Abstract
ETMRs are aggressive pediatric embryonal brain tumors with universally dismal outcome1. We collected 193 primary ETMRs and 23 matched relapses to investigate the genomic landscape of this distinct entity. We found that patients having tumors without C19MC amplification, the proposed driver3–5, frequently harbor DICER1 germline mutations or other miRNA-related aberrations including somatic miR-17–92 amplifications. Whole-genome sequencing revealed an overall low recurrence of SNVs, but prevalent R-loop-associated chromosomal instability, of which we show that this can be induced by loss of DICER1 function. Comparing primary tumors and matched relapses revealed a strong conservation of SVs but low conservation of SNVs. Moreover, many newly acquired SNVs are associated to a new cisplatin treatment related mutational signature. Finally, we show that targeting R-loops with topoisomerase and PARP inhibitors might be an effective treatment strategy for this deadly disease.
ETMR (Embryonal Tumor with Multilayered Rosettes) is a malignant type of brain tumor occurring almost exclusively in young children1. The tumors show diverse histological patterns described as EBL (Ependymoblastoma), MEPL (Medulloepithelioma) or ETANTR (Embryonal Tumor with Abundant Neuropil and True Rosettes), but together form one distinct biological entity termed ETMR1,2. Genetically, ETMRs are characterized by amplification and fusion of a microRNA cluster on chromosome 19 (C19MC) with TTYH1, present in ~90% of the tumors3–5. As current treatment options fail for nearly all patients, a better understanding of ETMR tumorigenesis is needed to develop new treatment strategies. Here, we have characterized the molecular landscape of a large cohort of primary and relapsed ETMRs to get more insight into the inter- and intra-tumor heterogeneity of ETMRs at diagnosis and relapse, and to identify tumor driving mechanisms that may lead to more effective treatment strategies.
Inter-tumor heterogeneity
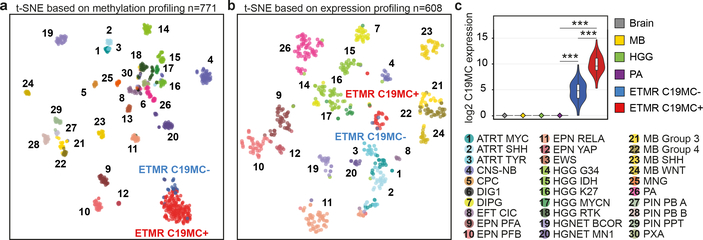
Cluster analysis of ETMR DNA methylation profiles (n=193; Supplementary Table 1) or mRNA data (n=28), confirmed that ETMRs are clearly distinct from other brain tumor entities (Fig. 1a–b)6,7. ETMRs without C19MC amplification (n=23) tend to cluster together at the edge of the main ETMR cluster but do not really separate, also not when clustering only ETMRs (Extended Data Fig. 1a). Additionally, miRNAs are expressed in a distinct pattern in ETMRs, but are similar between ETMRs with (n=7) and without (n=3) C19MC amplification (Extended Data Fig. 2). C19MC miRNAs are also expressed in ETMRs without C19MC amplification, albeit at a ~10-fold lower level, but not in normal brain or other brain tumors (Fig. 1c). Mature miRNAs specifically upregulated in ETMRs included all C19MC miRNAs and members of the miR-17–92 miRNA cluster, while several members of the let-7 family of miRNAs are specifically downregulated in ETMRs (Supplementary Table 2).
Figure 1. ETMRs regardless of C19MC amplification show high molecular similarity.
a-b, t-SNE clustering using either DNA methylation (a) or mRNA expression data (b). Colors represent different tumor entities classified as described previously7. c, Violin plot showing log2 transformed expression of C19MC miRNAs (n=56) in ETMRs and other tissues. P-values were calculated using two-sided Mann-Whitney U tests (***= P<0.0005). Boxplots show the median ± interquartile range, whiskers extend to 1.5x the interquartile range, violin plots depict kernel density estimates and represent the density distribution.
ETMRs occur throughout the brain, without any association to different histological variants, but tumors without C19MC amplification were significantly more often located in infratentorial regions (Fisher’s exact test, P-value: 8.80E-7). None of the other clinical annotations was associated to any molecular subgrouping (Extended Data Fig. 1a, b). Altogether, these data show that ETMRs are a molecularly distinct entity, with limited molecular inter-tumor heterogeneity despite diverse histopathological features, absence of C19MC amplification in a subset of tumors, and a variety of anatomic locations.
Intra-tumor heterogeneity
By comparing ETMR mRNA data against normal brain and other brain tumors (Supplementary Table 3), we identified several upregulated pathways related to development, including RNA processing, Hippo, WNT, NOTCH, and SHH signaling (Extended Data Fig. 3). Additionally, compared to other tumors, most ETMRs showed upregulation of DNA repair pathways, including base excision repair, Fanconi anemia and the p53 pathway (Extended Data Fig. 4a, Supplementary Table 4)8. However, a subset of tumors (10/28) lacked this upregulation. Since no distinct molecular subgroups were detected we investigated whether this was due to differences in intra-tumor heterogeneity. CIBERsort analysis was applied by comparing tumor expression data with single cell RNA-seq data of the developing pre-frontal cortex to delineate distinct tumor cell populations9,10. This resulted in the identification of stem cell-like, more differentiated oligodendrocyte precursor-like and astrocyte-like tumor cells (Extended Data Fig. 4b). A higher proportion of differentiated progeny corresponded to lower levels of stem cell markers LIN28A and HMGA2, lower levels of DNA damage checkpoint genes WEE1 and CHEK2, but higher levels of astrocyte markers AQP4 and GFAP (Extended Data Fig. 4c).
Interestingly, these observations correlated with the histology of the tumors. Tumors with more differentiated cells and lower expression of DNA repair genes tended to be more often diagnosed as ETANTRs, while tumors with less differentiated cells and higher expression of DNA repair genes were more likely described as EBL or MEPL (Extended Data Fig. 4d). These findings were validated by methylation profiling and RNA-sequencing of micro-dissected neuropil (more differentiated) and rosettes (less differentiated) of an ETMR diagnosed as ETANTR. While RNA-sequencing confirmed that expression of stem cell markers and astrocyte markers differed between the two cell populations, methylation profiles were highly similar and clustered together with other ETMRs (Extended data Fig. 4e–g) (Supplementary Table 3).
Previously, we reported that LIN28A is more widely expressed upon relapse and histology shifts towards a more EBL or MEPL phenotype1. To investigate whether this is due to outgrowth of the stem cell-like population after treatment, we compared the expression of LIN28A, HMGA2, WEE1, CHEK2, AQP4 and GFAP between a primary ETANTR and two matched recurrences and saw indeed that in both relapses the gene expression is shifting to be more stem cell-like (Extended Data Fig. 4h). Differences in expression of DNA repair genes and histology are therefore likely explained by the levels of differentiated progeny, while undifferentiated cells with high levels of DNA repair expression grow out upon relapse.
Recurrent aberrations
We sequenced (see methods) 82 tumors, including 16 cases without C19MC amplification and 12 recurrences (Supplementary Table 5, 6), to see whether there were any recurrent DNA aberrations other than C19MC amplification. Other recurrently mutated genes detected in the primary tumors included DICER1 (8/70), CTNNB1 (7/70), and TP53 (5/70), while mutations (apart from CTNNB1) in developmental pathways, previously suggested to play a role in ETMR tumorigenesis11, were detected only sporadically (Fig. 2a).
Figure 2. ETMRs without C19MC amplification recurrently harbor miRNA related aberrations.
a, Oncoplot showing somatic events occurring in ETMRs. b, Overview of identified DICER1 mutations. Alternating blue colors represent exons, yellow bars represent domains and pins represent the different aberrations found. c, Quantification of miRNA processing using the median ratio between 3p and 5p miRNAs (n=375), each bar representing one tumor. P-values were calculated using a one-sided Mann-Whitney U test (*** P<0.0005).
Interestingly, all DICER1 mutations occurred in cases that lacked the C19MC amplification. All eight cases harbored compound heterozygous mutations, including one somatic mutation (E1705K (3x), D1709N (2x), G1809R, E1813G and E1813K) in the hotspot region affecting the RNASE IIIb domain important for miRNA processing12. The second mutation was found in the germline (validated for 7/8 cases) and was in all cases deleterious (Fig. 2b). Sequencing mature miRNAs indeed showed a strongly increased ratio of 3’ over 5’ mature miRNAs in one of the DICER1 mutated cases (ET31) as compared to cases that have no DICER1 mutations (Fig. 2c).
Two ETMRs (ET68 and ET173, lacking C19MC amplification and DICER1 mutations) harbored amplifications of the miR-17–92 miRNA cluster at chromosome 13. In ET68 this was fused to a region at chromosome 11 to form a circular chromosome, confirmed by mate pair-sequencing (MP-seq), copy number profiles and FISH (Extended Data Fig. 5a, b). Genes on the circular chromosome, including the miR-17–92 miRNA cluster, were ~4-fold higher expressed compared to other ETMRs (Extended Data Fig. 5c). The miR-17–92 miRNA cluster amplification was also found once in an ETMR (ET85) with C19MC amplification.
Three other samples (ET89, ET160, and ET168, lacking C19MC amplification and DICER1 mutations) showed clustered breakpoints affecting the C19MC locus, suggesting that even though C19MC is not amplified its expression might be driven by structural rearrangements (Extended Data Fig. 5d). In ET160 we indeed found a fusion between C19MC and MYO9B, also located on chromosome 19. Alternative fusion partners to TTYH1 were not unique for cases lacking the C19MC amplification, as MP-seq also identified an alternative fusion partner, MIRLET7BHG, in ET20 with C19MC amplification.
Finally, we identified several cases throughout the cohort with severe chromosomal instability, including four cases without C19MC amplification (ET57, ET72, ET74, and ET104) (Extended Data Fig. 5e). Altogether, our data show that ETMRs harbor very few recurrently mutated genes, and are largely characterized by mutually exclusive aberrations affecting the miRNA pathway, including amplifications of the C19MC and miR-17–92 clusters and mutations in the miRNA processing gene DICER1.
Unlike SNVs and small indels, many copy number aberrations (CNAs) were recurrent throughout the cohort based on methylation array-derived copy number profiling (n=193). As shown previously, 90% of all ETMRs had C19MC amplification and chr2 gain was detected in 76% of the cases. Other recurrent CNAs included 6q loss (25%), and gain of 1q (26%), 17q (11%), 7 (12%), 3q (11%), and 11q (11%). However, no significant differences were found between ETMRs with or without C19MC amplification (Extended Data Fig. 6a–c). We also compared copy number profiles of 18 matched primary relapse pairs, identifying frequently acquired CNAs, including 6q loss (22%), and gains of 1q (33%), 17q (33%), and 7 (17%) (Extended Data Fig. 6d) (Supplementary Table 7). In addition, polyploidy was detected in 18% of the primary tumors, and acquired in 28% of the cases upon relapse (Extended Data Fig. 6d, e).
Furthermore, ETMRs sequenced by WGS were investigated to detect translocations, inversions, deletions and insertions (Supplementary Table 8). Many clustered breakpoints resembling found surrounding C19MC, but other chromosomes were also sporadically affected (Extended Data Fig. 6f). These breakpoints were also observed in copy number profiles: 16% of the cohort had visible alternating copy number states around C19MC. Together, the high number of CNAs, recurrent polyploidy, and recurrent complex rearrangements suggest that ETMR genomes are structurally instable.
Primary-relapse comparisons
To investigate which events are retained in relapses and therefore potentially tumor driving, we used WGS data of nine matched recurrences and analyzed allele frequencies of all detected SNVs in primary-relapse pairs (Supplementary Table 9). Overall, we found that only 51% (2138/4226) of all SNVs that are present at allele frequencies of at least 20% in the four matched primary tumors are also detectable at allele frequencies of 2% or more in the respective matching relapse (Fig. 3a). In addition, two out of four tumors did not have a single coding non-synonymous SNV detected in both primary tumor and first relapse, suggesting that acquiring somatic SNVs is not an early (driving) event (Extended Data Fig. 7).
Figure 3. Primary/relapse comparison reveals poor conservation of SNVs but high conservation of SVs.
a, Graph depicting allele frequencies of combined SNVs found between four primary tumors and matched initial relapses. Boxes represent SNVs that are gained, conserved and lost upon relapse. b, Analysis of mutational signatures of primary and relapsed tumors based on previously defined mutational signatures13,14. c, Radial plot depicting log2 fold changes of the somatic SNV burden between primary tumor (PRIM) and subsequent relapses (REC1 and REC2) colored by exposures shown in b. Number of mutations in primary tumor was set to 1 for visualization purposes. d, Venn diagram showing the overlap of breakpoints between a primary tumor and matched relapses. e, Circular representation of the genome of SVs and CNAs in a primary tumor with two matched recurrences shown in d. The outer rim represents, from outer to inner, the CNAs found in the primary tumor, the first recurrence and second recurrence. The middle part represents the different SVs found between primary tumors and recurrences. Chromosome 19 and X have been enlarged.
Relapsed tumors have a large increase in somatic SNVs as compared to primary tumors suggesting that new mutations may have been induced by treatment (Extended Data Fig. 8a). To investigate this, we compared mutational signatures between primary tumors and recurrences and observed a shift from mainly signature 1 and 16 (both associated to aging) in primary tumors to signature P1 in recurrences, which is a novel signature found in our pediatric pancancer cohort not associated to a mutagenesis process yet (Fig. 3b)13,14. To find the potential underlying mechanism we tested for mutational strand bias in somatic SNVs. No bias was detected in primary tumors, however, recurrences showed an increased bias of mainly C>A, C>T and T>A mutations towards the transcribed strand (Extended Data Fig. 8b), previously also observed in tumors treated with platinum based agents and cyclophosphamide15. Comparing matched primary tumors with multiple recurrences confirmed that the composition of SNVs changed upon treatment, but remained similar in subsequent relapses (Fig. 3c). Furthermore, the trinucleotide counts changed towards a signature highly similar to signature P1 and a cisplatin induced mutational signature derived from cisplatin treated cell lines16 (Extended Data Fig. 8c, d), suggesting that the majority of acquired mutations in relapsed ETMRs is induced by cisplatin treatment.
Despite the poor conservation of SNVs there was a strong conservation of breakpoints between primary tumors and recurrences. On average 73% (96%, 54%, 63%, 80%, respectively) of all breakpoints were conserved between primary tumors and relapses, including all breakpoints surrounding and forming C19MC aberrations (Extended Data Fig. 7). In the same case analyzed for shifts in mutational signatures, only one out of 110 breakpoints in the primary tumor was not detected in any recurrence (Fig. 3d, e). These data suggest that formation of SVs are early events in ETMR tumorigenesis. Further supporting this observation is an increased density of conserved mutations in close proximity to breakpoints, which were enriched for C>T and C>G mutations, previously described in association with chromothripsis and replication stress (Extended Data Fig. 9)17. The process of forming breakpoints continues upon relapse and relapses gain many new SVs as well, which may have a role in tumor progression (Extended Data Fig. 7). Therefore, even though we cannot fully exclude that acquisition of sporadic SNVs, also in non-coding regions of the genome, might play a role in tumor formation, the high conservation of breakpoints and recurrent chromosomal instability is more likely to be driving ETMRs than the poorly conserved SNVs
R-loop associated chromosomal instability
In recent years, multiple papers have described an association between the formation of R-loops, structures that form upon stalling of RNA polymerase resulting in a displaced non-template single stranded DNA loop, and DNA damage18. R-loops, which can form following disrupted helicase activity, can either facilitate or result from collision of transcription and replication and can potentially lead to chromosomal instability18,19. Interestingly, expression of DNA/RNA helicases and processes associated to helicase activity were highly upregulated in ETMRs suggesting a possible role for R-loops in the formation of breakpoints (Fig. 4a).
Figure 4. Breakpoint context reveals a possible role for R-loops in initiating ETMRs.
a,Schematic representation of GO-term enrichment. Circles represent GO-terms, sizes enrichment and colors groups based on similarity scores (Co-occurrence Association Score >0.05) (Supplementary Table 3). b, Enrichment in DRIP signal around C19MC (fold enrichment over input). c, Density of ETMR breakpoints (n=2301) overlapping DRIP-peaks (left; n=16002) and RLFS (right; n=85534) compared to random regions of the same size. P-values were calculated using a two-sided Chi-square test (***= P<0.0005). d, Enrichment of breakpoints overlapping RLFS compared to 10000 randomly generated sets of regions of the same size for ETMRs and other entities22. Boxes show the range (median, first and third quartile) of BH adjusted P-values calculated using one-sided Fisher’s exact tests, whiskers extend to upper and lower limits of the data (90% and 10% respectively). e, Immunohistochemistry pictures of full slides (left) and magnifications (right) for ETMRs (n=5) and a representative case of WNT (n=5) and Group4 MB (n=5). f, Immunohistochemistry of WT and DCR-KO cell lines stained for DAPI (blue), S9.6 (red) and y-H2AX (green). g, Quantification of signals shown in f, in mean signal per cell (n=255 WT cells, n=227 DCR-KO cells), normalized to DAPI signal. P-values were calculated using a two-sided Mann-Whitney U test (***= P<0.0005). Boxes show median, first and third quartile and whiskers extending to 1.5x the interquartile range. h, Dot blot of DNA-RNA hybrids extracted from WT and DCR-KO cells, ssDNA was used as loading control.
To investigate this, we applied prediction of R-loop forming sequences (RLFS)20 and performed DNA-RNA hybrid immunoprecipitation (DRIP) sequencing on the C19MC-amplified BT183 ETMR cell line. To compare how well the datasets matched we compared RLFS and the DRIP-seq data to published DRIP-seq data for Ewing sarcoma (EWS)21 and observed a similar genome-wide pattern (Extended Data Fig. 10a, b). R-loops in ETMRs were mostly observed in regions surrounding the TSS, CpG islands and g-quadruplex forming repeats and were also strongly enriched in peaks observed in EWS and RLFS (Extended Data Fig 10c). Overall, we observed a high density of R-loops on chromosome 19, but only in ETMRs we observed a large peak surrounding C19MC (Fig. 4b). Genome wide, many breakpoints overlapped with R-loops, and formation of breakpoints had a stronger association for regions that are enriched for R-loops, which was similar for breakpoints occurring in EWS but not for breakpoints occurring in other tumor entities (Extended Data Fig. 10d)22. Therefore, we compared the relative number of breakpoints in DRIP-seq peaks and RLFS as compared to regions outside R-loops and found a strong enrichment of all SV types, including those not forming the C19MC aberrations, in both DRIP-seq peaks and RLFS (Extended Data Fig. 10b, Fig. 4c). Compared to multiple other tumor entities the enrichment of breakpoints falling into R-loops or RLFS was specifically present in ETMRs (Fig. 4d). Furthermore, we observed an increased density of breakpoints in close proximity of R-loops and RLFS compared to other tumors which did not occur for other genomic elements (Extended Data Fig. 10e, f), most likely resulting from damage induced by stalled replication forks23. Finally, we also observed an increased density of SNVs falling into R-loops and RLFS, in line with the observed increased density of SNVs around breakpoints (Extended Data Fig. 10g). Together, these data suggest that R-loops may indeed play a role in generating breakpoints in ETMR, including the breakpoints observed around C19MC.
To validate the high levels of R-loops in ETMRs we stained five ETMRs for R-loops using the R-loop specific antibody S9.6 and compared them to staining in medulloblastomas (n=10) (Fig. 4e). ETMRs were highly positive for R-loops in the rosette structures while medulloblastomas were negative throughout the entire sections. Most ETMRs with or without C19MC amplification have miRNA related aberrations, and multiple miRNA processing factors have been associated with R-loop formation24,25, including DICER1 which has shown to be directly involved in DNA repair and contains helicase activity26. To test this, we generated dot blots and immunostainings for R-loops and double stranded breaks (DSBs) in a Dicer1 knockout (Dcr-KO) mouse stem cell line and its isogenic control. Indeed, both levels of R-loops and DSBs indicated by increased y-H2AX staining were increased in the Dcr-KO cells (Fig. 4f–h). In addition, sequencing detected chromothripsis events and many breakpoints in RLFS that resemble the phenotype in ETMRs around C19MC (Extended Data Fig. 10h, i). These data suggest that R-loops may cause the chromosomal instability in ETMRs and defective miRNA processing might play a role in generating the elevated R-loop levels.
Efficacy of PARP and TOP1 inhibition
Finally, we investigated whether R-loops could potentially be exploited for therapy. Previously, we reported that ETMRs are highly sensitive to inhibitors of topoisomerase, an enzyme that can resolve R-loops among other functions19,27. Other studies have shown that topotecan or irinotecan act as a TOP1 poison by covalently binding TOP1 to the DNA28, which can be further enhanced by trapping PARP1, as PARP1 is able to release TOP1 by parylation29. Therefore, we tested whether a combination treatment with PARP and TOP1 inhibitors would lead to a further increase in R-loops and increased response to therapy.
First, we tested whether there is synergy between PARP and TOP1 inhibition in ETMR cells. Topotecan alone is effective with IC50 values of ~5nM while PARP inhibitors are less effective with IC50 values of ~10μM for Pamiparib (BGB-290) and ~15μM for Veliparib. Both PARP inhibitors, however, act highly synergistically when used in combination with topotecan and lead to a larger decrease in viability than monotherapy (Fig. 5a). Synergy and cell death are only observed once concentrations of topotecan were higher than 1.25nM suggesting that the synergy is mostly dependent on sufficient levels of topoisomerase inhibition (Fig. 5b).
Figure 5. ETMR cells are sensitive to combination therapy with PARP and TOP1 inhibitors.
a, Dose response curves of ETMR cells treated with either Pamiparib or Veliparib, Topotecan and a combination of both drugs, P-values were calculated using two-way ANOVA. Error bars denote mean ± s.e.m. b, Calculation of synergy using the Chou-Talay method of drug treatments shown in a. c, Immunohistochemistry of ETMR cells stained for y-H2AX and S9.6. Cells were treated using IC50 concentrations of every drug or combination. d, Quantification of signal shown in c, in mean signal per cell (n=59 DMSO treated, n=158 Pamiparib treated, n=88 Topotecan treated, n=20 Combination treated cells), normalized by the total DAPI signal in the cell. P-values were calculated using two-sided Mann Whitney U tests (***= P<0.0005).
Next, we investigated whether topotecan alone or combined with PARP inhibitors is correlated with R-loop levels. Topotecan treatment (6h) increased the number of nuclear R-loops while Pamiparib had almost no effect on R-loops (Fig. 5c). Combination therapy induced R-loops at an even higher rate and caused more DNA damage as shown by y-H2AX staining. Quantification of the nuclear signals shows that y-H2AX and R-loop signals highly correlate, suggesting that an increase in R-loops increases the amount of DNA damage (Fig. 5d). Together, these data show that PARP and topoisomerase inhibitors act synergistically in ETMRs and combination treatment could, after thorough in vivo testing, potentially be used as treatment for ETMR patients.
Discussion
Here, we present the molecular landscape of ETMRs at diagnosis and relapse, resulting in a set of hallmarks describing the entity (Fig. 6). We show that all ETMRs are molecularly similar, despite differences in histology, the presence or absence of C19MC amplification and their location in the brain. C19MC amplification is still considered the main driver based on its strong conservation and high recurrence. Moreover, we have identified DICER1 as the first predisposition gene for ETMR. Our data give a better insight in what is driving these tumors and how they change after treatment, largely because of the cisplatin treatment.
Figure 6. Hallmarks of ETMR.
Schematic summary of ETMR characteristics
miRNA expression profiles correlate strongly between ETMRs even though the underlying mechanism deregulating miRNAs may be different. A possible explanation for this might be that tumors with C19MC or miR-17–92 amplification oversaturate the miRNA processing machinery. This could potentially mimic deregulated processing by DICER1 and also explains why RNA transport, vital for miRNA processing and shown to be affected by miRNA saturation, is highly upregulated in ETMRs (Extended Data Fig. 3)30,31. Oversaturation of the miRNA machinery could also explain why all ETMRs regardless of amplification status show many structural aberrations since several members of the miRNA processing machinery, including DICER1 and DROSHA, have been associated to R-loop formation as well24,25.
Other mechanisms causing chromosomal instability may play a role as well. Breakpoints were enriched in RLFS, which are an intrinsic property of the genome, suggesting that R-loops predispose certain loci to acquiring DNA damage. Especially since R-loops were found in close proximity of C19MC, there could be an event prior to C19MC amplification. Considering the early onset of the disease and strong conservation of SVs it is possible that there is a genetic predisposition that increases the levels of R-loops, which in turn causes breakage at sites that form R-loops, leading to C19MC amplification in ETMRs. Also, C19MC amplification itself might further increase the levels of R-loops causing the number of breakpoints to increase.
One such genetic predisposition is identified here through recurrent DICER1 germline mutations. With this finding, ETMR is added to the large spectrum of tumors that may arise in the context of the DICER1 syndrome32. The link between chromosomal instability, R-loops and DICER1 mutations in ETMRs may occur in other cancers as well, providing a rationale for the cancer susceptibility seen in those patients. Indeed, tumors associated to DICER1 syndrome seem to frequently acquire CNAs33,34, but whether this is related to R-loop formation remains to be elucidated. We recommend that ETMR patients and their families should consider genetic counseling for DICER1 syndrome, at least when C19MC is not amplified.
Presence of R-loops can also have therapeutic implications. A recent study in Ewing sarcoma has shown that R-loops may be associated with hyper transcription, which sequesters BRCA1 and prevents it from promoting double strand break repair21. That study explains why targeting these tumors with PARP inhibitors is successful. Here, we show that TOP1 inhibition can increase the levels of R-loops causing an increased amount of damage to cells already having high levels of R-loops. This mechanism may also be applicable to other drugs, providing potential novel treatment strategies.
Summarizing, by fully characterizing the events that drive ETMRs, we have provided a valuable resource for future ETMR research and potential novel therapies that could be further pursued to treat patients with this deadly tumor entity.
Methods
Experimental model and subject details
All patient samples included in this study were acquired under informed consent according to the ICGC (www.icgc.org) guidelines or INFORM (www.dkfz.de/en/inform/) guidelines (for cases ET85 and ET86) and included all relapses from those tumors. For all included cases consent was approved by the review board of the contributing centers before shipment. Primary tumors were derived from patients that did not undergo radiotherapy or chemotherapy prior to surgical removal of the tumor. Material from relapses was derived from patients that received therapy as described in Supplementary Table 1 (CHT=chemotherapy in all cases including cyclophosphamide and either cisplatin or carboplatin), (RT=radiotherapy).
Samples were included based on different ETMR histologies as described previously1 450k/EPIC classification7,35 and amplification of C19MC by visual inspection of copy number profiles. Since many tumors with CNS-PNET histologies could be reclassified as shown by Sturm et al.6, tumors with CNS-PNET and pineoblastoma histologies were also included if there was sufficient evidence based on 450k/EPIC clustering. The presence or absence of C19MC aberrations, based on copy numbers, were validated in approximately half of the cohort of primary tumors (90 tumors) using FISH and/or whole genome sequencing (WGS) and mate-pair sequencing (MP-seq) when available.
All experiments involving cell culture of ETMR cells were performed using the BT183 cell line that was obtained from J.A. Chan and the characteristics of this cell line have been described in the original publication36. Cells were cultured in low adhesion cell culture flasks as spheroid cultures as was described previously36. The cell line was derived from a primary tumor of a 2-year old patient and was acquired under informed consent. DICER1−/− and DICER1f/f mouse stem cells were obtained from ATCC (CRL-3220, CRL-3221) and cultured as specified by the manufacturer. All cell lines have been tested for mycoplasma contamination and were found to be negative.
Sample and library preparation for DNA sequencing
Out of the 70 primary tumors, we performed whole genome sequencing (WGS) for 20 cases, all with matched germline, and whole exome sequencing (WES) for 17 cases without germline to find (recurrent) genetic events that could potentially deregulate important pathways. In addition, we performed targeted sequencing for another 33 primary ETMRs and one matched recurrence, using a gene panel of 158 genes that were found recurrently affected in other brain tumors (Supplementary Table 4)37.
DNA was isolated from fresh frozen tumor material using either the Qiagen DNeasy Blood and Tissue kit or using the Promega Maxwell RSC Tissue DNA Kit (AS1610). DNA from the matching blood was extracted using the Qiagen Blood and Cell Culture Midi Kit or Promega Maxwell RSC Blood DNA Kit (AS1400) according to the provided protocol. For samples sequenced using targeted sequencing and methylation profiling of samples for which only FFPE material was available DNA was extracted using the Maxwell RSC FFPE kit (AS1450) following manufacturer’s instructions.
WGS libraries were prepared using the Illumina TruSeq Nano DNA LT Library Prep or TruSeq Nano DNA HT Library Prep Kit following the manufacturer’s instructions. Briefly, 100 ng of genomic DNA was fragmented to ~350 bp using a Covaris ultrasonicator (Covaris, Inc.). The fragmented DNA was then end-repaired, size-selected using magnetic beads, extended with an ‘A’ base on the 3′ end and ligated with TruSeq paired-end indexing adapters. The adapter-ligated fragment libraries were enriched using eight cycles of PCR and purified 1–2 times using magnetic beads. All generated libraries were validated using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) and using Agilent 2200 or 4200 TapeStation (Agilent Technologies). Libraries were sequenced on the Illumina HiSeq X (2×151 bp paired-end) according to the manufacturer’s protocol.
For samples ET1, ET12, ET16, ET19, ET20, ET30, ET31, ET66, ET68, ET71, ET72 and ET87 next generation sequencing libraries were created as described previously using Illumina, Inc. v2 protocols38. Libraries for those samples were subsequently sequenced using HiSeq 2000 v4 (Paired-End 125bp) instruments using three lanes on the machine achieving a comparable coverage to samples sequenced on the HiSeq X. The average sequencing coverage was 33.95X for tumor material and 34.99X for blood.
All samples sequenced for WGS were submitted to the DKFZ genomics and proteomics core facility and were only included for library preparation after passing all standard quality controls. Only samples with a DNA integrity (DIN) over 7 were included in the study.
Sequencing of samples using WES was performed by creating libraries using the IlluminaTruSeq exome enrichment kit following the manufacturer’s instructions after size selection. Size selection was performed by fractionation using a Covaris ultrasonicator and subsequent selection was performed using a 1.5% gel Pippin Prep cassette (Sage Science, Beverly, MA). Sequencing was performed at the Genome Quebec Innovation Centre, Montreal, Quebec using Illumina HiSeq 2000 instruments or at the ICGex NGS platform of the Institut Curie using HiSeq 2500 instruments for the two DICER1 mutated cases which were reported previously39.
Targeted sequencing was performed by creating libraries using the Agilent SureSelect XT technology. Libraries were sequenced using molecular barcode-indexed ligation-based sequencing at a NextSeq500 (Illumina) instrument19. For the targeted sequencing, we sequenced genes at an average coverage of roughly 100x for the genes listed in Supplementary Table 4.
DNA methylation array
DNA methylation profiling was performed as described previously6,7. DNA was extracted in the same manner as described for WGS using 500ng as input material for fresh frozen tissue and 250ng input material for FFPE tissue. Array data was created using the Infinium HumanMethylation450 BeadChip array (450k array) according to the manufacturer’s instructions (Illumina, San Diego, USA) at the DKFZ Genomics and Proteomics Core Facility (Heidelberg, Germany). For a subset of samples (described in Supplementary Table 1) Methylation BeadChip (EPIC) arrays were used. CpG probes that were used for the analysis were filtered based on presence of a common SNP within five bases of the probe, reads not mapping uniquely to the reference genome, probes mapping to the X and Y chromosome and reads not overlapping between 450K arrays and EPIC arrays.
Clustering was performed after correction of samples for the origin of the DNA (FFPE or Fresh frozen) using Surrogate Variable Analysis (sva)40 and only the 10000 most variable probes based on the full dataset after correction were selected for clustering. Distance was calculated using 1-Pearson correlation and linkage was calculated using average as measure. Subsequently, t-stochastic neighbourhood embedding (t-SNE) analysis using RTSNE (v0.13) was applied to generate the figures41.
Copy number profiling and analysis
Copy number profiles were created using the conumee package (v.1.3.0). For the analysis of copy numbers the average copy number change observed for chromosome 2 gains in annotated samples from Korshunov et al. was used as a cutoff to determine gains and losses1. Copy numbers were subsequently manually assessed to prevent false positives and false negatives due to differences in tumor cell content or ploidy. Calling of focal amplifications/deletions was performed similarly using three times the average copy number change for chromosome 2 gains followed by manual assessment. For samples with increased ploidy CNAs were filtered out manually that were likely due to ploidy changes to reduce the number of false positives.
Processing and alignment of DNA sequencing
Sequenced reads using WGS or WES approaches were aligned to the human genome version 19 (hg19) reference genome using bwa-0.7.8-r455 mem using default settings and T −0. Lanes were sorted using biobambam bamsort (version 0.0.148) and duplicate reads were removed using biobambam bammark duplicates. For data generated using the HiSeq X Picard (v1.125) (https://broadinstitute.github.io/picard/) was used to filter out duplicate mapping reads. Reads were removed if the phred-scaled base quality was below 25 taken over the length of the read. For targeted sequencing, reads were aligned to the human reference genome version 19 (hg19) using bwa mem v0.7.12-r1039. Duplicated reads were removed using Picard v1.113 and reads were sorted using SAMtools 0.1.17-r97342. Reads were removed if the phred-scaled base quality was below 25 taken over the length of the read.
Somatic SNV and indel calling
Detection of somatic SNVs and indels in WGS data was performed using our in-house pipeline Roddy (version 1.1.73). The pipeline is based on SAMtools (version 0.1.17-r973) mpileup and bcftools using parameter adjustments allowing for SNV calling38. Besides those filtering steps we applied additional filtering to remove low quality SNVs as described previously43. Variants were excluded that fell into ENCODE DAC blacklisted regions, Duke excluded regions, the hiSeqDepthTopPt1Pct track from UCSC genome browser or variants that had both the reference and altered allele annotated in DBsnp (version 135). In addition, we applied filtering criteria restricting the overlap of variants with features including tandem repeats, simple repeats, low complexity, satellite repeats, or segmental duplications to a maximum of two. Finally, all variants were filtered out that did not fulfill the heuristic criteria of having at least five reads in the sequenced tumor covering the position, a coverage of more than 10 reads but less than 300 reads in the germline control, more than 3% of the reads of the alternative variant in the germline, no bases other than wildtype or variant at the position, at least one read on every strand supporting the variant (or more than five reads in total) and a variant allele frequency at least over 10%. The final results were annotated using ANNOVAR (2016–02-01)44 using data derived from Interpro (https://www.ebi.ac.uk/interpro/ date: 2015–10-15), the CADD phred score45 (ljb26 date: 2014–09-25), cosmic (https://cancer.sanger.ac.uk/cosmic date: 2014–09-11), dbSNP (https://www.ncbi.nlm.nih.gov/SNP/ date: 2015–11-02) and clinvar (https://www.ncbi.nlm.nih.gov/clinvar/ date: 2016–03-02).
For targeted sequencing SNVs were detected using SAMtools mpileup and bcftools using a similar method described for WGS with few changes to increase stringency: variants were filtered out that did not have a minimum read depth of 20, a minimum RMS mapping quality of 30, a minimum of three reads supporting the variant, at least one read on every strand that supported the variant and an allele frequency of at least 10%. In addition, variants were filtered out that occurred in more than 0.1% of the 1000genomes population (http://www.internationalgenome.org date: 2015–08-24) or in more than 0.1% of the nonTCGA ExAc population (http://exac.broadinstitute.org date: 2016–04-23). All coding reported SNVs, SNVs and coding indels detected using panel sequencing were annotated using ANNOVAR and benign variants were filtered out.
For indel calling (< 50bp) in targeted sequencing both SAMtools mpileup and Platypus (version 0.8.1) was applied as described previously38.The same criteria were applied to indels that were applied to SNVs including read depth, mapping quality, reads supporting the variant allele frequency, allele frequency in general population, filtering of benign variants and manual reviewing. Indel calling and SNV calling in exome sequencing was performed in a similar manner as targeted sequencing with an extra filtering at alignment: as two germlines were available both were combined to form a pseudo-germline and used for filtering variants.
Germline SNV and indel calling
Germline SNV and indel calling was performed as described previously using freebayes (https://github.com/ekg/freebayes v1.1.0) applying the same settings and using the same filtering criteria46 using a panel of genes described in Supplementary Table 4. Genes not in the panel were excluded from analysis. In brief, raw variant predictions were filtered (QUAL>20, QUAL/AO>2, SAF>1, SAR>1, RPR >1 and RPL >1), and normalized across patients with vt (https://genome.sph.umich.edu/wiki/Vtv0.5).
Putative germline mutations were defined as frameshift, stop gain/loss, start loss, canonical splice site, exon/gene deletions, and damaging non-canonical splice site variants or pathogenic missense variants based on Clinvar annotation. In addition, homozygous missense variants, or missense variants having a somatic second hit that are present in a functional domain with a CADD score higher than 20 were annotated as putative germline mutation.
Putative germline mutations were filtered out if the minor allele frequency was higher than 0.1% in any continental population based on the non-TCGA ExAC database the 1000 genomes project or ESP (http://evs.gs.washington.edu/EVS/).
After annotation, filtered germline mutations were excluded from the analysis if annotated as benign in clinvar. In addition, variants with a reduction in allele frequency to less than 40% in the tumor were excluded. Finally, we only took along variants that were recurrent within the cohort. The context of all remaining variants was reviewed manually using IGV to prevent false positives.
For investigation of non-coding regions we included regions 100 bps preceding the TSS (based on known protein coding genes defined in gencode v19) and putative enhancers (based on H3K27ac data from cultured H9 neural progenitor cells, Encode, ENCSR449AXO, Bernstein lab)47.
Structural variant discovery using paired-end sequencing data
For structural variant (SV) discovery, aligned reads were processed with Delly (version 0.7.5) using paired-end mapping and split-read analysis21. Centromeric and telomeric regions of hg19 were excluded from analysis and to maximize specificity for the latter somatic filtering was run jointly on each pair of tumor sample and matched control. For germline SV discovery, we merged all SVs using the Delly merge command and genotyped all SV sites in all control samples. We then discarded SVs with an allele count of zero and filtered SVs using the germline mode of the Delly filter command. All germline SVs were then intersected with genes and protein truncating variants such as the DICER1 germline exon 19 deletion were manually inspected using IGV.
For somatic SV discovery, we first filtered each tumor sample against the matched control using the somatic mode of the Delly filter command. We then aggregated all somatic variants and genotyped them once more across all samples to fetch putative mismapping artifacts of high allele count. All somatic SVs that passed the second round of filtering against a panel of normal samples were collected for each tumor sample and overlayed on read-depth plots for manual inspection.
Mate-pair sequencing
Mate-pair DNA library preparation was carried out using the Illumina MP v2 reagents and protocol as described previously38. In brief, fragmentation of genomic DNA was performed using a Hydroshear device to an insert size of 4.5 kb followed by sequencing with Illumina HiSeq 2000 instruments. Alignment was performed using Eland (v2) retaining only uniquely aligned reads for downstream rearrangement analysis using Delly. Mate-pair sequencing was applied on complex rearrangements detected using WGS mainly as validation and reconstruction of the rearrangements but was not applied for discovery of structural alterations.
Oncoplots
Oncoplots were generated using custom scripts. Events (either germline or somatic) were only included if the minor allele frequency in the non-TCGA ExAC population was under 1% or unknown and the variant allele frequency was higher than 20%. In addition, every event had to fulfill one of the following criteria: the gene was either recurrently affected with at least one event present in the WGS cohort or a loss of function mutation within a recurrently affected pathway. Events were only included in the figure if events occurred in ETMRs without C19MC amplification, in multiple ETMRs with C19MC amplification or were loss of function (LOF) mutations in deregulated pathways. For the CNA oncoplot all CNAs were included after manual filtering for CNAs that were likely the result of increased ploidy.
Conservation between primary and relapse
Conservation of events between primary tumors and recurrences was performed using somatic calls of SNVs, indels and SVs. VAFs were recalculated at the defined positions using SAMtools mpileup followed by Varscan pileup2cns (version 2.3.9)48. Variants were only included if coverage in both germline and tumor was at least 10 reads, the coverage in both primary and recurrence was at least 10 reads, and no alleles other than wildtype or variant were detected in the germline, primary tumor or recurrence. In all sample combinations, more than 95% of SNVs had sufficient reads for further analysis. Retained events were defined as events having an allele frequency over 20% in both primary and recurrence, gained mutations in recurrences were defined as mutations that had an allele frequency less than 2% in the primary tumor and more than 10% in the recurrence and events that were lost in recurrence were defined as events with allele frequencies over 10% in primary tumors but less than 2% in recurrences.
Calculation of mutational signatures
Mutational signatures were calculated as described previously and compared to 30 signatures described by Alexandrov et al. and the novel P1 signature described by Gröbner et al.13,14. In brief, somatic mutations and mutation context (adjacent bases) were extracted from the called SNVs and used to construct a catalogue of every possible context for all samples. By using the formula M (mutational catalogue) = P (mutational signature) × E (exposures: contribution of mutational process within the mutational landscape) exposures were calculated.
Signatures were calculated over both primary tumors and recurrences with a mutation count over 200. Whole exomes and samples sequenced using targeted sequencing were therefore excluded from analysis. Exposures present under 5% in a sample were removed and exposures over 5% were reconstructed to form 100% of the sample to reduce the amount of noise. Changes in mutational signatures between primary tumors and recurrences were also calculated using only the 30 signatures described by Alexandrov et al.14 (data not shown). Signature calculation was also performed de novo and five signatures were called. Compared to the 30 signatures described by Alexandrov et al. one novel signature was found. This novel signature only had a cosine similarity over 0.85 for signature P1 therefore the P1 signature was included in the analysis instead of only the 30 classical signatures.
Calculation of transcriptional asymmetry
Transcriptional asymmetry was calculated using the MutationalPatterns (v1.4.3) R package based on methods described in Haradvala et al.49,50. Mutations were considered that overlapped gene bodies and were combined from either all primary tumors or all relapses. Substitutions were assigned as transcribed when present on the same strand as the gene definition (UCSC, hg19) and untranscribed if present on the opposite strand compared to the gene definition. When multiple genes in different directions overlapped, events were excluded in this region. Events were split in different substitution types and asymmetry was calculated separately for every substitution type for both primary and relapse.
RNA profiling
RNA was isolated from 28 ETMRs using the Qiagen RNeasy mini kit or the Maxwell RSC simplyRNA Tissue Kit (AS1340) using homogenized tumor tissue. All RNA isolations were performed according to the manufacturers protocol. QC was performed using an Agilent Bioanalyzer 2100 instrument and samples with an RNA integrity index (RIN) over 7 were included. Samples were profiled using Affymetrix GeneChip Human Genome U133 Plus2.0 arrays at the DKFZ genomics and proteomics core facility applying the manufacturers protocol for preparation, hybridization and QC.
Analysis of mRNA data
Samples were normalized and probe detection P-values were calculated using the MAS5.0 algorithm (Affymetrix), followed by QC of the percentage of present calls and manual inspection of GAPDH levels and 5’ to 3’ ratios. All probes were filtered to contain every gene once in the analysis. Analysis of ATRT51, MB52 and CNS-PNET6 data was performed in the same way for every sample included in the analysis.
Differential expression was calculated using anova using a P-value of 0.01 as a cut-off (corrected using false discovery rate). In addition, an absolute fold change of 2 and a minimum log2 gene expression value of 5 in at least one of the compared sets was used as criteria for the definition of differentially expressed genes. Heatmaps were generated using supervised clustering of expression of 450 DNA repair genes shown in (Supplementary Table 4) and pathways described by Pearl et al.8. All samples were z-score normalized and clustering was performed using Euclidean distance. All mRNA expression data was analyzed using the R2 platform (http://R2.amc.nl).
t-SNE clustering was performed by using z-score normalized gene expression using only representative probes by applying HugoOnce (http://R2.amc.nl). Gene expression data was derived from a set of 580 samples deposited in the R2 platform and 28 ETMRs which were all processed in the same way. Using these samples a 50-dim PCA was performed followed by t-SNE clustering using RTSNE.
GO and KEGG enrichment analysis
GO term enrichment and KEGG pathway enrichment was performed using TOPPgene53 and filtered using significant terms also found using the DAVID algorithm (v6.8)54. For all terms a P-value cut-off of 0.05 was used and P-values were corrected using False Discovery Rate (FDR), Bonferroni and Benjamini-Hochberg (BH). For GO term enrichment a gene limit of 200 was chosen to acquire more specificity in the results. Processes were defined using similarity between GO terms that was calculated using NaviGO55 using a minimum co-occurrence score of 0.05. For terms without similarity to any significant terms this was manually assessed.
Estimating cell populations from bulk mRNA sequencing
The Cibersort algorithm (v1.06)9 was applied to estimate the relative frequencies of different cell populations using normalized expression data. Signature genes were derived from the median gene expression levels of defined cell populations from the prefrontal cortex described by Zhong et al.10. Only informative genes were selected having a median expression higher than 1 in any of the subgroups. Both Relative and Absolute mode were applied using 500 permutations to estimate the relative abundance of each sample type.
Micro-dissection of FFPE material
Representative FFPE tissue sample of ET174 was histologically identified, targeted, and micro-dissected with a puncher for nucleic acid extraction. DNA was extracted in a similar manner as described for other FFPE material, RNA was extracted using the automated Maxwell system with the Maxwell 16 LEV RNA FFPE Kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions. In order to evaluate FFPE RNA quality we used the percentage of RNA fragments > 200 nt fragment determination value (DV200). Only RNA samples with DV200 > 70% were included for sequencing on a NextSeq 500 (Illumina). RNA-seq data was quantified using the quant option of kallisto (v0.43.0) using standard settings56.
miRNA sequencing and processing
Small RNAs were isolated as described previously57,58 from fresh frozen tumor material. In summary, total RNA is extracted using GITC/phenol extraction followed by 3’-adaptor ligation of barcoded adenylated adaptors. Samples were pooled in two sets of five samples. Subsequently, gel electrophoresis was used to isolate small RNAs (19–35nt) and purified using ethanol precipitation. Fragments were then amplified using standard PCR, isolated using gel electrophoresis and purified using ethanol precipitation. Samples were sequenced on a HiSeq 2000 v4 machine.
Small RNA-seq was aligned using Bowtie v1.00 (--seedmms 1 --maqerr 1000 --seedlen 21 --norc -M 1 --best --strata) to mirbase 18 after Reads were selected by cutting off adapters using cutadapt 9.5 (-e 0.1 -m 18 -M 34 -O 8 TCGTATGCCGTCTTCTGCTTG) and taking reads between 18 and 34 nucleotides. Subsequently reads were counted using SAMtools 0.1.17-r973 mpileup.
Analysis of miRNA data
Mature miRNA counts were quantified relatively to the total read count separately for both the 5’ (5p) and 3’ (3p) strand and the resulting RPM values were used in all subsequent analysis. Supervised clustering of miRNAs was performed using only significantly differentially expressed miRNAs between ETMRs with C19MC amplification and other samples (excluding ETMRs without C19MC amplification) using adjusted negative binomial testing with the DESeq2 package(1.18.1)59. Samples were normalized using z-score normalization and clustered using hierarchical clustering using average as method. Unsupervised clustering was performed after filtering for miRNAs that had an expression over 32 RPM in any included sample. Samples were normalized and clustered in the same way as the supervised analysis.
miRNA processing was quantified comparing the normalized levels of 3p mature miRNAs against the levels of 5p mature miRNAs using sequences provided by miRbase 1860. For every sample, RPM levels were compared for each 5p versus 3p miRNA and the mean value was calculated as processing ratio. miRNAs with multiple 5p or 3p variants were averaged. All miRNA data was compared against a set of 10 random samples per entity derived from unpublished ICGC data (Aichmuller et al., manuscript in preparation) that was processed in the same way as the ETMR data.
Analysis of R-loop levels
DNA:RNA hybrids were extracted from tissue derived from ETMR PDX models (BT183) that were treated using topotecan or saline as described in Schmidt et al.27. Tumors were subsequently frozen and pelleted using ultracentrifugation. DNA:RNA hybrids were extracted as described previously using the same protocol that is applied for cultured cells21. DNA was extracted using proteinase K followed by phenol/chloroform extraction and ethanol precipitation. Subsequently the DNA was fragmented using the restriction enzymes HindIII, EcoRI, BsrGI, XbaI and SspI (New England Biolabs). Digested DNA was subsequently incubated with anti-DNA:RNA hybrid antibody S9.6 (Merck, MABE1095) and immunoprecipitated using agarose beads. Bound DNA:RNA hybrids were eluted and incubated with proteinase K and cleaned with an additional phenol/chloroform/ethanol extraction. The DNA was subsequently sonicated and sequenced using a Hiseq 2000 machine with a 50bp-single-read protocol. Each treatment condition was performed in duplicate and both RNASE H and the input was included as negative control.
DRIP-seq analysis
DRIP-seq data was aligned using bwa using standard settings to hg19. Aligned reads were normalized to the input signal using bamCompare (deeptools 3.0.2) using log2 fold change increase as output for visualization purposes61. Peak calling was performed using MACS 2.0 (https://github.com/taoliu/MACS) using settings -g 2.7e9 -q 0.05, -B -m 2 30 and the input signal as background. Peak files were combined for each condition and each subsequent analysis was performed both for topotecan treated samples and untreated samples. Peak calling was compared to data from Ewing sarcoma21 and correlated to mRNA expression to ensure the quality of the dataset.
R-loop forming sequences (RLFS) were predicted using the qmRLFS finder (v1.5) tool20 by applying standard settings and using models m1 and m2 for both the human genome (hg19) and mouse genome (mm10) depending on the analysis. Both the RIZ and REZ were included in RLFS peaks.
Calculating R-loop enrichments
For the analysis comparing the overlap of SVs with R-loops, overlapping SVs were defined as either the start position of the SV, the end position of the SV or both positions falling in either an RLFS or DRIP peak depending on the analysis. Duplicate breakpoints were excluded when overlapping between matched primary tumor and relapse. The analysis comparing breakpoints in R-loops against the rest of the genome was performed by taking the total number of SVs falling in R-loop regions and by comparing this to 10000 randomly generated regions of the same size, with the same number of regions as the R-loop regions. For the comparison against other tumor entities 68018 somatic rearrangements were used from the study of Kloosterman et al.22 and locations were compared to either RLFS or R-loops derived from ETMRs. P-values were calculated using Fisher’s exact tests and corrected for multiple testing using BH correction. The same 10000 random regions were used for each entity and a P-value was calculated using Fisher’s exact test for every iteration and adjusted using Bonferroni correction. Only entities with more than 50 breakpoints were used for the comparison. For comparisons of R-loops with genomic regions ranges from both non-B-db 2.062 and repeatmasker 4.0.8. (http://www.repeatmasker.org/) were aggregated and the median enrichment over the input signal was calculated for 10000 randomly selected regions of every DNA element.
FISH analysis
Fluorescence in situ hybridization was applied as described previously to validate C19MC amplification63. Briefly, two probes corresponding to the 19q13.42 and 19q13 loci were applied, utilizing the 19q13 probe as a reference. The minichromosome formed between chromosome 11 and 13 was validated using probes at the loci 11q22.2 and 13q31.3. Ploidy of 28 different ETMRs was validated in a similar manner using FISH probes at the chromosome 9 and chromosome 11 centromeres as those chromosomes were found to be relatively stable in our cohort.
Detection of R-loop levels
DNA:RNA hybrids were extracted from cell pellets of DICER1 WT and KO (Dcr-KO) cells after lysis for 12–24 hours, with a solution containing TE, SDS and proteinase K as described previously64. DNA:RNA hybrids were subsequently extracted using Phenol/Choloform Isoamyl alcohol (Affymetrix, #75831) followed by phase-lock gel separation (Quantabio, 5prime phase lock gel light, #cat 2302820). DNA:RNA hybrids were subsequently purified using ethanol precipitation. DNA:RNA hybrids were diluted in 10x SSC buffer and loaded on a charged nylon membrane (Roche, 11209299001). Membranes were blocked in 1x TBS containing 5% skimmed milk and incubated overnight using an anti-DNA-RNA hybrid (S9.6) antibody (Merck, MABE1095) at a concentration of 1:1000. Subsequently an HRP linked secondary antibody (Santa cruz, sc-2005) was used followed by incubation with ECL (GE healthcare, RPN2232). Methylene blue staining (Sigma-Aldrich, M9140) supplemented with 0.3 mM NaOAc was used as a loading control for all dot blots. Dot blots were developed using a chemoluminescence imaging system (Intas, ECL chemostar). Experiments involving dot blots were performed five times to ensure reproducibility.
R-loop stainings for tumor material were performed on formalin fixed tissue slides of five ETMRs, five Group4 medulloblastomas and five WNT medulloblastomas using the same conditions as described by Gorthi et al.21 For all IHC slides representative pictures were taken in addition to pictures covering the entire slide.
In vitro drug response
Response to treatment with Pamiparib (BGB-290) (MedChemExpress, HY-104044) or Veliparib (Abbvie, S1004) and Topotecan (ApexBio, B4982) was evaluated using dose response curves. BT183 cells were plated in 96 well plates 24 hours before treatment. 2-fold increasing concentrations of the inhibitor were added with concentrations ranging from 0.08nM-20nM for Topotecan and 156,25nM- 40mM for Pamiparib and Veliparib. 72 hours after treatment cell viability was measured using an automated plate reader after using CellTiter-Glo (Promega, G7570). Graphpad Prism was used to generate dose response curves. For combination treatment, the IC20 was determined using the dose response curves of single inhibitors and added to increasing concentrations of the other drug. Synergism between inhibitors was calculated by applying the Chou-Talay method as described previously65.
Immunofluorescence
BT-183 cells were grown on sterilized glass coverslips coated with Human Laminin (Sigma, L2020) and Poly-L-Lysine (Sigma, P8920) for 24 hours before treatment with either DMSO (0.6%), Topotecan, Pamiparib or both Pamiparib and Topotecan. Before fixation using 4% PFA spheres were washed with 0.5% NP-40 for five minutes on ice to increase the permeability. Subsequently, cells were permeabilized using 0.3% Triton-X-100 for 10 minutes followed by one hour blocking with 10% donkey serum. Slides were incubated with primary antibody 1:100 for either y-H2AX (Abcam, ab11174) or S9.6 in 10% donkey serum overnight. Subsequently, coverslips were incubated with DAPI (1:1000) and Alexa Fluor 488/568 (1:500) (Life Technologies). Coverslips were mounted on glass slides using mountant (Invitrogen, P36930) and imaged at a confocal microscope (Zeiss LSM 800). Representative pictures were taken at 400x magnification and processed using Airyscan processing (included in the Zeiss LSM 800 software) and applying the middle of 21 Z-stacks. Red and green channels were normalized to the blue channel (DAPI) in all shown pictures.
Signals were quantified using ImageJ applying a custom-made macro over three random neurospheres having seven Z-stacks each for every treatment condition. Signal intensity was measured in separate regions by binarizing DAPI signal and using watershed image segmentation to generate regions that simulate cells within the neurosphere. Subsequently, background subtraction (rolling ball radius of 50 pixels) was applied on every Z-stack for every channel. For each channel the average signal per cell was calculated and the y-H2AX signal and S9.6 signal was normalized by DAPI signal per cell.
DICER1 WT and DICER1 KO cells were grown and mounted in the same way but were not grown on coated coverslips as cells were adherent. Representative pictures were taken at a 630x magnification using the middle of 11 Z-stacks. Quantification was performed at a 200x magnification using three representative regions with more than five cells using five different Z-stacks. Background correction, signal calculation and normalization was performed in the same manner as the neurospheres.
Statistics and Reproducibility
Statistical enrichment was calculated using Fisher’s exact tests for calculations of binary values and proportions unless stated otherwise. For comparison between continuous values Mann-Whitney U/Wilcoxon Rank sum tests were used unless otherwise stated in either the figure legend or method details. Multiple testing correction was performed using the Benjamini-Hochberg method unless otherwise stated in the figure legend. All statistical analysis was performed in R(v3.4.3) except for dose response curves which were calculated using Graphpad Prism 6. Unless stated otherwise, results from representative experiments were performed at least three times. Pictures shown in Fig. 4f, g are representative of three different regions each containing more than three cells, experiment is representative of three biological replicates. Dot blots in Fig 4h. are representative of five biological replicates. Dose response curves in Fig. 5a were based on three biological replicates for each concentration of every drug and combination of drugs. The experiment was repeated three times and a representative curve is given for each drug combination. Figures shown in Fig. 5c, d are representative of three different regions each containing more than three cells, experiment is representative of three biological replicates.
Extended Data
Extended Data Figure 1. Clinicopathological differences are not associated with molecular subgrouping.
a, t-SNE clustering analysis of DNA methylation profiles of 193 ETMRs. Samples were colored according to their clinical, histological or molecular annotation. b, Schematic representation of location (position of the circle), histological diagnosis (outer ring) and C19MC status (inner ring) of ETMRs. Circle size denotes the relative number of primary tumors that have been diagnosed in each part of the brain, each wedge representing one tumor. Tumors could be assigned to multiple locations depending on the diagnosis. Tumors were excluded for which no information on the site of occurrence was available.
Extended Data Figure 2. miRNA expression correlates strongly between ETMRs with or without C19MC amplification.
a, Supervised clustering of the 416 differentially expressed mature miRNAs (two-sided neg. binomial, BH adjusted p-value <0.05) between ETMRs (n=7) (excluding ETMRs without amplification) and other tissues (n=38). b, Unsupervised clustering of mature miRNAs with a minimum expression of 32 in at least one sample and a variance higher than 10 between all samples (n=294). Hierarchical clustering using average as distance measure was used to cluster the samples after values were z-score normalized. c-g, Regression of the median expression of mature miRNAs derived from ETMRs (n=7) against normal brain (n=8), other entities (n=10 for all entities) or ETMRs without C19MC amplification (n=3). miRNAs that had a median expression under 32 RPM in either of the compared entities were excluded. miRNAs that were differentially expressed between ETMRs (with and without C19MC amplification) against other entities (two-sided neg. binomial, BH adjusted p-value <0.05) were highlighted. For each comparison, the Pearson correlation was calculated (p-value <0.0005 for all comparisons).
Extended Data Figure 3. KEGG pathway enrichment in ETMRs.
a-b, Summary of KEGG pathway enrichment of ETMRs (n=28) against normal brain (n=38) (a) or 580 different brain tumors (b). Pathways are colored by similarity based on NaviGO co-occurrence scores55 and manual assessment. Significantly upregulated genes were calculated using ANOVA (FDR adjusted p-value <0.01).
Extended Data Figure 4. ETMRs consist of at least two distinct subtypes of cells.
a, Heatmap showing z-score normalized expression of 450 DNA repair genes and the corresponding pathways8 for 190 tumors of different entities including 28 ETMRs. Supervised clustering was used and samples were sorted by entity or C19MC amplification status. Entities include three ATRT subgroups, four MB subgroups, CNS EFT-CIC, CNS-NB FOXR2, HGNET-MN1, HGNET-BCOR, ETMRs with amplification of C19MC (red) and ETMRs without amplification (blue). ETMR subsets were manually assessed based on DNA repair pathway expression. b, Debulking of mRNA expression using CIBERsort by using the median expression of scRNA-seq data of the forebrain as gene signature10. The cumulative fraction of each cell type was calculated and samples were sorted according to the percentage of modeled neural stem cells. Samples were annotated based on the subsets derived from a. c, Boxplots showing expression of stem cell markers (HMGA2, LIN28A), astrocyte markers (AQP4, GFAP) and genes involved in the DNA damage response (WEE1, CHEK2) in ETMRs with high DNA repair expression (n=18) and low DNA repair expression (n=10). P-values were calculated using a two-sided Mann-Whitney U test (***= P<0.0005, **= P<0.005, *= P<0.05, NS= not significant). Boxes show the median, first and third quartile and whiskers extending to 1.5x the interquartile range. d, Distribution of histology annotation of 18 ETMRs for which these data were available divided into two subsets. The number of EBL phenotypes was significantly enriched in the high DNA repair expression group using a two-sided Fisher’s exact test (P-value = 3.7E-02). e, t-SNE clustering based on methylation profiles of a micro-dissected ETMR (ET174) (split in bulk, rosettes and neuropil) and 192 other ETMRs. f, Expression of LIN28A and AQP4 in rosette tissue and neuropil tissue of the same tumor. g, Copy number profiles of micro-dissected neuropil and rosettes from the same tumor. h, Fold change of expression of six markers in two matched recurrences normalized to the primary tumor.
Extended Data Figure 5. Recurrent events in ETMRs without C19MC amplification.
a, Schematic representation of the translocation / amplification of a region on chromosome 11 with the host-gene of the miR-17–92 miRNA cluster (MIR17HG) shown in red on chromosome 13. Regions were reconstructed using mate-pair sequencing. The actual amplified region is circular denoted by arrows on each end. b, Copy number profile of a tumor harboring the miR-17–92 cluster translocation / amplification. Copy numbers were derived from methylation array data with each dot representing a probe. Inset shows validation of both the chromosome 11 (YAP1; green) and chromosome 13 (MIR17HG; red) amplifications using FISH. c, Quantification of mature miRNAs in the miR-17–92 miRNA cluster (n=20) confirms that the ETMR (blue) with the chromosome 11 and chromosome 13 amplification/translocation has higher expression of miR-17–92 cluster miRNAs. Each bar represents one tumor corresponding to the given entity. P-values were calculated using a one-sided Mann-Whitney U test (*= P<0.05). d, Example of a copy number profile of a case showing clustered rearrangements around C19MC. This case did not have a C19MC amplification or DICER1 mutations. e, Copy number profile of an ETMR without C19MC amplification or DICER1 mutation showing an overall instable genome with many regions containing clustered breakpoints.
Extended Data Figure 6. ETMRs recurrently show genomic instability.
a, Oncoplot showing the co-occurrence of all CNVs separated by C19MC amplification status. b, Overview of copy number profiles of all ETMRs (n=193). Bars (gain, balanced, loss) add up to 100% for each chromosome arm. c, Overview of copy number profiles of all ETMRs with (n=170) or without (n=23) C19MC amplification. P-values were calculated using a two-sided Fisher’s exact tests and adjusted for multiple testing (BH) (*** P<0.0005, ** P<0.005, * P<0.05, NS= not significant). d, Overview of CNVs in matched primary tumor and recurrence pairs for the most variable CNVs. Events (copy number changes, clustered breakpoints or increases in ploidy) that were gained upon recurrence have a thicker outline. Percentages denote the percentage of matched samples acquiring a CNA or genome duplication. e, Example of a case in which polyploidy was validated using FISH (n=28 tested samples), the chromosome 9 and 11 centromeres were used as probes. f, Examples of cases showing clustered breakpoints on chromosome 19. Chromosome 19 is shown as a circular representation, translocations to other chromosomes were annotated as single positions. All SVs were detected using mate pair sequencing.
Extended Data Figure 7. Conservation of events for individual cases.
Summary of events occurring in seven matched primary tumors compared to recurrences (first second or third relapse) and two matched relapses. For every sample conservation of SNVs is given as a graph with the allele frequencies (AF) of the primary tumor on the x-axis and the recurrence on the y-axis. In the last panel two matched recurrences are shown with a recurrence on each axis. Boxes show events that are lost, conserved or gained. Each comparison has a table showing the total number of events in each quadrant (lost: AF primary >10% and AF recurrence < 2%, stable: AF primary >20% and AF recurrence >20% and gained: AF primary < 2% and AF recurrence >10%). Conservation of SVs is given as a circular representation of the genome having the CNVs from the primary tumor in the outer rim and the recurrence in the inner rim. SVs were colored by detection in either only the primary tumor (red), only in the relapse (grey) or in both (blue). Each combination also has a Venn diagram showing the total number of SVs that were detected in the primary tumor, the recurrence or both.
Extended Data Figure 8. Mutations in primary tumors and relapses.
a, Boxplots showing the total number of SNVs or indels in primary tumors (n=20) compared to relapses (n=12). Boxes show the median, first and third quartile and whiskers extending to 1.5x the interquartile range. We detected, on average, 1180 SNVs (range: 339–2544) and 468 indels (range: 299 – 1026) in primary tumors and 5162 SNVs (range: 2992–7773) and 847 indels (range: 554–1187) in relapsed tumors throughout the genome. In coding regions, there were on average 14 non-synonymous SNVs (range: 3 – 45) and two indels (range: 0 – 7) in primary tumors and 59 non-synonymous SNVs (range: 37 – 92) and six indels (range: 2 – 11) in relapsed tumors. b, Barchart showing the percentage of substitutions of either the combined primary tumors (n=20) or combined relapses (n=12) divided by substitution type and affected strand for SNVs residing in transcribed regions. Transcriptional asymmetry is defined as the difference between the amount of SNVs on the transcribed strand versus the untranscribed strand for each substitution type. Error bars denote mean ± s.e.m, P-values were calculated using two-sided Poisson tests (*** P<0.0005, ** P<0.005, * P<0.05, NS= not significant). c, Substitution type probability based on the 96 different trinucleotide contexts for a matched primary relapsed pair shown in d. compared to a cisplatin signature16 and new pediatric cancer signature (P1)13. d, Cosine similarity between the cisplatin signature and other signatures (n=36). P-values were calculated using an M-test after comparing all signatures pairwise (*** P<0.0005, ** P<0.005, * P<0.05, NS= not significant).
Extended Data Figure 9. ETMRs have dense and strongly conserved C>T and C>G mutations around breakpoints.
a, Rainfall plot showing an example of kataegis around C19MC. Every point represents a somatic SNV colored by substitution type, the x-axis represents the position in the genome and the position on the y-axis represents the density of SNVs. b, Lollipop plot showing SNVs per 1kb in a region of 10000 bp surrounding breakpoints for all ETMRs. Pins represent the percentage of substitution types of all SNVs within 1kb, while the height of the lollipops represents the substitutions per kb. c, Barchart showing the percentages of substitution types in regions 10kb around breakpoints (left, n= 543 SNVs) and the rest of the genome (right, n= 84991 SNVs). P-values were calculated using a one-sided Fisher’s exact test and annotated as *** P<0.0005, ** P<0.005, * P<0.05 or NS= not significant. d, Combined mutation density of four primary tumors colored by conservation in the matched recurrence (blue is conserved, grey is not conserved) as shown by a rainfall plot in the upper panel, density distribution is shown in the middle panel and breakpoint density is shown in the lower panel. e, Graph of allele frequencies (AF) of all primary (x-axis) versus relapse (y-axis). Boxes show conservation (lost: AF primary >10% and AF recurrence < 2%, conserved: AF primary >20% and AF recurrence >20% and gained: AF primary < 2% and AF recurrence >10%) (n=2100 SNVs over 20% allele frequency in the primary tumor). P-value was calculated using a two-sided Chi-square test f, Barchart showing the percentage of substitution types for SNVs in each quadrant (lost: AF primary >10% and AF recurrence < 2%, conserved: AF primary >20% and AF recurrence >20% and gained: AF primary < 2% and AF recurrence >10%). g, Graph showing the ratio of conserved SNVs against not conserved SNVs in regions around breakpoints with increasing sizes. Conservation is defined as SNVs with an allele frequency over 20% in the primary tumor and an allele frequency over 20% in the recurrence, SNVs with an allele frequency lower than 20% in the recurrence but higher than 20% in the primary tumor were defined as not conserved. P-value between 10kb around breakpoints and the rest of the genome using a two-sided Chi-square test (n=2100, p-value 5.4e-11).
Extended Data Figure 10. Context of R-loops and DNA damage in ETMRs and after Dicer1 KO.
a, Genome-wide density of R-loops in ETMRs, R-loops in Ewing sarcoma (EWS), RLFS and gene density. b, Representation of SVs genome-wide and their breakpoint context. Outer layers show the density of DRIP peaks (blue) or RLFS (red). The inner part shows all SVs from ETMRs sequenced using WGS, depicting SVs that fall in DRIP-seq peaks (blue) or RLFS (red). c, R-loop signal detected in genomic regions sorted by R-loop signal (including elements from non-B-DB62 and repeatmasker). R-loop signal was determined for 10000 randomly selected elements for every type of genomic feature (n=21). Violin plots depict kernel density estimates and represent the density distribution d, Genome-wide association of breakpoints with genomic regions sorted by R-loop signal shown in c. Genome-wide associations were calculated as distance to nearest element compared to a set of 10000 randomly generated breakpoints. Enrichments were calculated for EWS breakpoints66 and breakpoints from other entities22 (reference set). P-values were calculated using a two-sided Mann-Whitney U test and adjusted for multiple testing (BH). e, Density of distances between genomic regions and breakpoints detected in ETMR, EWS, random breakpoints and reference breakpoints. f, Total percentage of breakpoints within 1kb of genomic regions. g, Enrichment of SNVs (n=85534) in ETMR R-loops (n=16002 regions) and RLFS (n=85534 regions) compared to random regions of the same size. P-values were calculated using a two-sided Chi-square test (*** P<0.0005, ** P<0.005, * P<0.05, NS= not significant). h, Genome-wide distribution of mouse RLFS and breakpoints occurring in DICER1 KO cells compared to WT. The outer rim shows the genome wide density of mouse RLFS, the inner rim the CNAs that were found between WT and KO and the inner part shows the SVs that were detected between WT and KO. Breakpoints falling within RLFS are highlighted in red. i, Copy number profiles of an example of a translocation coupled to duplication in RLFS which were found in DICER1 KO compared to DICER1 WT cells. Red arrows depict the location of translocation/duplication.
Supplementary Material
Supplementary Table 8
Full list of identified somatic SVs in ETMRs.
Supplementary Table 9
Full list of somatic SNVs identified in primary tumors using WGS including non-coding regions, regions overlapping promotor regions and regions overlapping putative enhancers.
Supplementary Table 1
Information about ETMR samples included in the cohort.
Supplementary Table 2
Expression of mature miRNAs in ETMRs (n=10) and differential expression analysis of ETMRs (n=7) compared to other tissues (n=38). P-values were calculated using negative binomial testing and were BH adjusted.
Supplementary Table 3
Normalized expression values of ETMRs included in the paper (n=28), expression values of different regions that were micro-dissected and the KEGG and GO-term enrichments of ETMRs (n=28) compared to normal brain (n=38) or other brain tumors (n=580).
Supplementary Table 4
Lists of sequenced used for analysis using DNA repair genes and genes that were included for targeted sequencing.
Supplementary Table 5
Identified exonic somatic non-synonymous SNVs in primary tumors and relapsed tumors using WGS
Supplementary Table 6
Identified exonic non-synonymous SNVs using WES and targeted sequencing.
Supplementary Table 7
Copy number aberrations of the ETMR cohort and copy number changes between primary tumors and matched relapses.
Acknowledgements
We thank the DKFZ sequencing core facility for technical support, assistance with data generation and data management and we thank the DKFZ light microscopy facility for their assistance in generating microscopy images, and BeiGene for providing Pamiparib. This work was supported by the ICGC PedBrain Tumor Project, funded by the German Cancer Aid (109252) and by the German Federal Ministry of Education and Research: BMBF grants #01KU1201A (PedBrain Tumor) and #01KU1505A (ICGC-DE-MINING). Additional funding was awarded by the NIH (K22ES012264, 1R15ES019128, 1R01CA152063), Voelcker Fund Young Investigator Award and CPRIT (RP150445) to A.J.R.B.; CPRIT (RP101491), NCI T32 postdoctoral training grant (T32CA148724), NCATS TL1 (TL1TR002647) and the AACR-AstraZeneca Stimulating Therapeutic Advances through Research Training grant to A.G.; CPRIT (RP140105) to J.C.R.; and NCI (P30CA054174) to the sequencing core facility. S.L. and M.K. are supported by the Solving Kid’s Cancer foundation and the Bibi Fund for Childhood Cancer Research. A.K. is supported by the Helmholtz Association Research Grant (Germany). M.Ry. is supported by an RSF Research Grant (18–45-06012). J.O.K. was funded by an ERC starting grant.
Footnotes
Data availability statement
Data availability
Raw and processed 450K/EPIC methylation values and raw and processed expression data for all included ETMRs are deposited at the Gene Expression Omnibus (GEO) under accession number GSE122038. All NGS data is deposited at the European Genome-phenome Archive (EGA) under accession number EGAS00001003256.
Source Data
Source data are available for Fig. 1a–c, Fig. 2c, Fig. 3b, c, Fig. 4d, g, Fig. 5, a, b, d, Ext. Data Fig. 1a, Ext. Data Fig. 2a–g, Ext. Data Fig. 4b, c, h, Ext. Data Fig. 5c, Ext. Data Fig. 6b, c, Ext. Data Fig. 8a–d, Ext Data Fig. 9b, c, e, f, g, Ext Data Fig 10 g.
Code availability
All custom code used to generate the data in this study is available upon reasonable request.
Contact for reagent and resource sharing
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Marcel Kool (m.kool@kitz-heidelberg.de).
Declaration of Interests
The authors declare no competing interests
References
- 1.Korshunov A et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol 128, 279–289, doi: 10.1007/s00401-013-1228-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eberhart CG, Brat DJ, Cohen KJ & Burger PC Pediatric neuroblastic brain tumors containing abundant neuropil and true rosettes. Pediatr Dev Pathol 3, 346–352 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Pfister S et al. Novel genomic amplification targeting the microRNA cluster at 19q13.42 in a pediatric embryonal tumor with abundant neuropil and true rosettes. Acta Neuropathol 117, 457–464, doi: 10.1007/s00401-008-0467-y (2009). [DOI] [PubMed] [Google Scholar]
- 4.Li M et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16, 533–546, doi: 10.1016/j.ccr.2009.10.025 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleinman CL et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet 46, 39–44, doi: 10.1038/ng.2849 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Sturm D et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164, 1060–1072, doi: 10.1016/j.cell.2016.01.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capper D et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474, doi: 10.1038/nature26000 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearl LH, Schierz AC, Ward SE, Al-Lazikani B & Pearl FM Therapeutic opportunities within the DNA damage response. Nat Rev Cancer 15, 166–180, doi: 10.1038/nrc3891 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Newman AM et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12, 453–457, doi: 10.1038/nmeth.3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong S et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528, doi: 10.1038/nature25980 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Neumann JE et al. A mouse model for embryonal tumors with multilayered rosettes uncovers the therapeutic potential of Sonic-hedgehog inhibitors. Nat Med 23, 1191–1202, doi: 10.1038/nm.4402 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Anglesio MS et al. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol 229, 400–409, doi: 10.1002/path.4135 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Grobner SN et al. The landscape of genomic alterations across childhood cancers. Nature 555, 321–327, doi: 10.1038/nature25480 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421, doi: 10.1038/nature12477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szikriszt B et al. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol 17, 99, doi: 10.1186/s13059-016-0963-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boot A et al. In-depth characterization of the cisplatin mutational signature in human cell lines and in esophageal and liver tumors. Genome Res 28, 654–665, doi: 10.1101/gr.230219.117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maciejowski J, Li Y, Bosco N, Campbell PJ & de Lange T Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 163, 1641–1654, doi: 10.1016/j.cell.2015.11.054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos-Pereira JM & Aguilera A R loops: new modulators of genome dynamics and function. Nat Rev Genet 16, 583–597, doi: 10.1038/nrg3961 (2015). [DOI] [PubMed] [Google Scholar]
- 19.El Hage A, French SL, Beyer AL & Tollervey D Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24, 1546–1558, doi: 10.1101/gad.573310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenjaroenpun P, Wongsurawat T, Yenamandra SP & Kuznetsov VA QmRLFS-finder: a model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res 43, 10081, doi: 10.1093/nar/gkv974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorthi A et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 555, 387–391, doi: 10.1038/nature25748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloosterman WP et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep 1, 648–655, doi: 10.1016/j.celrep.2012.05.009 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Gan W et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25, 2041–2056, doi: 10.1101/gad.17010011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu WT et al. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat Commun 9, 532, doi: 10.1038/s41467-018-02893-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castel SE et al. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 159, 572–583, doi: 10.1016/j.cell.2014.09.031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francia S et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235, doi: 10.1038/nature11179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt C et al. Preclinical drug screen reveals topotecan, actinomycin D, and volasertib as potential new therapeutic candidates for ETMR brain tumor patients. Neuro Oncol 19, 1607–1617, doi: 10.1093/neuonc/nox093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staker BL et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci U S A 99, 15387–15392, doi: 10.1073/pnas.242259599 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das SK et al. Poly(ADP-ribose) polymers regulate DNA topoisomerase I (Top1) nuclear dynamics and camptothecin sensitivity in living cells. Nucleic Acids Res 44, 8363–8375, doi: 10.1093/nar/gkw665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennasser Y et al. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol 18, 323–327, doi: 10.1038/nsmb.1987 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm D et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541, doi: 10.1038/nature04791 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Schultz KAP et al. PTEN, DICER1, FH, and Their Associated Tumor Susceptibility Syndromes: Clinical Features, Genetics, and Surveillance Recommendations in Childhood. Clin Cancer Res 23, e76–e82, doi: 10.1158/1078-0432.CCR-17-0629 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Seki M et al. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res 74, 2742–2749, doi: 10.1158/0008-5472.CAN-13-2470 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Koelsche C et al. Primary intracranial spindle cell sarcoma with rhabdomyosarcoma-like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol 136, 327–337, doi: 10.1007/s00401-018-1871-6 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Hovestadt V et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125, 913–916, doi: 10.1007/s00401-013-1126-5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spence T et al. A novel C19MC amplified cell line links Lin28/let-7 to mTOR signaling in embryonal tumor with multilayered rosettes. Neuro Oncol 16, 62–71, doi: 10.1093/neuonc/not162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahm F et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol 131, 903–910, doi: 10.1007/s00401-015-1519-8 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Jones DT et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105, doi: 10.1038/nature11284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uro-Coste E et al. ETMR-like infantile cerebellar embryonal tumors in the extended morphologic spectrum of DICER1-related tumors. Acta Neuropathol, doi: 10.1007/s00401-018-1935-7 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883, doi: 10.1093/bioinformatics/bts034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Maaten L & Hinton G Visualizing Data using t-SNE. J Mach Learn Res 9, 2579–2605 (2008). [Google Scholar]
- 42.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kool M et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25, 393–405, doi: 10.1016/j.ccr.2014.02.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164, doi: 10.1093/nar/gkq603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kircher M et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–315, doi: 10.1038/ng.2892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waszak SM et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol 19, 785–798, doi: 10.1016/S1470-2045(18)30242-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koboldt DC et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285, doi: 10.1093/bioinformatics/btp373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haradhvala NJ et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell 164, 538–549, doi: 10.1016/j.cell.2015.12.050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blokzijl F, Janssen R, van Boxtel R & Cuppen E MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 10, 33, doi: 10.1186/s13073-018-0539-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johann PD et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 29, 379–393, doi: 10.1016/j.ccell.2016.02.001 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Northcott PA et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434, doi: 10.1038/nature13379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Bardes EE, Aronow BJ & Jegga AG ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311, doi: 10.1093/nar/gkp427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Wei Q, Khan IK, Ding Z, Yerneni S & Kihara D NaviGO: interactive tool for visualization and functional similarity and coherence analysis with gene ontology. BMC Bioinformatics 18, 177, doi: 10.1186/s12859-017-1600-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527, doi: 10.1038/nbt.3519 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Hovestadt V et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541, doi: 10.1038/nature13268 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Hafner M et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44, 3–12, doi: 10.1016/j.ymeth.2007.09.009 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S & Huber W Differential expression analysis for sequence count data. Genome Biol 11, R106, doi: 10.1186/gb-2010-11-10-r106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozomara A & Griffiths-Jones S miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42, D68–73, doi: 10.1093/nar/gkt1181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191, doi: 10.1093/nar/gku365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cer RZ et al. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res 41, D94–D100, doi: 10.1093/nar/gks955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korshunov A et al. Focal genomic amplification at 19q13.42 comprises a powerful diagnostic marker for embryonal tumors with ependymoblastic rosettes. Acta Neuropathol 120, 253–260, doi: 10.1007/s00401-010-0688-8 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Sanz LA et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell 63, 167–178, doi: 10.1016/j.molcel.2016.05.032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chou TC Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70, 440–446, doi: 10.1158/0008-5472.CAN-09-1947 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Anderson ND et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 361, doi: 10.1126/science.aam8419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 8
Full list of identified somatic SVs in ETMRs.
Supplementary Table 9
Full list of somatic SNVs identified in primary tumors using WGS including non-coding regions, regions overlapping promotor regions and regions overlapping putative enhancers.
Supplementary Table 1
Information about ETMR samples included in the cohort.
Supplementary Table 2
Expression of mature miRNAs in ETMRs (n=10) and differential expression analysis of ETMRs (n=7) compared to other tissues (n=38). P-values were calculated using negative binomial testing and were BH adjusted.
Supplementary Table 3
Normalized expression values of ETMRs included in the paper (n=28), expression values of different regions that were micro-dissected and the KEGG and GO-term enrichments of ETMRs (n=28) compared to normal brain (n=38) or other brain tumors (n=580).
Supplementary Table 4
Lists of sequenced used for analysis using DNA repair genes and genes that were included for targeted sequencing.
Supplementary Table 5
Identified exonic somatic non-synonymous SNVs in primary tumors and relapsed tumors using WGS
Supplementary Table 6
Identified exonic non-synonymous SNVs using WES and targeted sequencing.
Supplementary Table 7
Copy number aberrations of the ETMR cohort and copy number changes between primary tumors and matched relapses.