Abstract
Gigantopithecus blacki was a giant hominid that inhabited densely forested environments of Southeast Asia during the Pleistocene1. Its evolutionary relationships to other great ape species, and their divergence during the Middle and Late Miocene (16-5.3 Mya), remains disputed2,3. Hypotheses regarding relationships between Gigantopithecus and extinct and extant hominids are difficult to substantiate because of its highly derived dentognathic morphology and the absence of cranial and post-cranial remains1,3-6. Therefore, proposed hypotheses on the phylogenetic position of Gigantopithecus among hominids have been wide-ranging, but none have received independent molecular validation. We retrieved dental enamel proteome sequences from a 1.9 million years (Mya) old Gigantopithecus blacki molar found in Chuifeng Cave, China7,8. The thermal age of these protein sequences is approximately five times older than any previously published mammalian proteome or genome. We demonstrate that Gigantopithecus is a sister clade to orangutans (genus Pongo) with a common ancestor about 10-12 Mya, implying that the Gigantopithecus divergence from Pongo is part of the Miocene radiation of great apes. Additionally, we hypothesize that the expression of alpha-2-HS-glycoprotein (AHSG), which has not been observed in enamel proteomes previously, had a role in the biomineralization of the thick enamel crowns that characterize the large molars in the genus9,10. The survival of an Early Pleistocene dental enamel proteome in the subtropics further expands the scope of palaeoproteomic analysis into geographic areas and time periods previously considered incompatible with genetic preservation.
Gigantopithecus blacki is an extinct, potentially giant hominid species that once inhabited Asia. It was first discovered and identified by von Koenigswald in 1935 when he described an isolated tooth that he found in a Hong Kong drugstore11. The entire Gigantopithecus blacki fossil record, dated between the Early Pleistocene (~2.0 Mya) and the late Middle Pleistocene (~0.3 Mya12), includes thousands of teeth and four partial mandibles from subtropical Southeast Asia1,13,14. All the known Gigantopithecus blacki localities are situated in southern China, stretching from Longgupo Cave, just south of the Yangtze River, to the Xinchong Cave on Hainan Island, and, possibly, into northern Vietnam and Thailand15,16.
To address the evolutionary relationships between Gigantopithecus and extant hominoids, we performed protein extractions on dentine and enamel samples of a single molar (CF-B-16) found in Chuifeng Cave, China, that is morphologically assigned to Gigantopithecus blacki7,8. The site is dated using multiple approaches to 1.9±0.2 Mya (Extended Data Figs. 1, 2). Enamel and dentine samples were processed using recently established digestion-free protocols optimized for extremely degraded ancient proteomes17 (Methods). Enamel demineralization was replicated using two different acids, trifluoroacetic acid (TFA) and hydrochloric acid (HCl).
We identify no endogenous proteins from the dentine, but instead recover an ancient enamel proteome composed of 409 unique peptides matching to six endogenous proteins: amelogenin (AMELX), ameloblastin (AMBN), amelotin (AMTN), enamelin (ENAM), metalloproteinase-20 (MMP20) and alpha-2-HS-glycoprotein (AHSG, also known as FETUA; Extended Data Tab. 2). This observation extends the survival of ancient mammalian proteins to a thermal age, obtained by normalizing the chronological age to a constant temperature of 10°C, to approximately 11.8 Mya@10°C (Extended Data Tab. 1). Such a thermal age is well beyond the thermally oldest DNA (0.25 Mya@10°C, Sima de los Huesos – Spain18), collagen (0.22 Mya@10°C, Happisburgh – UK19) and enamel proteome (2.2 Mya@10°C, Dmanisi – Georgia17) reported to date. The Chuifeng Cave enamel proteome is thus, to the best of our knowledge, the oldest Cenozoic skeletal proteome currently reported (Fig. 1). The survival of a subtropical proteome at approximately 2 Mya suggests that chronologically older specimens from higher latitudes are likely to preserve ancient proteomes as well.
Figure 1. Thermal age of Chuifeng Cave, China, in the subtropics of Southeast Asia (red asterisks).
Chronological and thermal ages of other Cenozoic ancient genomes and proteomes are given for comparison. Inset: geographic location of Chuifeng Cave in the subtropics of Southeast Asia. Base map was generated using public domain data from www.naturalearthdata.com. The red asterisk also encloses the entire known geographic range of Gigantopithecus blacki fossils.
The content of the recovered enamel proteome is consistent with previously reported ancient enamel proteomes17,20,21, with the addition of several peptides deriving from a single region of AHSG. Peptide matches to these proteins cover a minimum of 43 informative single amino acid polymorphisms (SI Tab. 3). In addition, the retrieved protein regions largely fall within areas previously recovered from an Early Pleistocene Stephanorhinus enamel proteome from Dmanisi17 (SI Fig. 1). The absence of AMELY-specific peptides suggests that the sampled molar might have belonged to a female Gigantopithecus specimen. The endogenous peptide coverage of 456 amino acids is lower than the previously recovered sequence coverage for a Dmanisi Stephanorhinus specimen (875 amino acids17; SI Tab. 1). This observation is in agreement with the older thermal age for Chuifeng Cave, compared to Dmanisi17.
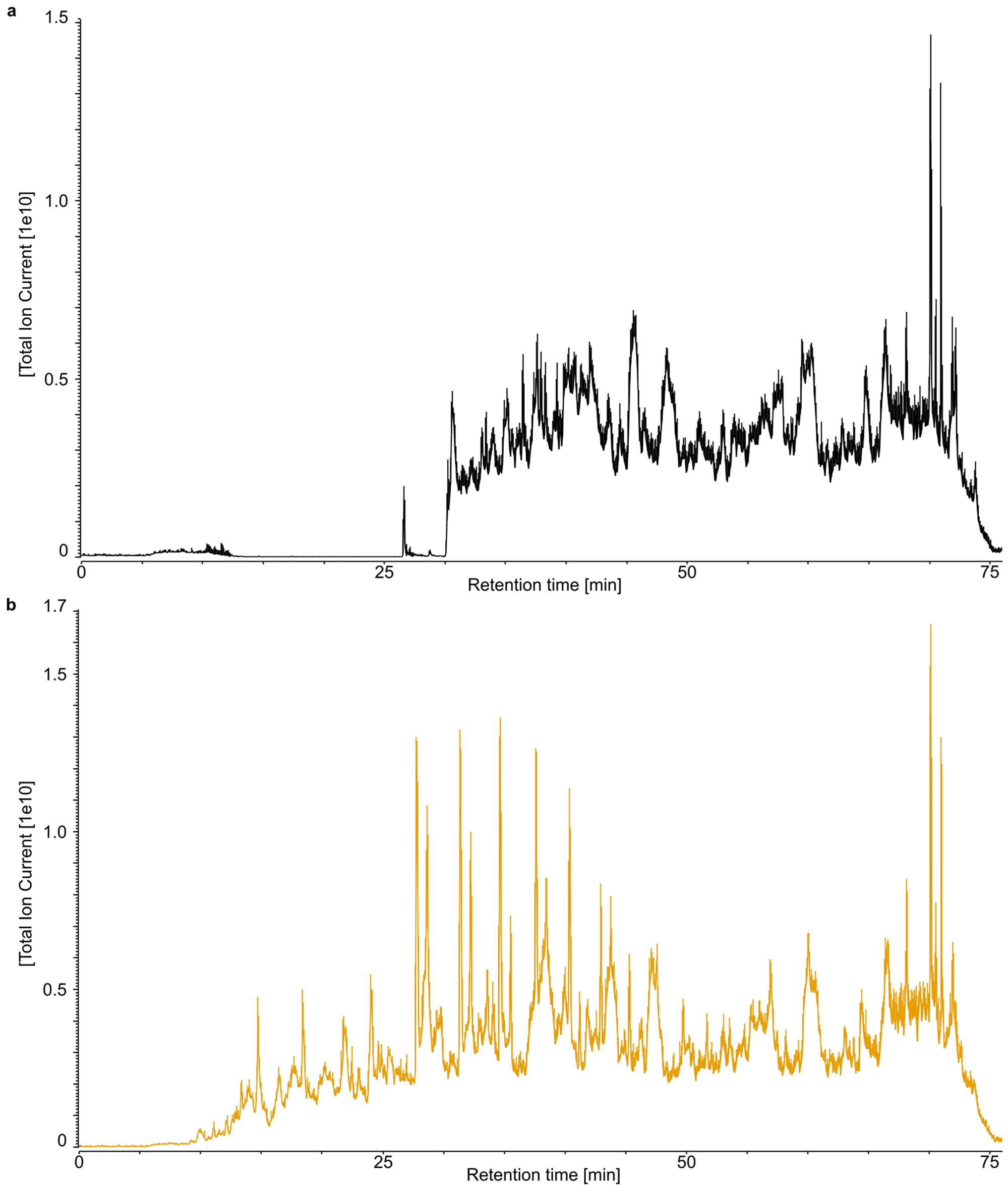
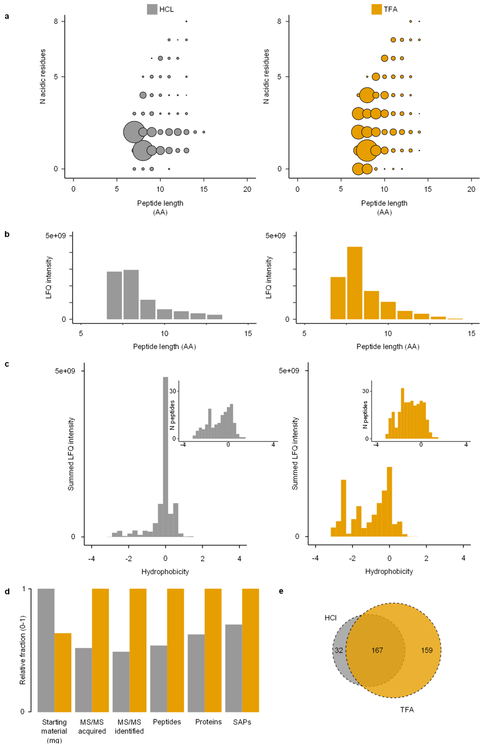
We replicated enamel demineralization using two different acids (TFA and HCl). When comparing the chromatograms of these two extracts, we observe that different peptide populations are released (Extended Data Fig. 3). Due to the partial acidic hydrolysis22, which potentially occurs alongside demineralization, peptide populations with a wider range of acidity (Extended Data Fig. 4a) and hydrophobicity (Extended Data Fig. 4c) are generated using TFA. We observe that the TFA-based demineralization returned 127 more unique non-overlapping peptide sequences compared to the HCl-based demineralization (Extended Data Fig. 4e). The TFA extract, therefore, outperformed the HCl-based extraction, despite a smaller amount of starting material17. Ultimately, the extended coverage of TFA-based demineralization increases the identification rate of informative single amino acid polymorphisms (SAPs), enhancing the phylogenetic information obtained (Extended Data Fig. 4d). Finally, we observe similar deamidation rates and average peptide lengths in the HCl- and TFA-demineralized samples (Extended Data Fig. 5), which indicate that the two acids release peptide populations modified to the same extend.
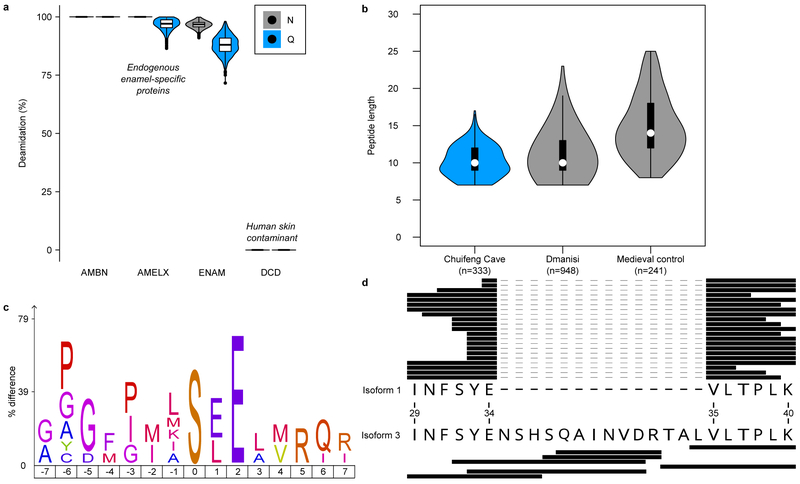
The Gigantopithecus enamel proteome is characterized by extensive diagenetic modifications, such as high rates of deamidation (Fig. 2a), and a high degree of degradation, as indicated by relatively short peptide lengths (Fig. 2b), as expected for an ancient proteome preserved in tropical conditions. When quantifying peptide intensities using label-free quantification (LFQ), implemented in MaxQuant23, we observe that summed and normalized MS1 spectral intensities are higher for shorter peptides compared to longer peptides (Extended Data Fig. 4b). Finally, the peptide lengths of the Chuifeng Cave enamel proteome are shorter than those identified in thermally younger enamel proteomes (Fig. 2b).
Figure 2. Gigantopithecus enamel proteome modifications and degradation.
a, Violin plots of asparagine (N) and glutamine (Q) deamidation for selected proteins (n=1,000 bootstrap replicates of intensity-based peptide deamidation32). Human dermcidin (DCD) is included as an example of a non-deamidated contaminant. For AMBN, all observed asparagines and glutamines are deamidated. For AMELX, all asparagines are deamidated. For DCD, no observed asparagines and glutamines are deamidated. b, Violin plots of peptide lengths for Gigantopithecus, an Early Pleistocene rhinoceros from Dmanisi, and a Medieval control sample17. c, Sequence-motif analysis of the over-representation of specific amino acids around the phosphorylated amino acid (position 0; n=14). d, Peptide coverage of AMELX protein isoforms. Matching peptides are indicated by black bars for isoform 1 (n=21) and isoform 3 (n=7). The latter includes an insertion due to alternative splicing between isoform 1, amino acid positions 34 and 35 (coordinates in reference to UniProt Accession number: Q99217-1 [AMELX_HUMAN]). a and b include data on AMELX, AMBN, ENAM, AMTN and MMP20 only. For a and b, boxplots define the range of the data (whiskers extending to 1.5 the interquartile range), 25th and 75th percentiles (boxes), and medians (dots).
Enamel-specific proteins are modified in vivo through protein phosphorylation, alternative splicing of AMELX, and MMP20- and KLK4-mediated proteolysis. Such modifications potentially survive in ancient proteomes. We detected evidence of surviving in vivo post-translational modifications, such as serine phosphorylation in the S-x-E/phS motif, recognised by the secreted kinase FAM20C (Fig. 2c). FAM20C kinase is known to regulate the phosphorylation of extracellular proteins involved in biomineralization24. Finally, we observe two alternative splicing-derived AMELX isoforms (Fig. 2d). These observations are similar to other Early Pleistocene enamel proteomes17. The Gigantopithecus enamel proteome therefore demonstrates that such in vivo modifications can likewise be recovered from hominid samples across the Pleistocene.
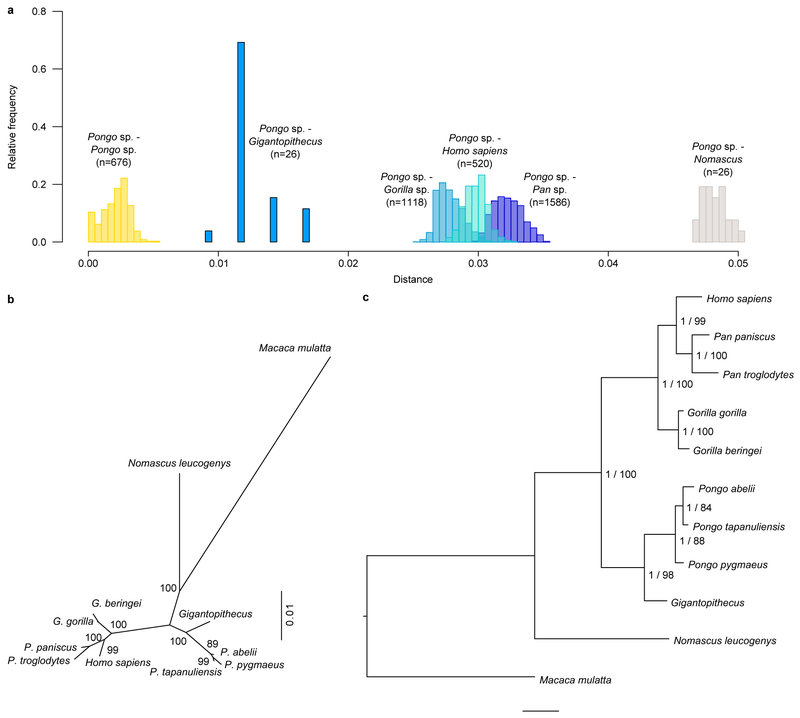
To achieve a protein-based phylogenetic placement of Gigantopithecus, we compared the enamel proteome sequences we retrieved with those of extant apes (Hominoidea). Publicly available whole-genome sequence data were used to predict enamel protein sequences from relevant species25,26 (SI Tab. 2, SI Figs. 2-12). Our results show that Gigantopithecus represents a sister taxon to all extant orangutans (Pongo sp.) forming a monophyletic group with extant pongines (Fig. 3a; Extended Data Figs. 6, 7). We then attempted to estimate the divergence time between Gigantopithecus and Pongo species using two approaches: (i) a pairwise distance approach and (ii) a Bayesian approach using MrBayes (Methods). While confidence intervals obtained for the divergence estimates of the Pongo-Gigantopithecus split are large, our results indicate that Gigantopithecus diverged from the extant Pongo species in the Middle or Late Miocene (~10 Mya and ~12 Mya using the Bayesian and pairwise distances approaches, respectively; Fig. 3b). This suggests that, despite an exclusively Pleistocene fossil record, Gigantopithecus is a member of an early radiation of pongines, whose diversity peaks during the Middle and Late Miocene (Fig. 3b). Our results thereby resolve the phylogenetic position of Gigantopithecus, but renew the debate on the evolutionary relationships between extant hominids and early hominids present in the fossil record2.
Figure 3. Bayesian phylogeny and divergence of Gigantopithecus among extant apes (Hominoidea).
a, Time-calibrated Bayesian phylogeny of Gigantopithecus. Circled nodes were fixed for topology (see Methods). Grey error bars represent the 95% highest posterior density (HPD) intervals for the divergence estimates. b, Distribution of probable and possible extinct pongines known from the fossil record, including Gigantopithecus (black bars). Grey bars represent the 95% HPD interval obtained from the Bayesian approach, and the 95% confidence interval obtained from the pairwise-distance based approach, of the Gigantopithecus-Pongo divergence.
The presence of AHSG in the Gigantopithecus proteome is intriguing, as this protein is not commonly observed in (modern) hominid enamel proteomes. All retrieved peptides derive from a single, highly conserved region that is bordered by disulfide cysteine bonds on either side (Extended Data Fig. 8). AHSG is highly glycosylated in vivo, but we observed no glycosylation during our bioinformatics analysis. The observed sequence contains regularly spaced aspartic acid residues that provide a suitable motif for binding to basic calcium phosphate lattices27. The notion that this specific peptide sequence is involved in biomineral binding is supported by the observation that this region is: (i) presented on the external surface of AHSG28, (ii) that such surfaces have been demonstrated to bind biominerals in other systems as well29, and (iii) that this type of binding enhances peptide preservation29. AHSG acts as a key component of bone and dentine mineralization processes through the inhibition of extrafibrillar mineralization of collagen type I helices30 and has previously been hypothesized to have a role in amelogenesis9. In our extracts, there are no endogenous plasma proteins present, such as serum albumin, or other common dentine proteins, such as collagen type I. We also do not identify any AHSG peptides in our dentine sample. We therefore exclude the possibility that the AHSG peptides derive from dentine. Gigantopithecus is known to have relatively long enamel formation times and thick enamel compared to several extant and extinct hominids, including its phylogenetically closest relatives10,31. We therefore hypothesize that Gigantopithecus has recruited AHSG as an additional molecular component to favour enamel biomineralization during prolonged amelogenesis, ultimately playing a role comparable to the one it has in bone and dentine mineralization9.
With our study, we are able to reveal the long-debated phylogenetic position of Gigantopithecus as an early diverging pongine. We demonstrate the ability to retrieve ancient enamel proteomes from Early Pleistocene samples preserved in subtropical conditions, well beyond the current limitations of biomolecular research in hominid and hominin evolution. In addition, the survival of an Early Pleistocene Gigantopithecus enamel proteome allows us to assess the presence of multiple forms of in vivo modifications. Finally, we demonstrate that palaeoproteomic analysis allowed revealing a hitherto unknown biological component of extinct hominid tooth formation. This finding suggests that the palaeoproteomic analysis of hominid enamel has great potential to provide a molecular perspective on human and great ape evolution.
METHODS
0. CHUIFENG CAVE
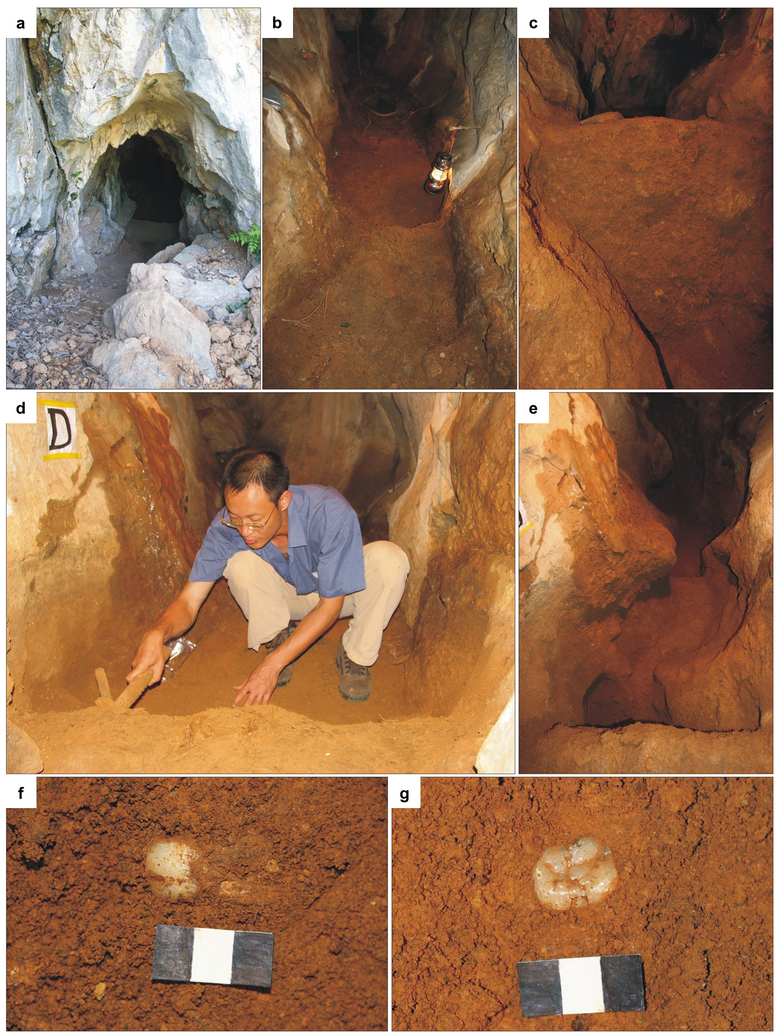
The Chuifeng Cave (23°34′27″N, 107°00′ 22″E) is one of the most representative sites for the Early Pleistocene Gigantopithecus blacki fauna8. The site is located in the Bubing Basin in the north-western part of the Guangxi Zhuang Autonomous Region, south China (Extended Data Fig. 1). The cave is 19 m in length, 0.5–2 m in width and 1.5–5 m in height, penetrating the limestone from southeast to northwest at a height of ~ 77 m above the local valley floor. A fossiliferous sandy-clay with a few limestone breccias fills most part of the cave, with an average depth of 1.3 m (Extended Data Fig. 2). Four excavation areas (A, B, C, and D) were excavated down to limestone bedrock in 10 cm intervals. Twenty-four large mammalian species, including 92 Gigantopithecus blacki teeth, were unearthed from the cave8. The Chuifeng Cave mammalian fauna is characterized by the occurrence of typical Early Pleistocene species, such as Hystrix magna, Sinomastodon sp., Stegodon preorientalis, Ailuropoda microta, Pachycrocuta licenti, Tapirus sanyuanensis, and Sus peii8. This mammalian fauna is comparable with other Gigantopithecus-containing faunas of the Early Pleistocene in southern China, such as Baikong33, Longgupo34, and Liucheng35. The mammalian fauna composition is consistent with the age results (~ 1.9 Mya) of combined ESR/U-series dating and sediment paleomagnetic studies36. In the present study, we collected one well-preserved Gigantopithecus blacki tooth (excavation number CF-B-16) for palaeoproteomic analysis. This tooth was excavated from area B at a depth of 90 cm from the sediment surface and, based on its stratigraphic position, is dated to ~ 1.9 Mya. No other samples were tested prior to CF-B-16, and no specific selection was made as to which Gigantopithecus tooth would be analysed.
1. THERMAL AGE
Thermal age was calculated to allow comparison with previously published ancient genomes, ancient proteomes, and collagen peptide mass fingerprinting studies, from other temporal and geographic localities. Temperature estimates for the hominin occupation of Dmanisi based on herpetological fauna suggest a temperature about 3.1 °C above current mean annual temperature, while the sea surface temperature record used 29 predicts a negative ΔT at the time of hominin occupation. Given this discrepancy and the widely different temperature estimates for the last glacial maximum (LGM) in the Caucasus, we conservatively use a scale factor of 0, correlating with a ΔT of approximately -0.2 °C, and a current mean annual temperature of 11.2 °C. Our thermal age prediction for Dmanisi (2.2 Myr@10 °C) should therefore be seen as conservative. Thermal age for Chuifeng Cave was calculated with a general lapse rate between mean annual temperature (MAT) and altitude of 5.0 °C/km, a scale factor of 0.7, and a ΔT at LGM of -3 °C. Again, actual ΔT at LGM might have been more pronounced, leading to a conservative estimate of thermal age for Chuifeng Cave as well. MAT was estimated based on the ten closest weather stations listed in publicly accessible World Meteorological Organization (WMO) data (Extended Data Table. 21). Thermal age calculations are, among other factors, altitude dependent, but only five out of these ten weather stations have altitude directly associated with them. We therefore estimated the altitude of the other five weather stations through an online resource (https://www.advancedconverter.com/map-tools/find-altitude-bycoordinates). The correlation between WMO altitude and estimated altitude was R = 0.99, providing sufficient validity to our estimated altitudes. The MATs for all weather stations were then averaged to obtain an approximate MAT for Chuifeng Cave. Next, thermal age was calculated for chronological ages of 1.7 Myr, 1.9 Myr and 2.1 Myr, giving estimates of the minimum (9.2 Myr@10 °C), maximum (15.0 Myr@10 °C), and mean (11.8 Myr@10 °C) thermal ages associated with the Chuifeng Cave fauna within a 95% confidence interval (Fig. 1). The Chuifeng Cave proteome is thereby substantially older than the oldest collagen peptide mass fingerprint (Ellesmere Island, 0.003 Myr@10 °C), oldest mammalian genome (Thistle Creek, 0.03 Myr@10 °C), oldest hominin genome (Sima de los Huesos, 0.25 Myr@10 °C), and oldest enamel proteome (Dmanisi, 2.2 Myr@10 °C) published to date29. Full thermal age calculations can be found in Supplementary Information File 3.
2. PROTEIN EXTRACTION
Ancient protein extractions took place in facilities at the Natural History Museum of Denmark dedicated to extracting ancient DNA and ancient proteins. These laboratories include clean rooms fitted with filtered ventilation and positive air pressure37. A negative extraction blank was processed alongside the ancient extractions, with the additional inclusion of injection blanks during MS/MS analysis to monitor potential protein contamination during all stages of analysis.
Two enamel (185 and 118 mg, respectively) and one dentine (192 mg) samples were removed from the same molar (CF-B-16), using a sterilized drill, and crushed to a rough powder. One enamel and the dentine sample were demineralized in 1.2 M HCl at 3°C for 24 hours, while the other enamel sample was demineralized at the same temperature and duration using 10% TFA. Subsequently, solubilized protein residues were cleaned, concentrated and immobilized on C18 Stage-Tips using previously published methods17. No other samples from Chuifeng Cave were analysed prior to or during the analysis of CF-B-16.
3. LC-MS/MS ANALYSIS
The extracts were analyzed by nanoflow liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) using a 15 cm capillary column (75 μm inner diameter, packed with 1.9 μm C18 beads (Reprosil-AQ Pur, Dr. Maisch)) on an EASY-nLC™ 1200 system (Proxeon, Odense, Denmark) connected to a Q-Exactive HF-X mass spectrometer (Thermo Scientific, Bremen, Germany). The nLC gradient and MS parameters followed a previously published Q-Exactive HF-X method32. System wash blanks were performed before and after every sample to hinder cross-contamination.
4. DATABASE CONSTRUCTION
We constructed a protein sequence database for Hominoidea proteins known to be present in enamel proteomes (SI Tab. 2), to which we added the homologous sequences from one Cercopithecoid (Macaca mulatta) as an outgroup for phylogenetic analysis. As few protein sequences are publicly available for Pongo pygmaeus, we predicted those sequences from publicly available genomic sequence data using the known gene coordinates of Pongo abelii homologous. Similarly, we generated de novo AMELY sequences for Pongo abelii and Pongo pygmaeus as well. Finally, we added common laboratory contaminants to allow spectra from such proteins to be confidently identified (file taken from the supplements of Hendy et al.37).
Ancestral Sequence Reconstruction
Previous research indicates that cross-species proteomic effects, observed during spectral identification, significantly reduce the identification of phylogenetically informative amino acid positions at large evolutionary distances38. We reasoned that this was likely to occur in the case of Gigantopithecus proteins39, and therefore reconstructed the ancestral protein sequences of enamel-specific proteins. Ancestral Sequence Recontruction (ASR) was conducted across the entire Hominoidea phylogeny using PhyloBot40. Input sequences were constrained phylogenetically to (Macaca,(Nomascus,((Pongo abelii, Pongo pygmaeus),Gorilla,(Homo,((Pan paniscus, Pan troglodytes)))))). We added those sequences to the reference protein database to account for them in the database search of PEAKS and MaxQuant.
Isoform variation
After obtaining complete protein sequences for all extant hominids, we added isoforms not present in UniProt or Genbank for the proteins AMELX, AMELY, AMBN, AMTN, KLK4, and TUFT1, including the reconstructed ASR sequences of these proteins, to the database. We assumed that the isoforms for these non-human hominids would result from identically placed alternative splicing across species and ancestral nodes (as also supported by all UniProt isoforms present for the studied proteins). Thus, we copied these alternative splicing sites onto the available reference sequences to create the missing isoforms. Database sequence names for these proteins were appended with “_ManIso2” or “_ManIso3”.
5. PROTEOMIC DATA ANALYSIS
Raw mass spectrometry data was searched per sample type (enamel, dentine, extraction blank and injection blanks) against a sequence database containing all common enamel proteins for all extant hominids (see above). We used PEAKS41 (v. 7.5) and MaxQuant23 (v. 1.6.2.6) software. The de novo and error-tolerant implementations of PEAKS, and the dependent peptide algorithm implemented in MaxQuant, were used to generate possible, additional, single-amino acid polymorphism (SAP) variation in enamel protein sequences. Such novel SAPs could represent unique amino acid substitutions on the Gigantopithecus lineage, which are not relevant to its phylogenetic placement but are relevant on dating the Pongo–Gigantopithecus divergence. Next, these potential sequence variants were added to a newly constructed sequence database and verified in separate searches in PEAKS and MaxQuant. We defined as variable modifications methionine oxidation, proline hydroxylation, glutamine and asparagine deamidation, pyro-glutamic acid from glutamic acid, pyro-glutamic acid from glutamine, and phosphorylation (STY). No fixed modifications were selected. We did not use an enzymatic protease during sample preparation, therefore the digestion mode was set to “unspecific”. For PEAKS, peptide spectrum matches were only accepted with an FDR ≤ 1.0%, and precursor mass tolerance was set to 10 ppm and fragment mass tolerance to 0.05 Da. For MaxQuant, peptide spectrum matches (PSM) and protein FDR were set at ≤ 1.0%, with a minimum Andromeda scores of 40 for all peptides. Protein matches were accepted with a minimum of two unique peptide sequences in at least one of the MaxQuant or PEAKS searches, including the removal of non-specific peptides after BLASTp searches of peptides matching to non-enamel proteins against UniProt and GenBank databases. Proteins that are retained after applying these criteria are listed in Extended Data Table 2. Examples of annotated MS/MS spectra after MaxQuant analysis can be found in Supplementary Figures S3 to S12.
Assessment of protein damage and degradation followed protocols explained elsewhere17,32,42 and included rates of deamidation and a comparison of observed peptide lengths. Peptide hydrophobicity was calculated using the R package “Peptides”, with the scale set to “KyteDoolittle”.
6. PHYLOGENETIC and DIVERGENCE ANALYSIS
Comparative reference dataset
We assembled a reference dataset with five protein sequences retrieved from the ancient sample (AMBN, AMELX, AMTN, ENAM and MMP20) and relevant extant species (SI Tab. 2). Protein sequences for human (Homo sapiens), common chimpanzee (Pan troglodytes), bonobo (Pan paniscus), Sumatran orangutan (Pongo abelii), Western gorilla (Gorilla gorilla), rhesus macaque (Macaca mulatta), and the white-cheeked gibbon (Nomascus leucogenys), were obtained from the UniProt database. Additionally, we expanded our dataset with protein sequences from publicly available whole-genome sequence data from present-day great apes, (in total 27 orangutans, 42 gorillas, 11 bonobos and 61 chimpanzees25,26,43), as well as 19 human individuals from the Simons Genome Diversity Project44. See the Supplementary Information for the human sample numbers taken from the SGDP dataset.
Reconstruction of protein sequences from whole-genome sequencing data
DNA sequence reads for reference samples used were mapped to human genome (version hg19) using BWA-MEM v0.7.5a-r405 (http://bio-bwa.sourceforge.net/bwa.shtml) with default parameters. PCR and optical duplicates were identified and removed using PICARD v1.91 (https://sourceforge.net/projects/picard/files/picard-tools/1.91/). Single nucleotide polymorphisms were called on the read alignments using the GATK UnifiedGenotyper: (https://software.broadinstitute.org/gatk/documentation/tooldocs/current/org_broadinstitute_gatk_tools_walkers_genotyper_UnifiedGenotyper.php).
To reconstruct the protein sequences from the genotype calls, we first created a consensus sequence for each of the five genes of interest and for each sample. Indels were not considered and a random allele was chosen at heterozygous positions. Next, we removed the intron sequences from each gene using the annotation of the reference human genome (hg19) available in the ENSEMBL database. For each of the in silico spliced genes, we performed a tblastn search45 using the human reference protein as the query. Finally, we obtained the translated protein sequences from the resulting alignments.
Assessing the phylogenetic position of Gigantopithecus blacki
We compared the Gigantopithecus blacki protein sequences with the corresponding homologous of the species in the reference panel. For each gene, we built two multiple sequence alignments using mafft46. The first incorporated all samples in the reference panel (n=164). The second incorporated only a single sample per species (SI Tab. 3). To account for isobaric amino acids (leucines=L and isoleucines=I), which cannot be distinguished in the ancient protein data, we changed all I to L at positions where the ancient sample carried either of those amino acids. To assess the phylogenetic position of the ancient sample, two inference approaches were used: a maximum-likelihood and a Bayesian inference.
Maximum-likelihood approach.
PhyML v. 3.147 was used to infer a maximum-likelihood tree, branch lengths and substitutions rates for each individual protein alignment (SI Fig. 2), and for the concatenated alignment. For each alignment, we started from three random trees (--n_rand_starts 3 -s BEST --rand_start), used the JTT model (-m JTT -f m), and obtained maximum likelihood estimates for the gamma distribution shape parameter (-a e) and the proportion of invariable sites (-v e). Support values were obtained for each bipartition based on 100 non-parametric bootstrap replicates. The bootstrap results per branch split are shown in Extended Data Figure 6b.
Bayesian approach.
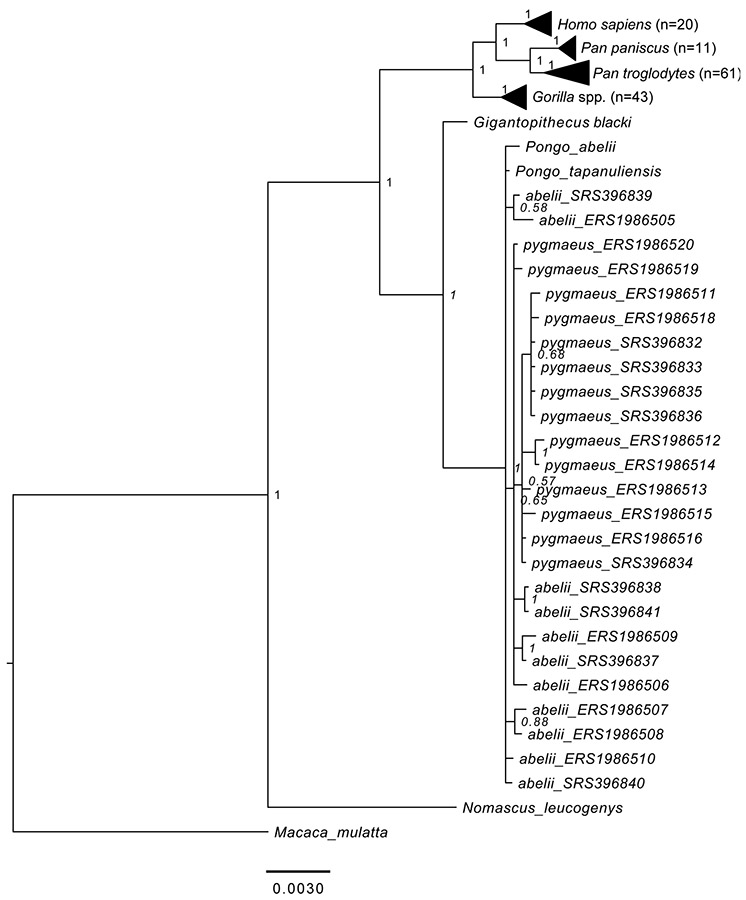
As a complementary approach, we used MrBayes48 and the concatenated alignment to infer the phylogenetic position of the ancient sample (Fig. 3, Extended Data Fig. 6b). We set an independent bipartition for each gene and estimated: substitution rates, across-site rate variation, and the proportion of invariable sites (unlink Statefreq=(all) Ratemultiplier=(all) Aamodel=(all) Shape=(all) Pinvar=(all)). MrBayes was executed using the CIPRES portal49. The MCMC algorithm was set to 5,000,000 cycles with 4 chains and a temperature parameter of 0.2. The convergence of the algorithm was assessed using Tracer v.1.6.0 after discarding 25% of the iterations as burn-in. MrBayes was run against the reference sequence for each species (Extended Data Fig. 6c) or against 162 great ape individuals, one hylobatid, and one cercopithecid (Extended Data Fig. 7). Both of these analyses, as well as the PHyML maximum likelihood approach, resulted in the same topology. The analysis utilizing a large number of individuals shows, however, that resolution within the genus Pongo is limited (Extended Data Fig. 7). Nevertheless, the placement of Gigantopithecus is fully supported.
Divergence time of Gigantopithecus
We estimated the divergence between Gigantopithecus and the Pongo branch first by using a distance-based approach. We used the alignment of the amino acid sequences of reference genome sequences for each species as well as diversity data (see above). A distance matrix was created from the concatenated protein sequences of all individuals using the function dist.ml from the R package phangorn50 under the LG amino acid substitution model51. We used pairwise exclusion to increase the amount of data for the present-day branches. We then calculated the mean difference of all orangutan sequences to all sequences from Homo, Pan, and Gorilla, and the mean difference of all orangutan sequences to Gigantopithecus (Extended Data Fig. 6a). We used the average distance between orangutan and the other extant great apes as a scaling factor, assuming a divergence time between these branches of 23.8 Mya52. Under this assumption, the molecular divergence of Gigantopithecus from the Pongo branch is 9.98 Mya. However, since Chuifeng Cave is dated to 1.9 Mya, this branch is likely underestimated and its age needs to be corrected to 11.88 Mya. We combine the 95% confidence interval of the distance matrix with the 95% confidence interval of the mutation rate estimate52, and add the upper and lower values of the 95% confidence interval for the Chuifeng Cave dating (1.7-2.1 Mya), and thereby suggest conservative upper and lower boundaries for the divergence time of 8.91 and 15.65 Mya, respectively.
If mutation rates did not substantially differ between extant Pongo and Gigantopithecus, this estimate should reflect the molecular evolution of their common branch. We calculated the divergence between the other great apes, taking into account the mutation rate differences on these lineages as scaling factors52. The resulting divergence time between Gorilla and the Homo/Pan branch is estimated at 10.27 Mya (7.9-13.25 Mya, 95% confidence interval), and the divergence between Homo and Pan at 8.72 Mya (8.06-13.81 Mya, 95% confidence interval). These values are in strong agreement with the estimates from Besenbacher et al.52, suggesting that these protein sequences represent well the known phylogeny of the great apes. Clearly, all divergence time estimates scale with assumptions on the mutation rates. We also caution that the small number of mutations in the peptide fragments in Gigantopithecus constitutes a severe limitation on the precision of these estimates on this branch. However, the phylogenetic position of Gigantopithecus as a sister clade to orangutans is also well supported in this analysis: a phylogenetic tree from a distance matrix of the reference sequences for these species (neighbor joining tree in phangorn; maximum likelihood computed with the pml function; 1,000 bootstrap replicates) separates Gigantopithecus from orangutans with 100% bootstrap support.
We used the program MrBayes48 to estimate divergence time estimates in a Bayesian framework using the reference genome sequences. We defined Macaca mulatta as outgroup, grouped Pan, Homo and Gorilla together as well as Pongo and Gigantopithecus, and set the divergence time of the two groups with a uniform distribution of 17.739-26.061 Mya, using the estimate from Besenbacher et al.52. Furthermore, we set the divergence time of the macaques and apes at 26.061-39.9 Mya (from the maximum divergence time of the hominids to a very high divergence time of the apes). We used a variable mutation rate and the VT amino acid substitution model53 in 5 million iterations. This results in a divergence time of Gigantopithecus–Pongo of 10.14 Mya (4.76-15.79 Mya, 95% HPD interval). The divergence of Gorilla from the Homo/Pan branch is estimated at 8.59 Mya (4.62-13.56 Mya, 95% HPD interval), and the divergence of Homo and Pan at 5.78 Mya (2.64-9.53 Mya, 95% HPD interval). These are largely consistent with, but somewhat younger than, previous estimates52,54, possibly due to a mutation slowdown on these lineages compared to the Pongo lineage, which is not taken into account here. However, they seem in agreement with the fossil record indicating the origin of hominins around 6-8 Mya and the dating of a possible early Gorillini (Chororapithecus) around 7-9 Mya54-58. Therefore, we conclude that the relative branch lengths of the tree (Fig. 3b) are concordant with the overall phylogeny and the estimates presented above.
DATA AVAILABILITY
All the mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD013838. Generated ancient protein consensus sequences for both hominins can be found in SI File 2.
Supplementary Material
Extended Data
Extended Data Figure 1. The current environment of Chuifeng Cave.
a, Landscape outside Chuifeng Cave. b, Elevated altitude of Chuifeng Cave (arrow points to the entrance). Photo credit: Wei Wang.
Extended Data Figure 2. Excavations in Chuifeng Cave.
a, Main entrance of the Chuifeng Cave. b, Well-preserved deposits before excavation. c, The stratigraphic profile (1.3 m in height) of Area D. d, W.W. excavating in Area D. e, Excavated channel. f and g, In situ Gigantopithecus blacki teeth (the scale bars are 3 cm). Photo credit: Wei Wang.
Extended Data Figure 3. Total ion current (TIC) chromatograms of the analysed samples.
a, HCl extract. b, TFA extract. Note differences in maximum TIC on the y-axis. Each extract was analysed only once.
Extended Data Figure 4. Comparison of HCl and TFA extraction protocols.
a, Summed and normalized peptide intensities for each combination of peptide length and number of acidic residues (D, E, deamidated N, deamidated Q). b, Summed and normalized intensities by peptide length. c, Summed and normalized peptide intensities across peptide hydrophobicity. Insets show peptide count distribution across peptide hydrophobicity. d, Extraction performance for various categories of the data. Values scaled to one and compared to the best-performing extraction method for each category independently. SAPs refer to those SAPs informative within Hominoidea. e, Proportional Venn diagram of unique peptide sequences identified in the two demineralization methods. All comparisons based on MaxQuant LFQ data only.
Extended Data Figure 5. Damage characteristics of HCl and TFA extraction protocols.
a, Comparison of mean peptide lengths, showing an identical distribution for the TFA (n=305) and HCl (n=191) extractions (two-sided t-test(394)=−0.599, p=0.5495). b, Comparison of asparagine deamidation. c, Comparison of glutamine deamidation. For b and c, violin plots describe the distribution of bootstrap replicates (n=1,000) of intensity-based peptide deamidation32. For some proteins, only deamidated asparagines or glutamines were observed (for example, AMBN), while DCD is included as an example of a non-deamidated contaminant. All comparisons based on MaxQuant data only. For a-c, boxplots define the range of the data (whiskers extending to 1.5 the interquartile range), outliers (beyond 1.5 the interquartile range) 25th and 75th percentiles (boxes), and medians (dots).
Extended Data Figure 6. Unconstrained phylogenetic analysis of Gigantopithecus blacki.
a, Pairwise distances between groups of selected Hominoids and Pongo estimated using the concatenated protein alignments and the phangorn R package. n=number of pairwise comparisons. b, Maximum-likelihood tree computed on a distance matrix using pml R function. Support values were obtained from 1,000 bootstrap replicates. c, Rooted phylogenetic tree obtained using MrBayes. For each bipartition, we show the posterior probability (0-1) obtained from the Bayesian approach and the support values obtained from 100 non-parametric bootstrap replicates in a PHyML maximum-likelihood (0-100) tree. PHyML and MrBayes recover the same topology. Both b and c are based on the same concatenated alignment of the five proteins retrieved from Gigantopithecus, and resulted in the same tree topology.
Extended Data Figure 7. Bayesian phylogenetic tree of Gigantopithecus blacki including 162 modern genomes of great apes, and a single hylobatid (Nomascus leucogenys).
Tree obtained from the concatenated alignment using MrBayes. Macaca mulatta was used as an outgroup. The internal nodes corresponding to Gorilla, Pan and Homo clades are collapsed for visualisation purposes. The number of individuals in each of these nodes is indicated in parentheses.
Extended Data Figure 8. Sequence conservation and structural relevance of retrieved AHSG peptides.
All AHSG-specific peptides, identified by PEAKS and MaxQuant, derive from a single sequence region bridging cystatin domain 1 and 2. The surviving sequence region is evolutionary conserved across Catarrhini. It contains a regular repeat of acidic amino acid (aspartic acids, D, on positions 133, 137, and 141) residues that enable binding of basic calcium phosphate (residues highlighted in green), similarly to a conserved region just N-terminal (glutamic acid, E, on position 111, and aspartic acids, D, on position 113 and 115). At the bottom, a fragment ion alignment is given of MaxQuant-identified AHSG peptides. The serine is phosphorylated in all matching spectra. The consensus sequence for Catarrhini is shown at the top for amino acid positions 100-149 (amino acid coordinates following UniProt accession P02765 FETUA_HUMAN).
Extended Data Table 1. Mean annual temperature (MAT) estimation at Chuifeng Cave.
The 10 geographically closest meteorological weather stations included in publicly available WMO data are used to estimate current MAT at Chuifeng Cave. Correlation of online altitude estimation and WMO provided altitude is R2=0.99 (Pearson correlation).
| WMO station ID |
WMO station name |
Longitude | Latitude | WMO altitude (m) |
Estimated altitude (m) |
First month |
Last month | MAT | Altitude source used |
Chuifeng Cave MAT |
|---|---|---|---|---|---|---|---|---|---|---|
| 5920901 | Ching His | 106.42E | 23.13N | 740 | 743 | February 1981 | October 1990 | 19.5 | WMO | 22.8 |
| 5698505 | Hekou | 103.95E | 22.5N | 137 | 114 | January 1961 | December 1970 | 22.6 | WMO | 22.1 |
| 5791601 | Tien-O | 107.17E | 25.0N | 305 | 245 | January 1981 | October 1990 | 19.7 | WMO | 20.0 |
| 5904602 | Ta Wan | 109.42E | 23.85N | 76 | 81 | January 1981 | October 1990 | 20.8 | WMO | 20.0 |
| 5963201 | Tung Hsing | 107.97E | 21.55N | 13 | 10 | February 1981 | October 1990 | 23.0 | WMO | 22.0 |
| 5921100 | Bose | 106.6E | 23.9N | na | 154 | January 1961 | October 1990 | 22.5 | Estimated altitude | 22.2 |
| 5920901 | Napo | 105.95E | 23.3N | na | 1214 | January 1981 | October 1990 | 19.6 | Estimated altitude | 24.5 |
| 5900700 | Guangnan | 105.07E | 24.07N | na | 1257 | January 1981 | October 1990 | 17.6 | Estimated altitude | 22.8 |
| 5943100 | Nanning | 108.35E | 22.82N | na | 81 | January 1922 | November 1993 | 22.0 | Estimated altitude | 21.2 |
| 5902300 | Hechi | 108.05E | 24.7N | na | 204 | January 1981 | October 1990 | 21.2 | Estimated altitude | 21.1 |
Extended Data Table 2. Enamel proteome sequence coverage.
Only proteins with 2 unique peptides in at least either MaxQuant or PEAKS searches were accepted. No protein matches in the dentine or blank extractions fulfilled this criterion. Primary entry refers to the Pongo abelii entry in UniProt for reference purposes. Protein sequence coverage in the final column indicates the coverage obtained after combining PEAKS and MaxQuant peptide recovery. For amino acid columns, numbers in brackets refer to amino acid positions uniquely identified in PEAKS or MaxQuant searches.
| Protein | Primary entry | Protein accession | MaxQuant peptides (all unique) |
MaxQuant amino acids |
PEAKS peptides (all unique) |
PEAKS amino acids |
Combined sequence coverage (%) |
|---|---|---|---|---|---|---|---|
| AMELX | H2PUX0_PONAB | H2PUX0 | 149 | 135 (4) | 270 | 141 (10) | 70.7 |
| AMBN | H2PDI5_PONAB | H2PDI5 | 55 | 105 (15) | 79 | 107 (11) | 27.5 |
| AMTN | H2PDI4_PONAB | H2PDI4 | 2 | 18 (0) | 2 | 18 (0) | 8.6 |
| ENAM | H2PDI6_PONAB | H2PDI6 | 125 | 129 (5) | 189 | 181 (57) | 16.3 |
| MMP20 | H2NF32_PONAB | H2NF32 | 2 | 9 (0) | 1 | 9 (0) | 1.9 |
| AHSG | H2PC98_PONAB | H2PC98 | 7 | 13 (0) | 12 | 13 (0) | 3.5 |
| ALB | ALBU_Bovin | P02769 | 2 | ||||
| DCD | DCD_human | P81605 | 3 | 8 | |||
| B2MG | B2MG_human | P61769 | 2 | ||||
| K1C9 | K1C9_human | P35527 | 3 |
Acknowledgements
EC and FW are supported by the VILLUM FONDEN (#17649) and by the European Commission through a Marie Skłodowska Curie (MSCA) Individual Fellowship (#795569). TMB is supported by BFU2017-86471-P (MINECO/FEDER, UE), NIHM U01 MH106874 grant, Howard Hughes International Early Career, Obra Social "La Caixa" and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880). E.C., J.C., J.V.O, D.S and P.G. are supported by the Marie Skłodowska-Curie European Training Network (ETN) TEMPERA, a project funded by the European Union’s EU Framework Program for Research and Innovation Horizon 2020 under Grant Agreement No. 722606. M.J.C. and M.M. are supported by the Danish National Research Foundation award PROTEIOS (DNRF128). Work at the Novo Nordisk Foundation Center for Protein Research is funded in part by a donation from the Novo Nordisk Foundation (#NNF14CC0001). Research at Chuifeng Cave is made possible by support from the National Natural Science Foundation of China (#41572023) and by a grant of the Bagui Scholar of Guangxi. MK was supported by a Deutsche Forschungsgemeinschaft (DFG) fellowship (KU 3467/1-1) and the Postdoctoral Junior Leader Fellowship Programme from “la Caixa” Banking Foundation (LCF/BQ/PR19/11700002). MEA is supported by the Independent Research Fund Denmark (#7027-00147B). The authors would like to thank Eske Willerslev for critical reading of the manuscript, scientific support and guidance.
Footnotes
Supplementary Information
Supplementary information is available in the online version of this article.
The authors declare no competing financial interests.
REFERENCES
- 1.Zhang Y & Harrison T Gigantopithecus blacki: a giant ape from the Pleistocene of Asia revisited. American Journal of Physical Anthropology 162, 153–177, doi: 10.1002/ajpa.23150 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Harrison T Apes among the tangled branches of human origins. Science 327, 532–534, doi: 10.1126/science.1184703 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Begun DR How to identify (as opposed to define) a homoplasy: Examples from fossil and living great apes. Journal of Human Evolution 52, 559–572, doi: 10.1016/j.jhevol.2006.11.017 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Kelley J in The Primate Fossil Record (ed Hartwig WC) 369–384 (Cambridge University Press, 2002). [Google Scholar]
- 5.Miller SF, White JL & Ciochon RL Assessing mandibular shape variation within Gigantopithecus using a geometric morphometric approach. American Journal of Physical Anthropology 137, 201–212, doi: 10.1002/ajpa.20856 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Grehan JR & Schwartz JH Evolution of the second orangutan: phylogeny and biogeography of hominid origins. Journal of Biogeography 36, 1823–1844, doi: 10.1111/j.1365-2699.2009.02141.x (2009). [DOI] [Google Scholar]
- 7.Shao Q et al. ESR, U-series and paleomagnetic dating of Gigantopithecus fauna from Chuifeng Cave, Guangxi, southern China. Quaternary Research 82, 270–280, doi: 10.1016/j.yqres.2014.04.009 (2014). [DOI] [Google Scholar]
- 8.Wang W New discoveries of Gigantopithecus blacki teeth from Chuifeng Cave in the Bubing Basin, Guangxi, south China. Journal of Human Evolution 57, 229–240, doi: 10.1016/j.jhevol.2009.05.004 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Bartlett JD et al. Protein–Protein Interactions of the Developing Enamel Matrix. Current Topics in Developmental Biology 74, 57–115, doi: 10.1016/S0070-2153(06)74003-0 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Dean MC & Schrenk F Enamel thickness and development in a third permanent molar of Gigantopithecus blacki. Journal of Human Evolution 45, 381–388, doi: 10.1016/j.jhevol.2003.08.009 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Von Koenigswald GHR Eine fossile Saugetierfauna mit Simia aus Sudchina. Proceedings van de Koninklijke Nederlandse Akademie van Wetenschappen 38, 872–879 (1935). [Google Scholar]
- 12.Zhang Y et al. New 400–320 ka Gigantopithecus blacki remains from Hejiang Cave, Chongzuo City, Guangxi, South China. Quaternary International 354, 35–45, doi: 10.1016/j.quaint.2013.12.008 (2014). [DOI] [Google Scholar]
- 13.Zhao LX & Zhang LZ New fossil evidence and diet analysis of Gigantopithecus blacki and its distribution and extinction in South China. Quaternary International 286, 69–74, doi: 10.1016/j.quaint.2011.12.016 (2013). [DOI] [Google Scholar]
- 14.Pei WC Excavation of Liucheng Gigantopithecus cave and exploration of other caves in Kwangsi. Memoir of the Institute of Vertebrate Palaeontology and Palaeoanthropology, Academia Sinica 7, 1–54 (1965). [Google Scholar]
- 15.Bocherens H et al. Flexibility of diet and habitat in Pleistocene South Asian mammals: Implications for the fate of the giant fossil ape Gigantopithecus. Quaternary International 434, 148–155, doi: 10.1016/j.quaint.2015.11.059 (2017). [DOI] [Google Scholar]
- 16.Ciochon R et al. Dated co-occurrence of Homo erectus and Gigantopithecus from Tham Khuyen Cave, Vietnam. Proceedings of the National Academy of Sciences 93, 3016–3020, doi: 10.1073/pnas.93.7.3016 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cappellini E et al. Early Pleistocene enamel proteome from Dmanisi resolves Stephanorhinus phylogeny. Nature, doi: 10.1038/s41586-019-1555-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer M et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507, doi: 10.1038/nature17405 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wadsworth C & Buckley M Proteome degradation in fossils: investigating the longevity of protein survival in ancient bone. Rapid Communications in Mass Spectrometry 28, 605–615, doi: 10.1002/rcm.6821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart NA et al. The identification of peptides by nanoLC-MS/MS from human surface tooth enamel following a simple acid etch extraction. RSC Advances 6, 61673–61679, doi: 10.1039/c6ra05120k (2016). [DOI] [Google Scholar]
- 21.Castiblanco GA et al. Identification of proteins from human permanent erupted enamel. European Journal of Oral Sciences 123, 390–395, doi: 10.1111/eos.12214 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Cristobal A et al. Toward an Optimized Workflow for Middle-Down Proteomics. Analytical Chemistry 89, 3318–3325, doi: 10.1021/acs.analchem.6b03756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology 26, 1367–1372, doi: 10.1038/nbt.1511 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Tagliabracci VS et al. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 336, 1150–1153, doi: 10.1126/science.1217817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prado-Martinez J et al. Great ape genetic diversity and population history. Nature 499, 471–475, doi: 10.1038/nature12228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nater A et al. Morphometric, Behavioral, and Genomic Evidence for a New Orangutan Species. Current Biology 27, 3487–3498, doi: 10.1016/j.cub.2017.11.020 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Tang N & Skibsted LH Calcium Binding to Amino Acids and Small Glycine Peptides in Aqueous Solution: Toward Peptide Design for Better Calcium Bioavailability. Journal of Agricultural and Food Chemistry 64, 4376–4389, doi: 10.1021/acs.jafc.6b01534 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Heiss A et al. Structural Basis of Calcification Inhibition by alpha sub(2)-HS Glycoprotein/Fetuin-A -- Formation Of Colloidal Calciprotein Particles. Journal of Biological Chemistry 278, 13333–13341, doi: 10.1074/jbc.M210868200 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Demarchi B et al. Protein sequences bound to mineral surfaces persist into deep time. eLife 5, e17092, doi: 10.7554/eLife.17092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price PA, Toroian D & Lim JE Mineralization by inhibitor exclusion: the calcification of collagen with fetuin. The Journal of biological chemistry 284, 17092–17101, doi: 10.1074/jbc.M109.007013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kono RT, Zhang Y, Jin C, Takai M & Suwa G A 3-dimensional assessment of molar enamel thickness and distribution pattern in Gigantopithecus blacki. Quaternary International 354, 46–51, doi: 10.1016/j.quaint.2014.02.012 (2014). [DOI] [Google Scholar]
- 32.Mackie M et al. Palaeoproteomic Profiling of Conservation Layers on a 14th Century Italian Wall Painting. Angewandte Chemie (International ed.) 57, 7369–7374, doi: 10.1002/anie.201713020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin C et al. Chronological sequence of the early Pleistocene Gigantopithecus faunas from cave sites in the Chongzuo, Zuojiang River area, South China. Quaternary International 354, 4–14, doi: 10.1016/j.quaint.2013.12.051 (2014). [DOI] [Google Scholar]
- 34.Huang WC,R; Gu Y; Larick R; Fang Q; Yonge C; de Vos J; Schwarcz HP; Rink WJ. Earliest hominids and artifacts from Asia: Longgupo Cave, Central China. Nature 378, 275–278 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Pei W Discovery of Gigantopithecus mandibles and other material in Liucheng district of central Kwangsi in South China. Vertebrata PalAsiatica 1, 65–71 (1957). [Google Scholar]
- 36.Sun L et al. Magnetochronological sequence of the Early Pleistocene Gigantopithecus faunas in Chongzuo, Guangxi, southern China. Quaternary International 354, 15–23, doi: 10.1016/j.quaint.2013.08.049 (2014). [DOI] [Google Scholar]
- 37.Hendy J et al. A guide to ancient protein studies. Nature Ecology & Evolution 2, 791–799, doi: 10.1038/s41559-018-0510-x (2018). [DOI] [PubMed] [Google Scholar]
- 38.Welker F Elucidation of cross-species proteomic effects in human and hominin bone proteome identification through a bioinformatics experiment. BMC Evolutionary Biology 18, 23, doi: 10.1186/s12862-018-1141-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welker F Palaeoproteomics for human evolution studies. Quaternary Science Reviews 190, 137–147, doi: 10.1016/j.quascirev.2018.04.033 (2018). [DOI] [Google Scholar]
- 40.Hanson-Smith V & Johnson A PhyloBot: A Web Portal for Automated Phylogenetics, Ancestral Sequence Reconstruction, and Exploration of Mutational Trajectories. PLoS computational biology 12, e1004976, doi: 10.1371/journal.pcbi.1004976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J et al. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Molecular and Cellular Proteomics 11, M111.010587, doi: 10.1074/mcp.M111.010587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welker F et al. Palaeoproteomic evidence identifies archaic hominins associated with the Châtelperronian at the Grotte du Renne. Proceedings of the National Academy of Sciences 113, 11162–11167, doi: 10.1073/pnas.1605834113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Manuel M et al. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science 354, 477–481, doi: 10.1126/science.aag2602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mallick S et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206, doi: 10.1038/nature18964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. Journal of molecular biology 215, 403–410, doi: 10.1006/enrs.2002.4406 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Katoh K & Frith MC Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28, 3144–3146, doi: 10.1093/bioinformatics/bts578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guindon S et al. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Systematic Biology 59, 307–321, doi: 10.1093/sysbio/syq010 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Ronquist F et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology 61, 539–542, doi: 10.1093/sysbio/sys029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller MA, Pfeiffer W & Schwartz T in Gateway Computing Environments Workshop (GCE) 1–8 (New Orleans, 2010). [Google Scholar]
- 50.Schliep KP phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593, doi: 10.1093/bioinformatics/btq706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le SQ & Gascuel O An Improved General Amino Acid Replacement Matrix. Molecular Biology and Evolution 25, 1307–1320, doi: 10.1093/molbev/msn067 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Besenbacher S, Hvilsom C, Marques-Bonet T, Mailund T & Schierup MH Direct estimation of mutations in great apes reconciles phylogenetic dating. Nature Ecology & Evolution 3, 286–292, doi: 10.1038/s41559-018-0778-x (2019). [DOI] [PubMed] [Google Scholar]
- 53.Müller T & Vingron M Modeling amino acid replacement. Journal of computational biology : a journal of computational molecular cell biology 7, 761–776, doi: 10.1089/10665270050514918 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Langergraber KE et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proceedings of the National Academy of Sciences 109, 15716–15721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh S et al. New geological and palaeontological age constraint for the gorilla–human lineage split. Nature 530, 215–218, doi: 10.1038/nature16510 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Senut B et al. First hominid from the Miocene (Lukeino Formation, Kenya). Comptes Rendus de l’Académie des Sciences - Series IIA - Earth and Planetary Science 332, 137–144, doi: 10.1016/S1251-8050(01)01529-4 (2001). [DOI] [Google Scholar]
- 57.Brunet M et al. A new hominid from the Upper Miocene of Chad, central Africa. Nature 418, 145–151, doi: 10.1038/nature00879 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Haile-Selassie Y Late Miocene hominids from the Middle Awash, Ethiopia. Nature 412, 178–181, doi: 10.1038/35084063 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD013838. Generated ancient protein consensus sequences for both hominins can be found in SI File 2.