Abstract
Genomic amplification of the oncogene MYCN is a major driver in the development of high-risk neuroblastoma, a pediatric cancer with poor prognosis. Given the challenge in targeting MYCN directly for therapy, we sought to identify MYCN-dependent metabolic vulnerabilities that can be targeted therapeutically. Here, we report that the gene encoding glycine decarboxylase (GLDC), which catalyzes the first and rate-limiting step in glycine breakdown with the production of the one-carbon unit 5,10-methylene-tetrahydrofolate, is a direct transcriptional target of MYCN. As a result, GLDC expression is markedly elevated in MYCN-amplified neuroblastoma tumors and cell lines. This transcriptional upregulation of GLDC expression is of functional significance, as GLDC depletion by RNA interference inhibits the proliferation and tumorigenicity of MYCN-amplified neuroblastoma cell lines by inducing G1 arrest. Metabolomic profiling reveals that GLDC knockdown disrupts purine and central carbon metabolism and reduces citrate production, leading to a decrease in the steady-state levels of cholesterol and fatty acids. Moreover, blocking purine or cholesterol synthesis recapitulates the growth inhibitory effect of GLDC knockdown. These findings reveal a critical role of GLDC in sustaining the proliferation of neuroblastoma cells with high-level GLDC expression and suggest that MYCN amplification is a biomarker for GLDC-based therapeutic strategies against high-risk neuroblastoma.
Keywords: Glycine cleavage, GLDC, MYCN, Neuroblastoma, Cancer metabolism, Serine-Glycine-One-Carbon metabolism
Introduction
Neuroblastoma is a malignant tumor of the sympathetic nervous system, originating from the sympathoadrenal lineage of neural crest cells during development [1, 2]. It is a cancer of young children and a leading cause of cancer-related deaths in children under 5 years of age. Most neuroblastoma tumors arise in the adrenal medulla and abdominal sympathetic ganglia [1, 3, 4]. Based on clinical and biological features, neuroblastoma is classified into low-, intermediate-, and high-risk categories [5]. Patients with low- or intermediate-risk neuroblastoma have an overall survival rate of more than 90% following minimum or standard treatment, whereas the overall survival rate for patients with high-risk neuroblastoma is less than 50% even after intensive, multimodal therapy [6, 7]. Thus, there is an urgent need to identify new therapeutic targets and strategies to tackle high-risk neuroblastoma. One approach is to identify metabolic vulnerabilities of high-risk neuroblastoma for therapeutic exploitation.
Genomic amplification of MYCN is a major cause of high-risk neuroblastoma and is strongly associated with poor outcome in neuroblastoma patients [5, 8–11]. MYCN is a member of the MYC family of oncogenic transcription factors that promote cell growth and proliferation through transcriptional regulation [12, 13]. There is a large body of evidence suggesting that transcriptional reprogramming of cellular metabolism by MYC oncoproteins is a key mechanism for their growth-promoting activity [14, 15]. This reprogramming, however, may also impose a dependence on otherwise dispensable metabolic pathways, which might be targeted therapeutically.
Increased activation of the serine-glycine-one-carbon (SGOC) metabolic pathway (Fig. 1a) is a common feature of metabolic reprogramming in various types of cancers. Genes encoding enzymes of this pathway are frequently overexpressed in cancers, as a result of genomic amplification [16, 17] or transcriptional activation [18–24]. The SGOC pathway links the glycolytic intermediate 3-phosphoglycerate to the production of serine, glycine, and the one-carbon carriers 5,10-methylene-tetrahydrofolate (5,10-MTHF) and 10-formyl-THF, which contribute carbon and nitrogen to purine nucleotide and thymidylate synthesis. In addition, 5,10-MTHF donates carbon for the synthesis of S-adenosylmethionine, the universal methyl donor for methylation reactions [18, 19, 25].
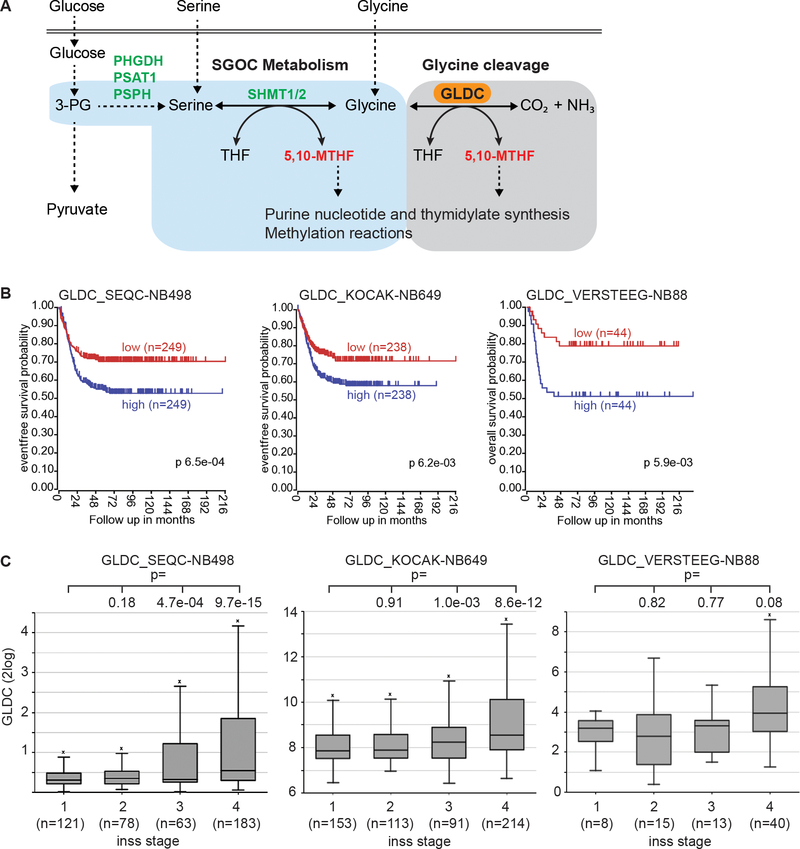
Fig. 1.
High GLDC expression is associated with poor prognosis and advanced stages of neuroblastoma. a Schematic of the SGOC pathway that provides one-carbon units for nucleotide biosynthesis and methylation reactions. 3-PG, 3-phosphoglycerate; PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; SHMT, serine hydroxymethyltransferase. b Kaplan-Meier survival curves for three independent cohorts of neuroblastoma patients based on GLDC mRNA expression. c Box plots of GLDC mRNA expression in relation to neuroblastoma stages using datasets from the three neuroblastoma cohorts.
As part of SGOC metabolism, mitochondrial glycine is catabolized by the glycine cleavage system (GCS), generating CO2, NH3, NADH, and the one-carbon unit 5,10-MTHF (Fig. 1a) [26, 27]. Stable isotope-tracing analysis in adult humans has provided evidence for a major role of glycine-derived one-carbon units in contributing to one-carbon metabolism and serine synthesis [28]. In addition, glycine clearance by the GCS helps to maintain the flux of the SGOC pathway [29] and to prevent the accumulation of glycine. Excess glycine is known to inhibit cell proliferation [30], probably by generating the toxic metabolites aminoacetone and methylglyoxal [29].
The GCS is a multi-enzyme complex, consisting of GLDC (P protein), aminomethyltransferase (T protein), dihydrolipoamide dehydrogenase (L protein), and the hydrogen carrier protein (H protein). GLDC catalyzes the first and rate limiting step in glycine breakdown. The physiological significance of GLDC in glycine clearance is underscored by genetic evidence: Mutations in GLDC cause glycine accumulation, leading to glycine encephalopathy (also known as non-ketotic hyperglycinemia, NKH) and neural tube defect (NTD) in mice and humans [31–33]. Moreover, in the mouse NTD model, supplementation of the one-carbon unit formate was able to rescue the NTD phenotype [33]. Further investigation revealed that embryonic neural tube closure depends on glycine cleavage as a key supply of one-carbon units to the folate cycle [34].
There is evidence supporting a role of GLDC in cancer. It has been reported that increased expression of GLDC in non-small cell lung cancer initiating cells is essential for their proliferation and tumorigenic potential by promoting pyrimidine biosynthesis, glycolysis, and sarcosine production [35]. Another study shows that in glioblastoma cell lines with high expression of SHMT2, which converts serine to glycine in the mitochondria (Fig. 1a), high GLDC expression is critical for the viability and growth of these cell lines by preventing alternative glycine catabolism that produces the toxic metabolites aminoacetone and methylglyoxal [29]. However, a study of the colon cancer cell line HCT116 suggests that glycine cleavage is not a significant source of one-carbon units for purine synthesis and cell proliferation [30]. Collectively, these findings suggest that GLDC may have a context-dependent oncogenic function. A molecular understanding of the underlying mechanism may identify biomarkers for selectively targeting GLDC-dependent cancers.
In this study, we investigated the function of GLDC in neuroblastoma. We found that GLDC expression is markedly upregulated in neuroblastoma tumors and cell lines with MYCN amplification. We identified GLDC as a direct transcriptional target of MYCN required for the proliferation and tumorigenicity of MYCN-amplified cell lines. Moreover, we obtained evidence for a critical role of GLDC in regulation of central carbon metabolism and lipid synthesis. Our findings suggest that targeting GLDC could be a selective therapeutic strategy against MYCN-amplified neuroblastoma.
Results
High GLDC expression is associated with poor prognosis and advanced stages of neuroblastoma
To investigate the relevance of GLDC in neuroblastoma, we examined GLDC expression in neuroblastoma tumors by analyzing published gene expression profiling data from three independent cohorts of neuroblastoma patients, including the SEQC [36], KOCAK [37] and VERSTEEG [38] cohorts (n =1235). Our analysis revealed that high levels of GLDC expression were significantly correlated with lower survival (Fig. 1b). Moreover, we found that higher GLDC expression was significantly associated with advanced stages of neuroblastoma tumors (Fig 1c). These findings provide genetic evidence for a potential role of high GLDC expression in neuroblastoma pathogenesis.
MYCN is a transcriptional activator of GLDC expression in neuroblastoma
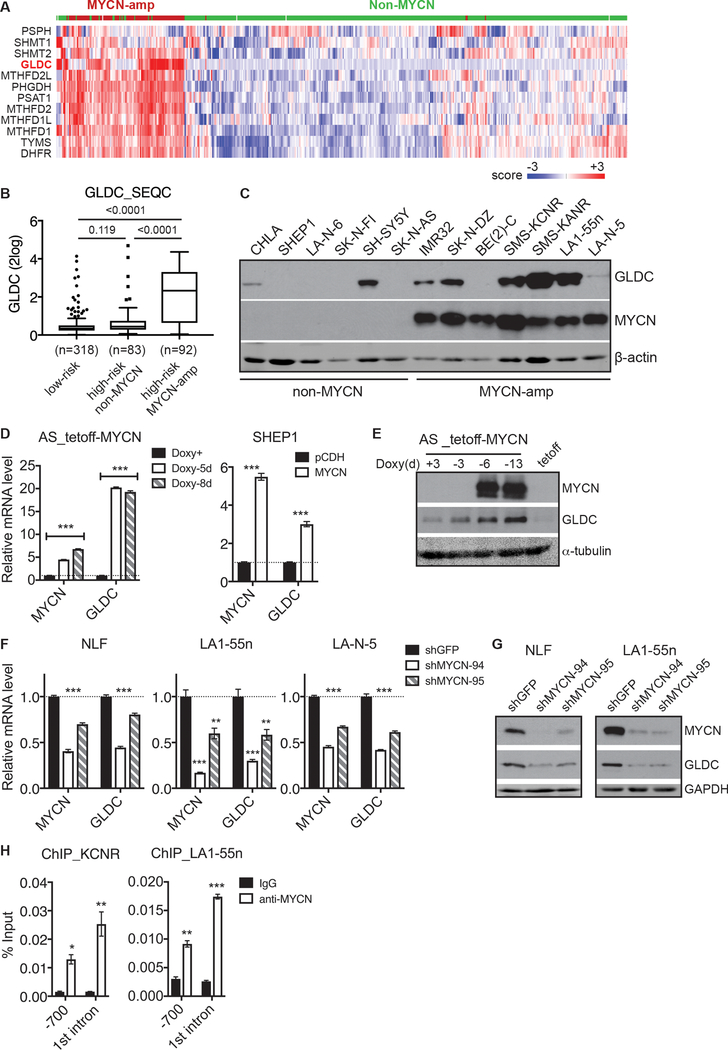
We sought to identify the transcription factor(s) responsible for the elevated GLDC expression in advanced neuroblastoma tumors. MYCN is a transcription factor and MYCN amplification is significantly associated with advanced stages of the disease [3–5]. We performed a hierarchical cluster analysis of the MYCN-amplification gene signature consisting of 143 genes upregulated in neuroblastoma tumors with MYCN amplification relative to those without [39]. This analysis revealed co-upregulation of GLDC and other SGOC pathway genes in MYCN-amplified neuroblastoma tumors (Fig. 2a). As a complementary approach, we analyzed the SEQC dataset for a correlation between GLDC mRNA expression and the MYCN amplification status. We found that GLDC expression was significantly higher in high-risk tumors with MYCN amplification compared to both low-risk tumors and to high-risk tumors without MYCN amplification (Fig. 2b). It is particularly interesting to note that there was no significant difference in GLDC mRNA expression levels between low-risk tumors and high-risk tumors without MYCN amplification (Fig. 2b). Similarly, we observed higher GLDC mRNA expression in MYCN-amplified neuroblastoma tumors from the KOCAK and VERSTEEG cohorts (Supplementary Figure 1A). In addition, we found a strong positive correlation in mRNA levels between GLDC and MYCN in tumor samples (Supplementary Figure 1B). In agreement with the findings from tumors, we found that six out of the seven MYCN-amplified neuroblastoma cell lines we examined showed detectable levels of GLDC protein, whereas only two out of six non-MYCN-amplified cell lines expressed GLDC (Fig. 2c). Moreover, quantitative analysis revealed a positive correlation in protein levels between GLDC and MYCN in these neuroblastoma cell lines (Supplementary Figure 1C). Together, these results indicate that high GLDC expression is associated with high MYCN expression in neuroblastoma tumors and cell lines.
Fig. 2.
GLDC is a direct transcriptional target of MYCN. a Heatmap showing coordinated upregulation of GLDC and other SGOC pathway genes in MYCN-amplified neuroblastoma tumors using the SEQC-498 dataset. b Box plot of GLDC mRNA expression in relation to neuroblastoma risk groups and MYCN amplification status using the SEQC dataset. c Immunoblotting of MYCN and GLDC in neuroblastoma cell lines with or without MYCN amplification. d-e qRT-PCR (d) and immunoblot (e) analyses of GLDC expression in non-MYCN-amplified SK-N-AS cells with inducible MYCN expression in the absence of doxycycline (Doxy) and SHEP1 cells with constitutive MYCN expression. f-g qRT-PCR (f) and immunoblot (g) analyses of GLDC expression in MYCN-amplified cell lines with MYCN knockdown. h ChIP-qPCR showing endogenous MYCN binding to the GLDC promoter and 1st intron in MYCN-amplified cell lines. Error bars (d, f, h) represent SEM (n = 2) and data were analyzed by unpaired, two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
We next investigated whether MYCN is able to induce GLDC expression. MYCN overexpression in non-MYCN-amplified SK-N-AS and SHEP1 neuroblastoma cell lines markedly increased GLDC mRNA (Fig. 2d) and protein expression (Fig. 2e). Conversely, knockdown of MYCN expression in MYCN-amplified NLF, LA1–55n, and LA-N-5 cell lines by two different shRNA constructs consistently downregulated the expression of GLDC mRNA (Fig. 2f) and protein (Fig. 2g). Thus, MYCN is a transcriptional activator of GLDC expression and is essential for maintaining GLDC expression in MYCN-amplified neuroblastoma cell lines.
MYCN and other members of the MYC family bind to the consensus E-box sequence CANNTG. Sequence examination revealed potential MYCN binding sites in the GLDC promoter region and first intron. To determine whether MYCN binds directly to the GLDC gene, we conducted chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) assays using MYCN-amplified SMS-KCNR and LA1–55n neuroblastoma cell lines. We detected significant levels of endogenous MYCN associated with the GLDC promoter and first intron (Fig. 2h), suggesting that GLDC is a direct transcriptional target of MYCN.
In light of above findings, we wondered if GLDC is also a target gene of MYC given the recent findings on MYC regulation of SGOC metabolism [14, 15]. We examined the P493–6 cell line, a lymphoid cell line with inducible MYC expression in the absence of doxycycline (tetoff-MYC) [40, 41]. Repression of MYC expression by doxycycline resulted in a marked reduction in GLDC mRNA expression (Supplementary Figure 2A), leading to decreased GLDC protein expression (Supplementary Figure 2B), whereas MYC induction by removing doxycycline resulted in a significant increase in GLDC protein expression (Supplementary Figure 2B). These findings suggest that GLDC is also a target gene of MYC.
High GLDC expression is essential for the proliferation and tumorigenicity of MYCN-amplified neuroblastoma cell lines
Since most of the MYCN-amplified cell lines we examined expressed high levels of GLDC protein (Fig. 2c) and MYCN directly activated GLDC transcription (Fig. 2d–h), we investigated the functional significance of high GLDC expression in MYCN-amplified cell lines by shRNA-mediated knockdown of GLDC expression. We tested five lentiviral constructs expressing shRNA sequences against different regions of the GLDC gene and selected two constructs (shGLDC-01 and shGLDC-99) that were most effective in silencing GLDC protein expression (Supplementary Figure 3A). GLDC knockdown by either shRNA construct significantly inhibited the proliferation of MYCN-amplified LA1–55n, NLF, and IMR5 neuroblastoma cell lines (Fig. 3a–b and Supplementary Figure 3B). We further investigated whether GLDC is required for the tumorigenicity of MYCN-amplified neuroblastoma cells using the tumorigenic SMS-KCNR cell line. NOD.SCID mice implanted with SMS-KCNR cells infected with control shGFP lentiviruses showed measurable tumor growth after 9 days, whereas no tumor growth was detected in the NOD.SCID mice implanted with shGLDC-expressing SMS-KCNR cells until 40 days after implantation (Fig. 3c–d). Moreover, GLDC knockdown markedly increased the survival of tumor-bearing mice (Fig. 3e). Interestingly, immunoblot analysis revealed that two of the three xenograft tumors derived from SMS-KCNR cells with GLDC knockdown regained the expression of GLDC protein at the levels comparable to those of xenograft tumors derived from control shGFP-expressing SMS-KCNR cells (Supplementary Figure 3C), suggesting a selective pressure for upregulation of GLDC expression during tumor growth, probably as a result of the outgrowth of a minor population of shGLDC cells that retained GLDC expression. Together, these findings indicate that high GLDC expression is required for the proliferation and tumorigenicity of MYCN-amplified neuroblastoma cell lines.
Fig. 3.
GLDC is required for the proliferation and tumorigenicity of MYCN-amplified neuroblastoma cell lines. a Cell growth assay of MYCN-amplified cell lines without (shGFP) or with GLDC knockdown (shGLDC). Error bars represent SEM (n = 5). b Microscopic image of IMR5 cells without or with GLDC knockdown. c-d Growth curves (c) and images (d) of SMS-KCNR xenografts without or with GLDC knockdown. Data were analyzed by multiple t test (Prism). e Survival curves for mice carrying SMS-KCNR xenografts without or with GLDC knockdown. Log-rank test p value is shown.
GLDC knockdown induces cell cycle arrest
To elucidate the mechanism underlying the growth-inhibitory effect of GLDC knockdown, we conducted microarray gene expression profiling of MYCN-amplified LA1–55n cells with inducible knockdown of GLDC expression in the presence of doxycycline (teton) (Supplementary Figure 4A). A total of 1094 genes showed significant changes in expression (≥ ±1.40 fold, p < 0.05) following GLDC knockdown, with 571 genes being upregulated and 523 genes downregulated (Supplementary Table 1). Gene ontology (GO) analysis of the upregulated genes revealed no significant enrichment of GO terms (FDR < 0.05), whereas GO analysis of the downregulated genes showed significant enrichment for GO terms associated with DNA replication, G1 to S transition, and cell proliferation (Fig. 4a and Supplementary Table 2). We obtained essential the same results with Gene Set Enrichment Analysis (GSEA) of the microarray data, which showed significant downregulation of gene sets involved in cell proliferation (Fig. 4b), including those of the MCM pathway (unwinding DNA prior to replication), DNA replication, cell cycle progression, and E2F targets (Fig. 4c). We confirmed the microarray data by qRT-PCR, showing that GLDC knockdown led to a significant reduction in mRNA expression of MCM5, cyclins (CCNs) and cyclin dependent kinases (CDKs) (Fig. 4d). Of note, GLDC knockdown had no significant effect on MYCN mRNA and protein expression (Supplementary Figures 4B–C), indicating that the downregulation of cyclins and CDK in cells with GLDC knockdown is not a result of nonspecific repression of MYCN expression.
Fig. 4.
GLDC knockdown reduces the expression of cell cycle genes. a-c GO (a) and GSEA (b-c) analyses of genes downregulated by GLDC knockdown showing significant enrichment of GO terms and gene sets for DNA replication and cell cycle progression. d qRT-PCR analysis of the indicated cell cycle genes in LA1–55n cells without (shGFP) or with GLDC knockown. Error bars represents SEM (n = 2) and data were analyzed by unpaired, two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
Based on the microarray data, we examined the effect of GLDC knockdown on cell cycle progression using the LA1–55n cell line with inducible GLDC knockdown. Culturing the cells in the presence of doxycycline resulted in time-dependent downregulation of GLDC protein expression (Supplementary Figure 4A), which was accompanied by a gradual increase in the fraction of cells in the G1 phase and a concomitant decrease in the S phase population (Fig. 5a–b). This change in cell cycle progression was most pronounced in the LA1–55n cells expressing shGLDC-99, which was most effective in silencing GLDC expression (Supplementary Figure 3A). Thus, both gene expression profiling and cell cycle analysis indicate that GLDC knockdown inhibits cell proliferation by inducing G1 arrest.
Fig. 5.
GLDC knockdown induces G1 phase arrest. a Time-course cell cycle analysis of LA1–55n cells without (teton-shGFP) or with inducible GLDC knockdown (teton-shGLDC-01 and shGLDC-99) in the presence of doxycycline (Dox). b Cell cycle distribution (mean ± SEM) of LA1–55n cells without or with inducible GLDC knockdown using data from (a).
In light of the findings that GLDC silencing downregulated cyclins and CDKs and induced G1 arrest, we further investigated whether a decrease in the expression or activity of cyclin-dependent CDKs is sufficient to induce G1 arrest. We focused our study on the cyclin E1-CDK2 complex given its major role in driving G1-S transition, DNA replication, and S-phase progression [42–44]. Treatment of LA1–55n and SMS-KCNR cells with CVT-313 [45], a specific inhibitor of CDK2, inhibited cell proliferation and arrested cells in the G1 phase (Supplementary Figure 5A–C). We obtained similar results with shRNA-mediated knockdown of cyclin E1 expression (Supplementary Figure 5D–E). These observations are consistent with the notion that downregulation of cyclins and CDKs is a key mechanism by which GLDC silencing induces G1 arrest.
GLDC knockdown alters amino acid and purine metabolism
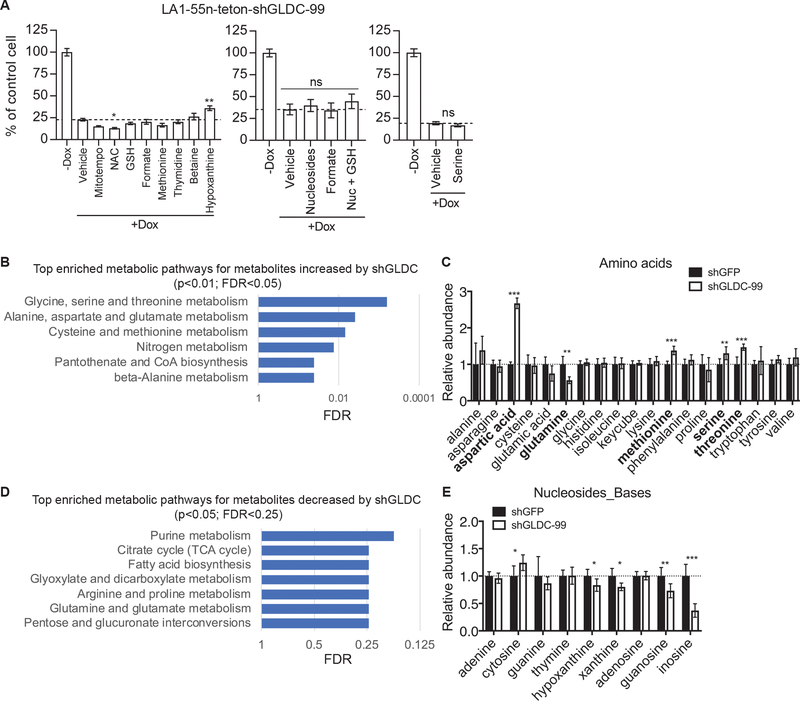
Given that glycine cleavage is part of SGOC metabolism, which generates serine, glycine, and one-carbon units for the production of proteins, nucleotides, and the antioxidant glutathione, we asked whether defects in SGOC metabolism are responsible for the growth-inhibitory effect of GLDC knockdown. We found that supplementation of serine, formate (one-carbon unit), nucleosides (adenosine, cytidine, guanosine, thymidine, and uridine) or antioxidants (N-acetyl-cysteine, MitoTEMPO or reduced glutathione) was unable to rescue the inhibitory effect of GLDC knockdown on cell proliferation, with the exception of hypoxanthine, which showed a small, but significant, rescuing effect (Fig. 6a). Hypoxanthine is a purine base that can be converted to inosine monophosphate (IMP), a precursor for AMP and GMP synthesis, by the enzyme hypoxanthine phosphoribosyltransferase (HPRT) through the salvage pathway of purine nucleotide synthesis [46], suggesting that GLDC knockdown might reduce the production of purine nucleotides.
Fig. 6.
GLDC regulates amino acid and purine metabolism. a Trypan blue exclusion assay of LA1–55n-teton-shGLDC-99 cells cultured in the absence or presence of 0.5 μg/ml doxycycline (Dox) for 9–11 days without (vehicle) or with supplementation of MitoTEMPO (10 μM), NAC (N-acetyl-L-cysteine, 100 μM), GSH (glutathion reduced, 250 μM), formate (2 mM), methionine (0.5 mM), thymidine (30 μM), betaine (10 mM), hypoxanthine (30 μM), nucleosides (30 μM each) or serine (0.5 mM). b Network-based pathway analysis of metabolites increased by GLDC knockdown. c Relative levels of amino acids in LA1–55n_teton-shGFP and -shGLDC-99 cells cultured in the presence of 1 μg/ml doxycycline for 9 days. d Network-based pathway analysis of metabolites decreased by GLDC knockdown. e Relative levels of nucleosides and bases in LA1–55n_teton-shGFP and -shGLDC-99 cells cultured in the presence of 1 μg/ml doxycycline for 9 days. Error bars represent SEM (a, n = 5) or SD (c and e, n = 6) and data were analyzed by unpaired, two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
To test this model and to determine the effect of GLDC knockdown on other metabolic processes, we performed metabolomic profiling of LA1–55n cells with inducible knockdown of GLDC expression using gas chromatography-mass spectrometry. The profiling revealed a total of 81 identified metabolites whose levels were significantly changed (p < 0.05) as a result of GLDC knockdown (Supplementary Table 3). Pathway analysis [47] of the increased metabolites showed significant enrichment (p < 0.01, FDR < 0.05) of metabolic pathways involved in serine, glycine and threonine metabolism, as well as other amino acid metabolism (Fig. 6b and Supplementary Table 4).
Although we observed no significant increase in the intracellular level of glycine, GLDC knockdown resulted in accumulation of serine, threonine, methionine, and aspartic acid (Fig. 6c). In addition to the direct link between serine and glycine, an analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [48] revealed metabolic pathways that connect glycine with threonine, aspartic acid, and methionine (Supplementary Figure 6A). Moreover, the levels of some intermediates of these pathways, including cystathionine and homoserine, were also significantly increased following GLDC knockdown (Supplementary Figure 6B). These findings suggest that interference with glycine breakdown by silencing GLDC expression might disrupt the flux of these metabolic pathways, leading to increased levels of these amino acids.
Pathway analysis of the decreased metabolites revealed that the most significantly enriched (p < 0.05, FDR < 0.25) metabolic pathways are related to purine metabolism, the TCA cycle, and fatty acid synthesis (Fig. 6d). The metabolomic profiling did not identify any nucleotides except for AMP, which showed a modest increase following GLDC knockdown (Supplementary Table 3). On the other hand, GLDC knockdown led to a significant decrease in the levels of the purine nucleosides guanosine and inosine, and of the purine bases hypoxanthine and xanthine (Fig. 6e). A primary source for these purine nucleosides and bases is the breakdown of purine nucleotides, including IMP, xanthosine monophosphate (XMP), and guanosine monophosphate (GMP). In turn, these nucleosides and bases can be used to synthesize IMP, XMP and GMP via nucleotide salvage pathways (Supplementary Figure 6C) [46]. Thus, their decrease could be a result of reduced levels of IMP, XMP, and GMP and/or an increase in their utilization for the synthesis of IMP, XMP, and GMP. Regardless of mechanistic details, the reduced levels of purine nucleosides and bases in LA1–55n cells with GLDC knockdown is consistent with the role of GLDC in producing one-carbon units for the purine backbone. The decrease in hypoxanthine levels also offers an explanation for the observed partial rescuing effect of supplemental hypoxanthine (Fig. 6a).
GLDC knockdown alters central carbon metabolism with reduced lipid production
Surprisingly, GLDC knockdown also had a significant impact on central carbon metabolism (glycolysis and the TCA cycle) and lipid (sterol and fatty acid) synthesis (Fig. 7a). Within the glycolysis pathway, the levels of glucose-6-phosphate (glucose-6-P), 3-phosphoglycerate (3-PG), and lactate were markedly decreased (Fig. 7b), suggesting that GLDC knockdown reduced glycolysis. Within the TCA cycle, GLDC knockdown significantly increased the levels of succinate, fumarate, and malate but decreased the levels of α-ketoglutarate (α-KG), aconitate, and citrate (Fig. 7c). The accumulation of succinate, fumarate, and malate could result from a block in the downstream reaction that catalyzes the condensation of acetyl-CoA with oxaloacetate to form citrate as suggested by the decreased citrate level and/or from an increase in malate production from aspartate via the malate-aspartate shuttle as suggested by the markedly increased aspartate level (Fig. 6c).
Fig. 7.
GLDC regulates central carbon metabolism. a Schematic of central carbon metabolism, glutaminolysis, and lipid synthesis. PPP, pentose phosphate pathway. b-d, Relative levels of metabolic intermediates in glycolysis (b), TCA cycle (c), and sterol-fatty acid synthesis (d). Error bars represent SD (n = 6) and data were analyzed by unpaired, two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
The marked reduction in the levels of α-KG, aconitate, citrate, and glutamine (Fig. 7c) suggests that GLDC knockdown might inhibit the glutamine-dependent reductive carboxylation pathway. Glutaminolysis and reductive carboxylation have a key role in supporting acetyl-CoA production for lipid synthesis in cancer cells [49–52]. Consistent with this model, we found that GLDC knockdown significantly reduced the levels of sterols and fatty acids (Fig. 7d).
Collectively, our metabolomic profiling revealed that GLDC knockdown resulted in alterations in multiple metabolic processes critical for supplying building blocks and energy to meet biosynthetic demands of cell proliferation.
Disruption of purine or cholesterol synthesis inhibits cell proliferation and represses the expression of cyclins and CDKs
The observations that GLDC knockdown disrupted multiple metabolic processes prompted us to investigate whether the metabolic alterations can account for the proliferation inhibitory effect of GLDC knockdown on neuroblastoma cells. We focused our study on purine and cholesterol synthesis, which was significantly reduced in cells with GLDC knockdown. The enzyme phosphoribosyl pyrophosphate amidotransferase (PPAT) catalyzes the first step of de novo purine biosynthetic pathway (Fig. 8a). We tested five lentiviral constructs expressing shRNA sequences against different regions of the PPAT gene and found two constructs (shPPAT-15 and shPPAT-17) that were most effective in silencing PPAT mRNA expression (Fig. 8b). PPAT knockdown by either shRNA construct significantly inhibited the proliferation of the MYCN-amplified neuroblastoma cell lines IMR32, LA1–55n, and SMS-KCNR (Fig. 8c). Moreover, PPAT knockdown significantly repressed the expression of CCNA2, CCNE1, CDK1 and CDK2 (Fig. 8d).
Fig. 8.
Disruption of purine or cholesterol synthesis inhibits cell proliferation and represses the expression of cyclins and CDKs. a Schematic of de novo purine synthesis and slavage pathways. b qRT-PCR analysis of PPAT mRNA expression in LA1–55n cells expressing shGFP or various shPPAT sequences, with HOXC9 mRNA expression as control. c Growth assay of the indicated neuroblastoma cell lines without (shGFP) or with PPAT knockdown (shPPAT) for 6 days. d qRT-PCR analysis of the indicated cell cycle genes in LA1–55n cells without (shGFP) or with PPAT knockdown (shPPAT). e Schematic of the mevalonate-cholesterol synthesis pathway. Statins inhibit the rate-limiting enzyme HMGCR (3-Hydroxy-3-Methylglutaryl-CoA Reductase). f Growth assay of LA1–55n cells treated with DMSO, mevalonate (Meval, 2 mM), simvastatin (Simva, 5 μM) or Meval plus Simva for 2 days. g qRT-PCR analysis of the indicated cell cycle genes in LA1–55n cells treated with DMSO, simvastatin (5 μM) or mevalonate (2 mM) plus simvastatin for 2 days. Error bars represent SEM (b, d and g, n = 3; and c and f, n = 4) and data were analyzed by unpaired, two-tailed Student’s t-test. **p < 0.01, ***p < 0.001.
The mevalonate pathway uses acetyl-CoA to produce cholesterol (Fig. 8e). We recently reported a critical role of the mevalonate-cholesterol synthesis pathway in sustaining neuroblastoma cell proliferation and tumorigenicity [23]. In agreement with our previous study, blocking the mevalonate-cholesterol synthesis pathway by simvastatin, a cholesterol-lowering drug that inhibits the rate-limiting enzyme HMGCR (Fig. 8e), inhibited the proliferation of LA1–55n cells (Fig. 8f). Addition of mevalonate, the metabolite immediately downstream of HMGCR (Fig. 8e), fully rescued the anti-proliferation effect of simvastatin (Fig. 8f). Importantly, simvastatin treatment significantly repressed the expression of CCNA2, CCNB1, CDK1 and CDK2, which could be fully reversed by supplemental mevalonate (Fig. 8g).
Collectively, our findings indicate that disruption of either purine or cholesterol synthesis is sufficient to repress the expression of cyclins and CDKs and to inhibit cell proliferation, recapitulating the growth inhibitory phenotype of GLDC knockdown in MYCN-amplified neuroblastoma cell lines.
Discussion
Our study presented in this report uncovers an important role of GLDC in MYCN-amplified neuroblastoma, a subgroup of high-risk neuroblastoma with extremely poor prognosis. We show that GLDC expression is elevated in neuroblastoma tumors and cell lines with MYCN-amplification. We further present evidence that MYCN directly targets the GLDC gene for transcriptional activation and that GLDC is a common transcriptional target of the MYC family of oncogenes. At the functional level, we show that high GLDC expression is required for the proliferation and tumorigenicity of MYCN-amplified cell lines and that GLDC depletion leads to G1 arrest.
At the mechanistic level, we show that GLDC knockdown induces broad changes in cellular metabolism, which may help to explain the inhibitory effect of GLDC knockdown on cell proliferation. GLDC is a mitochondrial enzyme that catalyzes the first and rate limiting step in glycine breakdown, generating CO2, NH3, NADH, and the one-carbon unit 5,10-MTHF [26, 27]. In mouse models and humans, GLDC mutations lead to accumulation of glycine and decreased supply of one-carbon units, causing glycine encephalopathy and neural tube defect [31–34]. Although in the neuroblastoma cell line (LA1–55n) examined, we did not observe a significant increase in glycine levels following GLDC knockdown, our metabolomic profiling data suggest that GLDC regulates SGOC metabolism in neuroblastoma cells. GLDC knockdown induced serine accumulation, which could be caused by a reduction in the conversion of serine to glycine as a result of decreased glycine breakdown. In the mitochondria, SHMT2 catalyzes the serine to glycine conversion with the production of 5,10-MTHF that, in turn, is converted to 10-formyl-THF for purine nucleotide synthesis. It is well established that SHMT2 and the mitochondrial one-carbon pathway have a major role in the conversion of serine to glycine and one-carbon unit production in the cell [18, 27, 53–58]. Importantly, It has been reported that high SHMT2 expression confers a survival advantage to glioma cells in ischemia but imposes a dependence on glycine clearance by GLDC [29], indicating a key role of GLDC in maintaining the flux of the SGOC metabolic pathway. In addition, GLDC knockdown might also reduce 5,10-MTHF production directly as a result of decreased glycine breakdown. Consistent with the model that GLDC modulates purine metabolism, GLDC knockdown significantly reduced the levels of purine nucleosides and bases in neuroblastoma cells and supplementation of hypoxanthine showed a small rescuing effect.
Perhaps more interestingly, we found that GLDC knockdown in neuroblastoma cells also altered central carbon metabolism with reduced glycolysis (as indicated by the lower level of lactate) and decreased citrate production. Glucose-derived acetyl-CoA is condensed with oxaloacetate to form citrate. In addition, glutamine-derived α-KG can generate citrate via either oxidation in which α-KG-derived oxaloacetate condenses with acetyl-CoA to form citrate or reductive carboxylation in which α-KG is reduced to citrate via NADPH consumption [59–64]. Our metabolomic profiling data are consistent with a decrease in reductive carboxylation of α-KG in neuroblastoma cells with GLDC knockdown as these cells showed increased levels of succinate, fumarate and malate but decreased levels of aconitate. Thus, the decrease in citrate levels in neuroblastoma cells with GLDC knockdown could be caused by a combination of reduced glycolysis and decreased α-KG reductive decarboxylation. Mitochondrial citrate is exported to the cytosol and cleaved by ATP citrate lyase to produce cytosolic acetyl-CoA for sterol and fatty acid synthesis. In agreement with the observed reduction in citrate production, neuroblastoma cells with GLDC knockdown had significantly lower levels of sterols (e.g., lanosterol and cholesterol) and fatty acids (e.g., palmate and myristic acid) in comparison with control cells. The molecular mechanism by which GLDC knockdown alters central carbon metabolism remains to be determined. In this regard, it is interesting to note that a recent study reports that inhibition of PHGDH, the first enzyme of the SGOC pathway (Fig. 1a), disrupts mass balance within central carbon metabolism. Thus, disruption of the carbon flux in the SGOC pathway can simultaneously alter the activity of multiple related metabolic pathways [65]. It is possible that GLDC knockdown acts through a similar mechanism.
GLDC knockdown results in downregulation of cyclins and CDKs and induces G1 arrest. We show that cyclin E1 knockdown or CDK2 inhibition is sufficient to inhibit cell proliferation and to induce G1 arrest in neuroblastoma cell lines. Moreover, we present evidence that blocking purine or cholesterol synthesis is sufficient to downregulate cyclins and CDKs and to inhibit cell proliferation. Taken together, these findings suggest that the alteration in cellular metabolism is likely a major mechanism for the growth arrest phenotype induced by GLDC knockdown in MYCN-amplified neuroblastoma cell lines.
Our findings also have therapeutic implications. MYCN has a major role in high-risk neuroblastoma development [5, 9, 66, 67]. However, targeting MYCN directly for therapy has proved to be challenging [68]. An alternative strategy is to identify MYCN-dependent metabolic vulnerabilities as therapeutic targets. We found that the GLDC gene is a direct transcriptional target of MYCN. As a result, neuroblastoma tumors and cell lines with MYCN amplification show a marked increase in GLDC expression, whereas non-MYCN-amplified neuroblastoma tumors and cell lines display low or undetectable levels of GLDC expression. Importantly, MYCN-amplified neuroblastoma cell lines require high levels of GLDC expression for their proliferation and tumorigenicity. This GLDC dependence may be a result of increased expression of SGOC pathway genes in MYCN-amplified neuroblastoma cells, which is expected to generate higher levels of glycine. Coordinated upregulation of GLDC expression could prevent alternative glycine catabolism that produces toxic metabolites [29]. Thus, we suggest that GLDC is a treatment target for high-risk neuroblastoma with MYCN amplification.
Materials and methods
Cell lines and culture
Neuroblastoma cell lines BE(2)-C (CRL-2268), IMR32 (CCL-127), SH-SY5Y (CRL2266), SK-N-AS (CRL-2137), SK-N-DZ (CRL-2149), and SK-N-FI (CRL-2141) were obtained from ATCC (Manassas, VA), LA1–55n (06041203) from Sigma-Aldrich (St. Louis, MO), and CHLA-90, LA-N-5, LA-N-6, SMS-KANR, and SMS-KCNR from Children’s Oncology Group Cell Culture and Xenograft Repository at Texas Tech University Health Sciences Center. IMR5 was a gift from J.K. Cowell (Augusta University), NLF from M.C. Simon (University of Pennsylvania), SHEP1 from V.P. Opipari (University of Michigan), and P493–6 from J. van Riggelen (Augusta University). All cell lines, except for IMR5 and P493–6, had been authenticated using short tandem repeat profiling (ATCC) and after authentication, large frozen stocks were made to ensure against contaminations by other cell lines. IMR5 was verified as an MYCN-amplified neuroblastoma cell line by high-level nuclear expression of MYCN and the specific neuroblastoma marker PHOX2B [69, 70], and P493–6 by inducible expression of MYC in the absence of doxycycline [40, 41]. All cell lines were used within 10 passages after reviving from the frozen stocks and were free of Mycoplasma contamination as determined by a LookOut Mycoplasma PCR kit (Sigma-Aldrich) and staining cells with DAPI every 3 months. IMR32, SHEP1, SH-SY5Y, and SK-N-AS were cultured in DMEM (HyClone SH30022, Thermo Fisher Scientific, Hampton, NH), BE(2)-C in DME/F-12 1:1 (HyClone SH30023), and all others in RPMI 1640 (HyClone SH30027). All culture media were supplemented with 10% FBS (Atlanta Biologicals S11050, Flowery Branch, GA) and 2 mM GlutaMAX (Gibco 35050–061, Thermo Fisher Scientific). CDK2 inhibitor III (CVT-313) was obtained from Sigma-Aldrich (238803), dissolved in DMSO, and stored at −800C until use. For cell rescue experiments, MitoTEMPO (Sigma-Aldrich SML0737), N-Acetyl-L-cysteine (Sigma-Aldrich A9165), L-glutathione reduced (Sigma-Aldrich G6013), sodium formate (Sigma-Aldrich 71539), thymidine (Sigma-Aldrich T1895), Hypoxanthine (Sigma Aldrich H9636), serine (Sigma-Aldrich S4311), and EmbryoMax Nucleosides (100X, Millipore Sigma ES-008-D, Sigma-Aldrich) were used at the indicated concentrations. Cell viability and proliferation were determined by trypan blue exclusion assay. Cell images were acquired using an EVOS FL imaging system (Thermo Fisher Scientific).
Patient data analysis
Patient data used in this study were described previously [36, 37, 71]. All analyses of the neuroblastoma patient data were conducted using R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl), and the resulting figures and p values were downloaded.
Overexpression and RNA interference
The Retro-X Tet-Off Advanced Inducible System (Clontech 632105, Takara Bio, Kusatsu, Shiga Prefecture, Japan) and the lentiviral vector pCDH-CMV-MCS-EF1-puro (CD510B-1, SBI System Biosciences, Palo Alto, CA) were used to generate neuroblastoma cell lines with inducible (in the absence of doxycycline) and constitutive MYCN expression, respectively. MYCN coding sequence was generated by PCR using pBabe-hygro/N-Myc [72] as a template. The sequence was verified by sequencing and subcloned into the expression vectors. Lentiviral pLKO.1 shRNA constructs shGLDC-00 (TRCN0000036600), shGLDC-01 (TRCN0000036601), shGLDC-02 (TRCN0000036602), shGLDC-71 (TRCN0000303371), shGLDC-99 (TRCN0000036599), shMYCN-94 (TRCN0000020694), shMYCN-95 (TRCN0000020695), shPPAT-13 (TRCN0000075213), shPPAT-14 (TRCN0000075214), shPPAT-15 (TRCN0000075215), shPPAT-16 (TRCN0000075216), and shPPAT-17 (TRCN0000075217) were purchased from Sigma-Aldrich, and shCCNE1–80 (RHS3979–200799580/TRCN0000045301) and shCCNE1–81 (RHS3979–200799581/ TRCN0000045302) were purchased from Open Biosystems (Dharmacon, Lafayette, CO). For inducible expression of shRNA against GLDC, the shGLDC-01 and shGLDC-99 sequences were cloned into the lentiviral pLKO-teton plasmid [73]. shRNA against GFP was used as control. Retroviruses were produced in 293FT cells using the packaging plasmids pHDM-G and pMD.MLVogp (gifts from R. Mulligan at MIT), and lentiviruses were produced in 293FT cells using the packaging plasmids pLP1, pLP2, and pLP/VSVG (Thermo Fisher Scientific K497500). Retroviral and lentiviral infections of cells were conducted according to standard procedures.
Quantitative reverse-transcription PCR (qRT-PCR)
Cells were lysed using TRIzol (Thermo Fisher Scientific 15596026) for total RNA isolation. Reverse transcription of RNA to cDNA was conducted using iScript Advanced cDNA Synthesis Kit (Bio-Rad 172–5038, Hercules, CA). qRT-PCR was done using a 2X SYBR green qPCR master mix (Bimake B21202, Houston, TX) on Real-Time PCR system CFX96 (Bio-Rad). Gene specific primers are described in Supplementary Table 5. qRT-PCR reactions were conducted in either duplicate or triplicate, presented as Mean ± SEM and normalized to β2 microglobulin (B2M) or GAPDH mRNA levels.
Immunoblotting
Cells were lysed using SDS sample buffer and protein concentrations were measured using Bio-Rad Protein Assay Dye (Bio-Rad 5000006). Proteins (~50 μg per sample) were separated on SDS-polyacrylamide gels, transferred to nitrocellulose membranes (926–31092, LI-COR Biosciences, Lincoln, NE), and probed with rabbit anti-cyclin E1 (ab33911, Abcam, Cambridge, MA), rabbit anti-GLDC (HPA002318, Sigma-Aldrich), mouse anti-MYCN (clone OP13, Millipore Sigma), rabbit anti-MYC (sc-764, Santa Cruz Biotech, Dallas, TX), rabbit anti-GAPDH (sc-25778, Santa Cruz Biotech), and mouse anti-α-tubulin (T5168, Sigma-Aldrich) antibodies. Horseradish Peroxidase conjugated goat anti-mouse (sc-2005, Santa Cruz Biotech) and goat anti-rabbit IgG (sc2004, Santa Cruz Biotech) were used as secondary antibodies to visualize proteins by chemiluminescence. For visualization with the Odyssey system (LI-COR Biosciences), Goat anti-mouse IRDye 800 (926–32210) or 680 (926–32220) and anti-rabbit IRDye 800 (926–32234) or 680 (926–68021) from LI-COR Biosciences were used as secondary antibodies.
ChIP-qPCR
Approximately 5×107 LA1–55n and SMS-KCNR neuroblastoma cells were collected for each ChIP assay according to the Young Lab protocol (http://younglab.wi.mit.edu/hESRegulation/ChIP.html). Briefly, following cross-linking, cells were lysed, chromatins sonicated, and DNA-protein complexes immunoprecipitated using Dynabeads protein G (Thermo Fisher Scientific 10004D). Mouse anti-MYCN (B8.4.B, Santa Cruz Biotech sc-53993) was used to capture MYCN associated genomic DNA fragments with mouse IgG (Santa Cruz Biotech sc2025) as control, followed by qPCR analysis using primers that cover the GLDC promoter 5’ region (−732 to −639; TSS +1) and the first intron (+578 to +654): −700 forward, TTTCCGAAAGTGTTGCGATTAC; −700 reverse, CTCCTCCTTCTTCTCACTCTCT; 1st intron forward, GTGGCGTGGAAGCAAATTC; 1st intron reverse, CTGGGTGCAGGAGAAAGTAAG.
Microarray
Total RNA was isolated using TRIzol from three independent samples of LA1–55n_teton-shGFP and -shGLDC-99 cells cultured in the presence of 0.5 μg/ml doxycycline for 7 days. RNA quality was analyzed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Affymetrix Human Gene 2.0 ST Array chips were used for microarray analysis (Affymetrix, Santa Clara, CA). Data were normalized, significance determined by ANOVA, and differential gene expression were calculated using the Partek Genomics Suite. Differentially expressed genes (≥ ±1.4-fold, p < 0.05) were subjected to GO analysis using DAVID [74]. GSEA [75] was conducted using GSEA desktop application software with annotated gene sets of Molecular Signature Database v6.2.
Cell cycle
LA1–55n_teton-shGFP and -shGLDC-99 cells were cultured in the presence of 0.5 μg/ml doxycycline for various times and LA1–55n cells were treated with DMSO or 5 μM CDK2 inhibitor III for 24 h. The cells were then collected, washed with PBS, and fixed in 70% cold ethanol for at least 30 minutes. After fixation, cells were treated with 100 μg/ml ribonuclease A (Qiagen 19101, Hilden, Germany) for 30 minutes at 370C and stained with 5 μg/ml propidium iodide (P3566, Thermo Fisher Scientific) for 10 minutes. Data were acquired using a FACSCanto system (BD Biosciences, San Jose, CA) and analyzed with FlowJo v10 software (FlowJo, Ashland, OR).
Xenograft assay
NOD.SCID mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Male and female mice of 6-week-old were randomly assigned to each group (n = 4 per group). SMS-KCNR cells expressing either shGFP or shGLDC-99 were suspended in 100 μl Hanks’ Balanced Salt Solution and injected subcutaneously into both flanks of the mice at 5 × 106 cells per injection site. Mice were examined every other day and tumor growth was monitored by measurement with a caliper. Tumor volume was calculated with the equation V = 1/2 × (length × width2). Mice were euthanized when their tumors reach ~2.0 cm in diameter or shows sign of necrosis. The experiment was terminated 9 weeks after injection. The animal study was approved by the Institutional Animal Care and Use Committee of Medical College of Georgia, Augusta University.
Metabolomics
LA1–55n_teton-shGFP and -shGLDC-99 cells were cultured in the presence of 1 μg/ml doxycycline for 9 days and collected by scraping and centrifugation. Cell pellets were washed once with ice-cold PBS, snap frozen in liquid nitrogen, and stored at −80oC until analysis. Six biological replicate samples (~5 × 106 cells/sample) were analyzed for each group. Metabolite extraction and metabolomic analysis were carried out in the NIH West Coast Metabolomics Center at the University of California Davis. Data were acquired using Agilent 6890 Gas Chromatography-Leco Pegasus IV Time of Flight Mass Spectrometry and processed using ChromaTOF software version 2.32 (Leco, St. Joseph, MI) and the BinBase algorithm as described previously [76].
Statistical analysis
Quantitative data from qRT-PCR and cell proliferation assays were presented as mean ± SEM and metabolomics as mean ± SD. The data were analyzed for statistical significance using unpaired, two-tailed Student’s t-test or two-way ANOVA with GraphPad Prism 7 software for Windows.
Supplementary Material
Acknowledgements
We thank John K. Cowell (Augusta University), Susan L. Cohn (University of Chicago), M. Celeste Simon (University of Pennsylvania), and C. Patrick Reynolds (Children’s Oncology Group, Texas Tech University) for providing cell lines, and Rogier Versteeg and the Department of Oncogenomics at the Academic Medical Center (Amsterdam, The Netherlands) for providing the R2 Genomics Analysis and Visualization Platform.
Funding
This work was supported by an NIH grant (R01 CA190429) to HFD.
Footnotes
Accession number
The NCBI Gene Expression Omnibus (GEO) accession number for the microarray data is GSE129807.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information is available at Oncogene’s website.
References:
- 1.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer 2013; 13: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall GM, Carter DR, Cheung BB, Liu T, Mateos MK, Meyerowitz JG et al. The prenatal origins of cancer. Nat Rev Cancer 2014; 14: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet 2007; 369: 2106–2120. [DOI] [PubMed] [Google Scholar]
- 4.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3: 203–216. [DOI] [PubMed] [Google Scholar]
- 5.Cohn SL, Pearson ADJ, London WB, Monclair T, Ambros PF, Brodeur GM et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J Clin Oncol 2009; 27: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK et al. Children’s Oncology Group’s 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer 2013; 60: 985–993. [DOI] [PubMed] [Google Scholar]
- 7.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J Clin Oncol 2015; 33: 3008–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwab M Amplification of N-myc as a prognostic marker for patients with neuroblastoma. Semin Cancer Biol 1993; 4: 13–18. [PubMed] [Google Scholar]
- 9.Campbell K, Gastier-Foster JM, Mann M, Naranjo AH, Van Ryn C, Bagatell R et al. Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: A report from the Children’s Oncology Group. Cancer 2017; 123: 4224–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984; 224: 1121–1124. [DOI] [PubMed] [Google Scholar]
- 11.Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med 1985; 313: 1111–1116. [DOI] [PubMed] [Google Scholar]
- 12.Dang CV. MYC on the path to cancer. Cell 2012; 149: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harbor perspectives in medicine 2013; 3: a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine 2013; 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J 2017; 36: 3409–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genet 2011; 43: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011; 476: 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab 2017; 25: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer 2016; 16: 650–662. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nature communications 2014; 5: 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J, Li T, Wang X, Zhao E, Choi JH, Yang L et al. The Histone H3 Methyltransferase G9A Epigenetically Activates the Serine-Glycine Synthesis Pathway to Sustain Cancer Cell Survival and Proliferation. Cell Metab 2013; 18: 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi JH et al. KDM4C and ATF4 Cooperate in Transcriptional Control of Amino Acid Metabolism. Cell Rep 2016; 14: 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu M, Xia Y, Ding J, Ye B, Zhao E, Choi JH et al. Transcriptional Profiling Reveals a Common Metabolic Program in High-Risk Human Neuroblastoma and Mouse Neuroblastoma Sphere-Forming Cells. Cell Rep 2016; 17: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia Y, Ye B, Ding J, Yu Y, Alptekin A, Thangaraju M et al. Metabolic Reprogramming by MYCN Confers Dependence on the Serine-Glycine-One-Carbon Biosynthetic Pathway. Cancer Res 2019; 79: 3837–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 2013; 13: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Wu Z, Dai Z, Yang Y, Wang J, Wu G. Glycine metabolism in animals and humans: implications for nutrition and health. Amino Acids 2013; 45: 463–477. [DOI] [PubMed] [Google Scholar]
- 27.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annual review of nutrition 2010; 30: 57–81. [DOI] [PubMed] [Google Scholar]
- 28.Lamers Y, Williamson J, Theriaque DW, Shuster JJ, Gilbert LR, Keeling C et al. Production of 1-carbon units from glycine is extensive in healthy men and women. J Nutr 2009; 139: 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature 2015; 520: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep 2014; 7: 1248–1258. [DOI] [PubMed] [Google Scholar]
- 31.Narisawa A, Komatsuzaki S, Kikuchi A, Niihori T, Aoki Y, Fujiwara K et al. Mutations in genes encoding the glycine cleavage system predispose to neural tube defects in mice and humans. Human molecular genetics 2012; 21: 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conter C, Rolland MO, Cheillan D, Bonnet V, Maire I, Froissart R. Genetic heterogeneity of the GLDC gene in 28 unrelated patients with glycine encephalopathy. Journal of inherited metabolic disease 2006; 29: 135–142. [DOI] [PubMed] [Google Scholar]
- 33.Pai YJ, Leung KY, Savery D, Hutchin T, Prunty H, Heales S et al. Glycine decarboxylase deficiency causes neural tube defects and features of non-ketotic hyperglycinemia in mice. Nature communications 2015; 6: 6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung KY, Pai YJ, Chen Q, Santos C, Calvani E, Sudiwala S et al. Partitioning of One-Carbon Units in Folate and Methionine Metabolism Is Essential for Neural Tube Closure. Cell Rep 2017; 21: 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012; 148: 259–272. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol 2015; 16: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis 2013; 4: e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012; 483: 589. [DOI] [PubMed] [Google Scholar]
- 39.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov 2013; 3: 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer 2000; 87: 787–793. [DOI] [PubMed] [Google Scholar]
- 41.Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol 1999; 9: 1255–1258. [DOI] [PubMed] [Google Scholar]
- 42.Tadesse S, Caldon EC, Tilley W, Wang S. Cyclin-Dependent Kinase 2 Inhibitors in Cancer Therapy: An Update. J Med Chem 2019; 62: 4233–4251. [DOI] [PubMed] [Google Scholar]
- 43.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9: 153–166. [DOI] [PubMed] [Google Scholar]
- 44.Mao L, Ding J, Perdue A, Yang L, Zha Y, Ren M et al. Cyclin E1 is a common target of BMI1 and MYCN and a prognostic marker for neuroblastoma progression. Oncogene 2012; 31: 3785–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks EE, Gray NS, Joly A, Kerwar SS, Lum R, Mackman RL et al. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J Biol Chem 1997; 272: 29207–29211. [DOI] [PubMed] [Google Scholar]
- 46.Manfredi JP, Holmes EW. Purine salvage pathways in myocardium. Annu Rev Physiol 1985; 47: 691–705. [DOI] [PubMed] [Google Scholar]
- 47.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 2011; 6: 743–760. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008; 7: 11–20. [DOI] [PubMed] [Google Scholar]
- 50.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016; 16: 619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J 2017; 36: 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012; 481: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stover PJ, Chen LH, Suh JR, Stover DM, Keyomarsi K, Shane B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J Biol Chem 1997; 272: 1842–1848. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida T, Kikuchi G. Major pathways of glycine and serine catabolism in rat liver. Arch Biochem Biophys 1970; 139: 380–392. [DOI] [PubMed] [Google Scholar]
- 55.Pfendner W, Pizer LI. The metabolism of serine and glycine in mutant lines of Chinese hamster ovary cells. Arch Biochem Biophys 1980; 200: 503–512. [DOI] [PubMed] [Google Scholar]
- 56.Narkewicz MR, Sauls SD, Tjoa SS, Teng C, Fennessey PV. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem J 1996; 313: 991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem 2002; 277: 38381–38389. [DOI] [PubMed] [Google Scholar]
- 58.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab 2016; 23: 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep 1985; 5: 393–400. [DOI] [PubMed] [Google Scholar]
- 60.Holleran AL, Briscoe DA, Fiskum G, Kelleher JK. Glutamine metabolism in AS-30D hepatoma cells. Evidence for its conversion into lipids via reductive carboxylation. Mol Cell Biochem 1995; 152: 95–101. [DOI] [PubMed] [Google Scholar]
- 61.D’Adamo AF Jr, Haft DE. An Alternate Pathway of Alpha-Ketoglutarate Catabolism in the Isolated, Perfused Rat Liver. I. Studies with Dl-Glutamate-2- and −5–14c. J Biol Chem 1965; 240: 613–617. [PubMed] [Google Scholar]
- 62.Des Rosiers C, Di Donato L, Comte B, Laplante A, Marcoux C, David F et al. Isotopomer analysis of citric acid cycle and gluconeogenesis in rat liver. Reversibility of isocitrate dehydrogenase and involvement of ATP-citrate lyase in gluconeogenesis. J Biol Chem 1995; 270: 10027–10036. [DOI] [PubMed] [Google Scholar]
- 63.Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 2008; 283: 20621–20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Comte B, Vincent G, Bouchard B, Benderdour M, Des Rosiers C. Reverse flux through cardiac NADP(+)-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol 2002; 283: H1505–1514. [DOI] [PubMed] [Google Scholar]
- 65.Reid MA, Allen AE, Liu S, Liberti MV, Liu P, Liu X et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nature communications 2018; 9: 5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teitz T, Stanke JJ, Federico S, Bradley CL, Brennan R, Zhang J et al. Preclinical models for neuroblastoma: establishing a baseline for treatment. PLoS One 2011; 6: e19133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J 1997; 16: 2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 2017; 17: 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alam G, Cui H, Shi H, Yang L, Ding J, Mao L et al. MYCN promotes the expansion of Phox2B-positive neuronal progenitors to drive neuroblastoma development. Am J Pathol 2009; 175: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017; 549: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci U S A 2012; 109: 19190–19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cui H, Li T, Ding HF. Linking of N-Myc to death receptor machinery in neuroblastoma cells. J Biol Chem 2005; 280: 9474–9481. [DOI] [PubMed] [Google Scholar]
- 73.Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell cycle (Georgetown, Tex 2009; 8: 498–504. [DOI] [PubMed] [Google Scholar]
- 74.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols 2008; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 75.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y et al. Quality control for plant metabolomics: reporting MSI-compliant studies. The Plant journal : for cell and molecular biology 2008; 53: 691–704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.