Abstract
To replicate in their hosts, viruses have to navigate the complexities of the mammalian cell, co-opting mechanisms of cellular physiology while defeating restriction factors that are dedicated to halting their progression. Primate lentiviruses devote a relatively large portion of their coding capacity to counteracting restriction factors by encoding accessory proteins dedicated to neutralizing the antiviral function of these intracellular inhibitors. Research into the roles of the accessory proteins has revealed the existence of previously undetected intrinsic defenses, provided insight into the evolution of primate lentiviruses as they adapt to new species and uncovered new targets for the development of therapeutics. This Review discusses the biology of the restriction factors APOBEC3, SAMHD1 and tetherin and the viral accessory proteins that counteract them.
When the nucleotide sequence of HIV-1 was determined in 1983, the complexity of its genome as compared to those of the murine and avian retroviruses became immediately apparent. In addition to the gag, pol and env genes that encode the structural proteins in all retroviruses, there are open reading frames in the 3′ portion of the genome that have no homologs in the genomes of the simpler animal retroviruses. In the years since, research into the regulatory and accessory proteins encoded by these open reading frames has provided fascinating insight not only into virus replication but into cell biology and immunology as well. The Tat and Rev regulatory proteins were found to be required for the replication of the virus in all cells, serving to induce transcription of the proviral DNA and to transport the unspliced and partially spliced viral RNA transcripts from the nucleus to cytoplasm, respectively. More mysterious were the accessory proteins that were found to be required for the virus to replicate in some, but not all, cell types (Vif, Vpx, Vpu, Nef and Vpr). As the roles of accessory proteins in virus replication were unraveled, the theme that has repeatedly emerged is that they serve as a means to counteract host antiviral defense mechanisms. It was through efforts to understand the roles of accessory proteins in virus replication that previously unimagined antiviral defense mechanisms and the ‘restriction factors’ that mediate them were identified. Viruses go to great lengths to counteract these restriction factors, devoting a substantial portion of their coding capacity to the task and devising complex splicing signals and overlapping reading frames to express accessory proteins. We review here the APOBEC3, SAMHD1 and tetherin (also known as BST2) restriction factors and the accessory proteins that the virus encodes to counteract them (Fig. 1).
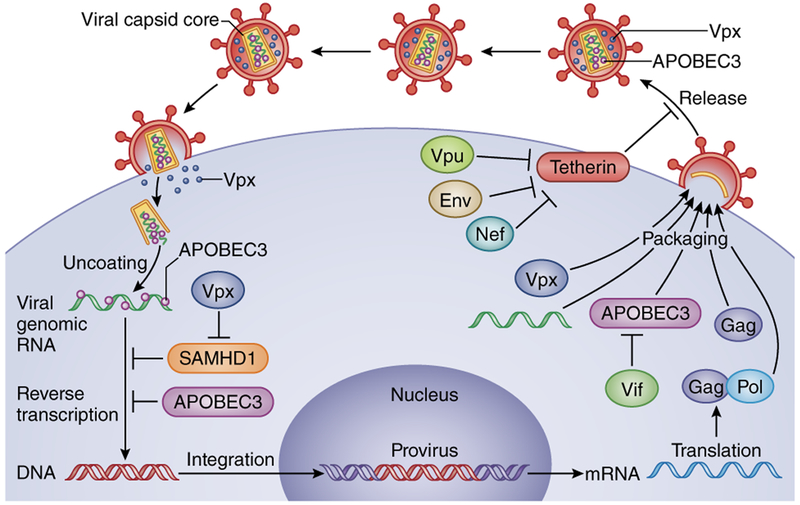
Figure 1.
The host restriction factors SAMHD1, APOBEC3 and tetherin, and the lentiviral accessory proteins that counteract them in the context of virus replication. After virus entry, SAMHD1 and APOBEC3 interfere with reverse transcription (SAMHD1) or modify the reverse-transcribed viral DNA (APOBEC3). Tetherin acts late in the replication cycle to prevent the release of virions. Vif counteracts APOBEC3-driven mutagenesis of the viral genome, by preventing its packaging into virions. Vpx relieves the SAMHD1-mediated block to reverse transcription. Vpu prevents tetherin from holding on to the virus at the plasma membrane. HIV-2 and SIVs that lack Vpu use Env and Nef, respectively, to counteract tetherin by sequestering it intracellularly.
Vif and the APOBEC3 cytidine deaminases
The virion infectivity factor (Vif) is encoded by all primate lentiviruses except equine infectious anemia virus1. Vif is required for lentiviral replication in nonpermissive cells, such as CD4+ T cells and monocyte-derived macrophages (MDMs)2, whereas in permissive T cell lines, Vif-deleted (Δvif) HIV-1 viruses replicate to high levels3,4. Moreover, Δvif HIV-1 virions produced in permissive cells are fully infectious and infect both permissive and nonpermissive cells4. Progress in understanding the role of Vif was stalled for several years until the elusive dominant inhibitor was identified as apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (A3G)5.
A3G belongs to a family of single-stranded DNA deaminases6,7. The APOBEC3 gene locus expanded from a single gene in rodents to seven in primates (encoding three single-domain deaminases, A3A, A3C, A3H and four double-domain deaminases, A3B, A3D, A3F, A3G)7. APOBEC3 genes have been subject to strong selective pressure resulting from mutations that provided resistance to lentivirus infection8,9. APOBEC3 proteins are expressed in many cell types, including CD4+ T cells and MDMs10. Work from many laboratories has elucidated the mode of action of APOBEC3 proteins and the mechanism by which they are degraded by Vif11,12. Initial studies focused on A3G, but A3B, A3D, A3F and A3H are also active against HIV-1.
APOBEC3 proteins, if not degraded by Vif, potently inhibit lentivirus replication through cytidine deamination of the viral genome. The antiviral activity requires that the proteins are packaged into the virion and is manifested only in the next cycle of infection (Fig. 2a)13,14. The reverse transcription products generated by the Δvif HIV-1 virions produced in cells that express APOBEC3 is characterized by the presence of numerous G→A mutations. These result from APOBEC3-mediated C→U deamination of the minus strand of the virus DNA14. Because U is read as T by the polymerase, synthesis of the plus strand results in G→A mutation. Depending on the degree of deamination, the reverse transcripts either are recognized as aberrant by the cell and degraded or become integrated into the genomic DNA but, because they contain translational termination codons and missense mutations, fail to produce infectious progeny14,15 (Fig. 2a).
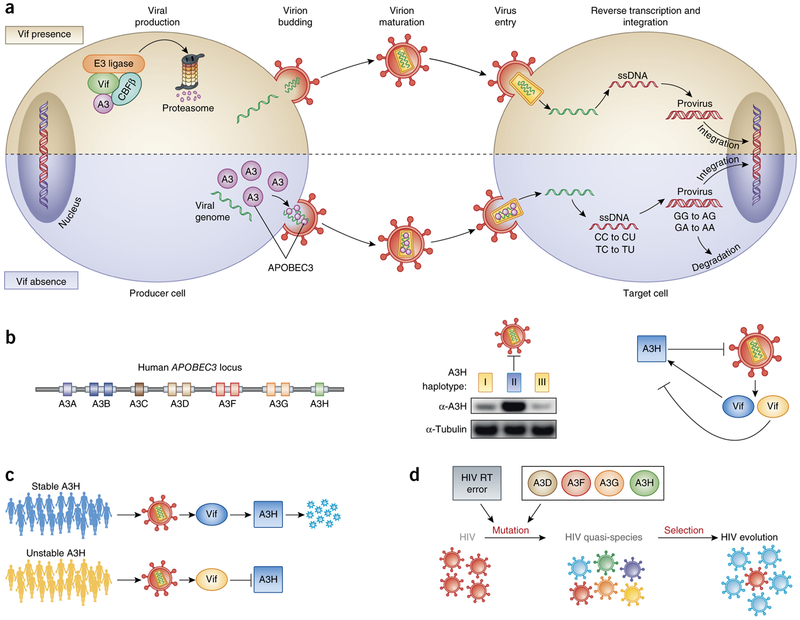
Figure 2.
Overview of the mode of APOBEC3 (A3) restriction and the implications of suboptimal Vif activity on HIV transmission and diversification. (a) APOBEC3 proteins are degraded in the producer cell in the presence of Vif, but in the absence of Vif they are packaged into the budding viral particle. In the next cycle of infection, the APOBEC3 proteins mutagenize the viral genome during reverse transcription by deaminating cytosines to uracils in the minus-strand DNA. Mutated viral DNA may be degraded by DNA repair enzymes or integrated into the host cell genome. (b) Left, the human APOBEC3 locus encodes seven different deaminases that carry either one (A3A, A3C, A3H) or two deaminase domains (A3B, A3D, A3F, A3G). Middle, A3H stands out among the APOBEC3 proteins because its haplotypes differ in protein stability and antiviral activity: haplotypes I and III (yellow) are unstable, whereas haplotype II (blue) is stable. Right, HIV Vif alleles differ in their ability to counteract the stable A3H haplotype II. (c) HIV Vif adapts to A3H haplotypes in vivo. A3H-resistant HIV (blue Vif) efficiently replicates in patients carrying an active A3H (blue individual). A3H-sensitive HIV (yellow Vif) replicates well in patients with unstable A3H (yellow individuals), but when it is transmitted to a new host who encodes stable A3H, its spread is limited. (d) HIV sequence diversification can be caused by reverse transcriptase (RT) errors and mutagenesis by different APOBEC3 proteins (A3D, A3F, A3G, A3H).
APOBEC3 proteins are packaged into virions as they assemble at the plasma membrane through interactions with several types of RNA (viral RNA, small noncoding RNAs, cellular mRNA) in a nucleocapsid-dependent manner16,17. Although virion packaging of APOBEC3 proteins is required for antiviral activity, it may not be sufficient. For example, A3A is a potent cytidine deaminase, but when packaged in virions it has little effect on infectivity. Packaging A3A in virions by fusing it to Vpr activates the ability of the deaminase to inhibit infectivity by altering its localization within the virion18.
APOBEC3-driven mutagenesis occurs over the entire viral genome, with a gradient of higher mutational frequency toward the 3′ end, an effect that is likely to result from this region of the viral genome remaining single-stranded for an extended time during reverse transcription19,20. Although many cytosine nucleotides are targeted for deamination, APOBEC3 proteins have preferences for specific dinucleotides in single-stranded DNA (minus strand: 5′-CC, 5′-TC11,20,21). The target-site preferences of the APOBEC3 proteins provide clues to the identity of the deaminase that left its footprint on a provirus: GGD (A3G, A3D), GAG (A3G) or GAD and GGA (A3F, A3H), where D is A, G or T11,21. Mutations in the A3G- or A3D-favored context (GG→GA) frequently produce translation termination codons (for example, TGG→TAG), enhancing the lethality of these deaminases11,12,21.
Although cytidine deamination is the primary mechanism by which APOBEC3 proteins restrict lentiviruses, there is evidence that they also exert deaminase-independent antiviral activity. Virion-packaged APOBEC3 can interfere with reverse transcription by preventing tRNA binding to the primer-binding site on the viral genomic RNA or by causing termination of minus-strand synthesis22,23. The contribution of deaminase-independent restriction remains to be determined in light of reports that catalytically inactive A3G mutants lack antiviral activity24,25.
To counteract the antiviral activity of APOBEC3 proteins, Vif induces their degradation before virion packaging (Fig. 2a). Vif binds to APOBEC3 molecules in the producer cell and recruits a cullin 5–based E3 ubiquitin ligase complex consisting of elongin B, elongin C and Rbx-1 (refs. 26,27). Vif expression is further stabilized by CBF-b, a transcription factor28–31. The complex ubiquitinates the bound APOBEC3 proteins, which are then rapidly degraded by proteasomes13,32. The crystal structure of the Vif-CBF-b E3 ligase pentameric complex33 shows Vif in an elongated cone-like shape with a larger and a smaller domain separated by a zinc-binding region33. The structure of Vif bound to APOBEC3 has not been determined but extensive evidence indicates that the N terminus of Vif interacts with the different APOBEC3 (refs. 11,12). The presence of A3G-, A3F- and A3H-specific amino acid motifs within Vif suggests that its activity against individual APOBEC3 proteins can adapt for optimal virus replication11,34–36.
Although lentiviruses go to great lengths to exclude APOBEC3 proteins from their virions, the packaging of the proteins in small amounts may provide a benefit to the virus. Nonlethal mutagenesis could provide a means by which the virus can generate sequence diversity to accelerate its evolution (Fig. 2d). The inability of Vif to fully prevent deamination is demonstrated by the hypermutated proviruses found in acutely and chronically infected patients and in vertically infected infants12,21. Several factors could contribute to incomplete counteraction of APOBEC3 by Vif. Interferon induction may cause the production of an amount of APOBEC3 sufficient to overwhelm the Vif produced by the virus; and viruses may encode Vif variants with reduced affinity for specific APOBEC3. The possibility that sublethal deamination may lead to beneficial genetic diversity has been demonstrated in cell culture by the appearance of antiretroviral drug–resistant virus as a result of APOBEC3-generated G→A mutation37,38. In support of this possibility in vivo, antiretroviral treatment failure has been found in some cases to be associated with viruses that bear partially defective Vif alleles39. In humanized mouse models, APOBEC3 increases the diversity of viral T cell epitopes, allowing escape from cytotoxic T lymphocytes40–42. APOBEC3 may also facilitate co-receptor switching by generating G→A mutations in env that encode amino acids in the V3 loop of gp120 (ref. 40).
Pandemic HIV-1 strains are resistant to multiple APOBEC3 proteins. Although these circulating viruses generally counteract A3G and A3F efficiently43–45, not all viral strains can target A3H for degradation45. The explanation for the differential susceptibility of HIV to A3H is found in the fact that not every individual expresses an active A3H haplotype46. Indeed, single-nucleotide polymorphisms in the human A3H coding region result either in stable variants that display potent antiviral activity or in unstable variants that are inactive46–50 (Fig. 2b). As the virus is transmitted and successfully spreads in the new host, its Vif has to evolve to counteract the recipient’s A3H haplotype36 (Fig. 2c). The failure of Vif to adapt could prevent transmission or attenuate HIV disease presentation in the case of an A3H-sensitive virus that encounters a host with stable A3H36,50 (Fig. 2c)—a scenario that is reminiscent of the role played by A3G in limiting the zoonotic transmission of lentiviruses from nonhuman primates. Because Vif-APOBEC3 interactions are species specific, they may determine whether a virus will be transmitted across species. A single charged amino acid at position 128 of A3G determines whether Vif will bind to human or nonhuman primate APOBEC3 proteins51–53. A successful jump across species requires that the nonhuman primate Vif be able to counteract the APOBEC3 proteins of the new host53,54. The APOBEC3-Vif axis is a key element of viral pathogenicity. It will be important in future studies to determine the degree of sensitivity of circulating viral strains to each APOBEC3 protein as a means of understanding how Vif diversity modulates AIDS progression.
SAMHD1 restriction of HIV-1 in non-dividing cells
The Vpr and Vpx accessory proteins arose by gene duplication during evolution and remain about 30% similar in amino acid sequence55,56. Both are virion packaged, a unique feature among the nonstructural HIV proteins. Both localize to the nucleus of the cell, yet lack a consensus nuclear localization sequence (NLS). Deletion of Vpx from SIVmac has no effect on the ability of the virus to infect activated CD4+ T cells, but renders the virus unable to infect myeloid cells such as dendritic cells (DCs) and MDMs57,58. Upon infection, few reverse transcripts are produced, suggesting a failure of reverse transcriptase to synthesize viral DNA. Although Vpx has no effect on the infection of activated T cells, it increases the ability of the virus to infect resting T cells59,60. Resting T cells do not support productive virus replication because of several blocks in the virus life-cycle. By relieving the block to reverse transcription in resting T cells, Vpx could allow the establishment of latently infected cells which upon subsequent activation would become producers of infectious virus. For HIV-2 and SIV, this effect could contribute to the pool of latently infected T cells that sustains chronic infection.
Vpr is encoded by all HIV and SIV lineages, whereas Vpx is present only in HIV-2 and some SIVs, notably those from macaques (SIVmac) and from red-capped (SIVrcm) and sooty mangabeys (SIVsm). Although HIV-1 does not encode Vpx, the virus is susceptible to the infectivity enhancement provided by the SIV accessory protein. Introducing Vpx from SIV into MDMs via Vpx-containing virus-like particles (VLPs) increases their susceptibility to HIV-1 by two orders of magnitude61. Moreover, HIV-1 that has been engineered to package SIV Vpx infects human DCs and MDMs with a similar increase in titer62,63, a feature that has been exploited as a means of producing lentiviral vectors that efficiently transduce primary myeloid cells64,65. Thus, it would appear advantageous for HIV-1 to encode Vpx, yet, curiously, it does not.
In cells, Vpx forms a complex with a cullin 4A–based E3 ubiquitin ligase, the components of which include DCAF1 and DDB1 (refs. 66,67). The complex regulates the degradation of a large number of cellular DNA repair proteins, replication enzymes and transcription factors. Its association with Vpx provided the first clue that Vpx might serve to neutralize a host restriction factor. Using a mass spectrometry–pull-down approach, the restriction factor was identified as SAM domain–and HD domain–containing protein 1 (SAMHD1)68,69. Vpx in virions or VLPs induces the degradation of SAMHD1, and knockdown of SAMHD1 in DCs relieves the block to infection by HIV-1 (refs. 68,69). To study the role of SAMHD1 in vivo, two groups generated SAMHD1-knockout mice70,71. MDMs from these SAMHD1-deficient mice are more susceptible to infection by HIV-1 (ref. 70) or an attenuated HIV-1 with a mutation in reverse transcriptase71. The deficiency did not increase susceptibility to mouse retroviruses, most likely because of additional blocks to retrovirus infection of myeloid cells70,71.
SAMHD1 consists of an N-terminal SAM (sterile alpha motif) domain and a C-terminal HD domain, the latter of which is characteristic of a class of enzymes with phosphodiesterase, phosphatase and nuclease activities. The enzyme contains an N-terminal consensus NLS and localizes to the nucleus72,73. Upon infection of MDM with SIV, HIV-2 or Vpx-containing engineered HIV-1, SAMHD1 is rapidly degraded. Its abundance starts to fall about an hour after infection and remains low for several days74,75. The deletion of amino acids 13–17 removes the NLS of SAMHD1, localizing it to the cytoplasm72,73. NLS-deficient SAMHD1 retains antiviral activity but cannot be degraded by Vpx, suggesting that Vpx seeks out SAMHD1 in the nucleus72,73. The need for Vpx to target SAMHD1 in the nucleus may explain its use of the DCAF1-DDB1 E3 ubiquitin ligase, a complex that is specialized in the degradation of nuclear proteins.
Recombinant SAMHD1 produced in Escherichia coli has dGTP-regulated phosphohydrolase activity that removes the triphosphate from deoxynucleotide triphosphates76–78. When expressed by lentiviral vector transduction in differentiated U937 cells, SAMHD1 causes a dramatic drop in the concentration of dNTPs78, suggesting a straightforward mechanism of restriction. After virus entry, reverse transcriptase generates a double-stranded DNA using the cellular pool of dNTPs. SAMHD1 could block HIV-1 infection by causing the concentration of dNTPs to fall to a level insufficient to support reverse transcription. In the case of HIV-2 or SIV infection, Vpx would be released from the virion, inducing the degradation of SAMHD1. As dNTPs levels were increased, reverse transcription would then proceed. In support of this mechanism, SAMHD1-mediated restriction is relieved by the addition of exogenous deoxynucleosides (dN) to DCs78. Moreover, such a mechanism could act more broadly. SAMHD1 blocks a wide range of retroviruses yet, interestingly, is inactive against foamy viruses, a class of retrovirus that is unusual for its ability to complete reverse transcription in the virion, before entry79.
Although this model made sense, recent findings have called into question whether dNTP pool depletion fully accounts for SAMHD1-mediated restriction. In activated cells, SAMHD1 is phosphorylated by specific cyclin-dependent kinases at amino acid Thr592. SAMHD1 with a T592E mutation that mimics the phosphorylated protein shows loss of antiviral activity80–83 yet, surprisingly, appears to retain dNTPase activity82. This finding would suggest that dNTP depletion is either not sufficient for antiviral activity or not required. If that is the case, then how does SAMHD1 restrict virus replication? Perhaps rather than preventing reverse transcription, it attacks the viral RNA or the newly synthesized viral DNA. In fact, E. coli–produced recombinant SAMHD1 was reported to have RNA and single-strand DNA exonuclease activity on synthetic substrates84. In an analysis of the relative contributions of the two catalytic activities to restriction, the dNTPase and RNase activities of SAMHD1 were genetically separated by point mutations85. A D137N mutation disabled the dNTPase but not the RNase activity yet had no effect on antiviral activity. Conversely, a Q548A mutation that inactivated the RNase but not the dNTPase activity caused the loss of antiviral activity, implicating the RNase and not the dNTPase activity as the mediator of virus restriction (Fig. 3). In spite of these findings, it is difficult to understand how SAMHD1 could degrade the viral genomic RNA or reverse transcription intermediates given that reverse transcription is generally thought to at least initiate in the cytoplasm and SAMHD1 is largely nuclear. In addition, Vpx-containing VLPs relieve the block to infection in DCs when added to cells several hours after infection86,87. Similarly, dNs added to DCs several hours after infection relieve the block to infection, and a temporary pharmacologic block to E3 ubiquitin ligase function stalls but does not prevent infection86. The reversibility of SAMHD1-mediated restriction is consistent with a lack of nucleotides, but hard to reconcile with a nucleolytic attack of the viral RNA.
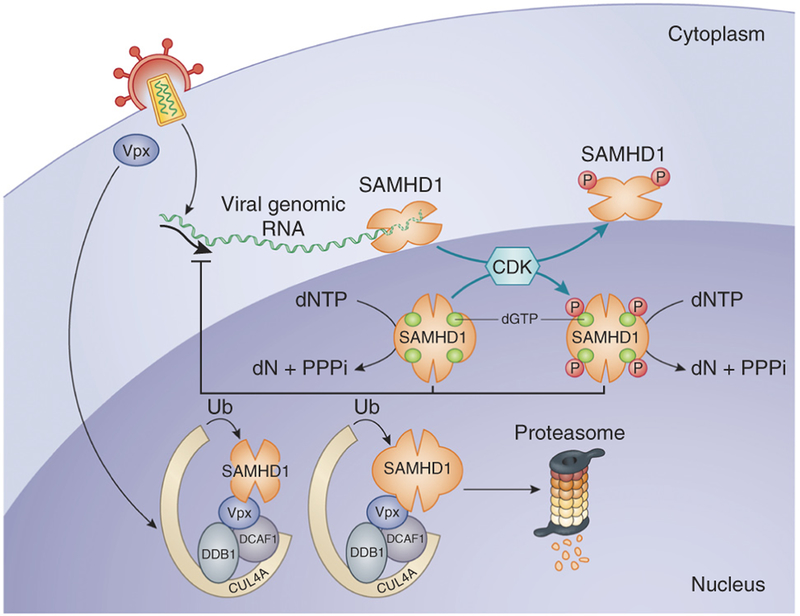
Figure 3.
Proposed models for SAMHD1-mediated restriction. HIV-2, SIV or engineered HIV-1 containing Vpx enters a myeloid cell. Vpx transits to the nucleus and binds to the DCAF1-DDB1-CUL4A E3 ubiquitin ligase complex and to SAMHD1, which is then degraded by proteasomes in the nucleus. If the virus lacks Vpx, SAMHD1 depletes the pool of dNTPs, blocking reverse transcription of the bound tRNA primer. An alternative model calls for SAMHD1 to degrade the viral genomic RNA as it is reverse transcribed in the cytoplasm. This activity is regulated by the phosphorylation of amino acid Thr592 by CDK-1, CDK-2 and CDK-6.
Another piece of the puzzle comes from an entirely different field of research. Aicardi-Goutières syndrome (AGS), a rare autosomal recessive neurological condition with early onset, is characterized by high expression of type I interferon (type I IFN) in the central nervous system and upregulation of IFN-stimulated genes. The disease can be caused by defects in the genes encoding SAMHD1, TREX1, RNase H2A, RNase H2B, RNase H2C, ADAR or IFIH1 (ref. 88), all of which are enzymes involved in nucleotide or nucleic acid metabolism. The 3′→5′ cytoplasmic single-strand DNA exonuclease TREX1 is a negative regulator of the interferon-stimulatory DNA response89 that digests endogenous cytoplasmic retroelement DNA, thereby preventing the activation of DNA sensors such as cGAS90. Such activation would trigger innate immune response pathways, leading to chronic inflammation similar to what is observed in patients with AGS. TREX1 is also thought to digest newly synthesized HIV-1 DNA, thereby limiting the activation of host antiviral defenses90,91. Similarly, SAMHD1 could control endogenous elements by degrading them or preventing their synthesis92, further blunting the innate immune response. In the absence of SAMHD1, the accumulation of cytoplasmic DNA would result in chronic inflammation. Consistent with this possibility, in SAMHD1-null mice, some interferon-stimulated genes are constitutively induced70,71. Such a model would offer a unified explanation as to how SAMHD1 restricts HIV-1 and causes AGS. However, evidence for increased amounts of endogenous retroelement RNA or DNA in SAMHD1-deficient mice or AGS patients is lacking.
How does HIV-1 survive without Vpx?
Although Vpx is the accessory protein most closely associated with inducing SAMHD1 degradation, it appears that it was Vpr that initially evolved for this purpose93. Consistent with this model, the SIV of African green monkeys (SIVagm) encodes Vpr but not Vpx, and its Vpr degrades SAMHD1. As the primate species evolved, mutations in the SAMHD1 gene that allowed for escape from SIV Vpx were selected An analysis of primate SAMHD1 sequences suggests that precisely those amino acids that are contacted by Vpx were under selective pressure93. The duplication of the Vpr open reading frame in the evolution of SIV allowed the two genes to specialize their functions, allowing Vpx the freedom to better adapt to amino acid sequence changes in SAMHD1. As the virus jumped from Old World monkeys to chimpanzees, before its transfer into humans, it deleted vpx, giving rise to the current SIV of chimpanzees (SIVcpz)94. Deletion of vpx disabled the ability of the virus to degrade SAMHD1 but freed vif, which overlaps with vpx, to alter its sequence, allowing it to evolve to better counteract chimpanzee APOBEC3.
The absence of Vpx in HIV-1 raises the question of how the virus replicates and induces pathogenesis without the benefit of an important accessory protein. One possibility is that the virus has assigned the role of Vpx to another viral protein. For example, HIV-1 reverse transcriptase has a higher affinity for dNTPs than that of HIV-2 or SIV, allowing it to synthesize DNA in low dNTP concentrations95. It has also been suggested that HIV-1 does induce SAMHD1 degradation, not through an accessory protein but by activating host cyclin L2 (ref. 96). Another possibility is simply that the virus, unlike HIV-2 or SIV, survives without the need to counteract the restriction. That HIV-1 largely fails to escape SAMHD1-mediated restriction is nicely demonstrated by the ability of Vpx to enhance the infectivity of the virus on DCs. HIV-1 could survive with little need to infect myeloid cells, replicating largely by T cell–to–T cell rather than MDM–to–T cell transmission. In contrast, HIV-2 and SIV may be dependent on MDM–to–T cell transfer of virus, an effective means of transmission in a setting in which there are few activated T cells, as is the case in the natural primate hosts. HIV-1 infection in humans may cause generalized T cell activation, providing a sufficient number of activated target cells to sustain the infection without the need for MDM–to–T cell transfer. In support of this model, rhesus macaques infected with Δvpx SIVmac have few infected myeloid lineage cells and low T cell viral burdens97. A lesson to be learned here is that even though HIV-1 has no Vpx, understanding this accessory protein has provided insight into AIDS pathogenesis.
The role of Vpu and tetherin in virus release
The Vpu accessory protein is a type I transmembrane protein that is expressed from a bicistronic viral mRNA and localizes to the ER. It is encoded by HIV-1 and some SIVs (those of macaques, mona monkeys and greater spot-nosed monkeys) but not by HIV-2 or most other SIVs. Δvpu HIV-1 has two distinct phenotypic traits that distinguish it from the wild-type virus: the virions have fewer envelope glycoprotein spikes, and they accumulate in massive clusters on the cell surface because they fail to detach from the producer cell98–100. Early studies showed that the decreased number of envelope glycoprotein spikes results from the interaction between CD4 and gp160 in the Golgi101, which prevents the transit of gp160 to the plasma membrane. Vpu acts on CD4 to prevent its interaction with gp160, and as such it frees gp160 to transit to the plasma membrane. The failure of Δvpu HIV-1 to release virions was more difficult to understand. The effects of Vpu on CD4 and on virion release were genetically separable by point mutations in Vpu that affected one function or the other. Moreover, the block to virion release was active only in certain cell types102,103 and could be enhanced by type I interferon104. The analysis of heterokaryons formed between permissive and nonpermissive cell types showed that the block in virion release was dominant, suggesting that it was caused by an inhibitor of this step105. A microarray screen to identify membrane-associated, interferon-inducible proteins specific to nonpermissive cell types identified the bone marrow stromal cell antigen BST2 (also known as CD317) (ref. 104). Transfection of BST2 into permissive cells blocked virion release by Δvpu HIV-1 but not wild-type HIV-1, confirming that BST2 is the Vpu target106,107. The protein was re-christened ‘tetherin’ for its role in tethering virus to the plasma membrane.
The interesting topology of tetherin immediately suggested how it might prevent the release of enveloped viruses. The protein is a disulfide-bonded homodimer with an extended coiled-coil alpha-helical extracellular domain flanked by a short cytoplasmic tail, an N-terminal transmembrane anchor and a C-terminal glycosyl-phosphatidylinositol (GPI) linkage108,109. If it were to insert one end into the viral lipid bilayer and the other into the cell plasma membrane, the virus would be unable to leave from the cell surface. In support of this model, the amino acid sequence of the central domain of tetherin is not critical for its function. Remarkably, an artificial tetherin could be constructed from the functional domains of unrelated proteins, demonstrating that it is the topology of the protein and not its sequence that endows it with antiviral activity110. Tetherin could presumably insert in either orientation in the viral and cell membranes. However, proteolytic treatment of tethered virions leaves an N-terminal fragment of tetherin in the plasma membrane, suggesting that this orientation is preferred102. To relieve the block to virus release, Vpu interacts with tetherin, sequestering it in the perinuclear region and causing its endocytosis and proteasomal degradation.
In addition to its role in preventing virion release, tetherin has an additional function as a sensor of viral infection (Fig. 4). As tetherin grabs onto budding virions, it clusters at the plasma membrane, becoming unlinked from the underlying cortical actin cytoskeleton and exposing dual tyrosines in the cytoplasmic domain that become targets for phosphorylation by the tyrosine kinase Syk105,111–113. The phosphorylated tetherin molecules recruit the signaling adaptors TRAF2, TRAF6 and the mitogen-activated protein kinase TAK-1; activates the transcription factor NF-kB111,113; and induces the expression of the proinflammatory cytokines CXCL10, IL-6 and type I IFN111. Tetherin activates NF-κB in HIV-1-infected cells, leading to the production of interferon, and this is prevented by Vpu114. Thus, Vpu antagonizes both the effect of tetherin on viral release and its role in viral sensing. During the course of chronic HIV-1 infection, as viral sequence variants accumulate, the ability of tetherin to downregulate CD4, sequester tetherin and prevent innate immune responses is preserved, indicating the importance of these functions in vivo115.
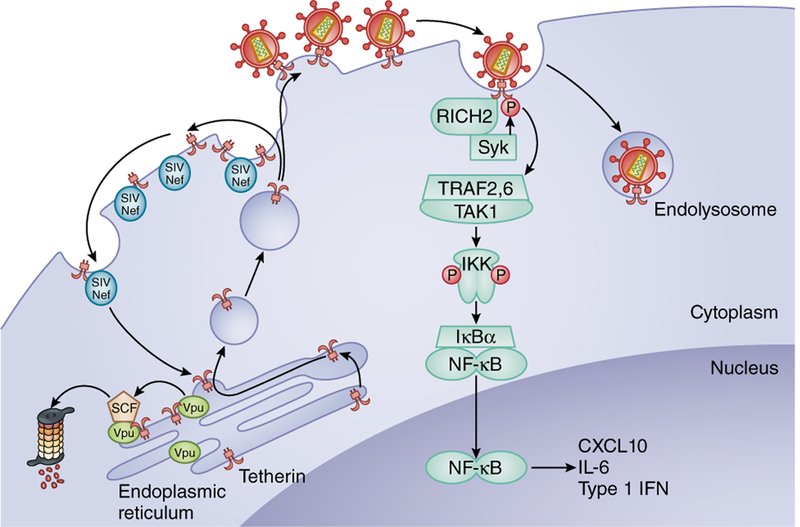
Figure 4.
Tetherin blocks virus release, activates an innate immune response and is counteracted by Vpu or Nef. Tetherin (BST2 or CD317) prevents virus release by inserting its N-terminal transmembrane domain in the plasma membrane and its GPI-linked C terminus in the virus envelope lipid bilayer. The tethered virus is then endocytosed. Tetherin contacts cortical actin through an interaction with RICH2. Tetherin is phosphorylated on tyrosines near its N terminus by Syk. Phosphorylated tetherin activates TRAF2, TRAF6 and TAK1. The complex phosphorylates IKK, causing the degradation of IκBα and thereby activating NF-κB, which induces the transcription of the proinflammatory cytokines CXCL10, IL-6 and IFN-β. Sequestration of tetherin by HIV-1 Vpu in the ER prevents its transit to the plasma membrane. Tetherin proteasomal degradation is induced by its interaction with the SCF-β-TRCP complex. In most SIVs, Nef binds to tetherin at the plasma membrane to induce its endocytosis through an AP2-dependent pathway.
How do HIV-2 and SIVs that lack Vpu escape restriction by tetherin? In SIVcpz SIVmac, SIVagm and SIVsm, this function is reassigned to Nef116–118. In these viruses, Nef binds to the cytoplasmic N terminus of tetherin, inducing its endocytosis in an AP-2-dependent mechanism. Human tetherin is not subject to antagonism by Nef because it has a deletion of the Nef binding site at amino acids 14–18 (ref. 119). Thus, zoonosis of SIVcpz into humans forced the virus to find another means to counteract tetherin. With just a few amino acid sequence changes, SIVcpz Vpu acquires the ability to target human tetherin120. The ease with which the species-specificity of Vpu could be altered is thought to have facilitated the ability of the virus to establish itself in humans. HIV-2 antagonizes tetherin with yet another viral protein. The virus, which is derived from SIVsm, lacks Vpu and instead uses its envelope glycoprotein protein for this purpose121,122. This evolutionary adaptation was recreated experimentally in Δnef SIV–infected rhesus macaques. During in vivo passage, the virus generated mutations in gp41 that allowed for escape from tetherin, mimicking HIV-2. Taken together, these findings again demonstrate the plasticity with which viral proteins mold to acquire different functions as they adapt to new species123.
Concluding remarks
This Review has focused on the three restriction factors that lentivirus have evolved accessory proteins to escape, but cells express other antiviral factors, some that have been identified and, most likely, others that remain to be discovered. In the case of antiviral proteins that are not counteracted by accessory proteins, the virus may have found an alternative means of escape. HIV-1 escapes TRIM5α by altering its capsid protein, for example. For antiviral proteins such as MxB, an interferon-induced protein that targets the viral capsid, the virus may simply live with a decreased ability to replicate.
Despite a plethora of sophisticated host antiviral defense mechanisms, viruses continue to ravage the human species. Viruses like HIV-1 are evolutionarily nimble, replicating small genomes with a generation time on the order of hours to produce huge numbers of genetically diverse progeny. HIV-1 has a remarkable degree of genetic plasticity that allows its proteins to interact with the altered host proteins it encounters during species transmission and that gives it the ability to swap the functionality of viral proteins when point mutations are insufficient. Although the virus may have found ways around antiviral host defenses, the restriction factors and viral accessory proteins provide targets for the development of new strategies by which the virus will be ultimately defeated. The study of the lentiviral accessory proteins has led to the discovery of new mechanisms of intrinsic immunity, continuing a long history of viruses teaching us about the inner working of cells.
ACKNOWLEDGMENTS
We thank M. Ooms and L.C.F. Mulder for critical reading of the manuscript. The work was supported by US National Institutes of Health/National Institute of Allergy and Infectious Disease grants to V.S. (AI064001, AI090935) and N.R.L. (AI067059, AI058864), the Vilcek Fellowship Endowment Fund to N.B. and the American Foundation for AIDS Research.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Strebel K et al. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature 328, 728–730 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Gabuzda DH et al. Role of Vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol 66, 6489–6495 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen BR HIV-1 Vif: counteracting innate antiretroviral defenses. Mol. Ther 8, 525–527 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Simon JH, Gaddis NC, Fouchier RA & Malim MH Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med 4, 1397–1400 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Sheehy AM, Gaddis N, Choi J & Malim M Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Harris RS, Petersen-Mahrt SK & Neuberger MS RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Jarmuz A et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Sawyer SL, Emerman M & Malik HS Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol 2, E275 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conticello SG, Thomas CJ, Petersen-Mahrt SK & Neuberger MS Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol 22, 367–377 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Refsland EW et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 38, 4274–4284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Refsland EW & Harris RS The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol 371, 1–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desimmie BA et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol 426, 1220–1245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariani R et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Mangeat B et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Bishop KN et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol 14, 1392–1396 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Apolonia L et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 11, e1004609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zennou V, Perez-Caballero D, Gottlinger H & Bieniasz PD APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol 78, 12058–12061 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguiar RS, Lovsin N, Tanuri A & Peterlin BM Vpr.A3A chimera inhibits HIV replication. J. Biol. Chem 283, 2518–2525 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Suspène R, Rusniok C, Vartanian JP & Wain-Hobson S Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res 34, 4677–4684 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol 11, 435–442 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Kim EY et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 10, e1004281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes RK, Malim MH & Bishop KN APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci 32, 118–128 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Bishop KN, Verma M, Kim EY, Wolinsky SM & Malim MH APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4, e1000231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Browne EP, Allers C & Landau NR Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology 387, 313–321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albin JS, Brown WL & Harris RS Catalytic activity of APOBEC3F is required for efficient restriction of Vif-deficient human immunodeficiency virus. Virology 450–451, 49–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Mehle A, Thomas ER, Rajendran KS & Gabuzda D A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem 281, 17259–17265 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Jäger S et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hultquist JF, Binka M, LaRue RS, Simon V & Harris RS Vif proteins of human and simian immunodeficiency viruses require cellular CBFbeta to degrade APOBEC3 restriction factors. J. Virol 86, 2874–2877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DY et al. CBFbeta stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell 49, 632–644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Du J, Evans SL, Yu Y & Yu XF T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Sheehy AM, Gaddis NC & Malim MH The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med 9, 1404–1407 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Guo Y et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Simon V et al. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1, e6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell RA & Pathak VK Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol 81, 8201–8210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ooms M et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 14, 411–421 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Mulder LC, Harari A & Simon V Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. USA 105, 5501–5506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim EY et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J. Virol 84, 10402–10405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fourati S et al. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. AIDS 24, 2313–2321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato K et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog 10, e1004453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krisko JF, Martinez-Torres F, Foster JL & Garcia JV HIV restriction by APOBEC3 in humanized mice. PLoS Pathog 9, e1003242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casartelli N et al. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. J. Exp. Med 207, 39–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwabu Y, Fujita H, Tanaka Y, Sata T & Tokunaga K Direct internalization of cell-surface BST-2/tetherin by the HIV-1 accessory protein Vpu. Commun. Integr. Biol 3, 366–369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwabu Y et al. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J. Biol. Chem 285, 35350–35358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binka M, Ooms M, Steward M & Simon V The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J. Virol 86, 49–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.OhAinle M, Kerns JA, Li MMH, Malik HS & Emerman M Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larue RS, Lengyel J, Jonsson SR, Andresdottir V & Harris RS Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol 84, 8193–8201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan L, Sarkis PT, Wang T, Tian C & Yu XF Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J 23, 279–287 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harari A, Ooms M, Mulder LC & Simon V Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol 83, 295–303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Refsland EW et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet 10, e1004761 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bogerd HP, Doehle BP, Wiegand HL & Cullen BR A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101, 3770–3774 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mangeat B, Turelli P, Liao S & Trono D A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem 279, 14481–14483 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Schröfelbauer B, Chen D & Landau NR A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101, 3927–3932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krupp A et al. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog 9, e1003641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tristem M, Marshall C, Karpas A, Petrik J & Hill F Origin of vpx in lentiviruses. Nature 347, 341–342 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Tristem M, Marshall C, Karpas A & Hill F Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J 11, 3405–3412 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guyader M, Emerman M, Montagnier L & Peden K VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. EMBO J 8, 1169–1175 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu XF, Yu QC, Essex M & Lee TH The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol 65, 5088–5091 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldauf HM et al. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med 18, 1682–1687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Descours B et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goujon C et al. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Ther 13, 991–994 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Sunseri N, O’Brien M, Bhardwaj N & Landau NR Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol 85, 6263–6274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bobadilla S, Sunseri N & Landau NR Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther 20, 514–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durand S et al. Tailored HIV-1 vectors for genetic modification of primary human dendritic cells and monocytes. J. Virol 87, 234–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Norton TD, Miller EA, Bhardwaj N & Landau NR Vpx-containing dendritic cell vaccine induces CTLs and reactivates latent HIV-1 in vitro. Gene Ther 22, 11–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le Rouzic E et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4–DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Srivastava S et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog 4, e1000059 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laguette N et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hrecka K et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Behrendt R et al. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Reports 4, 689–696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rehwinkel J et al. SAMHD1-dependent retroviral control and escape in mice. EMBO J 32, 2454–2462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hofmann H et al. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol 86, 12552–12560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandariz-Nuñez A et al. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim B, Nguyen LA, Daddacha W & Hollenbaugh JA Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem 287, 21570–21574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jáuregui P, Logue EC, Schultz ML, Fung S & Landau NR Degradation of SAMHD1 by Vpx is independent of uncoating. J. Virol (11 March 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goldstone DC et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Powell RD, Holland PJ, Hollis T & Perrino FW Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem 286, 43596–43600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lahouassa H et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol 13, 223–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gramberg T et al. Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pauls E et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J. Immunol 193, 1988–1997 (2014). [DOI] [PubMed] [Google Scholar]
- 81.St Gelais C et al. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol 88, 5834–5844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White TE et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cribier A, Descours B, Valadao AL, Laguette N & Benkirane M Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Reports 3, 1036–1043 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Beloglazova N et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem 288, 8101–8110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ryoo J et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat. Med 20, 936–941 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hofmann H et al. Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J. Virol 87, 11741–11750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mlcochova P, Watters SA, Towers GJ, Noursadeghi M & Gupta RK Vpx complementation of ‘non-macrophage tropic’ R5 viruses reveals robust entry of infectious HIV-1 cores into macrophages. Retrovirology 11, 25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crow YJ et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A 167, 296–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stetson DB, Ko JS, Heidmann T & Medzhitov R Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao D et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA & Lieberman J The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol 11, 1005–1013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao K et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep 4, 1108–1115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim ES et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Etienne L, Hahn BH, Sharp PM, Matsen FA & Emerman M Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 14, 85–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lenzi GM, Domaoal RA, Kim D, Schinazi RF & Kim B Kinetic variations between reverse transcriptases of viral protein X coding and noncoding lentiviruses. Retrovirology 11, 111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kyei GB, Cheng X, Ramani R & Ratner L Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe 17, 98–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Westmoreland SV et al. SIV vpx is essential for macrophage infection but not for development of AIDS. PLoS ONE 9, e84463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Strebel K, Klimkait T, Maldarelli F & Martin MA Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J. Virol 63, 3784–3791 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Terwilliger EF, Cohen EA, Lu YC, Sodroski JG & Haseltine WA Functional role of human immunodeficiency virus type 1 vpu. Proc. Natl. Acad. Sci. USA 86, 5163–5167 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klimkait T, Strebel K, Hoggan MD, Martin MA & Orenstein JM The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol 64, 621–629 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Willey RL, Maldarelli F, Martin MA & Strebel K Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160–CD4 complexes. J. Virol 66, 226–234 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neil SJ, Eastman SW, Jouvenet N & Bieniasz PD HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog 2, e39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sakai H, Tokunaga K, Kawamura M & Adachi A Function of human immunodeficiency virus type 1 Vpu protein in various cell types. J. Gen. Virol 76, 2717–2722 (1995). [DOI] [PubMed] [Google Scholar]
- 104.Neil SJ, Sandrin V, Sundquist WI & Bieniasz PD An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2, 193–203 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Varthakavi V, Smith RM, Bour SP, Strebel K & Spearman P Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 100, 15154–15159 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neil SJ, Zang T & Bieniasz PD Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Van Damme N et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hinz A et al. Structural basis of HIV-1 tethering to membranes by the BST-2/tetherin ectodomain. Cell Host Microbe 7, 314–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Venkatesh S & Bieniasz PD Mechanism of HIV-1 virion entrapment by tetherin. PLoS Pathog 9, e1003483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perez-Caballero D et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Galão RP, Le Tortorec A, Pickering S, Kueck T & Neil SJ Innate sensing of HIV-1 assembly by Tetherin induces NFkB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cocka LJ & Bates P Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog 8, e1002931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tokarev A et al. Stimulation of NF-kB activity by the HIV restriction factor BST2. J. Virol 87, 2046–2057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sauter D et al. Differential regulation of NF-kB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Reports 10, 586–599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pickering S et al. Preservation of tetherin and CD4 counter-activities in circulating Vpu alleles despite extensive sequence variation within HIV-1 infected individuals. PLoS Pathog 10, e1003895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jia B et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5, e1000429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang F et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lim ES, Malik HS & Emerman M Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol 84, 7124–7134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sauter D et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kluge SF et al. The transmembrane domain of HIV-1 Vpu is sufficient to confer anti-tetherin activity to SIVcpz and SIVgor Vpu proteins: cytoplasmic determinants of Vpu function. Retrovirology 10, 32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bour S & Strebel K The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol 70, 8285–8300 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bour S, Schubert U, Peden K & Strebel K The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J. Virol 70, 820–829 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Serra-Moreno R, Jia B, Breed M, Alvarez X & Evans DT Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9, 46–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]