Abstract
Alzheimer’s disease (AD) is a debilitating dementia with complex pathophysiological alterations including modifications to endogenous cysteine. S-nitrosylation (SNO) is a well-studied posttranslational modification (PTM) in the context of AD while S-glutathionylation (PSSG) remains less studied. Excess reactive oxygen and reactive nitrogen species (ROS/RNS) directly or indirectly generate SNO and PSSG. SNO is dysregulated in AD and plays a pervasive role in processes such as protein function, cell signaling, metabolism, and apoptosis. Despite some studies into the role of SNO in AD, multiple identified SNO proteins lack deep investigation and SNO modifications outside of brain tissues are limited, leaving the full role of SNO in AD to be elucidated. PSSG homeostasis is perturbed in AD and may affect a myriad of cellular processes. Here we overview the role of nitric oxide (NO•) in AD, discuss proteomic methodologies to investigate SNO and PSSG, and review SNO and PSSG in AD. A more thorough understanding of SNO, PSSG, and other cysteinyl PTMs in AD will be helpful for the development of novel therapeutics against neurodegenerative diseases.
1. Introduction
Alzheimer’s disease (AD) is the most prevalent form of human dementia, affecting over one third of persons aged 85 years or older and with over 5 million cases in the United States (Zolochevska, Bjorklund, Woltjer, Wiktorowicz, & Taglialatela, 2018). Characterized by extracellular amyloid-β (Aβ) plaques and intracellular neurofibrillary tangles, the disease involves the progressive loss of neurons and neural synapses amid complex pathophysiological alterations in the metabolism of neurons and glial cells (Combs, Hamel, & Kanaan, 2016; Selkoe, 2002). Implicated in such metabolic changes is the increased production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) including the gasotransmitter nitric oxide (NO•). Elevated levels of NO• in particular have been demonstrated to result in posttranslational modifications (PTMs) of proteins associated with mitochondrial dysfunction and bioenergetic compromise (Nakamura & Lipton, 2017).
Cysteine is an amino acid particularly vulnerable to modification in oxidative stress conditions, capable of undergoing a plethora of reversible modifications including S-nitrosylation (SNO) and S-glutathionylation (PSSG) (Butterfield, Gu, Di Domenico, & Robinson, 2014; Lennicke et al., 2016). Although it has a rare amino acid frequency of 2.6% in humans (Kozlowski, 2017), cysteine is known to be important to signaling and cellular redox status and is often found near the center of enzyme active sites (Baez, Reisz, & Furdui, 2015). SNO is believed to be specifically regulated and dysfunction of physiological SNO regulation can be critical to the pathogenesis of neurological disease (Foster, Hess, & Stamler, 2009; López-Sánchez, López-Pedrera, & Rodríguez-Ariza, 2014). Due to its low endogenous abundance and reversible nature, however, SNO presents challenges for in vitro analysis (Couvertier, Zhou, & Weerapana, 2014).
Proteomics, the study of the entire complement of proteins in cells, tissues, or organisms, offers techniques for the investigation of labile protein modifications such as SNO. Carefully designed high-throughput mass spectrometric methods allow cysteine modifications to be probed across hundreds and potentially thousands of proteins. Identified proteins may then be thoroughly investigated to elucidate the function of the cysteinyl modification, allowing a methodological approach in unraveling the roles of cysteine modifications in disease states. This review discusses the physiological function of NO• and SNO in the brain, common proteomic methodology for the analysis of SNO proteins, and findings of aberrant SNO in AD. Additionally PSSG, another cysteine PTM that is less studied in AD, will be presented.
2. Physiological and pathological NO
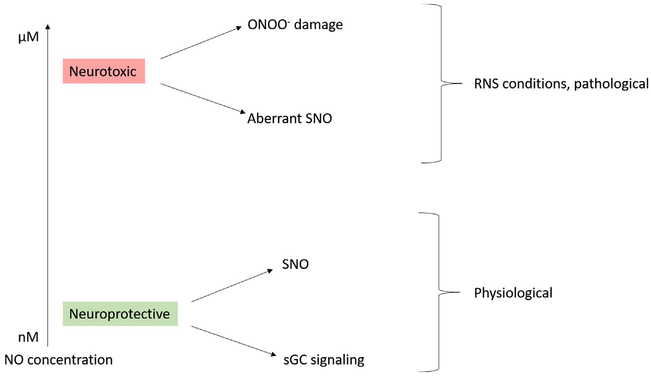
NO• is synthesized from L-arginine and oxygen via neuronal nitric oxide synthase (nNOS, NOS-1) with NADPH and calcium-calmodulin as cofactors within the brain. NO• also may be synthesized from inducible NOS (iNOS, NOS-2) or endothelial NOS (eNOS, NOS-3) under certain conditions (Förstermann & Sessa, 2012). Physiologically, NO• increases the activity of soluble guanylyl cyclase (sGC) to increase guanosine 3′,5′-cyclic monophosphate (cGMP) signaling and nitrosylating thiols of target proteins. Pathologically, NO• nitrosylates aberrant thiols and reacts with superoxide to form peroxynitrite leading to tyrosine nitration (El-Sehemy, Postovit, & Fu, 2016). The overall effect of NO• depends on the concentration of NO•. At low nanomolar concentrations, NO• provides pro-survival effects while at micromolar concentrations NO• exhibits pro-death effects due to unique radical reactivity of NO• and its capability to cross nonpolar membranes and access buried protein residues (Bradley & Steinert, 2016). Thus, NO• exhibits a dual role which may be neuroprotective under physiological conditions or neurodestructive under pathological conditions of increased RNS as highlighted in Fig. 1. A detailed description of NO• physiological function and its role in synaptic transmission and various diseases are reviewed elsewhere (Bradley & Steinert, 2016; Maher, Rahman, & Gad, 2017).
Fig. 1.
The dual effects of NO concentration on cell viability. At low concentrations NO• exhibits neuroprotective effects via soluble guanylyl cyclase (sGC) signaling and physiological SNO while at RNS concentrations NO exhibits neurotoxic effects via aberrant SNO and the generation of damaging peroxynitrite.
As neural NO• is produced largely by nNOS, Ca2+ levels are indirectly responsible for NO• levels. The nNOS isoform (as well as eNOS) requires greater than homeostatic Ca2+ levels in order to bind calmodulin for NO• synthesis. The iNOS isoform lacks the Ca2+ requirement and produces NO• at homeostatic conditions but is not known to be expressed in neurons at physiological conditions (Förstermann & Sessa, 2012; Mayer & Andrew, 1998). A primary source of short-term neural Ca2+ increase is the N-methyl-d-aspartate receptor (NMDAR), a nonselective cation channel which opens in response to glutamate binding. Stimulation of neurons by glutamate thus indirectly activates nNOS and increases NO• levels (Garthwaite, Charles, & Chess-Williams, 1988; Pierrefiche & Naassila, 2014). Under pathological AD conditions, Aβ1–42 oligomers stimulate α7 nicotinic acetylcholine receptors of neighboring astrocytes, resulting in a release of glutamate which in turn stimulates neuronal NMDAR’s. This flood of glutamate, known as excitotoxicity, disrupts neural Ca2+ homeostasis and generates RNS through the overproduction of NO• (Talantova et al., 2013).
3. S-nitrosylation
The putative formation of SNO results from the non-enzymatic reaction of a protein thiolate anion (RS−) with a nitrosonium cation (NO+), the latter formed from the metal-catalyzed oxidation of NO• (Lipton et al., 1993; Martínez-Ruiz, Cadenas, & Lamas, 2011; Nakamura et al., 2015). Radical recombination of protein thiyl and NO• radicals has additionally been proposed as a source of SNO (Smith & Marletta, 2012). Due to the non-enzymatic formation of SNO and contrasting endogenous SNO formation, several regulatory processes have been proposed: the subcellular environment, site specificity, and rate of denistrosylation. Random diffusion has been viewed as the primary transport mechanism and limits NO• distribution from generation sites. Thus, the formation of SNO may be regulated by the compartmentalization of SNO proteins in close proximity to NOS (Derakhshan, Hao, & Gross, 2007). The protein microenvironment of SNO residues influences the favorability of SNO formation, although variations in SNO proteins across variables such as pKa, hydrophobicity, solvent accessibility, charge of nearby residues, and presence or lack of metal coordination suggest an array of potential mechanisms for selective SNO formation (Doulias et al., 2010). Although labile, many SNO modifications do not spontaneously reverse in vivo and must be reduced enzymatically, such as by the thioredoxin or S-nitrosoglutathione (GSNO) reductase systems. The selective enzymatic reduction of physiologically stable SNO sites may thus provide an additional layer of SNO specificity (Benhar, Forrester, & Stamler, 2009).
Recently, the paradigm of non-enzymatic SNO formation has been challenged by a model of S-nitrosylases in which NO• enzymes generate proximity-based SNO proteins which then propagate the SNO modification throughout the cell via transnitrosylation (Seth et al., 2018). Knocked out genes in E. coli, that were induced during aerobic respiration on nitrate (ARN), resulted in variations in global SNO levels but not in NO• generation (Seth et al., 2018). The greatest reduction in SNO occurred during knockout of the hybrid cluster protein Hcp (Seth et al., 2018). Further analysis reinforced the S-nitrosylase role of Hcp, suggesting an ubiquitination-like model in which an NO• synthase (e.g., the nitrate reductase NarGHI) synthesizes NO•, NO• S-nitrosylates Hcp, then Hcp transnitrosylates proteins with transnitrosylase ability such as glyceraldehyde 3-phosphate dehydrogenase (GADPH). Due to the existence of mammalian transnitrosylases and S-nitrosylase metalloproteins, this S-nitrosylase framework may have applicability beyond bacterial systems (Seth et al., 2018).
Indeed, the propensity of certain SNO proteins to transfer the NO moiety to other proteins is well-established and has been proposed as a mechanism for the selectivity of S-nitrosylation (Anand & Stamler, 2012; Nakamura & Lipton, 2013; Stomberski, Hess, & Stamler, 2019). Chemically, this may be achieved due to the potential variability in bond character and differential stabilization of resonance structures based on the SNO microenvironment (Houk et al., 2003; Talipov & Timerghazin, 2013). Environments promoting nitrogen electrophilicity would favor a transnitrosylation reaction to another thiol while environments promoting sulfur electrophilicity would favor disulfide formation with another thiol such as glutathione (GSH) (Houk et al., 2003; Talipov & Timerghazin, 2013). Despite the establishment of transnitrosylases, human S-nitrosylases remain less investigated. Hemoglobin serves as an example of an established S-nitrosylase, capable of binding NO• to heme, S-nitrosylating Cysβ93 of hemoglobin, and transferring SNO to other protein or peptide thiols (Angelo, Singel, & Stamler, 2006). Another example raises the formation of a S-nitrosylation complex consisting of iNOS, S100A8, S100A9, and target proteins, whereby iNOS generated NO•, S100A8 and S100A9 directed target site-specificity, and S100A9 became S-nitrosylated and transnitrosylated 95 target proteins identified by proteomic analysis (Jia et al., 2014). The framework of enzymatic S-nitrosylation, however, requires broader evidence in human systems. Should the enzymatic SNO formation model hold, it may provide new scope into the investigation of S-nitrosylation in neurodegenerative diseases such as Alzheimer’s disease (AD).
Attempts to assign SNO motifs based on primary or secondary sequence have largely failed to account for all SNO sites identified in the corresponding studies (Chen et al., 2010; Doulias et al., 2010; Stamler, Toone, Lipton, & Sucher, 1997; Stomberski et al., 2019). Rather, the local microenvironment of target thiols accounting for tertiary and quaternary structure appears to best account for SNO site selectivity (Marino & Gladyshev, 2010). Indeed, computational tertiary structure analysis of 1250 known human SNO sites demonstrated ~86% of SNO sites were physically near charged residues without a primary sequence motif consensus (Chen et al., 2014; Stomberski et al., 2019). However, the specific local conditions of SNO formation remain to be fully elucidated and warrant further investigation.
In the context of AD, SNO proteins have been studied largely on a single to several protein basis with thorough investigations into the role each SNO protein plays in disease pathology. High-throughput proteome-wide studies of specific brain regions or structures have additionally been performed although to a much smaller extent than targeted investigations (Correani et al., 2015; Gu & Robinson, 2016; Riederer, Schiffrin, Kovari, Bouras, &Riederer, 2009; Seneviratne et al., 2016; Wang et al., 2011, 2012; Zareba-Koziol, Szwajda, Dadlez, Wyslouch-Cieszynska, & Lalowski, 2014). Taken as a whole, investigations into SNO proteins in AD present pervasive SNO dysregulation in AD brain tissue, suggesting the modification plays a critical role in disease pathogenesis. The specific effects of SNO in AD vary from protein to protein which is discussed in-depth in later sections. Also discussed are the findings of high-throughput studies, noting the full role of SNO in AD remains incomplete.
4. S-glutathionylation
Glutathione is a tripeptide consisting of glutamate, cysteine, and glycine; S-glutathionylation refers to the covalent attachment of glutathione to a protein at a cysteine residue via a disulfide bridge between the protein cysteine and glutathione cysteine. Glutathione exists in a cycle with glutathione disulfide (GSSG); GSH enzymatically reacts with PSSG to remove the S-glutathionylation modification and form GSSG which is then enzymatically reduced into two molecules of GSH. This allows glutathionylation to act as a regenerating cellular redox buffer against ROS (Johnson, Wilson-Delfosse, & Mieyal, 2012). Intracellular GSH can vary from 0.2–10mM and is found at 1–2mM for most cell types (Anderson, 1998). The ratio of GSH to GSSG meanwhile varies with cellular localization. Mitochondria for example typically exhibit a higher GSH:GSSG ratio than the cytosol, resulting in a more reducing environment (Wadey, Muyderman, Kwek, & Sims, 2009).
Historically, the glutathionylation/deglutathionylation cycle has been viewed as a process which acts mostly as a buffer against ROS/RNS via reducing aberrant cysteine modifications and thereby preventing the formation of damaging irreversible cysteine modifications. Recent perspectives suggest glutathionylation has a deeper role in cellular physiology with functions in cell signaling and alteration of protein activity (Zhang, Ye, Singh, Townsend, & Tew, 2018). Glutathionylation may play a deep role in the pathology of AD (Cordes, Bennett, Siford, & Hamel, 2009; Domenico et al., 2009; Lakunina, Petrushanko, Burnysheva, Mitkevich, & Makarov, 2017; Newman et al., 2007; Poulsen, Bahl, Simonsen, Hasselbalch, & Heegaard, 2014; Rani, Krishnan, & Rani Cathrine, 2017; Zhang, Kuo, Chiu, & Feng, 2012; Zhang, Rodriguez, Circu, Aw, & Feng, 2011).
5. Proteomic methods for SNO and PSSG protein identification
5.1. Shotgun proteomics
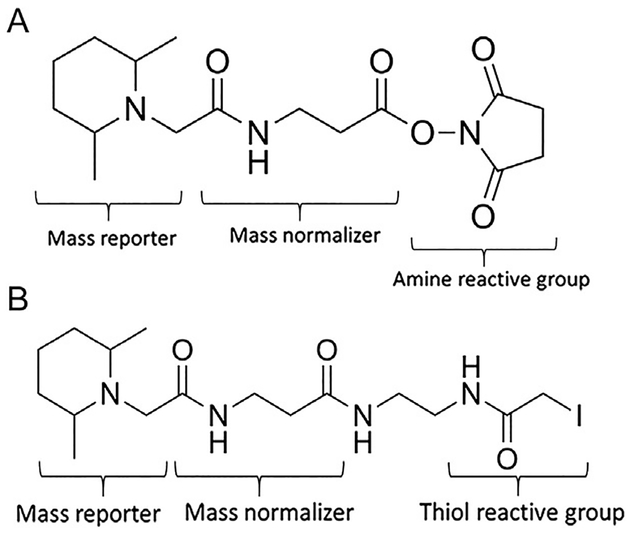
Shotgun proteomics is the analysis of complex peptide mixtures (Yates, 1998). A typical shotgun analysis entails experimental workup of the proteins of interest, separation by liquid chromatography (LC), analysis by tandem mass spectrometry (MS/MS), and data analysis with bioinformatic software. Typical MS and MS/MS analysis identifies full peptide mass to charge (m/z) ratios and m/z ratios of peptide fragments which allow determination of the peptide sequence (Zhang, Fonslow, Shan, Baek, & Yates, 2013). A variety of shotgun proteomics strategies have been developed for the relative quantification of cysteine PTMs. Isotopic labeling involves chemically modifying target peptides with reagents synthesized to contain light or heavy isotopes. As these labels are chemically identical but possess different masses, the mass difference can be detected within MS and thus distinguish experimental groups in a pooled sample (Hsu, Huang, Chow, & Chen, 2003). Isobaric tags such as tandem mass tag (TMT) involve the attachment of multiple isobaric chemical groups which produce unique mass ions during MS/MS analysis (Thompson et al., 2003). As displayed in Fig. 2A, TMT consists of three groups: a protein reactive group for attachment to primary amines (or protein thiols, shown in Fig. 2B), a mass reporter to produce a unique mass ion during MS analysis, and a mass normalizer to ensure each tag possesses the same exact mass (Thompson et al., 2003). TMT reagents are based on the location of heavy isotopes within its molecular structure. Only the mass reporter group is cleaved and detected during MS analysis, so each tag achieves a unique reporter ion mass by increasing the number of heavy isotopes in the mass reporter group while decreasing the number of heavy isotopes in the mass normalizer group (Dayon et al., 2008). Isotopic labeling and isobaric tagging thus allow relative quantification across multiple experimental groups in a single sample. Shotgun proteomic methods targeted to cysteine PTMs have incorporated these techniques.
Fig. 2.
Structure of tandem mass tags (TMT). TMT consists of a mass reporter, mass normalizer, and amine reactive group (A). Cys-reactive TMT reagents such as iodoTMT (B) replace the amine reactive group with a thiol reactive group.
5.2. SNO
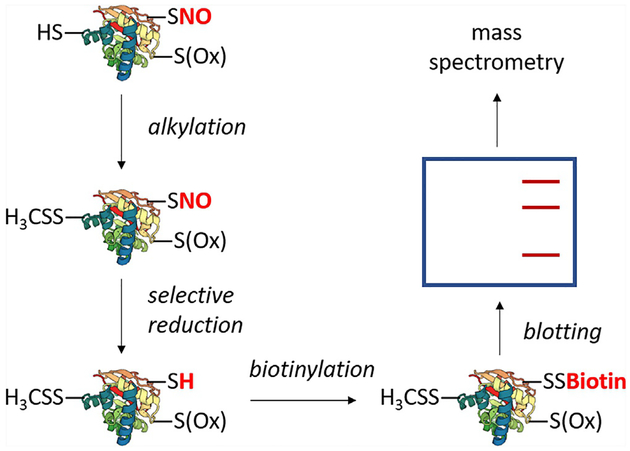
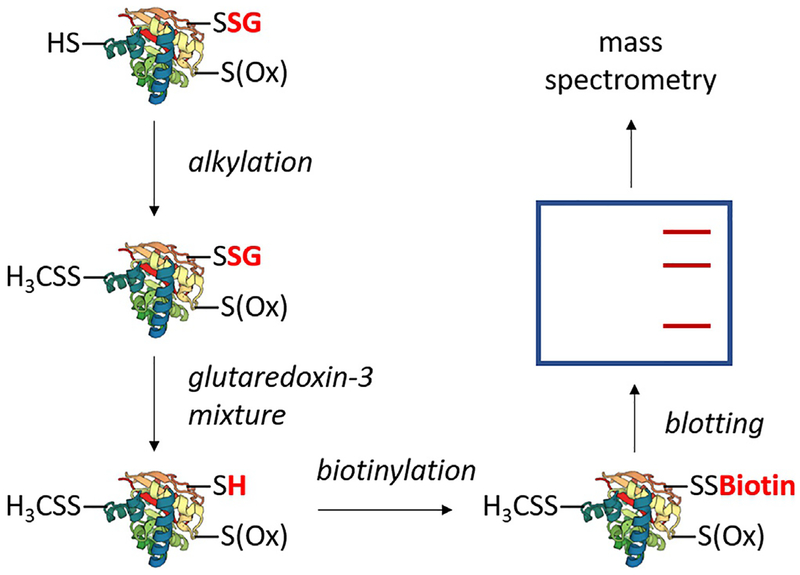
Various methods are aimed at the in vitro and in vivo study of SNO. In the context of AD, chief among these methods are variations of the biotin switch technique (BST), summarized in Fig. 3. The BST involves irreversibly alkylating all free cysteines, selectively reducing SNO, and biotinylating reduced SNO cysteines with a thiol-specific biotin reagent (Jaffrey, Erdjument-Bromage, Ferris, Tempst, & Snyder, 2001; Jaffrey & Snyder, 2001).
Fig. 3.
A generalized schematic of the biotin switch technique. The technique entails irreversible alkylation of free thiols, selective reduction of SNO, and biotinylation of SNO sites. Biotinylated proteins may be subject to blotting with anti-biotin or enriched with streptavidin followed by specific protein blotting. Bands can be excised, trypsinized, and subjected to mass spectrometry. Common variations include choice of thiol alkylating agent or replacement of biotin with an agent such as iodoTMT. Methods based on the biotin switch technique typically utilize modifications to the labeling and enrichment strategies, such as replacing biotin/streptavidin with a thiol-active resin (SNO-RAC) or direct tagging of SNO without prior selective reduction (SNOTRAP).
For samples with artificial induction of SNO, SNO proteins can be detected with anti-biotin immunoblotting but may not reflect in vivo SNO. For endogenous SNO, the low abundance of the modification typically requires streptavidin enrichment followed by Western blotting for specific proteins of interest (Jaffrey et al., 2001; Jaffrey & Snyder, 2001). Variations to the BST include different choices of cysteine alkylation agents (e.g., N-ethylmaleimide) (Correani et al., 2015; Kohr et al., 2011; Zahid et al., 2014), replacement of biotin with iodoTMT and anti-TMT Western blotting (Wang et al., 2017), and predominate use of MS analysis in place of blotting techniques. SNO Site Identification (SNOSID), a high-throughput proteomic modification to BST, added tryptic digestion after the biotin labeling step to allow selective pulldown of SNO peptides followed by LC-MS/MS analysis (Hao, Derakhshan, Shi, Campagne, & Gross, 2006). Another noteworthy modification to BST is SNO resin-assisted capture (SNO-RAC) (Forrester et al., 2009), an SNO-specific method more sensitive to high mass (>100kDa) proteins. SNO-RAC replaces biotin labeling and streptavidin enrichment with a thiol-specific resin, allowing the condensation of labeling and pulldown into a single step with protein analysis performed via Western blot or MS (Forrester, Thompson, et al., 2009). Drawbacks to the BST include many potential sources of error, generation of artifacts if not performed in darkness due to the nonspecific reaction of ascorbate with disulfides in light, and sensitivity to transition metals in solution due to nonspecific side reactions with absorbate (Forrester, Foster, Benhar, & Stamler, 2009). SNO-RAC suffers from similar drawbacks, remaining technically challenging despite a smaller number of sample preparation steps (Forrester, Thompson, et al., 2009). SNOSID has similar drawbacks but offers the ability to identify unknown endogenous SNO proteins, indirect identification of SNO sites, and is amenable to complex mixtures (López-Sánchez et al., 2014). Utilizing an anti-SNO antibody allows the direct confirmation of the presence of SNO proteins but the antibodies available are suitable for immunohistochemistry, not immunoprecipitation or Western blot (Torta, Usuelli, Malgaroli, & Bachi, 2008).
Several laboratories have developed other methods for studying SNO in AD (Gu & Robinson, 2016; Seneviratne et al., 2016; S. Wang et al., 2011) which we have recently reviewed (Gu & Robinson, 2016a). Capillary gel electrophoresis with laser induced fluorescent detection (CGE-LIF) is a method for the detection of SNO, replacing the biotin labeling step of BST with Dylight 488 maleimide to allow fluorescent detection with sensitivity in the picomolar range (Wang et al., 2011). CGE-LIF, however, lacks the ability to determine protein identities without use of a separate method such as BST (Wang et al., 2011). SNOTRAP is a technique similar to SNOSID, which follows free thiol alkylation with reaction of the SNO moiety with a triphenylphosphine thioester probe linked to biotin, allowing streptavidin enrichment followed by LC-MS/MS analysis. SNOTRAP offers more direct identification of SNO sites than SNOSID but remains technically challenging (Seneviratne et al., 2016).
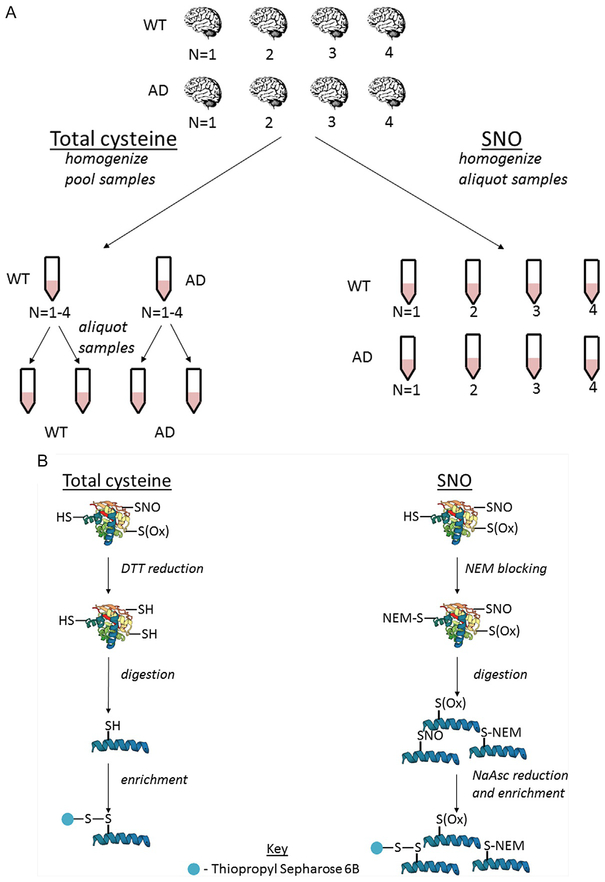
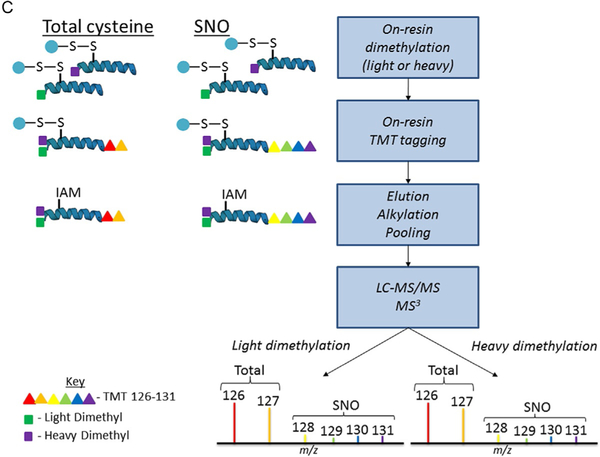
Our laboratory developed oxidized cysteine-selective combined precursor isotopic labeling and isobaric tagging (OxcyscPILOT), summarized in Fig. 4. As displayed in Fig. 4A, four samples of wild type (WT) and four samples of AD are selected for SNO analysis while pooled samples for WT and AD mice are generated for total cysteinyl analysis. As summarized in Fig. 4B, pooled samples are reduced, digested, then enriched while SNO samples are NEM blocked, digested, then simultaneously selectively reduced and enriched. As displayed in Fig. 4C, all samples are subjected to on-resin isotopic labeling and on-resin isobaric tagging to allow multiplexing during MS analysis. The use of light or heavy N-terminal dimethylation produces an ~8Da mass shift between light and heavy samples in the initial MS scan. The second MS scan analyzes peptide fragmentation while the third MS scan analyzes TMT reporter ion signal, producing a spectrum with TMT signals from the WT and AD pools and eight randomized SNO samples. OxcyscPILOT overcomes several limitations inherent in other proteomic SNO methods. First, the inclusion of a pooled WT and AD standard allows SNO levels to be normalized to the total cysteine proteome within individual spectra without the need to adjust for variability between scans. Second, multiplexing allows the statistical power of four biological replicates per WT and AD without increasing analysis time by a factor of 8. Third, the site occupancy of SNO can be calculated to determine the level at which a protein is SNO-modified versus unmodified. Although developed with TMT6-plex reagents, the use of TMT11-plex would allow the procedure’s multiplexing capability to increase to 18 samples or 9 biological replicates per WT and AD (Dyer et al., 2017; Gu & Robinson, 2016).
Fig. 4.
Schematic workflow for OxcyscPILOT. Panel (A) represents the cohort of WT/AD brain samples. Panel (B) shows the basic steps which include NEM-blocking (SNO)/reduction (total), tryptic digestion, selective reduction, and enrichment (SNO)/enrichment (total). Panel (C) represents the On-Resin dimethylation, TMT tagging, elution, alkylation, and LC-MS/MS and MS3 analysis.
5.3. OxcyscPILOT methodology
Our OxcyscPILOT methodology has been previously described in detail (Dyer, Gu, & Robinson).
5.3.1. Materials
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)
Acetic acid, glacial
Acetone
Acetonitrile, HPLC-grade (ACN)
Acetonitrile, MS-grade (ACN)
Ammonia
Ammonia acetate
BSA
Dithiothreitol (DTT)
Ethylenediaminetetraacetic acid (EDTA)
Formaldehyde, CH2O
Formic acid (FA)
Heavy formaldehyde, 13CD2O
Hydroxylamine
Iodoacetamide (IAM)
KCl
KH2PO4
Na2HPO4
NaCl
Nanopure water
NEM
NH4OH
Pierce BCA protein assay kit
Sodium ascorbate (NaAsc)
Sodium cyanoborodeuteride (NaBD3CN)
Sodium cyanoborohydride (NaBH3CN)
SDS
Triethylammonium Bicarbonate Buffer (TEAB)
Thiopropyl Sepharose 6B thiol-affinity resin
Tissue for analysis
TMT6 reagents
TPCK-treated trypsin from bovine pancreas
Trifluoroacetic acid (TFA)
Tris(hydroxylmethyl)aminomethane (Tris buffer)
Urea
Water, HPLC-grade
Water, MS-grade
5.3.2. Buffer recipes
Coupling buffer 1 (CB1): 50mM Tris, 21mM EDTA pH 7.5
Coupling buffer HEPES (CBH): 250mM HEPES, 1mM EDTA pH 7.7
Digestion buffer: 20mM Tris, 10mM CaCl2 pH 8.2
Enrichment washing buffers: Wash buffer (WB), 2M NaCl, 80% ACN with 0.1% TFA.
Fractionation buffers: 2, 4, 6, 8, 10, 12, 14, 16, 20, 25, 35, 50% MS-grade
ACN, adjusted to pH 10 with NH4OH
HLB clean-up buffers: HPLC-grade ACN with 0.1% FA, HPLC-grade
H2O with 0.1% FA, 40:60 H2O:ACN with 0.1% FA
NEM blocking buffer: 1 × PBS, 8M urea, 50mM NEM, 1% SDS pH 7.2
Reduction buffer: 50mM Tris, 8M urea, 10mM DTT pH 8.2
Resuspension buffer: 50mM Tris, 8M urea pH 8.2
SNO homogenization buffer: 1 × PBS, 8M urea, 5mM NEM, 5mM EDTA pH 7.2
Total cysteine homogenization buffer: 1 × PBS, 8M urea pH 7.2
WB: 50mM Tris, 1mM EDTA pH 8.0
5.3.3. Homogenization
Place equal mass portions of each brain and liver tissue to be analyzed for total cysteine and SNO quantification into homogenization tubes.
Add lysing matrix A and add 500μL of either total cysteine homogenization buffer or SNO homogenization buffer to the homogenization tube.
Homogenize tissues using the FastPrep-24 5G homogenizer with setting of 20s at 6m/s. Place the sample tubes on ice for 15min and remove supertant to a new 1.5mL eppendorf tube without collecting lysing matrix crystals.
Repeat step 2 (adding the appropriate homogenization buffer) and combine the new supertant with the previous collection.
Vortex the samples and centrifuge at 13,000 × g for 15min at 4°C. This final supernatant should be transferred to a new 1.5mL eppendorf tube.
Measure the protein concentration using BCA or other protein assay.
5.3.4. NEM-blocking and acetone precipitation (SNO samples ONLY)
Prepare a solution of 10% SDS (w/v) and a separate solution of 250mM NEM, both in 1 × PBS with 8M urea.
Generate two aliquots of SNO proteins (1mg) in 1.5mL Eppendorf tubes and add 25μL 10% SDS (1% final), 50μL 250mM NEM (50mM final), and complete with 1 × PBS with 8M urea to a final volume of 250μL per aliquot.
Keep the samples in the dark using aluminum foil and subject to low speed vortexing for 2h at room temperature.
Add 1mL of −20°C acetone (add to −20°C for a minimum of 3h prior) to the samples and incubate at −20°C overnight.
Centrifuge the samples at 13,000 × g for 10min at 4°C and discard the acetone. Allow the pellets to dry at room temperature for 20min.
Reconsitute the protein pellets in 125μL of resuspension buffer, vortexing frequently and then centrifuge at 1500 × g for 30s. After this step, the supernatants from the two individual aliquots can be combined and vortexed.
Determine the new protein concentration with BCA or other protein assay.
For further digestion steps, use 1mg of NEM-blocked protein.
5.3.5. Cysteinal reduction (total cysteine samples ONLY)
Prepare 1:40 protein/DTT ratio of 25mM DTT in 50mM Tris with 8M urea.
To each sample (100μg of protein) add 2.43μL of 25mM DTT and bring to a total volume of 100μL with 50mM Tris with 8M urea.
Incubate the samples in a 37°C water bath for 30min
5.3.6. Tryptic digestion (all samples)
Dilute the samples 10-fold with digestion buffer, add trypsin in a 4% w/w enzyme/protein ratio, and incubate overnight at 37°C.
Add one drop of pure FA to the samples to quench digestion. Test the pH of the samples with a pH test strip or a micro-electrode to ensure the pH is below 3. If the pH is above 3, adjust with FA.
Clean digested samples using C18 HLB cartridges (10mg) according to manufacturer’s protocol and dry to ~10μL.
5.3.7. Reduction and enrichment
Sonicate water, CB1, CBH, and WB for 30min to degas.
(For total cysteine only) Prepare 25mM DTT in CB1 and add 2.43μL of this solution to the samples. Next, add 20μL of CB1 to the samples and incubate at 37°C for 30min.
Place ~35mg of thiopropyl sepharose 6B thiol-affinity resin to a 1.5mL eppendorftube and add 1mL of water. Let resin sit at room temperature for 15min, aspirate several times with a 1mL pipette, and allow the resin to sit at room temperature for an additional 10min.
Transfer 500μL of resin to a 1mL spin column held in a 2mL centrifuge tube and centrifuge for 30s at 1000 × g.
Cap the bottom of the spin column and add 450μL of water. Cover the bottom with parafilm and vortex for 1min and centrifuge for 45s at 1000 × g.
Repeat step 5 two times with CB1 (for total cysteine) or CBH (for SNO). Cap the bottom of the spin column after the final wash.
For total cysteine, add CB1 to samples to a total volume of 100μL and transfer samples to resin. Cap the spin column and shake on a vortex at speed ~800rpm for 1h at room temperature.
For SNO, prepare 500mM NaAsc in CBH and add 4μL to samples (20mM final). Add CBH to samples to a volume of 100μL and transfer the samples to resin. Cap the spin column and shake on a vortex at ~800rpm for 2h to enrich at room temperature.
Place the spin column in a new 2mL tube and centrifuge at 1500 × g for 1min. If desired, store the unbound peptides in the −80°C.
Sequentially wash samples once with each of the following solutions (450μL) and in this order: WB, 2M NaCl, 80% ACN with 0.1% TFA, and 1% acetic acid. After adding the solution, cap both ends of the spin column, vortex for 1 min, and centrifuge at 1000 × g for 45s.
5.3.8. On-resin low pH dimethylation
Prepare solutions: 1% acetic acid, 4% formaldehyde, 0.6M sodium borohydride, 1% ammonia, and 100mM TEAB all in MS-grade water. For light dimethylation, prepare CH2O and NaBH3CN. For heavy dimethlyation, prepare 13CD2O and NaBD3CN.
Add 100μL of 1% acetic acid to samples, cap, and vortex. Ardd 8μL of 4% formaldehyde, recap, and vortex briefly. Immediately add 8μL of 0.6M sodium borohydride, cap, and vortex at ~800rpm for 10min at room temperature.
Add 16μL of 1% ammonia to samples and vortex at ~800 rpm for 3min. Add one drop of pure FA to the samples and let sit at room temperature for 1min.
Transfer the spin column to a new 2mL tube, and centrifuge at 1500 × g for 1min. Wash the samples with 450μL of 100mM TEAB, cap, and vortex for 1min and centrifuge at 1000 × g for 45s.
5.3.9. On-resin high pH TMT tagging
Reconstitute TMT reagents in MS-grade ACN to 0.02mg/μL and prepare 1% hydroxylamine.
Add 75μL of 100mM TEAB to samples. Add 20μL of TMT reagent to samples, cap, and vortex at ~800rpm for 1h at room temperature.
Add 8μL of 1% hydroxylamine and vortex at ~800rpm for 3min.
Transfer the spin column to a 2mL tube, centrifuge at 1500 × g for 1min. Wash the samples with 450μL of WB by vortex for 1min, and centrifuging at 1000 × g for 45s. Remember to cap before vortex or spinning steps.
5.3.10. Elution and free-thiol blocking
Prepare 25mM of DTT in WB and add 100μL of DTT to the samples, cap, and shake on a rocker at 300rpm for 30 min at room temperature. Transfer spin column to a new 2mL tube, and centrifuge at 1500 × g for 1min. Retain the eluent.
Repeat the above step twice with fresh DTT each time and use a 10-min shake time. Collect the eluent in the same tube. Repeat again once with 80% ACN in lieu of DTT.
Prepare 100mM IAM and add 300μL of 100mM IAM to samples. Incubate the samples in the dark on ice at room temperature for 1 h. Pool the samples and dry to ~100μL.
5.3.11. High pH reversed-phase fractionation
Prepare the fractionation buffers according to manufacturer’s protocol.
Reconstitute to 1550μL with 2% ACN, pH 10.
Activate a C18 HLB cartridge (30mg) with d1 mL of ACN with 0.1% FA (HLB clean-up buffer) and equilibrate the cartridge twice with 1mL of H2O with 0.1% FA (HLB clean-up buffer). Load the sample onto the HLB cartridge and wash once with 1mL H2O with 0.1% FA (HLB clean-up buffer).
Elute the sample into a clean 1.5mL Eppendorf tube with 500μL of 2% ACN, pH 10 buffer. Repeat elution with remaining fractionation buffers from 4 to 50% ACN, pH 10 into clean tubes each time. Dry the fractions to ~10μL and store in −80°C until ready for analysis.
5.3.12. LC-MS/MS and MS3 analysis
Reconstitute fractions in 258μL of MS-grade water with 0.1% FA and transfer to 0.65μm filters. Centrifuge at 12,000 × g for 1min and transfer samples to 250μL analysis vials. Transfer the sample vials to a nanoLC autosampler.
The LC gradient uses 3% ACN with 0.1% FA as buffer A and 100% ACN with 0.1% FA as buffer B. Set the gradient profile to: 0–5min, 10% mobile phase B; 5–40min, 10–15% B; 40–90min, 15–25% B; 90–115min, 25–30% B; 115–130min, 30–60% B; 130–135min, 60–80% B; 135–145min, 80% B; 145–150min, 80–10% B; 150–180min, 10% B.
Inject 5μL per fraction. Set the MS survey scan over m/z 375–1500 in the Orbitrap at 120,000 resolution, select the top mode 3 for each DDA cycle. Set precursor ions to be isolated with a width of 2.0 m/z, normalized collision energy to 35% in the Ion trap for MS/MS. For MS3 settings select the most intense CID fragment ion over m/z 375–1500 for HCD-MS3, the HCD fragment-ion isolation width to 2 m/z, and the normalized collision energy to 60%. Repeat injections for at least two technical replicates per fraction.
5.3.13. Database searching
Search light and heavy dimethylation RAW files separately with SEQUEST HT in Proteome Discoverer 2.3 against the most recent Uniprot database for the given species (i.e., mouse, human). Set the precursor mass tolerance to 15 ppm and fragment mass tolerance to 1Da. Select dynamic modifications as +28.0311Da (N-terminal) for light dimethylation or +36.076Da (N-terminal) for heavy dimethylation, +57.021Da (cysteine) for carbamidomethylation, +125.048Da (cysteine) for ethylmaleimide, +15.995Da (methionine) for methionine oxidation, and +229.163Da (lysine) for TMT-6plex. Enable decoy database searching with a 5% cutoff for false discovery rate. Set the enzyme to trypsin and allow up to two miscleavages. Identify reporter ions (126–131 m/z) with centroid using the smallest delta mass and 30 ppm mass tolerance settings.
5.4. PSSG
Several techniques have been utilized for the analysis of S-glutathionylation in the context of AD. Direct methods include Western blotting techniques utilizing an anti-GSH antibody and direct detection via MS by searching for a peptide mass shift corresponding to the attachment of GSH. The former suffers from low sensitivity while the latter is not easily amenable to complex mixtures (Zhang, Ye, et al., 2018). A notable indirect method summarized in Fig. 5 alkylates all free thiols followed by selective reduction of S-glutathionylation with a mixture of glutaredoxin-3, glutathione reductase, NADPH, and glutathione (Lind et al., 2002). The free thiols are then alkylated with a biotin-linked reagent, allowing streptavidin pull-down followed by gel separation and MS (Lind et al., 2002). Another CGE-LIF method replaces the biotinylation step with Dylight 488 maleimide to allow fluorescent detection of PSSG in the attomole range (Zhang et al., 2011, 2012). Both indirect methods, however, lack complete specificity due to glutaredoxin-3’s nonspecific reduction of non-PSSG disulfides, thus necessitating inclusion of positive and negative controls for high confidence in the identification of PSSG proteins (Zhang, Ye, et al., 2018).
Fig. 5.
A generalized schematic of indirect PSSG detection. The technique entails irreversible alkylation of free thiols, enzymatic reduction of PSSG, and biotinylation of PSSG sites. Biotinylated proteins are enriched with streptavidin followed by staining. Bands are then excised, trypsinized, and subjected to mass spectrometry.
6. SNO proteins in AD
A wide array of SNO proteins have been investigated in the context of AD, ranging from thorough studies of single proteins to high-throughput identification of multiple SNO proteins using proteomics. The majority of investigations have targeted single to several proteins, typically utilizing BST assay coupled with in-depth exploration of the function and effects of identified SNO proteins in cell models and/or animal models. These studies often combine in situ cell studies, animal model probing, and proteomic analysis of postmortem AD brain tissue (Akhtar et al., 2016; Bossy et al., 2010; Chakroborty, Kim, Schneider, West, & Stutzmann, 2015; Cho et al., 2009; Cordes et al., 2009; Kabiraj, Marin, Varela-Ramirez, & Narayan, 2016; Kwak et al., 2010; Kwak et al., 2011; Lacampagne et al., 2017; Molokanova et al., 2014; Nakamura, Cieplak, Cho, Godzik, & Lipton, 2010; Nakamura et al., 2010; Qu et al., 2011; Ryu, Lee, & Do, 2016; Seth et al., 2018; Uehara et al., 2006; Wang et al., 2018; Wang, Song, Tan, Albers, & Jia, 2012; Wang et al., 2017; Won et al., 2013; Zhang et al., 2018). On the other hand, high-throughput proteomics studies are typically performed in AD tissues, generating hundreds of SNO sites that will require deeper experimental investigation. Below, the findings of targeted studies for major proteins in AD are discussed. Additionally, the overall findings of high-throughput investigations are summarized in Table 1, and Fig. 6 represents possible crosstalk between the results of targeted studies.
Table 1.
List of selected SNO proteins in AD.
| Beta-secretase 1 | Amyloidogenic processing |
Loss of function | ↓ | Kwak et al. (2011) and Won et al. (2013) |
| Caspase 3 | Apoptosis | Transnitrosylation | N/A | Nakamura, Wang, et al. (2010) |
| Cyclin-dependent kinase 5 | Cell survival, proper synaptic spine density | Loss of synaptic spine density, transnitrosylation of Drp1 | ↑ | Molokanova et al. (2014), Qu et al. (2011), Wang et al. (2017) |
| Dexras 1 | Forms a complex with nNOS | Contributes to Aβ toxicity | N/A | Zhang, Zhu, et al. (2018) |
| Dynamin-related protein 1 | Initiator ofmitochondrial fission | Increased mitochondrial fission | ↑ | Akhtar et al. (2016), Cho et al. (2009), Gu and Robinson (2016), Molokanova et al. (2014), Nakamura, Cieplak, et al. (2010), Wang, Njoroge, et al. (2012), and Wang et al. (2017) |
| Glyceraldehyde 3-phosphate dehydrogenase | Glycolysis, apoptotic signaling | Increased apoptotic activity | ↑ | Sen, Saha, and Sen (2018), Wang et al. (2017), and Zareba-Koziol et al. (2014) |
| Insulin degrading enzyme | Degradation of insulin and Aβ | Reduction of function | ↑ | Akhtar et al. (2016) and Cordes et al. (2009) |
| O-linked N-acetylglucosaminyltransferase | Catalysis of Ser/Thr-linked O-GlcNAcylation | Reduction of function | N/A | Ryu et al. (2016) |
| Phosphate and tensin homolog | Negative regulation of P13k/Akt pathway | Increased PTEN degradation | ↑ | Kwak et al. (2010) |
| Protein disulfide isomerase | Protein folding | Loss of function | ↑ | Kabiraj et al. (2016), Uehara et al. (2006), and Wang et al. (2018) |
| Ryanodine receptor | Intracellular calcium release | Increased calcium leak | ↑ | Chakroborty et al. (2015) and Lacampagne et al. (2017) |
| X-linked inhibitor of apoptosis | Inhibition of caspases 3, 4, 7 | Loss of function | ↑ | Nakamura, Wang, et al. (2010) |
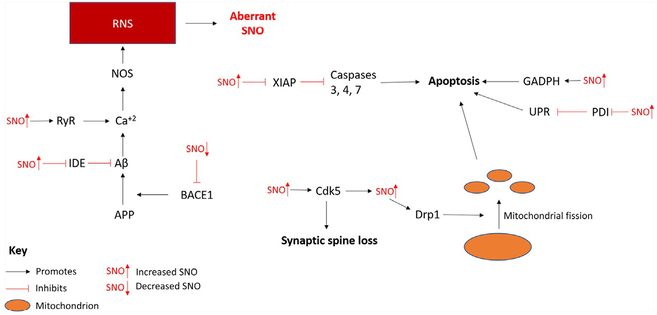
Fig. 6.
The effects of aberrant SNO in the AD brain. This is based on endogenous SNO protein quantification in mouse models or postmortem AD neural tissue. SNO may be inhibitory or enhance protein activity depending on the target protein. Abbreviations: APP, amyloid precursor protein; BACE1, beta-secretase 1; Cdk5, cyclin-dependent kinase 5; Drp1, dynamin-related protein 1; GADPH, glyceraldehyde 3-phosphate dehydrogenase; IDE, insulin degrading enzyme; NOS, nitric oxide synthase; PDI, protein disulfide isomerase; RyR, ryanodine receptor; UPR, unfolded protein response; XIAP, X-linked inhibitor of apoptosis.
6.1. Cyclin dependent kinase 5 and dynamin related protein 1
Cyclin dependent kinase 5 (Cdk5) is a neural protein involved in cell survival, synaptic plasticity, pain signaling, and drug addiction (Shah & Lahiri, 2014). For proper function, Cdk5 requires binding of the activator p35 (Shah & Lahiri, 2014). During neurotoxic conditions such as Aβ exposure, p35 may be cleaved into p25 and p10 which results in the hyperactivation of Cdk5 and neurotoxicity in AD (Shah & Lahiri, 2014). SNO of Cys83 or Cys157 of active Cdk5 enhanced activity in HEK293 cells and is likely a contributor to NMDAR-mediated dendritic spine loss (Qu et al., 2011). During Aβ exposure, neuronal cultures exhibited NMDAR-mediated SNO of Cdk5. Levels of SNO-Cdk5 were elevated in Tg2576 AD mouse brains. A further study demonstrated transfection of neural cultures with SNO-immune Cdk5 resulted in reduced neural spine loss when treated with Aβ (Qu et al., 2011). Additionally, SNO-Cdk5 was increased in postmortem human AD brain (Qu et al., 2011). NO• production and thus SNO-Cdk5 likely resulted primarily from extrasynaptic NMDARs activation rather than synaptic NMDARs activation (Molokanova et al., 2014). Further, inhibition of miRNA-132 (miR-132, known to be significantly downregulated in AD) in neural culture increased SNO-Cdk5 and nNOS with confirmation of decreased miR-132 expression in postmortem human AD temporal cortex (Wang et al., 2017). Interestingly, SNO-Cdk5 can favorably transnitrosylate dynamin related protein 1 (Drp1) (Qu et al., 2011).
Drp1 is a protein which participates in mitochondrial dynamics by initiating mitochondrial fission via the formation of complexes on the mitochondrial membrane, known to be induced by NO• (Barsoum et al., 2006; James, Parone, Mattenberger, & Martinou, 2003; Twig et al., 2008). Cerebrocortical neurons treated with Aβ25–35 induces pathological mitochondria fragmentation with SNO-Drp1 formation. Endogenous SNO-Drp1 in Tg2576 AD mice and postmortem AD human brains was elevated. SNO-Drp1 exhibits increased GTPase activity via a direct GTPase assay and mutant C644A (the endogenous SNO site) Drp1 suppressed NO•-induced mitochondrial fragmentation (Cho et al., 2009). A later study found no increase in GTPase activity of Drp1 due to SNO and detected no significant difference in SNO-Drp1 levels in postmortem human brain between AD and controls (Bossy et al., 2010). The methodology, however, may have introduced artifactual oxidation to endogenous Drp1 from ambient air (Nakamura, Cieplak, et al., 2010). Direct GTPase assays found increased GTPase activity from SNO in close Drp1 homologues (Nakamura, Cieplak, et al., 2010).
Further studies lend additional support for SNO-Drp1 in AD (Molokanova et al., 2014; Wang et al., 2017). SNO-Drp1 was increased and Drp1 was decreased in peripheral blood lymphocytes of patients with AD or mild cognitive impairment (MCI) (Wang, Song, et al., 2012). While more focused on insulin degrading enzyme (IDE, refer to following section), incubation of rat cortical cultures or rat cortico-hippocampal slices with Aβl-42, high glucose, or both, resulted in SNO-Drp1 formation at levels comparable to AD brains (Akhtar et al., 2016).
6.2. Protein disulfide isomerase
Protein disulfide isomerases (PDIs) are a family of endoplasmic reticulum (ER) chaperones which catalyze thiol-disulfide exchanges and rearrangement reactions for the proper folding of proteins via two catalytic-Cys-Gly-His-Cys- domains (Edman, Ellis, Blacher, Roth, & Rutter, 1985; Song & Wang, 1995; Vuori, Myllyla, Pihlajaniemi, & Kivirikko, 1992). SNO-PDI inhibits PDI (specifically PDIA1) activity and may be generated by NMDAR-mediated excitotoxicity, contributing to the unfolded protein response with sustained nitrosative stress. SNO-PDI was found in postmortem AD and Parkinson’s disease brains, suggesting SNO-PDI may contribute to neuronal cell death in neurodegenerative diseases (Uehara et al., 2006). Incubation of dopaminergic SH-SY5Y cells or primary cultured hippocampus neurons with Aβ25–35 resulted in elevated RNS and induced formation of SNO-PDI (Kabiraj et al., 2016; Wang et al., 2018) and reduced PDI activity which could be prevented with neohesperidin, an ROS-scavenger (Wang et al., 2018).
6.3. Glyceraldehyde 3-phosphate dehydrogenase and sirtuin 1
GADPH is a glycolytic enzyme with roles in activating apoptosis when S-nitrosylated (Bae et al., 2006; Hara et al., 2005; Li, Feng, Wu, & Zhang, 2012). Sirtuin 1 (SIRT1) is a nicotinamide adenosine dinucleotide-dependent class-III deacetylase that deacetylates Lys174 of tau protein and thus ameliorates tau aggregation (Min et al., 2018; Min, Sohn, Cho, Swanson, & Gan, 2013). Treatment of WT mice with Aβ1–42 induced the formation of SNO-GADPH which could transnitrosylate and deactivate SIRT1 which led to increased tau aggregation. Increased SNO-GADPH was present in postmortem AD cortical samples (Sen et al., 2018) consistent with other studies (Wang et al., 2017).
6.4. Insulin degrading enzyme
IDE is a zinc metalloendopeptidase responsible for the degradation of both insulin and Aβ1–42 (Duckworth, Bennett, & Hamel, 1998; Farris et al., 2003). NO• donors inhibit insulin degradation by IDE up to 70% and Aβ degradation by 25% with micromolar concentrations of NO• donors. Excess insulin almost fully inhibited IDE-mediated Aβ degradation and IDE was SNO-modified in situ (Cordes et al., 2009). Treatment of rat cortical cultures, rat hippocampal slices, or human neural cultures with glucose, Aβ1–42, or both elevated SNO-IDE due to NMDAR-mediated nitrosative stress. SNO inhibited the Aβ-degrading function of IDE and significant levels of SNO-IDE in postmortem AD brain were observed (Akhtar et al., 2016).
6.5. O-linked N-acetylglucosaminyltransferase
O-linked N-acetylglucosaminyltransferase (OGT) is a catalyst of Ser/Thr-linked O-GlcNAcylation (Hart, Housley, & Slawson, 2007). Aβ treatment of human neuroblastoma SK-N-MC cells elevated SNO-OGT and reduced global O-GlcNAcylation. OGT associated with nNOS after Aβ treatment while hyper O-GlcNAcylation appeared to prevent NMDAR-mediated calcium ion influx. SNO-OGT may contribute to tau hyperphosphorylation (Ryu et al., 2016).
6.6. Beta-secretase 1
Beta-secretase 1 (BACE1) is a membrane aspartyl protease responsible for the N-terminal cleavage of amyloid precursor protein (APP) in the amyloidogenic processing of APP (Cole & Vassar, 2007; Zhang & Xu, 2007). Treatment of rat primary cortical neurons with <100nM of NO• donors suppressed BACE1 transcription while 0.1–100μM treatment did not influence transcription but deactivated BACE1 via SNO (Kwak et al., 2011; Won et al., 2013). SNO-BACE1 was reduced in the entorhinal cortices of postmortem human AD brains (Kwak et al., 2011). Additionally, GSNO treatment of bEND3 cells induced the formation of SNO-dynamin 2, increasing Aβ uptake and clearance (Won et al., 2013).
6.7. X-linked inhibitor of apoptosis and caspase-3
X-linked inhibitor of apoptosis (XIAP) is an inhibitor of apoptosis via direct inhibition of caspases 3, 7, and 9 while caspase 9 is an inducer of apoptosis and caspases 3 and 7 are executors of apoptosis (Eckelman, Salvesen, & Scott, 2006). NO• donors can induce SNO of Cys450 of XIAP, inhibiting its E3-ligase and anti-apoptotic activity. SNO-XIAP in AD postmortem brains was increased and SNO-caspase-3 can transnitrosylate XIAP (Nakamura, Wang, et al., 2010).
6.8. Ryanodine receptor
Ryanodine receptors (RyRs) are endoplasmic reticulum (ER) channel proteins responsible for creating Ca2+ efflux from ER calcium stores into the cytosol (Verkhratsky, 2005). Blocking NO• synthesis increased synaptic depression in presymptomatic 3xTg-AD mice. NO• synthesis may result in SNO-RyR and increased RyR-mediated calcium release which may contribute to stress conditions in late AD (Chakroborty et al., 2015). Further investigation into RyR-mediated calcium leak found SNO as part of RyR remodeling in postmortem AD brain and the APP/presenilin 1 (PS1) AD mouse model (Lacampagne et al., 2017).
6.9. Dexamethasone-induced Ras-related protein 1
Dexamethasone-induced Ras-related protein 1 (Dexras1) is a small GTPase known to modulate signaling cascades important to neurogenesis in the hippocampus (Bouchard Cannon, Lowden, Trinh, & Cheng, 2018). SNO-Dexras1 formed on Cys11 in primary hippocampal neuron cultures after incubation with Aβ1–42 and in the hippocampus of 4-month APP/PS1 mice. Dexras1 mutated to lack the SNO site, resulted in reduced impairment in Aβ1–42 mice, suggesting SNO-Dexras1 contributes to the neurotoxicity of Aβ1–42 (Zhang, Zhu, et al., 2018).
6.10. Phosphatase and tensin homolog
Phosphatase and tensin homolog (PTEN) is a tumor suppressor phosphatase negative regulator of the P13K/Akt pathway (Maehama & Dixon, 1998; Myers et al., 1997). SNO-PTEN was found in the entorhinal cortexes of MCI and AD (Kwak et al., 2010). Both exogenous and endogenous NO• donors could induce stable SNO-PTEN in rat cortical cultures which resulted in ubiquination of PTEN and subsequent degradation by the ubiquitin-proteasome pathway. Due to PTEN’s neuroprotective effect, SNO-induced degradation of PTEN may contribute to neurodegenerative processes in AD (Kwak et al., 2010).
6.11. High-throughput SNO and modified cysteine studies
Numerous studies aimed at the proteome-wide identification of SNO proteins and other oxidative cysteine modifications have been performed. Forty-five endogenous SNO proteins were identified in AD hippocampus, substantia nigra, and cortex and are mostly involved in metabolism, signaling pathways, apoptosis, and redox regulation (Zahid et al., 2014). Endogenous SNO proteins of synaptosomes of WT and hAPP AD mice were profiled, identifying 138 SNO proteins of which 38 were detected only in the hAPP mice (Zareba-Koziol et al., 2014). These proteins were generally at higher levels of SNO and included pathways such as glycolysis/gluconeogenesis, calcium homeostasis, and ion and vesicle transport (Zareba-Koziol et al., 2014). SNO proteins were analyzed in the cortex, hippocampus, and cerebellum of CK-p25 AD mice, finding 313 endogenous SNO sites located on 251 proteins with 135 SNO-proteins unique to neurodegeneration (Seneviratne et al., 2016). The affected pathways included synapse function, metabolism, and AD pathology (Seneviratne et al., 2016) (Table 2).
Table 2.
SNO pathways identified in AD.
| Metabolism | Hippocampus, substantia nigra, cortex, cerebellum, whole brain | Gu and Robinson (2016), Seneviratne et al. (2016), and Zahid, Khan, Oellerich, Ahmed, and Asif (2014) |
| Signaling pathways | Hippocampus, substantia nigra, cortex, whole brain | Gu and Robinson (2016) and Zahid et al. (2014) |
| Apoptosis | Hippocampus, substantia nigra, cortex | Zahid et al. (2014) |
| Redox regulation | Hippocampus, substantia nigra, cortex | Zahid et al. (2014) |
| Glycolysis/gluconeogenesis | Synaptosomes | Zareba-Koziol et al. (2014) |
| Calcium homeostasis |
Synaptosomes | Zareba-Koziol et al. (2014) |
| Ion and vesicle transport | Synaptosomes | Zareba-Koziol et al. (2014) |
| Synapse function | Hippocampus, cortex, cerebellum | Seneviratne et al. (2016) |
| AD pathology | Hippocampus, cortex, cerebellum | Seneviratne et al. (2016) |
Our laboratory characterized the SNO proteome of the whole brain of the APP/PS1 mouse model, identifying 135 SNO proteins of which 11 proteins were differentially modified between WT and AD (Gu & Robinson, 2016). Significantly modified proteins previously identified in AD were ADP/ATP translocase 1 (Zareba-Koziol et al., 2014), ras-related C3 botulinum toxin substrate (Zareba-Koziol et al., 2014), 14-3-3 protein zeta/delta (Zahid et al., 2014), glutamine synthetase (Zahid et al., 2014; Zareba-Koziol et al., 2014), myelin proteolipid protein (Zareba-Koziol et al., 2014), citrate synthase (Zareba-Koziol et al., 2014), and 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (Zareba-Koziol et al., 2014) while newly discovered SNO proteins were septin 5, myc box-dependent-interacting protein 1, isocitrate dehydrogenase, and 14-3-3 protein gamma. Most site occupancies ofsignificant SNO proteins were below 1% with the exception of the 14-3-3 proteins which were 5–7% for 14-3-3 gamma and 28–54% for 14-3-3 zeta/delta (Gu & Robinson, 2016). Modified pathways consisted primarily of metabolism and signal transduction (Gu & Robinson, 2016). We additionally identified and quantified the cysteine proteome of 14-month APP/PS1 mouse livers, finding 2259 unique proteins with 65 proteins differentially expressed between WT and AD (Gu, Evans, & Robinson, 2015). We identified and quantified the reversibly oxidized cysteine proteome of 14-month APP/PS1 mouse livers by alkylating free thiols, reducing oxidized cysteines, and enriching previously oxidized cysteines followed by LC-MS/MS analysis (Gu & Robinson, 2016b). We identified 828 reversibly oxidized cysteine sites with 19 sites differentially expressed between WT and AD without identification of specific PTMs (Gu & Robinson, 2016b). Modified pathways consisted of amino acid metabolism, carbohydrate metabolism, lipid metabolism, citric acid cycle and respiratory electron transport, and biological oxidation (Gu & Robinson, 2016b).
The SNO proteome of the entorhinal brain tissue of postmortem AD patients was probed but there was no significant difference between AD and healthy controls, perhaps due to the sensitivity of the employed method (Riederer et al., 2009). Several studies targeted the SNO proteome without providing identifications for individual proteins. A highly sensitive SNO detection method was developed and demonstrated utility via quantifying the SNO protein amounts in the cerebrum of 5-month B6Cg-Tg AD mice compared to WT (Wang et al., 2011). A 2D micro-capillary gel electrophoresis fingerprint of the SNO proteome of 11-month B6Cg-Tg AD mouse brains was also later generated (Wang, Njoroge, et al., 2012). Proteins identified as significantly S-nitrosylated between AD and control across multiple high-throughput studies include GADPH (Seneviratne et al., 2016; Zareba-Koziol et al., 2014), glutamine synthase (Gu & Robinson, 2016; Seneviratne et al., 2016), ADP/ATP translocase 1 (Gu & Robinson, 2016; Zareba-Koziol et al., 2014), and 2′,3′ cyclic nucleotide 3′ phosphodiesterase (Gu & Robinson, 2016; Zareba-Koziol et al., 2014). BV2 microglia cells treated with Aβ25–35 for 24h changed cysteine redox status for 60 proteins in pathways such as cell death and survival, inflammatory response, or cellular growth and proliferation (Correani et al., 2015).
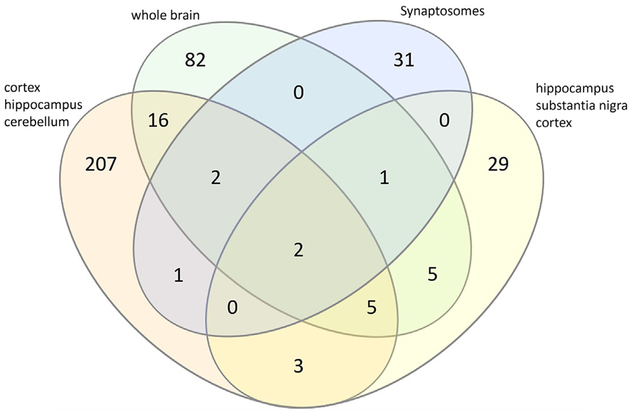
As displayed by Fig. 7, high-throughput SNO studies exhibit a small degree of overlap of SNO proteins and largely identify novel SNO proteins in AD. Given the heterogeneity of the brain, some difference between SNO proteomes is expected and illustrates the need to spatially investigate SNO in brain. The validity of shotgun proteomics methodology in probing SNO is shown in Fig. 7 because more than 350 SNO sites in AD have been determined. However, the largescale identification of unknown SNO proteins demonstrates the need for future studies to further understanding of SNO in AD.
Fig. 7.
Overlap between identified SNO proteins in AD in high-throughput proteomic studies. These studies targeted the cortex, hippocampus, and cerebellum (Seneviratne et al., 2016);whole brain (Gu & Robinson, 2016);synaptosomes (Zareba-Koziol et al., 2014);and hippocampus, substantia nigra, and cortex (Zahid et al., 2014). Diagram created with InteractiVenn (Heberle, Meirelles, da Silva, Telles, & Minghim, 2015).
7. PSSG proteins in AD
S-glutathionylation is less studied in the context of AD. Postmortem AD inferior parietal lobule (IPL) was probed, revealing significant increase of PSSG in deoxyhemoglobin, α-enolase, α-crystallin B, and GADPH (Newman et al., 2007). GADPH and α-enolase activity was reduced in AD IPL (Newman et al., 2007). Selective PSSG of the monomeric and dimeric forms of p53 were observed, suggesting that PSSG may prevent the formation of the more active p53 tetramer (Domenico et al., 2009). Like SNO-IDE; PSSG-IDE likely inhibits IDE degradation of Aβ (Cordes et al., 2009). Global PSSG levels in AD-Tg mice were compared to WT. PSSG levels were increased in AD cerebrum but decreased in AD hippocampus and whole blood (C. Zhang et al., 2011). Measurement of PSSG levels of 1, 5, and 11-month old AD-Tg mice were increased in the brains of all but 5-month old mice and global PSSG was increased in the blood of all age groups (Zhang et al., 2012). Principle component analysis could distinguish AD from WT with both brain PSSG and blood PSSG (Zhang et al., 2012). Blood collected from AD patients included a significant decrease in GSH/GSSG ratio which may affect glutathionylation (Rani et al., 2017). Transthyretin in cerebral spinal fluid from patients with AD, MCI, normal pressure hydrocephalus, or healthy controls possessed significant oxidation occurring on Cys10 in AD and MCI (Poulsen et al., 2014). Oxidation types included PSSG, S-cysteinylation, and S-cysteinylglycinylation (Poulsen et al., 2014). Incubation of SH-SY-5Y cells with Aβ42 increased PSSG of the α-subunit of Na,K-ATPase with enzyme inhibition, suggesting PSSG had an inhibitory effect (Lakunina et al., 2017). These findings are summarized in Table 3.
Table 3.
PSSG proteins and pathways identified in AD.
| α-enolase | Glycolysis, apoptotic signaling | Reduction of function | ↑ |
| α-crystallin B | Heat-inducible chaperone | Reduction of function | ↑ |
| Deoxyhemoglobin | Oxygen transport | Increased oxygen affinity | ↑ |
| Glyceraldehyde 3-phosphate dehydrogenase | Glycolysis, apoptotic signaling | Reduction of function | ↑ |
| Na,K-ATPase | Sodium-potassium pump | Reduction of function | N/A |
| Insulin degrading enzyme | Degradation of insulin and Aβ | Reduction of function | N/A |
| p53 | Tumor suppressor | Potential decrease in formation of active tetramer | ↑ |
| Transthyretin | Thyroxin/retinol transport | N/A | ↑ |
As PSSG remains relatively understudied in AD, further studies are required to better assess how PSSG may be involved in AD. With the advent of high-throughput proteomic studies and the development of indirect PSSG methods amenable to MS, a logical next step would be a high-throughput study aimed at a proteome-wide analysis of PSSG in AD.
8. Conclusions
Overall, shotgun proteomic studies offer a useful starting point for characterizing large numbers of SNO or PSSG proteins. Altered SNO levels of BACE1, IDE, and RyR likely contribute to impaired calcium ion homeostasis, indirectly increasing NO• production and maintaining RNS conditions whereas other studied SNO modifications likely contribute to the observed pathophysiology in AD. The full role of SNO in AD may thus be more fully elucidated with proper future study into different brain regions and neural cell types, across different disease stages to track alterations in SNO with disease progression, and into peripheral organs such as liver which may play a role in altered AD metabolism. Once SNO proteome-wide studies identify potential SNO dysregulation, targeted studies can determine the roles individual dysregulated SNO proteins play in disease pathology. Follow-up studies are required to expand upon the current results of high-throughput SNO experiments as proteins such as glutamine synthase and 2′,3′-cyclic-nucleotide 3′-phosphodiesterase have been repeatedly identified as significantly modified. While cell studies offer clues into the function of SNO proteins in vivo, such studies require endogenous confirmation of SNO proteins in AD tissue. Due to clear detrimental effects of SNO of proteins such as Cdk5 and amelioration of pathological features when prevented, SNO provides a potential avenue for novel drug development.
As a less studied modification in AD, PSSG may benefit greatly from high-throughput proteomic studies. Considering PSSG is viewed as a buffer against ROS/RNS stress and a mechanism by which aberrant cysteine modifications can be avoided, it would be interesting to discover if aberrant SNO might influence PSSG. Potential crosstalk between both modifications has not been investigated in AD and may reveal another layer of complexity to dysregulated cysteine modifications. Considering cysteine PTMs such as SNO and PSSG as potential drug targets in AD is an exciting direction for the field.
Acknowledgments
This review was supported by Vanderbilt University Start-up funds and NIH, NIGMS (R01GM117191) funds to RASR and supplement to support K.I.F.
References
- Akhtar MW, Sanz-Blasco S, Dolatabadi N, Parker J, Chon K, Lee MS,et al. (2016). Elevated glucose and oligomeric β-amyloid disrupt synapses via a common pathway of aberrant protein S-nitrosylation. Nature Communications, 7, 10242–10252. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4729876/pdf/ncomms10242.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P, & Stamler JS (2012). Enzymatic mechanisms regulating protein S-nitrosylation: Implications in health and disease. Journal of Molecular Medicine, 90(3), 233–244. 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson ME (1998). Glutathione: An overview of biosynthesis and modulation. Chemico-Biological Interactions, 111–112, 1–14. 10.1016/S0009-2797(97)00146-4. [DOI] [PubMed] [Google Scholar]
- Angelo M, Singel DJ, & Stamler JS (2006). An S-nitrosothiol (SNO) synthase function of hemoglobin that utilizes nitrite as a substrate. Proceedings of the National Academy of Sciences of the United States of America, 103(22), 8366–8371. 10.1073/pnas.0600942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B-I, Hara MR, Cascio MB, Wellington CL, Hayden MR, Ross CA, et al. (2006). Mutant Huntingtin: Nuclear translocation and cytotoxicity mediated by GAPDH. Proceedings of the National Academy of Sciences of the United States of America, 103(9), 3405–3409. 10.1073/pnas.0511316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez NO, Reisz JA, & Furdui CM (2015). Mass spectrometry in studies of protein thiol chemistry and signaling: Opportunities and caveats. Free Radical Biology & Medicine, 80, 191–211. 10.1016/j.freeradbiomed.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, et al. (2006). Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. The EMBO Journal, 25(16), 3900–3911. 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, & Stamler JS (2009). Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nature Reviews Molecular Cell Biology, 10, 721–732. 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- Bossy B, Petrilli A, Klinglmayr E, Chen J, Lütz-Meindl U, Knott AB, et al. (2010). S-nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer’s disease. Journal of Alzheimer’s Disease, 20(s2), S513–S526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard Cannon P, Lowden C, Trinh D, & Cheng H-Y (2018). Dexras1 is a homeostatic regulator of exercise-dependent proliferation and cell survival in the hippocampal neurogenic niche. Scientific Reports, 8, 5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley SA, & Steinert JR (2016). Nitric oxide-mediated posttranslational modifications: Impacts at the synapse. Oxidative Medicine and Cellular Longevity, 2016, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Gu L, Di Domenico F, & Robinson RA (2014). Mass spectrometry and redox proteomics: Applications in disease. Mass Spectrometry Reviews, 33(4), 277–301. 10.1002/mas.21374. [DOI] [PubMed] [Google Scholar]
- Chakroborty S, Kim J, Schneider C, West AR, & Stutzmann GE (2015). Nitric oxide signaling is recruited as a compensatory mechanism for sustaining synaptic plasticity in Alzheimer’s disease mice. Journal of Neuroscience, 35(17), 6893–6902. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4412902/pdf/zns6893.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Ku W-C, Lin P-Y, Chou H-C, Khoo K-H, & Chen Y-J (2010). S-alkylating labeling strategy for site-specific identification of the S-nitrosoproteome. Journal of Proteome Research, 9(12), 6417–6439. 10.1021/pr100680a. [DOI] [PubMed] [Google Scholar]
- Chen Y-J, Liao Y-C, Chen Y-J, Lu C-T, Yang H-H, Huang K-Y, et al. (2014). dbSNO 2.0: A resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Research, 43(D1), D503–D511. 10.1093/nar/gku1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, et al. (2009). S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science, 324(5923), 102–105. 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SL, & Vassar R (2007). The Alzheimer’s disease beta-secretase enzyme, BACE1. Molecular Neurodegeneration, 2, 22–46. 10.1186/1750-1326-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs B, Hamel C, & Kanaan NM (2016). Pathological conformations involving the amino terminus of tau occur early in Alzheimer’s disease and are differentially detected by monoclonal antibodies. Neurobiology of Disease, 94, 18–31. 10.1016/j.nbd.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes CM, Bennett RG, Siford GL, & Hamel FG (2009). Nitric oxide inhibits insulin-degrading enzyme activity and function through S-nitrosylation. Biochemical Pharmacology, 77(6), 1064–1073. Retrieved from https://ac.els-cdn.com/S0006295208008940/1-s2.0-S0006295208008940-main.pdf?_tid=b20c5ed1-24a7-4079-9fc0-7e29a7a92f8c&acdnat=1537206731_31d31afd323ca331ebcb6bafdaa70116. [DOI] [PubMed] [Google Scholar]
- Correani V, Di Francesco L, Cera I, Mignogna G, Giorgi A, Mazzanti M, et al. (2015). Reversible redox modifications in the microglial proteome challenged by beta amyloid. Molecular BioSystems, 11(6), 1584–1593. 10.1039/c4mb00703d. [DOI] [PubMed] [Google Scholar]
- Couvertier SM, Zhou Y, & Weerapana E (2014). Chemical-proteomic strategies to investigate cysteine posttranslational modifications. Biochimica et Biophysica Acta—Proteins and Proteomics, 1844(12), 2315–2330. 10.1016/j.bbapap.2014.09.024. [DOI] [PubMed] [Google Scholar]
- Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, et al. (2008). Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Analytical Chemistry, 80(8), 2921–2931. 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- Derakhshan B, Hao G, & Gross SS (2007). Balancing reactivity against selectivity: The evolution of protein S-nitrosylation as an effector of cell signaling by nitric oxide. Cardiovascular Research, 75(2), 210–219. 10.1016/j.cardiores.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenico FD, Cenini G, Sultana R, Perluigi M, Uberti D, Memo M, et al. (2009). Glutathionylation of the pro-apoptotic protein p53 in Alzheimer’s disease brain: Implications for AD pathogenesis. Neurochemical Research, 34(4), 727–733. 10.1007/s11064-009-9924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias P-T, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, et al. (2010). Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proceedings of the National Academy of Sciences of the United States of America, 107(39), 16958–16963. 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth WC, Bennett RG, & Hamel FG (1998). Insulin acts intracellularly on proteasomes through insulin-degrading enzyme. Biochemical and Biophysical Research Communications, 244(2), 390–394. 10.1006/bbrc.1998.8276. [DOI] [PubMed] [Google Scholar]
- Dyer RR, Gu L, & Robinson RAS (2017). S-nitrosylation in Alzheimer’s disease using oxidized cysteine-selective cPILOT In Santamana E & Fernández-Irigoyen J (Eds.), Current proteomic approaches applied to brain function (pp. 225–241). New York, NY: Springer New York. [Google Scholar]
- Eckelman BP, Salvesen GS, & Scott FL (2006). Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Reports, 7(10), 988–994. 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman JC, Ellis L, Blacher RW, Roth RA, & Rutter WJ (1985). Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature, 317, 267–270. 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- El-Sehemy A, Postovit LM, & Fu Y (2016). Nitric oxide signaling in human ovarian cancer: A potential therapeutic target. Nitric Oxide, 54, 30–37. 10.1016/j.niox.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, et al. (2003). Insulin-degrading enzyme regulates the levels of insulin, amyloid β-protein, and the β-amyloid precursor protein intracellular domain in vivo. Proceedings of the National Academy of Sciences of the United States of America, 100(7), 4162–4167. 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Foster MW, Benhar M, & Stamler JS (2009). Detection of protein S-nitrosylation with the biotin-switch technique. Free Radical Biology and Medicine, 46(2), 119–126. 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, & Stamler JS (2009). Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nature Biotechnology, 27(6), 557–559. 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstermann U, & Sessa WC (2012). Nitric oxide synthases: Regulation and function. European Heart Journal, 33(7), 829–837d. 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Hess DT, & Stamler JS (2009). Protein S-nitrosylation in health and disease: A current perspective. Trends in Molecular Medicine, 15(9), 391–404. 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthwaite J, Charles SL, & Chess-Williams R (1988). Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature, 336, 385–388. 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- Gu L, Evans AR, & Robinson RA (2015). Sample multiplexing with cysteine-selective approaches: cysDML and cPILOT. Journal of the American Society for Mass Spectrometry, 26(4), 615–630. 10.1007/s13361-014-1059-9. [DOI] [PubMed] [Google Scholar]
- Gu L, & Robinson RA (2016a). Proteomic approaches to quantify cysteine reversible modifications in aging and neurodegenerative diseases. Proteomics Clinical Applications, 10(12), 1159–1177. 10.1002/prca.201600015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, & Robinson RA (2016b). A simple isotopic labeling method to study cysteine oxidation in Alzheimer’s disease: Oxidized cysteine-selective dimethylation (OxcysDML). Analytical and Bioanalytical Chemistry, 408(11), 2993–3004. 10.1007/s00216-016-9307-4. [DOI] [PubMed] [Google Scholar]
- Gu L, & Robinson RAS (2016). High-throughput endogenous measurement of S-nitrosylation in Alzheimer’s disease using oxidized cysteine-selective cPILOT. Analyst, 141(12), 3904–3915. 10.1039/C6AN00417B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G, Derakhshan B, Shi L, Campagne F, & Gross SS (2006). SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proceedings of the National Academy of Sciences of the United States of America, 103(4), 1012–1017. 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature Cell Biology, 7, 665–674. 10.1038/ncb1268. https://www.nature.com/articles/ncb1268#supplementary-information. [DOI] [PubMed] [Google Scholar]
- Hart GW, Housley MP, & Slawson C (2007). Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature, 446, 1017–1022. 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, & Minghim R (2015). InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics, 16(1), 169–175. 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk KN, Hietbrink BN, Bartberger MD, McCarren PR, Choi BY, Voyksner RD, et al. (2003). Nitroxyl disulfides, novel intermediates in trans-nitrosation reactions. Journal of the American Chemical Society, 125(23), 6972–6976. 10.1021/ja029655l. [DOI] [PubMed] [Google Scholar]
- Hsu JL, Huang SY, Chow NH, & Chen SH (2003). Stable-isotope dimethyl labeling for quantitative proteomics. Analytical Chemistry, 75(24), 6843–6852. 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, & Snyder SH (2001). Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nature Cell Biology, 3, 193–197. 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, & Snyder SH (2001). The biotin switch method for the detection of S-nitrosylated proteins. Science’s STKE, 2001(86), pl1–pl9. 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, & Martinou JC (2003). hFis1, a novel component of the mammalian mitochondrial fission machinery. The Journal of Biological Chemistry, 278(38), 36373–36379. 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Jia J, Arif A, Terenzi F, Willard B, Plow EF, Hazen SL, et al. (2014). Target-selective protein S-nitrosylation by sequence motif recognition. Cell, 159(3), 623–634. 10.1016/j.cell.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WM, Wilson-Delfosse AL, & Mieyal JJ (2012). Dysregulation of glutathione homeostasisin neurodegenerative diseases. Nutrients, 4(10), 1399 Retrieved from http://www.mdpi.com/2072-6643/4/10/1399 (web archive link., 04 October 1399). http://www.mdpi.com/2072-6643/4/10/1399/pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabiraj P, Marin JE, Varela-Ramirez A, & Narayan M (2016). An 11-mer amyloid beta peptide fragment provokes chemical mutations and Parkinsonian biomarker aggregation in dopaminergic cells: A novel road map for “transfected” Parkinson’s. ACS Chemical Neuroscience, 7(11), 1519–1530. 10.1021/acschemneuro.6b00159. [DOI] [PubMed] [Google Scholar]
- Kohr MJ, Aponte AM, Sun J, Wang G, Murphy E, Gucek M, et al. (2011). Characterization of potential S-nitrosylation sites in the myocardium. American Journal of Physiology—Heart and Circulatory Physiology, 300(4), H1327–H1335. Retrieved from http://ajpheart.physiology.org/content/300/4/H1327.abstract. https://www.physiology.org/doi/pdf/10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski LP (2017). Proteome-pI: Proteome isoelectric point database. Nucleic Acids Research, 45(D1), D1112–D1116. 10.1093/nar/gkw978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y-D, Ma T, Diao S, Zhang X, Chen Y, Hsu J, et al. (2010). NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Molecular Neurodegeneration, 5(1), 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y-D, Wang R, Li JJ, Zhang Y-W, Xu H, & Liao F-F (2011). Differential regulation of BACE1 expression by oxidative and nitrosative signals. Molecular Neurodegeneration, 6(1), 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacampagne A, Liu X, Reiken S, Bussiere R, Meli AC, Lauritzen I, et al. (2017). Post-translational remodeling of ryanodine receptor induces calcium leak leading to Alzheimer’s disease-likepathologies and cognitive deficits. Acta Neuropathologica, 134(5), 749–767. 10.1007/s00401-017-1733-7. [DOI] [PubMed] [Google Scholar]
- Lakunina V, Petrushanko I, Burnysheva K, Mitkevich V, & Makarov A (2017). Alzheimer’s disease Aβ 42 peptide induces an increase in Na, K-ATPase glutathionylation. Doklady Biochemistry and Biophysics, 473(1), 114–117. Retrieved from https://link.springer.com/content/pdf/10.1134%2FS1607672917020077.pdf. [DOI] [PubMed] [Google Scholar]
- Lennicke C, Rahn J, Heimer N, Lichtenfels R, Wessjohann LA, & Seliger B (2016). Redox proteomics: Methods for the identification and enrichment of redox-modified proteins and their applications. Proteomics, 16(2), 197–213. 10.1002/pmic.201500268. [DOI] [PubMed] [Google Scholar]
- Li C, Feng J-J, Wu Y-P, & Zhang G-Y (2012). Cerebral ischemia-reperfusion induces GAPDH S-nitrosylation and nuclear translocation. Biochemistry (Moscow), 77(6), 671–678. 10.1134/S0006297912060156. [DOI] [PubMed] [Google Scholar]
- Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, et al. (2002). Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Archives of Biochemistry and Biophysics, 406(2), 229–240. Retrieved from https://ac.els-cdn.com/S000398610200468X/1-s2.0-S000398610200468X-main.pdf?_tid=925d5c31-3a40-4234-9900-d09746afb5a8&acdnat=1544752207_710cf362af708ac9ef46aa3fb6e6a4ed. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi Y-B, Pan Z-H, Lei SZ, Chen H-SV, Sucher NJ, et al. (1993). A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature, 364, 626–632. 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- López-Sánchez LM, López-Pedrera C, & Rodríguez-Ariza A (2014). Proteomic approaches to evaluate protein s-nitrosylation in disease. Mass Spectrometry Reviews, 33(1), 7–20. 10.1002/mas.21373. [DOI] [PubMed] [Google Scholar]
- Maehama T, & Dixon JE (1998). The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of Biological Chemistry, 273(22), 13375–13378. Retrieved from http://www.jbc.org/content/273/22/13375.full.pdf. [DOI] [PubMed] [Google Scholar]
- Maher A, Rahman MFA, & Gad MZ (2017). The role of nitric oxide from neurological disease to cancer In El-Khamisy S (Ed.), Personalised medicine (pp. 71–88). Springer. [DOI] [PubMed] [Google Scholar]
- Marino SM, & Gladyshev VN (2010). Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. Journal of Molecular Biology, 395(4), 844–859. 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ruiz A, Cadenas S, & Lamas S (2011). Nitric oxide signaling: Classical, less classical, and nonclassical mechanisms. Free Radical Biology and Medicine, 51(1), 17–29. 10.1016/j.freeradbiomed.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Mayer B, & Andrew P (1998). Nitric oxide synthases: Catalytic function and progress towards selective inhibition. Naunyn-Schmiedeberg’s Archives of Pharmacology, 358(1), 127–133. [DOI] [PubMed] [Google Scholar]
- Min S-W, Sohn PD, Li Y, Devidze N, Johnson JR, Krogan NJ, et al. (2018). SIRT1 deacetylates tau and reduces pathogenic tau spread in a mouse model of tauopathy. The Journal of Neuroscience, 38(15), 3680–3688. 10.1523/jneurosci.2369-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SW, Sohn PD, Cho SH, Swanson RA, & Gan L (2013). Sirtuins in neurodegenerative diseases: An update on potential mechanisms. Frontiers in Aging Neuroscience, 5, 53–61. 10.3389/fnagi.2013.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molokanova E, Akhtar MW, Sanz-Blasco S, Tu S, Pina-Crespo JC, McKercher SR, et al. (2014). Differential effects of synaptic and extrasynaptic NMDA receptors on Aβ-induced nitric oxide production in cerebrocortical neurons. Journal of Neuroscience, 34(14), 5023–5028. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3972726/pdf/zns5023.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, et al. (1997). P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proceedings of the National Academy of Sciences of the United States of America, 94(17), 9052–9057. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9256433. https://www.ncbi.nlm.nih.gov/pmc/PMC23024/. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC23024/pdf/pq009052.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Cieplak P, Cho D-H, Godzik A, & Lipton SA (2010). S-nitrosylation of Drp1 links excessive mitochondrial fission to neuronal injury in neurodegeneration. Mitochondrion, 10(5), 573–578. Retrieved from https://ac.els-cdn.com/S156772491000070X/1-s2.0-S156772491000070X-main.pdf?_tid=f3c25002-8753-4d43-9922-b1e8768d19ab&acdnat=1537204261_bb74b0bb46413ed0e99f321af990f0b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, & Lipton SA (2013). Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxidants & Redox Signaling, 18(3), 239–249. 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, & Lipton SA (2017). ‘SNO’-storms compromise protein activity and mitochondrial metabolism in neurodegenerative disorders. Trends in Endocrinology and Metabolism, 28(12), 879–892. 10.1016/j.tem.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Prikhodko OA, Pirie E, Nagar S, Akhtar MW, Oh C-K, et al. (2015). Aberrant protein S-nitrosylation contributes to the pathophysiology of neurodegenerative diseases. Neurobiology of Disease, 84, 99–108. Retrieved from https://ac.els-cdn.com/S0969996115000893/1-s2.0-S0969996115000893-main.pdf?_tid=75bbe070-8f90-48fa-be71-392958e17cc6&acdnat=1536860884_5ed2e04d364eaf9a29f86479e84ece8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, et al. (2010). Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Molecular Cell, 39(2), 184–195. Retrieved from https://ac.els-cdn.com/S1097276510005241/1-s2.0-S1097276510005241-main.pdf?_tid=6148f140-1b5d-4032-a5da-1755bdad4703&acdnat=1537204602_c0a1d129062efdb9801eb2533f4944f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, et al. (2007). An increase in S-glutathionylated proteins in the Alzheimer’s disease inferior parietal lobule, a proteomics approach. Journal of Neuroscience Research, 85(7), 1506–1514. 10.1002/jnr.21275. [DOI] [PubMed] [Google Scholar]
- Pierrefiche O, & Naassila M (2014). Endogenous nitric oxide but not exogenous no-donor S-nitroprussiate facilitates NMDA excitation in spontaneous rhythmic neonatal rat brainstem slice. Brain Research, 1543, 9–16. 10.1016/j.brainres.2013.10.042. [DOI] [PubMed] [Google Scholar]
- Poulsen K, Bahl JM, Simonsen AH, Hasselbalch SG, & Heegaard NH (2014). Distinct transthyretin oxidation isoform profile in spinal fluid from patients with Alzheimer’s disease and mild cognitive impairment. Clinical Proteomics, 11(1), 12–20. 10.1186/1559-0275-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Nakamura T, Cao G, Holland EA, McKercher SR, & Lipton SA (2011). S-nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proceedings of the National Academy of Sciences of the United States of America, 108(34), 14330–14335. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3161554/pdf/pnas.201105172.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani P, Krishnan S, & Rani Cathrine C (2017). Study on analysis of peripheral biomarkers for Alzheimer’s disease diagnosis. Frontiers in Neurology, 8, 328 10.3389/fneur.2017.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer IM, Schiffrin M, Kovari E, Bouras C, & Riederer BM (2009). Ubiquitination and cysteine nitrosylation during aging and Alzheimer’s disease. Brain Research Bulletin, 80(4–5), 233–241. 10.1016/j.brainresbull.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Ryu I-H, Lee K-Y, & Do S-I (2016). Aβ-affected pathogenic induction of S-nitrosylation of OGT and identification of Cys-NO linkage triplet. Biochimica et Biophysica Acta Proteins and Proteomics, 1864(5), 609–621. Retrieved from https://ac.els-cdn.com/S1570963916300085/1-s2.0-S1570963916300085-main.pdf?_tid=7ce3104d-94d8-4b43-bcb9-7434487cbf3c&acdnat=1536618446_275b213a9ea80851973f0e11daa58ad6. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ (2002). Alzheimer’s disease is a synaptic failure. Science, 298(5594), 789–791. 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Sen T, Saha P, & Sen N (2018). Nitrosylation of GAPDH augments pathological tau acetylation upon exposure to amyloid-β. Science Signaling, 11(522), 1–21. 10.1126/scisignal.aao6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne U, Nott A, Bhat VB, Ravindra KC, Wishnok JS, Tsai LH, et al. (2016). S-nitrosation of proteins relevant to Alzheimer’s disease during early stages of neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America, 113(15), 4152–4157. 10.1073/pnas.1521318113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth D, Hess DT, Hausladen A, Wang L, Wang Y-J, & Stamler JS (2018). A multiplex enzymatic machinery for cellular protein S-nitrosylation. Molecular Cell, 69(3), 451–464.e456. 10.1016/j.molcel.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, & Lahiri DK (2014). Cdk5 activity in the brain—multiple paths of regulation. Journal of Cell Science, 127(11), 2391–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, & Marletta MA (2012). Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Current Opinion in Chemical Biology, 16(5), 498–506. 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J-L, & Wang C-C (1995). Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. European Journal of Biochemistry, 231(2), 312–316. 10.1111/j.1432-1033.1995.0312e.x. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, & Sucher NJ (1997). (S)NO signals: Translocation, regulation, and a consensus motif. Neuron, 18(5), 691–696. 10.1016/S0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Stomberski CT, Hess DT, & Stamler JS (2019). Protein S-nitrosylation: Determinants of specificity and enzymatic regulation of S-nitrosothiol-based signaling. Antioxidants & Redox Signaling, 30(10), 1131–1351. 10.1089/ars.2017.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talantova M, Sanz-Blasco S, Zhang X, Xia P, Akhtar MW, Okamoto S-I, et al. (2013). Aβ induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss. Proceedings of the National Academy of Sciences of the United States of America, 110(27), E2518–E2527. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3704025/pdf/pnas.201306832.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talipov MR, & Timerghazin QK (2013). Protein control of S-nitrosothiol reactivity: Interplay of antagonistic resonance structures. The Journal of Physical Chemistry B, 117(6), 1827–1837. 10.1021/jp310664z. [DOI] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. (2003). Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry, 75(8), 1895–1904. [DOI] [PubMed] [Google Scholar]
- Torta F, Usuelli V, Malgaroli A, & Bachi A (2008). Proteomic analysis of protein S-nitrosylation. Proteomics, 8(21), 4484–4494. 10.1002/pmic.200800089. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, et al. (2008). Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO Journal, 27(2), 433–446. 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi Z-Q, Gu Z, Ma Y, et al. (2006). S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature, 441(7092), 513–517. Retrieved from https://www.nature.com/articles/nature04782.pdf. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A (2005). Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiological Reviews, 85(1), 201–279. 10.1152/physrev.00004.2004. [DOI] [PubMed] [Google Scholar]
- Vuori K, Myllyla R, Pihlajaniemi T, & Kivirikko KI (1992). Expression and site-directed mutagenesis of human protein disulfide isomerase in Escherichia coli. This multifunctional polypeptide has two independently acting catalytic sites for the isomerase activity. The Journal of Biological Chemistry, 267(11), 7211–7214. Retrieved from http://www.jbc.org/content/267/11/7211.full.pdf. [PubMed] [Google Scholar]
- Wadey AL, Muyderman H, Kwek PT, & Sims NR (2009). Mitochondrial glutathione uptake: Characterization in isolated brain mitochondria and astrocytes in culture. Journal of Neurochemistry, 109(Suppl. 1), 101–108. 10.1111/j.1471-4159.2009.05936.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Yuan Y, Zhang P, Zhang H, Liu X, & Zhang Y (2018). Neohesperidin prevents Aβ25–35-induced apoptosis in primary cultured hippocampal neurons by blocking the S-nitrosylation of protein-disulphide isomerase. Neurochemical Research, 43(9), 1736–1744. 10.1007/s11064-018-2589-5. [DOI] [PubMed] [Google Scholar]
- Wang S, Circu ML, Zhou H, Figeys D, Aw TY, & Feng J (2011). Highly sensitive detection of S-nitrosylated proteins by capillary gel electrophoresis with laser induced fluorescence. Journal of Chromatography A, 1218(38), 6756–6762. 10.1016/j.chroma.2011.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Njoroge SK, Battle K, Zhang C, Hollins BC, Soper SA, et al. (2012). Two-dimensional nitrosylated protein fingerprinting by using poly (methyl methacrylate) microchips. Lab on a Chip, 12(18), 3362–3369. 10.1039/c2lc40132k. [DOI] [PubMed] [Google Scholar]
- Wang S, Song J, Tan M, Albers K, & Jia J (2012). Mitochondrial fission proteins in peripheral blood lymphocytes are potential biomarkers for Alzheimer’s disease. European Journal of Neurology, 19(7), 1015–1022. Retrieved from https://onlinelibrary.wiley.com/doi/pdf/10.1111/j.1468-1331.2012.03670.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Veremeyko T, Wong AH-K, El Fatimy R, Wei Z, Cai W, et al. (2017). Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiology of Aging, 51, 156–166. Retrieved from https://ac.els-cdn.com/S0197458016303281/1-s2.0-S0197458016303281-main.pdf?_tid=371a060d-276d-46ed-9ba7-3d1c8a5a1080&acdnat=1536603699_25b39ce188aef958e1e71b80a34ca48f. [DOI] [PubMed] [Google Scholar]
- Won J-S, Kim J, Annamalai B, Shunmugavel A, Singh I, & Singh AK (2013). Protective role of S-nitrosoglutathione (GSNO) against cognitive impairment in rat model of chronic cerebral hypoperfusion. Journal of Alzheimer’s Disease, 34(3), 621–635. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4040220/pdf/nihms569802.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR 3rd. (1998). Mass spectrometry and the age of the proteome. Journal of Mass Spectrometry, 33(1), 1–19. . [DOI] [PubMed] [Google Scholar]
- Zahid S, Khan R, Oellerich M, Ahmed N, & Asif A (2014). Differential S-nitrosylation of proteins in Alzheimer’s disease. Neuroscience, 256, 126–136. Retrieved from https://ac.els-cdn.com/S0306452213008798/1-s2.0-S0306452213008798-main.pdf?_tid=55f32e3d-c0b3-48db-a3e0-f3d333b279a6&acdnat=1536877816_b2bbbdf4421741e70b6af2712b376360. [DOI] [PubMed] [Google Scholar]
- Zareba-Koziol M, Szwajda A, Dadlez M, Wyslouch-Cieszynska A, & Lalowski M (2014). Global analysis of S-nitrosylation sites in the wild type (APP) transgenic mouse brain-clues for synaptic pathology. Molecular & Cellular Proteomics, 13(9), 2288–2305. 10.1074/mcp.M113.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kuo C-C, Chiu AWL, & Feng J (2012). Prediction of S-glutathionylated proteins progression in Alzheimer’s transgenic mouse model using principle component analysis. Journal of Alzheimer’s Disease, 30(4), 919–934. 10.3233/JAD-2012-120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rodriguez C, Circu ML, Aw TY, & Feng J (2011). S-glutathionyl quantification in the attomole range using glutaredoxin-3-catalyzed cysteine derivatization and capillary gel electrophoresis with laser-induced fluorescence detection. Analytical and Bioanalytical Chemistry, 401(7), 2165–2175. Retrieved from https://link.springer.com/content/pdf/10.1007%2Fs00216-011-5311-x.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ye Z-W, Singh S, Townsend DM, & Tew KD (2018). An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radical Biology and Medicine, 120, 204–216. 10.1016/j.freeradbiomed.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fonslow BR, Shan B, Baek M-C, & Yates JR (2013). Protein analysis by shotgun/bottom-up proteomics. Chemical Reviews, 113(4), 2343–2394. 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu Z, Liang H-Y, Zhang L, Zhou Q-G, Ni H-Y, et al. (2018). nNOS–CAPON interaction mediates amyloid-β-induced neurotoxicity, especially in the early stages. Aging Cell, 17(3), e12754–e12765. 10.1111/acel.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, & Xu H (2007). Molecular and cellular mechanisms for Alzheimer’s disease: Understanding APP metabolism. Current Molecular Medicine, 7(7), 687–696. Retrieved from http://www.eurekaselect.com/65935/article. [DOI] [PubMed] [Google Scholar]
- Zolochevska O, Bjorklund N, Woltjer R, Wiktorowicz JE, & Taglialatela G (2018). Postsynaptic proteome of non-demented individuals with Alzheimer’s disease neuropathology. Journal of Alzheimer’s Disease, 65(2), 659–682[Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]