Abstract
In this study, we develop a reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for rapid and easy detection of goose astroviruses (GAstVs) in clinical samples. The specific LAMP primer sets were designed targeting the ORF2 gene of GAstV. The conditions of LAMP amplification were optimized in terms of reaction time and temperature. The optimal conditions are 60 min in a 60 °C water bath. No cross-reactivity was noted with fowl adenovirus serotype 4 (FAdV-4), duck tembusu virus (DTMUV), goose parvovirus (GPV), avian infectious bronchitis virus (IBV), or chicken anemia virus (CAV). The proposed RT-LAMP method was compared with conventional RT polymerase chain reaction (RT-PCR) and with nested RT-PCR. The results showed that the sensitivity of the proposed method was comparable to that of nested RT-PCR and tenfold higher than that of the conventional RT-PCR. Clinical samples (N = 129) of the liver and kidney from sick geese collected from six commercial goose farms were tested. The positive rate was 39.5% (51/129), 38.8% (50/129), and 34.9% (45/129) using RT-LAMP, nested RT-PCR and conventional RT-PCR, respectively. The developed RT-LAMP diagnostic method is not only simple, rapid, and highly specific, but also portable for use on the field. It may be used in epidemiological investigation to detect GAstVs.
Keywords: GAstVs, Reverse-transcription loop-mediated isothermal amplification (RT-LAMP), Rapid diagnosis
Introduction
Astroviruses (AstVs) are non-enveloped, single-strand, positive-sense RNA viruses that belong to the Astrovirus family and have a genome length of approximately 6.2–7.7 Kb (Cortez et al. 2017; De Benedictis et al. 2011). According to the hosts of infection, the Astrovirus family is classified into two genera—Mamastrovirus, which includes human, pig, cattle, cat, dog, rodent, rabbit, mouse, and bat AstVs, and Avastrovirus, which includes guineafowl, chicken, duck, pigeon, and turkey AstVs (Donato and Vijaykrishna 2017; Krishnan 2014). Their viral genomes consist of a short 5′-untranslated region (UTR), three open reading frames (ORFs 1a, 1b, and 2), and a 3′-UTR (Zhang et al. 2017b). It is noteworthy that ORF2 encodes capsid proteins, which are required for virion formation and contain a highly conserved N-terminus and highly mutated C-terminus (Zhang et al. 2017b).
In 2015, an outbreak of gout was reported in 1-week-old goslings on a goose farm in Anhui Province, China. By 2017, the disease spread to Jiangsu, Shandong, Guangdong, and Fujian Provinces (Zhang et al. 2018b), causing significant economic loss within the goose industry. The main clinical symptoms of the disease are visceral urate deposition and swelling on the surfaces of the heart, liver, and kidneys (Zhang et al. 2018a). Several researchers have used high-throughput sequencing technology and the rapid amplification of cDNA ends to prove that the disease is caused by a new virus called the “goose AstV” (GAstV) (Niu et al. 2018; Yang et al. 2018; Zhang et al. 2018b).
The ability to quickly and accurately detect GAstV is essential for controlling the disease. Isolation of the virus from cell cultures (LMH or GEK cell), reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and next-generation sequencing have been used to identify GAstVs (Niu et al. 2018; Yuan et al. 2018; Zhang et al. 2018b); however, these experimental methods are time-consuming and require high-quality, expensive equipment. Moreover, they are not suitable for field situations or primitive clinical laboratories.
Loop-mediated isothermal amplification (LAMP) is a diagnostic method that depends on Bst DNA polymerase and requires four to six primers targeting four to six specific sequences. LAMP has been used to detect and identify pathogens (Guo et al. 2018; Mansour et al. 2015) and does not need expensive equipment or lengthy experimental steps. A constant-temperature water bath or heating block for a duration of 60 min is the only equipment needed for the samples to be assayed using the LAMP method. In addition, the experimental results can be observed with the naked eye on adding fluorescent dyes. This method has good practicability, rapidity, and sensitivity (Venkatesanet al. 2012). In this study, a RT-LAMP method targeting the ORF2 of GAstV is successfully established, compared with conventional RT-PCR and nested RT-PCR, and its detection specificity is evaluated using clinical specimens.
Materials and methods
Virus strains and clinical samples
A new goose AstV strain, AH/2018 (accession number: MN099162), was isolated from a sick goose in Anhui Province, China. FAdV-4, DTMUV, GPV, IBV and CAV were stored at − 80 °C and preserved in our laboratory. One hundred twenty-nine tissue samples (livers and kidneys) were collected from sick geese from different locations in Anhui Province of China between 2018 and 2019.
Design of primers for RT-PCR, nested RT-PCR, and RT-LAMP
ORF2 genome sequences of several strains of GAstV were obtained from GenBank (accession numbers: MH052598.1; MH807626.1; MH410610.1; MF772821.1; MG934571.1). The sequences were analyzed using MegAlign (DNASTAR, Inc., Madison, WI, USA). Primers for RT-LAMP were designed with online software, Primer Explorer ver. 4 (https://primerexplorer.jp/e/). The primer targeted for the highly conserved ORF2 gene sequences of the goose Astrovirus strain (accession number: MH052598.1). The primer design of RT-PCR and nested RT-PCR using software Primer Premier version 5.0 (PREMIER Biosoft International, Palo Alto, California, USA) were custom synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) with polyacrylamide gel purification. Detailed information on the primers is provided in Table 1.
Table 1.
Primers and reaction conditions used in this study
| Test | Primer name | Type | Length (bp) | Sequence | Optimal reaction conditions |
|---|---|---|---|---|---|
| RT-LAMP | GAstV-F3 | Forward outer | 20 | GCTCTCACTCATGACCAGTG | 60 °C for 60 min, 80 °C for 10 min to terminate amplification |
| GAstV-B3 | Reverse outer | 20 | CCACTTCTGTGGAAACAGCC | ||
| GAstV-FIP | Forward inner | 42 | TCTGCAATGCGCCACTAACTGT-TTCAGCATCAGGGAAAACGG | ||
| GAstV-BIP | Reverse inner | 40 | GAGAAGGAGCAACACAGGCGC-AAAACACCTCACCGACACC | ||
| GAstV-LF | Forward loop | 23 | ATCCTGGGTATTTACAAGGGTTG | ||
| GAstV-LB | Reverse loop | 20 | GTCTCAAATGCGCAGACCTC | ||
| RT-PCR | GAstV-NF | Forward | 16 | AGGAAAATGGGCAAAC | 95 °C 5 min; 94 °C 30 s, 56 °C 30 s, 72 °C 35 s, 30 cycles; 72 °C 10 min |
| GAstV-NR | Reverse | 16 | AGGAAAATGGGCAAAC | ||
| Nested RT-PCR | GAstV-OF | Forward outer | 20 | ACAAAATGACTACCACGATA | 95 °C 5 min; 94 °C 30 s, 54 °C 30 s, 72 °C 30 s, 30 cycles; 72 °C 10 min |
| GAstV-OR | Reverse outer | 17 | AACACCTCACCGACACC | ||
| GAstV-IF | Forward inner | 16 | AGGAAAATGGGCAAAC | ||
| GAstV-IR | Reverse inner | 15 | AAGGGTTGCCGTTTT | ||
| Standard control | GAstV-ORF2-F | Forward | 18 | ATGGCAGACAGGGCGGTG | 95 °C 5 min; 94 °C 30 s, 56 °C 30 s, 72 °C 40 s, 30 cycles; 72 °C 10 min |
| GAstV-ORF2-R | Reverse | 19 | ATGGCAGACAGGGCGGTG |
Nucleic acid extraction
All tissue samples were homogenized in 1 mL of phosphate-buffered saline and centrifuged for 5 min at 12,000 rpm after being frozen and thawed three times. Viral DNA was extracted using the TIANamp Virus DNA/RNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Viral RNA was extracted from the supernatants using the TIANamp Virus DNA/RNA Kit (TIANGEN) according to the manufacturer’s instructions. Reverse transcription of 100 ng viral RNA sample was conducted using the FastQuant cDNA Kit (TIANGEN) according to the manufacturer’s instructions. The extracted cDNA was stored in − 20 °C.
Construction of a standard plasmid containing ORF2
The RNA genome of GAstV was reverse transcribed into cDNA, and ORF2 was amplified using PCR with GAstV-ORF2-F and GAstV-ORF2-R (Table 1). The PCR products were purified using a DNA purification kit (TIANGEN), cloned into the pMD19-T vector (TaKaRa, Dalian, China), and transfected into Escherichia coli DH5α cells for amplification. The recombinant plasmid was extracted using the TIANprep Mini Plasmid Kit (TIANGEN) and sent to Sangon Biotech Shanghai Co, Ltd. for sequencing. The concentration of plasmids was quantified using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). The copy number of the recombinant plasmid was calculated using the following formula: amount (copies/μL) = (6.02 × 1023) × (ng/μL × 10–9)/(DNA length × 660).
Reaction and optimizations of RT-LAMP
Referring to the basic reaction system of LAMP, the concentrations of following ingredients were optimized in a total reaction volume of 25 μL: primers, deoxynucleoside triphosphate (dNTP), and MgSO4. After determining the optimal reaction system, the temperature and time were optimized in the experiment. First, the RT-LAMP reactions were observed for 60 min using optimal reaction system at 58, 60, 62, 64, and 66 °C, each. After selecting the optimum temperature, the reaction time was optimized. At the optimum temperature, the reaction times were 50, 55, 60, 65, and 70 min. Finally, the reaction was terminated in a water bath at 80 °C for 10 min. The reaction products were analyzed using 1.5% agarose gel electrophoresis. Finally, 1 μL 10,000 × SYBR Green I dye (Solarbio, Beijing, China) was added to the reaction product, and the result was observed with the naked eye. The optimal reaction conditions and times for the optimization of RT-PCR and nested RT-PCR are provided in Table 1.
Conventional RT-PCR and nested RT-PCR
The total volume of the conventional RT-PCR is 25 μL, and its composition was as follows: 1 × Premix Taq® Version 2.0 (TaKaRa), 1 μL of template, 0.4 μM GAstV-NF and GAstV-NR primers. The composition of the nested RT-PCR system in a total of 25 μL was as follows: 1 × Premix Taq® Version 2.0 (TaKaRa), 1 μL of template, 0.4 μM GAstV-OF and GAstV-OR primers. One microliter of initial PCR product was used as a template in PCR reaction with GAstV-IF and GAstV-IR primers. The primers and reaction conditions are shown in Table 1.
Sensitivity and specificity test of the RT-LAMP assay
The recombinant plasmid was serially diluted ten times (1 × 109 to 1 × 101 copies) according to the known concentration and used as a template for RT-LAMP, nested RT-PCR, and conventional RT-PCR. Before the specificity evaluation, the concentration of each virus DNA or cDNA was adjusted and unified at 100 ng/μL. Genomic RNAs/DNAs of avian pathogenic viruses FAdV-4, DTMUV, GPV, IBV, and CAV were used as templates to verify the specificity of the RT-LAMP assay. GAstV was used as a positive control and the blank template was used as a negative control. All reactions were conducted in triplicate. The amplified products were electrophoresed on 1.5% agarose gel with GoldView II (Solarbio) and visually inspected using SYBR Green I dye under daylight conditions (Fig. 1).
Fig. 1.
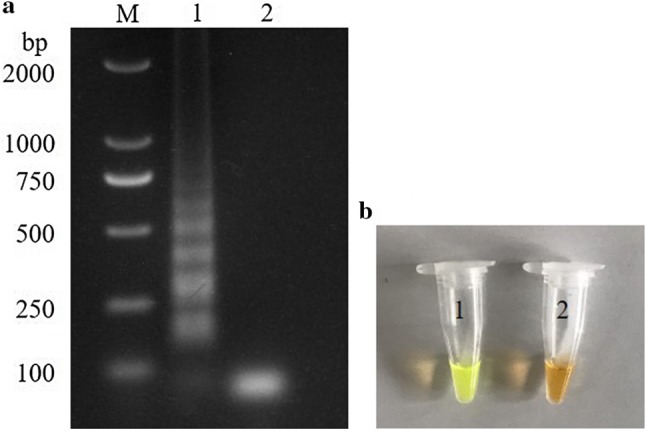
Analysis of LAMP products. LAMP amplification products were analyzed using agarose gel electrophoresis (a) M, DL2000 DNA marker; lane 1, positive control; lane 2, negative control; and visually inspected with SYBR Green I dye under daylight conditions (b) tube 1, positive control and tube 2, negative control
Evaluation of the RT-LAMP assay in clinical samples
The 129 tissue samples (liver and kidney) were tested using RT-LAMP, nested RT-PCR, and conventional RT-PCR assays, and the positive detection rates were simultaneously recorded.
Results
Reaction mixtures and optimal conditions
After optimization of various parameters, the final optimized in a total reaction volume of 25 μL contained 1.6 μM (each) of forward and backward inner primers, 0.2 μM (each) of outer primers F3 and B3, 6 mM MgSO4, 1.4 mM dNTP mix (TaKaRa), 1 × Thermo Pol buffer (20 mM Tris–HCl, 50 mM KCl, 10 mM (NH4)2 SO4, 2 mM MgSO4, 0.1% Tween-20), 8 U Bst DNA polymerase (New England Biolabs, Ipswich, MA, USA), and 1 μL template. The products of RT-LAMP showed multiple characteristic ladder bands at different temperatures. At 60 °C, the bands of light were strongest. The optimal reaction time was 60 min.
Sensitivity and specificity
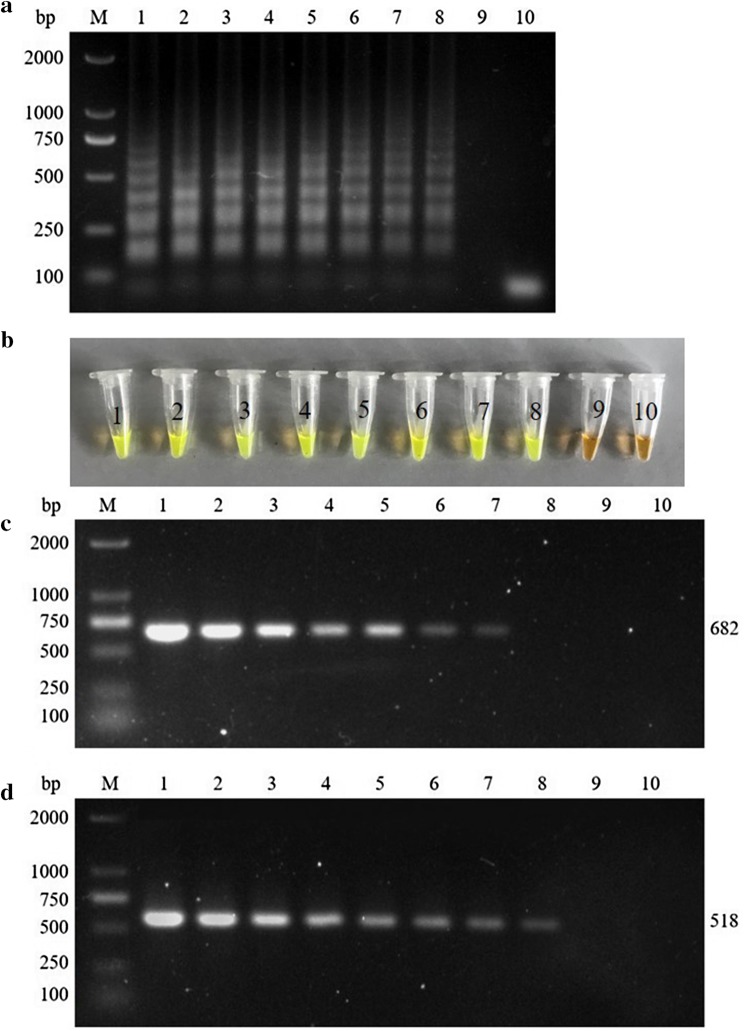
Using 1 × 109 to 1 × 101 copies of the GAstV-ORF2 recombinant plasmid as templates, RT-LAMP, nested RT-PCR and conventional RT-PCR assays were conducted to determine their detections limits. The results showed that RT-LAMP can detect a minimum of 1 × 102 plasmid copies. Moreover, the nested RT-PCR and conventional RT-PCR have detection limits of 1 × 102 and 1 × 103 copies, respectively (Fig. 2). These results suggested that RT-LAMP was ten times more sensitive than conventional RT-PCR and that its sensitivity was similar to that of nested RT-PCR.
Fig. 2.
Comparative sensitivity of RT-LAMP, nested RT-PCR and conventional RT-PCR for GAstVs. a Electrophoretic analysis of LAMP amplified products. b Visually inspected with SYBR Green I dye under daylight conditions visual observation of LAMP amplified products. c Electrophoretic analysis of RT-PCR amplified products. d Electrophoretic analysis of nested RT-PCR amplified products. M, DL2000 DNA marker; lane 1, 1 × 109 copies/tube; lane 2, 1 × 108 copies/tube; lane 3, 1 × 107 copies/tube; lane 4, 1 × 106 copies/tube; lane 5, 1 × 105 copies/tube; lane 6, 1 × 104 copies/tube; lane 7, 1 × 103 copies/tube; lane 8, 1 × 102 copies/tube; lane 9, 1 × 101 copies/tube; lane 10, negative control
The results of specificity determination showed that only GAstV and positive recombinant plasmid templates exhibited a color reaction. Not surprisingly, the other viruses used in the experiment showed no color reaction, which was consistent with the negative control (Fig. 3). This demonstrated the high specificity of RT-LAMP used in our study.
Fig. 3.

Analysis of RT-LAMP specificity for GAstV. M, DL2000 DNA marker; specificity of the RT-LAMP assay determined by electrophoresis analysis (a) and SYBR Green I staining (b); lane 1, GAstV; lane 2, FAdV-4; lane 3, DTMUV; lane 4, GPV; lane 5, IBV; lane 6, CAV; lane 7, negative control
Evaluation of the RT-LAMP assay
The practicability of the established RT-LAMP method was evaluated using the 129 collected samples. Nested RT-PCR and conventional RT-PCR were also used in the detection processes. As shown in Table 2, the positive detection rates of RT-LAMP, nested RT-PCR and conventional RT-PCR were 39.5% (51/129), 38.8% (50/129), and 34.9% (45/129), respectively. The comparison of the positive detection rates showed that the RT-LAMP method had a slight advantage over the other two methods and was portable enough to be useful for disease detection in the field.
Table 2.
Performance of conventional RT-PCR, nested RT-PCR and RT-LAMP assays for detection of GAstVs in clinical samples
| Samples | Total | Conventional RT-PCR | Nested RT-PCR | RT-LAMP | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| Liver | 72 | 26 | 46 | 29 | 43 | 29 | 43 |
| Kidney | 57 | 19 | 38 | 21 | 36 | 22 | 35 |
| Total | 129 | 45 | 84 | 50 | 79 | 51 | 78 |
Discussion
Since it was detected in China in 2017, the disease associated with goose gout has increased gosling mortality and caused heavy losses in the poultry industry (Zhang et al. 2018b). This disease was later confirmed to be caused by GAstV. To effectively monitor and prevent it, a portable detection method was vital. A variety of methods are used to detect viral infections, such as conventional RT-PCR and nested RT-PCR (Yuan et al. 2018); however, these methods require highly accurate and expensive equipment, which make them much less suitable for clinical surveillance of animal epidemics in developing countries, such as China. RT-LAMP was recently developed to detect viruses and has proved to be a simple and reliable method. It has the advantages such as being faster and simpler when compared with conventional RT-PCR and nested RT-PCR (Zhao et al. 2014; Zhang et al. 2017a). Conducting an assay under this method requires only a water bath under isothermal conditions for the samples. In our study, three pairs of primers were designed for the ORF2 of GAstV to establish a simple, rapid, and highly specific RT-LAMP assay. We also concurrently evaluated RT-LAMP, nested RT-PCR, and conventional RT-PCR assay. The results of sensitivity showed that the detection limit of RT-LAMP at 1 × 102 copies was better than that of conventional RT-PCR and comparable to that of nested RT-PCR. According to previous studies, a real-time fluorescence qPCR method based on TaqMan can detect a minimum of 52.5 copies (Yuan et al. 2018). Similar results were observed with RT-LAMP, which is a simpler, faster (i.e., only 60 min), and less expensive method. The test environment was strictly partitioned to avoid false-positive contamination. To evaluate the feasibility of RT-LAMP in clinical applications, 129 clinical samples were tested. The results showed that GAstV was widely distributed in the Anhui Province of China, consistent with the results of previous studies (Zhang et al. 2018b).
In summary, the established RT-LAMP could detect GAstV, and it exhibits simplicity, rapidity, and high specificity. This assay could be used in epidemiological investigation to detect GAstV.
Acknowledgements
The authors express gratitude to editage for the help with revising this manuscript.
Author contributions
ZY and DZ conceived of the study, carried out the experiment and drafted the manuscript, contributed equally to this work. KY and CB participated in the data collection and analysis. YL involved in drafting of the manuscript. JL and SJ participated in statistical analysis. YW conceived of the study, revising the manuscript critically. All authors have read and approved the final manuscript.
Funding
This work was supported financially by the National Natural Science Foundation of China (no. 31602063), National key research and development program (no. 2018YFD0502006), Anhui Key Research and Development Program (no. 201904f06020030). Accession number: A new GAstV strain (no. MN099162), FAdV-4 strain (no. KY379035), DTMUV strain (no. KY623435), GPV strain (no. MK333463), IBV strain (no. MH020185) and CAV strain (no. KU645509), was uploaded to GenBanK database.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Zhaorong Yu and Da Zhang contributed equally to this work.
References
- Cortez V, Meliopoulos VA, Karlsson EA, Hargest V, Johnson C, Schultz-Cherry S. Astrovirus biology and pathogenesis. Annu Rev Virol. 2017;4(1):327–348. doi: 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol. 2011;11(7):1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato C, Vijaykrishna D. The broad host range and genetic diversity of mammalian and avian astroviruses. Viruses. 2017;9(5):102–119. doi: 10.3390/v9050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XG, Zhou YZ, Li Q, Wang W, Wen JZ, Zheng L, Wang Q. Rapid and reliable diagnostic method to detect Zika virus by real-time fluorescence reverse transcription loop-mediated isothermal amplification. AMB Express. 2018;8(1):60–67. doi: 10.1186/s13568-018-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan T. Novel human astroviruses: challenges for developing countries. Virus Dis. 2014;25(2):208–214. doi: 10.1007/s13337-014-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SM, Ali H, Chase CC, Cepica A. Loop-mediated isothermal amplification for diagnosis of 18 World Organization for Animal Health (OIE) notifiable viral diseases of ruminants, swine and poultry. Anim Health Res Rev. 2015;16(2):89–106. doi: 10.1017/S1466252315000018. [DOI] [PubMed] [Google Scholar]
- Niu X, Tian J, Yang J, Jiang X, Wang H, Chen H, Yi T, Diao Y. Novel goose astrovirus associated gout in Gosling, China. Vet Microbiol. 2018;220:53–56. doi: 10.1016/j.vetmic.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Venkatesan G, Bhanuprakash V, Balamurugan V, Singh RK, Pandey AB. Development of loop-mediated isothermal amplification assay for specific and rapid detection of camelpox virus in clinical samples. J Virol Methods. 2012;183(1):34–39. doi: 10.1016/j.jviromet.2012.03.019. [DOI] [PubMed] [Google Scholar]
- Yang J, Tian J, Tang Y, Diao Y. Isolation and genomic characterization of gosling gout caused by a novel goose astrovirus. Transbound Emerg Dis. 2018;65(6):1689–1696. doi: 10.1111/tbed.12928. [DOI] [PubMed] [Google Scholar]
- Yuan X, Meng K, Zhang Y, Qi L, Ai W, Wang Y. Establishment and application of rapid diagnosis for reverse transcription-quantitative PCR of newly emerging goose-origin nephrotic astrovirus in China. mSphere. 2018;3(6):e00380-18. doi: 10.1128/mSphere.00380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ye Y, Song D, Guo N, Peng Q, Li A, Zhou X, Chen Y, Zhang M, Huang D, Tang Y. A simple and rapid identification method for newly emerged porcine deltacoronavirus with loop-mediated isothermal amplification. Biol Res. 2017;50(1):30–36. doi: 10.1186/s40659-017-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang F, Liu N, Yang L, Zhang D. Complete genome sequence of a novel avastrovirus in goose. Arch Virol. 2017;162(7):2135–2139. doi: 10.1007/s00705-017-3297-1. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Cao Y, Wang J, Fu G, Sun M, Zhang L, Meng L, Cui G, Huang Y, Hu X, Su J. Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg Microbes Infect. 2018;7(1):71–81. doi: 10.1038/s41426-018-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren D, Li T, Zhou H, Liu X, Wang X, Lu H, Gao W, Wang Y, Zou X, Sun H, Ye J. An emerging novel goose astrovirus associated with gosling gout disease, China. Emerg Microbes Infect. 2018;7(1):152–159. doi: 10.1038/s41426-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fan B, Wu G, Yan X, Li Y, Zhou X, Yue H, Dai X, Zhu H, Tian B, Li J, Zhang Q. Development of loop-mediated isothermal amplification assay for specific and rapid detection of differential goat pox virus and sheep pox virus. BMC Microbiol. 2014;14(1):10–19. doi: 10.1186/1471-2180-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]