Abstract
The past decade has shown exponential growth in the field of RNA nanotechnology. The rapid advances of using RNA nanoparticles for biomedical applications, especially targeted cancer therapy, suggest its potential as a new generation of drug. After the first milestone of small molecule drugs and the second milestone of antibody drugs, it was predicted that RNA drugs, either RNA itself or chemicals/ligands that target RNA, will be the third milestone in drug development. Thus, a comprehensive assessment of the current therapeutic RNA nanoparticles is urgently needed to meet the drug evaluation criteria. Specifically, the pharmacological and immunological profiles of RNA nanoparticles need to be systematically studied to provide insights in rational design of RNA-based therapeutics. By virtue of its programmability and biocompatibility, RNA molecules can be designed to construct sophisticated nanoparticles with versatile functions/applications and highly tunable physicochemical properties. This intrinsic characteristic allows the systemic study of the effects of various properties of RNA nanoparticles on their in vivo behaviors such as cancer targeting and immune responses. This review will focus on the recent progress of RNA nanoparticles in cancer targeting, and summarize the effects of common physicochemical properties such as size and shape on the RNA nanoparticles’ bio-distribution and immunostimulation profiles.
Keywords: biodistribution, immune response, immunomodulation, immunostimulation, nanobiotechnology, RNA nanoparticle, RNA nanostructure, RNA nanotechnology
1 |. INTRODUCTION
Nowadays, nanotechnology-based platforms such as lipid-based particles (Puri et al., 2009), nucleic acids nanoparticles (Andersen et al., 2009; Guo, Tschammer, Mohammed, & Guo, 2005), viral nanoparticles (Singh et al., 2007), and synthetic inorganic and polymeric particles (Astruc, 2012) are finding ever-increasing applications in various fields, including nanomedicine. Nevertheless, one challenge in clinical development of nanomedicine is the lack of sufficient evidence regarding their safety, immunological, and pharmacological profiles. Inadvertent recognition of nanomaterials as invaders by the immune systems frequently results in varying levels of immunostimulation or immunosuppression, which consequently leads to toxicity and reduced therapeutic efficacy. Particularly, the common challenges in the immunotoxicology assessment of nanomaterials have been summarized by the NCI Nanotechnology Characterization Laboratory, highlighting four key areas (Chemistry, Efficacy, Pharmacology and Toxicology, and Hematology and Immunology) requiring thorough characterization to efficiently translate nanoparticle-formulated drugs toward the clinic (Dobrovolskaia, 2015). Typically, their physicochemical characterization needs to be well-assessed, and they are expected to display favorable pharmacokinetics (PK), pharmacodynamics (PD), and strong safety profiles while retaining potent drug efficacy against the targeted disease. Some strategies, for example, poly (ethylene glycol) (PEG) coating on cationic lipid-based nanocarriers (Suk, Xu, Kim, Hanes, & Ensign, 2016), have been proposed to engineer nanoparticles that minimize unwanted immunotoxicity while improving their in vivo performance. However, additional side effects induced by PEG-specific antibodies were also reported (Zhang, Sun, Liu, & Jiang, 2016). Refining nanoparticle size is another approach proposed to diminish immune response because phagocytic cells in the immune systems tend to pick up larger nanoparticles. Unfortunately, some nanoparticles were formulated with unpredictable or heterogeneous size distributions, making consistent assembly particularly difficult (Desai, 2012). While immune responses are vital, the favorable in vivo biodistribution of a nanoplatform to achieve specific cancer targeting is another key factor for efficacious drug delivery. With active targeting ligands, nanoparticles exhibit distinct advantages over traditional small molecule therapeutics for overcoming PK limitations by virtue of their prolonged circulation time and extended accumulation in the tumors (Singh & Lillard Jr., 2009). Upon systemic administration, nanoparticles are exposed to the physiological environment rich with proteins, cells and tissues. Size and shape have both been reported to play important roles on the PK profile and biodistribution of nanoparticles (Hoshyar, Gray, Han, & Bao, 2016; Toy, Peiris, Ghaghada, & Karathanasis, 2014). Nanoparticles smaller than 5 nm undergo significant filtration by the kidneys and are mainly excreted in urine. Larger particles, ranging from 20 to 100 nm, are easily engulfed by macrophages or sequestered in healthy tissues, thus causing nonspecific and undesirable accumulation in healthy organs (Gustafson, Holt-Casper, Grainger, & Ghandehari, 2015). Nanoparticles with distinct shapes also exhibit different hemorheological dynamics and cellular uptake. For instance, Discher et al reported prolonged circulation time of filamentous polymer micelles compared to spherical counterparts (Geng et al., 2007). Additionally, the surface characteristics of nanoparticles such as hydrophobicity and surface charge can greatly influence protein adsorption and cellular membrane interactions that are important to the in vivo performance of nanoparticles (Albanese, Tang, & Chan, 2012). Adsorption to specific opsonin proteins such as the complement proteins, IgG and laminin will facilitate the recognition and uptake by immune cells, such as macrophages. As a result of this nanoparticle uptake, macrophages secrete cytokines, possibly leading to immunostimulatory responses. Thus, notwithstanding the achievement of nanotechnology has shown in drug delivery, the translation of synthetic or biological nanoparticles into clinical trials will mandate extensive investigations in accordance with strict criteria.
As a naturally-occurring biopolymer, RNA is unique compared to other nanomaterials. RNA molecules possess diverse sequences, secondary structures, tertiary and quaternary interactions by nature (Hendrix, Brenner, & Holbrook, 2005). Unlike other biomacromolecules (e.g., DNA and protein), RNA is structurally more flexible and functionally more versatile (Guo, 2010; Jasinski, Haque, Binzel, & Guo, 2017). Taking advantage of both canonical Watson-Crick (A-U, G-C) and non-canonical (e.g., G-U) base pairing, a substantial number of natural RNA motifs and self-folding RNAs have been discovered. These include, but are not limited to: kink-turns (Huang & Lilley, 2013; Huang & Lilley, 2016), kissing hairpins (Bindewald, Hayes, Yingling, Kasprzak, & Shapiro, 2008), paranemic motifs (Afonin, Cieply, & Leontis, 2008), pseudoknot (Bindewald, Afonin, Jaeger, & Shapiro, 2011), three-way (Shu, Shu, Haque, Abdelmawla, & Guo, 2011; Zhang et al., 2013) and multi-helix junctions (Laing, Jung, Iqbal, & Schlick, 2009), bulges and loops (Zacharias & Hagerman, 1995). As a result of the highly influential research in previous traditional RNA biology, the field of RNA nanotechnology has experienced an exponential growth in the past decade (Jasinski et al., 2017; Li et al., 2015). Myriads of sophisticated RNA architectures with highly-ordered structures and multivalent functionalities were assembled using RNA as building blocks, such as RNA polygons (Boerneke, Dibrov, & Hermann, 2016; Bui et al., 2017; Dibrov, McLean, Parsons, & Hermann, 2011; Huang & Lilley, 2016; Jasinski, Khisamutdinov, Lyubchenko, & Guo, 2014; Khisamutdinov et al., 2014; Ohno et al., 2011; Severcan, Geary, Verzemnieks, Chworos, & Jaeger, 2009), RNA polyhedrons (Afonin et al., 2010; Hao et al., 2014; Khisamutdinov et al., 2016; Li et al., 2016; Severcan et al., 2010; Xu et al., 2019; Yu, Liu, Jiang, Wang, & Mao, 2015), RNA rings (Geary, Chworos, Verzemnieks, Voss, & Jaeger, 2017; Grabow et al., 2011; Shu et al., 2013), RNA dendrimers (Sharma et al., 2015), jigsaw puzzles (Chworos et al., 2004), and RNA filaments (Nasalean, Baudrey, Leontis, & Jaeger, 2006) (Figure 1).
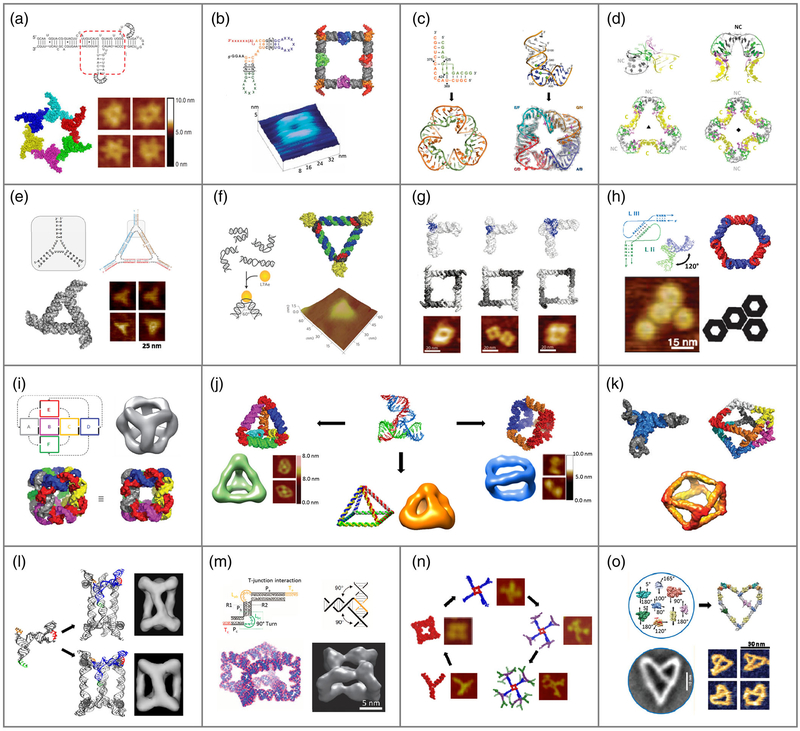
FIGURE 1.
Modular design and construction of RNA nanostructures. (a) pRNA hexamer from phi29 DNA packaging motor (Reprinted with permission from Shu, Haque, et al. (2013). Copyright 2013 RNA society); (b) RNA square with tectoRNA motif (Reprinted with permission from Chworos et al. (2004). Copyright 2004 The American Association for the Advancement of Science); (c) RNA triangle with IIa motif from SVV IRES (Reprinted with permission from Boerneke et al. (2016)). Copyright 2016 John Wiley & Sons, Inc.) & RNA square with IIa-1 motif from HCV (Reprinted with permission from Dibrov et al. (2011). Copyright 2011 National Academy of Sciences; (d) RNA triangle and square with k-turn motif (Reprinted with permission from Huang and Lilley (2016). Copyright 2016 The Royal Society of Chemistry); (e) RNA triangle with tetra-U motif (Reprinted with permission from Bui et al. (2017). Copyright 2017 Elsevier Inc).; (f) RNA-protein triangle with k-turn motif (Reprinted with permission from Ohno et al. (2011). Copyright 2011 Nature Publishing Group); (g) RNA tectosquare with RA, 3WJ, and tRNA motifs (Reprinted with permission from Severcan et al. (2009). Copyright 2009 American Chemical Society); (h) RNA nanoring based on RNAI/II inverse kissing complex (Reprinted with permission from Grabow et al. (2011). Copyright 2011 American Chemical Society); (i) RNA cube designed in silico (Reprinted with permission from Afonin et al. (2010). Copyright 2010 Macmillan Publishers Limited); (j) pRNA-3WJ motif-based RNA tetrahedron (Reprinted with permission from Li et al. (2016). Copyright 2016 John Wiley & Sons, Inc.), pyramid (Reprinted with permission from Xu C et al. (2018). Copyright 2018 Springer Nature), & nanoprism (Reprinted with permission from Khisamutdinov et al. (2016)). Copyright 2016 John Wiley & Sons, Inc.); (k) RNA polyhedron made of tRNA subunit (Reprinted with permission from Severcan et al. (2010). Copyright 2010 Macmillan Publishers Limited); (l) triangular and tetragonal RNA prism from re-engineered pRNA (Reprinted with permission from Hao et al. (2014). Copyright 2014 Macmillan Publishers Limited); (m) homo-octameric RNA prism with T-junction RNA tile (Reprinted with permission from Yu et al. (2015). Copyright 2015 Macmillan Publishers Limited); (n) RNA dendrimers from pRNA-3WJ motif (Reprinted with permission from Sharma et al. (2015). Copyright 2015 Elsevier Inc.); (o) RNA nanoheart from a syntax of RNA modules (Reprinted with permission from Geary et al. (2017)). Copyright 2017 American Chemical Society
Recently, the three-way junction (3WJ) motif of packaging RNA (pRNA) originated from phi29 DNA packaging motor has been exploited as a scaffold for fabrication of multi-functional and thermodynamically stable RNA nanoparticles (Shu et al., 2011). Different components including therapeutic (miRNA, siRNA, chemical drugs), targeting (RNA aptamers or chemical ligands), and imaging (fluorophores or fluorogenic aptamers) modules were incorporated to the 3WJ motif without affecting the authentic folding and original functions of modules (Shu, Khisamutdinov, Zhang, & Guo, 2013). Tremendous efforts have been made to improve the enzymatic stability of RNA by chemical modifications such as 2′-fluorine (2’-F) on pyrimidines, making them dramatically resistant to RNase degradation (Corey, 2007). These RNA nanoparticles are highly programmable, namely their physicochemical properties can be easily engineered to favor in vivo therapeutic delivery (Guo et al., 2017; Jasinski et al., 2014; Jasinski, Li, & Guo, 2018; Jasinski, Yin, Li, & Guo, 2018; Khisamutdinov, Li, et al., 2014; Pi et al., 2018; Shu et al., 2018). Upon systemic injection in tumor-bearing mice, the RNA nanoparticles displayed favored accumulation at the tumor site, with little to no accumulation in vital organs (Binzel et al., 2016; Cui et al., 2015; Lee et al., 2015; Pi et al., 2018; Rychahou et al., 2015; Shu et al., 2015; Shu et al., 2018; Shu, Haque, et al., 2013). The negatively charged backbone of RNA disallows nonspecific interaction with negatively charged cell membranes. The pRNA-based nanoparticles showed favorable pharmacological profiles and induced negligible interferon or cytokine secretion in vivo (Abdelmawla et al., 2011; Guo et al., 2017; Khisamutdinov, Li, et al., 2014; Shu et al., 2018). Thus, the significant progress of therapeutic RNA nanoparticles has revealed its potential to pioneer a new generation of drug for the future market. It was predicted that RNA drugs, either RNA itself or chemicals that target RNA, will become the third milestone in drug development, following the first generation of small molecule drugs and the second generation of antibody drugs (Shu et al., 2014).
In order to develop therapeutic RNA nanoparticles into a new drug, they must meet all the FDA’s drug evaluation criteria and overcome the obstacles limiting their clinical advancement. To date, many important barriers (e.g., chemical stability and thermodynamic stability) in the field of RNA nanotechnology have been successfully overcome (Corey, 2007; Khisamutdinov, Jasinski, & Guo, 2014; Shu et al., 2011). The progress in these aspects has been discussed in previous reviews (Jasinski et al., 2017; Li et al., 2015; Xu et al., 2018). Recently, however, more studies have been reported on understanding the broad mechanisms that define the pharmacological and immunological profile of RNA nanoparticles. A variety of their physicochemical properties were recognized to be associated with their in vivo behaviors, including size, shape, stoichiometry/module density, surface chemistry, composition, hydrophobicity, and elasticity (Abdelmawla et al., 2011; Afonin et al., 2014; Guo et al., 2017; Halman et al., 2017; Hong et al., 2018; Jasinski, Li, & Guo, 2018; Jasinski, Yin, et al., 2018; Johnson et al., 2017; Ke et al., 2018; Khisamutdinov, Li, et al., 2014; Lee, Urban, Xu, Sullenger, & Lee, 2016; Pi et al., 2018; Shi et al., 2018; Shu et al., 2018). Here, this brief review aims to integrate the recent progress seen in the study of RNA nanoparticle on biodistribution and immunostimulation, while focusing on the defining aspects of their design and physiochemical properties precisely tuned to treat different cancers.
2 |. DEFINITION OF RNA NANOTECHNOLOGY
RNA nanotechnology is a relatively new field distinctive from traditional RNA biology. The concept of RNA nanotechnology is defined as the study on the design, construction, and application of RNA nanoparticles at the nanometer scale that are primarily composed of RNA via bottom-up self-assembly (Guo, 2010). The major frame of RNA nanoparticles can include scaffolds, targeting ligands, regulatory moieties, and therapeutic modules. The field focuses on the inter-RNA interactions and quaternary interactions of small RNA motifs, which is different from classical RNA research focusing on intra-RNA interactions and 2D/3D structure–function relationships. Nevertheless, the broad knowledge gained from classical RNA biology research has laid a solid foundation for RNA nanotechnology. Since the introduction of the concept describing RNA nanoarchitectures in 1996 and 1998 by RNA tectonics and reengineered pRNA molecules respectively (Guo, Zhang, Chen, Trottier, & Garver, 1998; Jaeger, Westhof, & Leontis, 2001), RNA nanotechnology has grown rapidly as a platform with extensive applicational studies in nanomedicine over the past decade (Jasinski et al., 2017; Li et al., 2015).
3 |. ADVANTAGES OF RNA NANOTECHNOLOGY FOR CANCER TARGETING AND IMMUNOMODULATION
3.1 |. RNA nanoparticles are distinct from traditional therapeutic RNAs
Traditional therapeutic RNAs, such as siRNA (Elbashir et al., 2001), miRNA (Bartel, 2004), anti-miRNA (Elmen et al., 2008), mRNA (Sahin, Kariko, & Tureci, 2014), viral immunostimulatory RNA (isRNA) (Bourquin et al., 2007), and ribozymes (Sarver et al., 1990), have a long history of fundamental research. Some of these small RNAs were reported to be immunogenic (Berger et al., 2009; Bourquin et al., 2007; Heil et al., 2004; Hornung et al., 2005; Judge et al., 2005). Cellular uptake of these RNAs might stimulate RNA-sensing pattern recognition receptors (PRRs) (Mogensen, 2009). RNA nanoparticles are distinguishable from traditional therapeutic RNAi and other small functional RNAs, though they share the similar chemical property of ribonucleotide composition. The advantages of RNA nanotechnology include: (a) therapeutic RNAi components or small non-coding RNA can serve as both scaffolds and functional moieties to construct larger scale nanostructures through a bottom-up self-assembly (Figure 1); (b) the nanoscale size offers favorable PK and PD profiles, enhanced permeability and retention (EPR) effect (Yin, Wang, Li, Shu, & Guo, 2019), minimal liver accumulation (Binzel et al., 2016; Shu et al., 2015), and undetectable toxicity in vivo (Abdelmawla et al., 2011; Yin et al., 2019); (c) RNA nanoparticles are generally assembled with a defined size, shape, and structure, and these morphologies are easily tunable (Bui et al., 2017; Jasinski et al., 2014; Khisamutdinov, Li, et al., 2014; Monferrer, Zhang, Lushnikov, & Hermann, 2019); (d) Unlike small RNAs, RNA nanoparticles can harbor multi-valent functionalities such as a targeting ligand and various drugs for combination therapy (Afonin et al., 2014; Zhang et al., 2017); (e) RNA nanoparticles display thermal, chemical and metabolic stability in biological matrices. In a more comprehensive pharmacological characterization study of pRNA nanoparticles (Abdelmawla et al., 2011), various secondary PK parameters have been evaluated (Table 1). The normalized volume of distribution Vd (1.2 L/kg) of pRNA nanoparticles suggested the distribution of a significant fraction to peripheral tissues (outside vascular or extravasation), particularly in tumor. The relatively small clearance (Cl) value (significantly below the kidney filtration rate) suggests that the nanoparticles were not efficiently filtered out by the kidneys. By taking advantage of these features, traditional small therapeutic RNAs can be conveniently incorporated into RNA nanoparticles to achieve specific targeted delivery, an extended in vivo half-life, and higher therapeutic efficacy (Jasinski et al., 2017; Xu, Haque, et al., 2018).
TABLE 1.
Secondary PK parameters of pRNA nanoparticles (Reprinted with permission from Abdelmawla et al. (2011). Copyright 2011 Elsevier Inc.)
| Key parameters | 2’-F-pRNA | siRNA |
|---|---|---|
| AUClast (hour·ng/ml) | 2 × 105 | 1.1 × 103 |
| T1/2 (hours) | 5–10 | 0.25 |
| Vd (l/kg) | 1.2 | 0.36 |
| Cl (l/kg/hour) | 0.13 | 1.0 |
| Dose (mg/kg) | 24 | 1.2 |
Abbreviations: AUC, area under the curve; Cl, clearance; PK, pharmacokinetic; T1/2, half-life; Vd, volume of distribution.
3.2 |. RNA nanoparticles show favorable cancer targeting and minimal organ accumulation
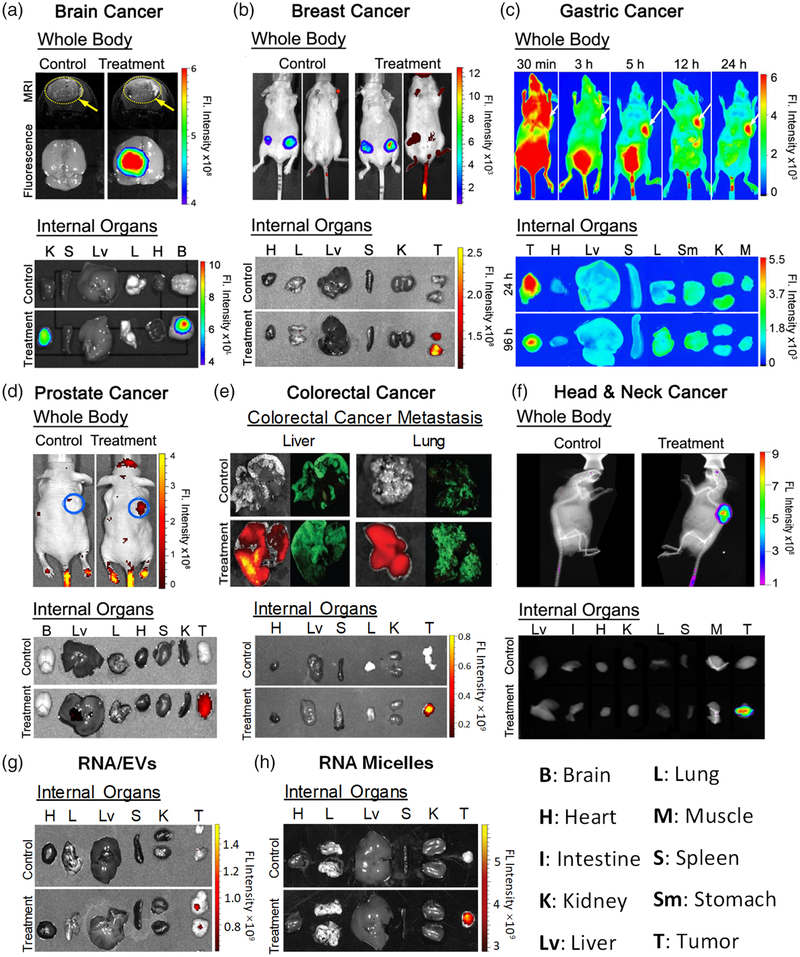
One of the critical challenges to overcome in cancer nanotechnology is the non-specific accumulation of administered nanoparticles in healthy organs including the liver, lungs, kidneys, and spleen (Shi, Kantoff, Wooster, & Farokhzad, 2017). This non-specificity not only distracts nanoparticles from transporting to tumors, but also causes toxicity and unwanted side effects. To address this challenge, Guo Lab has developed a series of pRNA-3WJ based RNA nanoparticles incorporated with targeting ligands (Binzel et al., 2016; Cui et al., 2015; Lee et al., 2015; Pi et al., 2018; Rychahou et al., 2015; Shu et al., 2015; Shu et al., 2018; Shu, Haque, et al., 2013; Xu et al., 2018). Upon systemic administration in tumor-bearing mice, these RNA nanoparticles specifically transport to tumors within about 4 hours(h), and successfully remain at the tumor site for over 24 h. No or minimal organ accumulation was detected several hours post-injection. Consistent results were observed in several common cancer models (glioblastoma, breast, gastric, prostate, colorectal, and head and neck cancers) (Figure 2), in which the incorporated targeting ligands were changed to bind specific receptors overexpressed on different cancer cell surfaces. Meanwhile, therapeutics such as anti-miRNA21 can be delivered into cancer cells with the aid of RNA nanoparticles via receptor-mediated endocytosis, thus improving the therapeutic effects (Binzel et al., 2016; Shu et al., 2015). Another novel study reported that a globular RNA micelle can efficiently deliver anti-miRNA21 to tumors without a targeting ligand (Guo et al., 2017; Khisamutdinov et al., 2016; Shu et al., 2018; Yin et al., 2019). It is suggested the negatively charged nature of RNA also plays an important role in targeting specificity because it minimizes nonspecific interactions with the negatively charged cell membranes. Furthermore, the strong elasticity and branched ratchet-like shape of pRNA-3WJ based nanoparticles confer higher tumor penetration and improve EPR effects. Collectively, these results demonstrate that pRNA-3WJ based nanoparticles can be conveniently engineered with active targeting ligands to achieve specific cancer targeting with low accumulation in healthy organs. This favorable biodistribution is an important indication of RNA nanoparticles’ pharmacological profiles.
FIGURE 2.
Specific cancer targeting in vivo of RNA nanoparticles to (a) brain cancer (Reprinted with permission from Lee et al., 2015). Copyright 2015 Impact Journals; (b) breast cancer (Reprinted with permission from Shu et al. (2015). Copyright 2015 American Chemical Society); (c) gastric cancer (Reprinted with permission from Cui et al. (2015). Copyright 2015 Macmillan Publishers Limited); (d) prostate cancer (Reprinted with permission from Binzel et al. (2016). Copyright 2016 Elsevier Inc.); (e) colorectal cancer (Reprinted with permission from Rychahou et al. (2015); Xu, Pang, et al. (2018). Copyright 2015 American Chemical Society & Elsevier B.V).; (f) Head & Neck cancer (Reprinted with permission from Shu, Haque, et al. (2013). Copyright 2013 RNA society); (g) specific cancer targeting of RNA/EVs (Reprinted with permission from Pi et al. (2018). Copyright 2018 Springer Nature Publishing), and (h) RNA micelles (Reprinted with permission from Shu et al. (2018)). Copyright 2018 Elsevier Inc)
3.3 |. RNA nanoparticles intrinsically display immunologically inert property and non-toxicity
Due to the lack of a universal nomenclature to categorize traditional therapeutic RNAs and RNA nanoparticles, the literatures on RNA immunogenicity have been controversial. Though the immunogenicity of traditional small RNAs has been widely investigated, only a limited number of studies focused on the immunogenicity of RNA nanoparticles. Recent studies revealed that pRNA-based RNA nanoparticles intrinsically display immunologically inert properties (Abdelmawla et al., 2011; Guo et al., 2017; Khisamutdinov, Li, et al., 2014; Shu et al., 2018). Specifically, no or negligible cytokine induction including TNF-α (Tumor Necrosis Factor-α), IL-6 (Interleukin-6) and IFN-α (Interferon-α) has been observed following treatment with pRNA nanoparticles in vitro and in vivo. TNF-α is a cytokine involved in systemic inflammation and the acute phase reaction. IL-6 is an interleukin secreted to stimulate immune responses during infection. IFN-α, which belongs to type I interferon, is also a cytokine involved in pro-inflammatory reactions released in response to the presence of viral pathogens. Besides, no stimulation of TLRs (Toll-like receptors) pathway, and no damage to normal tissue and organs was detected in multiple cell types and mice (Cui et al., 2015; Zhang et al., 2017). Similarly, RNA polygons (RNA triangle, square, and pentagon) constructed from the thermodynamically stable pRNA-3WJ were studied (Guo et al., 2017; Khisamutdinov, Li, et al., 2014). These RNA polygons have been considered nonimmunogenic and nontoxic because undetectable or negligible cytokine induction and cytotoxicity were observed in vitro and in vivo, compared to positive controls. Consistent results were found in three dimensional pRNA-based nanoparticles, including the RNA tetrahedron, RNA nanoprism, and RNA micelles (Guo et al., 2017; Khisamutdinov et al., 2016; Shu et al., 2018; Yin et al., 2019). Additionally, RNA aptamers, a family of RNA oligonucleotides commonly incorporated into RNA nanoparticles as targeted ligands to enhance binding specificity, have been reported to go unrecognized by the host immune system in various animal studies (Song, Lee, & Ban, 2012). These findings demonstrate that RNA nanoparticles equipped with targeting ligands can serve as safe delivery vectors in therapeutic interventions. Studies by Afonin Lab using a different system have also shown no immune response detection upon treatment with RNA nanoparticles in human peripheral blood mononuclear cells (PBMCs) from healthy donors (Hong et al., 2018). Interestingly, only complexation with a delivery carrier such as lipofectamine 2000 induced immunorecognition by PBMCs.
4 |. PHYSICOCHEMICAL PROPERTIES OF RNA NANOPARTICLES AFFECT IN VIVO BIODISTRIBUTION AND IMMUNE RESPONSE
Nanotechnology offers a substantial number of benefits over traditional routes for drug delivery, but unfavorable immune responses and liver accumulation have also been reported (Buzea, Pacheco, & Robbie, 2007; Zolnik, Gonzalez-Fernandez, Sadrieh, & Dobrovolskaia, 2010). It has been suggested that the adverse effects were elicited by numerous physicochemical characteristics, including size, shape, surface chemistry, or hydrophobicity (Dobrovolskaia, 2015; Dobrovolskaia, Shurin, & Shvedova, 2016). Engineering these properties with precision and homogeneity is a common strategy to improve the in vivo performance of nanomaterials. One of the advantages of RNA nanotechnology is its high programmability. In other words, their physicochemical properties are easily tunable (Figure 3), and the production process is highly consistent. Therefore, the effects of these properties can simply be studied as a result of reproducible nanoparticle assembly. The following subsections will focus on the main physicochemical properties of RNA nanoparticles and the corresponding effects on their immunostimulation and biodistribution.
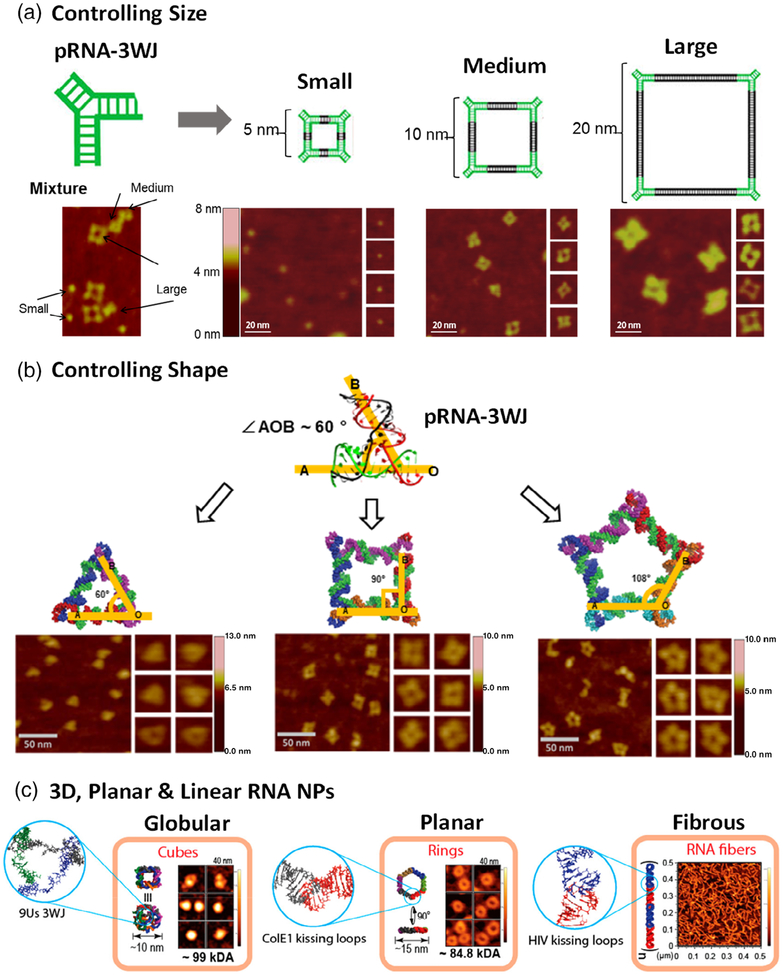
FIGURE 3.
Construction of RNA nanostructures with tunable properties. (a) RNA squares with small, medium, and large size by tuning the length of the connecting helix (Reprinted with permission from Jasinski et al. (2014). Copyright 2014 American Chemical Society). (b) RNA triangle, square and pentagon by tuning the interior pRNA-3WJ angle (Reprinted with permission from Khisamutdinov, Li, et al. (2014). Copyright 2014 American Chemical Society). (c) 3D RNA cube, planar RNA nanoring, and linear RNA fiber by different connectivity (Reprinted with permission from Hong et al. (2018). Copyright 2018 American Chemical Society)
4.1 |. Nanoparticle size
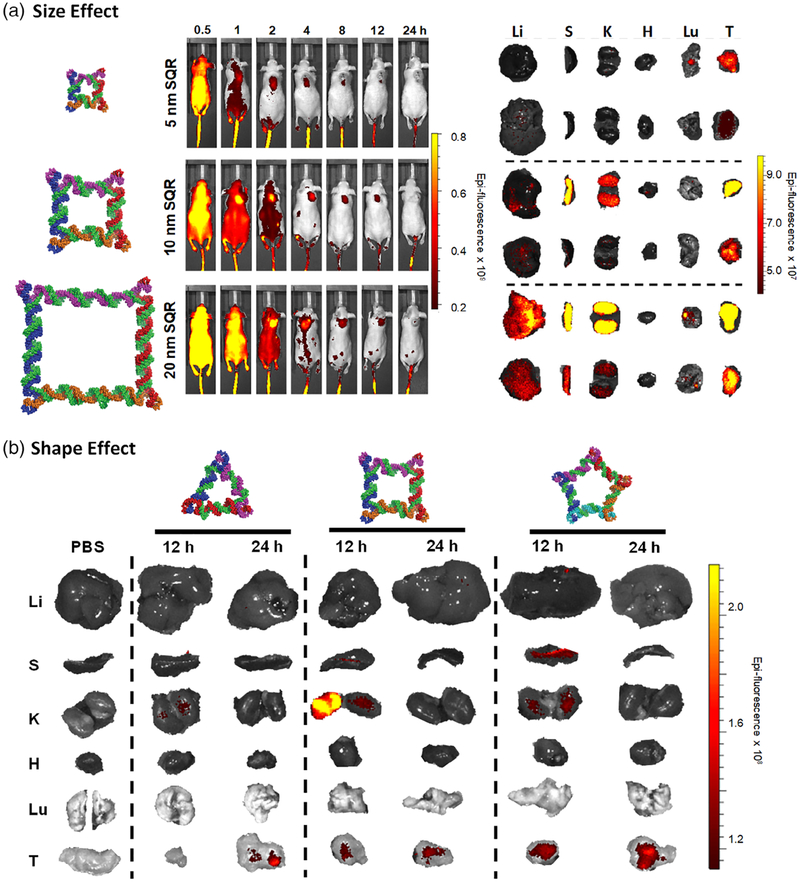
Nanoparticle size represents one of the most critical considerations in the design and construction of RNA nanoparticles. It was found that size significantly dictates nanoparticle performance at the nano-bio interface, including vascular transportation, plasma protein binding, and cellular membrane interaction (Albanese et al., 2012; Hoshyar et al., 2016). Large particles (>100 nm) tend to be trapped in the liver and spleen as a result of the stronger recognition by the mononuclear phagocytic system (MPS) in these organs (Gustafson et al., 2015). Particles with a small diameter (<10 nm) are more likely to have a faster renal clearance, thus leading to a shorter half-life in vivo (Longmire, Choyke, & Kobayashi, 2008). In a systemic in vivo biodistribution study, the effect of RNA nanoparticle size on their circulation time and accumulation in healthy organs and tumors has been evaluated (Jasinski, Li, & Guo, 2018). Specifically, RNA squares of three different sizes (5 nm, 10 nm, 20 nm) were intravenously administered into tumor-bearing mice. Internal organ imaging at 12 and 24 h time points showed the rapid elimination of 5 nm RNA squares from vital organs with significant accumulation in the tumor after 12 h (Figure 4a). It was suggested that renal excretion is the primary excretion route for the nanoparticles (Piao, Wang, Binzel, & Guo, 2018). For the 10 and 20 nm RNA squares, stronger interaction with macrophages and slower metabolism in the liver was observed, which is possibly caused by the different protein binding profiles of large nanoparticles compared to that of small ones. The correlation of size with the biodistribution profile should be considered from the perspective of its effects on renal clearance and macrophage uptake. Particularly, smaller RNA nanoparticles exhibited less uptake by macrophages of the MPS due to less serum protein binding. However, they are more rapidly excreted by kidney filtration, while the opposite trend is seen with the larger RNA nanoparticles. Thus, the final in vivo fate of RNA nanoparticles will take a balance between these two size-dependent elimination pathways (i.e., macrophages and urinary excretion).
FIGURE 4.
Effects of RNA nanoparticle size and shape onin vivo biodistribution. (a) RNA squares with identical shape but varying size, and (b) RNA polygons with identical size but varying shape show different circulation times and tumor accumulation in vivo. (Reprinted with permission from Jasinski, Li, and Guo (2018). Copyright 2018 Elsevier Inc.)
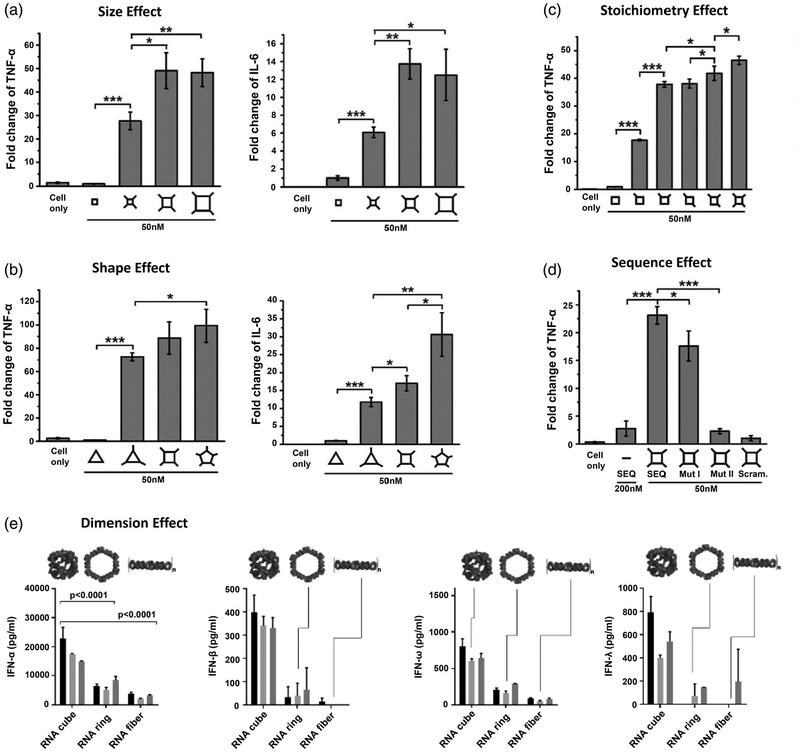
The effect of varied sizes will manifest itself in the interaction between particles and immune system as well. Small sizes will benefit from not being recognized by the bulky opsonins in complement cascade, a key component of the immune system, due to the inadequate accommodation on particle surfaces (Ventola, 2012). RNA nanoparticles have been deliberately constructed in a size range from 10 to 40 nm, making them advantageous for drug delivery. Recently, the effect of size on their immunostimulation has been studied (Guo et al., 2017). RNA nanoparticles with identical square shape but varying size were used as the model (Figure 5a). After extending the single-stranded sequence at the vertexes, RNA squares were endowed with immunostimulatory activity in a size-dependent fashion. Small RNA squares (7.10 nm) elevated the immunomodulation to some extent, while stronger responses were observed with medium (12.31 nm) and large (21.15 nm) RNA squares. This can likely be attributed to the engulfing behavior of phagocytes. Larger nanoparticle sizes show greater sensitivity to phagocytosis than the corresponding smaller ones. A similar finding was reported when using varying sizes of RNA polygons, in which RNA hexagons induced more IFNs than any smaller RNA polygons (Hong et al., 2018). These findings answered important questions regarding size in the rational design of RNA nanoparticles for favorable cancer accumulation and immune-interactions.
FIGURE 5.
Effects of RNA nanoparticles’ physicochemical properties on immunostimulation. RNA nanoparticles with (a) varying size, (b) varying shape, (c) different stoichiometry, (d) different sequence, and (e) different dimension induced cytokines and interferons secretion to various levels. (figures A-D and E reprinted with permission from Guo et al. (2017). Copyright 2018 Elsevier Inc). and (Reprinted with permission from Hong et al. (2018). Copyright 2018 American Chemical Society, respectively)
4.2 |. Nanoparticle shape
Shape is another critical design parameter in nanotechnology. However, detailed immunological and pharmacokinetic studies of shape effects are limited; most of these studies require complex construction procedures to produce nanoparticles of varying shapes while maintaining uniform size and composition (Toy et al., 2014). In contrast, RNA nanotechnology exceptionally enables the controlled self-assembly of nanostructures with custom size or shape independently, resulting in a true shape comparison. For example, different RNA polygons (triangle, square, and pentagon) were all constructed with identical size, benefiting from the inherent flexibility of the pRNA-3WJ scaffold (Guo et al., 2017; Khisamutdinov, Li, et al., 2014). Intriguingly, when immunostimulatory oligonucleotides were implemented on RNA polygons, the RNA pentagon appeared to be the most potent inducer of pro-inflammatory cytokines while the triangular counterparts remained the least among the polygonal structures studied (Figure 5b), implying the shape-dependent immunomodulation of RNA nanoparticles. Regarding RNA polygons, another interesting trend was observed: number of sides. The RNA triangle and pentagon, having an odd number of sides, tended to stimulate more IFN-β secretion than the even-sided RNA square and hexagon (Johnson et al., 2017). Additionally, a significant difference was found in immune responses between RNA nanoparticles with varying dimensional structures. A 3D RNA tetrahedron carrying immunostimulatory oligonucleotides exhibited stronger immunostimulatory activity than the planar triangular counterpart, when size and payload stoichiometry were controlled to be equivalent (Guo et al., 2017). Likewise, globular RNA cubes induced stronger immunostimulation compared to planar hexameric RNA rings, which were more immunostimulatory than RNA fibers (Figure 5e) (Hong et al., 2018). These findings suggest a trend of increasing immunostimulatory properties of RNA nanoparticles from linear, to planar, and to 3D structure. One interpretation may be that the increased surface area intrinsic to 3D structures provides a spacious surface for complement opsonins assembly and deposition, while a greater proportion of opsonins released into the surrounding medium with linear and planar RNA structures.
The shape of nanoparticles has been shown to dictate the interactions that occur with cell membranes and circulating serum proteins. For instance, studies have suggested that oblate-shaped nanoparticles with discoidal geometries are more likely to migrate toward blood vessel walls and establish greater interactions with endothelial cells of blood vessels in comparison to spherical nanoparticles (Muller, Fedosov, & Gompper, 2014). The biodistribution profiles of RNA polygons of different shape but uniform size were compared in tumor-bearing mouse models after systemic administration (Jasinski, Li, & Guo, 2018). Different retention in organs were observed at 12 h time point as nanosquares showed high fluorescent signal intensity while triangle nanoparticles showed none and the pentagon very little. In the spleen, pentagon nanoparticles exhibited the highest fluorescence. A similar biodistribution in organs was found among the particles after 24 h (Figure 4b). Therefore, the protein corona formation on the RNA nanoparticles may drastically change in response to nanoparticle shapes, which will further impact their elimination pathways. Additionally, the cellular interactions with nanoparticles as well as internalization are also closely related to the size and shape. Cell receptors that mediate the endocytosis are of various sizes and shapes, so it will provide beneficial information on rational design of nanoparticles that possess favorable binding to the receptors, thus enhancing nanoparticle recognition and cellular uptake. Considering the controllable size, shape and other physicochemical properties, RNA nanoparticles could potentially be designed with enhanced tumor cell uptake and retention.
4.3 |. Sequence signature and modular stoichiometry
As a biocompatible nanomaterial, RNA nanoparticles are immunologically inert. In contrast, some special RNA sequences have been reported to trigger immune responses, named isRNAs, due to the specific recognition by toll-like receptors (TLRs) or cytosolic sensors (PKR, RIG-1, and MDA-5) in immune cells (Berger et al., 2009; Bourquin et al., 2007; Heil et al., 2004; Hornung et al., 2005; Judge et al., 2005). Incorporation of these isRNA sequences can turn immunologically inert RNA nanoparticles to immunologically active, or even enhance the immune response associated with an incorporated module. In a study from Guo Lab, a specific RNA SEQ was extended to the vertexes of RNA squares and dramatically engendered the production of pro-inflammatory cytokines in vitro and in vivo (Figure 5d) (Guo et al., 2017). The immune responses were in direct proportion to the stoichiometry of single-stranded RNA extensions. RNA squares with increasing copies of payload induced stronger cytokine levels (Figure 5c). Conversely, mutation or complementary blockage of the extension sequence resulted in reduced immune responses, while scrambling the extension sequence led to complete abrogation of immune response. This study affords a new sight in the design and construction of RNA nanoparticles—they can be constructed to serve as safe therapeutic nanocarriers with non-immunogenicity, or deliberately trigger a strong immune response for immunotherapy.
4.4 |. Surface chemistry
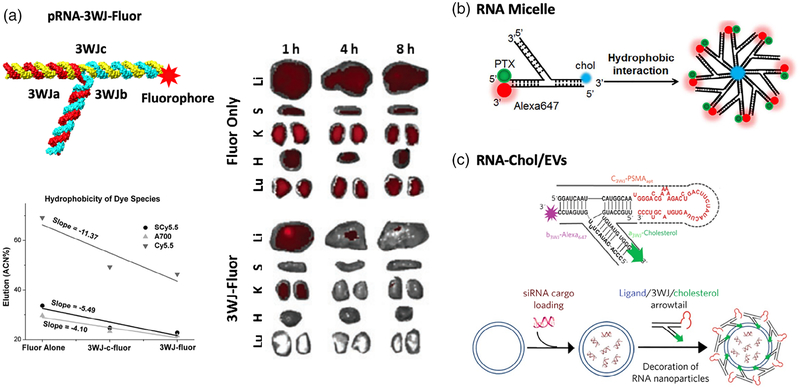
Surface characteristics have been shown to be a significant parameter in the pharmacokinetics and pharmacodynamics of many nanomaterials, as well as influencing their immune system interactions (Albanese et al., 2012). Some cationic nanoparticles, such as polyethyleneimines, can easily interact with cell membranes, causing nonspecific cytotoxicity and giving rise to complement system activation (Merkel et al., 2011). In contrast, RNA nanoparticles have consistently exhibited the advantage of causing no or undetectable cytotoxicity in many studies due to their polyanionic nature (Shu et al., 2014). Surface-projected composition is another factor that greatly defines the in vivo fate of nanoparticles (Merkel et al., 2011). As a naturally aqueous-soluble biopolymer, RNA nanoparticles are distinct from many synthetic nanomaterials in that they do not require surface modifications, such as polyethylene glycol (PEG) grafting (Suk et al., 2016), to increase aqueous-solubility and consequently limit their immune response and increase in vivo circulation. This advantage provides RNA nanoparticles the flexibility to incorporate various functional modules, including RNAi therapeutics, chemical drugs, fluorophores, and targeting ligands, to achieve multi-functionality. Particularly, some of these surface compositions, especially drugs, fluorophores, or other hydrophobic compounds, might be important factors in determining how RNA nanoparticles communicate with cells or proteins in vivo. Nanoparticles decorated with more hydrophobic reagents will result in greater plasma protein binding, and therefore greater accumulation in the liver or other organs. Jasinski et al. reported that different chemicals incorporated to RNA nanoparticles altered the RNA hydrophobicity to varying degrees (Figure 6a) (Jasinski, Yin, et al., 2018). The changes in vital organ accumulation as a function of hydrophobicity variation was investigated. Weaker organ accumulation was detected for RNA nanoparticles (3WJ-Fluor) containing hydrophobic fluorophores (Cyanine5.5, Sulfonated-Cyanine 5.5, and AlexaFluor700) than these fluorophores alone (Figure 6a–c), clearly indicating the capacity of RNA nanoparticles to solubilize hydrophobic compounds. In another study, paclitaxel (PTX), an antitumor chemo-drug with poor aqueous-solubility, was conjugated to micellar RNA nanoparticle (Figure 6b) (Shu et al., 2018). Consequently, the RNA micelle/PTX complex showed significantly enhanced water-solubility and efficient cancer targeting in vivo. Additionally, RNA nanoparticles conjugated with a cholesterol molecule were used to control the ligand-displaying on extracellular vesicle (EVs) membranes. As a result of orientational control enabled by RNA nanoparticles, EVs decorated with multi-functional RNA nanoparticles were able to provide specific delivery of siRNA (Figure 6c) (Li et al., 2018; Pi et al., 2018). As shown here, the biological implications of RNA nanoparticles following systemic administration are closely related to their chemical modifications. While changes in the protein binding activity of RNA nanoparticles might be difficult to predict, understanding the effects of hydrophobic drugs conjugation on its solubility, and in turn its in vivo biodistribution, can help researchers develop RNA-based therapeutics for future clinical settings.
FIGURE 6.
Construction of RNA nanoparticles with various surface characteristics. (a) pRNA-3WJ nanoparticles conjugated with hydrophobic fluorophores and their effects on in vivo biodistribution (Reprinted with permission from Jasinski, Yin, et al. (2018). Copyright 2018, Mary Ann Liebert, Inc). (b) RNA micelles assembled from pRNA-3WJ conjugated with cholesterol, paclitaxel and fluorophore (Reprinted with permission from Shu et al. (2018). Copyright 2018 Elsevier Inc). (c) Ligand-displaying extracellular vesicles by pRNA-3WJ conjugated with cholesterol, ligand aptamer, and fluorophore (Reprinted with permission from Pi et al. (2018). Copyright 2018 Springer Nature Publishing)
4.5 |. Other factors
In addition to the common physicochemical properties discussed above, other parameters can also make their own unique contributions to RNA nanoparticle’s immunostimulation or biodistribution profiles. Recent studies have shown that some chemical modifications not only improve the enzymatic stability of RNA, but also play vital roles in RNA immunostimulation (Ge et al., 2010; Peacock et al., 2011). For example, 2′-fluorine(2’-F) modified siRNA displayed immune-inert behavior in human peripheral blood mononuclear cells, whereas the unmodified siRNA induced the production of TNF-α and IFN-α (Lee et al., 2016). It is suggested that this property can be consistently applied to RNA nanoparticles when siRNA is incorporated, because most RNA nanoparticles used in cancer targeting or drug delivery were 2’-F modified. Besides, 2’-O-methyl (2’-O-Me) modification has been found to prevent the recognition of siRNAs by TLR7/8 and RIG-I receptor, thus reducing induction of TNF-α and IFN-β in human fibroblast MRC-5 cells (Ge et al., 2010). In addition, it has been reported that unmodified galactosidase (GAL) siRNA transiently induced the expression of TNF-α, IL-6, IL-10, IFN-β and IFN-sensitive gene in vivo, whereas a formulation of 2′-O-Me-Luciferase (LUC) siRNA had no such effects (Broering et al., 2014). Thus, 2’-O-Me is potentially another facile approach capable of fine-tuning the properties of RNA to limit or enhance immune and inflammatory responses, depending on the therapeutic objective.
Interestingly, the intra- and intermolecular connectivity appeared to be another factor that affects the immunostimulation of RNA nanoparticles. Afonin Lab reported that RNA rings, assembled from pre-folded monomers via intermolecular interaction (kissing loops), exhibited less immunostimulatory activity than RNA cubes formed via intramolecular hydrogen bonds (Hong et al., 2018). This result suggests that RNA connectivity may play a role, but this mechanism needs further investigation.
Additionally, it has been reported that nanoparticle elasticity influences vascular transport, biodistribution, cellular internalization, and immunostimulation (Anselmo et al., 2015). RNA as a biopolymer has shown rubber-like elastic property (Chiu et al., 2014; Jacobson, McIntosh, Stevens, Rubinstein, & Saleh, 2017), allowing RNA nanoparticles to “squeeze” through vasculatures of the tumor microenvironment by blood pressure without altering its thermodynamic stability. Meanwhile, many RNA nanoparticles were constructed to be ratchet-shaped after incorporating multiple modules (Haque et al., 2012; Shu et al., 2011), preventing them from returning to blood circulation. Therefore, the effects of these properties favor the transport of RNA nanoparticles toward tumors and enhance the EPR effect.
5 |. PERSPECTIVES
5.1 |. RNA nanotechnology for potential immunotherapy
One of the most important recent breakthroughs in cancer research is cancer immunotherapy (Couzin-Frankel, 2013; McNutt, 2013). Extensive studies revealed important information regarding the complicated cancer-immune system relationship, and many researchers are now looking to summon the self-defense system in the hosts to kill cancer. The most common immunotherapies include chimeric antigen receptors (CAR) T-cell therapy (June, O’Connor, Kawalekar, Ghassemi, & Milone, 2018), immune checkpoint blockade (Pardoll, 2012), monoclonal antibodies (mAb) (Weiner, Dhodapkar, & Ferrone, 2009), and cancer vaccines and adjuvants (Temizoz, Kuroda, & Ishii, 2016), just to name a few. Particularly, nucleic acid aptamers have emerged as a new type of therapeutic for immunotherapy (Pastor et al., 2018). Aptamers are short nucleic acid oligomers (12–80 nt) selected from SELEX (systematic evolution of ligands by exponential enrichment) and are capable of binding targets specifically and tightly. Various kinds of co-stimulatory molecules belonging to the B7/CD28 family have been selected to trigger cell mediated immune responses. For instance, CTLA-4 RNA aptamer developed by Santilli-Marotto et al. can bind CTLA-4 with high affinity, inhibit CTLA-4 function, and enhance tumor immunity in mice (Santulli-Marotto, Nair, Rusconi, Sullenger, & Gilboa, 2003). Furthermore, aptamers targeting the TNF/TNFR family which are involved in the later phase of T-cells activation have been developed, including OX40 and 4–1BB aptamers (McNamara et al., 2008). In order to take advantage of the RNA nanoparticle platforms, nucleic acid aptamers can be incorporated into the RNA scaffold to achieve stronger immunotherapeutic effects. Meanwhile, the multivalent property allows the additional incorporation of RNAi therapeutics or chemotherapeutic drugs to realize combination therapy.
Although immunotherapy has shown success in various cancers, some clinical challenges remain, such as the safety and efficacy concerns derived from the systemic dosing of immunomodulatory agents (Whiteside, Demaria, Rodriguez-Ruiz, Zarour, & Melero, 2016). RNA nanotechnology, as a safe and efficient drug delivery platform, can potentially enhance the efficacy as well as reduce the side effects of such immunotherapies by improving the delivery, retention, and release of immunomodulatory agents in targeted cell populations and organs. RNA, as a biomacromolecule, can be successfully recognized by the immune system as a self-entity, and thus RNA nanoparticles intrinsically display immunologically inert property. As described above, though naturally inert, RNA nanoparticles can be manually designed using their tunable and programmable properties to exhibit no, low, or high immunostimulation, thus allowing them to be employed as safe therapeutic carriers without triggering an immune response, or as potential immunomodulators for cancer immunotherapy.
5.2 |. Understanding the interactions of RNA nanoparticles at the nano-bio interface
Upon introduction into biological environment, nanoparticles interacting with proteins, membranes, cells and organs establish a series of nanoparticle/biological interactions at the interfaces which govern the in vivo fate of nanoparticles (Cheng, Jiang, Wang, Chen, & Liu, 2013; Nel et al., 2009). As little is known about the interactions of RNA nanoparticles with biological components, a better understanding at the nano-bio interface will be essential to the rational design of RNA nanoparticles capable of targeted delivery (Xu, Haque, et al., 2018). At the molecular level, protein corona formation around nanoparticles drastically influences their in vivo behavior, resulting in different elimination pathways (Kim, Faix, & Schnitzer, 2017; Tenzer et al., 2013). Moreover, the component in the protein corona, such as the complement proteins, can potentially lead to altered immune responses (Chen et al., 2017). Previous studies on serum protein binding of RNA polygon nanoparticles showed that the size and shape play critical roles in protein binding (Jasinski, Li, & Guo, 2018). However, the composition of protein corona formation on RNA nanoparticles has not been comprehensively determined. At the cellular lever, it appears the frequency of RNA nanoparticle uptake into macrophages is closely related to their morphology and size, as these factors impact the engulfing process (Guo et al., 2017). The internalization of RNA nanoparticles with targeting ligands into cancer cells is proposed to be receptor-mediated pathway (Shu et al., 2014). However, the impacts of physicochemical properties on the intracellular trafficking of RNA nanoparticles are still not completely understood and await more investigation.
6 |. CONCLUSION
RNA nanotechnology is growing exponentially, though its inception lags behind other nano-delivery systems. As shown within this review, RNA nanoparticles display many advantages in biomedicine. As drug carriers, RNA nanoparticles have repeatedly shown immunologically inert behavior, while can be concomitantly manipulated to exhibit controlled immunostimulation. RNA nanoparticles possess a variety of advantageous physicochemical properties over other nanomaterials, including the capacity to be precisely programmed. As a result, RNA nanoparticles can be rationally designed, optimized, and constructed for specialized in vivo applications. As a biocompatible nanomaterial, RNA nanoparticles show favorable tumor targeting proficiency, as evidenced in various pre-clinical cancer models. The extensive research conducted in order to understand the safety, immunological, and pharmacological profiles of RNA nanoparticles have positively paved a path toward clinical trials. Evidently, RNA nanotechnology is bespeaking a bright future in cancer therapy.
ACKNOWLEDGMENTS
The research in P.G.’s lab was supported by NIH grants R01EB019036 and U01CA207946. The authors would like to thank Lora E. McBride for her constructive comments and revisions on the article. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation.
Funding information
National Cancer Institute, Grant/Award Numbers: R01EB019036, U01CA207946; CM Chen Foundation
Footnotes
CONFLICT OF INTEREST
P.G. is the consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd, as well as the cofounder of Shenzhen P&Z Bio-medical Co. Ltd and its subsidiary US P&Z Biological Technology LLC, as well as ExonanoRNA, LLC and its subsidiary ExonanoRNA (Foshan) Biomedicine Co., Ltd.
REFERENCES
- Abdelmawla S, Guo S, Zhang L, Pulukuri SM, Patankar P, Conley P, … Li QX (2011). Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery. Molecular Therapy, 19, 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, & Jaeger L (2010). In vitro assembly of cubic RNA-based scaffolds designed in silico. Nature Nanotechnology, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Cieply DJ, & Leontis NB (2008). Specific RNA self-assembly with minimal paranemic motifs. Journal of the American Chemical Society, 130, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Viard M, Kagiampakis I, Case CL, Dobrovolskaia MA, Hofmann J, et al. (2014). Triggering of RNA interference with RNA-RNA, RNA-DNA, and DNA-RNA nanoparticles. ACS Nano, 9, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, et al. (2014). Multifunctional RNA nanoparticles. Nano Letters, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese A, Tang PS, & Chan WC (2012). The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annual Review of Biomedical Engineering, 14, 1–16. [DOI] [PubMed] [Google Scholar]
- Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, … Kjems J (2009). Self-assembly of a nanoscale DNA box with a controllable lid. Nature, 459, 73–76. [DOI] [PubMed] [Google Scholar]
- Anselmo AC, Zhang M, Kumar S, Vogus DR, Menegatti S, Helgeson ME, & Mitragotri S (2015). Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano, 9, 3169–3177. [DOI] [PubMed] [Google Scholar]
- Astruc D (2012). Electron-transfer processes in dendrimers and their implication in biology, catalysis, sensing and nanotechnology. Nature Chemistry, 4, 255–267. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Berger M, Ablasser A, Kim S, Bekeredjian-Ding I, Giese T, Endres S, … Hartmann G (2009). TLR8-driven IL-12-dependent reciprocal and synergistic activation of NK cells and monocytes by immunostimulatory RNA. Journal of Immunotherapy, 32, 262–271. [DOI] [PubMed] [Google Scholar]
- Bindewald E, Afonin K, Jaeger L, & Shapiro BA (2011). Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS Nano, 5, 9542–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E, Hayes R, Yingling YG, Kasprzak W, & Shapiro BA (2008). RNAJunction: A database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Research, 36, D392–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzel D, Shu Y, Li H, Sun M, Zhang Q, Shu D, et al. (2016). Specific delivery of MiRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Molecular Therapy, 24, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerneke MA, Dibrov SM, & Hermann T (2016). Crystal-structure-guided Design of Self-Assembling RNA Nanotriangles. Angewandte Chemie (International Ed. in English), 55, 4097–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin C, Schmidt L, Hornung V, Wurzenberger C, Anz D, Sandholzer N, … Endres S (2007). Immunostimulatory RNA oligonucleotides trigger an antigen-specific cytotoxic T-cell and IgG2a response. Blood, 109, 2953–2960. [DOI] [PubMed] [Google Scholar]
- Broering R, Real CI, John MJ, Jahn-Hofmann K, Ickenstein LM, Kleinehr K, … Schlaak JF (2014). Chemical modifications on siRNAs avoid toll-like-receptor-mediated activation of the hepatic immune system in vivo and in vitro. International Immunology, 26, 35–46. [DOI] [PubMed] [Google Scholar]
- Bui MN, Brittany JM, Viard M, Satterwhite E, Martins AN, Li Z, et al. (2017). Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine, 13, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzea C, Pacheco II, & Robbie K (2007). Nanomaterials and nanoparticles: Sources and toxicity. Biointerphases, 2, MR17–MR71. [DOI] [PubMed] [Google Scholar]
- Chen F, Wang G, Griffin JI, Brenneman B, Banda NK, Holers VM, … Simberg D (2017). Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nature Nanotechnology, 12, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Jiang X, Wang J, Chen C, & Liu RS (2013). Nano-bio effects: Interaction of nanomaterials with cells. Nanoscale, 5, 3547–3569. [DOI] [PubMed] [Google Scholar]
- Chiu HC, Koh K, Evich M, Lesiak A, Germann MW, Bongiorno A, et al. (2014). RNA intrusions change DNA elastic properties and structure. Nanoscale, 6, 10009–10017. [DOI] [PubMed] [Google Scholar]
- Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, & Jaeger L (2004). Building programmable jigsaw puzzles with RNA. Science, 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- Corey DR (2007). Chemical modification: The key to clinical application of RNA interference? Journal of Clinical Investigation, 117, 3615–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin-Frankel J (2013). Breakthrough of the year 2013.Cancer immunotherapy. Science, 342, 1432–1433. [DOI] [PubMed] [Google Scholar]
- Cui D, Zhang C, Liu B, Shu Y, Du T, Shu D, et al. (2015). Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Scientific Reports, 5, 10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N (2012). Challenges in development of nanoparticle-based therapeutics. The AAPS JournalAAPS, 14, 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov SM, McLean J, Parsons J, & Hermann T (2011). Self-assembling RNA square. Proceedings of the National Academy of Sciences of the United States of America, 108, 6405–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaia MA (2015). Pre-clinical immunotoxicity studies of nanotechnology-formulated drugs: Challenges, considerations and strategy. Journal of Controlled ReleaseJ, 220, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaia MA, Shurin M, & Shvedova AA (2016). Current understanding of interactions between nanoparticles and the immune system. Toxicology and Applied Pharmacology, 299, 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, & Tuschl T (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature, 452, 896–899. [DOI] [PubMed] [Google Scholar]
- Ge Q, Dallas A, Ilves H, Shorenstein J, Behlke MA, & Johnston BH (2010). Effects of chemical modification on the potency, serum stability, and immunostimulatory properties of short shRNAs. RNA, 16, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary C, Chworos A, Verzemnieks E, Voss NR, & Jaeger L (2017). Composing RNA nanostructures from a syntax of RNA structural modules. Nano Letters, 17, 7095–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, & Discher DE (2007). Shape effects of filaments versus spherical particles in flow and drug delivery. Nature Nanotechnology, 2, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, & Jaeger L (2011). Self-assembling RNA Nanorings based on RNAI/-II inverse kissing complexes. Nano Letters, 11, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P (2010). The emerging field of RNA nanotechnology. Nature Nanotechnology, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, & Garver K (1998). Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Molecular Cell, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- Guo S, Li H, Ma M, Fu J, Dong Y, & Guo P (2017). Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles. Molecular Therapy--Nucleic Acids, 9, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Tschammer N, Mohammed S, & Guo P (2005). Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Human Gene Therapy, 16, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson HH, Holt-Casper D, Grainger DW, & Ghandehari H (2015). Nanoparticle uptake: The phagocyte problem. Nano Today, 10, 487–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halman JR, Satterwhite E, Roark B, Chandler M, Viard M, Ivanina A, … Afonin KA (2017). Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Research, 45, 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C, Li X, Tian C, Jiang W, Wang G, & Mao C (2014). Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nature Communications, 5, 3890. [DOI] [PubMed] [Google Scholar]
- Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, et al. (2012). Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science, 303, 1526–1529. [DOI] [PubMed] [Google Scholar]
- Hendrix DK, Brenner SE, & Holbrook SR (2005). RNA structural motifs: Building blocks of a modular biomolecule. Quarterly Reviews of Biophysics, 38, 221–243. [DOI] [PubMed] [Google Scholar]
- Hong E, Halman JR, Shah AB, Khisamutdinov EF, Dobrovolskaia MA, & Afonin KA (2018). Structure and composition define Immunorecognition of nucleic acid nanoparticles. Nano Letters, 18, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, … Hartmann G (2005). Sequence-specific potent induction of IFN-[alpha] by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Medicine, 11, 263–270. [DOI] [PubMed] [Google Scholar]
- Hoshyar N, Gray S, Han H, & Bao G (2016). The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine (London, England), 11, 673–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, & Lilley DM (2013). The molecular recognition of kink-turn structure by the L7Ae class of proteins. RNA, 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, & Lilley DM (2016). A quasi-cyclic RNA nano-scale molecular object constructed using kink turns. Nanoscale, 8, 15189–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DR, McIntosh DB, Stevens MJ, Rubinstein M, & Saleh OA (2017). Single-stranded nucleic acid elasticity arises from internal electrostatic tension. Proceedings of the National Academy of Sciences of the United States of America, 114, 5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L, Westhof E, & Leontis NB (2001). TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Research, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski D, Haque F, Binzel DW, & Guo P (2017). Advancement of the emerging field of RNA nanotechnology. ACS Nano, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski D, Khisamutdinov EF, Lyubchenko YL, & Guo P (2014). Physicochemically tunable poly-functionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano, 8, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski DL, Li H, & Guo P (2018). The effect of size and shape of RNA nanoparticles on biodistribution. Molecular Therapy, 26, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski DL, Yin H, Li Z, & Guo P (2018). Hydrophobic effect from conjugated chemicals or drugs on in vivo biodistribution of RNA nanoparticles. Human Gene Therapy, 29, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Halman JR, Satterwhite E, Zakharov AV, Bui MN, Benkato K, et al. (2017). Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling. Small, 13, 1701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, & MacLachlan I (2005). Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnology, 23, 457–462. [DOI] [PubMed] [Google Scholar]
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, & Milone MC (2018). CAR T cell immunotherapy for human cancer. Science, 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Ke W, Hong E, Saito RF, Rangel MC, Wang J, Viard M, et al. (2018). RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic Acids Research, 47, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov E, Li H, Jasinski D, Chen J, Fu J, & Guo P (2014). Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square, and pentagon nanovehicles. Nucleic Acids Research, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov EF, Jasinski DL, & Guo P (2014). RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano, 8, 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov EF, Jasinski DL, Li H, Zhang K, Chiu W, & Guo P (2016). Fabrication of RNA 3D nanoprism for loading and protection of small RNAs and model drugs. Advanced Materials, 28, 100079–100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Faix PH, & Schnitzer JE (2017). Overcoming key biological barriers to cancer drug delivery and efficacy. Journal of Controlled ReleaseJ, 267, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing C, Jung S, Iqbal A, & Schlick T (2009). Tertiary motifs revealed in analyses of higher-order RNA junctions. Journal of Molecular Biology, 393, 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Haque F, Shu D, Yoo JY, Li H, Yokel RA, et al. (2015). RNA nanoparticles as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget, 6, 14766–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Urban JH, Xu L, Sullenger BA, & Lee J (2016). 2’Fluoro modification differentially modulates the ability of RNAs to activate pattern recognition receptors. Nucleic Acid Therapeutics, 26, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, … Guo P (2015). RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today, 10, 631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang K, Pi F, Guo S, Shlyakhtenko L, Chiu W, … Guo P (2016). Controllable self-assembly of RNA tetrahedrons with precise shape and size for cancer targeting. Advanced Materials, 28, 7501–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang H, Yin H, Bennett C, Zhang HG, & Guo P (2018). Arrowtail RNA for ligand display on ginger exosome-like nanovesicles to systemic deliver siRNA for cancer suppression. Scientific Reports, 8, 14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire M, Choyke PL, & Kobayashi H (2008). Clearance properties of nano-sized particles and molecules as imaging agents: Considerations and caveats. Nanomedicine (London, England), 3, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, et al. (2008). Multivalent 4–1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. Journal of Clinical Investigation, 118, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt M (2013). Cancer immunotherapy. Science, 342, 1417. [DOI] [PubMed] [Google Scholar]
- Merkel OM, Urbanics R, Bedocs P, Rozsnyay Z, Rosivall L, Toth M, et al. (2011). In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials, 32, 4936–4942. [DOI] [PubMed] [Google Scholar]
- Mogensen TH (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clinical Microbiology Reviews, 22, 240–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monferrer A, Zhang D, Lushnikov AJ, & Hermann T (2019). Versatile kit of robust nanoshapes self-assembling from RNA and DNA modules. Nature Communications, 10, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, Fedosov DA, & Gompper G (2014). Margination of micro- and nano-particles in blood flow and its effect on drug delivery. Scientific Reports, 4, 4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasalean L, Baudrey S, Leontis NB, & Jaeger L (2006). Controlling RNA self-assembly to form filaments. Nucleic Acids Research, 34, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, et al. (2009). Understanding biophysicochemical interactions at the nano-bio interface. Nature Materials, 8, 543–557. [DOI] [PubMed] [Google Scholar]
- Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, … Saito H (2011). Synthetic RNA-protein complex shaped like an equilateral triangle. Nature Nanotechnology, 6, 116–120. [DOI] [PubMed] [Google Scholar]
- Pardoll DM (2012). The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer, 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor F, Berraondo P, Etxeberria I, Frederick J, Sahin U, Gilboa E, & Melero I (2018). An RNA toolbox for cancer immunotherapy. Nature Reviews Drug Discovery, 17, 751–767. [DOI] [PubMed] [Google Scholar]
- Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, et al. (2011). Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. Journal of the American Chemical Society, 133, 9200–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, … Guo P (2018). Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nature Nanotechnology, 13, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao X, Wang H, Binzel DW, & Guo P (2018). Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2’-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA, 24, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri A, Loomis K, Smith B, Lee JH, Yavlovich A, Heldman E, & Blumenthal R (2009). Lipid-based nanoparticles as pharmaceutical drug carriers: From concepts to clinic. Critical Reviews in Therapeutic Drug Carrier Systems, 26, 523–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, … Evers BM (2015). Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano, 9, 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Kariko K, & Tureci O (2014). mRNA-based therapeutics—Developing a new class of drugs. Nature Reviews Drug Discovery, 13, 759–780. [DOI] [PubMed] [Google Scholar]
- Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, & Gilboa E (2003). Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Research, 63, 7483–7489. [PubMed] [Google Scholar]
- Sarver NA, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, et al. (1990). Ribozymes as potential anti-HIV-1 therapeutic agents. Science, 24, 1222–1225. [DOI] [PubMed] [Google Scholar]
- Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, & Jaeger L (2010). A polyhedron made of tRNAs. Nature Chemistry, 2, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severcan I, Geary C, Verzemnieks E, Chworos A, & Jaeger L (2009). Square-shaped RNA particles from different RNA folds. Nano Letters, 9, 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Haque F, Pi F, Shlyakhtenko L, Evers BM, & Guo P (2015). Controllable self-assembly of RNA dendrimers. Nanomedicine: Nanotechnology, Biology and Medicine, 12, 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Kantoff PW, Wooster R, & Farokhzad OC (2017). Cancer nanomedicine: Progress, challenges and opportunities. Nature Reviews Cancer, 17, 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Li SK, Charoenputtakun P, Liu CY, Jasinski D, & Guo P (2018). RNA nanoparticle distribution and clearance in the eye after sub-conjunctival injection with and without thermosensitive hydrogels. Journal of Controlled ReleaseJ, 270, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Khisamutdinov E, Zhang L, & Guo P (2013). Programmable folding of fusion RNA complex driven by the 3WJ motif of phi29 motor pRNA. Nucleic Acids Research, 42, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, et al. (2015). Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano, 9, 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, & Guo P (2011). Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nature Nanotechnology, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, et al. (2013). Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA, 19, 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, et al. (2014). Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Advanced Drug Delivery Reviews, 66C, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Yin H, Rajabi M, Li H, Vieweger M, Guo S, … Guo P (2018). RNA-based micelles: A novel platform for paclitaxel loading and delivery. Journal of Controlled ReleaseJ, 276, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, … Manchester M (2007). Bio-distribution, toxicity and pathology of cow-pea mosaic virus nanoparticles in vivo. Journal of Controlled Release, 120, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, & Lillard JW Jr. (2009). Nanoparticle-based targeted drug delivery. Experimental and Molecular Pathology, 86, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KM, Lee S, & Ban C (2012). Aptamers and their biological applications. Sensors (Basel), 12, 612–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk JS, Xu Q, Kim N, Hanes J, & Ensign LM (2016). PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Advanced Drug Delivery Reviews, 99, 28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temizoz B, Kuroda E, & Ishii KJ (2016). Vaccine adjuvants as potential cancer immunotherapeutics. International Immunology, 28, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, … Stauber RH (2013). Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nature Nanotechnology, 8, 772–781. [DOI] [PubMed] [Google Scholar]
- Toy R, Peiris PM, Ghaghada KB, & Karathanasis E (2014). Shaping cancer nanomedicine: The effect of particle shape on the in vivo journey of nanoparticles. Nanomedicine (London, England), 9, 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola CL (2012). The nanomedicine revolution: Part 1: Emerging concepts. PT, 37, 512–525. [PMC free article] [PubMed] [Google Scholar]
- Weiner LM, Dhodapkar MV, & Ferrone S (2009). Monoclonal antibodies for cancer immunotherapy. Lancet, 373, 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside TL, Demaria S, Rodriguez-Ruiz ME, Zarour HM, & Melero I (2016). Emerging opportunities and challenges in cancer immunotherapy. Clinical Cancer Research, 22, 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Haque F, Jasinski DL, Binzel DW, Shu D, & Guo P (2018). Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Letters, 414, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Li H, Zhang K, Binzel DW, Yin H, Chiu W, & Guo P (2019). Photo-controlled release of paclitaxel and model drugs from RNA pyramids. Nano Research, 12, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Pang L, Wang H, Xu C, Shah H, Guo P, et al. (2018). Specific delivery of delta-5-desaturase siRNA via RNA nanoparticles supplemented with dihomo-gamma-linolenic acid for colon cancer suppression. Redox Biology, 21, 101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Wang H, Li Z, Shu D, & Guo P (2019). RNA micelles for systemic delivery of anti-miRNA for cancer targeting and inhibition without ligand. ACS Nano, 13, 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P, & Shu D (2019). Delivery of anti-miRNA for triple-negative breast cancer therapy using RNA nanoparticles targeting stem cell marker CD133. Molecular Therapy, 27, 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Liu ZY, Jiang W, Wang GS, & Mao CD (2015). De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism. Nature Communications, 6, 5724–5729. [DOI] [PubMed] [Google Scholar]
- Zacharias M, & Hagerman PJ (1995). Bulge-induced bends in RNA: Quantification by transient electric birefringence. Journal of Molecular Biology, 247, 486–500. [DOI] [PubMed] [Google Scholar]
- Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, … Chi YI (2013). Crystal structure of 3WJ Core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA, 19, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Sun F, Liu S, & Jiang S (2016). Anti-PEG antibodies in the clinic: Current issues and beyond PEGylation. Journal of Controlled ReleaseJ, 244, 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Leonard M, Shu Y, Yang Y, Shu D, Guo P, & Zhang X (2017). Overcoming Tamoxifen resistance of human breast cancer by targeted gene silencing using multifunctional pRNA nanoparticles. ACS Nano, 11, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnik BS, Gonzalez-Fernandez A, Sadrieh N, & Dobrovolskaia MA (2010). Nanoparticles and the immune system. Endocrinology, 151, 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]