and MTT assays (Life Technologies). HEK293 (ATCC) cells were transfected with 100ng pCMV6 plasmid encoding untagged APOL1 using Lipofectamine2000 (Life Technologies). Stable T-Rex 293 cells generated using the Flp-In T-Rex system (Life Technologies) were induced with 100ng/mL tetracycline. Cytotoxicity measurements for stable T-Rex 293 cells were normalized to no-tetracycline controls. Detailed experimental methods and gene sequences are provided in the Supplement.
Statistics
We used ANOVA with post-testing (Dunnett) for comparisons involving multiple groups or standard two-tailed t-tests for comparisons involving only two groups. P < 0.05 after post-testing was considered statistically significant.
Supplementary Material
Acknowledgments:
We are grateful to funding from the National Institutes of Health (NIMHD), Department of Defense, the Doris Duke Charitable Foundation, and NephCure for this work.
Support: National Institutes of Health (NIMHD), Department of Defense, the Doris Duke Charitable Foundation, NephCure
Disclosure: DJF and MRP are named on patents filed by BIDMC related to APOL1 diagnostics and therapeutics; hold equity in Apolo1bio; and receive research support from Vertex.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary information is available at the Kidney International website.
References:
- 1.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010; 329: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 2010; 128: 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman DJ, Pollak MR. Apolipoprotein L1 and Kidney Disease in African Americans. Trends Endocrinol Metab 2016; 27: 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp JB, Nelson GW, Sampath K, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 2011; 22: 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Toole JF, Schilling W, Kunze D, et al. ApoL1 Overexpression Drives Variant-Independent Cytotoxicity. J Am Soc Nephrol 2018; 29: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckerman P, Bi-Karchin J, Park AS, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med 2017; 23: 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Zhu JY, Richman A, et al. APOL1-G1 in Nephrocytes Induces Hypertrophy and Accelerates Cell Death. J Am Soc Nephrol 2017; 28: 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan X, Jhaveri A, Cheng K, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. American journal of physiology Renal physiology 2014; 307: F326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan X, Wen H, Saleem MA, et al. Vascular smooth muscle cells contribute to APOL1-induced podocyte injury in HIV milieu. Exp Mol Pathol 2015; 98: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, Chou JW, Snipes JA, et al. APOL1 Renal-Risk Variants Induce Mitochondrial Dysfunction. J Am Soc Nephrol 2017; 28: 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols B, Jog P, Lee JH, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int 2015; 87: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olabisi OA, Zhang JY, VerPlank L, et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A 2016; 113: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen H, Kumar V, Lan X, et al. APOL1 Risk Variants Cause Podocytes Injury through Enhancing Endoplasmic Reticulum Stress. Biosci Rep 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruzel-Davila E, Shemer R, Ofir A, et al. APOL1-Mediated Cell Injury Involves Disruption of Conserved Trafficking Processes. J Am Soc Nephrol 2017; 28: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayek SS, Koh KH, Grams ME, et al. A tripartite complex of suPAR, APOL1 risk variants and alphavbeta3 integrin on podocytes mediates chronic kidney disease. Nat Med 2017; 23: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granado D, Muller D, Krausel V, et al. Intracellular APOL1 Risk Variants Cause Cytotoxicity Accompanied by Energy Depletion. J Am Soc Nephrol 2017; 28: 3227–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson R, Genovese G, Canon C, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A 2014; 111: E2130–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuypers B, Lecordier L, Meehan CJ, et al. Apolipoprotein L1 Variant Associated with Increased Susceptibility to Trypanosome Infection. MBio 2016; 7: e02198–02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.