Abstract
Rapid identification and characterisation of drug-resistant bacterial pathogens have an important role in diagnostic and antimicrobial stewardship. Response time in the diagnosis of not only the etiological agent but also in antimicrobial susceptibility results is of utmost importance in patient treatment. In this study, matrix-assisted laser desorption ionisation–time of flight (MALDI-TOF) mass spectrometry (MS) was used to screen for biomarkers of ESCAPE (vancomycin-resistant Enterococcus faecium, methicillin-resistant Staphylococcus aureus, hypervirulent NAP1/ribotype 027 Clostridioides [Clostridium] difficile, multidrug resistant Acinetobacter baumannii, multidrug resistant Pseudomonas aeruginosa, and carbapenem-resistant Enterobacteriaceae) pathogens to predict antimicrobial resistance or hypervirulence. Several biomarkers of drug-resistant genotypes in S. aureus, A. baumannii, P. aeruginosa, and K. pneumoniae, as well as hypervirulence in C. difficile, were detected. The fastest possible susceptibility testing with MALDI-TOF MS is simultaneous detection of a characteristic drug-resistant peak and species identification in the same spectra generated in routine processing. According to our approach, resistance or virulence biomarker peaks can be identified while performing routine microbiology analysis, and no additional assays nor prolonged incubation time is needed. Outstanding biomarker peaks detected in our study should be further analysed by additional methods to identify the specific proteins involved.
Subject terms: Mass spectrometry, Clinical microbiology, Infectious-disease diagnostics, Pathogens
Introduction
The emergence of drug-resistant bacterial strains has been accelerated by both antibiotic overuse and reduced infection control. In 2013, the Centers for Disease Control and Prevention issued a report with the most concerning drug-resistant threats in the United States1. Of those mentioned, the most notable are pathogens predominately associated with healthcare-associated infections (HAI)2.
Reports by the Infectious Diseases Society of America have highlighted the importance of specific nosocomial drug-resistant pathogens3. Designated by the acronym ESKAPE, this group comprises the following pathogens: vancomycin-resistant Enterococcus faecium (VREfm), methicillin-resistant Staphylococcus aureus (MRSA), carbapenem-resistant Klebsiella pneumoniae, multidrug resistant (MDR) Acinetobacter baumannii, MDR Pseudomonas aeruginosa, and carbapenem-resistant Enterobacter cloacae3,4. These pathogens are of considerable concern due to their high frequency in HAIs5,6.
Clostridioides [Clostridium] difficile infection is one of the most common HAIs in hospitalised patients receiving antimicrobial therapy. In particular, the infection of hypervirulent C. difficile NAP1/ribotype 027 is associated with severe disease and high mortality rates7,8. In recent years, hospitals have reported a rise in C. difficile infection rates9. Thus, researchers have suggested amending the ESKAPE group by replacing K. pneumoniae with C. difficile and adding the entire Enterobacteriaceae family (instead of only K. pneumoniae and E. cloacae), changing the acronym to ESCAPE10,11.
Rapid identification and characterisation of drug-resistant bacterial pathogens have an important role in diagnostic and antimicrobial stewardship. The mission of the Antibacterial Resistance Leadership Group is to reduce the public health threat of antibacterial resistance12, which can be achieved by rapid detection of MDR isolates in clinical diagnostic laboratories. Matrix-assisted laser desorption ionisation–time of flight (MALDI-TOF) mass spectrometry (MS) is a fast and reliable technique that has been implemented in clinical microbiology laboratories to replace or complement conventional phenotypic identification for most isolated bacterial strains13. Although the report of antibiotic susceptibilities in the laboratory is necessary, the rapid determination of antimicrobial resistance through the use of MALDI-TOF MS is not yet available14. Response time in the diagnosis of not only the etiological agent but also in antimicrobial susceptibility results is of utmost importance in patient treatment. The fastest possible susceptibility testing with MALDI-TOF MS is simultaneous detection of a characteristic drug-resistant peak and species identification in the same spectra generated in routine processing15. Therefore, this study aimed to screen for biomarkers using MALDI-TOF MS in ESCAPE pathogens to predict antimicrobial resistance or hypervirulence.
Results
Selected genes and virulence factors
Among the E. faecium isolates, 53.1% (34/64) were vancomycin-resistant, and 46.9% (30/64) were vancomycin-susceptible (Table 1). The vanA gene was detected in all (34/34) VREfm isolates, whereas the vanB gene was not detected in any of the strains.
Table 1.
Distribution of potential peaks detected, protein/peptide assignment and diagnostic utility.
| Species, virulence/resistance phenotype (n) | Theoretical peaks (p ≤ 0.05) | Potential biomarkers | Protein | Mascot score | Protein sequence coverage (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | N | m/z | AUC | p | ||||||||
| E. faecium, vancomycin-resistant (34) vs vancomycin-susceptible (30) | 70 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| MRSA (36) vs MSSA (31) | 45 | 1 | 4,594 (P) | 0.90 | <0.001 | 50 S ribosomal protein L28 | 26 | 64 | 83.3 | 96.8 | 96.8 | 83.3 |
| C. difficile, 027 ribotype (57) vs non-027 (36) | 76 | 2 | 6,654 (A) | 0.96 | <0.001 | 30 S ribosomal protein S20 | 31 | 67 | 100.0 | 91.7 | 95.0 | 100.0 |
| 6,712 (P) | 0.98 | <0.001 | 30S ribosomal protein S21 | 34 | 98 | 100.0 | 91.7 | 95.0 | 100.0 | |||
| A. baumannii, bla OXA-24 or bla OXA -58 (33) vs | ||||||||||||
| non-blaOXA-24 or bla OXA -58 (35) | 2 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| A. baumannii, blaOXA-24 (26) vs bla OXA -58 (7) | 6 | 2 | 6,304 (P) | 0.99 | <0.001 | NADH-quinone oxidoreductase subunit K | 29 | 56 | 85.7 | 100.0 | 100.0 | 96.3 |
| 6,332 (A) | 0.99 | <0.001 | NADH-quinone oxidoreductase subunit K | 29 | 56 | 100.0 | 100.0 | 100.0 | 100.0 | |||
| P. aeruginosa, MDR (27) vs non-MDR (28) | 61 | 3 | 2,726 (P) | 0.81 | <0.001 | UPF0270 protein Pfl01_4103 | 20 | 32 | 75.0 | 66.6 | 70.0 | 72.0 |
| 5,455 (P) | 0.81 | <0.001 | UPF0391 membrane protein Patl_1732 | 32 | 98 | 82.1 | 51.9 | 67.7 | 66.7 | |||
| 5,742 (P) | 0.84 | <0.001 | Not determined | NA | NA | 85.7 | 40.7 | 60.0 | 73.0 | |||
| 2,726, 5,455, and 5,742 (P) | NA | NA | NA | 75.0 | 74.1 | 75.0 | 74.1 | |||||
| K. pneumoniae, carbapenem-resistant (49) vs carbapenem-susceptible (20) | 96 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| K. pneumoniae, carbapenem- and colistin-resistant (16) vs carbapenem- and colistin-susceptible (20) | 75 | 1 | 6,100 (A) | 0.86 | <0.001 | Not determined | NA | NA | 93.8 | 55.0 | 62.5 | 91.7 |
A: absence; AUC: area under the curve; MDR: multidrug resistant; MRSA: methicillin-resistant S. aureus; MSSA: methicillin-susceptible S. aureus; NPV: negative predictive value; P: presence, PPV: positive predictive value.
Of the S. aureus isolates, 53.7% (36/67) were MRSA (cefoxitin-resistant) and mecA positive; 46.3% (31/67) were methicillin-susceptible S. aureus (MSSA) and mecA negative (Table 1).
For C. difficile isolates, 61.3% (57/93) were ribotypes 027 and 38.7% (36/93) were non-027 ribotypes (Table 1). The following non-027 ribotypes were identified: 001 (n = 17; 47.2%), 106 (n = 4; 11.1%), 003 (n = 3; 8.3%), 176 (n = 3; 8.3%), 002 (n = 1; 2.8%), 012 (n = 1; 2.8%), 014 (n = 1; 2.8%), 017 (n = 1; 2.8%), 019 (n = 1; 2.8%), 020 (n = 1; 2.8%), 076 (n = 1; 2.8%), 220 (n = 1; 2.8%), and 353 (n = 1; 2.8%).
For A. baumannii isolates, 45.6% (31/68) were carbapenem resistant. Class D blaOXA carbapenemase genes blaOXA-24 and blaOXA-58 were detected in 26/68 and 7/68 isolates, respectively. In addition, blaOXA-58 was detected in two carbapenem-susceptible isolates. As expected, the blaOXA-51 gene was detected in all (68/68) strains. Neither class D blaOXA-23, nor encoding metallo-β-lactamases genes blaNDM, blaVIM, and blaIMP genes were detected.
Among P. aeruginosa isolates, 49.1% (27/55) were MDR, and 50.9% (28/55) were non-MDR. The blaVIM and blaIMP genes were not detected.
For K. pneumoniae isolates, 72.6% (49/69) were carbapenem resistant, and 27.4% (20/69) were carbapenem susceptible. Of the carbapenem-resistant isolates, 16/49 (32.7%) were colistin-resistant; none of the carbapenem-susceptible isolates presented colistin resistance. The blaNDM gene was detected in all the carbapenem-resistant isolates. Neither blaKPC, blaIMP, blaVIM, nor blaOXA-48 were detected.
Potential biomarkers
The generated protein profile of each isolate spot had mass-charge ratio (m/z) values between 2,000 and 20,000. A MS spectrum sample of each species is shown in Supplementary Fig. S1. Classification models were generated based on the GA, SNN, and QC algorithms, in which at least one of them yielded adequate cross-validation and recognition capability values for vancomycin-resistant vs vancomycin-susceptible E. faecium (90% and 98%, respectively), C. difficile ribotype 027 vs non-027 (90% and 96%, respectively), mecA positive vs mecA negative (87% and 98%, respectively), carbapenem-resistant vs carbapenem-susceptible K. pneumoniae (78% and 90%, respectively), MDR vs non-MDR P. aeruginosa (76% and 89%, respectively), carbapenem/colistin-resistant vs carbapenem/colistin-susceptible K. pneumoniae (67% and 83%, respectively), blaOXA-24 vs blaOXA-58 carbapenem-resistant A. baumannii (59% and 51%, respectively), and blaOXA-24/58 positive vs blaOXA-24/58 negative A. baumannii (50% and 72%, respectively). A complete description of all classification analyses is shown in Supplementary Table S2. The ability of each algorithm to classify spectra from the same set of samples was used to perform an external validation. At least one of the models was able to allocate most of the spectra of the validation set to their corresponding phenotype/genotype profile (Table S2).
In the initial screening and after group comparisons (phenotype/genotype absent or present), statistically significant peaks (p ≤ 0.05) were detected for carbapenem-resistant K. pneumoniae (n = 96), C. difficile ribotype 027 (n = 76), carbapenem and colistin-resistant K. pneumoniae (n = 75), VREfm (n = 70), MDR P. aeruginosa (n = 61), MRSA (n = 45), blaOXA-58 A. baumannii (n = 6), and carbapenem-resistant A. baumannii (blaOXA-24 or blaOXA-58; n = 2) (Table 1). The complete list describing all significant peaks is shown in Supplementary Dataset S3.
The distribution of each strain after PCA based analysis per phenotype/genotype of all species is shown in Supplementary Fig. S4. The distinguishing capability between drug-resistance (or virulence) phenotype/genotype profiles in the isolates based on their peptide mass fingerprints was highly variable.
These statistically significant peaks were selected for further analysis which were the analysis of the peak area/intensity of the spectra, the coefficient of variation and the value of area under the curve [AUC] of each peak. When all three criteria were met, these peaks were considered as “potential biomarkers peaks”.
When analysis VREfm, no potential biomarkers were finally selected.
For S. aureus isolates, one peak with a m/z of 4,594 (with a value of area under the receiver operating characteristics curve [AUC-ROC] of 0.82; p < 0.001) was selected (Table 1). This peak was present as a singlet in the MSSA isolates; however, in the MRSA isolates, it displayed a splitting pattern as a doublet (Fig. 1). The presence of the peak m/z 4,594 as a doublet had a sensitivity of 83.3% and a specificity of 96.8% for the detection of methicillin-resistant phenotype (Table 1). Furthermore, a peak previously reported as a potential biomarker (m/z 2,415) was detected in 63.9% (23/36) of the MRSA isolates16.
Figure 1.
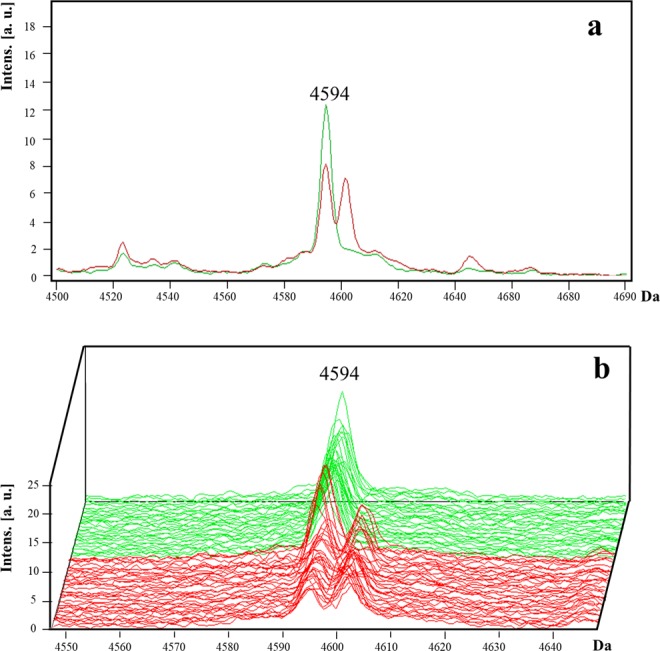
Representative mass spectra of peak m/z 4,594 in S. aureus. The peak m/z 4,594 is presented as a doublet in MRSA isolates (red line) and as a singlet in MSSA isolates (green line). The average spectrum of each group of peak m/z 4,594 (a) and the spectra of all the analysed isolates (b) are shown.
For C. difficile isolates, peaks were selected by strain: the peak at m/z 6,654 (AUC = 0.96) was present only in non-027 strains, except for the 176 ribotype; and the peak at m/z 6,712 (AUC = 0.99; p < 0.001) was present only in 027 and 176 strains (Fig. 2). To detect the hypervirulent 027 ribotype, the absence of peak m/z 6,654 and presence of peak m/z 6,712 presented a sensitivity of 100%, specificity of 91.7%, and PPV of 95%.
Figure 2.
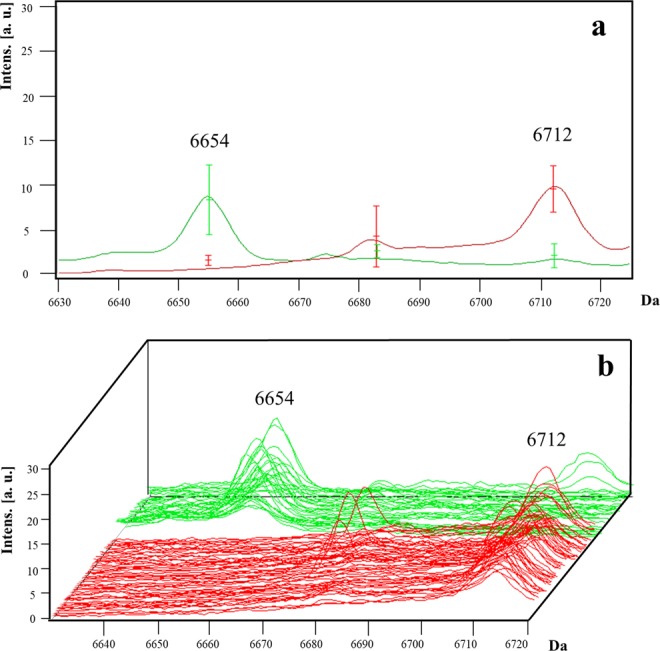
Representative mass spectra of peaks m/z 6,654 and 6,712 in C. difficile. The peak m/z 6,654 is present in non-027 ribotype isolates (green line) and is absent in 027 and 176 ribotype (red line). By contrast, the peak m/z 6,712 is present in 027 and ribotypes but absent in non-027 ribotypes. The average spectrum of each group of peaks m/z 6,654 and 6,712 (a) and the spectra of all the analysed isolates (b) are shown.
A. baumannii blaOXA-24 were compared with blaOXA-58 isolates, and two peaks were selected: the peak at m/z 6,304 (AUC = 0.99; p < 0.001) was present only in blaOXA-58-positive isolates (specificity, and positive predictive value [PPV] of 100%), and the peak at m/z 6,332 (AUC = 0.99, p < 0.001) was present only in blaOXA-24-positive isolates (sensitivity, specificity, PPV, and negative predictive value [NPV] of 100%) (Table 1) (Fig. 3). The comparison of blaOXA-58-positive with blaOXA-58- plus blaoxa-24-negative isolates yielded a statistically significant peak at m/z 6,332 (p = 0.016). The comparison of isolates that harboured either blaOXA-24 or blaOXA-58 with isolates that harboured exclusively blaOXA-51 genes showed no potential biomarkers.
Figure 3.
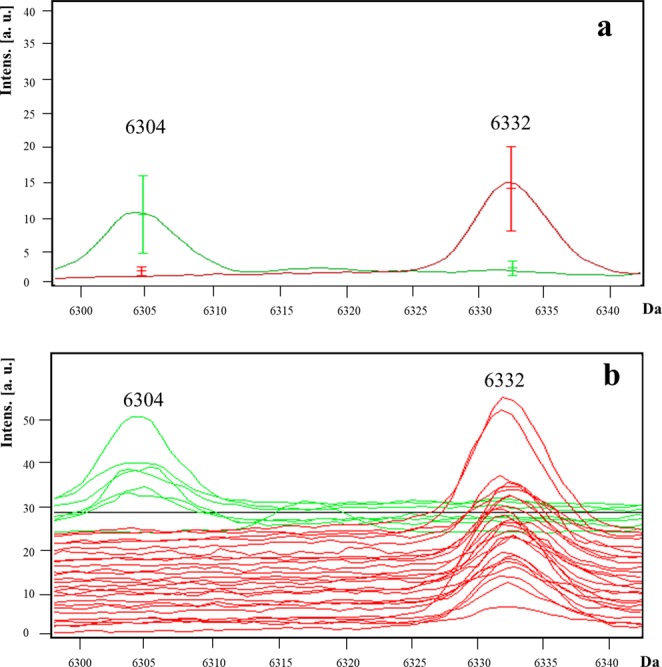
Representative mass spectra of peaks m/z 6,304 and 6,332 in A. baumannii. The peak m/z 6,304 is present in blaOXA-58 isolates (green line) and is absent in blaOXA-24 isolates (red line). By contrast, the peak m/z 6,332 is present in blaOXA-24 but absent in blaOXA-58 isolates. The average spectrum of each group of peaks m/z 6,304 and 6,332 (a) and the spectra of all the analysed isolates (b) are shown.
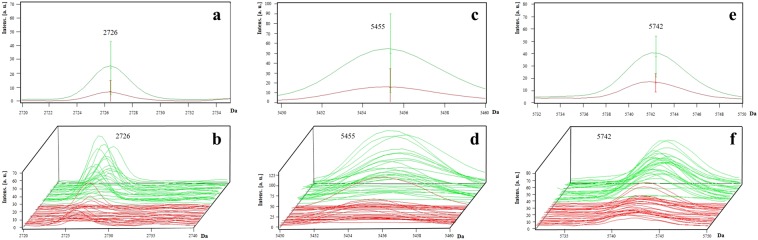
For P. aeruginosa isolates, three potential biomarkers were detected at peaks m/z 2,726 (AUC = 0.81; p < 0.001), 5,455 (AUC = 0.81; p < 0.001), and 5,742 (AUC = 0.84; p < 0.001), all of which predominated in the non-MDR isolates (Fig. 4). The presence of all three peaks increased the specificity to 74.1%, PPV to 75%, and NPV to 74.1% for the detection of non-MDR isolates (Table 1).
Figure 4.
Representative mass spectra of peaks m/z 2,726, 5,455 and 5,742 in P. aeruginosa. Three peaks m/z 2,726, 5,455, and 5,742 were present in the majority of non-MDR isolates (green line) and at higher intensities than those in MDR isolates (red line). The average spectrum of each group of peaks m/z 2,726 (a), 5,455 (c), and 5,742 (e) and the spectra of all the analysed isolates for peaks m/z 2,726 (b), 5,455 (d), and 5,742 (f) are shown.
No potential biomarkers were detected for the carbapenem-resistant K. pneumoniae isolates. However, the absence of peak m/z 6100 (AUC = 0.88) had a sensitivity of 93.8% and a NPV of 91.7% for the detection of carbapenem and colistin-resistant K. pneumoniae (Fig. 5) (Table 1).
Figure 5.
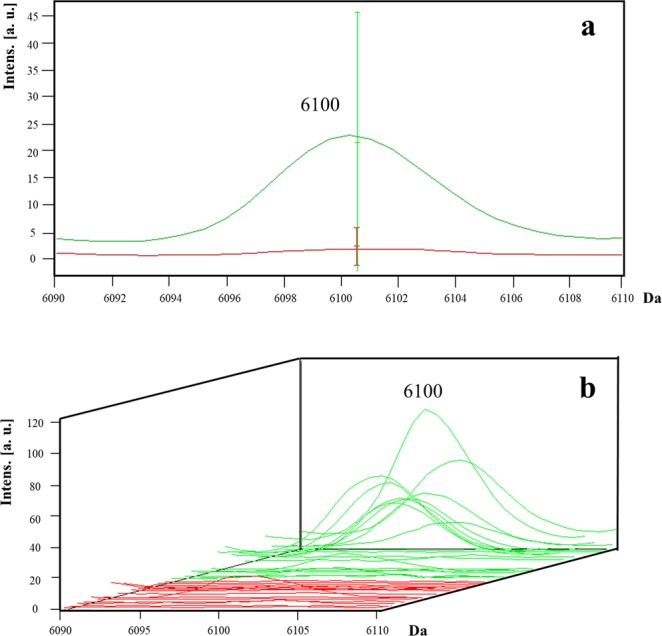
Representative mass spectra of peaks m/z 6,100 in K. pneumoniae. The peak m/z 6,100 is absent in the carbapenem- and colistin-resistant isolates (red line) and is present in the majority of the carbapenem- and colistin-susceptible isolates (green line). The average spectrum of each group of peak m/z 6,100 (a) and the spectra of all the analysed isolates (b) are shown.
Peptide/protein identity assignment
Assignment of possible peptide/protein identity of all the identified potential marker peaks is shown in Table 1. The m/z 4,594 peak of S. aureus was assigned to the 50 S ribosomal protein L28 with MASCOT score of 26 and protein sequence coverage of 64% Both the m/z 6,654 and 6,712 peaks of C. difficile were assigned to the 30 S ribosomal protein (S20 and S21, respectively), with 31–34 score and 67–98% protein sequence coverage. Both the m/z 6,304 and the m/z 6,332 peak of A. baumannii was assigned to a NADH-quinone oxidoreductase subunit K with a score of 29 and 56% of protein sequence coverage for each peak. In P. aeruginosa, the m/z 2,726 peak was assigned to a UPF0270 protein Pfl01_4103 with a score of 20 and 32% of protein sequence coverage, and the m/z 5,455 peak was assigned to a UPF0391 membrane protein Patl_1732 with a score of 32 and 98% of protein sequence coverage. The m/z 5,742 peak of P. aeruginosa and the m/z 6,100 peak of K. pneumoniae could not be determined.
Discussion
MALDI-TOF MS is now widely used in clinical microbiology for bacterial identification, taxonomy, and strain typing17. One application in development is the detection of antibiotic resistance in bacteria. Thus far, several approaches have been proposed in MALDI-TOF MS to detect antimicrobial resistance such as the detection of the entire cell profile, enzymatic activity by antibiotic hydrolysis, or resistance proteins within the cell18–21. In this study, we identified several potential biomarkers of drug-resistant genotypes in S. aureus, A. baumannii, P. aeruginosa, and K. pneumoniae, as well as hypervirulence in C. difficile, using a direct approach.
Regarding C. difficile, 2 out of 73 peaks reached statistical significance, wherein isolates were divided into 027 and non-027 ribotype groups. The peak m/z 6,654 was absent in 027 and 176 ribotypes, whereas the peak m/z 6,712 was present in both ribotypes. These findings complicate the differentiation of hypervirulent strains (027) from non-hypervirulent strains (176). However, ribotype 176 shares many similarities with 027, such as the presence of binary toxin genes and a single-base-pair deletion at nucleotide 117 in the gene that encodes a negative regulator of toxin production22, in such way that is actually difficult to differentiate both ribotypes in most methodologies23,24. Nevertheless, severe disease outcomes and mortality are associated with ribotype 17625 and in clinical epidemiology, hypervirulent strains should be notified24. In our work, 027 strains were differentiated from other non-027 strains, such as 001, 002, 003, 012, 014, 017, 019, 020, 076, 106, 220, and 353 ribotypes. It is highly likely (98% of protein sequence coverage) that 30S ribosomal protein was assigned to both peaks, which binds directly to 16S ribosomal RNA. In addition, both peaks had excellent AUC values (0.96–0.99), sensitivity, specificity, PPV, and NPV, all of which indicate a good performance of a biomarker.
MRSA is one of the most studied pathogens of the ESCAPE group regarding MALDI-TOF analysis15,19,21,26–28. A number of studies have used MALDI-TOF MS to discriminate clonal complexes of MRSA (CC5, CC8, CC22, CC30, and CC45)29. In the present study, the peak m/z 4,594 had a high specificity of 96.8% to detect MRSA isolates. The 50S ribosomal protein was most probably assigned to this peak. Different protein synthesis inhibitors, such as macrolides, ketolides, lincosamides, and B-type streptogramins, prevent the 50S particle from being formed in growing cells30. To our knowledge, this is the first report of a potential biomarker peak for MRSA isolates. In addition, a m/z 2,415 peak was detected in the MRSA isolates (not shown). This peak was previously reported in MRSA isolates with the class A mec gene complex16, though in our study the characterisation beyond the mecA gene detection was not performed.
Few reports have included the analysis of drug-resistant A. baumannii through MALDI-TOF MS, and although promising, their results were obtained using the carbapenem hydrolysis assay, not the direct approach15. In our study, two potential biomarker peaks were detected for MDR A. baumannii: m/z 6,304 and m/z 6,332. Both peaks had high AUC values (0.99) and were present in blaOXA-58- and blaOXA-24-positive isolates, respectively. A NADH-quinone oxidoreductase subunit was likely assigned to both peaks. Type II NADH-quinone oxidoreductases of Gram-negative bacteria can be inhibited by polymyxin B and colistin31. The performance of peak m/z 6,332 as a biomarker improved when comparing blaOXA-58-positive with blaOXA-58-negative and blaOXA-24 isolates. Our results indicate the discriminatory power of MALDI-TOF MS to differentiate blaOXA-58 from blaOXA-24 isolates, and this may be applied directly in clinical microbiology.
A signal loss of three peaks was detected for MDR P. aeruginosa isolates in comparison to non-MDR isolates. All three peaks had acceptable individual AUC values (0.81–0.84), and interestingly, the inclusion of all three peaks increased their potential as biomarker peaks for the detection of non-MDR isolates (sensitivity, 75%; specificity, 74.1%; PPV, 75%; and NPV, 74.1%). Two of the peaks were likely assigned to hypotheticals proteins, such as UPF0270 protein Pfl01_4103 (m/z 2,726 peak) and UPF0391 membrane protein Patl_1732 (m/z 5,455 peak). In case the identified peaks were proteins, the signal loss could be due to a mutation introducing a premature stop codon or a frame-shift in a gene. As it is, we cannot assume they are indeed associated to proteins and other explanations must be considered. Growth conditions and sample preparation may also contribute to signal loss. Therefore, loss of a signal does not alone give distinctive information about the strain genotype and cannot be used as a reliable biomarker peak29.
One potential biomarker for carbapenem and colistin resistance in K. pneumoniae was identified at peak m/z 6,100, which could be used to detect this phenotype, giving its NPV (91.7%). The use and the resistance to polymyxins have increased in carbapenemase-producing Enterobacteriaceae. A MALDI-TOF MS–based method that detects polymyxin resistance-related modifications to lipid A (i.e., phosphoethanolamine addition) was previously developed in E. coli. This methodology can be performed in 15 min and simultaneously discriminates between chromosome- and plasmid-encoded resistance32,33. This methodology has an advantage over our approach, both in specificity and processing time.
In addition, we did not assess the frequency of the colistin resistance-associated mcr gene in our Enterobacteriaceae isolates. Our study population contained the blaNDM gene only in the carbapenem-resistant K. pneumoniae isolates. Because the blaKPC K. pneumoniae genotype has been previously identified by MALDI-TOF34, future studies should analyse a higher number of isolates and a wider diversity of drug resistance–associated genes of K. pneumoniae.
Previous studies have compared VRE mass spectra among vanA or vanB positive and negative groups, specifically on E. faecium isolates15,21. A number of peaks were detected among vanA-positive isolates35,36 as well as vanB-positive isolates37. In our study, we detected 70 potential peaks with significant difference (p ≤ 0.05), but no final potential biomarkers were detected. Failure to identify lineage-specific biomarker peaks in Gram-positive pathogens such as E. faecium has been reported previously38.
Changes in the distribution of peaks among strains suggest a peak shift, possibly corresponding to a slight protein weight modification due to an amino acid substitution after gene mutations. This shift can be used as a trustable biomarker to identify the strain genotype (either related to virulence or drug resistance), which was the approach used in this study29. According to our approach, resistance or virulence biomarker peaks can be identified while performing routine microbiology analysis, and no additional assays nor prolonged incubation time is needed.
The primary limitation of this study was the lack of validation studies. Predictive biomarkers for MALDI-TOF should be first identified with a training set of data and afterward should be validated on an independent set of data. This validation set should include isolates with geographic, temporal, and clonal diversity to ensure the validity of the peak biomarkers29. In addition, outstanding potential biomarker peaks detected in our study should be further analysed by additional methods to identify which proteins/peptide each peak corresponds to.
In conclusion, we identified several potential biomarker peaks suggestive of drug resistance in S. aureus, A. baumannii, P. aeruginosa, and K. pneumoniae, as well as hypervirulence in C. difficile using MALDI-TOF MS.
Methods
Clinical isolates
Selected ESCAPE clinical isolates obtained between January 2007 and December 2017 from the Hospital Civil de Guadalajara “Fray Antonio Alcalde” (Jalisco, Mexico) were included. The VITEK 2 system (Biomérieux, Craponne, France) was used according to the manufacturer’s instructions, and either identification card for Gram positive or negative were used for identification. Bacteria were grown on plates containing Trypticase soy agar with 5% sheep blood and incubated overnight at 35 °C. A suspension of bacteria from pure cultures was performed in a 0.45% sodium chloride solution. This suspension was adjusted to a McFarland standard of 0.5 and along with the ID card, introduced into the system, which automatically inoculated the card and incubated at 35 °C until the end of the test.
A. baumannii species identification was confirmed by polymerase chain reaction (PCR). Primers and PCR conditions were used as described previously39,40. First, rapid DNA extraction was performed, in which three to five bacterial colonies were suspended in 100 µl of sterile distilled water and heated at 95 °C for 15 min. After centrifugation at 13,500 rpm for 5 min, the supernatant was collected. A pair of primers, forward (F): 5′-CATTATCACGGTAATTAGTG-3′ and reverse (R): 5′-AGAGCACTGTGCACTTAAG-3′, was used for the amplification of an internal fragment from the 16S-23S rRNA intergenic spacer region of A. baumannii. A second pair of primers, F5′-CCTGAATCTTCTGGTAAAAC-3′ and R5′-GTTTCTGGGCTGCCAAACATTAC-3′, were used for the amplification of a highly conserved region of the recA gene of Acinetobacter genus. The reaction mixture contained 1X PCR buffer, 2 mM MgCl2, 0.2 mM of each dNTP, 200 nM of each primer, 1 U of AmpliTaq polymerase and 2 µl of DNA. PCR was initiated by denaturation for 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 55 °C and 30 s at 72 °C with final extension for 7 min at 72 °C and a holding step at 4 °C until analysis. Next, 5 µl of the PCR products were electrophoresed on a 2.0% agarose gel prepared with 0.5x TBE buffer for 1 h at 130 V. The gel was stained with 10 µM ethidium bromide, and the PCR product bands were visualized using a UV transilluminator. Expected PCR products for the 16S-23S rRNA intergenic spacer region and recA gene were 208 and 425 bp, respectively.
Selected clinical isolates included E. faecium (n = 64), S. aureus (n = 67), C. difficile (n = 93), A. baumannii (n = 68), P. aeruginosa (n = 55), and K. pneumoniae isolates (n = 69).
Antimicrobial susceptibility and phenotypic tests
Antimicrobial susceptibility was determined using the broth microdilution method according to the 2019 Clinical and Laboratory Standards Institute41. The following strains were evaluated for resistance to the corresponding drugs: E. faecium, vancomycin; S. aureus, cefoxitin and oxacillin; A. baumannii, imipenem and meropenem; P. aeruginosa, gentamicin, cefepime, ciprofloxacin, imipenem, and meropenem; K. pneumoniae, ertapenem, imipenem, meropenem and colistin. Strains were classified as MDR as described elsewhere, i.e. resistant to three or more antimicrobial classes42.
Genotyping
E. faecium isolates were screened as previously described for the presence of vanA and vanB genes after rapid DNA extraction. Primers F5′-CATGAATAGAATAAAAGTTGCAATA-3′ and R5′-CCCCTTTAACGCTAATACGATCAA-3′ were used for the amplification of vanA gene (1,030 bp); and F5′-GTGACAAACCGGAGGCGAGGA-3′ and R5′-CCOGCCATCCTCCTGCAAAAAA-3′, for the vanB gene (433 bp)43. PCR conditions were 10 min at 95 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 58 °C and 30 s at 72 °C with final extension for 10 min at 72 °C.
The mecA gene (147 bp) was screened in all S. aureus isolates, using primers F5′- GTGAAGATATACCAAGTGATT-3′ and R5′- ATGCGCTATAGATTGAAAGGAT-3′44. PCR conditions were 5 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 1 min at 50 °C and 2 min at 72 °C with final extension for 10 min at 72 °C.
A. baumannii isolates were screened for class D blaOXA carbapenemases, including blaOXA-23 (F5′-GATCGGATTGGAGAACCAGA-3′ and R5′-ATTTCTGACCGCATTTCCAT-3′, 501 bp), blaOXA-24 (F5′-GGTTAGTTGGCCCCCTTAAA-3′ and R5′-AGTTGAGCGAAAAGGGGATT-3′, 246 bp), blaOXA-51 (F5′-TAATGCTTTGATCGGCCTTG-3′ and R5′-TGGATTGCACTTCATCTTGG-3′, 353 bp), and blaOXA-58 (F5′-AAGTATTGGGGCTTGTGCTG-3′ and R5′-CCCCTCTGCGCTCTACATAC-3′, 599 bp)45, and metallo-β-lactamases genes including blaVIM (F5′-ATGGTGTTTGGTCGCATATC-′3 and R5′-TGGGCCATTCAGCCAGATC-′3, 510 bp)46, blaIMP (F5′-GGAATAGAGTGGCTTAAYTCTC-′3 and R5′-CCAAACYACTASGTTATCT-′3, 188 bp)47, and blaNDM (F5′-GGAAACTGGCGACCAACG-3′ and R5′-ATGCGGGCCGTATGAGTGA-3′, 678 bp)48. P. aeruginosa isolates were screened for blaVIM, and blaIMP genes. K. pneumoniae isolates were screened for blaKPC (F5′-GCAGCGGCAGCAGTTTGTTGATT-3′ and R5′-GTAGACGGCCAACACAATAGGTGC-3′, 184 bp)49, blaVIM, blaIMP, blaNDM, and blaOXA-48 (F5′-TTCGGCCACGGAGCAAATCAG-3′ and R5′-GATGTGGGCATATCCATATTCATCGCA-3′, 240 bp)49. Conditions for multiplex PCR (blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58) were 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 55 °C and 30 s at 72 °C, and 6 min at 72 °C. For blaNDM and blaVIM, PCR conditions were 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 60 s at 60 °C and 45 s at 72 °C, and 5 min at 72 °C. For blaIMP, PCR conditions were 5 min at 95 °C, 30 cycles of 30 s at 95 °C, 40 s at 52 °C and 50 s at 72 °C, and 5 min at 72 °C. For multiplex PCR blaOXA-48 and blaKPC, conditions were 5 min at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 60 °C and 30 s at 72 °C, and 5 min at 72 °C.
Ribotyping of C. difficile isolates included the amplification of the 16S-23S rRNA intergenic spacer region using primers 5′-GTGCGGCTGGATCACCTCCT-3′ and 5′-CCCTGCACCCTTAATAACTTGACC-3′50. PCR conditions were 95 °C for 15 min, followed by 24 cycles at 95 °C for 1 min, 57 °C for 1 min and 72 °C for 1 min, and 72 °C for 30 min. Capillary electrophoresis was performed at the C. difficile Ribotyping Network Reference Laboratory at Leeds Teaching Hospitals Trust (Leeds, UK).
Mass spectra analysis
All selected strains were prepared for MALDI-TOF MS (Microflex LT system, Bruker Daltonics, Bremen, Germany) according to the manufacturer’s recommendation. For the direct colony method, an overnight culture grown on blood agar at 37 °C under aerobic conditions of each bacterial strain or from a culture grown on C. difficile agar (Neogen Corporation, MI) in anaerobic conditions for up to 24 h, were included. One colony of each bacteria was applied using sterile wooden toothpicks on a 96-spot stainless steel target plate (Bruker Daltonics, Bremen, Germany). After drying, 1 μl 70% formic acid was added and air-dried prior to adding 1 μl of a matrix solution, which was composed of a saturated solution of α-cyano-4-hydroxycinnamic acid 10 mg/mL in 50% acetonitrile (Sigma-Aldrich, Toluca, Mexico) and 2.5% trifluoroacetic acid (Sigma-Aldrich), which enables measurement of peptides and proteins from 0.7 to 20 kDa. Finally, spots were allowed to dry completely, and the target plate was introduced into the equipment. Samples were analysed by MALDI Biotyper 3.0 software for spectra profile match in the database.
All the isolates were classified according to manufacturer’s recommended score identification criteria. A score of 2.0 to 3.0 indicated reliable species-level identification, a score of 1.7 to 1.9 indicated reliable genus identification but questionable species-level identification, and a score of <1.7 indicated an unreliable identification.
Potential marker peaks
The MS spectrum of each spot was analysed by the MALDI Biotyper V.3.1.66 with the most updated spectra library, V.7.0 (7,311 spectra). High-quality spectra were captured using the flexControl V.3.4 software (Bruker Daltonics). These spectra were then imported into the ClinProTools V.3.0 software (Bruker Daltonics) for recognition of mass spectra patterns between groups and preprocessed using the default parameters. Spectra were smoothed with 10 cycles of the Savitzky/Golay algorithm for 10 cycles with a width of 2 m/z. Baseline subtraction was performed with the Top Hat algorithm. Peak picking was performed on the average spectrum from each group, with a signal to noise threshold of 5. Peak selection was performed using the P-value T-test/ANOVA sort mode.
Group selection was based on the presence or absence of drug-resistance genotypes, except for C. difficile in which hypervirulent ribotypes determination was considered.
Statistical analyses
The selection of potential biomarker peaks of drug-resistant phenotypes was performed using statistical analyses within the algorithms included in the ClinProTools software. Classification models were generated using the genetic algorithm (GA), supervised neural network (SNN), and quick classifier (QC) algorithms. The parameters were maximum of 5 peaks, maximum of 50 generations, k-nearest neighbor classification with 3 neighbors for GA; 1–25 peaks automatically detected for SNN, and QC (automatic peak detection 1–25 peaks, p-value T-test/ANOVA). The recognition capability and cross validation values were calculated to demonstrate the reliability and accuracy of the model.
The Anderson-Darling test was used to determine the distribution of the population (≤0.05: not normally distributed, >0.05, normally distributed). According to the results, either the t-test (normally distributed data) or the Wilcoxon test (not normally distributed data) were used to confirm significant differences between two classes (on the presence or absence of drug-resistance or virulence phenotype/genotypes). As the p-value provides a measure of the strength of an association, the lower it is, the better the respective peak signal can be used to discriminate the groups. If the p-value for the t-test or the Wilcoxon test was ≤0.05, the peak was confirmed to be significantly different.
Peaks that obtained statistical significance were selected for further analysis, which included evaluating the peak area/intensity of the spectra and the coefficient of variation of each peak; this analysis allowed the selection of potential peaks. ClinProTools also calculates a Receiver Operating Characteristic (ROC) curve for each peak. The ROC curve provides an evaluation of the discrimination quality of a peak. In addition, the area under the curve (AUC) was determined and only peaks with AUC ≥ 0.80 values were selected. The principal component analysis (PCA) was used to evaluate the distinguishing capability between drug-resistance (or virulence) phenotype/genotype profiles in the isolates based on their peptide mass fingerprints.
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for the potential biomarker peaks.
Peptide/protein identity assignment
MS spectra were first processed using flexAnalysis software (Bruker Daltonics) in which top-hat baseline subtraction and spectra smoothing were performed. The spectra were then exported to the Biotools software (Bruker Daltonics), in which protein/peptide identification was performed by latter submitting the data to the Mascot Server (Matrix Science, Boston, USA) and run against the SwissProt database for peptide/protein assignment.
Supplementary information
Author contributions
S.F.T. and E.G.G. contributed equally to this work. S.F.T., E.G.G., S.M.O. and P.B.I.: conceived and designed the experiments; P.B.I. and S.M.O.: performed the experiments; S.F.T., and P.B.I. performed the statistical analysis; S.M.O., A.M.M., R.M.O., A.C.O., and E.R.N. revised the data; S.F.T., E.G.G. and P.B.I. wrote the paper, E.G.G. and P.B.I. supervised the project; and E.G.G. and R.M.O.: provided grants, samples and reagents.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Samantha Flores-Treviño and Elvira Garza-González.
Supplementary information
is available for this paper at 10.1038/s41598-019-55430-1.
References
- 1.Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, Available from, http://www.cdc.gov/drugresistance/threat-report-2013/, Accesed on October 28, 2019 (2013).
- 2.Weiner LM, et al. Vital Signs: Preventing Antibiotic-Resistant Infections in Hospitals - United States, 2014. MMWR Morb Mortal Wkly Rep. 2016;65:235–241. doi: 10.15585/mmwr.mm6509e1. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol. 2010;31(Suppl 1):S7–10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 5.Llaca-Diaz JM, Mendoza-Olazaran S, Camacho-Ortiz A, Flores S, Garza-Gonzalez E. One-year surveillance of ESKAPE pathogens in an intensive care unit of Monterrey, Mexico. Chemotherapy. 2012;58:475–481. doi: 10.1159/000346352. [DOI] [PubMed] [Google Scholar]
- 6.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 7.Camacho-Ortiz A, et al. First report of Clostridium difficile NAP1/027 in a Mexican hospital. PLoS One. 2015;10:e0122627. doi: 10.1371/journal.pone.0122627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morfin-Otero R, et al. Clostridium difficile outbreak caused by NAP1/BI/027 strain and non-027 strains in a Mexican hospital. Braz J Infect Dis. 2016;20:8–13. doi: 10.1016/j.bjid.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davila LP, et al. Increasing rates of Clostridium difficile infection in Mexican hospitals. Braz J Infect Dis. 2017;21:530–534. doi: 10.1016/j.bjid.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rosa FG, Corcione S, Pagani N, Di Perri G. From ESKAPE to ESCAPE, from KPC to CCC. Clin Infect Dis. 2015;60:1289–1290. doi: 10.1093/cid/ciu1170. [DOI] [PubMed] [Google Scholar]
- 11.Peterson LR. Bad bugs, no drugs: no ESCAPE revisited. Clin Infect Dis. 2009;49:992–993. doi: 10.1086/605539. [DOI] [PubMed] [Google Scholar]
- 12.Manca C, et al. Leading Antibacterial Laboratory Research by Integrating Conventional and Innovative Approaches: The Laboratory Center of the Antibacterial Resistance Leadership Group. Clin Infect Dis. 2017;64:S13–S17. doi: 10.1093/cid/ciw826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect. 2010;16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 14.Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burckhardt I, Zimmermann S. Susceptibility Testing of Bacteria Using Maldi-Tof Mass Spectrometry. Front Microbiol. 2018;9:1744. doi: 10.3389/fmicb.2018.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhoads DD, Wang H, Karichu J, Richter SS. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn Microbiol Infect Dis. 2016;86:257–261. doi: 10.1016/j.diagmicrobio.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Llarena FJ, Bou G. Proteomics As a Tool for Studying Bacterial Virulence and Antimicrobial Resistance. Front Microbiol. 2016;7:410. doi: 10.3389/fmicb.2016.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrabak J, Chudackova E, Walkova R. Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev. 2013;26:103–114. doi: 10.1128/CMR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostrzewa M, Sparbier K, Maier T, Schubert S. MALDI-TOF MS: an upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteomics Clin Appl. 2013;7:767–778. doi: 10.1002/prca.201300042. [DOI] [PubMed] [Google Scholar]
- 21.Vrioni G, et al. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: current achievements and future perspectives. Ann Transl Med. 2018;6:240. doi: 10.21037/atm.2018.06.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyc O, Pituch H, Matejkova J, Obuch-Woszczatynski P, Kuijper EJ. Clostridium difficile PCR ribotype 176 in the Czech Republic and Poland. Lancet. 2011;377:1407. doi: 10.1016/S0140-6736(11)60575-8. [DOI] [PubMed] [Google Scholar]
- 23.Krutova M, Matejkova J, Nyc O. C. difficile ribotype 027 or 176? Folia Microbiol (Praha) 2014;59:523–526. doi: 10.1007/s12223-014-0323-5. [DOI] [PubMed] [Google Scholar]
- 24.Mentula S, Laakso S, Lyytikainen O, Kirveskari J. Differentiating virulent 027 and non-027 Clostridium difficile strains by molecular methods. Expert Rev Mol Diagn. 2015;15:1225–1229. doi: 10.1586/14737159.2015.1069710. [DOI] [PubMed] [Google Scholar]
- 25.Polivkova S, Krutova M, Petrlova K, Benes J, Nyc O. Clostridium difficile ribotype 176 - A predictor for high mortality and risk of nosocomial spread? Anaerobe. 2016;40:35–40. doi: 10.1016/j.anaerobe.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Wang HY, et al. A new scheme for strain typing of methicillin-resistant Staphylococcus aureus on the basis of matrix-assisted laser desorption ionization time-of-flight mass spectrometry by using machine learning approach. PLoS One. 2018;13:e0194289. doi: 10.1371/journal.pone.0194289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, et al. Analysis of methicillin-resistant Staphylococcus aureus major clonal lineages by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) J Microbiol Methods. 2015;117:122–127. doi: 10.1016/j.mimet.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Camoez M, et al. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2016;22:161 e161–161 e167. doi: 10.1016/j.cmi.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Sauget M, Valot B, Bertrand X, Hocquet D. Can MALDI-TOF Mass Spectrometry Reasonably Type Bacteria? Trends Microbiol. 2017;25:447–455. doi: 10.1016/j.tim.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Champney WS. Bacterial ribosomal subunit synthesis: a novel antibiotic target. Curr Drug Targets Infect Disord. 2001;1:19–36. doi: 10.2174/1568005013343281. [DOI] [PubMed] [Google Scholar]
- 31.Deris ZZ, et al. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J Antibiot (Tokyo) 2014;67:147–151. doi: 10.1038/ja.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dortet L, et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J Antimicrob Chemother. 2018;73:3359–3367. doi: 10.1093/jac/dky330. [DOI] [PubMed] [Google Scholar]
- 33.Oviano, M. & Bou, G. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for the Rapid Detection of Antimicrobial Resistance Mechanisms and Beyond. Clin Microbiol Rev32, 10.1128/CMR.00037-18 (2019). [DOI] [PMC free article] [PubMed]
- 34.Angeletti S, et al. MALDI-TOF mass spectrometry and blakpc gene phylogenetic analysis of an outbreak of carbapenem-resistant K. pneumoniae strains. New Microbiol. 2015;38:541–550. [PubMed] [Google Scholar]
- 35.Nakano S, et al. Differentiation of vanA-positive Enterococcus faecium from vanA-negative E. faecium by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Int J Antimicrob Agents. 2014;44:256–259. doi: 10.1016/j.ijantimicag.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Wang LJ, Lu XX, Wu W, Sui WJ, Zhang G. Application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry in the screening of vanA-positive Enterococcus faecium. Eur J Mass Spectrom (Chichester) 2014;20:461–465. doi: 10.1255/ejms.1298. [DOI] [PubMed] [Google Scholar]
- 37.Griffin PM, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J Clin Microbiol. 2012;50:2918–2931. doi: 10.1128/JCM.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasch P, et al. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J Microbiol Methods. 2014;100:58–69. doi: 10.1016/j.mimet.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Bocanegra-Ibarias P, et al. Genetic characterisation of drug resistance and clonal dynamics of Acinetobacter baumannii in a hospital setting in Mexico. Int J Antimicrob Agents. 2015;45:309–313. doi: 10.1016/j.ijantimicag.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 40.Chen TL, et al. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13:801–806. doi: 10.1111/j.1469-0691.2007.01744.x. [DOI] [PubMed] [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement M100-S29 (2019).
- 42.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 43.Bocanegra-Ibarias P, et al. Phenotypic and genotypic characterization of vancomycin-resistant Enterococcus faecium clinical isolates from two hospitals in Mexico: First detection of VanB phenotype-vanA genotype. Enferm Infecc Microbiol Clin. 2016;34:415–421. doi: 10.1016/j.eimc.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodford N, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Nordmann P, Poirel L. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect. 2002;8:321–331. doi: 10.1046/j.1469-0691.2002.00401.x. [DOI] [PubMed] [Google Scholar]
- 47.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 48.Baroud M, et al. Underlying mechanisms of carbapenem resistance in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates at a tertiary care centre in Lebanon: role of OXA-48 and NDM-1 carbapenemases. Int J Antimicrob Agents. 2013;41:75–79. doi: 10.1016/j.ijantimicag.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Swayne RL, et al. for rapid detection of genes encoding five types of non-metallo- (class A and D) carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents. 2011;38:35–38. doi: 10.1016/j.ijantimicag.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS microbiology letters. 1999;175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.