Molecular detection of biological agents in the field has traditionally relied on the use of quantitative real-time PCR (qPCR), which now includes commercially available instruments that can be used in the laboratory or field. Adapting this technology for field-forward applications necessitated innovation to minimize size, weight, and power requirements.
KEYWORDS: detection, real-time PCR, diagnostics, genomic sequencing, field laboratory
ABSTRACT
Molecular detection of biological agents in the field has traditionally relied on the use of quantitative real-time PCR (qPCR), which now includes commercially available instruments that can be used in the laboratory or field. Adapting this technology for field-forward applications necessitated innovation to minimize size, weight, and power requirements. Rugged, portable instruments, efficient power sources, freeze-dried reagents, data communications, and standard operating procedures for minimally trained users are some examples of limitations that have been overcome to allow qPCR-based data to be generated at the point of need. Despite the high specificity and sensitivity of qPCR, the assays require a priori sequence-based knowledge of the etiological agent to design and produce specific targeted assays with primers and probes. However, in many cases the etiological agent may not be known and pathogen identification must rely on the use of an untargeted screening method. By extracting, preparing, and sequencing all of the genomic material in a particular sample at once, known as metagenomics, a less biased view of the biological entities in that sample can be ascertained. Using metagenomics methods in the field requires the development and optimization of straightforward sample preparation, sequencing, and bioinformatics workflows reminiscent of the challenges faced during the development of field-forward qPCR 15 years ago. To review the state of qPCR and sequencing in the field, we summarized a panel discussion from the 2019 ASM Biothreats Conference. Our discussion focused on the development, evolution, and comparison of molecular methods for biological agents and their utility in the field.
Deployment of a laboratory capability outside the clinical laboratory walls is the necessary response to diseases that readily cross international borders. The Laboratory Without Walls is our answer to diseases without borders.
—Timothy J. J. Inglis (1)
Detection, identification, and characterization of biological agents, especially in the field, is a constant challenge due to the complex and variable nature of biological material. Traditionally, the most sensitive methods require the use of culture-based approaches, which allow for pathogen identification through demonstration of viability. Culture-based methods require specific equipment and laboratory personnel, and early PCR relied on a priori knowledge of the suspected pathogen. In addition, culture-based pathogen detection requires high turnaround times (TAT), from days to months, depending on the organism. Therefore, a variety of culture-independent diagnostic tests (CIDTs), especially those employing nucleic acid amplification and next-generation sequencing (NGS), have been developed for rapid pathogen detection and are better suited for field applications (2). By coupling sample triage via molecular methods at the point of need (PON) with expedient sample transport to a fixed laboratory setting, the TAT for pathogen detection is greatly reduced. Many organizations, including military defense, intergovernmental-international, public and veterinary health, and law enforcement, deploy field laboratories in various applications, such as detection of weapons of mass destruction (WMD), infectious disease outbreaks, and treaty verification (3). Since targeted assays limit pathogen detection to the number of targets included in the assay panel, further efforts are needed to fully develop unbiased sample analysis in the field. These efforts should focus on the development of rugged laboratory instruments, especially size, weight, and power (SWaP), reagents without the need for cold storage, and simple standard operating procedures (1).
The most common field-forward technology relies on using PCR, which was discovered by the late Kary Mullis in 1985 and revolutionized molecular biology (4). This discovery earned K. Mullis the Nobel Prize in Chemistry in 1993 and has since evolved to enable real-time analysis and quantitation of target nucleic acids through the use of real-time or qPCR instruments and corresponding chemistries (5). Real-time detection of target nucleic acids is based on the utilization of fluorescent, intercalating dyes or dually labeled probes containing a fluorophore and quencher, such as TaqMan hydrolysis probes or single-labeled fluorescent resonance energy transfer (FRET) hybridization probes (6–8). The most commonly used method, TaqMan, utilizes Taq polymerase for the amplification of target DNA and hydrolysis of the dually labeled probe, resulting in an increase in fluorescence that is detected by the instrument. Alternatively, the use of FRET probes or intercalating dyes enables both the detection of target DNA and melting curve analysis (sample fluorescence versus temperature) where post-PCR amplification yields melting peaks (first negative derivative of sample fluorescence versus temperature) according to single-nucleotide sequences (9).
The availability of targeted qPCR assays, especially those designed and validated for commercial use, has increased the advantages of this technology, since the suspected target agent can be detected within a few hours with minimal hands-on time. Real-time thermal cyclers have evolved to enable the simultaneous analysis of up to 384 samples in a single run (10–12). In addition, the use of multiplex assays allows for testing of more than one target and the incorporation of internal controls in a single sample. Alternatively, field-forward qPCR instruments are smaller, lightweight, rugged instruments with lower throughput to enable sample analysis in the field (13).
Since the confirmation of the three-dimensional structure of DNA by Watson and Crick, there has been a significant amount of research and progress made toward developing methods for reading the genetic code inscribed in DNA molecules. The ability to read DNA sequences was revolutionized by the advent of Fred Sanger’s chain termination method (14, 15). This breakthrough Sanger sequencing method established the first generation of DNA sequencing and serves as the gold-standard method for DNA analysis (16). The second generation of sequencing, often referred to as NGS, was significant for its introduction of massively parallel sequencing, allowing for higher throughput of sequencing millions of reads per run (17). Currently, the third generation of DNA sequencing, in its nascent stages, is focused on the sequencing of single DNA molecules (18). Long-read sequencing technologies are currently available to perform this innovative sequencing method (19, 20). Advancements in NGS protocols and instrumentation have enabled faster sample analysis with reduced cost. The use of NGS-based approaches offers the advantage of unbiased, metagenomic analysis of a sample without the need for any a priori knowledge of the suspected etiological agent. NGS-based sample analysis is beneficial for the detection of polymicrobial infections, detection of antimicrobial resistance, surveillance, and identification of novel pathogens in a single assay. In addition, this method allows for pathogen identification where other targeted approaches may fail. Despite the advantages, there has not been widespread adaptation of these methods in the field due to the need for cold storage, use of large instrumentation, and a trained bioinformatician. In recent years, commercially available, handheld devices capable of sequencing in the field have enabled pathogen detection in <6 h (21–23). Meeting operational constraints in the field, such as remote power, eliminating cold storage, and incorporating simple, intuitive product design, have allowed minimally trained users to run tests and report results (24). However, further development for sample preparation and data analysis is required to enable the implementation of sequencing for routine sample analysis.
In general, as the technology for qPCR and NGS continue to advance, the utility of NGS in the field is less mature than current qPCR products and their concepts of operation. Currently, qPCR still maintains higher analytical sensitivity, specificity, reproducibility, and ease of use over NGS. In addition, qPCR has an advantage in turnaround time for actionable results and lower cost per test (2). However, NGS retains the advantage of discovery power and scalability in the number of markers that can be analyzed simultaneously (25, 26). Significant progress has been made to address SWaP and the portability and ruggedness of performing qPCR and NGS in field-forward settings (27, 28). Our workshop, “Detection of Biological Agents: Then and Now,” focused on the development, evolution, and comparison of qPCR and sequencing as related to the challenge and these associated issues (Fig. 1).
FIG 1.
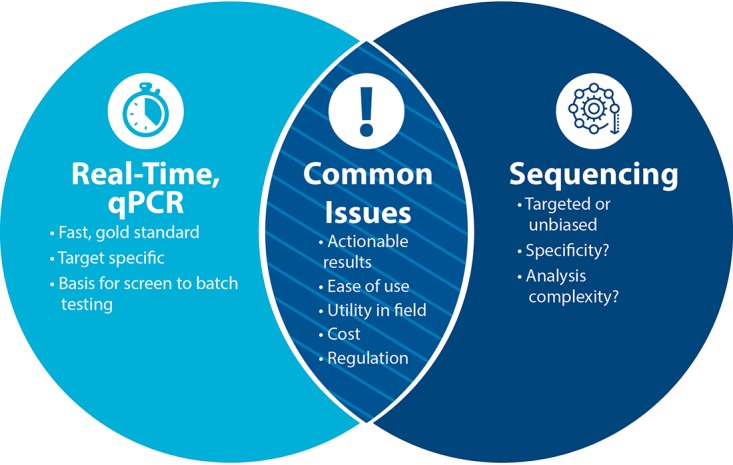
Comparison of qPCR and sequencing-based molecular methods. This Venn diagram illustrates the advantages of each technology and the common issues. (Courtesy of Christina May, reproduced with permission.)
WORKSHOP DESIGN, STRUCTURE, AND TOPICS
The panel discussion, moderated by Kenneth Yeh, included Matt Scullion, Bryan Gnade, Anthony Jones, Kay Mereish, Joe Russell, and Chris Whittier. Our objective was to craft a relevant discussion on two convergent areas of technical and operational concepts that would be moderated among expert panelists. Before the panel discussion, the moderator and team worked with the panelists to define topics, objectives, and questions. Our objective also included collaborating to develop and publish the findings of our panel discussion to further reinforce the value of presenting relevant topics and issues at scientific conferences, especially those presented through professional organizations such as the American Society for Microbiology (ASM).
DISCUSSION
First movers and fast followers.
The products and related systems developed for field-forward applications, including technologies based on qPCR and NGS, come from companies and performers that can be termed “first movers and fast followers,” which was derived from an industrial economics model that has been applied to marketing technology (29). Often first movers are considered the pack leaders, pioneers, and first to market by performing as the tip of the spear. They have successfully applied novel technology through patenting research and development concepts, accelerated their product development, especially with early end user champions, and leveraged strategic sales, marketing with business, and successful market entry (30). Simon Sinek coined the Law of Diffusion, which is a theory on how, why, and how fast new ideas and technology spread where first movers and fast followers are at the forefront of this ecosystem. Although second to market, fast followers have first-mover characteristics and are still competitive and agile.
Earlier, the military and defense focused on capabilities and requirements for detecting biological agents in the field, which the 2001 anthrax attacks in the U.S. amplified, resulting in broader efforts across the U.S. government, thereby creating a biodefense enterprise along with related security applications. In order for qPCR and NGS products to be successful in the field, operational discriminators, including ease of use, portability, ruggedness, speed, sensitivity, specificity, reproducibility, discovery power, and throughput, need to be balanced. Based on our literature search (25–28, 31, 32) and the panel’s observations, we present a notional qualitative comparison of the two technologies that can serve as the basis for a further questionnaire to collect data and feedback from end users (Fig. 2).
FIG 2.
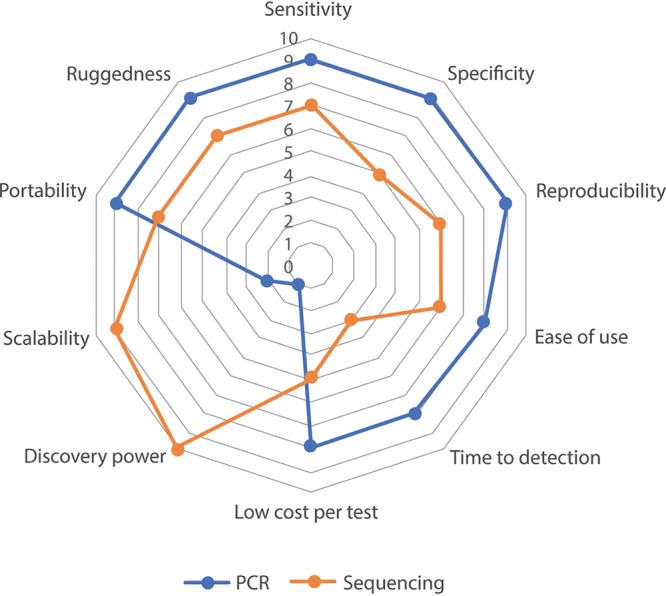
Comparison of the state the art of qPCR and sequencing. The qualitative analysis is based on our observations and literature search based on these ten criteria.
Costs, in terms of consumables per test, life cycle, instrument maintenance/warranties, and operator training, are also important. Our panel discussion included expert panelists representing examples of first movers and fast followers in the field.
(i) qPCR. One first mover for molecular detection of biological agents in the field was BioFire, formerly Idaho Technology, Inc. BioFire optimized an existing laboratory-based qPCR platform, called the R.A.P.I.D., for field-forward applications. During the mid-1990s to the early 2000s, this optimization focused on making a rugged instrument and developing freeze-dried reagents and automated software analysis to aid in field logistics and improve ease of use.
BioFire also fielded three different military field PCR systems (RAZOR, JBAIDS, and FilmArray) for biologic identification with FDA clearances, including a clinical laboratory improvement amendments (CLIA)-waived system. In the mid-2000s, they developed the field portable system RAZOR, working with the U.S. Special Operations Command. They incorporated the features from the previous generation of field-ready qPCR systems (rugged, freeze-dried reagents, and autoanalysis), minimized the system footprint, and added battery power. The most important advancement they made was in its ease of use by eliminating pipetting and minimizing the number of steps required to generate results. In the last decade, BioFire further integrated their systems with sample preparation and increased their multiplex capacity, all while improving the ease of use and reducing the number of steps to obtain results.
Future technology includes “Extreme PCR,” which is a phrase coined by its inventor, Carl Wittwer (one of Idaho Technology’s and BioFire’s founders), and refers to techniques that can shorten qPCR runs from tens of minutes to tens of seconds. While the current time to results (TTR) for a multiplexed CLIA-waived test of 45 min is acceptable, it does not fully satisfy the needs of the point-of-care market.
(ii) Sequencing. The sequencing space has seen several iterations of first movers and fast followers, many of which transitioned into first casualties and fast burials. The most notable and dramatic instances of the latter were primarily seen in second-generation sequencing. Pyrosequencing (e.g., 454), semiconductor sequencing (e.g., Ion Torrent), and sequencing by ligation (e.g., SOLiD) all brought unique advantages with their technology and enjoyed brief periods of substantial market penetration. However, their advantages could not eclipse the quality and reliability of first-generation chain-terminator sequencing (which some will argue still maintains gold-standard status) or compete with the throughput, read length, and low per-base cost of third-generation platforms, such as sequencing by synthesis (i.e., Illumina), single-molecule sequencing (i.e., PacBio), or nanopore sequencing (i.e., Oxford Nanopore Technologies [ONT]). However, even among the third-generation offerings dominating the market for the past 5 years, critical differentiating factors are emerging that indicate where the sequencing market is moving and how sequencing data will be leveraged in the next 5 to 10 years. Among the leading third-generation platforms mentioned above, ONT can be considered a first mover in field-forward sequencing, or sequencing outside of a traditional brick-and-mortar laboratory at or near the location of sampling. It is the first and, as of now, only sequencing platform that can be reasonably considered to be portable. The paradigm shift associated with portable sequencing is centered on the potential for sequence-based information being generated at the point of need rather than traditional hospitals, advanced clinical settings, or research laboratories. In other words, the benefits of positive health outcomes, biosphere management, and natural product discovery associated with genomic sequencing will no longer be limited to those fortunate enough to live in close proximity to more traditional research facilities. The option of bringing the laboratory to the sample, rather than the other way around, is now feasible. Therein, however, lies the remaining development challenges in order to fully realize the small form factor of emerging sequencing hardware: you still need to bring the laboratory. Front-end sample preparation (i.e., nucleic acid extraction, target molecule enrichment, library preparation, etc.), as well as back-end data analysis (i.e., bioinformatics, taxonomy assignment, sequence assembly, functional annotation, phylogeny, etc.) lag behind the sequencing hardware as related to portability and usability. The full, end-to-end process of portable sequencing has a much larger footprint than the hardware itself. To effectively support and execute the full process, one needs (i) a robust portable power source to support (ii) substantial computational capacity needed for basecalling and analyzing sequence data, (iii) cold-storage solutions for flow cells and sequencing reagents, (iv) an ergonomic approximation of a laboratory workbench to perform manual workflow steps and access different pipettes, pipette tips, and other consumables, (v) biosafety and biohazard storage/disposal options, and (vi) accessory hardware/devices for library preparation (e.g., thermocycler, minicentrifuge, etc.), at a minimum. ONT has developed some accessory hardware to address some of these issues, namely, the Voltrax device (an automated library preparation device) and the MinIT (a GPU-based computational device that integrates the MinION operating software and rapid basecalling). Recently, ONT has developed a field sequencing kit comprised of lyophilized reagents that do not require cold storage. Third-party developers have begun to innovate further solutions, such as FPGA-mediated basecalling that improves on the scalability of the MinIT (33), optimized algorithms to more efficiently analyze nanopore data (34, 35), and mobile laboratory structures that arrange logistical requirements into single-person portable configurations (e.g., http://www.mriglobal.org/2018/11/08/mercury-labs/). Fast followers to ONT’s first-mover advantage include integrated device manufacturers such as Ontera, makers of a nanopore-based molecular detection platform in a portable form factor. While not explicitly a sequencing technology, this platform offers a novel method of detecting molecular signatures from a range of targets, including DNA, RNA, and protein, in point-of-need settings.
Significant innovation potential remains in streamlined, push-button bioinformatics software with graphical user interfaces (GUIs) and standardized reporting nomenclature, as well as automation of wet-lab processing steps (e.g., DNA/RNA extraction from various sample matrices, molecular enrichment, sequencing library preparation, etc.). Such innovation will be necessary to move the technology into routine biosurveillance and clinical settings subject to regulatory frameworks. Regulatory frameworks themselves will need to evolve to accommodate emerging markets of validated sequence-based point-of-care diagnostic tests, point-of-sampling food safety and agriculture surveillance tests, point-of-entry supply chain authentication tests, and forensic human identification, to name a few examples. Ethical policy related to human DNA testing has not kept pace with the utility of field-forward sequencing. Recent DNA testing at the U.S. and Mexico border and consumer DNA testing at large have highlighted the need for established norms on consent, acceptable procurement, and legal use of a person’s genomic data.
Given the diversity of sample matrices, genome-based maladies, functional utility of genomic information in biosurveillance, and lack of regulatory and policy consensus on the generation and use of genomic information in novel settings, a robust emerging market addressing these concepts is poised to rapidly grow over the coming decade.
Retrospective example in the field: issue of weaponized anthrax.
After the first and second Gulf Wars, the United Nations formed two special commissions: United Nations Special Commission (UNSCOM) and then United Nations Monitoring, Verification, and Inspection Commission (UNMOVIC) to inspect, verify, destroy, and monitor weapons of mass destruction (WMD) of Iraq. Iraq produced several biological agents and weaponized them by filling certain munitions, such as the R400 bomb. The inspectors’ approach to this task was similar to that of a forensic scientist, thereby accounting for and verifying what Iraq declared for production, weaponization, and unilateral destruction. Although the UNSCOM’s work often rests on UN Security Council decisions and related diplomatic and political factors, their experts in the field were very much fast followers when adapting new technology. In 2002, UNMOVIC fielded a mobile laboratory that had chemical and biological analysis capabilities. In the biological analysis, the laboratory heavily relied on qPCR (Idaho Technology), immunoassay techniques, and others funded by different governments.
In 2002, UNMOVIC was permitted to excavate a site declared to be where anthrax-filled R400 bombs were destroyed. The inspectors unearthed 8 intact bombs, filled with liquid, which were suspected to contain Clostridium botulinum, aflatoxin, or anthrax, because those agents were declared to fill R400 bombs. The verification of the R400 bombs’ contents followed a primary, secondary, and tertiary process (Fig. 3).
FIG 3.

Sampling from R400 bomb body that was filled with liquid agent. (Photo taken by Kay Mereish.)
The primary verification process started with safe drilling to discern if the R400s were filled with chemical warfare agents, and then the samples were taken to the field laboratory and screened for chemical warfare agents and enzyme-linked immunosorbent assay (ELISA) and PCR technology to identify the biological agent (Fig. 4). In this case, potassium permanganate, which the Iraqis used to deactivate the biological agent-filled bombs, presented a challenge as a chemical interferent to the PCR analysis. Besides chemical interferents, earlier issues included performing DNA extraction, which was limited by the current quality of kits used in the field.
FIG 4.

Excavated R400 bombs from Al Azizia site. On the left of the photo is the drilling prototype device called MONICA. (Photo taken by Kay Mereish.)
The secondary and tertiary verification process was done at the reference laboratory by following standard chain-of-custody procedures, while the primary process was done by the UNMOVIC’s inspectors on-site. Two international reference laboratories tested the samples for chemical warfare and biological warfare agents, and the results confirmed the presence of potassium permanganate and Bacillus anthracis DNA. The results of the reference laboratory analysis were reported to the Security Council. The genome of Bacillus anthracis was identified using a multilocus variable number of tandem repeat analysis (ML-VNTR), published in 2000 (36).
Retrospectively, the UNMOVIC field laboratory deployed for testing biological warfare agents was the first of its kind, and it was successful in making analyses in a field-forward setting where samples were confidently shipped to reference laboratories for confirmation. Back in 2003, UNMOVIC proposed to apply qPCR analysis at the Al Hakam facility, which was the Iraqi disposal site for bulk agent. Although inspectors could not convince the UN to validate their proposed protocol for field use, their results from the field laboratory in conjunction with the results from the reference laboratories could have been used as a model to support the ongoing negotiations to establish a verification protocol for the Biological Weapons Convention (BWC) by an ad hoc group of experts. Currently, there are no further negotiations or plans to establish a verification protocol for the BWC similar to that of the Chemical Weapons Convention.
Other experiences from the field.
In addition to the treaty verification example, which applied qPCR in the field and confirmatory sequencing at a reference laboratory, our expert panelists described their experiences as fast followers in the field. Examples for military (clinical diagnostics) and wildlife health applications were described. Their experiences, which mainly used qPCR platforms, reflect the need and objective for rapid identification of biological agents in addition to the use of mature products, such as detection assays, to meet PON.
(i) 2014–2016 Outbreak of Ebola. When the U.S. military deployed to West Africa in response to the outbreak of Ebola in 2014, they faced a significant challenge to meet a workload that would eventually total over 32,000 samples tested from 2014 to 2016. The EZ1 Ebola Zaire assay, developed by the Diagnostic Systems Division at USAMRIID, was utilized by a team of Army, NIAID, and local national laboratorians working at the Liberian Institute for Biomedical Research to meet this need. Thanks to some forward thinkers, the EZ1 assay had been prepositioned with the FDA as a “Pre-EUA” test for the BioFire JBAIDS, Roche LightCycler, and ABI 7500 Fast DX systems. It was quickly updated and subsequently authorized for emergency use (EUA) by the FDA in August 2014, making it the first EUA Ebola assay available for U.S. personnel in the affected region. The ability to develop content on open platforms was critical to the success of the EZ1 assay. This demonstrated the obvious benefit to working with more open platforms that are more usable, and in a resource-constrained environment this development philosophy complements other efforts seeking fully cleared in vitro diagnostic (IVD) devices. The vision of the DOD’s diagnostics development leadership more than a decade ago, along with the large number of personnel willing to deploy to the region and committed local laboratory staff, succeeded in meeting the challenge of the 2014–2016 Ebola outbreak.
(ii) Afghanistan. U.S. DOD doctrine lists five roles, or levels, of medical care patients may receive across first care: Role 1 (unit level), Role 2 (emergency trauma care), Role 3 (medical treatment facility), and Role 4 (hospitals). Another example of military and defense experience was related to a deployment in Afghanistan, which was at a Role 3 medical treatment facility in Afghanistan. The priority of the Role 3 medical treatment facility was on blood bank and blood chemistry, which can be measured using handheld devices, such as the Abbott Piccolo and i-STAT. The BioFire JBAIDS allowed us to have a field diagnostic system. The first challenge was making space in a mobile Conex-type laboratory to account for microbiology and molecular biology, which required clean and dirty areas.
The next challenge in the deployment was being familiar with the technology and associated concept of operations, i.e., organizing and quickly setting all of the necessary components and material. Completing simple operational tasks, such as managing computer passwords and routine communication issues that were not easily resolved, became very time-consuming in a demanding and unforgiving environment. Two JBAIDS instruments were used: one was functional and the other was for spare parts. Overall the system was compact, but material such as extraction kits and assorted consumable boxes were still larger than those used by our contemporaries in biochemistry. Cold-chain storage was unreliable and virtually nonexistent. Despite the operational challenges, the technology helped us get results relatively quickly but required technical expertise.
(iii) Understanding disease transmission in wildlife. Examining wildlife health in remote places was the third experience mentioned by one of the panelists. The example of testing wild mountain gorillas for Campylobacter, which is difficult to culture in the field, showed that qPCR technologies can be useful and practical for early detection of a pathogen and containing a potential outbreak (37). As noted, the ability to perform qPCR and sequencing in the field reduces TAT of results, which also benefits wildlife by rapidly informing treatment, potentially even while the animal is still anesthetized. Similar challenges for performing qPCR in the field, such as instrument power and lack of cold-chain storage, were also noted. When animal samples can be reliably obtained and analyzed in situ by qPCR, the timely results are valuable and often lower or remove the need for complicated export and import of animal samples that might be subject to various international trade regulations, such as CITES.
One important challenge, noted by a panelist who learned it as a Ph.D. student and is still challenged by it as a Ph.D. professor, was the expense of the laboratory instrumentation, whether it is a qPCR or sequencing instrument. This cost remains a budget obstacle for most academics working with wildlife. He underscored the importance of loaner and demo programs allowing investigators to borrow otherwise costly instruments in order to expand the applications of these technologies and advance diagnostics on wildlife and the understanding of how pathogens interact with their hosts. The panelist also reminded participants that these technologies fit well with the One Health approach of recognizing the shared health issues between animals, humans, plants, and the greater environment by better enabling discovery and investigation of shared infectious disease agents and their natural ecology.
WORKSHOP FINDINGS
The purpose of our panel was 2-fold: catalyzing a discussion and creating awareness on (i) the technical aspect of qPCR and sequencing, which have similar direct and targeted approaches, with NGS offering greater discovery power, and (ii) the operational aspect of performing these methods in the field, which are parallel, and qPCR arguably is more mature. From our discussion, which included audience questions, our panel identified the following points related to utility and future challenges.
Current qPCR and sequencing methods are complementary and interdependent.
While orthogonal technologies such as immuno-based assays have value, qPCR systems designed for field-based applications are the most robust due to significant investment in optimization of the chemistry and instrumentation (SWaP). Technologies for qPCR and sequencing are related and intertwined, for example, strains need to be sequenced in order to develop new and improved qPCR assays. In addition, robust qPCR and sequencing at the PON offset obstacles for sending sample material out of countries where permission issues and sensitivities exist.
The advent of funding for biodefense research and development attracted scientists, especially those in the life sciences industry, and created an enterprise (38). Related industries, such as environmental testing, also shifted to meet the needs in biodefense. Since lower cost allows for greater end user adoption, manufacturers still require consistent funding opportunities and incentives in order to sustain their research and development in otherwise reactive markets to bridge the “valley of death” gap for their products. At the opposite end of the spectrum, one panelist noted the importance of prioritizing underrepresented research topics, such as environment and wildlife biology. The funding opportunities are few and tend to get grouped in larger clinical opportunities.
Maintaining effective qPCR assays is a continuing challenge.
The effect of signature erosion is when a given qPCR assay has reduced sensitivity and specificity when tested against new strains. The quality of a qPCR assay is dependent on continual testing and evaluation against novel isolates and/or known strains that have mutated in nature. This requires time and resources in terms of a reach-back laboratory with high-throughput sequencing capacity. In addition to sample inhibition mentioned in the UNMOVIC example, sample volume imposes limitations on testing, and as a result nondestructive analysis methods are also needed to accommodate the spectrum of different biological and chemical methods.
As an example from the clinical diagnostic perspective, BioFire Defense is developing a tropical disease panel that can detect approximately 20 pathogens simultaneously, and the development requires significant time for testing, validation, an institutional review board (IRB), and clinical trials around the world. This process is lengthy, and the intended use of the diagnostic assay is carefully defined. As outbreaks happen, sequence data are reviewed to see if the assay will be efficacious in detecting the causative agent. While this is part of product maintenance, there is often a lag between an outbreak and the availability of sequence data or samples. This delay is compounded by the geopolitical pressures around sample and information sharing and concerns over reportable diseases. As qPCR and sequencing platforms become easier to use, people will begin testing in creative ways and in various locations. There is often no standard policy in place for when an unexpected disease is identified. Subsequently, an interesting problem has resulted where customers often request to “hide” results for targets on our panels when they have no policy for dealing with a positive result.
In closing, the panel wholly agreed that greater scientific transparency is mutually beneficial. While exchanging sequencing data through shared transfers is more convenient, the need for better exchange of isolate material in goodwill is still important to continue fulfilling scientific transparency. As a whole, this starts with working collaborations and building trust in a consistent, bilateral manner. Examples include partners doing research while actively engaging with partner country governments and allowing them to have a voice and ownership. This was exemplified in Liberia following the 2014–2015 outbreak of ebolavirus. Together, we should all advocate for continued scientific transparency.
ACKNOWLEDGMENTS
We thank the audience for attending our panel discussion at the 2019 ASM Biothreats Conference. Special thanks to the organizing committee for the opportunity to develop and present this important topic and case examples. We also thank Christina May for her graphic design work and the reviewers and editors for their critique.
The views and opinions expressed in this paper are those of the authors and not necessarily the views and opinions of the United States Department of Defense and United States Department of Homeland Security.
REFERENCES
- 1.Inglis TJ. 2013. The lab without walls: a deployable approach to tropical infectious diseases. Am J Trop Med Hyg 88:614–618. doi: 10.4269/ajtmh.12-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doggett NA, Mukundan H, Lefkowitz EJ, Slezak TR, Chain PS, Morse S, Anderson K, Hodge DR, Pillai S. 2016. Culture-independent diagnostics for health security. Health Secur 14:122–142. doi: 10.1089/hs.2015.0074. [DOI] [PubMed] [Google Scholar]
- 3.Parsons A, Matero P, Adams M, Yeh K. 2018. Examining the utility and readiness of mobile and field transportable laboratories for biodefence and global health security-related purposes. Glob Secur Health Sci Policy 3:1–13. doi: 10.1080/23779497.2018.1480403. [DOI] [Google Scholar]
- 4.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 5.Wittwer CT, Ririe KM, Andrew RV, David DA, Gundry RA, Balis UJ. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques 22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi R, Fockler C, Dollinger G, Watson R. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology 11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 7.Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res 6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 8.De Silva D, Reiser A, Herrmann M, Tabiti K, Wittwer C. 1998. Rapid genotyping and quantification on the LightCycler with hybridization probes. Biochemica 2:12–15. [Google Scholar]
- 9.Lyon E. 2001. Mutation detection using fluorescent hybridization probes and melting curve analysis. Expert Rev Mol Diagn 1:92–101. doi: 10.1586/14737159.1.1.92. [DOI] [PubMed] [Google Scholar]
- 10.Schaad NW, Opgenorth D, Gaush P. 2002. Real-time polymerase chain reaction for one-hour on-site diagnosis of Pierce’s disease of grape in early season asymptomatic vines. Phytopathology 92:721–728. doi: 10.1094/PHYTO.2002.92.7.721. [DOI] [PubMed] [Google Scholar]
- 11.Pierce KE, Mistry R, Reid SM, Bharya S, Dukes JP, Hartshorn C, King DP, Wangh LJ. 2010. Design and optimization of a novel reverse transcription linear-after-the-exponential PCR for the detection of foot-and-mouth disease virus. J Appl Microbiol 109:180–189. doi: 10.1111/j.1365-2672.2009.04640.x. [DOI] [PubMed] [Google Scholar]
- 12.Matero P, Hemmila H, Tomaso H, Piiparinen H, Rantakokko-Jalava K, Nuotio L, Nikkari S. 2011. Rapid field detection assays for Bacillus anthracis, Brucella spp., Francisella tularensis and Yersinia pestis. Clin Microbiol Infect 17:34–43. doi: 10.1111/j.1469-0691.2010.03178.x. [DOI] [PubMed] [Google Scholar]
- 13.Ozanich RM, Colburn HA, Victry KD, Bartholomew RA, Arce JS, Heredia-Langner A, Jarman K, Kreuzer HW, Bruckner-Lea CJ. 2017. Evaluation of PCR systems for field screening of Bacillus anthracis. Health Secur 15:70–80. doi: 10.1089/hs.2016.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanger F, Coulson AR. 1975. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heather JM, Chain B. 2016. The sequence of sequencers: the history of sequencing DNA. Genomics 107:1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voelkerding KV, Dames SA, Durtschi JD. 2009. Next-generation sequencing: from basic research to diagnostics. Clin Chem 55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Sun W, Liu X, Wang X, Xiao Y, Bi D, Yin J, Shi D. 2016. Third-generation sequencing and analysis of four complete pig liver esterase gene sequences in clones identified by screening BAC library. PLoS One 11:e0163295. doi: 10.1371/journal.pone.0163295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Gurtowski J, Yoo S, Nattestad M, Marcus S, Goodwin S, McCombie W, Schatz MC. 2016. Third-generation sequencing and the future of genomics. BioRxiv doi: 10.1101/048603. [DOI]
- 20.Schadt EE, Turner S, Kasarskis A. 2010. A window into third-generation sequencing. Hum Mol Genet 19:R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 21.Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Stryke D, Bouquet J, Somasekar S, Linnen JM, Dodd R, Mulembakani P, Schneider BS, Muyembe-Tamfum JJ, Stramer SL, Chiu CY. 2015. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, Khan AM, Woodford N, Saunders NJ, Wain J, O'Grady J, Livermore DM. 2017. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 72:104–114. doi: 10.1093/jac/dkw397. [DOI] [PubMed] [Google Scholar]
- 24.Greninger AL. 2018. The challenge of diagnostic metagenomics. Expert Rev Mol Diagn 18:605–615. doi: 10.1080/14737159.2018.1487292. [DOI] [PubMed] [Google Scholar]
- 25.Khodakov D, Wang C, Zhang DY. 2016. Diagnostics based on nucleic acid sequence variant profiling: PCR, hybridization, and NGS approaches. Adv Drug Deliv Rev 105:3–19. doi: 10.1016/j.addr.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Lecuit M, Eloit M. 2014. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol 4:25. doi: 10.3389/fcimb.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, Ouédraogo N, Afrough B, Bah A, Baum JH, Becker-Ziaja B, Boettcher J-P, Cabeza-Cabrerizo M, Camino-Sanchez A, Carter LL, Doerrbecker J, Enkirch T, Dorival IGG, Hetzelt N, Hinzmann J, Holm T, Kafetzopoulou LE, Koropogui M, Kosgey A, Kuisma E, Logue CH, Mazzarelli A, Meisel S, Mertens M, Michel J, Ngabo D, Nitzsche K, Pallash E, Patrono LV, Portmann J, Repits JG, Rickett NY, Sachse A, Singethan K, Vitoriano I, Yemanaberhan RL, Zekeng EG, Trina R, Bello A, Sall AA, Faye O, Faye O, Magassouba N, Williams CV, Amburgey V, Winona L, Davis E, Gerlach J, Washington F, Monteil V, Jourdain M, Bererd M, Camara A, Somlare H, Camara A, Gerard M, Bado G, Baillet B, Delaune D, Nebie KY, Diarra A, Savane Y, Pallawo RB, Gutierrez GJ, Milhano N, Roger I, Williams CJ, Yattara F, Lewandowski K, Taylor J, Rachwal P, Turner D, Pollakis G, Hiscox JA, Matthews DA, O'Shea MK, Johnston AM, Wilson D, Hutley E, Smit E, Di Caro A, Woelfel R, Stoecker K, Fleischmann E, Gabriel M, Weller SA, Koivogui L, Diallo B, Keita S, Rambaut A, Formenty P, Gunther S, Carroll MW. 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaulding UK, Christensen CJ, Crisp RJ, Vaughn MB, Trauscht RC, Gardner JR, Thatcher SA, Clemens KM, Teng DH, Bird A, Ota IM, Hadfield T, Ryan V, Brunelle SL. 2012. RAZOR EX anthrax air detection system. J AOAC Int 95:860–891. doi: 10.5740/jaoacint.11-521. [DOI] [PubMed] [Google Scholar]
- 29.Liberman MB, Montgomery DB. 1988. First-mover advantages. Strateg Manage J 9:41–58. doi: 10.1002/smj.4250090706. [DOI] [Google Scholar]
- 30.Liberman MB, Montgomery DB. 1998. First-mover (dis)advantages: retrospective and link with the resource-based view. Strateg Manage J 19:1111–1125. doi:. [DOI] [Google Scholar]
- 31.Qiagen. 2017. miRNA-seq bs qPCR in 2017: an update. Qiagen, Venlo, Netherlands: http://biomarkerinsights.qiagen.com/2017/04/26/ngs-mirna-ngs-vs-qpcr-2017/. Accessed 31 October 2019. [Google Scholar]
- 32.Illumina. 2019. Advantages of next-generation sequencing vs. qPCR. Illumina, San Diego, CA: https://emea.illumina.com/science/technology/next-generation-sequencing/ngs-vs-qpcr.html?langsel=/se/. Accessed 31 October 2019. [Google Scholar]
- 33.Wu Z H K, Mittmann R, Magierowski S, Ghafar-Zadeh E, Zhong X. 2018. FPGA-based DNA basecalling hardware acceleration. Abstr IEEE 61st Int Midwest Symp Circuits Syst (MWSCAS), abstr 18412603. [Google Scholar]
- 34.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 35.Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, Okinaka R, Jackson PJ, Hugh-Jones ME. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol 182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittier CA, Cranfield MR, Stoskopf MK. 2010. Real-time PCR detection of Campylobacter spp. in free-ranging mountain gorillas (Gorilla beringei beringei). J Wildl Dis 46:791–802. doi: 10.7589/0090-3558-46.3.791. [DOI] [PubMed] [Google Scholar]
- 38.Bill Kelly RR, Zemlo T. 2006. Current market opportunities in biodefense research. Industrial Biotechnol 2:32–35. doi: 10.1089/ind.2006.2.32. [DOI] [Google Scholar]


