Abstract
The sustained generation of genomic data in the last decade has increased the knowledge on the causal mutations of a large number of diseases, especially for highly penetrant Mendelian diseases, typically caused by a unique or a few genes. However, the discovery of causal genes in complex diseases has been far less successful. Many complex diseases are actually a consequence of the failure of complex biological modules, composed by interrelated proteins, which can happen in many different ways, which conferring a multigenic nature to the condition that can hardly be attributed to one or a few genes. We present a mechanistic model, Hipathia, implemented in a web server that allows estimating the effect that mutations, or changes in the expression of genes, have over the whole system of human signaling and the corresponding functional consequences. We show several use cases where we demonstrate how different the ultimate impact of mutations with similar loss-of-function potential can be and how the potential pathological role of a damaged gene can be inferred within the context of a signaling network. The use of systems biology-based approaches, such as mechanistic models, allows estimating the potential impact of loss-of-function mutations occurring in proteins that are part of complex biological interaction networks, such as signaling pathways. This holistic approach provides an elegant alternative to gene-centric approaches that can open new avenues in the interpretation of the genomic variability in complex diseases.
Subject terms: Cellular signalling networks, Systems analysis
Introduction
The extraordinarily fast increase in throughput of sequencing technologies in the last decade1,2 has fostered different international collaborative projects3–5 that resulted in an unprecedented increase in our knowledge of the mutational spectrum of diseases, especially thosewith significant morbidity and mortality and caused by highly penetrant (typically protein-coding) variants6,7. However, in addition to the expected pathogenic variation, these projects have revealed an unanticipated amount of variation at genome level in apparently normal, healthy individuals. Actually, putative loss-of-function (pLoF) variants, with a potential severe effect on the function of human protein-coding genes8 seems to be surprisingly pervasive, according to reports from different genome sequencing projects3,4,9. Conservative estimates suggest that there are more than 250 pLoF variants predicted to be highly damaging10 per sequenced genome, in protein coding regions8, as well as other non-coding regions, such as miRNAs11, transcription factor binding sites12 and others13. Therefore, a better understanding on the contribution of pLoF to disease is critical for clinical applications of genomic data14.
From a historical perspective, the application of sequential heuristic filters, in a process called prioritization, has demonstrated to be a useful tool for the clinical interpretation of genomic variation in rare Mendelian diseases6,7. Thus, extensively used filtering criteria are: (i) the potential impact of the variant in the resulting gene product, estimated by different indexes that predict the potential pathologic effect of an amino acid substitution (e.g. Polyphen15, SIFT16, SNPeffect17, PMUT18, etc.) that can be combined with allelic frequency, conservation, etc. (e.g. PROVEAN19, PupaSNP20, CONDEL21, VAAST22, MutationTaster23, etc.); (ii) variant population frequencies, given that variants with a relatively high frequency in the population are unlikely to be causative of many hereditary disorders (obtained from different repositories such as the 1000 genomes3, the Exome Aggregation Consortium24, the gnomAD25, or also from local population repositories, which have demonstrated to be useful for this purpose26); (iii) evolutionary conservation (e.g. PhyloP27, GERP28, etc.); (iv) compendiums of different criteria, such as CADD29 or, more recently, based on artificial intelligence30. These filters can also be used in combination with knowledge on functional labels31, syndromes and phenotypes32, or diseases33–35, previously associated to the most likely candidate genes, as implemented in tools such as Phen-Gen36, eXtasy37, PhenIX38, Exomiser39, etc. Different computer applications, such as Annovar40, the Variant Effect Predictor41 or the CellBase42, collect all this information that is subsequently used by different web interfaces that allow carry out this prioritization interactively, likeOVA43, BiERapp44, QueryOR45 or Mutation Distiller46, etc.
Despite success of gene centric clinical interpretation of variation in finding disease genes in single gene Mendelian disorders7,47,48, the application of these concepts to complex diseases has produced more modest results49. Contrarily to the case of RDs, complex diseases are characterized by phenotypic heterogeneity, that is, patients with similar presentations often have different underlying disease mechanisms, and incomplete penetrance, as a consequence of its multigenic nature and the significant role of the environment50–52. However, the most widely used tools for the interpretation of genetic variation all focus on monogenic genetic models or, on oligogenic ones as much.
As a matter of fact, complex, multigenic diseases can be better understood as failures of functional modules caused by different combinations of perturbed gene activities rather than by the failure of a unique gene53. Actually, the idea of cell functionality as a result of the complex interactions between their molecular components is not new54 and was proposed almost two decades ago in the context of systems biology55. These interacting components define operational entities or modules to which different elementary functions can be attributed. This modularity, extensively described in numerous reports56,57, suggests that causative genes for the same disease often reside in the same biological module, which can be a protein complex or any type of biological network58,59. Currently, a detailed recapitulation of the knowledge on biological networks that account for cell functionality, metabolism and other cell processes is available in different pathway repositories such as the Kyoto Encyclopedia of Genes and Genomes (KEGG)60, Reactome61, Pathway Commons62, Wikipathways63 and others, including pathways with specific, curated descriptions of disease mechanisms64. Since pathways describe how proteins interact among them to trigger cell functionalities or to generate different molecules and metabolites, mathematical models of the activity of such pathways constitute ultimately mechanistic descriptors of the behavior of the cell. In fact, recent reports demonstrate that mechanistic models of the activity of metabolic or signaling pathways, render highly precise predictions of complex phenotypes, such as patient survival65,66, drug response67, etc.
An interesting property of mechanistic models is that, in addition to study molecular mechanisms of disease in a particular condition, they can be used to predict the potential consequences that perturbations (mutations or changes in the expression) of the proteins that compose the pathway can have over the individual circuits that trigger cell actions or the production of metabolites68,69. This mechanistic view of the effects of a change in the integrity or the activity of one or several proteins within the context of signaling66 or metabolic70 pathways can be used to understand the functional consequences of pLoF mutations and/or gene expression perturbations, thus providing a clinical interpretation for these variations in complex scenarios that takes into account the whole context of the impact of the perturbation from a functional angle.
Here we present a web server that implements a version of the Hipathia66, an algorithm that has recently been shown that outperforms other current competing algorithms71, and demonstrate the advantages of mechanistic models in the interpretation of complex variability in two scenarios of different complexity: a rare disease, Fanconi anemia (FA) and a common disease, diabetes.
Implementation
Overview
The general idea for the interpretation of complex variability is based on the use of an algorithm that uses gene expression (as a proxy of the presence of the corresponding protein within a pathway) to model the activity of signaling circuits defined within signaling pathways, which ultimately provide a hint on functional cell activity. A functional assessment of the differences in functional cell behavior between two conditions can be achieved by comparing the corresponding signaling circuit activity profiles to detect what circuits behave differently. Since the model only requires gene expression and the topology of the signaling circuits, it is easy to simulate a “mutated” condition just by reducing or setting to zero the expression of one or several genes (in the assumption that the effect in signal transduction of an inactive protein is equivalent to its absence) and constructing an artificial gene expression profile for the simulated mutated condition. Then, the original condition can be compared to the simulated condition to detect the circuits and the corresponding functions affected by the LoF of the genes.
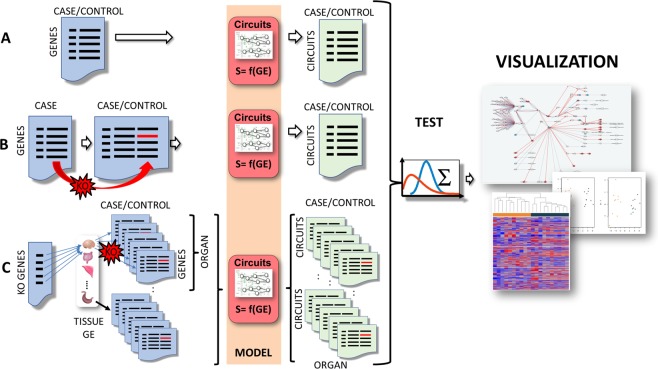
Figure 1 shows the schema of the three analysis scenarios implemented in the Hipathia web server. The simplest scenario, Differential Signaling, (Fig. 1A) represents a conventional transcriptomics case/control study that can be transformed into a differential signaling circuit activity contrast. Gene expression values are used to define circuit activities (see below for details) and a Wilcoxon test is used to detect circuits that are differentially activated between cases and controls. In the second scenario, the Perturbation effect, the effect of a LoF mutation is interactively simulated in one or several genes (Fig. 1B). To carry out this simulation the gene expression corresponding to a specific condition is uploaded. The graphical interface allows the user to select one or several genes and decide the mutation effect by modulating the expression value of the selected genes (setting the value to 0 would simulate a total loss of function mutation and raising the value would simulate an over expression). Then, a fold change between the original and the simulated conditions is used to detect effects of the perturbation over the different circuits. The third scenario, Variant interpreter, (Fig. 1C) allows estimating the potential effect of a list of mutations over different human tissues. To achieve so, the GTEx gene expression matrices corresponding to 30 different tissues (see Additional Table 1), containing each a variable number of individuals, are used as controls and then, equivalent matrices of cases are generated by simulating the mutations as previously described on all the individuals. Then a case/control contrast with a Wilcoxon test is carried out for each tissue, which would reveal whether some of the mutations in the list have a significant impact on one or several tissues and the functional nature of such impact.
Figure 1.
Schematic representation of the three analysis scenarios implemented in the Hipathia web server. (A) Conventional transcriptomics case/control study transformed by Hipathia into a differential signaling circuit activity contrast. (B) Interactive simulation of the effect of a mutation over the signaling circuits. (C) Estimation of the potential effect of a list of mutations over the different signaling circuits in a number of selected human tissues. Icons for organs were taken from the Reactome Icon Library111
The web interface
The input of the program consists of normalized gene expression matrices in CSV format for the two first options of Differential signaling activity and the Perturbation effect (Fig. 1A,B) and also optionally for the Variant interpreter option that explores the effect of mutations across tissues (Fig. 1C), as user defined tissue. Expression may have been measured with any sequencing or microarray technology. The gene expression matrix must include samples as columns and genes as rows. Gene names must be Entrez or HUGO IDs.
For the Variant Interpreter option, a list of Entrez or HUGO gene names can be provided.
Graphical representation of the results
Different analysis types are carried out on the circuit’s activities calculated, which include two-class comparisons and PCA, with the corresponding visualizations as heatmaps and PCA plots. Graphical representation of the circuits significantly up- or down-activated, including the individual node expression change, are also provided (see Fig. 1 right). An interactive graphical output in which the pathways analyzed are displayed with the possible ways in which the signal can be transmitted from receptor proteins to the corresponding effector proteins, highlighting those in which significant changes in signaling are found. In this visual representation, disruptions or activations in the signal transduction caused by gene perturbations (mutations or expression changes) can be easily visualized and understood in terms of their consequences on cell signaling and their ultimate effect over the corresponding functions triggered by the effectors.
The client of the web application has been implemented in JavaScript using the HTML5 and SVG standards and uses CellMaps72 libraries for interactive visual representation of pathways.
Mechanistic model of cell functionality triggered by signaling
The Hipathia (acronym for High-throughput pathway interpretation and analysis) is a mechanistic model of signaling circuit activities previously described66. In brief, circuits that connect receptor proteins to specific effector proteins, which ultimately trigger cell activities, are defined using KEGG pathways60. Such circuits represent the sequence of activation (and inhibition) steps that mediates the transduction of the signal from the receptor to the effector protein. The method assumptions are that, in order to transduce the signal, all the proteins that connect the receptor with the effector should be present and the higher the amount of these proteins the stronger will be the signal. Measurements of mRNA levels are taken as proxies of the amount of the corresponding proteins (a quite common assumption73–78). Then, in order to quantify the intensity of signal transduction, the following steps are taken: normalized gene expression values, rescaled to a value in the range [0,1], obtained as explained above, are used as proxies of the protein activities (activations or inhibitions in the transmission chain)73,75,79. Thus, the intensity value of signal transduced along a circuit that reaches the effector is estimated by starting with initial signal intensity with the maximum value of 1 in the receptor, which is propagated along the nodes of the signaling circuits according the recursive formula:
| 1 |
where Sn is the signal intensity for the current node n, vn is its normalized gene expression value, A is the set of activation signals (sa), arriving to the node n from the corresponding activation edges, I is the set of inhibitory signals (si) arriving to the node from inhibition edges66. Like normalized gene expression values, circuit activity values are measurements with no absolute meaning by themselves but rather in a comparison.
The application of this formula to all the circuits defined in all the pathways allows transforming a gene expression profile into the corresponding signaling circuit activity profile for any sample studied. If two conditions are compared, a Wilcoxon test can used to assess differences in signaling circuit activity between both types of samples.
Estimation of the impact of a mutation over cell functionality
The effect of a mutation is dependent on the context which includes the activity (gene expression status) and the integrity (mutational status) of the rest of proteins involved in the pathways that trigger functionalities relevant to the disease analyzed (disease hallmarks). The effect of one or several simultaneous mutations in a specific tissue can easily be predicted using the mechanistic model68,69. The reference or control dataset is taken from the tissue of interest in GTEx80. Then, an affected dataset is simulated from the control dataset by drastically reducing the expression of the gene(s) with a pLoF mutation by multiplying their expression values by 0.01 in all the control samples. This simulates either an inactive gene or a non-functional gene product. Then, the circuit activities are recalculated in the affected dataset and it is compared to the reference dataset. Although not completely realistic, given that the model does not have information on the way in which the diseased tissue will transcriptionally react to the perturbation induced by the mutated genes, the results will certainly point with precision to those cell functions affected in first instance.
Data Sources
In the current version of HiPathia more than 8000 circuits have been identified and modeled within a total of more than 150 pathways downloaded from KEGG60 corresponding to three species (human 145, mouse 141 and rat 141).
Gene expression data from 30 non-diseased tissue sites (See Additional Table 1) used in the third option were taken from the GTEx Portal80 (GTEx Analysis V7; dbGaP Accession phs000424.v7.p2).
Data and methods for the examples
Gene expression for bone marrow, which is not present in GTEx, was downloaded from the Gene Expression Omnibus (GEO) database (GSE16334)81.
Gene expression microarray study that compares human islets gene expression from 54 non-diabetic and 9 type 2 diabetic donors82 was downloaded from GEO (GSE38642).
Data on natural variability of different populations, which comprises over 88 million variants of 2,504 individuals from 26 populations, was obtained from the 1000 Genomes project portal3,83.
In order to assess the impact of the natural variation found in genes of healthy population, variants located within gene regions were annotated using CADD29. As proposed by CADD developers, a gene was considered to carry a pLoF mutation when the CADD score is over the threshold of 2084. A gene is considered to be affected by pLoF in a recessive scenario, when the two alternative alleles are present.
Transcriptomics data processing
Gene expression data from microarrays were summarized and normalized by quantiles with the Robust Multiarray Analysis method using affy R package85. Probes were mapped to the corresponding genes using BiomaRt86. Gene expression values are estimated as the 90 percentile of probe expression values. Probes that mapped in more than one gene were discarded (except in the case that they were the unique probes mapping on the gene, that the median value of intensities was taken.)
RNA-seq gene expression data were normalized with the Trimmed mean of M values (TMM) normalization method using the edgeR package87.
Then, the Hipathia66 algorithm requires some extra steps for the calculation of the signal intensities. Thus, a logarithm transformation (apply log(matrix + 1)) followed by a truncation by the quantile 0.99 (all values greater than quantile 0.99 are truncated to this upper value, all values lower than quantile 0.01 are truncated to this lower value) were applied to the normalized gene expression values. Finally, in both cases, quantiles normalization using the preprocessCoreR package88 was carried out.
Results
We demonstrate the possibilities that mechanistic models offer for the interpretation of genomic variability in two different scenarios. The first one is the case of Fanconi Anemia (ORPHA:84), a rare disease with well-known hallmarks (chromosomal instability caused by failures in the repair machinery) which have been mapped in the FA pathway. Therefore, variants having an impact in the pathway, in the absence of other considerations, are potential genetic disease drivers. However, if they affect a large number of pathways as well, then they are probably lethal genes rather than disease driver genes.
A second scenario considered a complex disease, such as type 2 diabetes, where their disease hallmarks are not so well defined. Some general cell processes could potentially be associated with the disease phenotype but, in many cases, these are too general to be considered a clear causative agent of diabetes. Then, the number of cell processes potentially related to the disease can be reduced by considering only those that display a significantly differential behavior in a case-control comparison.
Fanconi anemia, a case example of a rare disease
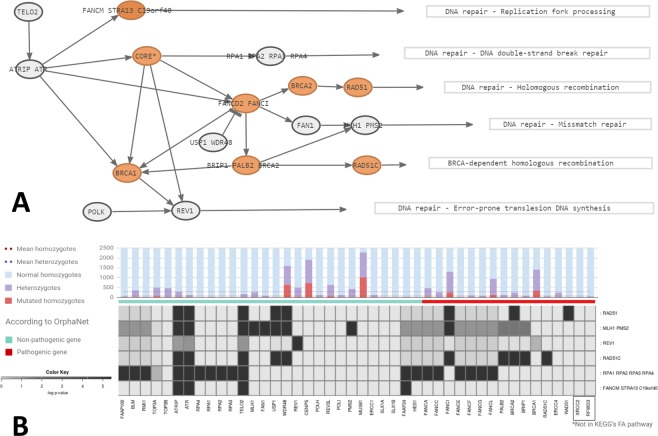
A detailed map of FA signaling is available in KEGG (ID: 03460) and is already implemented in the corresponding mechanistic model in Hipathia. Figure 2A shows the basic map of the signal transduction chain used to relate gene expression to the activity of signaling circuits within the FA pathway, which ultimately trigger cell activities related to FA hallmarks.
Figure 2.
Fanconi anemia simulation. (A) The FA pathway. Nodes in orange contain known FA pathogenic genes. (B) The result of the LoF simulations over the different FA circuits. Over each gene column, the frequencies at which these genes have been found in the normal population (1000 genomes) with a pLoF mutation is represented.
Since the main tissue affected by FA, bone marrow, is not present in GTEx, we uploaded to Hipathia data from an experiment available in the GEO database (GSE16334) in which bone marrow from healthy donors was analyzed81.
The Loss of function (LoF) mutations in all the genes in the pathway was simulated and the results are shown in Fig. 2B. As expected, known disease genes produced more impact over the FA pathway than most of the genes that have not been described as disease-causing (to date). Other genes not identified as causal genetic drivers of FA produce a devastating effect on the FA circuits, but also affect many other signaling circuits in other pathways (see Additional Table 2), and the LoF mutations in them were almost absent in the healthy population (data taken from 1000 genomes), which suggest that these genes are lethal and, consequently, unrelated with the disease.
Diabetes as a common disease case example
Diabetes is a highly prevalent disease, affecting to more than 8% of the population and being a major cause of blindness, kidney failure, heart attacks, stroke and lower limb amputation89. Inflammation is one of the most prominent phenotypic consequences of diabetes90. However, inflammation is a too general concept that can be associated to many conditions and, consequently, focusing on all the circuits that directly or indirectly trigger inflammation could result in the detection of activations unrelated to the disease. Therefore, a diabetes gene expression dataset that compares human islets gene expression of healthy donors with type 2 diabetic patients82 was used to determine which, among the 30 inflammation-related circuits, were specifically deregulated in the disease. Additional Table 3 shows three inflammation-related circuits significantly deregulated in diabetic human islets with respect their normal counterparts (FDR-adjusted p-value < 0.05)91, which are Rap1 signaling pathway:PRKCI-PARD6A-PARD3, NF-kappa B signaling pathway: CCL19 and NF-kappa B signaling pathway: CCL21.
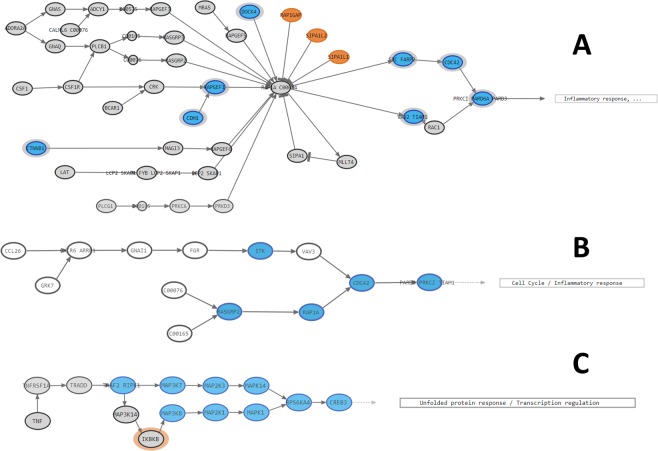
In order to demonstrate the suitability of mechanistic models for the interpretation of complex genomic variation we have focused on three circuits that represent three different scenarios. Firstly, the circuit Rap1 signaling pathway:PRKCI-PARD6A-PARD3 (Fig. 3A) that triggers inflammation and displays one of the highest differences in activity between diabetic and non-diabetic samples. Secondly, the signaling circuit Chemokynes pathway:PARD3-PRKCZ-TIAM1 (Fig. 3B), which also triggers inflammation but does not display any significant difference in the comparison (see Additional Table 3), which would represent a circuit with a disease hallmark but unrelated with the disease. And finally, the signaling circuit TNF signaling pathway:CREB3 (Fig. 3C), which neither presents a different activity in the comparison nor the functions triggered are likely to be directly related with diabetes (see Additional Table 4).
Figure 3.
Signaling circuits analyzed in the diabetes case study. Nodes in orange contain genes whose LoF causes an upregulation of the circuit and nodes in blue contain genes whose inactivation causes circuit downregulation. (A) Rap1 signaling pathway:PRKCI PARD6A PARD3, (B) Chemokynes pathway:PARD3-PRKCZ-TIAM1 and (C) TNF signaling pathway:CREB3 circuits.
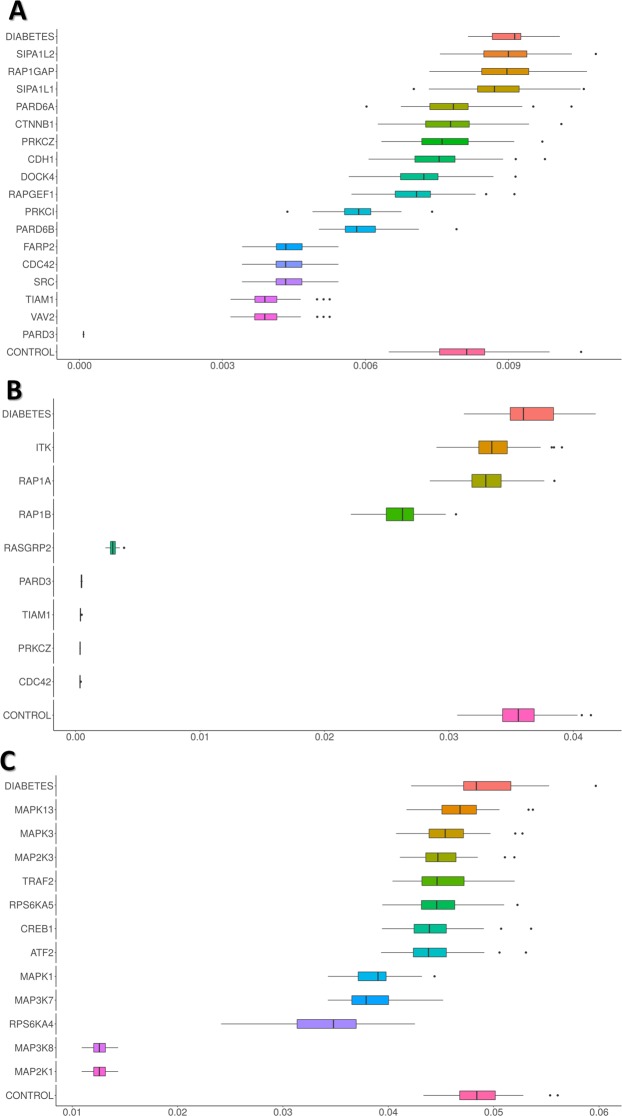
Then we used the Hipathia functionality to check the effect of genes with LoF mutations using the pancreatic islet tissue as user-defined tissue, and comparing the results of the resulting pathway activity after the simulations with those displayed by both the normal and the type 2 diabetes tissues. Figure 4 shows the results on the LoF simulations in the three above mentioned signaling circuits. Three genes, SIPA1L2, RAP1GAP and SIPA1L1, produce in the Rap1 signaling pathway:PRKCI-PARD6A-PARD3 circuit a level of activity in the control quite similar to the observed in the diabetic tissue. However, neither in the Chemokynes pathway:PARD3-PRKCZ-TIAM1 nor in the TNF signaling pathway:CREB3 circuits this trend is observed.
Figure 4.
Effect of genes with LoF mutations using the pancreatic islet tissue as used defined tissue, and comparing the resulting pathway activity after the simulations with those displayed by both the normal and the type 2 diabetes tissues. (A) Rap1 signaling pathway:PRKCI PARD6A PARD3, (B) Chemokynes pathway:PARD3-PRKCZ-TIAM1 and (C) TNF signaling pathway:CREB3 circuits.
Interestingly, when the differential activity caused by simulating LoF mutations in the three circuits is compared with the frequency at which these genes appear in the healthy population with a homozygous LoF mutation, it is clear that genes with relatively high LoF variation frequencies cause no effect in the pathway activity as a general rule (see Additional Fig. 1).
Discussion
Mechanistic models of pathway activity can be considered (complete or partial) representations of disease mechanisms and have been successfully used to uncover details on the molecular mechanisms behind different cancers65,66,92–94, common diseases95,96, rare diseases97 or the mechanisms of action of drugs67,98. Moreover, mechanistic models have been used in other biologically interesting scenarios such as the discovery of molecular mechanisms that explain how stress-induced activation of brown adipose tissue prevents obesity99 or the mechanisms of death and the post-mortem ischemia of a tissue100. Therefore, mechanistic models provide a holistic and accurate framework for the interpretation of the consequences of the genetic variation found in whole exome or genome sequencing, especially in the case of complex diseases, where the effect can be highly dependent on the specific condition of the patient.
However, even in rare Mendelian diseases, typically caused by one or a few highly penetrant variants6,7, the scenario is sometimes far away from the expected simplicity, as the example with FA reveals. When the LoF mutations were simulated, WDR48 LoF displayed a surprisingly high impact over RAD51, MLH1/PMS2 and RAD51C circuits. However, a large number of healthy individuals carry potential deleterious mutations in this gene, as shown in Fig. 2. Protein WDR48 acts as a complex together with USP1 to inhibit FANCD2/FANCI complex (Fig. 2A), indeed the coupling of WDR48 to the distal end of USP1 dramatically enhances USP1 activity, which catalyzes the deubiquitination of FANCD2. Thus, is USP1 that plays the catalytic role, whilst WDR48 is only required to join USP1101,102. Therefore, WDR48 gene may accumulate mutations that do not affect WD repeat domain structure, and that maintain its union with USP1intact, mutations that would be predicted as deleterious in other genes103.
A similar situation was observed with FANCI, that also showed an impact over RAD51, MLH1/PMS2 and RAD51C signaling circuits, but we found several individuals harboring homozygous mutations in this gene. Protein complex FANCI/FANCD2 is required to DNA repair function104, however, FANCI appears to be more naturally mutated in normal population than FANCD2 (Additional Table 2, data on LoF simulation are not available since FANCD2 expression was not present in the analyzed dataset), suggesting that FANCD2 may play a more determinant role than FANCI in the DNA repair-related function of the complex.
LoF simulations of ATRIP, ATR and TELO2 have an important impact over FA pathway, given that affects 5 out of 6 sub-pathways. Moreover, all three genes have a low frequency of mutations in 1000 genomes healthy population, which point to them as good candidates. However, none of these genes are categorized as pathogenic by Orphanet. Nevertheless, ATR LoF simulation affects not only FA pathway, but also to 38 more circuits, most of them belonging to p53 signaling pathway. This fact suggests that ATRIP/ATR complex is relevant to many other cellular processes, so, while detected as good candidates with high impact over the cell, its impact might not be specific of FA disease. Protein TELO2 functions as an S-phase checkpoint protein in the cell cycle, aside of its role in DNA repair, indeed mutations in this gene result in severe developmental diseases. Therefore, it is not misguided to present this gene as a candidate gene to further study.
Interestingly, only 12.29% of LoF mutations categorized as pathogenic by Orphanet have no impact over FA pathway, while 35.71% of the non-pathogenic genes have no impact on the pathway according to our model (Additional Table 2). This suggests that our model can be a useful tool to provide variant interpretation, especially in those cases where no variant is found in the disease-associated known genes.
As for Rap1 signaling pathway:PRKCI-PARD6A-PARD3 LoF simulation analysis related to diabetes, we found up to nodes disturbing the activation of the sub-pathway: CTNNB1, CDH1, RAPGEF1, DOCK4, RAP1GAP, SIPA1L2, SIPA1L1, SRC/FARP2, CDC42, VAV2/TIAM1 and PKKCI/PARD6A/PARD3.Among them, the LoF of nodes RAP1GAP, SIPA1L2 and SIPA1L1 were the only ones showing diabetes-like activation values, therefore resulting in an overactivation of the Rap1 signaling pathway:PRKCI-PARD6A-PARD3 circuit, as found in the analysis performed with the diabetes dataset. As seen in Fig. 3A, all three genes code for proteins that inactivate the RAS-related RAP1 protein, which is involved in many cellular processes, including inflammatory response, cell proliferation and adhesion and Thyroid Stimulating Hormone (TSH) signaling pathway.
The relationship between inflammation and type 2 diabetes has long been established. The proposed mechanisms to explain impaired insulin secretion include oxidative stress, endoplasmic reticulum stress, ectopic lipid deposition in muscle, liver and pancreas, lipotoxicity and glucotoxicity, all processes that may cause an inflammatory response105,106.
Focusing on RAP1 inactivation, a recent study reported the association between inhibition of RAP1 (via miRNAs) and hyperglycemia in patients with Type 1 Diabetes107. Moreover, other authors have suggested its relation with diabetic cardiomyopathy and with immune-mediated diabetes108,109. Therefore, it is reasonable to think that LoF mutations in those genes inactivating RAP1 protein would result in an over-activation of inflammatory response and thus, an increased risk of developing immune-mediated diabetes.
Beyond signaling, which can be mechanistically linked to cell functionalities71, other cellular processes such as metabolism, can also be used for a detailed interpretation of the consequences of variation over other relevant cell biological processes. Actually, it has recently been demonstrated that mutations with an impact over several metabolic pathways have a clear connection with certain cancer processes and could be easily related to patient phenotypes such as survival70,92.
Mechanistic models like the one used in this work rely on the accuracy of the pathways used to derive circuit topologies71. Although in general pathways from the most known repositories (KEGG60, Reactome61, Pathway Commons62, Wikipathways63, etc.) represent curated biological knowledge and are expected to be quite accurate, they are not absent of errors. Moreover, in many cases pathways represent incomplete biological knowledge. Errors or missing links will define erroneous or incomplete circuits with unpredictable (but most likely erroneous) behavior. Recently, we have described some constitutively inactive circuits that were incorrectly annotated in the Fanconi Anemia pathway that, after literature exploration, could be fixed and recovered the expected behaviour110.
Conclusions
By transforming the current variant-centric and gene-centric data into function-centric measurements a more holistic and context-dependent approach to the interpretation of complex variability, especially useful for common diseases, can be attained. Mechanistic models of cell functionality will open new avenues for understanding the complex relationships among genes that ultimately shape the phenotype.
Availability and requirements
Project name: HiPathia. Project home page: http://hipathia.babelomics.org. Operating system(s): Platform independent. Programming language: HTML/JavaScript in the frontend; Java, R and web services in Node and Express for the backend. Other requirements: Modern browsers. License: Does not apply. Any restrictions to use by non-academics: no
Supplementary information
Acknowledgements
This work is supported by grants SAF2017-88908-R from the Spanish Ministry of Economy and Competitiveness and “Plataforma de Recursos Biomoleculares y Bioinformáticos” PT17/0009/0006 from the ISCIII, both co-funded with European Regional Development Funds (ERDF) as well as H2020 Programme of the European Union grants Marie Curie Innovative Training Network “Machine Learning Frontiers in Precision Medicine” (MLFPM) (GA 813533) and “ELIXIR-EXCELERATE fast-track ELIXIR implementation and drive early user exploitation across the life sciences” (GA 676559). The project was also supported by grant “Large-scale drug repurposing in rare diseases by genomic Big Data analysis with machine learning methods - AYUDAS FUNDACIÓN BBVA A EQUIPOS DE INVESTIGACION CIENTIFICA 2018” and by CIBERER intramural grant ACCI2018/29.
Author contributions
M.E.M. has performed the data collection and the analysis, M.P.C. has collaborated in the analysis of the data and participated in writing the manuscript, M.M.F. has participated in data collection and analysis, K.R. has developed the web-based tool, M.H. has developed the Hipathia algorithm, C.L. has participated in the development of the Hipathia tool and J.D. has conceived the work and wrote the manuscript.
Data availability
The datasets analyzed during the current study are available in the GEO repository (accession: GSE16334) [https://www.ncbi.nlm.nih.gov/geo/], GTEx portal (dbGaP accession phs000424.v7.p2) [https://www.gtexportal.org/home/].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55454-7.
References
- 1.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER. DNA sequencing technologies: 2006–2016. Nature protocols. 2017;12:213. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- 3.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boycott KM, et al. International cooperation to enable the diagnosis of all rare genetic diseases. The American Journal of Human Genetics. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nature Reviews Genetics. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 8.MacArthur DG, Tyler-Smith C. Loss-of-function variants in the genomes of healthy humans. Hum Mol Genet. 2010;19:R125–130. doi: 10.1093/hmg/ddq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue Y, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carbonell J, et al. A map of human microRNA variation uncovers unexpectedly high levels of variability. Genome Med. 2012;4:62. doi: 10.1186/gm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spivakov M, et al. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 2012;13:R49. doi: 10.1186/gb-2012-13-9-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston JJ, et al. Individualized Iterative Phenotyping for Genome-wide Analysis of Loss-of-Function Mutations. The American Journal of Human Genetics. 2015;96:913–925. doi: 10.1016/j.ajhg.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 17.Reumers J, et al. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrer-Costa C, et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 19.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLOS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conde L, et al. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu H, et al. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;37:622–634. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11:361. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 24.Karczewski KJ, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Research. 2016;45:D840–D845. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dopazo J, et al. 267 Spanish exomes reveal population-specific differences in disease-related genetic variation. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome research. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS computational biology. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundaram L, et al. Predicting the clinical impact of human mutation with deep neural networks. Nature Genetics. 2018;50:1161–1170. doi: 10.1038/s41588-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler S, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Research. 2013;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenson, P. D. et al. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics Chapter.1, Unit1 13, 10.1002/0471250953.bi0113s39 (2012). [DOI] [PubMed]
- 34.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The_Uniprot_Consortium. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res42, D191-198, 10.1093/nar/gkt1140 (2014). [DOI] [PMC free article] [PubMed]
- 36.Javed A, Agrawal S, Ng PC. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nature methods. 2014;11:935. doi: 10.1038/nmeth.3046. [DOI] [PubMed] [Google Scholar]
- 37.Sifrim A, et al. eXtasy: variant prioritization by genomic data fusion. Nat Methods. 2013;10:1083–1084. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 38.Zemojtel T, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Science translational medicine. 2014;6:252ra123–252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson PN, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome research. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLaren W, et al. The ensembl variant effect predictor. Genome Biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bleda M, et al. CellBase, a comprehensive collection of RESTful web services for retrieving relevant biological information from heterogeneous sources. Nucleic Acids Res. 2012;40:W609–614. doi: 10.1093/nar/gks575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antanaviciute, A. et al. OVA: integrating molecular and physical phenotype data from multiple biomedical domain ontologies with variant filtering for enhanced variant prioritization. Bioinformatics, btv473 (2015). [DOI] [PMC free article] [PubMed]
- 44.Aleman A, Garcia-Garcia F, Salavert F, Medina I, Dopazo J. A web-based interactive framework to assist in the prioritization of disease candidate genes in whole-exome sequencing studies. Nucleic Acids Res. 2014;42:W88–93. doi: 10.1093/nar/gku407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertoldi L, et al. QueryOR: a comprehensive web platform for genetic variant analysis and prioritization. BMC Bioinformatics. 2017;18:225. doi: 10.1186/s12859-017-1654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hombach, D. et al. MutationDistiller: user-driven identification of pathogenic DNA variants. Nucleic Acids Research, 10.1093/nar/gkz330%J Nucleic Acids Research (2019). [DOI] [PMC free article] [PubMed]
- 47.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 48.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156:872–877. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gustafsson M, et al. Modules, networks and systems medicine for understanding disease and aiding diagnosis. Genome Medicine. 2014;6:82. doi: 10.1186/s13073-014-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsanis N. The continuum of causality in human genetic disorders. Genome Biology. 2016;17:233. doi: 10.1186/s13059-016-1107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oti M, Brunner HG. The modular nature of genetic diseases. Clin Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 54.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 55.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 56.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitra K, Carvunis AR, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nat Rev Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Alonso L, et al. The role of the interactome in the maintenance of deleterious variability in human populations. Mol Syst Biol. 2014;10:752. doi: 10.15252/msb.20145222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research. 2016;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic acids research. 2017;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2010;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Slenter, D. N. et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic acids research (2017). [DOI] [PMC free article] [PubMed]
- 64.Mazein A, et al. Systems medicine disease maps: community-driven comprehensive representation of disease mechanisms. npj Systems Biology and Applications. 2018;4:21. doi: 10.1038/s41540-018-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fey D, et al. Signaling pathway models as biomarkers: Patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal. 2015;8:ra130. doi: 10.1126/scisignal.aab0990. [DOI] [PubMed] [Google Scholar]
- 66.Hidalgo MR, et al. High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget. 2017;8:5160–5178. doi: 10.18632/oncotarget.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amadoz A, Sebastian-Leon P, Vidal E, Salavert F, Dopazo J. Using activation status of signaling pathways as mechanism-based biomarkers to predict drug sensitivity. Scientific reports. 2015;5:18494. doi: 10.1038/srep18494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salavert F, et al. Actionable pathways: interactive discovery of therapeutic targets using signaling pathway models. Nucleic Acids Res. 2016;44:W212–216. doi: 10.1093/nar/gkw369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hernansaiz-Ballesteros RD, et al. Assessing the impact of mutations found in next generation sequencing data over human signaling pathways. Nucleic Acids Res. 2015;43:W270–275. doi: 10.1093/nar/gkv349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Çubuk C, et al. Differential metabolic activity and discovery of therapeutic targets using summarized metabolic pathway models. NPJ Systems Biology. 2019;5:7. doi: 10.1038/s41540-019-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amadoz, A., Hidalgo, M. R., Çubuk, C., Carbonell-Caballero, J. & Dopazo, J. A comparison of mechanistic signaling pathway activity analysis methods. Briefings in bioinformatics Advanced publication (2018). [DOI] [PMC free article] [PubMed]
- 72.Salavert F, et al. Web-based network analysis and visualization using CellMaps. Bioinformatics. 2016;32:3041–3043. doi: 10.1093/bioinformatics/btw332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Efroni S, Schaefer CF, Buetow KH. Identification of key processes underlying cancer phenotypes using biologic pathway analysis. PLoS ONE. 2007;2:e425. doi: 10.1371/journal.pone.0000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sebastian-Leon P, et al. Inferring the functional effect of gene expression changes in signaling pathways. Nucleic Acids Res. 2013;41:W213–217. doi: 10.1093/nar/gkt451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sebastian-Leon P, et al. Understanding disease mechanisms with models of signaling pathway activities. BMC Syst Biol. 2014;8:121. doi: 10.1186/s12918-014-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jacob L, Neuvial P, Dudoit S. More power via graph-structured tests for differential expression of gene networks. Ann. Appl. Stat. 2012;6:561–600. doi: 10.1214/11-AOAS528. [DOI] [Google Scholar]
- 77.Martini P, Sales G, Massa MS, Chiogna M, Romualdi C. Along signal paths: an empirical gene set approach exploiting pathway topology. Nucleic Acids Res. 2013;41:e19. doi: 10.1093/nar/gks866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitsos A, et al. Identifying drug effects via pathway alterations using an integer linear programming optimization formulation on phosphoproteomic data. PLoS Comput Biol. 2009;5:e1000591. doi: 10.1371/journal.pcbi.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montaner D, Minguez P, Al-Shahrour F, Dopazo J. Gene set internal coherence in the context of functional profiling. BMC Genomics. 2009;10:197. doi: 10.1186/1471-2164-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lonsdale J, et al. The genotype-tissue expression (GTEx) project. Nature genetics. 2013;45:580. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanderwerf SM, et al. TLR8-dependent TNF-(alpha) overexpression in Fanconi anemia group C cells. Blood. 2009;114:5290–5298. doi: 10.1182/blood-2009-05-222414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taneera J, et al. A Systems Genetics Approach Identifies Genes and Pathways for Type 2 Diabetes in Human Islets. Cell metabolism. 2012;16:122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 83.The 1000 genomes project, http://www.1000genomes.org/
- 84.Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research47, D886-D894, https://doi.org/10.1093/nar/gky1016%J Nucleic Acids Research (2018). [DOI] [PMC free article] [PubMed]
- 85.Gautier L, Cope L, Bolstad BM, Irizarry R. A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 86.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 87.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 89.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 90.Dandona P, Aljada A, Chaudhuri A, Mohanty P, Garg R. Metabolic syndrome: a comprehensive perspective based on interactions between obesity, diabetes, and inflammation. Circulation. 2005;111:1448–1454. doi: 10.1161/01.CIR.0000158483.13093.9D. [DOI] [PubMed] [Google Scholar]
- 91.Benjamini Y, Yekutieli D. The control of false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 92.Cubuk C, et al. Gene expression integration into pathway modules reveals a pan-cancer metabolic landscape. Cancer research. 2018;78:6059–6072. doi: 10.1158/0008-5472.CAN-17-2705. [DOI] [PubMed] [Google Scholar]
- 93.Iorio, F. et al. Pathway-based dissection of the genomic heterogeneity of cancer hallmarks’ acquisition with SLAPenrich. Scientific reports8 (2018). [DOI] [PMC free article] [PubMed]
- 94.Hidalgo MR, Amadoz A, Cubuk C, Carbonell-Caballero J, Dopazo J. Models of cell signaling uncover molecular mechanisms of high-risk neuroblastoma and predict disease outcome. Biology direct. 2018;13:16. doi: 10.1186/s13062-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh, V. et al. Computational systems biology approach for the study of rheumatoid arthritis: From a molecular map to a dynamical model. Genomics computational biology4 (2018). [DOI] [PMC free article] [PubMed]
- 96.Niarakis, A. et al. In Fc Receptors Computational modeling of the main signaling pathways involved in mast cell activation 69–93 (Springer, (2014). [DOI] [PubMed]
- 97.Chacón-Solano E, et al. Fibroblasts activation and abnormal extracellular matrix remodelling as common hallmarks in three cancer-prone genodermatoses. J British Journal of Dermatology. 2019;181:512–522. doi: 10.1111/bjd.17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frieboes HB, et al. Prediction of drug response in breast cancer using integrative experimental/computational modeling. Cancer research. 2009;69:4484–4492. doi: 10.1158/0008-5472.CAN-08-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Razzoli M, et al. Stress-induced activation of brown adipose tissue prevents obesity in conditions of low adaptive thermogenesis. Molecular metabolism. 2016;5:19–33. doi: 10.1016/j.molmet.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ferreira PG, et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nature communications. 2018;9:490. doi: 10.1038/s41467-017-02772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cohn MA, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Molecular cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 102.Yin J, et al. Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46. Structure. 2015;23:2043–2054. doi: 10.1016/j.str.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 103.LiD R. WD-repeatproteins: Structurecharacteristics, biologicalfunction, andtheirinvolvementinhumandiseases. CellMolLifeSci. 2001;58:2085–2097. [Google Scholar]
- 104.Liang C-C, et al. The FANCD2–FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nature Communications. 2016;7:12124. doi: 10.1038/ncomms12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. The Journal of Clinical Investigation. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nature Reviews Immunology. 2011;11:98. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 107.Satake E, et al. Circulating miRNA Profiles Associated With Hyperglycemia in Patients With Type 1 Diabetes. Diabetes. 2018;67:1013. doi: 10.2337/db17-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cai Y, et al. Decoding telomere protein Rap1: Its telomeric and nontelomeric functions and potential implications in diabetic cardiomyopathy. Cell Cycle. 2017;16:1765–1773. doi: 10.1080/15384101.2017.1371886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lagarrigue F, Gertler FB, Ginsberg MH, Cantor JM. Cutting Edge: Loss of T Cell RIAM Precludes Conjugate Formation with APC and Prevents Immune-Mediated Diabetes. The Journal of Immunology. 2017;198:3410. doi: 10.4049/jimmunol.1601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Esteban-Medina M, Peña-Chilet M, Loucera C, Dopazo J. Exploring the druggable space around the Fanconi anemia pathway using machine learning and mechanistic models. BMC Bioinformatics. 2019;20:370. doi: 10.1186/s12859-019-2969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sidiropoulos K, et al. Reactome enhanced pathway visualization. Bioinformatics. 2017;33:3461–3467. doi: 10.1093/bioinformatics/btx441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the GEO repository (accession: GSE16334) [https://www.ncbi.nlm.nih.gov/geo/], GTEx portal (dbGaP accession phs000424.v7.p2) [https://www.gtexportal.org/home/].