Abstract
The taxonomic composition of microbial communities in animals varies among animal species, but the contribution of interspecific differences in filtering of the microbial pool by the animal host to this variation is uncertain. Here, we demonstrate significant interspecific variation in microbial community composition among laboratory-reared Drosophila species that was not related to host phylogeny. Complementary reciprocal transfer experiments yielded different microbial communities for a single microbiota administered to homologous and heterologous hosts (i.e., the same and different Drosophila species from which the microbiota was derived), indicative of among-host species differences in traits that shape microbiota composition. The difference in microbiota composition between homologous and heterologous hosts was not greater for distantly related than for closely related host species pairs. Furthermore, Drosophila survival to adulthood was significantly reduced in heterologous associations relative to homologous associations and microbiologically sterile flies, suggesting that microbial taxa that are advantageous for their homologous host species can be deleterious for other host species. We conclude that drosophilid flies display robust among-host species variation in host controls over microbiota composition that has diversified in response to selection pressures which are not tracked by host phylogeny.
Subject terms: Microbiome, Molecular ecology
Introduction
Animals host diverse microbial communities, and there is increasing evidence that variation in the composition of these communities can influence the health and fitness of the host [1, 2]. As our knowledge of microbiota effects on host phenotype increases, so does interest in applying this knowledge to achieve specific outcomes, for example to optimize microbial therapies for human health [3, 4] and to design microbial strategies for improved control of insect pests [5–7]. The success of these applications will rely on a fundamental understanding of the processes that shape the composition of these microbial communities.
The conditions and resources in the host habitat, e.g., pH or redox conditions of the gut, can influence microbiota assembly by acting as ecological filters determining which of the microbes the host encounters can colonize the host environment. However, quantifying the contribution of filtering by the host environment is a challenge as many other processes also contribute to the assembly of the host microbiota, including transmission of microbes among hosts and from the external environment to hosts, among-microbe interactions, and ecological drift [8, 9]. Variation in the host environment is often cited as an explanation for observed differences in microbiota composition among host species. However, interspecific variation in the niche, social behavior, and geographic range of the host can generate differences in gut microbiota composition among species by altering the pool of microbial species that the host encounters and patterns of microbial transmission [10–13]. Furthermore, differences in host diet can generate variation in composition of the gut microbiota both by altering the resources in the gut and by acting as a source of microbes [14–18].
The drosophilid flies offer an excellent system to investigate the role of the host environment in generating among-species variation in gut microbiota composition because multiple species can be maintained under uniform conditions in the laboratory. Previous studies have indicated that, while some microbial taxa are common in the gut microbiome of multiple Drosophila species (e.g., Acetobacteraceae, Lactobacillaceae and Enterobacteriaceae), gut microbiota composition varies among host species [19–21]. This interspecific variation in microbiota composition may be most evident under uniform conditions in the laboratory, e.g., refs. [19, 20] and can be masked in field populations by other processes influencing microbiota assembly. In particular, neutral processes (i.e., ecological drift and passive dispersal) contribute to the assembly of the drosophilid gut microbiota [22, 23], and the large-scale variation in microbiota composition with time and site of collection [21, 22, 24, 25], suggests that the pool of microbes in the environment may also play an important role in the shaping the composition of the gut microbiota. However, interspecific variation in microbiota composition has only been quantified under controlled laboratory conditions for a small number of Drosophila species, e.g., refs. [19, 20], and the ecological processes that underlie this variation have not yet been addressed.
In this study, we first investigated gut microbiota composition in Drosophila species that were maintained under uniform conditions, using two independent sets of fly species that were cultured and processed for analysis by different methods. Then, we tested the role of host filtering experimentally by making reciprocal transfers of microbiota between two pairs of Drosophila species, one closely related and one distantly related, such that each host species was colonized with microbiota from the same species (homologous association) and a different species (heterologous association). We also scored survival to adulthood, as an index of the fitness of homologous and heterologous associations, following evidence that heterologous associations can reduce host performance in both mice of the genus Peromyscus and Nasonia jewel wasps [19, 26].
Materials and methods
The flies
The experimental material comprises two different sets of Drosophila species (Table 1). Set-1 consisted of eight species maintained at 25 °C, 36% relative humidity, and 12:12 h (L:D) at Cornell University, reared on a sterile diet of 10% inactive Brewer’s yeast (MP Biomedicals, Santa Ana, CA, USA), 10% glucose (Sigma-Aldrich, St. Louis, MO, USA), 1.2% agar (Apex Bio, Houston, TX), and preservatives (0.04% phosphoric acid and 0.42% propionic acid; Sigma-Aldrich). Set-2 consisted of 14 species maintained at 21.5 °C, 60% relative humidity, and 13:11 h (L:D) at the University of Western Ontario, reared on a non-sterile diet of 13.75% organic bananas, 4.75% corn syrup, 3% liquid malt, 2.75% active yeast (Fleischmann’s active dry yeast, ADY, Canada), 0.725% agar (Apex Bioresearch Products; Genesee Drosophila agar type II), and the preservatives 0.2% methylparaben (Sigma-Aldrich) and 0.3% propionic acid (Sigma-Aldrich) [27]. The diet for two species (D. mojavensis, D. persimilis) was supplemented with 0.213% Opuntia ficus-indicta powder (OroVerde Export, Morelos, Mexico). Set-2 Drosophila had been treated with 0.03% dietary tetracycline hydrochloride (Sigma-Aldrich) for two consecutive generations to eliminate Wolbachia endosymbionts at least five generations prior to collection for this study.
Table 1.
The Drosophila species source information and diet type
| Species | Subgenus | Species group | Sourcea | Strain/Stock no. | Collection location |
|---|---|---|---|---|---|
| Set-1: reared at Cornell University on yeast/glucose diet | |||||
| D. ananassae | Sophophora | Melanogaster | DSSC | 14024-0371.13 | Hawaii |
| D. melanogaster | Sophophora | Melanogaster | DSSC | Canton-S | Not known |
| D. pseudoobscura | Sophophora | Obscura | DSSC | SD02 | Not known |
| D. santomea | Sophophora | Melanogaster | DSSC | 1402-0271.01 | San Tome and Principe Island |
| D. sechellia | Sophophora | Melanogaster | DSSC | 14021-0248.03 | Cousin Island, Seychelles |
| D. simulans | Sophophora | Melanogaster | DSSC | 14021-0251.001 | Georgetown, Guyana |
| D. willistoni | Sophophora | Willistoni | DSSC | 14030-0811.24 | Guadaloupe Island, Caribbean |
| D. yakuba | Sophophora | Melanogaster | DSSC | 14021-0261.01 | Liberia |
| Set-2: reared at Western University on banana diet | |||||
| D. ananassae | Sophophora | Melanogaster | EU | E-11002 | Kagoshima, Japan |
| D. bipectinata | Sophophora | Melanogaster | DSSC | 14024-0381.20 | Kep Province, Cambodia |
| D. borealis | Drosophila | Virilis | DSSC | 15010-0961.05 | Lytton, Quebec, Canada |
| D. hydei | Drosophila | Repleta | Field | NA | London, Ontario, Canada |
| D. immigrans | Drosophila | Immigrans | DSSC | 15111-1731.03 | Patan, Nepal |
| D. melanogaster | Sophophora | Melanogaster | Field | NA | London, Ontario, Canada |
| D. mercatorum | Drosophila | Repleta | DSSC | 15082-1521.25 | Argoin, Bahia, Brazil |
| D. mojavensis | Drosophila | Repleta | DSSC | 15081-1352.22 | Catalina Island, California, USA |
| D. persimilis | Sophophora | Obscura | DSSC | 14011-0111.49 | Mt. St. Helena, California, USA |
| D. simulans | Sophophora | Melanogaster | Field | NA | Florida City, USA |
| D. suzukii | Sophophora | Melanogaster | Field | NA | South-Ontario, Canada |
| D. tropicalis | Sophophora | Willistoni | DSSC | 14030-0801.00 | San Salvador, El Salvador |
| D. virilis | Drosophila | Virilis | EU | E-15601 | Hokkaido, Japan |
| D. willistoni | Sophophora | Willistoni | DSSC | 14030-0811.24 | Guadaloupe Island, Caribbean |
aDSSC = National Drosophila Species Stock Center (University of California, San Diego); EU = Drosophila Stocks of Ehime University (Japan); Field = field collection
DNA extraction and amplicon sequencing
For the samples in Set-1, flies were individually surface-washed in sterile water, and then dissected with fine pins to isolate the gut (comprising the crop, midgut, and hindgut, but excluding the Malpighian tubules). For each species and sex, two samples of 30 dissected guts were collected. DNA was extracted in phenol:chloroform:isoamyl alcohol, following the protocol in ref. [24], designed specifically to isolate DNA from both Gram-positive and Gram-negative bacteria. 16S ribosomal RNA (rRNA) genes were amplified with the primers 319F and 806R that target the V3-V4 variable region, using the dual-indexing strategy of [28]. Briefly, each 25 μl reaction comprises 0.25 μl Phusion polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 5 μl HF buffer, 0.5 μl 10 mM dNTPs, 0.75 μl DMSO (dimethyl sulfoxide), 8.5 μl water, 2.5 μl each 1 μM primer, and 5 μl template DNA. The cycling conditions were: 98 °C for 60 s, followed by 35 cycles of 98 °C for 15 s, 58 °C for 15 s, and 72 °C for 15 s, and a final elongation of 72 °C for 1 min. Amplicons were cleaned and normalized using the SequalPrep 96-well plate kit (Life Technologies, Carlsbad, CA, USA), pooled and concentrated with the AMPure kit (Beckman Coulter Inc., Brea, CA, USA), according to manufacturer’s instructions. Multiplexed, paired-end sequencing was performed on the Illumina MiSeq platform (2 × 300 bp reads) at the University of Rochester Genomics Research Center.
For Set 2, three replicate samples of whole flies were generated for both sexes of each Drosophila species by combining flies from multiple rearing vials. Each replicate consisted of either 15–20 male or 10 female adult flies at 5 days post-eclosion. The pooled flies were washed three times in sterile phosphate-buffered saline (PBS) to remove food and fecal debris. Washed flies were stored at −20 °C prior to DNA extraction. Flies were homogenized in 180 μl enzymatic lysis buffer (20 mM Tris·Cl, pH 8.0; 2 mM sodium EDTA; 1.2% Triton® X-100; 20 mg/ml lysozyme) with a sterile pestle, then DNA was isolated using the DNeasy Blood & Tissue kit with pre-treatment for Gram-positive bacteria (Qiagen, Germantown, MD, USA). The V3-V4 region of bacterial 16S rRNA genes were amplified with the primers 341f (5′-CCTACGGGAGGCAGCAG-3′) and 806r (5′-GGACTACHVGGGTWTCTAAT-3′) utilizing a dual-indexing strategy modified from [29]. PCR reactions contained 1X KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA), 0.3 μM each primer, and 20 ng template DNA. Amplicons were generated with the cycling protocol: 3 min at 95 °C, 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final extension of 5 min at 72 °C. Following purification with Agencourt Ampure XP beads (Beckman Coulter, Indianapolis, Indiana, USA), PCR products were quantified with a Qubit 2.0 fluorimeter (Invitrogen), and pooled in equimolar concentrations. The pooled amplicons were quality checked with a Fragment Analyzer (Advanced Analytical Technologies, Ames, IA, USA), and sequenced on a MiSeq instrument (2 × 250 bp; Illumina, San Diego, CA, USA) at the Cornell Biotechnology Resource Center. Both sequencing runs included template-free controls, and the ASV read counts for these negative controls are provided in Dataset S1a and S1c.
Microbiota transfer experiments
Microbiota slurries were obtained from adult flies of D. melanogaster, D. simulans, and D. willistoni strains from Set-1 (Table 1) that had been reared in the laboratory at Cornell University on sterile diet of 10% inactive Brewer’s yeast, 10% glucose, 1.2% agar, and preservatives (0.04% phosphoric acid and 0.42% propionic acid; Sigma-Aldrich). For each slurry, 60 adult flies (30 male and 30 female, 5–10 days post-eclosion, and collected from multiple vials) were pooled, washed 3x in sterile PBS to remove external debris, and crushed with a sterile pestle. The homogenate was resuspended in 3 ml sterile PBS and centrifuged at 2880 × g for 1 min. Glycerol was added to each supernatant to a final concentration of 20% and the microbiota-glycerol stocks were stored at −80 °C until required.
Axenic (i.e., microbiologically sterile) D. melanogaster, D. simulans, and D. willistoni were generated by egg dechorionation [30]. Eggs ( < 18 h old) were collected from yeast/glucose/grape juice agar plates (D. melanogaster), yeast/glucose/grape juice agar plates covered with inactive Brewer’s yeast paste (D. willistoni), and plates of Formula 4–24® Instant Drosophila Medium (Carolina Biological Supply Company, Burlington, North Carolina, USA) rehydrated in grape juice (D. simulans). The eggs of D. melanogaster and D. willistoni were gently removed from agar plates with a paintbrush, and D. simulans eggs were floated from the medium in 20% sucrose solution. Eggs of all species were surface-sterilized by three washes in 0.6% hypochlorite, followed by three rinses in sterile water.
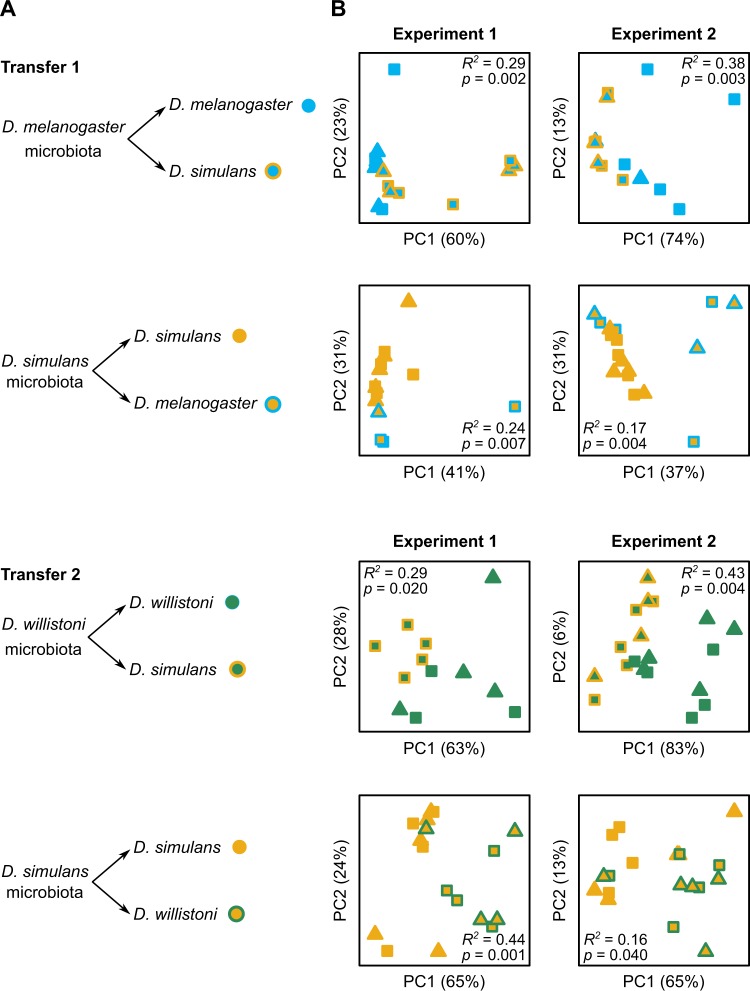
The reciprocal microbiota transfer experiments comprised two independent transfers, each conducted between two pairs of species: D. simulans and D. melanogaster (closely related), and D. simulans and D. willistoni (more distantly related) (Fig. 2a). Approximately 60 axenic eggs were transferred aseptically to sterile 50 ml Falcon centrifuge tubes containing 7.5 ml autoclaved yeast/glucose diet (described above), followed by an inoculum of 35 μl microbiota slurry that had been thawed on ice. Each reciprocal transfer comprised five replicate vials of four treatments, i.e., two heterologous associations (between insects and microbiota of different Drosophila species) and two homologous associations (between insects and microbiota of the same Drosophila species), together with five vials administered with 35 μl sterile PBS, as a check for the microbiological sterility of eggs. The insects were reared at 25 °C and 36% humidity in a 12:12 h (L:D) cycle. The number of pupae present was counted at 7 or 8 days after inoculation, and the number of surviving flies of each sex in each vial was determined at 5 days post-eclosion. Three flies of each sex in each vial were stored at −20 °C, and then processed using the DNA extraction method for Set-1 (above) and 16S rRNA gene amplicon sequencing by the procedure for Set-2. In parallel, two male and female flies per vial were individually checked for microbiological sterility by the method of [30]. Briefly, each fly was homogenized in 100 μl sterile PBS for 30 s using a FastPrep-24 instrument (MP Biomedicals). The homogenate was diluted to 1 ml in PBS, plated onto modified MRS agar medium (1.25% vegetable peptone (Becton Dickinson), 0.75% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% dipotassium hydrogen phosphate, 0.2% triammonium citrate, 0.02% magnesium sulfate heptahydrate, 0.005% manganese sulfate tetrahydrate, 1.2% agar) using the WASP-2 apparatus (Microbiology International) and incubated for 1–2 days at 30 °C. Colony-forming units (CFUs) were counted using a Protocol 3 colony counter (Microbiology International). Homogenates from most flies derived from the vials administered PBS contained no CFUs, with a maximum of four CFUs per fly, indicating that microbial contamination of the experiments was minimal. The abundance of 16S rRNA gene copies in the DNA extractions from homologous and heterologous associations was quantified with the quantitative PCR (qPCR) protocol described in ref. [31]. To test whether compounds from the flies themselves present in the homogenates could influence the survival of the recipient flies, we conducted transfers 1 and 2 (Fig. 2a) with homogenates from axenic flies generated as described above. The number of flies at 5 days post-eclosion was assessed.
Fig. 2.
Microbiota composition in Drosophila differs significantly between microbial slurries administered to homologous and heterologous host species. a Design of reciprocal transfer experiments between closely related species (Transfer-1) and distantly related species (Transfer-2). b Principal coordinates analysis plots for bacterial communities in two replicate experiments (Experiment-1 and Experiment-2) for each Transfer design. The percentage of variation explained by each axis is given, and the results of PERMANOVA tests for differences in community composition between host species are shown on each plot. Male flies are represented by triangles, and female flies by squares
Processing of 16S rRNA gene amplicon sequences
The QIIME2 v2018.4 [32] microbiome bioinformatics platform was used to resolve amplicon sequence variants (ASVs) from paired-end reads with the DADA2 pipeline [32, 33]. Taxonomic assignments were made with a naive Bayes classifier trained on the Silva 128 99% operational taxonomic units (OTUs) reference sequences trimmed with the 341f and 806r primer sequences [34, 35]. Amplicon sequence variants that were not assigned to the domain Bacteria were omitted from analysis. Read counts for the remaining sequence variants are provided in Dataset S1a–c for Set-1, Set-2, and the reciprocal transfer experiments, respectively. Read counts for ASVs assigned to Wolbachia are highlighted in Datasets S1a–c, and they are excluded from subsequent analysis, as are ASVs detected in only one sample. The raw reads from all datasets are deposited in the NCBI Short Read Archive under BioProject PRJNA515926.
Statistical analysis
All statistical analyses were conducted in R [36] with functions from the vegan package [37], except where specified. Prior to analysis, reads from Set-1 samples were subsampled to 90,900 reads per sample, Set 2 to 3950 reads per sample, and transfer experiment samples to 1800 reads per sample. Sequence coverage plots generated with the iNEXT package [38] confirmed that these sequencing depths were sufficient to cover the ASV diversity present (Supplementary Fig. S1).
Relationships among microbiome samples were visualized with principal coordinate analysis (PCoA) plots of Bray–Curtis or Jaccard dissimilarity calculated from the relative abundance or presence/absence of ASVs, respectively. To test for differences in microbiome composition between Drosophila species, we conducted one-way PERMANOVA analyses [39] of Bray–Curtis and Jaccard dissimilarities. For samples from Set-2, we tested whether microbiome composition differed between the subgenera Drosophila and Sophophora with a nested PERMANOVA (host species nested within subgenus) using the nested.npmanova function from the BiodiversityR package [40]. We tested the influences of host species and rearing laboratory for the species common to both sets of species with a two-way PERMANOVA based on the relative abundance of bacterial genera. For Acetobacter and Lactobacillus, we tested for differences between rearing laboratory in each species common to both sets with linear models of logit transformed relative abundances. The relationship between host phylogeny and microbiota composition for each set of host species was assessed in two complementary ways [41]: congruence in topology of host phylogenetic trees and microbiota dendrograms, determined with the normalized Robinson-Foulds metric [42]; and significance of the correlation between host phylogenetic distance and similarity in microbiome composition determined with Mantel tests based on Pearson’s product-moment correlations. Host phylogenies were constructed from ref. [43] (see legend to Fig. 1 for detail). Microbiota composition dendrograms were constructed by averaging the microbiota composition of all samples for each species, followed by hierarchical clustering of Bray–Curtis and Jaccard dissimilarities by average linkage.
Fig. 1.
Microbiota composition does not track phylogeny of Drosophila species. Principal coordinates plots for Set 1 (a) and Set 2 (b) with % variation explained by each axis shown in paretheses; and comparison of host phylogeny with dendrogram of similarity in microbiota composition for Set 1 (c) and Set 2 (d). For a, b male (triangles) and female (squares), and the color code for species is provided in c, d, respectively
For the reciprocal transfer experiments, microbiome composition and characteristics were compared between homologous and heterologous associations for each of the eight tests. Relationships among samples were visualized with PCoA ordination plots of Bray–Curtis dissimilarities. Differences in microbiome composition between association type were tested with PERMANOVA tests and p-values were adjusted with the false-discovery rate procedure of ref. [44]. To determine the microbial taxa that differed in relative abundance between homologous and heterologous associations in each test, we used the linear discriminant analysis effect size method (LEfSe) [45] with α = 0.05 for the initial Kruskal–Wallis test and applying a linear discriminant analysis (LDA) score threshold of 2.0. To determine whether association type, host species, host sex, microbiota source species, and experiment influenced microbiome characteristics in the reciprocal transfer experiments, we ran linear models. For the number of ASVs and genera present (i.e., richness), we performed generalized linear models with quasipoisson error distributions and likelihood ratio chi-square tests. For dissimilarity-based measures (i.e., distance to microbiome inoculum and multivariate dispersion), we applied a logit transformation and conducted F-tests. A generalized linear model with quasipossion error distribution and likelihood ratio chi-square tests were used to determine the influence of association type, host species, microbiota source species, and experiment on the number of surviving flies at 5 days post-eclosion.
Results
Microbiota composition varies with host species but does not reflect host phylogeny
Our first analysis examined whether composition of the gut bacterial communities varied among drosophilid species maintained under uniform conditions. To check for the generality of our findings, this analysis was conducted on two sets of Drosophila species that had been reared on different diets in different laboratories, and processed for 16S rRNA gene amplicon sequences by different procedures. The microbiota in every drosophilid species from both sets was dominated by two families of bacteria, the Acetobacteraceae (α-proteobacteria) and Lactobacillaceae (Firmicutes) (Supplementary Fig. S2). However, at the level of amplicon sequence variants (ASVs), microbiome composition differed significantly among Drosophila species in both Set-1 (species, R2 = 0.42, p = 0.002; sex, R2 = 0.01, p = 0.860; Fig. 1a) and Set-2 (species, R2 = 0.70, p < 0.001; sex, R2 = 0.004, p = 0.44; Fig. 1b).
Four Drosophila species (D. ananassae, D. melanogaster, D. simulans, and D. willistoni) were represented in both sets, and for these species, “Set” explained more of the variation in microbiome composition based on relative abundance of bacterial genera, than Drosophila species (Set, R2 = 0.33, p < 0.001; Drosophila species, R2 = 0.22, p = 0.004). The effect of “Set” is primarily due to a higher relative abundance of Acetobacter in Set-1 than Set-2, and the reverse pattern for Lactobacillus, particularly in D. ananassae and D. simulans (Suplementary Fig. S3). An equivalent analysis of ASVs was not conducted because differences in the methods for generating amplicons between the two sets would confound interpretation.
We investigated how the pattern of variation in microbiota composition related to host phylogeny using two complementary statistical methods of normalized Robinson-Foulds (nRF) index of dendrogram similarity and Mantel test for correlation between host phylogenetic distance and dissimilarity in microbial community composition. For the relative abundance data for both species sets (Fig. 1c, d), host phylogeny and microbiota dendrograms were completely incongruent (nRF = 1.0) and Mantel tests using Pearson’s r were not significant (Set-1, r = 0.41, p = 0.07; Set-2, r = −0.08, p = 0.74). Significant phylogenetic signal was also not obtained for the presence/absence of ASVs (Set-1: nRF = 0.6; r = 0.39, p = 0.13; Set-2: nRF = 1.0; r = −0.03, p = 0.61), nor for bacterial OTUs generated with 97%, 95% and 90% sequence identity cutoffs (Supplementary Table S1).
We conducted a supplementary test for Set-2, which included Drosophila species of two subgenera (Drosophila and Sophophora) (Table 1). Pairs of species from the same subgenus did not have more similar microbiome composition than pairs of species from different subgenera (Supplementary Fig. S4A, Mann–Whitney U-test: = 0.87), and subgenus did not significantly contribute to variation in microbiota composition among samples (Supplementary Fig. S4B; PERMANOVA: subgenus, p = 0.613; species nested within subgenus, p < 0.001).
Our results provide clear evidence that microbiome composition varied among the Drosophila species. We reasoned that among species differences in compatibility with various bacterial taxa present in each laboratory may make an important contribution to this variation. Alternatively, the patterns may be the legacy of differences in the microbiological history of individual fly cultures, for example at the point of acquisition from the field or during subsequent maintenance in the laboratory. To test between these alternative explanations, we conducted a set of reciprocal transfer experiments, described next.
Reciprocal transfer experiments reveal differences in microbiota composition between host species administered the same microbial inoculum
We conducted two reciprocal microbiota transfers: between D. simulans and, first, the closely related species D. melanogaster (Transfer 1) and, second, the more distantly related species D. willistoni (Transfer 2) (Fig. 2a). As the microbiota composition of laboratory cultures of drosophilid flies can vary over time [20, 21], each transfer was conducted twice (Experiment 1 and Experiment 2) with different microbial inocula yielding eight tests. Specifically, a microbial inoculum from one Drosophila species was administered to eggs of the same species (homologous host) and different species (heterologous host), and the microbiota in the resultant adult flies was determined at 5 days post-eclosion. Significant differences in microbiome composition between the homologous and heterologous hosts would be indicative of among-host species differences in compatibility with microorganisms present in the inoculum.
Microbiome composition, quantified as the relative abundance of ASVs, differed between the two Drosophila species administered the same microbial inoculum in all eight tests (Fig. 2b). For presence/absence of ASVs, the microbiomes of D. simulans and D. melanogaster differed significantly in three of the four tests while there were no differences in the microbiomes of D. simulans and D. willistoni for this metric (Supplementary Table S2). Our data did not indicate that the magnitude of the differences was greater for distantly related host species as the amount of variation explained by host species did not differ between Transfer 1 and Transfer 2 (t-test for differences in PERMANOVA R2 values; t = 0.75, p = 0.48).
In transfers between D. melanogaster and D. simulans (Transfer 1), differences in microbiota composition were attributed primarily to the genera Acetobacter and Lactobacillus, which dominated the microbiota of these flies. Lactobacillus were more abundant in D. melanogaster than in D. simulans for all four tests, while the opposite pattern was observed for Acetobacter (Table 2 and Supplementary Fig. S5). In addition, three lines of evidence suggest that D. melanogaster is compatible with a wider range of microbial taxa than D. simulans. First, significantly more genera were detected in D. melanogaster than D. simulans (Supplementary Fig. S6; likelihood ratio χ2 = 8.44, p = 0.004). Second, low abundance genera (i.e., genera other than Acetobacter and Lactobacillus) were more abundant in D. melanogaster than D. simulans (Table 2). Finally, many more bacterial ASVs were enriched in the microbiota of D. melanogaster relative to D. simulans than the reverse (Table 3).
Table 2.
Linear discriminant analysis effect sizes (LEFSe) for bacterial genera that differed significantly in relative abundance between D. melanogaster and D. simulans in microbiome transfer experiments
 |
Values are LDA scores (log 10) with p-values for Kruskal–Wallis tests in parentheses. Cells are shaded to indicate the Drosophila species in which the genus was more abundant (blue = D. melanogaster, yellow = D. simulans). There were no significant differences between fly species for any bacterial genera in the microbiome transfer experiments between D. willistoni and D. simulans
Table 3.
Linear discriminant analysis effect sizes (LEFSe) for bacterial ASVs that differed significantly in relative abundance between host species in microbiome transfer experiments
 |
Values are LDA scores (log 10) with p-values for Kruskal–Wallis tests in parentheses. Cells are shaded to indicate the Drosophila species in which the ASV was more abundant (blue = D. melanogaster, yellow = D. simulans, green = D. willistoni)
For the reciprocal transfer experiments between D. simulans and D. willistoni, Acetobacter ASVs dominated the microbiota (Supplementary Fig. S7). Three of these Acetobacter ASVs differed significantly between the host species (Table 3). The two most abundant ASVs, #387 and #402 (Supplementry Fig. S7), showed differing responses to the type of association. Specifically, ASV #387 tended to be more abundant in homologous associations while ASV #402 was more abundant in heterologous associations (Table 3). The responses of these ASVs to association type are consistent with results from the microbiota transfers between D. simulans and D. melanogaster. A further ASV, #388, tended to be more abundant in D. willistoni than D. simulans (significantly so for two of the four tests), and more abundant in D. simulans than D. melanogaster for one of the four tests in Transfer 1 (Table 3). Differences between host species in the absolute abundance of ASVs generally reflected those based on the relative abundance of ASVs (Supplementary Table S3). Altogether, these results are indicative of species-specific determinants of microbiota community composition, and suggest that D. melanogaster is a more permissive host than D. simulans or D. willistoni.
Further analyses revealed no consistent pattern of differences between homologous and heterologous associations in ASV richness (Supplementary Fig. S8), similarity to composition of the inoculum microbiota (Supplementary Fig. S9), or microbiota variation among replicate vials (i.e., multivariate dispersion) (Supplementary Fig. S10). Constraining definitive interpretation of these results, all these indices are influenced by the composition of the microbiota inoculum (Table 4), which differed significantly in dissimilarity to the composition of the fly microbiome between Experiment-1 and Experiment-2 (Table 4). Host species also influenced ASV richness (Table 4) and pair-wise post-hoc analysis showed that ASV richness was greatest in D. melanogaster, intermediate in D. simulans, and lowest in D. willistoni (p < 0.05 for all).
Table 4.
ANOVA tables for generalized linear models of microbiota characteristics
| ASV richness | Dissimilarity to inoculum | Multivariate dispersion | |||||
|---|---|---|---|---|---|---|---|
| df | χ2 | p | F | P | F | p | |
| Inoculum microbiota | 2 | 8.73 | 0.013 | 89.97 | <0.001 | 3.66 | 0.028 |
| Host species | 2 | 45.49 | <0.001 | 0.84 | 0.435 | 1.60 | 0.206 |
| Association type | 1 | 0.83 | 0.362 | 0.59 | 0.445 | 0.05 | 0.827 |
| Host sex | 1 | 2.07 | 0.150 | 0.03 | 0.856 | 0.07 | 0.797 |
| Experiment | 1 | 3.28 | 0.070 | 118.12 | <0.001 | 1.00 | 0.320 |
| Residuals | 122 | ||||||
Significant values are shown in bold (α = 0.05)
Host survival to adulthood is lower in heterologous associations than in homologous associations
As a measure of host fitness in homologous and heterologous associations, the number of flies at harvest (5 days post-eclosion) of the reciprocal transfer experiments was scored, with Drosophila reared under microbiologically sterile conditions (axenic flies) as controls (Supplementary Fig. S11). Survival to adulthood varied significantly with association type (χ2 = 10.8, p = 0.001), host species (χ2 = 12.8, p = 0.002) and microbiota inoculum (χ2 = 19.1, p < 0.001). However, following standardization of the data by host species and experiment, the overall survival in homologous associations was significantly greater than in heterologous associations (F2,116 = 12.7, p = 0.001; Fig. 3). Post-hoc tests revealed no significant difference between the standardized survival of flies in homologous associations and axenic flies, a result that confirms and extends the evidence that axenic status does not significantly affect survivorship to adulthood of D. melanogaster under laboratory conditions on nutritionally adequate diets [46]. However, host survivorship was significantly reduced in heterologous associations, indicating that the microbiota from different species can be deleterious for Drosophila.
Fig. 3.
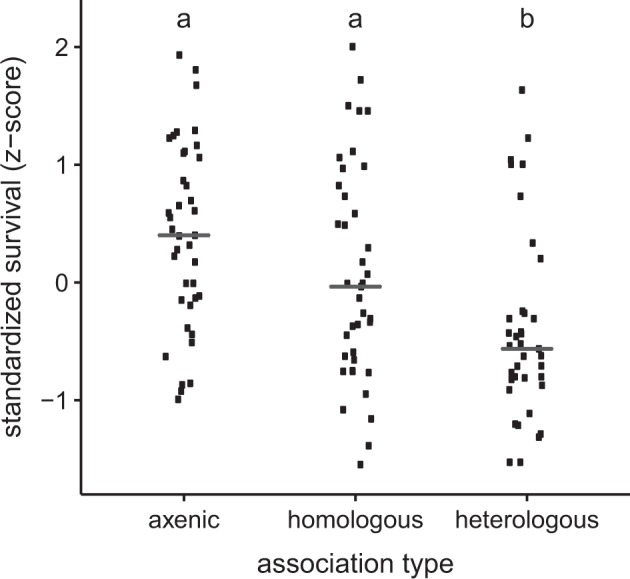
Host survival is significantly greater in homologous associations than heterologous associations. The number of flies in each vial at 5 days post-eclosion was standardized within each host species and experiment
As a check for the possible deleterious effects of insect material in the microbial slurries on the survival of heterologous species, we administered homogenates from axenic flies of the same and different species. Fly mortality did not differ significantly between treatments with axenic homogenates of the same or different species (Supplementary Fig. S12; p > 0.05 for all comparisons). Furthermore, our index of bacterial abundance (16S rRNA gene copy number, determined by qPCR with general bacterial primers) did not differ significantly between homologous and heterologous associations (Supplementary Fig. S13). 16S copy number also did not vary with microbiota source species or experiment, but the effect of host species was significant, with significantly lower abundance in D. simulans than D. melanogaster (Supplementary Fig. S13).
Discussion
This study has yielded two key results. First, the analysis of microbial communities in 18 Drosophila species (Fig. 1) reveals that microbiota composition varies among Drosophila species maintained under uniform laboratory conditions. Second, the reciprocal transfer experiments demonstrate that two drosophilid species administered with the same microorganisms yield microbial communities of significantly different composition, and that host survival to adulthood is significantly depressed in associations with microbiota derived from a different host species. Specifically, these results indicate that variation in microbiota composition can be explained by differences among host species in compatibility with different microbial taxa, rather than their microbiological history; and that these differences are important for host fitness.
The fate of microorganisms that enter the gut is determined by multiple host factors [47], as well as among-microbe competition [48, 49]. Current understanding of host filtering by the gut environment in drosophilids is dominated by research on D. melanogaster, with evidence that some ingested microorganisms persist and others are lost, either by dying in the gut or by elimination with the bulk flow of food through the gut [50–52]. Candidate host determinants of the fate of microbial cells include the profile of antimicrobial peptides expressed in the gut [53, 54], the patterns of gut epithelial turnover [55–57], and the pH of the acidic region of the gut [58]. How these host traits vary among Drosophila species is largely unknown, beyond the genome-scale evidence for extensive evolutionary diversification of the repertoire of canonical immune genes among Drosophila species [59]. Previous studies have found that D. melanogaster is relatively permissive, for example capable of associating with many bacterial taxa administered individually [60]. The finding from the reciprocal transfer experiments in this study that D. melanogaster supports a higher taxonomic diversity of microorganisms than both D. simulans and D. willistoni indicates that host filtering may be more restrictive in some drosophilid species than in D. melanogaster.
It is also important to consider the microbial taxonomic resolution of host filtering. Our results concur with the growing evidence that host–microbe compatibility can vary widely among closely related bacterial strains, with the implication that biologically relevant variation can be undetected in studies that use of 97% sequence identity of 16S gene amplicons as the cutoff for taxon identification [61]. Illustrating the importance of fine-scale variation, we found that the relative abundance of some individual ASVs of Acetobacter displayed the reverse of the general pattern that Acetobacter is promoted in D. simulans relative to D. melanogaster (Table 3). A further layer of complexity is indicated by two Acetobacter ASVs that displayed the intriguing pattern of elevated abundance in either homologous or heterologous hosts (ASV #387 and #402, respectively, in Table 3) rather than in one host species. The reduced survival to adulthood of heterologous hosts raises the possibility that this pattern reflects processes, which favor some taxa in healthy hosts, while other taxa flourish in unhealthy hosts, potentially contributing to dysbiosis [53]. One specific scenario is that flies have inadequate controls over certain bacterial taxa derived from different host species, and the resultant high abundance of these bacteria is deleterious for the host. Future research can test this hypothesis by identifying the bacterial functional traits that dictate compatibility with different Drosophila species, and investigating how compatibility both influences, and is influenced by, the physiological condition of the host.
The evidence from this study that the composition of microbial communities varies among drosophilid species is overlain by the lack of evidence that this variation tracks host phylogeny. There is, however, one systematic caveat in the approach of this and other studies [19, 20] of among-species variation in the microbiota of drosophilids: that the analyses are conducted on long-term laboratory cultures. The microbiota in these cultures is of lower diversity and differs in both taxonomic composition and functional traits from natural populations [20, 21, 25, 62], raising the possibility that the microbial taxa which have been eliminated are precisely those that are controlled by host traits that track host phylogeny. Tempering this concern, a robust relationship between host phylogeny and microbiota composition is detected in mammals maintained in captivity [19, 63], even though the microbiota in these animals can be depauperate and divergent from field individuals [64, 65].
Weak or undetectable correspondence between host phylogeny and microbiota composition has been reported for various systems other than drosophilid flies, notably marine nematodes [66], passerine birds [67], and possibly bats [63] (but see [68, 69]). To date, the factors that may influence the host phylogenetic pattern of microbiota composition in different groups has received limited consideration. It has been argued [19] that the poor phylogenetic signal for drosophilids can be attributed to the variability in the microbiota composition (also argued by ref. [20]), the rapid transit of some bacterial taxa through the Drosophila gut, and the dominance of the microbiota by Acetobacter. However, these explanations are contradicted by other data. Specifically, there is excellent evidence that some bacterial taxa can persist for extended periods in the Drosophila gut [50, 51] and that bacterial taxa other than Acetobacteraceae can dominate the gut microbiota, varying with diet and Drosophila species [23, 24, 70]. Furthermore, no detectable phylogenetic pattern is obtained in analyses of greater phylogenetic scope (including representatives of subgenus Drosophila in Set 2 of this study), even though the reciprocal transfer experiments in this study show that host factors are an important driver of among-species variation in microbiota. Taken together, these considerations indicate that the lack of phylogenetic signal does not arise from a dearth of among-host species variation in controls over the microbiota. Instead, the host controls may have diversified in response to ever-shifting selection pressures that preclude simple correspondence with host phylogeny.
Further insights come from the finding of this study that the survival to adulthood of Drosophila species is significantly depressed by association with microbiota from different species, suggesting that some microbial taxa that are advantageous for one host species may be deleterious for a related species. Similar results have been obtained for Nasonia jewel wasps [19, 26]. This effect is likely mediated predominantly by ineffective filtering by the heterologous host of bacteria that are specifically deleterious in that host. Additionally, filtering by the donor host may exclude bacterial taxa that are specifically advantageous to the recipient host from the microbiota inocula generated in this study. Among-host species variation in traits that influence microbiota composition can arise from selection for optimal microbial partners, which may vary among host species in relation to ecological circumstance (especially diet) and physiological differences in metabolism, etc. However, some relevant host traits may be adaptations for diet processing in the gut or resistance to gut pathogens, with incidental consequences for the microbiota composition. For example, the expression of antimicrobial peptides, which influence gut microbiota composition (see above), is strongly diet-dependent in some insects [71], and pH of the acidic region of the D. melanogaster gut influences susceptibility to the bacterial pathogen Pseudomonas entomophila, as well as gut microbiota abundance [58]. As a consequence, among-host species variation in compatible microorganisms may be dictated, at least in part, by interspecific differences in the natural diet and the ever-changing selection pressures exerted by pathogens.
In conclusion, this study reinforces the growing appreciation that multi-species approaches provide valuable insights into microbial community assembly in animal hosts [19, 63, 72]. To date, most research on host–microbial compatibility has focused on the synthesis of symbioses with single microbial taxa, e.g., rhizobia in legumes [73], bioluminescent Vibrio in squid [74]. However, these systems are increasingly being complemented by studies of animals with complex microbiomes, including the zebrafish [75, 76], Peromyscus mice [69, 77], and Drosophila [23, 78]. This study demonstrates the technical tractability of Drosophila system for multi-species experiments, offering the opportunity to identify host traits shaping microbiota composition and the evolutionary processes dictating among-host species variation.
Supplementary information
Acknowledgements
We thank Marita Wilson (Cornell University) and Dina Mostafa, Adrian Ng and Peter Denezis (University of Western Ontario) for technical assistance, and Brent J. Sinclair (University of Western Ontario) and Andrew Moeller (Cornell University) for helpful discussions. This study was funded by NSF grant Bio 1241099 to AED, and a Natural Sciences and Engineering Council of Canada Discovery Grant to BJS. SK and RK were supported by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant Agreement No. 661122, and additionally SK was supported by the Emil Aaltonen Foundation and RK by the Finnish Cultural Foundation Grant for sabbatical research leave.
Author contributions
KLA, AB, and AED designed the study; AB and VGM prepared the samples for set-1; SK, RK, and GPS prepared the samples for set-2; KLA, EB, AB, and GPS conducted the reciprocal transfer experiments; KLA, AB, and VGM processed the sequence data; KLA conducted the statistical analyses; KLA and AED wrote the manuscript; all authors contributed to manuscript revisions and agreed to the final version.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0532-7) contains supplementary material, which is available to authorized users.
References
- 1.Douglas AE. Fundamentals of microbiome science: how microbes shape animal biology. Princeton, NJ, USA: Princeton University Press; 2018. [Google Scholar]
- 2.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krumbeck JA, Walter J, Hutkins RW. Synbiotics for improved human health: Recent developments, challenges, and opportunities. Annu Rev Food Sci Technol. 2018;9:451–79. doi: 10.1146/annurev-food-030117-012757. [DOI] [PubMed] [Google Scholar]
- 4.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora AK, Douglas AE. Hype or opportunity? Using microbial symbionts in novel strategies for insect pest control. J Insect Physiol. 2017;103:10–17. doi: 10.1016/j.jinsphys.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Gressel J. Microbiome facilitated pest resistance: potential problems and uses. Pest Manag Sci. 2018;74:511–5. doi: 10.1002/ps.4777. [DOI] [PubMed] [Google Scholar]
- 7.Berasategui A, Shukla S, Salem H, Kaltenpoth M. Potential applications of insect symbionts in biotechnology. Appl Microbiol Biotechnol. 2016;100:1567–77. doi: 10.1007/s00253-015-7186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–62. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adair KL, Douglas AE. Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol. 2017;35:23–9. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, et al. Social networks predict gut microbiome composition in wild baboons. eLife. 2015;4:e05224. doi: 10.7554/eLife.05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc Natl Acad Sci USA. 2017;201700122. [DOI] [PMC free article] [PubMed]
- 12.Perofsky AC, Lewis RJ, Meyers LA. Terrestriality and bacterial transfer: a comparative study of gut microbiomes in sympatric Malagasy mammals. ISMEJ. 2019;13:50–63. doi: 10.1038/s41396-018-0251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moeller AH, Peeters M, Ndjango J-B, Li Y, Hahn BH, Ochman H. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 2013;23:1715–20. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Knight R, Caporaso JG, et al. Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch) Ecol Lett. 2014;17:979–87. doi: 10.1111/ele.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–5. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CCR, Snowberg LK, Caporaso GJ, Knight R, Bolnick DI. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 2015;9:1–12. doi: 10.1038/ismej.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016;14:e2000225. doi: 10.1371/journal.pbio.2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong AC-N, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–32. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandler JA, Morgan Lang J, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinson VG, Douglas AE, Jaenike J. Community structure of the gut microbiota in sympatric species of wild Drosophila. Ecol Lett. 2017;20:629–39. doi: 10.1111/ele.12761. [DOI] [PubMed] [Google Scholar]
- 23.Adair KL, Wilson M, Bost A, Douglas AE. Microbial community assembly in wild populations of the fruit fly Drosophila melanogaster. ISME J. 2018;12:959–72. doi: 10.1038/s41396-017-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bost A, Martinson VG, Franzenburg S, Adair KL, Albasi A, Wells MT, et al. Functional variation in the gut microbiome of wild Drosophila populations. Mol Ecol. 2018;27:2834–45. doi: 10.1111/mec.14728. [DOI] [PubMed] [Google Scholar]
- 25.Staubach F, Baines JF, Künzel S, Bik EM, Petrov DA. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS ONE. 2013;8:e70749. doi: 10.1371/journal.pone.0070749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brucker RM, Bordenstein SR. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science. 2013;341:667–9. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 27.Markow TA, O’Grady PM. Dietary considerations. In: TA Markow, PM OʼGrady (eds). Drosophila: a guide to species identification and use. Elsevier, London. 2006, p. 215–26.
- 28.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the Miseq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koyle ML, Veloz M, Judd AM, Wong AC-N, Newell PD, Douglas AE, et al. Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J Vis Exp. 2016;e54219. [DOI] [PMC free article] [PubMed]
- 31.Wong AC-N, Luo Y, Jing X, Franzenburg S, Bost A, Douglas AE. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl Environ Microbiol. 2015;81:6232–40. doi: 10.1128/AEM.01442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Epub ahead of print 2018. 10.7287/peerj.preprints.27295v2. [DOI] [PMC free article] [PubMed]
- 33.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2012;12:2825–30. [Google Scholar]
- 35.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 37.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. Vegan: community ecology package. R package version 2.3-4.
- 38.Hsieh TC, Ma KH, Chao A. iNEXT: iNterpolation and EXTrapolation for species diversity. R package version 2.0.8.
- 39.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 40.Kindt R, Coe R. A manual and software for common statistical methods for ecological and biodiversity studies. Nairobi, Kenya: World Agroforestry Centre (ICRAF); 2005. Tree diversity analysis. [Google Scholar]
- 41.Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems. 2018;3:e00097–18. doi: 10.1128/mSystems.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53:131–47. [Google Scholar]
- 43.Van Der Linde K, Houle D, Spicer GS, Steppan SJ. A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet Res. 2010;92:25–38. doi: 10.1017/S001667231000008X. [DOI] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 45.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong AC, Dobson AJ, Douglas AE. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol. 2014;217:1894–901. doi: 10.1242/jeb.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miguel-Aliaga I, Jasper H, Lemaitre B. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics. 2018;210:357–96. doi: 10.1534/genetics.118.300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350:663–6. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 49.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inamine H, Ellner SP, Newell PD, Luo Y, Buchon N, Douglas AE. Spatiotemporally heterogeneous population dynamics of gut bacteria inferred from fecal time series data. mBio. 2018;9:e01453–17. doi: 10.1128/mBio.01453-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pais IS, Valente RS, Sporniak M, Teixeira L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 2018;16:e2005710. doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blum JE, Fischer CN, Miles J, Handelsman J. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio. 2013;4:e00860–13-e00860-13. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu J-H, Kim S-H, Lee H-Y, Bai JY, Nam YD, Bae JW, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–82. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 54.Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio. 2014;5:e01117–14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, et al. Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep. 2015;12:1656–67. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–22. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–44. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overend G, Luo Y, Henderson L, Douglas AE, Davies SA, Dow JAT. Molecular mechanism and functional significance of acid generation in the Drosophila midgut. Sci Rep. 2016;6:27242. doi: 10.1038/srep27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–8. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 60.Chaston JM, Newell PD, Douglas AE. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio. 2014;5:1–12. doi: 10.1128/mBio.01631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segata N. On the road to strain-resolved comparative metagenomics. mSystems. 2018;3:e00190–17. doi: 10.1128/mSystems.00190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winans NJ, Walter A, Chouaia B, Chaston JM, Douglas AE, Newell PD. A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Mol Ecol. 2017;26:4536–50. doi: 10.1111/mec.14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishida AH, Ochman H. Rates of gut microbiome divergence in mammals. Mol Ecol. 2018;27:1884–97. doi: 10.1111/mec.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA. 2016;113:10376–81. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ericsson AC, Montonye DR, Smith CR, Franklin CL. Modeling a superorganism – considerations regarding the use of “dirty” mice in biomedical research. Yale J Biol Med. 2017;90:361–71. [PMC free article] [PubMed] [Google Scholar]
- 66.Schuelke T, Pereira TJ, Hardy SM, Bik HM. Nematode-associated microbial taxa do not correlate with host phylogeny, geographic region or feeding morphology in marine sediment habitats. Mol Ecol. 2018;27:1930–51. doi: 10.1111/mec.14539. [DOI] [PubMed] [Google Scholar]
- 67.Hird SM, Sánchez C, Carstens BC, Brumfield RT. Comparative gut microbiota of 59 neotropical bird species. Front Microbiol. 2015;6:1403. doi: 10.3389/fmicb.2015.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ingala MR, Simmons NB, Wultsch C, Krampis K, Speer KA, Perkins SL. Comparing microbiome sampling methods in a wild mammal: Fecal and intestinal samples record different signals of host ecology, evolution. Front Microbiol. 2018;9:803. doi: 10.3389/fmicb.2018.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips C, Phelan G, Dowd S, McDonough MM, Ferguson AW, Delton Hanson J, et al. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol. 2012;21:2617–27. doi: 10.1111/j.1365-294X.2012.05568.x. [DOI] [PubMed] [Google Scholar]
- 70.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, et al. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017;41:S27–48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 71.Vogel H, Müller A, Heckel DG, Gutzeit H, Vilcinskas A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev Comp Immunol. 2018;78:141–8. doi: 10.1016/j.dci.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 72.Kohl KD, Dearing MD, Bordenstein SR. Microbial communities exhibit host species distinguishability and phylosymbiosis along the length of the gastrointestinal tract. Mol Ecol. 2018;27:1874–83. doi: 10.1111/mec.14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oldroyd GEDD, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–44. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 74.McFall-Ngai M. Divining the essence of symbiosis: Insights from the squid-Vibrio model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stagaman K, Burns AR, Guillemin K, Bohannan BJM. The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J. 2017;11:1630–9. doi: 10.1038/ismej.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016;10:655–64. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baxter NT, Wan JJ, Schubert AM, Jenior ML, Myers P, Schloss PD. Intra- and interindividual variations mask interspecies variation in the microbiota of sympatric Peromyscus populations. Appl Environ Microbiol. 2015;81:396–404. doi: 10.1128/AEM.02303-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Obadia B, Güvener ZT, Zhang V, Ceja-Navarro JA, Brodie EL, Ja WW, et al. Probabilistic invasion underlies natural gut microbiome stability. Curr Biol. 2017;27:1999–2006.e8. doi: 10.1016/j.cub.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.