Abstract
Loci discovered by genome-wide association studies (GWAS) predominantly map outside protein-coding genes. The interpretation of the functional consequences of non-coding variants can be greatly enhanced by catalogues of regulatory genomic regions in cell lines and primary tissues. However, robust and readily applicable methods to systematically evaluate the contribution of these regions to genetic variation implicated in diseases or quantitative traits are still lacking. Here we propose a novel approach that leverages GWAS findings with regulatory or functional annotations to classify features relevant to a phenotype of interest. Within our framework, we account for major sources of confounding that current methods do not offer. We further assess enrichment for 29 GWAS traits within ENCODE and Roadmap derived regulatory regions. We characterize unique enrichment patterns for traits and annotations, driving novel biological insights. The method is implemented in standalone software and an R package to facilitate its application by the research community.
Introduction
Genome-wide association studies (GWAS) in humans have discovered susceptibility variants for complex diseases and biomedical quantitative traits, with over 75,000 associations found to date 1,2, representing a large investment in resources, time and organization by the worldwide research community. The majority (~90%) of implicated variants are classified as intronic or intergenic 3 and thus cannot be readily assigned to an underlying cellular or molecular mechanism. This has prompted a number of efforts to annotate the putative functional consequences of variants in cell-specific contexts from experimentally derived regulatory regions (e.g. regions marked by histone modifications, transcription factor binding 3–6), principally as a means to inform and accelerate functional validation.
The robust identification of the combinations of annotations for these regulatory regions (henceforth referred to generically as ‘regulatory annotations’) and cell types that are biologically most informative for a given disease or quantitative trait of interest (henceforth referred generically to as 'phenotype') requires that one can confidently distinguish correlations driven by biology from those arising by chance. Regulatory annotations may cover a large proportion of the genome, and thus many disease-associated variants will map within them by chance. In addition, the heterogeneous distribution of genetic variants and functional regions in the genome may result in their non-random association with genomic features such as genes 7,8, which in turn may drive spurious correlations that confound correct interpretation of these correlation patterns.
Functional enrichment methods assess the relative contribution of regulatory annotations to a phenotype of interest. In their simplest implementation, they estimate enrichment of association p-values (or z-scores) based on comparisons of the full set of genome-wide association (GWA) variants 9–13, or on subsets of highly associated variants (e.g. genome-wide significant variants) 14–16. These approaches have identified many biologically plausible patterns of enrichment and can be broadly used for ranking the relative contribution of features. For instance, variants associated with lipid traits and Crohn’s disease are enriched in open chromatin derived from liver and immune cells, respectively 13, reflecting biological functions. However, there is currently little confidence in interpreting unexpected enrichments, because of various statistical concerns. First, overly simplistic models that do not account for known confounders such as local linkage disequilibrium (LD) and local gene density can lead to spurious enrichment patterns 14. Second, tests based on subsets of variants typically probe a limited number of genomic features, whereas evidence of enrichment occurs well below genome-wide significance 11,12. Due to the large number of annotations now available, a third problem has emerged of prioritizing the most informative set from a large number of often correlated functional annotations. Methodological improvements are thus needed to increase the accuracy of inference, and to realize the full potential of those costly experiments in focused analysis.
Here we present a novel statistical approach that leverages GWAS findings with functional (i.e. regulatory or protein-coding) annotations to find features relevant to a phenotype of interest. This method accounts for LD, matched genotyping variants and local gene density with the application of logistic regression to derive statistical significance. We name our method GARFIELD, which stands for GWAS Analysis of Regulatory or Functional Information Enrichment with LD correction. We use GARFIELD to analyze the enrichment patterns of publicly available GWAS summary statistics using regulatory maps from the ENCODE 3 and Roadmap Epigenomics 5 projects. We describe expected and novel enrichments that illustrate the molecular and cellular basis of well-studied traits, which we expect to help drive novel biological insights and enhance efforts to prioritize variants for focused functional exploration. Finally, we developed new software to facilitate the application of our approach by the research community, and tools for effective visualization of enrichment results that scale to thousands of potential functional elements.
Results
Method Overview
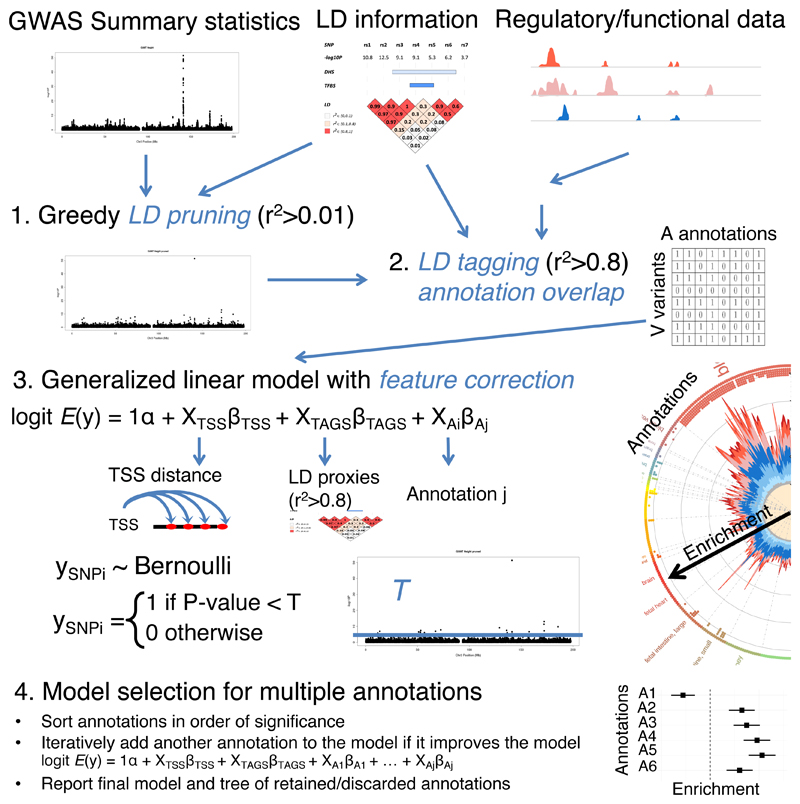
The analysis workflow implemented in GARFIELD is summarized in Figure 1 and Online Methods. The method requires four inputs: (i) a set of genome-wide genetic variant association p-values with a phenotype of interest; (ii) genome-wide genomic coordinates for regulatory annotations of interest; (iii) lists of LD tags for each variant (r2 ≥ 0.01 and r2 ≥ 0.8 within 1-MB windows) from a reference population of interest (e.g. Caucasian) and (iv) the distance of each variant to the nearest transcription start site (TSS). Given these inputs, the method first uses a greedy procedure to extract a set of independent variants from the genome-wide genetic variants, using LD (r2 ≥ 0.01) and distance information (‘LD pruning step’). Second, it annotates each variant with a regulatory annotation if either the variant, or a correlated variant (r2 ≥ 0.8), overlaps the feature (‘LD tagging annotation step’). Third, it calculates odds ratios (OR) and enrichment p-values at different GWAS p-value thresholds (denoted as ‘T’) for each annotation using a logistic regression model with ‘feature matching’ (Online Methods) on variants by distance to the nearest TSS and number of LD proxies (r2 ≥ 0.8) (used as categorical covariates). This pruning strategy is conservative, as a potential loss of the true causal variant at a small fraction of the loci due to pruning will be offset by the analysis of genome-wide enrichment patterns. We thus believe this is a conservative but sound approach for identifying annotations that harbor more GWAS variants (at a given threshold T) than expected by chance. To correct for multiple testing on the number of different annotations, we further estimate the effective number of independent annotations by using the eigenvalues of the correlation matrix of the binary annotation overlap matrix from Figure 1 (adapted from Galwey et al. 17) (Online Methods) and then apply a Bonferroni correction at the 95% significance level. This takes into account the tissue-selective components of regulatory data, namely that closely related cell types and tissues are more similar to each other than different ones. Our single annotation approach can be viewed as an extension of Maurano et al. 11 (see also Supplementary Table 1) with two critical improvements. First, we account for the effect of local variant correlations by restricting enrichment calculations to sets of independent variants (LD pruning step). Second, we employ a testing procedure that accounts for systematic differences in gene distance and number of proxies in the variant set.
Figure 1. Outline of the GARFIELD method.
Top panel: three inputs (annotation, p-value and linkage disequilibrium (LD) data) are used for the first two analytical steps (LD pruning and variant functional annotation), which result in a binary annotation overlap matrix of V pruned variants and A annotations. Middle panel: a logistic regression approach is used for testing for enrichment at a GWAS significance P-value threshold T while controlling for confounding features such as TSS distance and number of LD proxies. Bottom panel: model selection procedure for multiple annotations.
Additionally, we implement a heuristic procedure to combine the biological signal contained in correlated annotations, which allows us to identify conditionally independent sets of regulatory annotations underpinning the enrichment signals. To reduce the computational burden of searching through all possible combinations of available annotations, we first obtain enrichment p-values for all annotations separately, using the default single-annotation GARFIELD model. We then rank all statistically significant annotations by their enrichment p-value and iteratively add each such annotation to the model if it significantly improves the model fit compared to the model not including the annotation (analysis of deviance using a chi-squared test).
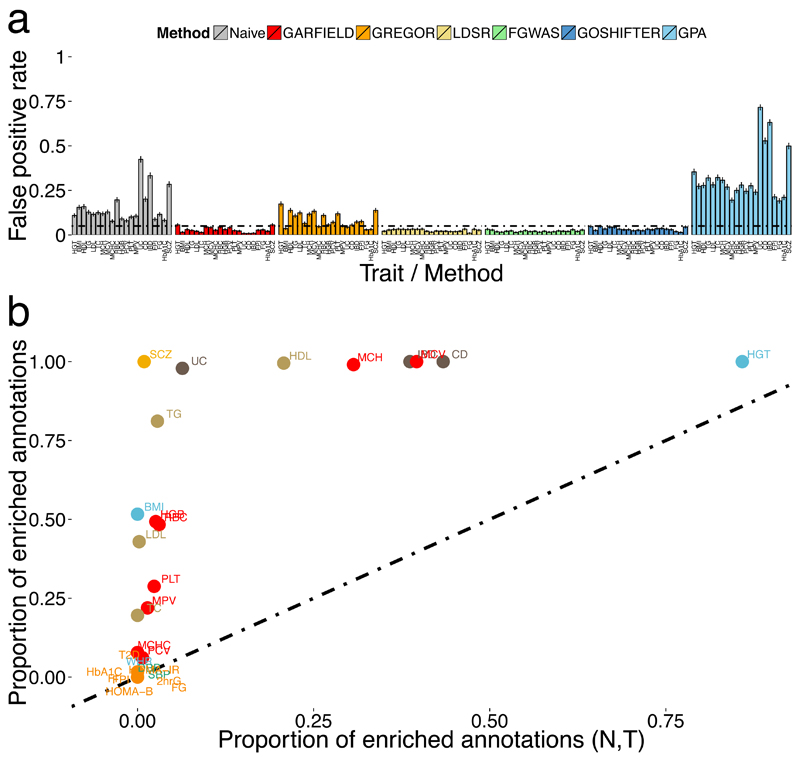
We compared GARFIELD to five widely used alternative methods (LDSC 10, fgwas 13, GoShifter 14, GREGOR 16 and GPA 9), while noting that benchmarking of methods is typically best done by investigators independent of the method developers. To estimate the false positive rate (FPR), we used 21 real disease or quantitative trait GWASs with the required summary statistics for all methods and greater than five independent genetic variants at the T < 10-8 threshold (Online Methods). We assessed the enrichment of each trait against 1,000 peak region annotations, simulated to match observed peak lengths and between peak distances for DNaseI hypersensitive sites (DHS) in HepG2 cells (ENCODE). We compared GARFIELD to the five alternative methods and to a naive model, where enrichment ORs are calculated without accounting for LD or other features. FPRs were estimated by the observed proportion of significantly enriched annotations per phenotype (Online Methods). At the 5% significance level, methods not modelling LD produced significantly inflated FPRs (0.15 and 0.33 on average for Naïve and GPA, respectively) (Figure 2a). GARFIELD, fgwas, LDSC and GoShifter preserved the FPR for all traits, while GREGOR yielded more false positive results than expected (average FPR 0.09). Further assessment of GARFIELD for a set of 29 traits showed that FPRs are also preserved when lowering the threshold from T < 10-8 to T < 10-5 (Supplementary Figure 1a).
Figure 2. Method assessment.
(a) Estimated false positive rate (FPR) from 21 publicly available disease or quantitative traits and n = 1,000 simulated independent annotations. The black horizontal line denotes the 5% FPR threshold. Error bars denote standard errors. (b) Comparison between the proportion of significant annotations (GARFIELD enrichment p-value < 2.6 × 10-4 for multiple testing correction) found from models accounting for number of proxies (N) and distance to nearest TSS (T) respectively (x-axis), to a model not accounting for any feature (y-axis), for each of 29 publicly available GWA studies and n = 424 DNaseI hypersensitive site annotations. Key of trait name labels is shown in Supplementary Table 3.
To assess the value of feature matching in significance testing, we employed GARFIELD with and without feature correction to 424 open chromatin annotations in 29 phenotypes at the T < 10-8 threshold. As expected, we found that feature matching controls for biases in enrichment analysis by significantly reducing the number of observed enrichments (Wilcoxon signed rank test proportion median = 0.46, p-value = 1.4 × 10-4) (Figure 2b). We further explored the relative contribution of each feature by comparing the number of significant enrichments detected in a feature-corrected model compared to the uncorrected model. We found median proportion reduction estimates of enrichments of 0.34 (p-value = 1.4 × 10-4) and 0.10 (p-value = 1.1 × 10-3) for the number of LD proxies and TSS distance, respectively (Supplementary Figure 1b-c). Estimates were concordant between GWAS p-value thresholds (Supplementary Figure 1d). These tests suggest that LD proxy number is the single most important confounder, although not sufficient to correct for individually when compared to the model correcting for both features together.
Enrichment in open chromatin regions
To assess the relative enrichment of phenotype-genotype associations in different cell types, we first applied GARFIELD to a generic regulatory annotation denoting open chromatin (DNaseI hypersensitive sites) in 424 cell lines and primary cell types from ENCODE 3 and Roadmap Epigenomics 5 (Supplementary Table 2). We considered five diseases and 24 quantitative traits with publicly available GWAS summary statistics. For each trait-annotation pair we derived enrichment estimates at eight GWAS P-value thresholds (T < 10-1 to T < 10-8). At the most stringent cut-off (T < 10-8), there were a median of 21 independent variants per trait after LD pruning (range 0-117, Table 1 and Supplementary Table 3), while at a more permissive threshold (T < 10-5) there were a median of 76 variants per trait (range 11-619).
Table 1. Summary of GARFIELD enrichment analyses in DNaseI hypersensitive sites, histone modifications and genomic segmentations per phenotype.
Columns denote (A) phenotype category and (B) its index, (C) trait full name and (D) abbreviation, (E) total number of variants after LD pruning, (F) number of independent SNPs at GWAS p-value threshold 10-8, (G) number of enriched cell types in open chromatin marks at GWAS p-value threshold 10-8, (H) number of conditionally independent cell types in open chromatin marks at GWAS p-value threshold 10-8, (I) number of enriched cell types in H3K27ac at GWAS p-value threshold 10-8, (J) number of enriched cell types in H3K4me3 at GWAS p-value threshold 10-8, (K) number of enriched cell types/segmentation states at GWAS p-value threshold 10-8, (L) number of depleted cell types/segmentation states at GWAS p-value threshold 10-8, (M) number of independent SNPs at GWAS p-value threshold 10-5, (N) number of enriched cell types in open chromatin marks at GWAS p-value threshold 10-5, (O) number of conditionally independent cell types in open chromatin marks at GWAS p-value threshold 10-5, (P) number of enriched cell types in H3K27ac at GWAS p-value threshold 10-5, (Q) number of enriched cell types in H3K4me3 at GWAS p-value threshold 10-5, (R) number of enriched cell types/segmentation states at GWAS p-value threshold 10-5, (S) number of depleted cell types/segmentation states at GWAS p-value threshold 10-5, (T) Tissues showing the largest enrichment in open chromatin states per trait category, (U) Tissues/histone modifications showing the largest enrichment per trait category, (V) Tissues/segmentation states showing the largest enrichment per trait category. Total number of GWAS variants (n) per trait can be found in Supplementary Table 3.
| Broad category | Similarity group | Trait | Abbreviation | Total independent SNPs genome-wide | T<10-8 | T<10-5 | Top enriched categories | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs at threshold | Enriched DHS/Cells | DHS/Cells from multiple annotation approach | H3K27ac enrichments | H3K4me3 enrichments | Enriched Seg. States/Cells | Depleted Seg. States/Cells | SNPs at threshold | Enriched DHS/Cells | DHS/Cells from multiple annotation approach | H3K27ac enrichments | H3K4me3 enrichments | Enriched Seg. States/Cells | Depleted Seg. States/Cells | DHSs | Histone Modifications | Segmentations | |||||
| Anthropometric | 1 | Body mass index | BMI | 50,166 | 16 | 0 | - | 0 | 1 | 14 | 0 | 62 | 0 | - | 0 | 0 | 14 | 0 | foetal_muscle* | Connective/epithelial/bone | active enhancers, tissues including adipose and skeletal muscle* |
| 1 | Height | HGT | 49,909 | 114 | 364 | 7 | 65 | 106 | 297 | 0 | 290 | 395 | 10 | 88 | 124 | 806 | 28 | ||||
| 1 | Waist hip ratio adjusted for BMI | WHR | 50,500 | 7 | 0 | - | 0 | 0 | 0 | 0 | 34 | 19 | 2 | 2 | 0 | 27 | 0 | ||||
| Glycaemic | 2 | 2hr Glucose adjusted for BMI | 2hrG | 46,691 | 1 | 0 | - | 0 | 0 | 0 | 0 | 11 | 0 | - | 0 | 0 | 1 | 0 | blood and foetal intestine | foetal intestine, spleen, adipose, pancreas | poised promoters in pancreatic islets and stomach mucosa, active enhancers in pancreatic islets and transcription regulation in blood |
| 2 | HbA1C | HbA1C | 52,079 | 8 | 0 | - | 0 | 0 | 0 | 0 | 45 | 2 | 2 | 7 | 0 | 2 | 0 | ||||
| 2 | Fasting Proinsulin | FPI | 56,429 | 10 | 0 | - | 0 | 0 | 22 | 0 | 30 | 1 | 1 | 1 | 0 | 0 | 5 | ||||
| 2 | Fasting Glucose | FG | 51,578 | 13 | 0 | - | 0 | 0 | 0 | 0 | 43 | 11 | 3 | 1 | 2 | 7 | 0 | ||||
| 2 | Fasting Insulin | FI | 50,132 | 0 | 0 | - | 0 | 0 | 0 | 0 | 12 | 0 | - | 1 | 0 | 0 | 0 | ||||
| 2 | Type 2 Diabetes | T2D | 47,928 | 10 | 0 | - | 0 | 0 | 1 | 0 | 65 | 5 | 3 | 3 | 5 | 3 | 0 | ||||
| 2 | HOMA-IR | HOMA-IR | 49,450 | 0 | 0 | - | 0 | 0 | 0 | 0 | 14 | 0 | - | 0 | 0 | 1 | 0 | ||||
| 2 | HOMA-B | HOMA-B | 49,249 | 4 | 0 | - | 0 | 0 | 0 | 0 | 20 | 0 | - | 0 | 0 | 2 | 0 | ||||
| Blood pressure | 3 | Diastolic blood pressure | DBP | 52,446 | 7 | 0 | - | 0 | 0 | 0 | 0 | 43 | 0 | - | 1 | 0 | 2 | 0 | lung | left ventricle | transcription enhancers in blood and left ventricle |
| 3 | Systolic blood pressure | SBP | 52,592 | 7 | 0 | - | 0 | 0 | 4 | 0 | 48 | 1 | 1 | 3 | 0 | 9 | 0 | ||||
| Lipids | 4 | High-density lipoprotein | HDL | 61,981 | 66 | 88 | 3 | 22 | 6 | 135 | 26 | 150 | 154 | 3 | 65 | 35 | 483 | 119 | several (liver, blood, foetal intestine, colon, foetal thymus) | liver, blood, gastrointestinal and adipose | active enhancers in liver, transcription enhancers in foetal intestine and blood |
| 4 | Total cholesterol | TC | 62,029 | 64 | 0 | - | 2 | 1 | 8 | 6 | 131 | 15 | 3 | 20 | 16 | 122 | 14 | ||||
| 4 | Triglyceride | TG | 61,999 | 36 | 12 | 2 | 6 | 1 | 2 | 0 | 95 | 43 | 3 | 29 | 32 | 41 | 0 | ||||
| 4 | Low-density lipoprotein | LDL | 61,933 | 49 | 1 | 1 | 7 | 6 | 12 | 2 | 115 | 75 | 3 | 25 | 23 | 45 | 27 | ||||
| Haematological | 5 | Haemoglobin count | HGB | 49,658 | 21 | 11 | 3 | 10 | 2 | 141 | 0 | 84 | 20 | 3 | 15 | 4 | 97 | 0 | blood | HSC/Blood/Immune | transcriptional regulation and active enhancers in blood |
| 5 | Mean corpuscular volume | MCV | 49,387 | 45 | 167 | 8 | 71 | 103 | 187 | 0 | 132 | 247 | 5 | 87 | 120 | 831 | 1 | ||||
| 5 | Red blood cell count | RBC | 49,414 | 25 | 12 | 2 | 9 | 2 | 11 | 0 | 103 | 91 | 4 | 47 | 35 | 357 | 0 | ||||
| 5 | Mean corpuscular haemoglobin concentration | MCHC | 49,466 | 13 | 0 | - | 2 | 0 | 3 | 0 | 45 | 6 | 2 | 3 | 0 | 10 | 0 | ||||
| 5 | Mean corpuscular haemoglobin | MCH | 49,239 | 39 | 129 | 5 | 54 | 106 | 134 | 0 | 116 | 200 | 5 | 72 | 122 | 323 | 0 | ||||
| 5 | Packed cell volume | PCV | 49,236 | 16 | 3 | 2 | 2 | 0 | 31 | 0 | 63 | 69 | 4 | 47 | 40 | 254 | 0 | ||||
| 5 | Mean platelet volume | MPV | 56,683 | 27 | 6 | 2 | 3 | 1 | 4 | 0 | 76 | 18 | 3 | 17 | 3 | 37 | 1 | ||||
| 5 | Platelet count | PLT | 58,181 | 35 | 10 | 2 | 15 | 9 | 19 | 0 | 111 | 66 | 4 | 74 | 95 | 352 | 2 | ||||
| Autoimmune disease | 6 | Crohn's Disease | CD | 347,359 | 65 | 183 | 4 | 28 | 35 | 54 | 22 | 215 | 187 | 5 | 40 | 67 | 221 | 28 | blood, foetal intestine, foetal thymus | blood immune and gastrointestinal cells | active enhancers in blood immune cells and thymus |
| 6 | Ulcerative colitis | UC | 356,248 | 67 | 27 | 2 | 13 | 11 | 49 | 3 | 218 | 150 | 3 | 39 | 45 | 183 | 22 | ||||
| 6 | Inflammatory Bowel Disease | IBD | 393,352 | 94 | 164 | 4 | 35 | 17 | 155 | 15 | 283 | 168 | 7 | 40 | 37 | 263 | 38 | ||||
| Psychiatric disorder | 7 | Schizophrenia | SCZ | 170,825 | 117 | 4 | 1 | 17 | 15 | 98 | 0 | 619 | 2 | 2 | 8 | 1 | 29 | 6 | blood, foetal brain | CNS and HSC/Blood/Immune | transcription/bivalent promoters in foetal brain |
Features = Total independent SNPs significant in GWAS for a given T
excluding height
Total number of GWAS variants (n) per trait can be found in Supplementary Table 3.
We found statistically significant enrichments (p < 2.6 × 10-4; Online methods, Supplementary Note) for the majority of traits considered, highlighting clear differences in enrichment patterns between traits (Supplementary Table 4). As clearly visible from enrichment wheel plots, some traits displayed relatively ubiquitous enrichment (e.g. height, Figure 3a), while others showed relatively narrow enrichment (e.g. ulcerative colitis, Figure 3b, see also Supplementary Figure 2). Blood cells were overall the most enriched tissue type in hematological traits and autoimmune diseases, but provided little to no enrichment for glycemic, blood pressure and anthropometric traits (except height which was enriched in nearly all tissues). As predicted, incorporating sub-threshold associations (T < 10-5) increased the resolution of enrichment patterns across traits (Table 1). For instance, at T < 10-8 there were no annotations enriched for waist-to-hip ratio (WHR), while at T < 10-5 there were 19 significant enrichments, 18 of which coming from muscle or fetal muscle tissue. For HbA1C and fasting glucose again there were no enrichments at T < 10-8, while at T < 10-5 we uncovered links to blood, fetal stomach and fetal intestine tissues. Additionally, for low density lipoprotein (LDL) cholesterol we found a single enrichment in colon at T < 10-8, while the permissive threshold allowed us to detect much larger number of relevant annotations (75), including liver, blood and fetal intestine cell types. Overall, 89% of the enrichments at the T < 10-8 threshold were also identified at T < 10-5 (between-threshold log10 enrichment p-value correlation = 0.85) (Supplementary Figure 3) showing high degree of agreement between thresholds.
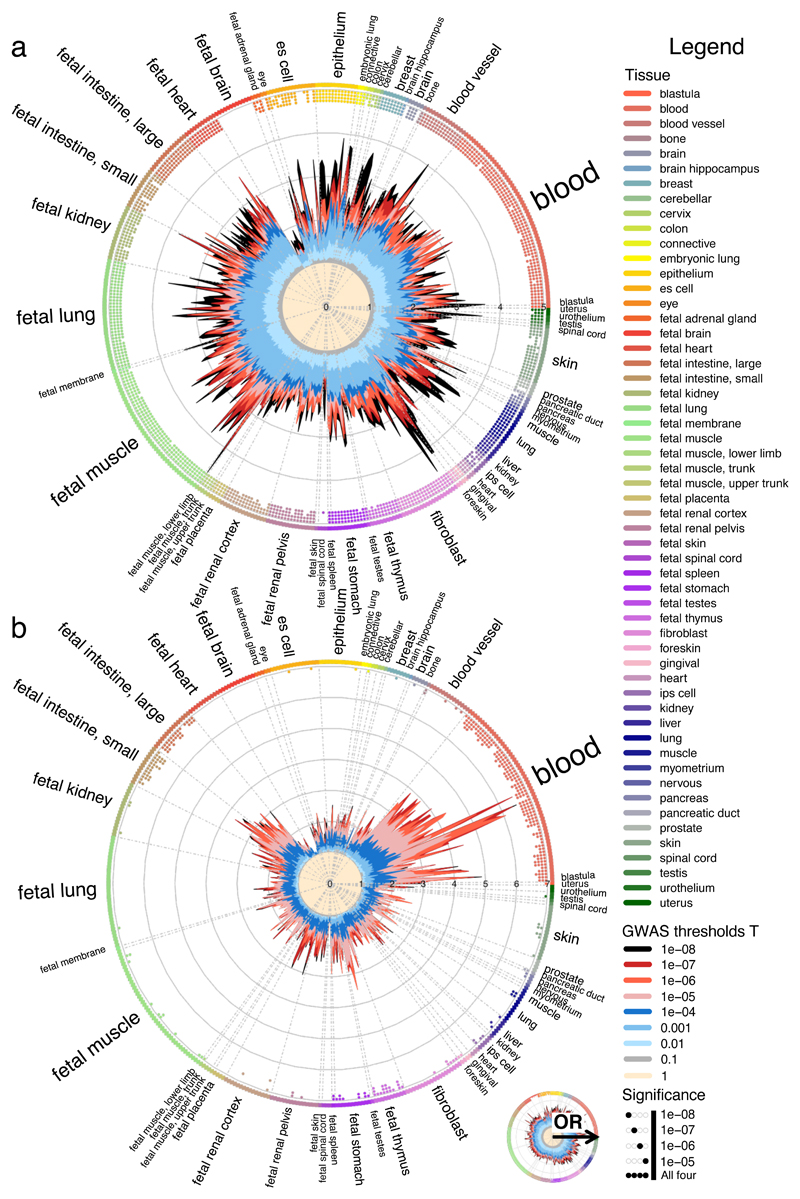
Figure 3. Enrichment of genome-wide association analysis p-values in DNaseI hypersensitive sites (hotspots).
(a) Height (HGT) (n = 2,468,982 GWAS variants). (b) Ulcerative colitis (UC) (n = 11,113,952 GWAS variants). Radial lines show odds ratio values at eight GWAS P-value thresholds (T) for all ENCODE and Roadmap Epigenomics DHS cell lines, sorted by tissue on the outer circle. Dots in the inner ring of the outer circle denote significant GARFIELD enrichment (if present) at T < 10-5 (outermost) to T < 10-8 (innermost) after multiple testing correction for the number of effective annotations and are coloured with respect to the tissue of the cell type they test. Font size of tissue labels reflects the number of cell types from that tissue. Crohn’s disease shows predominant enrichment in blood, fetal thymus and fetal intestine tissues whereas height exhibits an overall enrichment. OR, odd’s ratio.
The observed enrichments reflect current understanding of key cellular types for disease, augmented with novel observations. In the former category were enrichments of lipid traits in blood, liver, fetal intestine and fetal thymus cell types; of hematological traits in blood, and of autoimmune diseases (ulcerative colitis, Crohn’s disease, inflammatory bowel disease (IBD)) in blood and fetal intestine 11,13,18. Potentially interesting examples of the latter category include the enrichment of Caco-2 (a well-established gut epithelia cellular model) elements for LDL cholesterol, the enrichment of (fetal) muscle and placenta cell type elements in high density lipoprotein (HDL) cholesterol and foetal intestine in Hemoglobin (HGB). For each trait, we also employed GARFIELD’s heuristic multiple annotation approach illustrated earlier to further prioritize a parsimonious set of non-correlated cell types from those with significant enrichment. Only a small proportion of enriched annotations detected under univariate settings were retained in the multiple annotation model (proportion median = 17%, range 2-100%; median number of annotations retained = 2, range 1-8; Table 1 and Supplementary Figure 4). For instance, in height we narrow down the annotations from 364 to 7 (2%). These findings are suggestive of a high degree in redundancy between annotations, while also highlighting that in the majority of cases biological enrichments are driven by more than a single annotation. For instance, for HDL cholesterol we obtain conditionally independent signals coming from blood and liver cell types.
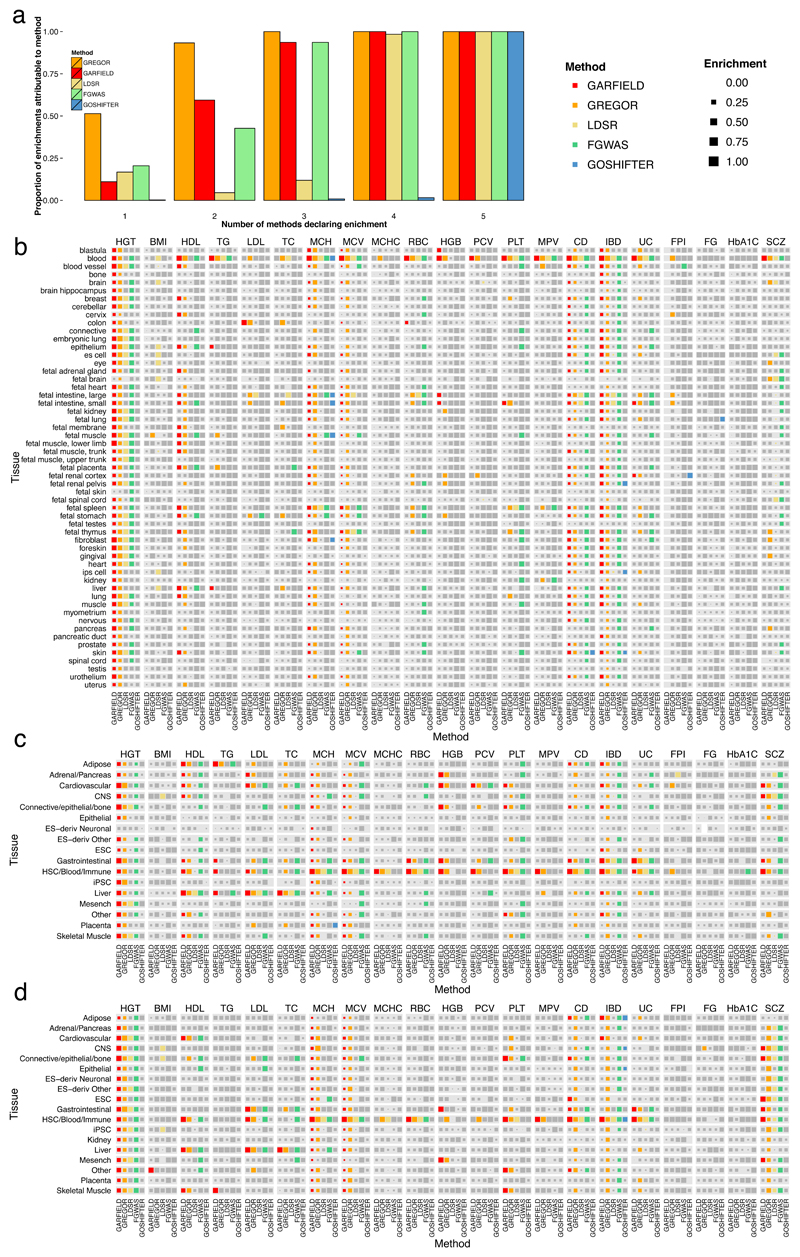
Next, we sought to evaluate GARFIELD against alternative enrichment methods when considering empirical phenotypes and DHS data. We performed enrichment analysis for each of the 21 traits from the simulation study in each of the 424 cell types using each of the five methods (GARFIELD, GoShifter, fgwas, GREGOR and LDSC) shown previously to preserve (or nearly preserve) FPR in simulations (Online Methods; Supplementary Table 5). GREGOR yielded the largest number of enrichments (median = 24, max = 398), followed by GARFIELD (median = 10, max = 364). Fgwas and LDSC yielded intermediate levels of enrichment (median = 5, max = 327; median = 5, max = 144, respectively), while GoShifter was very conservative (median = 0, max = 5). Stratification of the enrichments to groups according to the number of methods supporting them further showed that GREGOR identified the largest number of enrichments found by at least one other method. GARFIELD closely followed GREGOR, whereas fgwas, LDSC and GoShifter showed much lower between-method concordance rates. GREGOR also identified the largest number of method-specific enrichments, however, the inflated FPR indicates that more enrichments are discovered at the cost of also reporting more false positives, making utility of GREGOR alone less desirable in practice (Figure 4a). In the absence of a truth set, the observation that GARFIELD captures a large proportion of enrichments consistent with other methods, while preserving the FPR, provides an indirect assessment of the robustness of our approach. Overall, enrichments of blood cell traits with blood cell regulatory annotations tended to be highly consistent between most methods (supported by GARFIELD for 7 traits; GREGOR in 8; fgwas and LDSC in 5 and GoShifter in 1; Figure 4b), as expected given their clear biological relevance. Likewise, we observed highly consistent results for height in the majority of cell types; schizophrenia (SCZ) in blood and fetal brain; HDL cholesterol in liver, blood and fetal placenta (supported by GARFIELD, GREGOR and fgwas); triglycerides in blood (GARFIELD, GREGOR, fgwas and LDSC); mean corpuscular volume (MCV) in fetal stomach, fetal spleen and fetal thymus, mean corpuscular hemoglobin (MCH) in fetal intestine, fetal stomach and fetal spleen, all of which were supported by at least three methods.
Figure 4. Method comparison for 21 GWAS datasets in DNaseI hypersensitive sites (hotspots) and histone modifications (H3K27ac and H3K4me3) at the T < 10-8 GWAS significance threshold.
(a) Proportion of enriched cell types in DNaseI hypersensitive sites identified by each method, where enrichments are stratified by the number of methods that support them. GARFIELD, fgwas and LDSC are restricted to positive enrichments only so as to be comparable to GREGOR and GoShifter. (b) Summary of significant enrichments per tissue and per method for DNaseI hypersensitive data. A colored box is present if the corresponding method has found at least one significantly enriched cell type for that tissue after multiple testing correction. Colors correspond to the different methods and are the same as in panel a. A grey box denotes that the enrichment did not reach significance. Additionally, the size of each box represents the relative magnitude of the enrichment. Since each method uses a different enrichment statistic, we have scaled each of them separately per method and per trait (e.g. for GARFIELD we scaled the ORs for all cell types for HDL so that 1 denotes the cell type with the highest enrichment found and 0 the lowest one). (c) Summary of significant enrichments per tissue and per method for H3K27ac data. (d) Summary of significant enrichments per tissue and per method for H3K4me3 data. (b-d) Sample sizes n per trait (and trait name labels) can be found in Supplementary Table 3 denoted by the number of variants in each GWAS study.
Finally, we compared the average CPU time used per method, trait and annotation based on the analysis of 21 traits and 424 annotations. GARFIELD was faster than all other methods with an average of 0.64 mins needed, compared to 2.32 mins for GoShifter, 6.70 mins for LDSC, 16 mins for fgwas and 0.96 mins for GREGOR (Online Methods). It has to be noted however that LDSC is fast to run but had a substantial computational burden of generating the necessary input files for our custom data (Supplementary Table 6).
Enrichment in promoter and enhancer marks
In light of the current knowledge of relevant links between cell types and complex traits based on promoter and enhancer activity, we also sought to evaluate GARFIELD against alternative enrichment methods when considering empirical phenotypes and marks of active enhancer (H3K27ac) and active promoter (H3K4me3) activity in 127 cell types, similarly to DHS comparisons presented earlier.
We found statistically significant enrichments (p < 5 × 10-4; Online methods) that confirm known biology for both H3K27ac and H3K4me3 (Supplementary Table 5). Namely, height was enriched in the majority of tissues for both regulatory marks; SCZ showed predominantly enriched in central nervous system (CNS) tissue; blood cell traits were enriched in HSC/Blood/Immune cell types and lipids traits in liver tissues for both marks. Overall results also show fewer and more specific enrichments in H3K27ac in comparison to H3K4me3 (mean 17, range [0-72]; and mean 20, range [0-106] number of enrichments, respectively) consistent with higher cell type specificity found in active enhancers versus active promoter regions.
Enrichment in genomic segmentations
We additionally sought to compare the relative enrichment of different types of functional genomic marks, using ChromHMM 15 data on genomic segmentations for 127 cell types (Supplementary Table 7). For each segmentation state and cell type, we analyzed our 29 phenotypes at two different GWAS p-value thresholds (T < 10-5 and T < 10-8). Overall, when considering only significantly enriched trait-annotation pairs (p < 3.3 × 10-5; Supplementary Table 8), we found higher levels of enrichment for promoters (median OR = 3.4, range [2.0-10.9] for T < 10-5) and enhancers (median OR = 3.8, range [1.9-68.0]) compared to transcribed regions (median OR = 2.6, range [1.8-13.8]), and depletion in quiescent regions (Figure 5a) (similar patterns were obtained for T < 10-8, Supplementary Figure 5). Given that transcriptional states mainly mark active genes, it is unsurprising to see the contrast of enrichment in transcriptional regions compared to the depletion in quiescent regions. Interestingly the enhancer states consistently had stronger enrichments than transcribed regions, an observation in agreement with enrichments of hematological traits in cell-matched regulatory states from the BLUEPRINT project 19. To confirm these patterns, while controlling for the effect of annotation density on the number of enrichments found, we sought to compare only ORs for cell types enriched in both transcribed and enhancer pair states (and promoter and transcribed states). Similarly to our previous observations, results showed on average greater ORs for enrichment for enhancers when compared to transcribed regions (Figure 5b) (with a similar but weaker effect for promoters), which provides further evidence that our observation is not due to difference in power for enrichment detection between annotations of different density but due to their biological relevance to the studied traits.
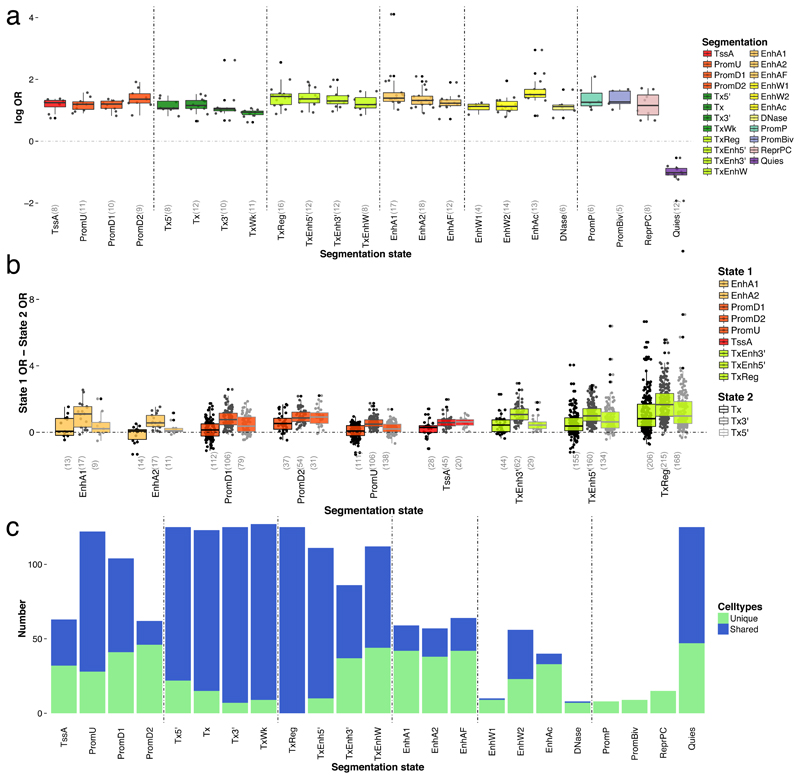
Figure 5. Enrichment levels (log OR) and extent of sharing between traits for 25-state chromatin segmentations of the NIH Roadmap and ENCODE projects at the T < 10-5 GWAS significance threshold.
(a) Distribution of significant (log) OR values across the 29 traits considered, split by segmentation state and coloured to highlight predicted functional elements (Supplementary Table 9). Number of points n is shown on the x-axis below each category. (b) Distribution of the pairwise difference between ORs from all enhancer, promoter and transcriptional enhancers and transcriptional regulatory states tested (‘state 1’) to ORs from transcription states for significant enrichments only (‘state 2’; e.g. measuring ORc,tEnhA1-ORc,tTx for all cell types c and traits t for which p-valuec,tEnhA1 and p-valuec,tTx are both significant). Number of points n is shown on the x-axis below each category. Boxplots show the median (center line); upper and lower quartiles (box limits), whiskers, furthest away point less than l.5x interquartile range (whiskers); points in the distribution(grey points) and outliers (black points). (c) Sharing of significantly enriched (or depleted) annotations (n=127 cell types) across 27 phenotypes (excluding Crohn’s disease (CD) and Ulcerative colitis (UC) as categories of IBD). The barplot displays the number of cell types where an annotation is uniquely enriched/depleted in a trait or shared between traits.
When considering cell-type specificity, again the trait height was the most ubiquitously enriched phenotype. In general, we found the largest ORs for anthropometric traits in active enhancers in adipose and skeletal muscle tissues; glycemic traits in active enhancers in pancreatic islets, poised promoters in pancreatic islets and stomach mucosa and transcription regulation in blood; lipid traits in active enhancers in liver, transcription enhancers in blood and fetal intestine tissue; autoimmune diseases and blood traits in active enhancers in tissues including blood and thymus; psychiatric disorder in transcription and bivalent promoters in fetal brain. As expected, incorporating sub-threshold associations again greatly increased the resolution of enrichment patterns across different traits (Table 1). For example, we found no significant enrichment at T < 10-8 for the glycemic indices β-cell activity index (HOMA-B), glycated hemoglobin (HbA1C) and fasting glucose (FG), whereas at T < 10-5 HOMA-B was predominantly enriched in active enhancers in pancreatic islets and ES-I3 cells, HbA1C in active enhancers in psoas muscle and fasting glucose in poised promoters in pancreatic islets and stomach mucosa.
Finally, we assessed the extent to which traits shared significantly enriched annotations, by comparing the number of cell types per segmentation state that were found to be significantly enriched (or depleted) for a single trait compared to multiple traits (Figure 5c and Supplementary Figure 5). Our results confirmed patterns of higher cell type specificity for enhancer states, with a median of 67% of cell types in enhancer states that were unique to a single trait compared to only 45% for promoter regions at T<10-5 (76% and 50% at T<10-8, respectively). This confirms enhancer states as prime regions of interest 19 when seeking to investigate gene function underlying complex trait and disease associations.
Software implementation
Many GWA studies seek to explore functional enrichment patterns, but often rely on customized, in-house pipelines. We implemented GARFIELD as a standalone tool in C++ in order to facilitate use by the research community (Online Methods). The software allows for enrichment analysis of any user-provided trait with variant GWAS p-values and GRCh37 genomic coordinates. We provide over 1000 GENCODE 20, ENCODE 3 and Roadmap Epigenomics 5 pre-compiled annotations, UK10K sequence LD data and TSS distance information for a ready to use package. Furthermore, custom annotation data can be easily accommodated when provided in a simple bed format. In addition, we have also developed a Bioconductor package for the R statistical framework to further increase usability.
Discussion
Large-scale efforts 3–6 have been devoted to systematically mapping molecular traits associated with genomic regulatory regions. They have greatly enhanced the annotation of putative functional consequences of non-coding variants in cell-specific contexts, and have further shown to provide links to disease association. However, current methods that aim to evaluate the contribution of such regions to genetic variation in disease cannot always do so robustly or are not readily applicable for systematic analysis and comparison of broad sets of features. In particular, it has been shown that LD and gene density can confound enrichment analysis results 14. Here we further estimated the relative effect of each of those features and identified LD as the largest confounder. Additionally, because of their design, different genotyping platforms (and imputation strategies) can create different biases (e.g. number of variants, genomic location distribution). GARFIELD accounts for all those features, by obtaining independent signals, expansion to relevant annotations using a population scale LD reference and feature matching, and to the best of our knowledge there is no other method that can do so without making extremely restrictive assumptions (e.g. Pickrell et al. 13 assume at most one causal variant at a given genomic region). Furthermore, many available approaches use variants that reach genome-wide significance from association analysis (T < 5 × 10-8) although there has been evidence of enrichment occurring well below that level 11,12. To capture these effects, GARFIELD allows for parallel enrichment analyses at multiple p-value sub-thresholds, which improves power to detect statistically significant enrichment patterns by increasing the number of variants tested, thus enabling its application to traits with underpowered GWA studies. Finally, we provide a flexible software platform with effective visualization to enable researchers to carry out simultaneous enrichment analysis for thousands of annotations at multiple association thresholds.
In our own application of GARFIELD on existing GWAS and functional datasets we identified a broad set of largely expected or previously identified enrichments, for example lipids traits in open chromatin in liver, hematological traits in blood and anthropometric traits in active enhancers in adipose tissue. A number of GWAS hits do not show significant enrichments even with established cell types when using higher thresholds, but GARFIELD’s stepwise, stratified approach uncovers these more nuanced enrichments, shown in the case of pancreatic islets with fasting HOMA-B. By analyzing large-scale genome segmentation data, we assessed the relative contribution of each segmentation state to the phenotypic traits. We discovered a larger number of enrichments coming from transcription states as opposed to promoter and enhancer states together with a larger number of shared cell types between traits. These findings may be biologically relevant, or could also be a result of statistically larger power for enrichment detection for broader region annotations. Here we show that study power differences are not responsible for larger OR values for significant enrichments in promoter and enhancer regions when compared to transcribed regions, highlighting them as much more relevant for trait associated variants.
Robust, usable and modular methods are critical in the modern large-scale analysis arena, where we expect many discoveries to come from principled combinations of heterogeneous datasets. In our hands, GARFIELD provided the greatest number of enrichments on real data among methods with full control of FPR in simulated data and was among the fastest methods. However, we acknowledge that as authors of this method we are not the right group to provide unbiased benchmarking of these methods and look forward to independent analysis of these methods. We have already deployed GARFIELD in a number of association study settings both in house and more broadly in the community. Our aim in developing it has been to provide the most robust statistical framework for analyzing functional enrichments coupled with practical ease of use and visualization, and we hope the community will continue to exploit this tool to provide more insights into disease mechanisms.
URLs
Association Summary Statistics
http://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium GIANT BMI 21, height 22 and waist hip ratio adjusted for BMI 23
http://www.magicinvestigators.org/downloads MAGIC BMI adjusted 2hr glucose 24, HOMA B, HOMA IR, fasting glucose, fasting insulin 25, fasting proinsulin 26 and HbA1C 27
http://www.sph.umich.edu/csg/abecasis/public/lipids2010/ Global lipid GWAS summary statistics for LDL, HDL, TC and TG 28
http://www.ibdgenetics.org/downloads.html IIBDGC data on Crohn’s disease, ulcerative colitis and Inflammatory Bowel Disease 29
http://www.georgehretlab.org/icbp_088023401234-9812599.html ICBP data on SBP and DBP 30
http://diagram-consortium.org/downloads.html DIAGRAM Type 2 diabetes 31 GWAS summary statistics
http://www.med.unc.edu/pgc/ PGC data on Schizophrenia.
DHS data
http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=hub_4607_uniformDnase&hubUrl=http://ftp.ebi.ac.uk/pub/databases/ensembl/encode/integration_data_jan2011/hub.txt ENCODE DNaseI hypersensitive sites
http://www.genboree.org/EdaccData/Current-Release/experiment-sample/Chromatin_Accessibility/ NIH Roadmap Epigenomics Mapping
H3K27ac, H3K4me3 and epigenome segmentation data
http://egg2.wustl.edu/roadmap/web_portal/ Processed NarrowPeak consolidated epigenome data and 25 state chromatin state model on imputed data for 12 marks.
Software
http://www.ebi.ac.uk/birney-srv/GARFIELD/ GARFIELD standalone tool; Code availability
http://bioconductor.org/packages/release/bioc/html/garfield.html/ GARFIELD R-package
Online Methods
Association Summary Statistics Data
GWAS summary statistics from the analysis of 29 disease and quantitative phenotypes were obtained from a number of sources (see URLs). From GIANT we downloaded large studies on BMI 21, Height 22 and Waist hip ratio adjusted for BMI 23. From MAGIC we downloaded data on BMI adjusted 2hr glucose 24, HOMA B, HOMA IR, Fasting glucose, Fasting insulin 25, Fasting proinsulin 26 and HbA1C 27. Global lipid GWAS summary statistics for LDL, HDL, TC and TG we obtained from 28. Crohn’s disease, Ulcerative colitis and Inflammatory Bowel Disease 29 data was obtained from IIBDGC. SBP and DBP 30 data was downloaded ICBP. Type 2 diabetes 31 GWAS summary statistics were downloaded from DIAGRAM. Schizophrenia data from 32 was further obtained and analysed. Blood trait data on HGB, MCH, MCV, MCHC, RBC and PCV was additionally obtained from the authors of van der Harst et al 33 and MPV and PLT data from the authors of Gieger et. al. 34 (Supplementary Table 3).
DHS data
DNaseI hypersensitive sites (hotspots) were obtained from ENCODE and the NIH Roadmap Epigenomics Mapping (see URLs) on all available cell types. DHS data was processed following DHSs data processing protocol described in an ENCODE study 4. Further information on the data can be found in Supplementary Table 2.
H3K27ac and H3K4me3 data
Processed NarrowPeak consolidated epigenome data was downloaded from the Roadmap Epigenomics portal (see URLs) for all available cell types for H3K27ac and H3K4me3 marks (98 and 127 cell types, respectively). Cell line information can be found in Supplementary Tables 7.
Epigenome segmentation data
Data from a chromatin state model with 25 states based on imputed data for 12 marks (H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac, H4K20me1, H3K79me2, H3K36me3, H3K9me3, H3K27me3, H2A.Z, and DNase) across 111 Roadmap Epigenomics 15 and 16 ENCODE reference epigenomes was downloaded from the Roadmap Epigenomics portal (see URLs). State and cell line information can be found in Supplementary Tables 9 and 7.
LD data
LD information (proxies) was calculated using PLINK 35(v1.7) and the --tag-r2 0.01 --tag-kb 500 (and --tag-r2 0.8 --tag-kb 500) flags in order to find all proxies within a 1Mb window around each variant at R-squared thresholds of 0.01 and 0.8. We computed these from the UK10K 36 sequence data on 3621 samples from two population cohorts (TwinsUK and ALSPAC) (data described elsewhere 36). Variants that were not observed in the UK10K data were excluded from our analysis.
Data processing
Given a genome-wide distribution of p-values for association with a given disease or quantitative trait, we perform the following pre-processing steps in order to calculate the level of enrichment and its significance for an annotation of interest. To remove possible biases due to linkage disequilibrium (LD) or dependence between variants we compute the r2 between all SNPs within 1-Mb windows and consider r2 of less than 0.01 between two variants to mean (approximate) independence. Next, from the full set of genetic variants for each phenotype, we create an independent set of SNPs where in order to keep all possible GWAS signals we sequentially find and retain the next most significant (lowest P-value) variant independent of all other variants in our independence set. After LD pruning an average of 2.2% (with range 1.9-3.4%) of genome-wide variants remained in our independence set for enrichment analysis (Supplementary Table 3). Next, we annotate each independent SNP and consider it as overlapping a functional element if (i) the SNP itself resides in such a genomic region or (ii) at least one of its proxies in LD (r2 ≥ 0.8) and within 500 Kb with it does. We include the latter as the association of a SNP in GWAS potentially tags the effect of other variants, which could underlie the observed association signal. The advantage of our greedy pruning over a P-value independent pruning is that we retain larger proportion of potentially causal variants (or tags of such SNPs). This is particularly advantageous for GWA studies with low power and more pronounced at more stringent pruning thresholds.
Quantifying enrichment and statistical significance
To find the enrichment of GWAS signals within a given functional annotation at a genome-wide significance P-value threshold T, we use the following logistic regression model
where yi = 1 if SNP i has GWAS P-value < T, and yi = 0 otherwise. 1 denotes an intercept term (a vector of 1’s) and XAj denotes a binary annotation covariate for annotation j. XTSS and XTAGS are categorical covariates denoting which quantile bin of distance to nearest TSS and number of LD proxies (r2≥0.8) a variant falls in (by default we use 5 quantiles for TSS distance and 15 for number of LD proxies). These terms are added to account for possible biases in the analysis due to the GWAS P-value distribution correlating to them, which may also non-randomly associate with functional data. Due to the discreteness of the number of proxies and the skewness of their distribution in the pruned data, exact quantile binning is not always possible, in which case we create a stepwise binning in which we iteratively find the first (Q-q)’th quantile from the remaining variants after having already created q (out of Q) bins and removed those variants from consideration. We calculate ORs and test for their significance at T=10-1, 10-2, …, 10-8 for all traits at each given threshold.
Testing for significant association between an annotation and GWAS SNP status means testing for βAj=0 vs βAj∈ℜ. If, additionally, βAj>0, this denotes enrichment, otherwise we consider it to be a depletion. OR statistic is then calculated via the following equation
Model selection for multiple annotations is done by (i) sorting annotations in order of significance from single annotation model; (ii) iteratively trying to add an annotation to the model if it significantly improved the model fit (p<0.05) given all other annotations in the model using the following model:
And (iii) reporting the final model and tree or retained/discarded annotations.
Multiple testing
To account for multiple testing in the number of annotations used, we apply a Bonferroni correction for the number of independent tests carried out. Due to the nature of the data, annotations need not be (and are not in general) independent (e.g. biological replicates of the same cell types). Thus, correcting for all annotations by assuming independence would be extremely stringent in practice. Instead, we estimate the effective number of independent tests performed similarly to Galwey, 2009 17. More specifically, we take an independent subsample of SNPs and find the eigenvalues of the correlation matrix between all considered annotations and then find the effective number of independent test from equation 16 in Galwey, 2009. This results in at most 194 independent annotations out of a total of 424 for the DHS data (for the 29 phenotypes considered), to which we apply Bonferroni correction (p~2.6x10-4). Further details can be found in the Supplementary Note. Similarly, for the segmentation data a total of 25x127=3175 annotations were used, which resulted in p~3.3x10-5 after correcting for multiple testing on the effective number of independent annotations at the 5% significance level. Finally, for the histone modification data we used a threshold of p~4.7 × 10-4.
False positive rate
To get an estimate of GARFIELD’s false positive rate, we simulated 1000 random annotations by mimicking the peak lengths and between peak distances from the ENCODE HepG2 DHS cell line. We then performed enrichment analysis for each annotation-trait pair from the 1,000 simulated annotations and 29 publicly available disease or quantitative trait GWAS studies. We estimated the false positive rate as the proportion of cell types showing significant enrichment for a given trait and further compared GARFIELD to each of six other tools for a subset of 21 of the traits with the necessary summary statistics for running all other approaches.
Analysis with other software
For the method comparison analysis, we used threshold of T<10-8 for GARFIELD, GREGOR and GoShifter and no threshold for fgwas and LDSC. Enrichment was defined as p<2.6x10-4 and an effect with positive direction (ORGARFIELD>1, EnrichmentLDSC>1, Estimatefgwas>1; GREGOR and GoShifter only test for enrichment and not for depletion so they were used without this constraint).
fgwas
We used full GWAS summary statistics (no LD pruning or tagging) against each annotation at a time as recommended by the fgwas user manual. Enrichment was defined by p-value<0.05 for the false positive rate estimation and p-value<2.6x10-4 for the real data analysis to correct for multiple testing.
LD-score regression (LDSC)
For each annotation we prepared .ldscore files. Then for each annotation/trait pair we run LD-score regression accounting for the baseline model. We obtained enrichment p-values based on the resulting regression coefficients as per the software documentation. Analysis was restricted to hapmap3 SNPs again as per the user manual recommendation. Enrichment was defined by p-value<0.05 for the false positive rate estimation and p-value<2.6x10-4 for the real data analysis to correct for multiple testing.
GoShifter
We restricted the variants to those from the 1000 genomes project due to LD tagging in GoShifter using the same panel. For each study, we selected variants with GWAS P-value less than 10-8 and pruned them similarly to GARFIELD according to LD r2≥0.01. Testing was done using r2≥0.8 for LD tagging and 10,000 permutations. Enrichment was defined by p-value<0.05 for the false positive rate estimation and p-value < 2.6 × 10-4 for the real data analysis to correct for multiple testing. The p-value of enrichment was calculated as the proportion of permutations producing at least as extreme overlap as the observed SNP-annotation overlap.
GREGOR
We restricted the variants to those from the 1000 genomes project due to LD tagging in GREGOR using the same panel. For each study, we selected variants with GWAS P-value less than 10-8 and pruned them similarly to GARFIELD according to LD r2≥0.01. Testing was done using r2≥0.8 and 500 minimum neighbouring SNPs for each tested variant. Enrichment was defined by p-value<0.05 for the false positive rate estimation and p-value<2.6x10-4 for the real data analysis to correct for multiple testing.
GPA
We used full GWAS summary statistics, with no LD pruning or tagging and used a maximum of 10,000 EM iterations. Enrichment was defined by p-value<0.05 and q1>q0 for the false positive rate estimation.
CPU time estimates
We compared total CPU usage times between different methods for the analyses of 21 traits and 424 annotations and the respective average CPU times for a single trait/annotation pair. Analyses for each trait/annotation were run separately (whenever possible) on a compute cluster containing machines with the following architecture: Linux (x86-64) and 2x2.1 Ghz 16 core AMD 6378. Then cumulative run time over all traits/annotations and average run time for a single trait/annotation pair was reported (Supplementary Table 6).
Segmentation OR distribution and between trait sharing
From all significantly enriched cell types per trait and segmentation state, we calculated the median OR and then plotted its distribution (on a log scale) across traits in order to estimate the per-state OR. Additionally, we took all significantly enriched cell types for pairs of annotations in order to remove the effect of power for annotation density and looked at the distribution of ORs for enhancer and promoter states versus those of transcription states. Finally, we counted the number of cell types per feature that were found to be significantly enriched (or depleted) in a single trait or shared between multiple traits.
Software
GARFIELD is available as a standalone tool and an R-package (see URLs). The tool consists of two main parts: (i) pruning and annotation of the GWA study of interest and (ii) calculating odds ratios and significance of the observed enrichment. Additionally, we provide scripts for further prioritization of annotations by iteratively adding annotations in a joint model if they improve the model fit (Chi-squared test).
Supplementary Material
Editorial summary.
GARFIELD is a new approach that classifies genomic features related to phenotypes based on integrating GWAS signals with functional annotations. GARFIELD is used to characterize enrichment patterns for 29 traits integrated with ENCODE and Roadmap Epigenomics annotations.
Acknowledgements and grant support
This study makes use of data generated by the UK10K Consortium. A full list of the investigators who contributed to the generation of the data is available from www.UK10K.org. Funding for UK10K was provided by the Wellcome Trust under award WT091310. Nicole Soranzo's research is supported by the Wellcome Trust (Grant Codes WT098051 and WT091310). NJT is a Wellcome Trust Investigator (202802/Z/16/Z), is the PI of the Avon Longitudinal Study of Parents and Children (MRC & WT 102215/2/13/2), is supported by the University of Bristol NIHR Biomedical Research Centre (BRC-1215-20011), the MRC Integrative Epidemiology Unit (MC_UU_12013/3) and works within the CRUK Integrative Cancer Epidemiology Programme (C18281/A19169). EMBL-WTSI postdoctoral fellowships (G.R.S. Ritchie, V. Iotchkova).
Footnotes
Reporting Summary
Further information on research design in available in the Life Sciences Reporting Summary linked to this article.
Data availability
Web links for publicly available GWAS datasets and regulatory information databases are included in URLs section. Restriction of availability apply to blood cell indices GWASs from van der Harst et al. 33 and Gieger et al. 34, which have been obtained through the manuscripts authors. Any other data that supports the findings of this study is available from the corresponding author upon request.
Code availability
Custom code can be found at http://www.ebi.ac.uk/birney-srv/GARFIELD/
Author Contributions
Contributed data or materials: G.R.S.R., J.L.M., K.W., N.J.T., I.D., N.S.; Developed the method: E.B., G.R.S.R., I.D., J.L.M., N.S., V.I.; Analysed the data: V.I.; Provided critical interpretation of results: E.B., I.D., N.J.T., N.S., V.I.; Designed tools: M.G., S.M.; Wrote the manuscript: E.B., N.S., V.I.; Evaluated the manuscript: E.B., G.R.S.R., I.D., J.L.M., K.W., M.G., N.J.T., N.S., S.M., V.I.; Designed and managed the project: E.B., N.S.
Competing interests
The authors declare no competing interests.
References
- 1.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein BE, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 7.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen H, et al. Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians. PLoS ONE. 2013;8:e59494. doi: 10.1371/journal.pone.0059494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. doi: 10.1371/journal.pgen.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schork AJ, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trynka G, et al. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am J Hum Genet. 2015;97:139–152. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt EM, et al. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galwey NW. A new measure of the effective number of tests, a practical tool for comparing families of non-independent significance tests. Genet Epidemiol. 2009;33:559–568. doi: 10.1002/gepi.20408. [DOI] [PubMed] [Google Scholar]
- 18.Dunham I, Kulesha E, Iotchkova V, Morganella S, Birney E. FORGE: A tool to discover cell specific enrichments of GWAS associated SNPs in regulatory regions [version 1; referees: 2 approved with reservations] F1000Res. 2015;4 [Google Scholar]
- 19.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strawbridge RJ, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soranzo N, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Consortium for Blood Pressure Genome-Wide Association Studies et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gieger C, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.UK10K Consortium et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.