Abstract
Most autotrophs use the Calvin–Benson–Bassham (CBB) cycle for carbon fixation. In contrast, all currently described autotrophs from the Campylobacterota (previously Epsilonproteobacteria) use the reductive tricarboxylic acid cycle (rTCA) instead. We discovered campylobacterotal epibionts (“Candidatus Thiobarba”) of deep-sea mussels that have acquired a complete CBB cycle and may have lost most key genes of the rTCA cycle. Intriguingly, the phylogenies of campylobacterotal CBB cycle genes suggest they were acquired in multiple transfers from Gammaproteobacteria closely related to sulfur-oxidizing endosymbionts associated with the mussels, as well as from Betaproteobacteria. We hypothesize that “Ca. Thiobarba” switched from the rTCA cycle to a fully functional CBB cycle during its evolution, by acquiring genes from multiple sources, including co-occurring symbionts. We also found key CBB cycle genes in free-living Campylobacterota, suggesting that the CBB cycle may be more widespread in this phylum than previously known. Metatranscriptomics and metaproteomics confirmed high expression of CBB cycle genes in mussel-associated “Ca. Thiobarba”. Direct stable isotope fingerprinting showed that “Ca. Thiobarba” has typical CBB signatures, suggesting that it uses this cycle for carbon fixation. Our discovery calls into question current assumptions about the distribution of carbon fixation pathways in microbial lineages, and the interpretation of stable isotope measurements in the environment.
Subject terms: Water microbiology, Microbial ecology, Bacterial genetics, Molecular evolution, Metagenomics
Introduction
All life on Earth is based on carbon fixation, and its molecular machinery is increasingly becoming a focus of biotechnology and geo-engineering efforts due to its potential to improve crop yields and sequester carbon dioxide from the atmosphere [1]. Seven carbon fixation pathways have evolved in nature, and one purely synthetic pathway runs in vitro [2–4]. Of the seven natural pathways, the Calvin–Benson–Bassham (CBB) cycle was the first discovered, and is believed to be the most widespread [5–7]. The CBB cycle is used by a diverse array of organisms throughout the tree of life, including plants and algae, cyanobacteria, and autotrophic members of the Alpha-, Beta-, and Gammaproteobacteria. Its key enzyme, the ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is thought to be the most abundant, as well as one of the most ancient enzymes on Earth [8, 9].
The reductive tricarboxylic acid (rTCA) cycle was the second described carbon fixation pathway [10]. In short, it is a reversal of the energy-generating oxidative TCA cycle. Instead of oxidizing acetyl-CoA and generating ATP and reducing equivalents, it reduces CO2 at the expense of ATP and reducing equivalents [2, 7, 10]. Most of the enzymes are shared with the TCA cycle, except for those that catalyze irreversible reactions in the TCA, such as citrate synthase, which is catalyzed by ATP citrate lyase in the rTCA cycle. However, given sufficiently high reactant to product ratios and enzyme concentrations, the citrate synthase reaction can be reversed to run the TCA cycle reductively, without any additional enzymes [11, 12]. The rTCA pathway is widely distributed in nature, and has been described in diverse lineages of anaerobes and microaerobes, such as the Chlorobi, Aquificae, Nitrospirae, and is also commonly observed among the Proteobacteria, including the Deltaproteobacteria and the Campylobacterota, (formerly Epsilonproteobacteria) [13, 14]. It is particularly prominent in the Campylobacterota, as all previously described autotrophic members of this class use the rTCA pathway for CO2 fixation [2, 13].
Carbon fixation by chemoautotrophic microorganisms forms the basis of entire ecosystems at deep-sea hydrothermal vents and cold seeps [15, 16]. Most of this carbon is fixed either via the CBB cycle, used by many gammaproteobacterial autotrophs, or the rTCA cycle, used by campylobacterotal autotrophs. This difference is reflected by the different niches colonized by these organisms at hydrothermal vents and seeps, with Gammaproteobacteria typically dominating habitats with higher oxygen and lower sulfide concentrations where the CBB cycle would be more efficient, and Campylobacterota typically thriving at lower oxygen and higher sulfide concentrations where the rTCA cycle could provide a selective advantage [17–23]. Experimental studies have linked substrate preferences in cultured Gammaproteobacteria and Campylobacterota to these ecological distributions [24–26]. Symbiotic invertebrates at hydrothermal vents and cold seeps associate with either gammaproteobacterial or campylobacterotal endosymbionts, which they rely on for most of their nutrition [27, 28]. Some vent and seep invertebrates associate with both gammaproteobacterial and campylobacterotal symbionts simultaneously, which raises the question of how these co-occurring symbionts with differing habitat preferences can both be provided with suitable conditions [27, 29, 30].
Bathymodiolin mussels, a subfamily of mytilid bivalves, are found worldwide at hydrothermal vents and cold seeps [31]. They have evolved symbiotic relationships with chemosynthetic bacteria, allowing them to colonize these extreme environments. Inside their gills, they host intracellular gammaproteobacterial endosymbionts in epithelial cells called bacteriocytes. The dominant endosymbionts are sulfur- and methane-oxidizing bacteria, often co-occurring in the same mussel species. Some sulfur-oxidizing symbionts also use hydrogen as an energy source, and some mussel species host additional symbionts that gain energy from short-chain alkanes [32, 33]. In addition to these dominant endosymbionts, Assié et al. recently discovered epibionts that colonize bathymodiolin mussels from around the world [34]. In contrast to the gammaproteobacterial endosymbionts of bathymodiolins, these epibionts belong to the Campylobacterota. They are filamentous and colonize the surfaces of the gill epithelia in dense patches in the extracellular spaces between the gill filaments, through which the mussel pumps the inflow of oxygenated seawater (Fig. S1). The nature of the association between the epibiotic Campylobacterota and their mussel hosts is not clear. Similar associations in other deep-sea invertebrates, such as Kiwa crabs [35], gastropods [36], and shallow-water nematodes [37] are thought to be beneficial or commensal.
In this study, we used a multi-omics approach to investigate the metabolism of the Campylobacterota epibionts in two bathymodiolin mussels species, “Bathymodiolus” childressi from cold seeps in the Gulf of Mexico, which have only methane oxidizers as their dominant endosymbiont, and Bathymodiolus azoricus from the Mid-Atlantic Ridge, which host both a sulfur- and a methane-oxidizing endosymbiont [27]. Unexpectedly, the epibionts had, and expressed, all genes required for the CBB cycle but appeared to lack key genes of the rTCA cycle. These CBB cycle genes were most likely acquired by horizontal gene transfer (HGT) from diverse sources. With a recently developed, highly sensitive, direct stable isotope fingerprinting technique [38], we show that the proteins of these epibionts had an isotopic signature typical of the CBB cycle, further demonstrating its importance for the metabolism of these epibionts. The discovery of Campylobacterota that use the CBB cycle for CO2 fixation has implications for understanding the evolution of carbon fixation pathways, and for interpreting stable isotope values in environmental samples.
Results and discussion
Genome assemblies and annotations
We assembled Campylobacterota draft genomes from gill metagenomes of two mussel species: “B.” childressi and B. azoricus. The draft genome from “B.” childressi was 2.2 Mb, and estimated to be 95% complete. It was composed of 354 contigs (longer than 900 bp) with an N50 of 6367 bp, and had 30% GC content, 2204 predicted protein-coding genes and 31 tRNA-encoding genes. The draft genome from B. azoricus was estimated to be 92% complete at 2.3 Mb. It was composed of 523 contigs (longer than 900 bp) with an N50 of 4446 bp. It had 30% GC content, 2155 predicted protein-coding genes and 37 tRNAs (Supplementary Table S1). The draft genomes had an average nucleotide sequence identity (ANI) of 83.1%, indicating that they represent different species likely belonging to the same genus [39–42]. The low GC content of both genomes falls within the range found in other Campylobacterota genera, such as Arcobacter with 27%, and Sulfurovum with 41% GC content.
Sequence analysis of 16S ribosomal RNA genes (16S rRNA, Fig. S2) identified these epibionts as a novel family-level, deep-branching, sister group of the Sulfurovum clade within the Campylobacterota [34]. The Campylobacterota draft genome from “B.” childressi contained a partial 16S rRNA sequence (586 bp) that was 100% identical to the epibiont sequence previously published [34]. The metagenomic 16S rRNA sequences and ANI information allowed us to link the two draft genomes to the previously described epibiont [34].
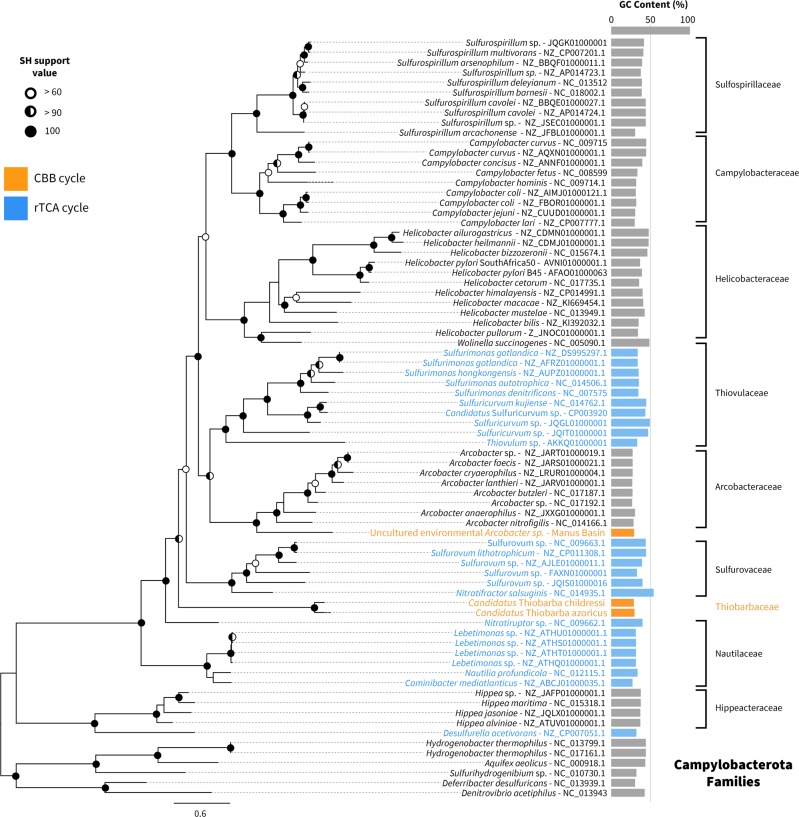
To better resolve the relationships of the mussel epibionts to other Campylobacterota, we analyzed a set of 18 conserved marker genes from the two epibiont draft genomes and other publicly available Campylobacterota genomes (Fig. 1). In contrast to the 16S rRNA based phylogeny (Fig. S2), our analysis placed the mussel epibionts on a long branch, basal to the main Campylobacterota families. The long-branch formation for the genomes presented in this study is likely related to low amino acid sequence identity (AAI) values between these and the Campylobacterota representative genomes. It is unlikely caused by long-branch attraction artifacts due to major differences in GC content, because the genes and genomes compared have similar GC contents (Fig. 1). AAI values were below 48% when comparing the Campylobacterota bins found in our bathymodiolin samples with their closest relatives Sulfurospirillum arcachonense and Arcobacter anaerophilus (Supplementary Table S2). According to the guidelines of Rodriguez and Konstantinidis [42], organisms with AAI values higher than 30% and lower than 55–60% likely belong to the same division, but not the same genus. In summary, both 16S rRNA and concatenated marker gene phylogenies indicated that the epibionts belong to a novel family of Campylobacterota.
Fig. 1.
Phylogenomic tree of representatives of Campylobacterota. The 18 single copy genes used in this analysis were chosen based on the AMPHORA2 marker database [90]. Five deltaproteobacterial species were used to root the tree. In blue are genomes with rTCA cycle genes and in orange genomes with CBB cycle genes. The right column indicates GC content of each genome, the dotted line indicates 50% GC content
We therefore propose the new Candidatus family “Thiobarbaceae” (Campylobacterales, Campylobacterota), with the name composed of “Thio-” from the Greek word θεῖον, theîon, for sulfur and “barba” from the Latin word for beard. The proposed family includes the novel Candidatus genus “Thiobarba” with two Candidatus species “Ca. T. azoricus” and “Ca. T. childressi”, for the two epibiont species in reference to their respective hosts, B. azoricus and B. childressi. For more details on the etiology see SI Appendix note 1.
Unexpected carbon fixation pathways of “Candidatus Thiobarba spp.”
Considering their phylogenetic relationship to free-living chemolithoautotrophic and mixotrophic Campylobacterota and their presence in sulfide-rich environments, we searched the epibiont draft genomes for metabolic pathways indicative of heterotrophy, autotrophy, and sulfur oxidation. Both “Ca. Thiobarba” genomes encoded all the genes for the SOX multi-enzyme pathway of sulfur oxidation and are thus capable of lithotrophy using reduced sulfur compounds as electron donors (Fig. 2). Like other sulfur-oxidizing Campylobacterota, they also appear capable of heterotrophic growth as their genomes contained an oxidative TCA cycle, a partial glycogenesis/glycolysis pathway and numerous ABC-like and TRAP transporters (SI Appendix note 2 and Fig. S3).
Fig. 2.
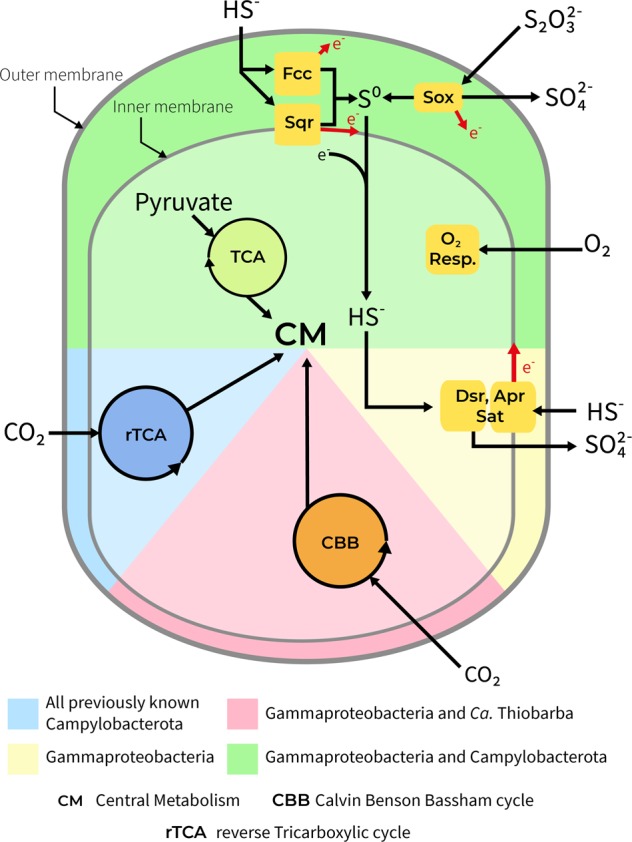
“Ca. Thiobarba spp.” share metabolic features of Gammaproteobacteria and Campylobacterota. Figure shows overview of the main metabolic pathways for energy generation and carbon fixation in known chemosynthetic Gammaproteobacteria and Campylobacterota compared with the metabolism of “Ca. Thiobarba spp.”
All previously described sulfur-oxidizing Campylobacterota use the reverse TCA cycle for carbon fixation [2]. All of these bacteria have genes encoding the enzymes for this cycle including the pyruvate: ferredoxin oxidoreductase genes porABCD, the 2-oxoglutarate oxidoreductase genes oorABDG, and the ATP citrate lyase genes aclAB. Unexpectedly, we could not find most of these genes in the “Ca. Thiobarba” genomes. The “Ca. T. childressi” draft genome contained only the porAB genes, and the “Ca T. azoricus” draft genome contained porABCD and aclA, but not the oorABDG genes. In “Ca. T. childressi” the aclA genes was the only gene present on a 1 kb-long contig. To confirm that these genes were not missing because of errors in assembly, binning or annotation, we performed two different analyses: we searched the genomes and unbinned metagenome assemblies with BLAST, and we mapped the unassembled reads to a database we created from hundreds of published sequences for the key rTCA cycle genes, oor, por, and acl (SI Appendix note 3 and Table S9). No additional rTCA cycle genes were found in the draft genomes or in the “B.” childressi metagenome assembly. The absence of rTCA cycle genes suggests that either (a) the epibionts never had a complete rTCA cycle, (b) it was lost over evolution, or (c) as the “Ca. Thiobarba” genomes are not yet complete, it is possible that the missing rTCA cycle genes could still be present in the epibiont genomes but were not recovered. The por genes are also part of other metabolic pathways, such as pyruvate fermentation, and are widely distributed across the Campylobacterota, including non-chemotrophic members such as Helicobacter pylori [43]. This could explain why the por genes were present in both lineages [44].
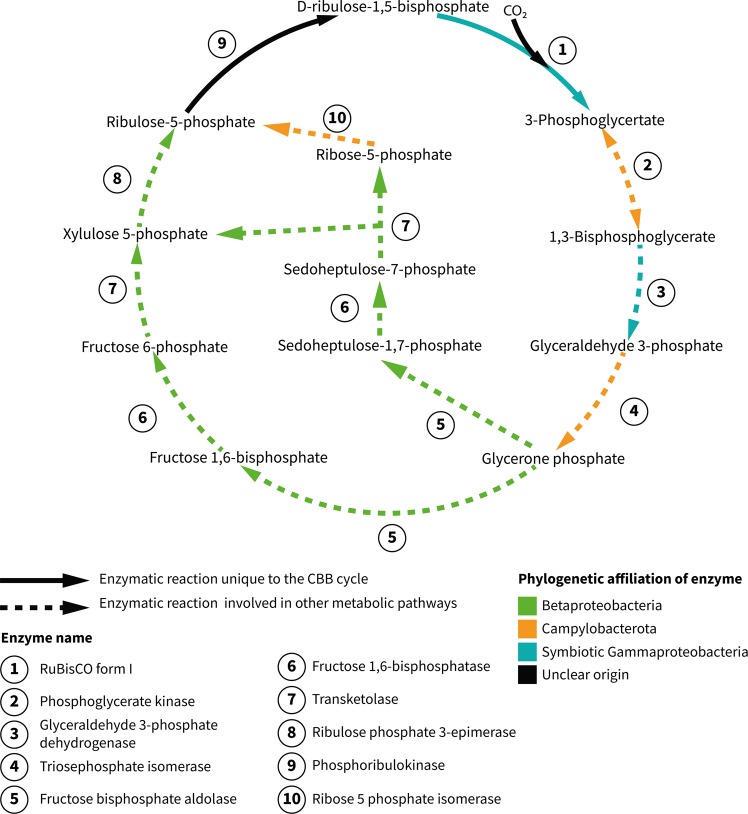
Although the rTCA cycles were incomplete, both “Ca. Thiobarba” genomes contained all the genes required for carbon fixation via the CBB cycle (Fig. 2). Most CBB cycle enzymes are used in other metabolic pathways, and are thus also found in heterotrophic bacteria, but two enzymes are unique to the cycle: phosphoribulokinase (PRK) and ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) [2]. In both “Ca. Thiobarba” species, 9 out of the 12 genes encoding PRK, RuBisCO, and accessory proteins were grouped in two clusters, while three additional genes for the CBB cycle were scattered on separate contigs (Fig. S4). The first cluster consisted of the RuBisCO Form I large and small subunits (rbcL and rbcS), a conserved hypothetical protein, and the RuBisCO activation protein cbbQ. The order of these genes was conserved in both epibionts (Fig. S5). “Ca. T. childressi” had an additional gene encoding the RuBisCO activation protein cbbO in this first cluster. In the “Ca. T. azoricus” genome, this gene was located on a separate contig. The second cluster included the genes coding for fructose-1,6-bisphosphatase, PRK, transketolase, phosphoglycolate phosphatase, fructose-bisphosphate aldolase, and ribulose-phosphate 3-epimerase (Fig. S4). The order of the second gene cluster was consistent in both epibiont genomes, but the gene neighborhoods surrounding this cluster differed (Fig. S5). As the genomes of “Ca. Thiobarba” are not closed, we analyzed the assemblies further to rule out that the CBB cycle genes originated from assembly error. Multiple lines of evidence support our interpretation. Firstly, identical gene cluster structures, for both RuBisCO and accessory genes, have been reconstructed independently from two separate host species. Secondly, analysis of the sequence coverage of these clusters did not show deviation from the rest of the genome, which would be an indicator for an assembly artifact [45]. Furthermore, “B.” childressi does not host a sulfur-oxidizing endosymbiont from which a RuBisCO sequence could originate, further reducing the likelihood of a metagenomic miss-assembly. Because of this and considering the higher quality of the draft genome assembly, we focused our further analyses on “B.” childressi.
CBB cycle expression in “Ca. Thiobarba childressi”
To confirm expression of the CBB cycle by the epibionts, we analyzed the metatranscriptomes and -proteomes of “B.” childressi, the mussel species with the highest abundance of these epibionts [34]. We found that all CBB cycle genes were expressed in the transcriptomes, including the rbcL and rbcS, which were among the most highly expressed genes of this epibiont (Table 1). “Ca T. childressi” was present in relatively low abundance in the metaproteome samples (~0.5% of the total sample protein, calculated according to [46]), thus, only the most abundantly expressed proteins could be detected. The RuBisCO small and large subunits were among those “Ca T. childressi” proteins detected, further indicating high expression levels. The abundance of CBB cycle transcripts and proteins highlights their importance in the metabolism of “Ca. T. childressi” (for full transcription and expression information, see Supplementary Tables S3 and S4). In contrast, the porABCD genes were transcribed, but not detectable in the proteome suggesting a less important role of these genes.
Table 1.
Transcription and translation ranks for the detectable genes involved in the CBB cycle of “Ca. T. childressi”
| Gene_ID | Name | Transcription rank | Translation rank |
|---|---|---|---|
| BCM6EPS_1532 | Ribulose 1,5-bisphosphate carboxylase large chain - EC 4.1.1.39 | 14 | 18 |
| BCM6EPS_1531 | Ribulose 1,5-bisphosphate carboxylase small chain - EC 4.1.1.39 | 17 | 19 |
| BCM6EPS_1455 | NAD-dependent glyceraldehyde-3-phosphate dehydrogenase - EC 1.2.1.12 | 75 | Not detected |
| BCM6EPS_1028 | Transketolase - EC 2.2.1.1 | 80 | Not detected |
| BCM6EPS_1030 | Fructose-bisphosphate aldolase class II - EC 4.1.2.13 | 90 | Not detected |
| BCM6EPS_126 | Pyruvate-ferredoxin oxidoreductase- delta subunit -EC 1.2.7.1- CDS | 129 | Not detected |
| BCM6EPS_1031 | Ribulose-phosphate 3-epimerase - EC 5.1.3.1 | 150 | Not detected |
| BCM6EPS_1027 | Phosphoribulokinase - EC 2.7.1.19 | 160 | Not detected |
| BCM6EPS_1026 | Fructose-1-6-bisphosphatase- type I - EC 3.1.3.11 | 182 | Not detected |
| BCM6EPS_125 | Pyruvate-ferredoxin oxidoreductase- alpha subunit -EC 1.2.7.1- CDS | 217 | Not detected |
| BCM6EPS_1029 | Phosphoglycolate phosphatase - EC 3.1.3.18 | 236 | Not detected |
| BCM6EPS_124 | Pyruvate-ferredoxin oxidoreductase- beta subunit -EC 1.2.7.1- CDS | 256 | Not detected |
| BCM6EPS_1456 | Phosphoglycerate kinase - EC 2.7.2.3 | 440 | Not detected |
| BCM6EPS_514 | Triosephosphate isomerase - EC 5.3.1.1 | 461 | Not detected |
| BCM6EPS_127 | Pyruvate-ferredoxin oxidoreductase- gamma subunit -EC 1.2.7.1- CDS | 720 | Not detected |
Direct stable isotope fingerprinting suggests a CBB signature for “Ca. Thiobarba childressi”
The stable carbon isotope signatures of an environmental sample reflect the pathway that dominates inorganic carbon fixation in the chemoautotrophic members of the bacterial community [47]. Due to differences in kinetic isotope effects, the enzymes involved in the different carbon fixation pathways vary in the degree to which they discriminate against the heavier 13C. This leads to a shift in the 12C/13C ratio between the inorganic carbon source and the generated biomass that is characteristic for the carbon fixation pathway. The CBB cycle generates a −13 to −26‰ shift of the δ13C ratio, while the rTCA cycle leads to a much smaller −3 to −13‰ shift [47].
The average δ13C value of bulk “B.” childressi gill tissues was −47.1 ± 2.6‰, (Supplementary Table S5). However, these values reflect the stable isotope composition of all members of the symbiotic community. As most of the biomass is from the host animal or the highly abundant methane-oxidizing gammaproteobacterial endosymbiont, the signal of the epibiont was greatly diluted [48]. To overcome this limitation and to distinguish between the stable carbon isotope values of the symbiotic partners, we employed the recently-developed direct Protein-SIF method (SIF = stable isotope fingerprinting) on our metaproteomic data [38]. Direct Protein-SIF quantifies the stable isotopic composition of uncultivated members of a mixed community for which genomes or transcriptomes are available. Peptides from the methane-oxidizing symbionts had a δ13C of −38.8 ± 0.7‰, and host peptides had −44.2 ± 0.6‰. These values are similar to those of the methane gas at this cold seep site. Thus, the methane-oxidizing symbionts likely obtain most of their carbon from methane [49]. The host values were similar to those of bulk measurements. However, they were unexpectedly light compared with the methane-oxidizing symbionts, considering that these mussels are thought to gain most of their nutrition from their methane-oxidizing symbionts, and would therefore be expected to have similar δ13C values. As “B.” childressi is known to be capable of filter-feeding [50], these values possibly reflect nutritional supplementation from filter-feeding on microorganisms with even lighter δ13C values than the methane-oxidizing symbionts, that is from the seep environment (as phototrophic microorganisms from the surface would have heavier δ13C values).
“Ca. T. childressi” had a much lower abundance in the metaproteomic dataset compared with the host and the methane-oxidizing symbionts, yet we detected 49 peptides that were unique to “Ca. T. childressi”. This allowed us to estimate the epibionts natural δ13C value, which was relatively light at −66.6 ± 12.5‰. There are two possible inorganic carbon sources for this epibiont: (1) ambient seawater inorganic carbon, which has a δ13C value of + 3‰ [49], and (2) inorganic carbon produced as an end product of methane oxidation by methane-oxidizing bacteria or respiration by the host, which we expect to be around −39‰ for a gas hydrate site, similar to our collection site [48, 49]. We calculated the expected values of biomass generated if either of these carbon sources were fixed through the rTCA cycle or the CBB cycle (Fig. 3). Regardless of the inorganic carbon source, the δ13C values of “Ca. T. childressi” peptides were far lighter than would be expected if they used the rTCA cycle. They were, however, consistent with the expected values for carbon fixation using the CBB cycle, with inorganic carbon derived from symbiont methane oxidation or host respiration.
Fig. 3.
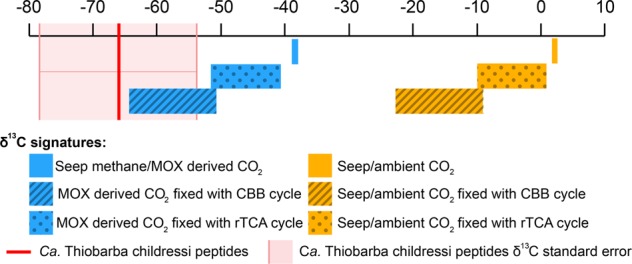
Stable carbon isotope values of “Ca. Thiobarba childressi” are consistent with carbon fixation via the CBB cycle. Model of δ13C values of deep-sea carbon and the predicted influence of different inorganic fixation pathways on these values. The δ13C values of CO2 originating from ambient seawater are shown in yellow, and the expected δ13C values of CO2 originating from methane oxidation (MOX) are shown in blue. The red line represents the average δ13C value measured for “Ca. Thiobarba” peptides using direct Protein-SIF. Reference δ13C values for “Seep methane” and “Seep/ambient CO2” are based on Macavoy et al. [48] and Sassen et al. [49]. Transformations of δ13C values for each metabolic pathway are estimated based on Pearson et al. [47]
Calvin cycle genes in free-living Campylobacterota
After discovering the CBB cycle in the mussel epibionts, we asked if other members of the Campylobacterota might also have acquired these genes. We discovered key CBB cycle genes in a Campylobacterota draft genome binned from a metagenomic library from diffuse hydrothermal fluids collected in the Manus Basin (Western Pacific) [23]. This draft genome was composed of 60 contigs with 29.1% GC content, and based on CheckM, was 92.2% complete [51]. Phylogenomic reconstruction placed this organism on a deep branch basal to the Arcobacteraceae family. AAI values showed between 55 and 58% similarity with the Arcobacteraceae, suggesting that this Campylobacterota bin might belong to a new genus within the Arcobacteraceae (Supplementary Table S2). Our phylogenetic analyses and AAI values indicate that this environmental bin belongs to a Campylobacteraota family distinct from “Ca. Thiobarba” (Fig. 1). Although a full rTCA cycle was present in the draft genome, we also found genes coding for a RuBisCO form I enzyme, a hypothetical gene and CbbQ in one cluster. This cluster shared the same gene order, as well as 84% nucleotide sequence identity, with the CBB cycle cluster we found in “Ca. Thiobarba” (Fig. S5). The high sequence identity between these clusters suggests a similar origin for both. If these genes and the enzymes they encode are active in the Manus Basin bacterium, then free-living Campylobacterota may also be able to use the CBB cycle to fix carbon. These bacteria could use either or both cycles depending on the environmental setting, as suggested for the sulfur-oxidizing gammaproteobacterial symbionts of vestimentiferan tubeworms found at hydrothermal vents [52–54], the large sulfur bacteria Beggiatoa and Thiomargarita spp. [55–57], and recently, the cultivable sulfur oxidizer Thioflavicoccus mobilis [58]. The tubeworm symbiont “Ca. E. persephone” expressed both the CBB and the rTCA cycle in the same host individual, but it is still unclear how these two cycles are coordinated at the level of individual symbiont cells, or over time [53, 54].
Possible evolutionary origins of Campylobacterota CBB cycle genes
Considering the lack of CBB cycle genes in all Campylobacterota investigated prior to this study, it is most likely that this carbon fixation pathway was acquired by “Ca. Thiobarba” and free-living Campylobacterota through HGT, rather than being an ancestral pathway in this phylum. We investigated the evolutionary origins of the genes coding for CBB enzymes, including those with additional roles in other metabolic pathways, using BLAST analyses of nucleotide and protein sequences, and phylogenetic reconstruction of protein sequences. BLAST analyses revealed that only two of the “Ca. Thiobarba” CBB cycle genes were affiliated with genes from other Campylobacterota. Of the other ten, five had best hits to Gammaproteobacteria, and five had best hits to Betaproteobacteria (Supplementary Table S6).
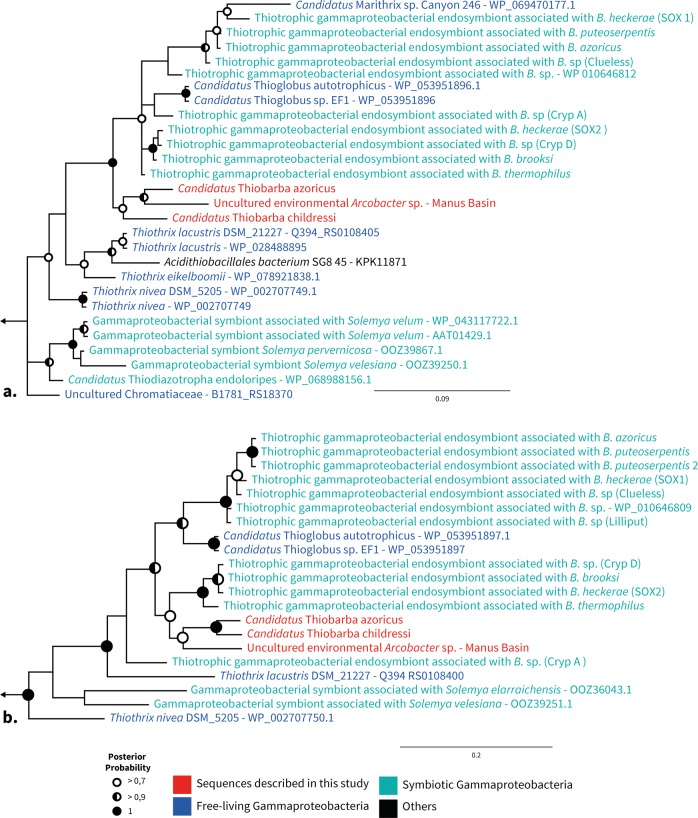
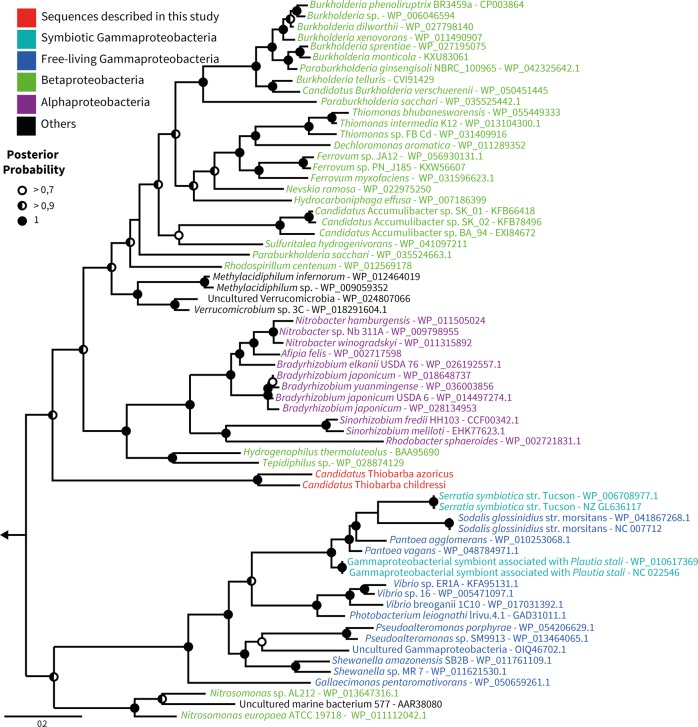
Phylogenetic reconstruction further supported our hypothesis that the “Ca. Thiobarba” CBB cycle is a ‘patchwork’ of genes with evolutionary origins in the Betaproteobacteria, Gammaproteobacteria, and Campylobacterota (Fig. 4). The RuBisCO large and small subunits rbcL and rbcS, their accessory proteins cbbQ and cbbO, as well as the glyceraldehyde-3-phosphate dehydrogenase proteins clustered with a clade of gammaproteobacterial sulfur-oxidizing chemolithoautotrophs. Many of the related sequences belonged to free-living sulfur oxidizers such as “Ca. Thioglobus autotrophicus” and the gammaproteobacterial sulfur-oxidizing endosymbionts of bathymodiolin mussels (Fig. 5). Phylogenetic analysis of “Ca. Thiobarba” PRK proteins placed these on a long branch between gamma-, alpha- and betaproteobacterial clades, but this placement did not have high support (Fig. 6). This could indicate that the “Ca. Thiobarba” PRK proteins truly belong to a Campylobacterota gene family, and because these are the first sequences available from this family, their phylogenetic placement is currently not well supported. Further sampling may help to clarify their evolutionary history. Four “Ca. Thiobarba” CBB cycle proteins consistently belonged to a sister branch to betaproteobacterial sequences (fructose-1,6-bisphophatase, 1,6-bisphophate aldolase, transketolase, and ribulose-phosphate 3-epimerase). Only two proteins were phylogenetically related to those from other Campylobacterota (phosphoglycerate kinase and triose phosphate isomerase) (Figs. S6–S25).
Fig. 4.
“Ca. Thiobarba” genomes encode a CBB cycle with genes affiliated to at least three phylogenetically distinct classes. The solid arrows indicate enzymatic reactions that are unique to the CBB cycle, while the dashed arrows indicate that the enzymes are also involved in other metabolic pathways. Enzyme names are shown in bold and the colors represent their phylogenetic affiliations
Fig. 5.
“Ca. Thiobarba” RuBisCO proteins cluster with gammaproteobacterial sequences. Bayesian inference trees of RuBisCO large (a) and small (b) subunit amino acid sequences under an LG model with Gamma-distributed rates of evolution. Analyses were performed with 6 million generations using two parallel Monte Carlo Markov chains. Sample trees were taken every 25,000 generations. Left arrows indicate truncated tree, tree roots were built from Prochlorococcus and Synechococcus sequences for (a) and Planktothrix and Synechococcus sequences for (b). Full trees are displayed as Figs. S14 and S15
Fig. 6.
“Ca. Thiobarba” phosphoribulokinases are loosely affiliated with those from Betaproteobacteria, Alphaproteobacteria, and Verrucomicrobia. Bayesian inference tree of phosphoribulokinase amino acid sequences under an LG model with Gamma-distributed rates of evolution and a proportion of invariant sites. Analyses were performed with 6 million generations using two parallel Monte Carlo Markov chains. Sample trees were taken every 25,000 generations. Left arrow indicates truncated root, the root is built from distant Prochlorococcus and Synechococcus sequences. Full tree is displayed as SI appendix Fig. S23
The CBB cycle genes consistently fell into three phylogenetic groups: three CBB enzymes were encoded by genes most closely related to those of other Campylobacterota, two were encoded by genes closely related to gammaproteobacterial genes, and seven steps were encoded by genes related to those from Betaproteobacteria. The “Ca. Thiobarba” CBB cycle genes that fell within the Gammaproteobacteria were organized in the same order as genes with which they were most closely related, such as those from “Ca. Thioglobus autotrophicus” and the endosymbionts of bathymodiolin mussels (Fig. S5 and SI Appendix note 4), including the same hypothetical genes placed between the rbcS and cbbQ genes. Furthermore, the genes that were most closely related to Betaproteobacteria had a similar organization to genes found in free-living Betaproteobacteria, such as Paraburkholderia xenovorans (NC_007651) and Dechloromonas aromatica (NC_007298) (Fig. S5), the difference being that “Ca. Thiobarba” operons contain a phosphoglycolate phosphatase gene absent in the free-living genomes. This similarity further supports the hypothesis that these CBB cycle genes were acquired by “Ca. Thiobarba” at least twice, in independent HGT events, with one possibly originating from Gammaproteobacteria, and another possibly from Betaproteobacteria. Alternatively, it is also possible that the Betaproteobacteria-like genes clustering on long branches, such as the PRK, are Campylobacterota genes that have not previously been sequenced. Regardless of the number of HGT events, the acquisition of these genes presumably happened in a common ancestor to the “Ca. Thiobarbaceae”.
Codon usage of horizontally acquired genes is initially expected to reflect the codon usage of the original donor’s genome. Over time, codon usage will evolve to match that of its new host [59]. Some of the “Ca. Thiobarbaceae” CBB cycle genes that we hypothesize were acquired via HGT have, when compared with the closest relative belonging to a different class, a highly similar amino acid identity but differ strongly at the nucleotide level. For example, the RuBisCO small and large subunit amino acid sequences were 98% identical to the sequence from the sulfur-oxidizing gammaproteobacterial endosymbiont, although the nucleotide sequences shared only 47% identity (more details are available in Supplementary Table S6). In addition, codon usage analysis of all “Ca. Thiobarba” CBB cycle genes showed that these had a codon usage similar to that of the “Ca. Thiobarbaceae” core genome (Fig. S26). These results suggest that if these CBB cycle genes were acquired horizontally from donors that had different codon usage patterns to “Ca. Thiobarbaceae”, this was not a recent event.
Gene order within each of the two CBB clusters was identical in “Ca. T. azoricus” and “Ca. T. childressi”. This further supports our hypothesis of a single acquisition event for each cluster in a common ancient ancestor. This synteny also highlights the tendency of these clusters to resist genomic rearrangements. In contrast, the genomic neighborhoods of the CBB clusters differed between the two “Ca. Thiobarba”, indicating that subsequent genome rearrangements occurred since the divergence of these two epibionts. Mobile element genes and transposases were the most highly expressed genes in “Ca. T. childressi” based on our transcriptomes, which, if active, could explain these rearrangements (Supplementary Table S3) [60].
Evolutionary advantages of the CBB cycle
Members of the Campylobacterota occupy remarkably diverse habitats, and have a range of different lifestyles and metabolic capabilities, from chemolithoautotrophs that use a suite of electron donors and acceptors, to heterotrophic symbionts and pathogens of humans and other animals [13, 61]. Evolutionary studies suggest that Campylobacterota emerged in deep-sea habitats, subsequently colonizing and diversifying across terrestrial and human-associated environments [13, 62]. Considering the distribution of chemosynthetic potential within the Campylobacterota, it has been hypothesized that they evolved from an autotrophic common ancestor that first used the Wood–Ljungdahl pathway before switching to a more flexible rTCA cycle [13, 17, 63]. We hypothesize that in the symbiotic “Ca. Thiobarba” lineage, the rTCA cycle was replaced by yet another carbon fixation pathway, the CBB cycle.
Several environmental and genomic factors provide important clues as to why the CBB cycle may be selected for over the rTCA cycle in “Ca. Thiobarba”. Both the CBB and rTCA cycles serve the same purpose, the fixation of inorganic carbon to provide building blocks for cell biomass. But a major difference between the two known carbon fixation pathways is their energy requirements. For example, if one molecule of pyruvate is synthesized from CO2 via the CBB cycle seven molecules of ATP are used, while the rTCA cycle only requires two ATPs [2]. From an evolutionary point of view, exchanging a more energy-efficient carbon fixation pathway with a costlier one could best be explained if it comes with an additional advantage such as oxygen tolerance. The rTCA cycle relies on ferredoxin-based enzymes, which are quickly oxidized by oxygen, and as a result, most organisms with an rTCA cycle are anaerobes or microaerobes [64, 65]. In contrast, CBB cycle enzymes are less affected by oxygen [66]. “Ca. Thiobarba” colonizes the space between the individual gill filaments (Fig. S1), a gas exchange organ that is exposed to oxygen and is typically dominated by gammaproteobacterial endosymbionts. Oxygen dissolved in seawater is therefore first encountered by “Ca. Thiobarba”, before the host or the endosymbionts. Considering that the gills are constantly pumping water through the gills and the inter-filament space, the epibionts will experience oxygen concentrations as high as those in the surrounding seawater. The close phylogenetic relationship between some “Ca. Thiobarba” CBB cycle genes with those from the sulfur-oxidizing gammaproteobacterial endosymbionts of bathymodiolin mussels suggests that either (i) both symbionts acquired CBB cycle genes from the same source or (ii) “Ca. Thiobarba” acquired key genes from the gammaproteobacterial endosymbionts already adapted to the mussel gill niche.
Many free-living, deep-sea Campylobacterota colonize abiotic and biotic surfaces [29, 67, 68]. Thus, the ancestor of “Ca. Thiobarba” might have colonized mussel gills prior to acquiring the CBB cycle. Colonizing mussel gills would bring “Ca. Thiobarba” into close proximity to the mussel’s gammaproteobacterial endosymbionts. Sharing a niche has been shown to be a stronger predictor of HGT than phylogenetic relatedness [69]. Moreover, Campylobacterota have remarkably flexible genomes, with rampant genomic rearrangement and DNA uptake [70–72]. This affinity for foreign DNA uptake, and the physical proximity of mussel epibionts and endosymbionts support scenario (ii) above. The acquisition of the CBB carbon fixation pathway may have enabled “Ca. Thiobarba” to thrive attached to an animal host, leading to the complete reliance on the CBB cycle for carbon fixation and potentially to the gradual loss of the rTCA cycle.
Evolving a Calvin cycle in nature and the laboratory
The complex metabolic network that links carbon fixation and central carbon metabolism poses a massive challenge to switching carbon fixation pathways, either in nature or in the laboratory. These links are usually specific to each pathway and to each organism [66]. Efforts to introduce nonnative carbon fixation pathways have mainly focused on the CBB cycle because theoretically, only two additional enzymes are needed to run this cycle, even in heterotrophs, such as Escherichia coli [73, 74]. However, a number of challenges must be overcome to express ‘foreign’ carbon fixation pathways in new organisms. In addition to the challenges inherent in expressing horizontally acquired genes, such as nonnative promoter and codon usage, and the need for chaperones and biosynthesis enzymes, gene expression must be tightly regulated to balance the production and consumption of intermediates and end products. Because of this, to run the CBB cycle in engineered E. coli, the CBB cycle had to be synthetically decoupled from gluconeogenesis by deleting the phosphoglycerate mutase gene [73]. Switching from one carbon fixation pathway to another may be simpler in chemolithoautotrophs than rewiring a chemoorganoheterotroph, such as E. coli to use the CBB cycle. In a chemolithoautotroph, production of energy and reducing equivalents are already decoupled from carbon fixation, as they are generated through oxidation of reduced compounds such as sulfur. Nevertheless, switching from the rTCA to the CBB cycle is a major shift in cellular metabolism, requiring adaptation of diverse biosynthetic pathways linked to carbon fixation. As far as we are aware, this has not yet been observed in nature, but in the laboratory, E. coli required extensive fine-tuning of metabolic enzymes beyond the CBB cycle through experimental evolution to run a fully functional CBB cycle [3, 73].
Conclusions
The environment is a potent driving force in structuring symbiotic and free-living microbial communities [27, 30, 75]. The distribution of gammaproteobacterial and campylobacterotal sulfur oxidizers is a typical example of adaptation to geochemical niches in a range of environments from hydrothermal vents [23] and cold seeps [21] to oxygen minimum zones [76] and coastal sediments [77]. Gammaproteobacteria are usually associated with low-sulfide, high-oxygen environments, and Campylobacterota with high-sulfide, low-oxygen environments. The horizontal acquisition of the CBB cycle genes may have allowed campylobacterotal “Ca. Thiobarba” to establish a symbiotic relationship in a niche that is usually dominated by Gammaproteobacteria.
The diverse origins of “Ca. Thiobarba’s” CBB cycle genes showcases the modularity [78] of bacterial metabolism and demonstrates that in principle, fully functional metabolic cycles can be pieced together with enzymes from different organisms, both in the laboratory [3] and in nature. In addition to acquiring the two genes theoretically required by a heterotroph to encode a full CBB cycle, “Ca. Thiobarba” seems to have replaced an extensive set of additional CBB cycle genes. This suggests that similar to laboratory models, this natural metabolic switch required ‘tweaking’ of further enzymes of this pathway, and possibly other pathways that siphon off intermediates. Metabolic modularity is considered one of the main factors organizing biological networks [78]. Understanding genome evolution in “Ca. Thiobarba” will shed light on the complex interplay between gene acquisition, expression and the selection that caused the evolution of this major metabolic shift. Our findings highlight the central role that HGT plays in metabolic modularity and environmental adaptation.
Carbon isotope signatures are routinely used to assess the relative importance of the CBB and rTCA cycles in contemporary and past natural environments, and to infer the key organisms responsible for primary production [79–82]. Although stable isotope signatures may accurately reflect the relative importance of distinct carbon fixation pathways in environmental samples, our study shows that assigning these key ecological functions to particular microbial groups requires a deeper understanding of how the underlying metabolic pathways are distributed in nature.
Material and methods
Sample collection
“B.” childressi individuals were collected at cold seeps in the northern Gulf of Mexico at the GC246 and GC234 sites during the R/V Atlantis AT26-13 cruise in April 2014, Nautilus cruise NA044 in July 2014 and Nautilus NA058 cruise in May 2015. The B. azoricus individual was collected at the Lucky Strike hydrothermal vent field on the North Mid-Atlantic Ridge during the Biobaz cruise in 2013. All mussels were recovered in ambient seawater in insulated containers to maintain the water temperature at their collection site. A list of samples and fixation details are summarized in Supplementary Table S7.
DNA and RNA extraction
DNA was extracted from mussel gill tissue according to Zhou et al. [83] with the following modifications: an initial overnight incubation step was performed at 37 °C in 360 µl of extraction buffer (100 mM Tris-HCl [pH 8.0], 100 mM sodium EDTA [pH 8.0], 100 mM sodium phosphate [pH 8.0], 1.5 M NaCl, 1% CTAB), and 40 µl of proteinase K (10 mg/ml). For transcriptome sequencing, RNA was extracted with an Allprep® DNA/RNA micro kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Concentrations of DNA and RNA were measured with a Qubit® 2.0 Fluorometer (Invitrogen, Eugen, USA).
Metagenome sequencing and assembly
DNA extracted from gill tissues of one “B.” childressi individual was sequenced at the Center for Biotechnology at the University of Bielefeld (Bielefeld, Germany). A total of 471,459,598 paired-end reads (150 bp) and 7,739,150 paired-end reads (250 bp long) were generated on Illumina HiSeq 1500 and MiSeq machines, respectively. DNA extracted from gill tissues of one “B.” and one B. azoricus individual was sequenced by the Max Planck Genome Center (Cologne, Germany) and generated, respectively, 57,172,785 and 159,408,731 paired-end reads (150 bp long) on an Illumina HiSeq 2500.
We screened the metagenomic and metatranscriptomic libraries for the presence of campylobacterotal 16S rRNA sequences. The PhyloFlash 2.0 suite (https://github.com/HRGV/phyloFlash) was used to perform RNA small subunit (SSU) screening and reconstructions.
Metagenome assembly was performed as follows: First the raw reads were quality trimmed (Q = 2) and Illumina adapters were removed using BBduk (BBmap suite v37.9 from Bushnell B. - sourceforge.net/projects/bbmap/). An initial assembly was performed with Megahit [84] using default settings. The resulting assembly file was then analyzed with metawatt V2.0 binning tools [85], and draft genome bins were generated by analyzing contig tetranucleotide frequency, differential coverage and GC content. Contigs belonging to bins with a Campylobacterota taxonomic signature were extracted. The quality-trimmed metagenomic reads were then mapped against the Campylobacterota contigs using Bbmap (BBmap suite v37.9), filtering reads with a minimum identity of 98%. The mapped reads were then used for a new assembly using SPAdes 3.4.2 [86] with default settings. Additional details on the assembly of “Ca. T. azoricus” are described in SI appendix note 3. The bin of the free-living Campylobacterota carrying CBB cycle genes was obtained from the Manus Basin metagenome “NSu-F5” as described in [23] with three rounds of read-mapping, reassembly and binning for final bin completion of 92 and 11.7% contamination.
Bin quality was checked with CheckM [51] and a new iteration of taxonomic binning, mapping, and assembly was performed until no contamination from other bacterial strains or host remained in the assembly. Contigs smaller than 900 bp were included in BLAST analysis but excluded from subsequent analyses because they were unlikely to have any relevant genetic information. Genomes were annotated with RAST and cross-checked with IMG ER web servers [87–89]. Genome ANI and AAI were calculated using the AAI and ANI calculator from the enveomics collection [90] with the default settings. The specific coverage for genomes and gene was calculated using BBmap.
Raw data were uploaded to the European Nucleotide Archive under the accession numbers: PRJEB19882, PRJEB23284, and PRJEB23286.
Transcriptome sequencing and processing
Transcriptomes of three “B.” childressi individuals were sequenced at the Max Planck Genome Center (Cologne, Germany); details are in Supplementary Table S7. Transcriptome reads were processed as in Rubin-Blum et al. [33]. Briefly, raw reads were mapped against the “Ca. T. childressi” draft genome with BBmap (BBmap suite v.37.09): reads were quality trimmed (Q = 2), Illumina adapters removed and a minimum similarity of 98% used to map to the reference genome. The number of transcriptome reads mapping to each gene was estimated with featureCounts v1.5.2 [91]. To compare the transcriptome libraries of each individual, a normalization factor was estimated with calcNormFactors based on the trimmed mean of M-values (TMM) implemented in the edgeR version 3.16.5 [92]. The TMM normalized read counts were converted to reads per kilobases of exon per million reads mapped (RPKM) with edgeR (http://www.bioconductor.org).
rTCA cycle gene screening
To confirm presence or absence of the rTCA cycle in the metagenomic and transcriptomic libraries, we created a BLAST database containing published amino acid sequences of Campylobacterota rTCA key genes, citrate lyase, 2-oxoglutarate ferredoxin oxidoreductase, and pyruvate ferredoxin kinase. The first metagenomic assembly iterations, as well as the final Campylobacterota bins, were screened using BLASTX against the respective database to detect the presence of potential rTCA cycle genes.
Phylogenomic reconstruction
Phylogenomic trees were calculated using Phylogenomics-tools (Brandon Seah, https://github.com/kbseah/phylogenomics-tools). The draft genomes “Ca. T. childressi” and “Ca. T. azoricus” and the free-living Campylobacterotum from Manus basin were compared with the genomes of 41 Campylobacterota representatives. Five Deltaproteobacteria genomes were used as outgroup. Universal marker proteins conserved across all bacteria were screened using Amphora2 [93]. Genes present in one copy in every draft genome were selected for the phylogenomic reconstruction (rpsI, rplT, rpsB, rplM, rpsS, rplK, rplL, frr, rplP, rplA, rplB, pyrG,rpsM, smpB). Each gene set was aligned using MUSCLE [94]. The concat_align.pl script (phylogenomics-tools) was used for determining the best protein substitution model of each marker alignment (rpsI::LG, rplT::LG, rpsB::LG, rplM::LG, rpsS::LG, rplK::RTREV, rplL::LG, frr::LG, rplP::LG, rplA::LG, rplB::LG, pyrG::LG,rpsM::LG, smpB::LG). To calculate the multi-gene phylogeny, the marker genes from each genome were concatenated. The best tree with SH-like aLRT support value was calculated with RAxML [95] using the tree_calculations.pl script (phylogenomics-tools).
Phylogenetic analysis
The IMG ER pipeline detected genes with a gammaproteobacterial signature based on homologies to sequences in its database. We extracted and analyzed these sequences with the Geneious software version v 9.1.8 [96] (http://www.geneious.com). Genes predicted by automated annotations were manually verified and curated using the public databases NCBI, Uniprot and Swissprot. Sequences of interest were compared with the NCBI nucleotide and amino acid databases using nucleotide- and amino acid-BLAST. We retrieved closely related sequences from the BLASTX results on the NCBI nonredundant database. In addition, other reference sequences were included in the analysis and all sequences were aligned using MUSCLE (v3.6.) [94]. To detect the best substitution model to use for phylogenetic reconstruction, we used the ProtTest3 package [97] (Model summarized in Supplementary Table S8). Phylogenetic analyses were then performed using Bayesian and Maximum likelihood analyses. Bayesian analysis was performed with MrBayes (v3.2) [98] under a General Time Reversible model with the best-fitted substitution model. Analyses were performed for 2 million generations using four parallel Monte Carlo Markov chains. Sample trees were taken every 1000 generations. Maximum likelihood trees were calculated with PHYML [99] using the best-fitted substitution model. We used 1000 bootstraps as support values for nodes in the trees.
The 16S rRNA phylogeny (Fig. S2) includes the full and partial 16SrRNA sequences identified in the genomes of the two “Ca. Thiobarbaceae”, the free-living Arcobacter, as well as the same 41 Campylobacterota representatives used for phylogenomic analyses. The phylogeny was calculated using Bayesian inference, the analysis was performed with MrBayes (v3.2) [100, 101] under a General Time Reversible model with Gamma-distributed rates of evolution and a proportion of invariant sites. Analyses were performed for 10 million generations using four parallel Monte Carlo Markov chains. Sample trees were taken every 5000 generations. Posterior probabilities calculated with PHYML using a GTR substitution model with 5000 bootstraps were used as support values for nodes in the tree.
Codon usage analysis
The codon usage of “Ca. T. azoricus” and “Ca. T. childressi” genes was determined with CodonW [59] using default parameters. The principal component analysis was plotted with R (version 3.4.0).
Bulk isotope analysis
Parts of “B.” childressi gill tissues were used for bulk stable isotope analysis. Tissue pieces were oven-dried overnight and ground to a fine powder. The dried tissue was weighed and samples (0.3–0.7 mg dry weight) were packaged in tin capsules for mass spectrometry, and analyzed using a Costech (Valencia, CA USA) elemental analyzer interfaced with a continuous flow Micromass (Manchester, UK) Isoprime isotope ratio mass spectrometer (EA-IRMS) for 15N/14N and 13C/12C ratios. Measurements are reported in δ notation [per mil (‰) units] and ovalbumin was used as a routine standard. Precision for δ13C and δ15N was ± 0.2 and ± 0.4‰.
Protein extraction and peptide preparation
Parts of the gills (see SI appendix Supplementary Table S7) of three “B.” childressi specimen were used to prepare tryptic digests following the filter-aided sample preparation (FASP) protocol of Wisniewski et al. [102] with minor modifications [61]. Prior to FASP, cells were disrupted by beat-beating samples in SDT lysis buffer (4% (w/v) SDS, 100 mM Tris-HCl [pH 7.6], 0.1 M DTT) using lysing matrix D tubes (MP Biomedicals) before heating to 95 °C for 10 min.
To allow binding of peptides to the SCX column for 2D-LC methods, peptides were desalted using Sep-Pak C18 Plus Light Cartridges (Waters) according to the manufacturer’s instructions. A centrifugal vacuum concentrator was used to exchange acetonitrile after peptide elution with 0.2% (v/v) formic acid. The Pierce Micro BCA assay (Thermo Scientific) was used to determine peptide concentrations, following the manufacturer’s instructions.
1D- and 2D-LC-MS/MS
All three samples were analyzed by 1D-LC-MS/MS and 2D-LC-MS/MS as described in Kleiner et al. [46]. Briefly, sample analysis via 1D-LC-MS/MS was run twice. An UltiMateTM 3000 RSLCnano Liquid Chromatograph (Thermo Fisher Scientific) was used to load 1.5–3 μg peptide with loading solvent A (2% acetonitrile, 0.05% trifluoroacetic acid) onto a 5 mm, 300 µm ID C18 Acclaim® PepMap100 pre-column (Thermo Fisher Scientific). Peptides were eluted from the pre-column onto a 50 cm × 75 µm analytical EASY-Spray column packed with PepMap RSLC C18, 2 µm material (Thermo Fisher Scientific) heated to 45 °C. An Easy-Spray source connected the analytical column to a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Separation of peptides on the analytical column was achieved at a flow rate of 225 nl min−1 using a 460 min gradient going from 98% buffer A (0.1% formic acid) to 31% buffer B (0.1% formic acid, 80% acetonitrile) in 363 min, then to 50% B in 70 min, to 99% B in 1 min and ending with 99% B. Electrospray ionization was used to ionize eluting peptides. Carryover was reduced by two wash runs (injection of 20 µl acetonitrile, 99% eluent B) and one blank run between samples. Data acquisition with the Q Exactive Plus were done as in [103].
The 2D-LC-MS/MS experiments were performed as described by Kleiner et al. [46] with the modification that pH plugs instead of NaCl salt plugs were used for peptide elution from the SCX column. Briefly, 4.5 μg of peptide was loaded with loading solvent B (2% acetonitrile, 0.5% formic acid) onto a 10 cm, 300 μm ID Poros 10 S SCX column (Thermo Fisher Scientific) at a flow rate of 5 µl min−1 using the same LC as for 1D-LC-MS/MS. Peptides that did not bind to the SCX column were captured by the C18 pre-column (same as for 1D-LC), which was in-line downstream of the SCX column. The C18 pre-column was then switched in-line with the 50 cm × 75 μm analytical column (same as for 1D) and the breakthrough separated using a gradient of eluent A and B (2–31% B in 82 min, 50% B in 10 min, 99% B in 1 min, holding 99% B for 7 min, back to 2% B in 1 min, holding 2% B for 19 min). Peptides were eluted stepwise from the SCX to the C18 pre-column by injecting 20 μl of pH buffers with increasing pH ([pH 2.5-pH 8], CTIBiphase buffers, Column Technology Inc.) from the autosampler. After each pH plug, the C18 pre-column was again switched in-line with the analytical column and peptides separated as above. Between samples, the SCX column was washed twice (injection of 20 µl 4 M NaCl in loading solvent B, 100% eluent B), the RP column once (injection of 20 µl acetonitrile, 99% eluent B) and a blank run was done to reduce carryover. Data were acquired with the Q Exactive Plus as in [103].
Protein identification and quantification
A database containing protein sequences predicted from the metatranscriptomic and -genomic data of the “B.” childressi symbiosis generated in this study was used for protein identification as described in the ‘Metagenome assembly’ section above. The cRAP protein sequence database (http://www.thegpm.org/crap/), which contains sequences of common lab contaminants, was appended to the database. The final database contained 38,418 protein sequences. For protein identification, MS/MS spectra were searched against this database using the Sequest HT node in Proteome Discoverer version 2.0.0.802 (Thermo Fisher Scientific) as in [38].
To quantify proteins, normalized spectral abundance factors (NSAFs) [104] were calculated per species and multiplied by 100, to give the relative protein abundance in %. For biomass calculations, the method described by Kleiner et al. [46] was used (see Supplementary Table S4). Calculations of NSAFs and biomass for each sample were based on the combined data from both 1D-LC-MS/MS runs and the one 2D-LC-MS/MS run.
Direct Protein-SIF
Stable carbon isotope fingerprints (SIFs) for “B.” childressi and its symbionts were determined as described by Kleiner et al. [38]. Human hair with a known δ13C value was used as a reference to correct for instrument fractionation. A tryptic digest of the reference material was prepared as described above and with the same 1D-LC-MS/MS method as the samples. Due to the low abundance of the “Ca. Thiobarba” symbiont in terms of biomass, the six 1D-LC-MS/MS datasets (technical replicate runs of three gill samples) were combined in one peptide identification search to obtain enough peptides for SIF estimation. For peptide identification, MS/MS spectra were searched against the database using the Sequest HT node in Proteome Discoverer version 2.0.0.802 (Thermo Fisher Scientific) and peptide spectral matches were filtered using the Percolator node as described by Petersen et al. [103]. The peptide-spectrum match (PSM) files generated by Proteome Discoverer were exported in tab-delimited text format. The 1D-LC-MS/MS raw files were converted to mzML format using the MSConvertGUI available in the ProteoWizard tool suite [105]. Only the MS1 spectra were retained in the mzML files and the spectra were converted to centroided data by Vendor algorithm peak picking. The PSM and mzML files were used as input for the Calis-p software (https://sourceforge.net/projects/calis-p/) to extract peptide isotope distributions and to compute the direct Protein-SIF δ13C value for each species [38]. The direct Protein-SIF δ13C values were corrected for instrument fragmentation by applying the offset determined by comparing the direct Protein-SIF δ13C value of the reference material with its known δ13C value.
Electron microscopy
One “B.” childressi mussel, retrieved from location GC 234, was dissected and gill pieces were fixed for 12 h with 2.5% glutaraldehyde in 1.5X PHEM buffer (containing 90 mM PIPES, 37.5 mM HEPES, 15 mM EGTA, and 3 mM MgCl2) and 9% sucrose at 4 °C after Montanaro et al. [106]. Samples were then washed in 1.5X PHEM with 9% sucrose three times. Gill pieces were dehydrated in a stepwise ethanol series (30–100% in 10% increments), transferred into pure acetone and infiltrated with resin with a stepwise resin series (25–100% in 25% steps) using centrifugation embedding [107]. In short, the sample was transferred to a 2 ml tube filled with resin and centrifuged for 30 s in a benchtop centrifuge at 2000 g. After the second pure resin step, samples were transferred into fresh resin in an embedding mold and polymerized at 60 °C for 24 h. Seventy nanometers of sections were cut with an Ultracut UC7, picked up on formvar coated copper grids and stained with 0.5% aqueous uranyl acetate for 20 min and 2% lead citrate for 6 min. Ultrathin sections were imaged at 30 kV with a Quanta FEG 250 scanning electron microscope equipped with a STEM detector using the xT microscope control software ver. 6.2.6.3123.
Supplementary information
Acknowledgements
We would like to thank the crew and captains of the scientific vessels of the following cruises: Atlantis cruise AT26-13, Nautilus cruises NA044 and NA058 and the “Pourquoi Pas?” Biobaz cruise. We also thank the Max Planck Genome Center in Cologne for the genome and transcriptome sequencing. We are grateful to Lizbeth Sayavedra for transcriptome assembly advice and support. We also thank Brandon Seah for his help with the naming of “Ca. Thiobarba”. We thank Marc Strous for access to proteomics equipment. The purchase of the proteomics equipment was supported by a grant of the Canadian Foundation for Innovation to Marc Strous. This work was funded by the Max Planck Society, the DFG Cluster of Excellence ‘The Ocean in the Earth System’ at MARUM (University of Bremen), a European Research Council Advanced Grant (BathyBiome, Grant 340535) and a Gordon and Betty Moore Foundation Marine Microbiology Initiative Investigator Award through Grant GBMF3811 to ND, the European Union (EU) Marie Curie Actions Initial Training Network (ITN) SYMBIOMICS (contract number 264774) and an NSF award #0801741 to SJ to fund the AT26-13 expedition to the Gulf of Mexico. JMP was supported by the Vienna Science and Technology Fund (WWTF) through project VRG14-021. MK was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada through a Banting fellowship and the NC State Chancellor’s Faculty Excellence Program Cluster on Microbiomes and Complex Microbial Communities. TH was supported through a fellowship of the German Academic Exchange Service DAAD.
Author contributions
AA, NL, JMP, and ND conceived and developed the study. AA and NL analyzed data. AA and HGV performed the metagenomic assemblies. AA performed transcriptome and genome analyses, as well as phylogenetic reconstructions. MK and TH performed proteomic analyses. MK developed and performed isotopic fingerprinting method. NL and AA provided bulk Isotope analysis. NL provided key support for isotope work and interpretations. NL performed the elecron microscopy and analyzed the data. AM and DVM provided environmental genomic bin. HET performed genomic sequencing. SJ and MS provided key biological samples. AA and NL wrote the paper with support from JMP and ND. All authors discussed the results and contributed to the final paper.
Data availability
The metagenomic and metatranscriptomic raw reads are available in the European Nucleotide Archive under Study Accession Number: PRJEB23286, PRJEB23284, and PRJEB19882. The mass spectrometry metaproteomics data, direct Protein-SIF relevant files, and protein sequence database were deposited in the ProteomeXchange Consortium via the PRIDE [108] partner repository with the dataset identifier PXD008089.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Adrien Assié, Nikolaus Leisch
Contributor Information
Adrien Assié, Phone: +1-713-1819156, Email: adrien.assie@bcm.edu.
Nicole Dubilier, Phone: +49-421-2028-932, Email: ndubilie@mpi-bremen.de.
Jillian M. Petersen, Phone: +43-1-4277-91206, Email: petersen@microbial-ecology.net
Supplementary information
The online version of this article (10.1038/s41396-019-0508-7) contains supplementary material, which is available to authorized users.
References
- 1.Rosgaard L, de Porcellinis AJ, Jacobsen JH, Frigaard N-U, Sakuragi Y. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J Biotechnol. 2012;162:134–47. doi: 10.1016/j.jbiotec.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Hügler M, Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci. 2011;3:261–89. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 3.Schwander T, Schada von Borzyskowski L, Burgener S, Cortina NS, Erb TJ. A synthetic pathway for the fixation of carbon dioxide in vitro. Science. 2016;354:900–4. doi: 10.1126/science.aah5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa IA, Barnum TP, Somasekhar PY, Carlström CI, Engelbrektson AL, Coates JD. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc Natl Acad Sci. 2018;115:E92–101. doi: 10.1073/pnas.1715549114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raven J. Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat Micro Ecol. 2009;56:177–92. [Google Scholar]
- 6.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive earth’s biogeochemical cycles. Science. 2008;320:1034–9. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 7.Bassham JA, Benson AA, Kay LD, Harris AZ, Wilson AT, Calvin M. The path of carbon in photosynthesis. XXI. The cyclic regeneration of carbon dioxide acceptor 1. J Am Chem Soc. 1954;76:1760–70. [Google Scholar]
- 8.Raven JA. Rubisco: still the most abundant protein of Earth? New Phytol. 2013;198:1–3. doi: 10.1111/nph.12197. [DOI] [PubMed] [Google Scholar]
- 9.Erb TJ, Zarzycki J. A short history of RubisCO: the rise and fall (?) of Nature’s predominant CO2 fixing enzyme. Curr Opin Biotechnol. 2018;49:100–7. doi: 10.1016/j.copbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans MC, Buchanan BB, Arnon DI. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci. 1966;55:928–34. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mall A, Sobotta J, Huber C, Tschirner C, Kowarschik S, Bačnik K, et al. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science. 2018;359:563–7. doi: 10.1126/science.aao2410. [DOI] [PubMed] [Google Scholar]
- 12.Nunoura T, Chikaraishi Y, Izaki R, Suwa T, Sato T, Harada T, et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science. 2018;359:559–63. doi: 10.1126/science.aao3407. [DOI] [PubMed] [Google Scholar]
- 13.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.) Front Microbiol. 2017;8:682. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, et al. Addendum: comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.) Front Microbiol. 2018;9:772. doi: 10.3389/fmicb.2018.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith C. Chemosynthesis in the deep-sea: life without the sun. Biogeosci Discuss. 2012;9:17037–52. [Google Scholar]
- 16.Van Dover CL. The Ecology of deep-sea hydrothermal vents. Princeton university press, Princeton. 2000.
- 17.Campbell BJ, Engel AS, Porter ML, Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–68. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol. 2008;65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- 19.Sievert S, Vetriani C. Chemoautotrophy at deep-sea vents: past, present, and future. Oceanography. 2012;25:218–33. [Google Scholar]
- 20.Reeves EP, Yoshinaga MY, Pjevac P, Goldenstein NI, Peplies J, Meyerdierks A, et al. Microbial lipids reveal carbon assimilation patterns on hydrothermal sulfide chimneys. Environ Microbiol. 2014;16:3515–32. doi: 10.1111/1462-2920.12525. [DOI] [PubMed] [Google Scholar]
- 21.Pop Ristova P, Wenzhöfer F, Ramette A, Felden J, Boetius A. Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea) ISME J. 2014;9:1–13. doi: 10.1038/ismej.2014.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sievert SM, Hügler M, Taylor CD, Wirsen CO. Sulfur Oxidation at Deep-Sea Hydrothermal Vents. Microbial Sulfur Metabolism. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008. pp. 238–58. [Google Scholar]
- 23.Meier DV, Pjevac P, Bach W, Hourdez S, Girguis PR, Vidoudez C, et al. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545–58. doi: 10.1038/ismej.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall KT, Morris RM. Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J. 2013;7:452–5. doi: 10.1038/ismej.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagaki F, Takai K, Nealson KH, Horikoshi K. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the E-Proteobacteria isolated from Okinawa trough hydrothermal sediments. Int J Syst Evol Microbiol. 2004;54:1477–82. doi: 10.1099/ijs.0.03042-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto M, Takai K. Sulfur metabolisms in Epsilon- and Gamma-Proteobacteria in deep-sea hydrothermal fields. Front Microbiol. 2011;2:192. doi: 10.3389/fmicb.2011.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–40. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 28.Galkin SV. Structure of Hydrothermal Vent Communities. In: Galkin SV, Demina LL, editors. Trace Metal Biogeochemistry and Ecology of Deep-Sea Hydrothermal Vent Systems. Switzerland: Springer International Publishing; 2016. pp. 41–53. [Google Scholar]
- 29.Petersen JM, Ramette A, Lott C, Cambon-Bonavita M-A, Zbinden M, Dubilier N. Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environ Microbiol. 2010;12:2204–18. doi: 10.1111/j.1462-2920.2009.02129.x. [DOI] [PubMed] [Google Scholar]
- 30.Beinart RA, Sanders JG, Faure B, Sylva SP, Lee RW, Becker EL, et al. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc Natl Acad Sci. 2012;109:E3241–50. doi: 10.1073/pnas.1202690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duperron S, Lorion J, Samadi S, Gros O, Gaill F. Symbioses between deep-sea mussels (Mytilidae: Bathymodiolinae) and chemosynthetic bacteria: diversity, function and evolution. C R Biol. 2009;332:298–310. doi: 10.1016/j.crvi.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–80. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 33.Rubin-Blum M, Antony CP, Borowski C, Sayavedra L, Pape T, Sahling H, et al. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat Microbiol. 2017;2:17093. doi: 10.1038/nmicrobiol.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Assié A, Borowski C, van der Heijden K, Raggi L, Geier B, Leisch N, et al. A specific and widespread association between deep-sea Bathymodiolus mussels and a novel family of Epsilonproteobacteria. Environ Microbiol Rep. 2016;8:805–13. doi: 10.1111/1758-2229.12442. [DOI] [PubMed] [Google Scholar]
- 35.Thurber AR, Jones WJ, Schnabel K. Dancing for food in the deep sea: bacterial farming by a new species of Yeti crab. PLoS One. 2011;6:1–12. doi: 10.1371/journal.pone.0026243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zbinden M, Marqué L, Gaudron SM, Ravaux J, Léger N, Duperron S. Epsilonproteobacteria as gill epibionts of the hydrothermal vent gastropod Cyathermia naticoides (North East-Pacific Rise) Mar Biol. 2015;162:435–48. [Google Scholar]
- 37.Bellec L, Bonavita M-AC, Hourdez S, Jebbar M, Tasiemski A, Durand L, et al. Chemosynthetic ectosymbionts associated with a shallow-water marine nematode. Sci Rep. 2019;9:7019. doi: 10.1038/s41598-019-43517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleiner M, Dong X, Hinzke T, Wippler J, Thorson E, Mayer B, et al. Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities. Proc Natl Acad Sci. 2018;115:E5576–84. doi: 10.1073/pnas.1722325115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goris J, Konstantinidis KT, Klappenbach JA, Vandamme P, Coenye T, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 40.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–64. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–45. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-R LM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe. 2014;9:111–8. [Google Scholar]
- 43.Marais A, Mendz GL, Hazell SL, Mégraud F. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol Mol Biol Rev. 1999;63:642–74. doi: 10.1128/mmbr.63.3.642-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenthal B, Mai Z, Caplivski D, Ghosh S, De La Vega H, Graf T, et al. Evidence for the bacterial origin of genes encoding fermentation enzymes of the amitochondriate protozoan parasite Entamoeba histolytica. J Bacteriol. 1997;179:3736–45. doi: 10.1128/jb.179.11.3736-3745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillippy AM, Schatz MC, Pop M. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol. 2008;9:R55. doi: 10.1186/gb-2008-9-3-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, Li C, et al. Assessing species biomass contributions in microbial communities via metaproteomics. Nat Commun. 2017;8:1558. doi: 10.1038/s41467-017-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson A. Pathways of Carbon Assimilation and Their Impact on Organic Matter Values δ13C. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. pp. 143–56. [Google Scholar]
- 48.MacAvoy SE, Carney RS, Morgan E, Macko SA. Stable isotope variation among the mussel Bathymodiolus childressi and associated heterotrophic fauna at four cold-seep communities in the Gulf of Mexico. J Shellfish Res. 2008;27:147–51. [Google Scholar]
- 49.Sassen R, Joye S, Sweet ST, DeFreitas DA, Milkov AV, MacDonald IR. Thermogenic gas hydrates and hydrocarbon gases in complex chemosynthetic communities, Gulf of Mexico continental slope. Org Geochem. 1999;30:485–97. [Google Scholar]
- 50.Riekenberg P, Carney R, Fry B. Trophic plasticity of the methanotrophic mussel Bathymodiolus childressi in the Gulf of Mexico. Mar Ecol Prog Ser. 2016;547:91–106. [Google Scholar]
- 51.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Liles MR, Halanych KM. Endosymbiont genomes yield clues of tubeworm success. ISME J. 2018;12:2785–95. doi: 10.1038/s41396-018-0220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robidart JC, Bench SR, Feldman RA, Novoradovsky A, Podell SB, Gaasterland T, et al. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ Microbiol. 2008;10:727–37. doi: 10.1111/j.1462-2920.2007.01496.x. [DOI] [PubMed] [Google Scholar]
- 54.Markert S, Arndt C, Felbeck H, Becher DD, Sievert SM, Hugler M, et al. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science. 2007;315:247–50. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- 55.Winkel M, Salman-Carvalho V, Woyke T, Richter M, Schulz-Vogt HN, Flood BE, et al. Single-cell sequencing of thiomargarita reveals genomic flexibility for adaptation to dynamic redox conditions. Front Microbiol. 2016;7:964. doi: 10.3389/fmicb.2016.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flood BE, Fliss P, Jones DS, Dick GJ, Jain S, Kaster A-K, et al. Single-Cell (Meta-)Genomics of a Dimorphic Candidatus Thiomargarita nelsonii reveals genomic plasticity. Front Microbiol. 2016;7:603. doi: 10.3389/fmicb.2016.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacGregor BJ, Biddle JF, Harbort C, Matthysse AG, Teske A. Sulfide oxidation, nitrate respiration, carbon acquisition, and electron transport pathways suggested by the draft genome of a single orange Guaymas Basin Beggiatoa (Cand. Maribeggiatoa) sp. filament. Mar Genomics. 2013;11:53–65. doi: 10.1016/j.margen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Rubin-Blum M, Dubilier N, Kleiner M. Genetic evidence for two carbon fixation pathways (the Calvin–Benson–Bassham Cycle and the Reverse Tricarboxylic Acid Cycle) in symbiotic and FRee-living Bacteria. mSphere. 2019;4:pii: e00394–18. doi: 10.1128/mSphere.00394-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peden JF. Analysis of codon usage. Biosystems. 2011;106:45–50. doi: 10.1016/j.biosystems.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleiner M, Young JC, Shah M, Verberkmoes NC, Dubilier N. Metaproteomics reveals abundant transposase expression in mutualistic endosymbionts. MBio. 2013;4:e00223–13. doi: 10.1128/mBio.00223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamann E, Gruber-Vodicka H, Kleiner M, Tegetmeyer HE, Riedel D, Littmann S, et al. Environmental breviatea harbour mutualistic Arcobacter epibionts. Nature. 2016;534:254–8. doi: 10.1038/nature18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Sievert SM. Pan-genome analyses identify lineage- and niche-specific markers of evolution and adaptation in Epsilonproteobacteria. Front Microbiol. 2014;5:110. doi: 10.3389/fmicb.2014.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hugler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ε-subdivision of proteobacteria. J Bacteriol. 2005;187:3020–7. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ragsdale SW. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem Rev. 2003;103:2333–46. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- 65.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–82. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 66.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pjevac P, Meier DV, Markert S, Hentschker C, Schweder T, Becher D, et al. Metaproteogenomic profiling of microbial communities colonizing actively venting hydrothermal chimneys. Front Microbiol. 2018;9:680. doi: 10.3389/fmicb.2018.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalenitchenko D, Dupraz M, Le Bris N, Petetin C, Rose C, West NJ, et al. Ecological succession leads to chemosynthesis in mats colonizing wood in sea water. ISME J. 2016;10:2246–58. doi: 10.1038/ismej.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 70.Gilbreath JJ, Cody WL, Merrell DS, Hendrixson DR. Change is good: variations in common biological mechanisms in the Epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol Mol Biol Rev. 2011;75:84–32. doi: 10.1128/MMBR.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Porcelli I, Reuter M, Pearson BM, Wilhelm T, van Vliet AHM. Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria. BMC Genomics. 2013;14:616. doi: 10.1186/1471-2164-14-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anderson RE, Reveillaud J, Reddington E, Delmont TO, Eren AM, McDermott JM, et al. Genomic variation in microbial populations inhabiting the marine subseafloor at deep-sea hydrothermal vents. Nat Commun. 2017;8:1114. doi: 10.1038/s41467-017-01228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antonovsky N, Gleizer S, Noor E, Zohar Y, Herz E, Barenholz U, et al. Sugar synthesis from CO2 in Escherichia coli. Cell. 2016;166:115–25. doi: 10.1016/j.cell.2016.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bar-Even A, Noor E, Lewis NE, Milo R. Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci. 2010;107:8889–94. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwirglmaier K, Reid WDK, Heywood J, Sweeting CJ, Wigham BD, Polunin NVC, et al. Linking regional variation of epibiotic bacterial diversity and trophic ecology in a new species of Kiwaidae (Decapoda, Anomura) from East Scotia Ridge (Antarctica) hydrothermal vents. Microbiologyopen. 2015;4:136–50. doi: 10.1002/mbo3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rogge A, Vogts A, Voss M, Jürgens K, Jost G, Labrenz M. Success of chemolithoautotrophic SUP05 and Sulfurimonas GD17 cells in pelagic Baltic Sea redox zones is facilitated by their lifestyles as K- and r -strategists. Environ Microbiol. 2017;19:2495–506. doi: 10.1111/1462-2920.13783. [DOI] [PubMed] [Google Scholar]
- 77.Pjevac P, Kamyshny A, Dyksma S, Mußmann M. Microbial consumption of zero-valence sulfur in marine benthic habitats. Environ Microbiol. 2014;16:3416–30. doi: 10.1111/1462-2920.12410. [DOI] [PubMed] [Google Scholar]
- 78.Kreimer A, Borenstein E, Gophna U, Ruppin E. The evolution of modularity in bacterial metabolic networks. Proc Natl Acad Sci. 2008;105:6976–81. doi: 10.1073/pnas.0712149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levin LA, Mendoza GF, Konotchick T, Lee R. Macrobenthos community structure and trophic relationships within active and inactive Pacific hydrothermal sediments. Deep Sea Res Part II Top Stud Oceanogr. 2009;56:1632–48. [Google Scholar]
- 80.Zhang CL, Fouke BW, Bonheyo GT, Peacock AD, White DC, Huang Y, et al. Lipid biomarkers and carbon-isotopes of modern travertine deposits (Yellowstone National Park, USA): Implications for biogeochemical dynamics in hot-spring systems. Geochim Cosmochim Acta. 2004;68:3157–69. [Google Scholar]
- 81.Kelley CA, Coffin RB, Cifuentes LA. Stable isotope evidence for alternative bacterial carbon sources in the Gulf of Mexico. Limnol Oceanogr. 1998;43:1962–9. [Google Scholar]
- 82.Fry B, Sherr EB. δ13C Measurements as Indicators of Carbon Flow in Marine and Freshwater Ecosystems. In: Rundel PW, Ehleringer NKA Jr., editors. Stable Isotopes in Ecological Research. New York, NY: Springer; 1989. [Google Scholar]
- 83.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–22. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2014;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 85.Strous M, Kraft B, Bisdorf R, Tegetmeyer HE. The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol. 2012;3:1–11. doi: 10.3389/fmicb.2012.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bankevich A, Nurk S, Antipov D, Gurevich Aa, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyer F, Paarmann D, D’Souza M, Olson R, Glass E, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–7. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. Peer J Prepr. 2016;4:e1900v1. [Google Scholar]
- 91.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 92.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–4. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 94.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and Model Choice Across a Large Model Space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 100.Ronquist, F., J. P. Huelsenbeck, M. Teslenko, and J. A. A Nylander. MrBayes version 3.2 manual: tutorials and model summaries. Distributed with the software from https://github.com/NBISweden/MrBayes, 2019.
- 101.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–4. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 102.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 103.Petersen JM, Kemper A, Gruber-Vodicka H, Cardini U, van der Geest M, Kleiner M, et al. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat Microbiol. 2016;2:16195. doi: 10.1038/nmicrobiol.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–47. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 105.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Montanaro J, Gruber D, Leisch N. Improved ultrastructure of marine invertebrates using non-toxic buffers. PeerJ. 2016;4:e1860. doi: 10.7717/peerj.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McDonald KL. Rapid embedding methods into epoxy and LR white resins for morphological and immunological analysis of cryofixed biological specimens. Microsc Microanal. 2014;20:152–63. doi: 10.1017/S1431927613013846. [DOI] [PubMed] [Google Scholar]
- 108.Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metagenomic and metatranscriptomic raw reads are available in the European Nucleotide Archive under Study Accession Number: PRJEB23286, PRJEB23284, and PRJEB19882. The mass spectrometry metaproteomics data, direct Protein-SIF relevant files, and protein sequence database were deposited in the ProteomeXchange Consortium via the PRIDE [108] partner repository with the dataset identifier PXD008089.