Abstract
Picornaviridae family comprises single-stranded, positive-sense RNA viruses distributed into forty-seven genera. Picornaviruses have a broad host range and geographic distribution in all continents. In this study, we applied a high-throughput sequencing approach to examine the presence of picornaviruses in penguins from King George Island, Antarctica. We discovered and characterized a novel picornavirus from cloacal swab samples of gentoo penguins (Pygoscelis papua), which we tentatively named Pingu virus. Also, using RT-PCR we detected this virus in 12.9 per cent of cloacal swabs derived from P. papua, but not in samples from adélie penguins (Pygoscelis adeliae) or chinstrap penguins (Pygoscelis antarcticus). Attempts to isolate the virus in a chicken cell line and in embryonated chicken eggs were unsuccessful. Our results expand the viral diversity, host range, and geographical distribution of the Picornaviridae.
Keywords: picornavirus, penguins, Antarctica, virus discovery, metagenomics
1. Introduction
Picornaviruses are single-stranded, positive-sense RNA viruses with a genome of 6.7–10.1 kb in length (Zell et al. 2017). The Picornaviridae family contains ∼110 species classified into forty-seven genera, and forty-nine unassigned picornaviruses (Zell et al. 2017; Zell 2018). The majority of picornavirus infections are asymptomatic, although some of these viruses may cause important human and animal diseases, which can involve the central nervous system (e.g. poliovirus), heart (e.g. encephalomyocarditis virus), liver (e.g. hepatitis A virus), skin (e.g. coxsackie A virus), gastrointestinal tract (e.g. human parechovirus), or upper respiratory tract (e.g. human rhinovirus 89) (Zell 2018). Picornaviruses are mainly transmitted via the fecal–oral and respiratory routes (Zell et al. 2017).
Picornaviruses have been identified in a wide range of hosts, including all classes of vertebrates, such as mammals, birds, fish, amphibians, and reptiles (Zell et al. 2017; Shi et al. 2018; Zell 2018). In the last decade, our knowledge about the host range, diversity, and geographic distribution of picornaviruses has dramatically increased (Shi et al. 2018; Zell 2018). Avian picornaviruses can infect wild and domestic birds, including Anseriformes (i.e. ducks, goose), Galliformes (i.e. chicken, domestic turkey, pheasant, common quail), Columbiformes (e.g. feral pigeon), and Passeriformes (oriental magpie robin, Japanese thrush, grey-backed thrush, blackbird, and pale thrush), causing important diseases such as hemorrhagic hepatitis, runting–stunting enteritis, avian encephalomyelitis, catarrhal enteritis, and malabsorption syndrome (Boros, Pankovics, and Reuter 2014). Recent studies have increased the diversity of picornaviruses in apparently healthy birds, including waterfowl and shorebird populations (Shi et al. 2018; Wille et al. 2019b).
The Antarctic and sub-Antarctic islands provide a unique environment for the existence of seabirds and marine mammals, particularly during their breeding seasons (Smeele, Ainley, and Varsani 2018). In the past few years, our understanding of microbial diversity in the Antarctic continent has increased (Cavicchioli 2015; Lopez-Bueno et al. 2015; Smeele, Ainley, and Varsani 2018). However, information on viruses associated with animals within this region is still very limited. Previous studies based on serology and molecular assays have shown the presence of viruses of the Birnaviridae, Flaviviridae, Herpesviridae, Orthomyxoviridae, and Paramyxoviridae families associated with various animal species including penguins, skua, leopard seal, crabeater seal, Ross seal, Weddell seal, and seabirds (Smeele, Ainley, and Varsani 2018). Antarctic penguins appear to play a key role in the epidemiology of some pathogenic viruses (e.g. influenza viruses), which can potentially infect other avian hosts (Barriga et al. 2016; Neira et al. 2017; Wille et al. 2019a). Our aim was to identify and characterize novel viruses from penguins in King George Island, Antarctica. To this end, we applied a high-throughput sequencing (HTS) approach to nucleic acids derived from blood and cloacal samples collected from this animal species.
2. Materials and methods
2.1 Penguin samples
Blood and cloacal swab samples were collected from ninety-three healthy penguins from a beach near to three penguin colonies in Keller Peninsula, King George Island (61.9882°S 58.0196°W) between 20 November and 1 December 2006. King George Island is located in the Southern Ocean, 120 km off the coast of Antarctica (Fig. 1) and is the largest of the Antarctic islands in South Shetland. Penguins were classified into three different species: gentoo (Pygoscelis papua), adélie (Pygoscelis adeliae), and chinstrap (Pygoscelis antarcticus). Each penguin was sampled by cloacal insertion of a sterile swab, and samples were kept in 500 μl of viral transport medium (glycerol 5% in physiological saline with penicillin 2000 U/ml and fungizone 2000 U/ml, 6 μg of gentamicin, 2.5 μl of streptomycin, 1% bovine serum albumin, 20% glycerol). Blood was collected from the medial metatarsal vein. The volume of blood obtained was up to 1 per cent relative to the weight of the bird (e.g. 1 ml for a bird to 100 g). Blood samples were centrifuged at 800×g for 7 min and serum was separated and stored in microtubes. Both sera and cloacal samples were stored in liquid nitrogen (−200 °C) until arrival at the laboratories at University of São Paulo in Brazil, where samples were stored at −80 °C.
Figure 1.
Map of the collection site in the King George Island, Antarctica. Red dot represents the place of collection.
Sample collection and handling procedures were approved by the Ethics Committee on Animal Experiments of the Institute of Biomedical Science at University of São Paulo (No. 122/04), license 201/2006 of CGFAU/IBAMA. This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use by The National Council Scientific and Technological Development (CNPq) and the Brazilian Antarctic Program (PROANTAR). Field activities are part of the Projects CNPQ/PROANTAR 550040/2007-2 and 558837/2005.
2.2 Viral extraction, sequencing, and assembly
Samples were pooled based on penguin species and sample type (Table 1), and prepared as previously described in de Souza et al. (2018). Subsequently, viral RNA of pooled samples was extracted using the QIAamp viral RNA mini kit (Qiagen, Hilden, USA). Nucleic acids were analyzed using a Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA) to evaluate the quantity and quality of the extracted RNA, respectively. cDNA synthesis was performed using SuperScript II and random hexamer primers (Invitrogen, Carlsbad, USA) according to the manufacturer’s recommendations. Sequencing was performed using the TruSeq RNA sample preparation kit in an Illumina HiSeq 2500 instrument (Illumina, San Diego, USA) with a paired-end and 150-base-read protocol using a RAPID module. All reads generated in this project are available at the NCBI Short Read Archive (SRA) under accessions SAMN12324479–SAMN12324487 (BioProject ID: PRJNA555609). Sequencing reads were assembled de novo using the metaViC pipeline (https://github.com/sejmodha/MetaViC). Briefly, adapters and short reads (<80 nucleotides) were removed using Trim Galore, and ribosomal RNA sequences were removed using RiboPicker (Schmieder, Lim, and Edwards 2012). After this step, each read was run against the RefSeq protein database using DIAMOND (Buchfink, Xie, and Huson 2015). We kept sequences that matched viruses, environmental sequences and sequences that did not match anything in the database. Furthermore, sequence reads were de novo assembled using SPAdes and IDBA-UD with the k-mer values of 31, 55, 77, and 99 for SPAdes and a range of the k-mer values between 31 and 99 for IDBA-UD (Bankevich et al. 2012; Peng et al. 2012). Assembled contigs obtained with both tools were merged using GARM (Soto-Jimenez, Estrada, and Sanchez-Flores 2014). Contigs longer than 200 nucleotides and supercontigs generated by GARM were merged and classified using DIAMOND as described above. Krona was used to visualize the BLAST tabular outputs. We also performed sequence alignments to reference viral genomes using Bowtie2 (Langdon 2015) as previously described in Souza et al. (2018).
Table 1.
Information of sample pools used in this study.
| Pool | Species | Sample type | N 1 | Reads | Q30 | SRA |
|---|---|---|---|---|---|---|
| 05 | P. papua | Serum | 11 | 35,828,584 | 69.51 | SAMN12324479 |
| 06 | P. papua | Cloacal swab | 12 | 53,129,208 | 78.83 | SAMN12324480 |
| 07 | P. adeliae | Serum | 17 | 31,249,794 | 67.57 | SAMN12324481 |
| 08 | P. adeliae | Cloacal swab | 13 | 10,716,114 | 90.64 | SAMN12324482 |
| 09 | P. adeliae | Serum | 12 | 8,392,766 | 81.80 | SAMN12324483 |
| 10 | P. adeliae | Cloacal swab | 11 | 10,690,302 | 84.40 | SAMN12324484 |
| 11 | P. papua | Cloacal swab | 19 | 16,660,310 | 88.70 | SAMN12324485 |
| 12 | P. adeliae | Cloacal swab | 24 | 14,634,546 | 88.55 | SAMN12324486 |
| 13 | P. antarcticus | Cloacal swab | 8 | 8,518,380 | 79.08 | SAMN12324487 |
N 1, number of individual samples per pool.
2.3 Viral genome characterization
To characterize the genome of Pingu picornavirus (PingPcV), we first estimated the genome size and then performed ORF prediction using Geneious 9.1.2 (Biomatters, Auckland, New Zealand). Putative viral proteins were confirmed using BLASTX. Protein domains were screened using InterProScan (Finn et al. 2017) and manually compared with those of related picornaviruses. The nucleotide sequence of PingPcV was deposited in GenBank (accession number MH255796).
2.4 Phylogenetic and p-distance analysis
A maximum likelihood (ML) phylogenetic tree was inferred using a protein alignment of 435 amino acids of the 3Dpol protein (from nucleotide position 6,112 to 7,463 of the PingPcV genome). The alignment included fifty-seven sequences of representative members of the Picornaviridae family (Zell et al. 2017), was generated using PROMALS3D and edited using Geneious (Pei, Tang, and Grishin 2008). The ML tree was inferred using IQ-TREE (version 1.6.0) using the LG+I+G4 substitution model with 1,000 ultrafast bootstraps (Nguyen et al. 2015). The best-fit model was determined based on 144 reversible amino acids substitution models using the Bayesian Information Criterion in ModelFinder (Kalyaanamoorthy et al. 2017). Statistical support for individual nodes of the phylogenetic tree was estimated using 1,000 bootstraps. The phylogenetic tree was visualized using FigTree v.1.4.2.
To determine the sequence variability of the 2Chel, 3Cpro, and 3Dpol proteins of PingPcV and other picornaviruses, we calculated the amino acid p-distance values. All ambiguous positions were removed for each sequence pair. Standard error estimations were calculated by bootstrapping (n = 1,000) using MEGA v.7 (Kumar, Stecher, and Tamura 2016) and results were converted to percentages.
2.5 RT-PCR for novel picornavirus
We used RT-PCR to determine the prevalence of PingPcV. Primers were designed to amplify a fragment of ∼700 nt of the 3Dpol gene (forward primer: 5′-GCCGGTCGGATGCTCTTTGGTGG-3′—position 6,590–6,612; reverse primer: 5′-CTCCACCGCGAGACCACATGATC-3′—position 7,267–7,289 nt). cDNA was generated using Superscript III (Invitrogen, Carlsbad, USA) with random hexamers (Invitrogen, Carlsbad, USA) following the manufacturer’s instructions. PCR was performed using Platinum Taq DNA polymerase high fidelity (Thermo Fisher Scientific, Waltham, USA) following the manufacturer's instructions. Cycling conditions were as follows: 94 °C for 30 s, followed by 35 cycles at 94 °C for 15 s, 66 °C for 30 s and 68 °C for 1 min (Supplementary Information S1). Amplicons were visualized by gel electrophoresis in 2 per cent agarose gels. All PCR products were sequenced using the Sanger method in an ABI 3730 genetic analyzer (Applied Biosystems, Foster City, USA). PingPcV partial sequences are available in Supplementary Information S2.
2.6 Virus isolation
Virus-positive samples were filtered through a 0.22-μm filter, and 250 μl was inoculated onto UMNSAH/DF cell monolayers in T25 flasks. Flasks were gently rocked for 1 h at 37 °C before adding 7 ml of D-MEM containing 2 per cent fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml streptomycin. Inoculated cultures were kept at 37 °C with 5 per cent CO2 for 6 days. Viruses were blind passaged three times, and for each passage, RNA was extracted from both cells and supernatant. In addition, 9-day-old specific-pathogen-free embryonated chicken eggs were inoculated with 100 μl of material obtained from PCR-positive samples. Inoculated eggs were incubated for 5 days at 37 °C and 50 per cent humidity and observed daily for viability. Embryos that died during the first 24 h were discarded. Embryos that died after 24 h post-inoculation and those that survived the 5-day incubation period were subject to necropsy as previously described for other avian picornaviruses (Tannock and Shafren 1994; MacLachlan and Dubovi 2016). In addition, RNA was extracted from the allantoic and amniotic fluid and subject to RT-PCR as described above. This process was repeated in four independent experiments.
3. Results and discussion
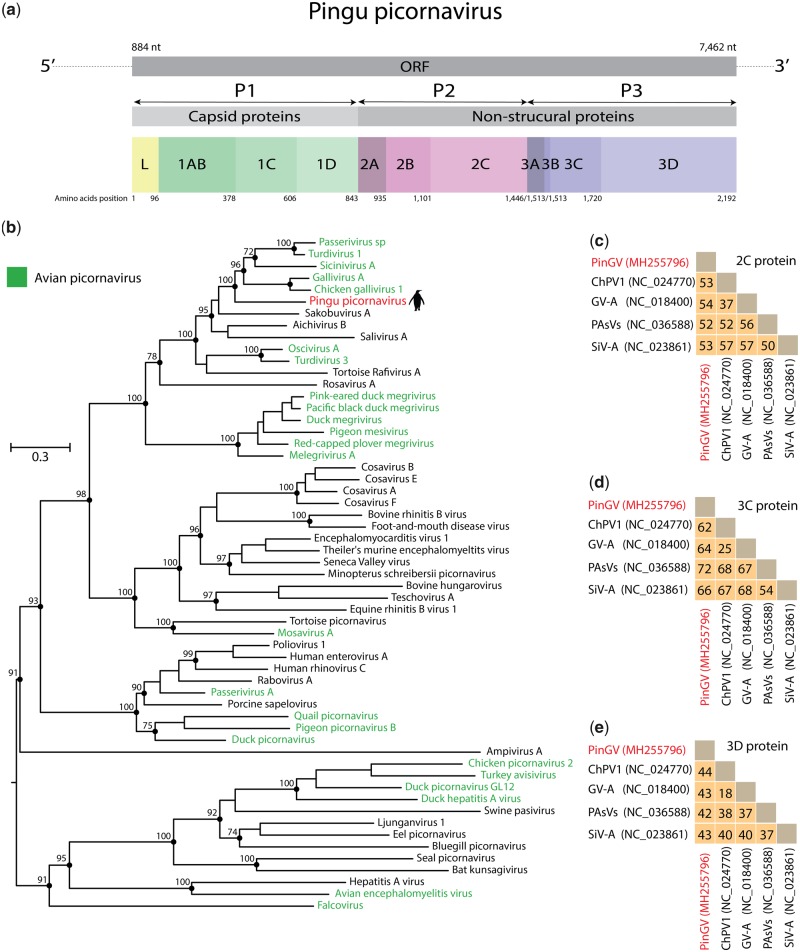
Analysis of three blood pools and six cloacal pools generated a total of 8,518,380–53,129,208 paired-end reads per pool with 67.57–90.64 per cent of bases ≥Q30 and a base call accuracy of 99.9 per cent (Table 1). We identified a nearly complete genome of a novel picornavirus from a pool of cloacal swabs (Pool 11—Table 1) derived from gentoo penguins (P. papua), which was tentatively named as PingPcV (Fig. 2a). The PingPcV genome was assembled using 4,593 reads and exhibited a mean coverage of 271×. Our metagenomic approach did not identify any other sequences of putative vertebrate viruses.
Figure 2.
(a) Genome organization of the complete coding sequence of PingPcV. (b) ML phylogenetic tree showing the evolutionary relationships of viruses identified in our study with representatives members of the Picornaviridae family. Phylogenies are midpoint rooted for clarity of presentation. The scale bar indicates evolutionary distance in numbers of substitutions per amino acid site. Bootstrap values of 1,000 replicates are shown in principal nodes. PingPcV is shown in red with a penguin silhouette. Amino acid identity of PingPcV and representative picornaviruses related of 2Chel (c), 3Cpro (d), and 3Dpol (e) proteins.
The nearly complete sequence of PingPcV is 7,601 nt long and exhibits a genome organization similar to those of the Galivirus, Kobuvirus, Passerivirus, Sabokovirus, Salivirus, and Sinicivirus genera (Jiang et al. 2014; Zell et al. 2017). The viral genome contains a single open reading frame encoding a polyprotein of 2,192 amino acids (238.59 kDa), which is predicted to be cleaved in eleven proteins (Fig. 2a). Also, we obtained partial sequences of the 5′ (717 nucleotides) and 3′ (672 nucleotides) untranslated regions. Based on BLASTX analysis, PingPcV displays 45 per cent identity with Passerivirus A1 (also known as Turdivirus 1), which was described in a dead grey-backed thrush from Hong Kong (Woo et al. 2010). In addition, PingPcV shared ∼38 per cent amino acid identity with Penguin megrivirus, a recent picornavirus identified in cloacal swabs of Adélie penguins also from King George Island (Yinda et al. 2019).
Phylogenetic analysis using the 3Dpol protein showed that PingPcV grouped with high bootstrap values with picornaviruses of the Gallivirus, Passerivirus, and Sicinivirus genera (Fig. 2b). All the viruses within this clade were identified in birds, reinforcing that PingPcV may infect penguins. Almost all picornaviruses phylogenetically related to PingPcV have been described in poultry without any apparent symptoms of disease in the USA, Ireland, Hong Kong, and Hungary (Boros et al. 2012; Bullman et al. 2014; Lau et al. 2014). However, it should be noted that a passerivirus sp. (unclassified picornavirus) was associated with an enteric outbreak of home-reared estrildid finches in Hungary (Pankovics et al. 2018). However, all samples used in this study were collected from penguins apparently without symptoms or disease, which suggests this virus might be endemic and non-pathogenic in penguins.
We examined the level of amino acid identity of the 2Chel, 3Cpro, and 3Dpol proteins of PingPcV and picornaviruses. As shown in Fig. 2c–e, 2Chel and 3Dpol of PingPcV are more similar to Chicken gallivirus 1, whereas 3Cpro is more similar to that of Sicinivirus 1. Both viruses were identified in chicken in Hong Kong and Ireland, respectively (Bullman et al. 2014; Lau et al. 2014). Based on our analysis and the species demarcation criteria of the Picornaviridae family by the International Committee on Taxonomy of Viruses (Zell et al. 2017), we propose that PingPcV should constitute the first member of a novel genus within the Picornaviridae.
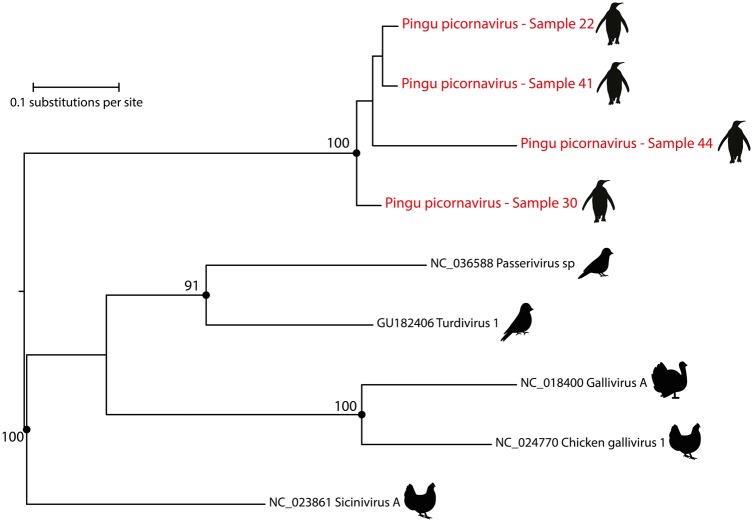
Individual samples derived from P. papua were screened by RT-PCR. Four out of 31 (12.9%) samples were positive to PingPcV. Obtained PCR products were 85–98 per cent identical at the nucleotide level to the consensus sequence of the pool (Fig. 3). Two samples (22 and 20) were collected on 21 November, another (sample 41) on 23 November, and the last one (sample 44) on 27 November, (Supplementary Information S2). This suggests that different lineages of PingPcV are circulating among penguins in King George Island. Also, as expected, positive samples were identified in the pool of cloacal swabs only (consistent with HTS data), suggesting a potential transmission route of this virus to the environment. PingPcV was not detected in P. adeliae and P. antarcticus. Collectively, our results suggest that PingPcV may infect penguins and that transmission may occur via the fecal–oral route, as reported for other picornaviruses (Zell et al. 2017). Our attempts to isolate virus in chicken cell lines (UMNSAH/DF) and embryonated chicken eggs were unsuccessful. It is feasible to think that PingPcV infects specifically P. papua and not P. adeliae or P. antarcticus as we did not detect PingPcV RNA in samples derived from the latter two species. Future serological studies might confirm these findings.
Figure 3.
ML phylogenetic tree showing the evolutionary relationships of four partial genomes of PingPcV with related avian picornaviruses. The phylogeny was inferred with IQ-TREE (TPM3u+F+I nucleotide substitution model) using an alignment of 417 nucleotides derived from the RdRp gene. Phylogenies are midpoint rooted for clarity of presentation. The scale bar indicates evolutionary distance in numbers of substitutions per nucleotide site. Bootstrap values of 1,000 replicates are shown in principal nodes. PingPcV sequences are shown in red with a penguin silhouette.
Previous studies identified viruses in penguins of the following families: Paramyxoviridae (Thomazelli et al. 2010; Neira et al. 2017), Herpesviridae (Pfaff et al. 2017), Orthomyxoviridae (Barriga et al. 2016), Adenoviridae (Lee et al. 2016), Polyomaviridae (Varsani et al. 2015), Poxviridae (Offerman et al. 2014), Papillomaviridae (Varsani et al. 2014), and Birnaviridae (Jackwood, Gough, and Sommer 2005), and recently Picornaviridae (Yinda et al. 2019). Therefore, our study increased the known picornaviral diversity of penguins in Antarctica.
Supplementary Material
Acknowledgements
We thank the National Council Scientific and Technological Development (CNPq) and the Prof. Vivian Helena Pellizari, coordinator Project Micropolar of Brazilian Antarctic Program (PROANTAR). The authors acknowledge the support from the Brazilian Ministries of Science, Technology, and Innovation (MCTI), of Interministerial Commission for Sea Resources (SECIRM). Also, we would like to thank the Brazilian Navy Ary Rongel H-44 by logistic support of the Antarctica Expedition.
Data availability
All sequence reads generated in this project are available at the Short Read Archive (SRA) under accessions SAMN12324479–SAMN12324487 (BioProject ID: PRJNA555609) and the consensus virus genome sequence was deposited in GenBank (accession number: MH255796).
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil (Grant no. 13/14929-1, and Scholarships nos. 17/13981-0; 12/24150-9; 15/05778-5; 14/20851-8; 16/01414-1; 06/00572-0). P.R.M. was supported by the Medical Research Council of the UK (Grant no. MC_UU_120/14/9).
Conflict of interest: None declared.
References
- Bankevich A. et al. (2012) ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-cell Sequencing’, Journal of Computational Biology, 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga G. P. et al. (2016) ‘Avian Influenza Virus H5 Strain with North American and Eurasian Lineage Genes in an Antarctic Penguin’, Emerging Infectious Diseases, 22: 2221–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A. et al. (2012) ‘Identification and Complete Genome Characterization of a Novel Picornavirus in turkey (Meleagris gallopavo)’, The Journal of General Virology, 93: 2171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A., Pankovics P., Reuter G. (2014) ‘Avian Picornaviruses: Molecular Evolution, Genome Diversity and Unusual Genome Features of a Rapidly Expanding Group of Viruses in Birds’, Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 28: 151–66. [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Bullman S. et al. (2014) ‘Identification and Genetic Characterization of a Novel Picornavirus from Chickens’, Journal of General Virology, 95: 1094–103. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. (2015) ‘Microbial Ecology of Antarctic Aquatic Systems’, Nature Reviews Microbiology, 13: 691–706. [DOI] [PubMed] [Google Scholar]
- de Souza W. M. et al. (2018) ‘Discovery of Novel Anelloviruses in Small Mammals Expands the Host Range and Diversity of the Anelloviridae’, Virology, 514: 9–17. [DOI] [PubMed] [Google Scholar]
- Finn R. D. et al. (2017) ‘InterPro in 2017-beyond Protein Family and Domain Annotations’, Nucleic Acids Research, 45: D190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood D. J., Gough R. E., Sommer S. E. (2005) ‘Nucleotide and Amino Acid Sequence Analysis of a Birnavirus Isolated from Penguins’, Veterinary Record, 156: 550–2. [DOI] [PubMed] [Google Scholar]
- Jiang P. et al. (2014) ‘Picornavirus Morphogenesis’, Microbiology and Molecular Biology Reviews, 78: 418–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S. et al. (2017) ‘ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates’, Nature Methods, 14: 587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016) ‘MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets’, Molecular Biology and Evolution, 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon W. B. (2015) ‘Performance of Genetic Programming Optimised Bowtie2 on Genome Comparison and Analytic Testing (GCAT) Benchmarks’, BioData Mining, 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. et al. (2014) ‘Chickens Host Diverse Picornaviruses Originated from Potential Interspecies Transmission with Recombination’, Journal of General Virology, 95: 1929–44. [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. (2016) ‘Genetic and Molecular Epidemiological Characterization of a Novel Adenovirus in Antarctic Penguins Collected Between 2008 and 2013’, PLoS One, 11: e0157032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bueno A. et al. (2015) ‘Ecological Connectivity Shapes Quasispecies Structure of RNA Viruses in an Antarctic Lake’, Molecular Ecology, 24: 4812–25. [DOI] [PubMed] [Google Scholar]
- MacLachlan N. J., Dubovi E. J. (2016) Chapter 26—Picornaviridae, Fenner's Veterinary Virology, 5th edn San Diego, CA, USA: Academic Press, p. 602. [Google Scholar]
- Neira V. et al. (2017) ‘Novel Avulaviruses in Penguins, Antarctica’, Emerging Infectious Diseases, 23: 1212–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offerman K. et al. (2014) ‘The Complete Genome Sequences of Poxviruses Isolated from a Penguin and a Pigeon in South Africa and Comparison to Other Sequenced Avipoxviruses’, BMC Genomics, 15: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankovics P. et al. (2018) ‘A Novel Passerivirus (Family Picornaviridae) in an Outbreak of Enteritis with High Mortality in Estrildid Finches (Uraeginthus sp.)’, Archives of Virology, 163: 1063–71. [DOI] [PubMed] [Google Scholar]
- Pei J., Tang M., Grishin N. V. (2008) ‘PROMALS3D Web Server for Accurate Multiple Protein Sequence and Structure Alignments’, Nucleic Acids Research, 36: W30–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. (2012) ‘IDBA-UD: A De Novo Assembler for Single-cell and Metagenomic Sequencing Data with Highly Uneven Depth’, Bioinformatics, 28: 1420–8. [DOI] [PubMed] [Google Scholar]
- Pfaff F. et al. (2017) ‘A Novel Alphaherpesvirus Associated with Fatal Diseases in Banded Penguins’, Journal of General Virology, 98: 89–95. [DOI] [PubMed] [Google Scholar]
- Schmieder R., Lim Y. W., Edwards R. (2012) ‘Identification and Removal of Ribosomal RNA Sequences from Metatranscriptomes’, Bioinformatics, 28: 433–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M. et al. (2018) ‘The Evolutionary History of Vertebrate RNA Viruses’, Nature, 556: 197–202. [DOI] [PubMed] [Google Scholar]
- Smeele Z. E., Ainley D. G., Varsani A. (2018) ‘Viruses Associated with Antarctic Wildlife: From Serology Based Detection to Identification of Genomes Using High Throughput Sequencing’, Virus Research, 243: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Jimenez L. M., Estrada K., Sanchez-Flores A. (2014) ‘GARM: Genome Assembly, Reconciliation and Merging Pipeline’, Current Topics in Medicinal Chemistry, 14: 418–24. [DOI] [PubMed] [Google Scholar]
- Souza W. M. et al. (2018) ‘Viral Diversity of Rhipicephalus microplus Parasitizing Cattle in Southern Brazil’, Scientific Reports, 8: 16315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G. A., Shafren D. R. (1994) ‘Avian Encephalomyelitis: A Review’, Avian Pathology, 23: 603–20. [DOI] [PubMed] [Google Scholar]
- Thomazelli L. M. et al. (2010) ‘Newcastle Disease Virus in Penguins from King George Island on the Antarctic Region’, Veterinary Microbiology, 146: 155–60. [DOI] [PubMed] [Google Scholar]
- Varsani A. et al. (2014) ‘A Novel Papillomavirus in Adelie Penguin (Pygoscelis adeliae) Faeces Sampled at the Cape Crozier Colony, Antarctica’, Journal of General Virology, 95: 1352–65. [DOI] [PubMed] [Google Scholar]
- Varsani A. et al. (2015) ‘Identification of an Avian Polyomavirus Associated with Adelie Penguins (Pygoscelis adeliae)’, Journal of General Virology, 96: 851–7. [DOI] [PubMed] [Google Scholar]
- Wille M. et al. (2019a) ‘Antarctic Penguins as Reservoirs of Diversity for Avian Avulaviruses’, Journal of Virology, 93: e00271–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M. et al. (2019b) ‘Virome Heterogeneity and Connectivity in Waterfowl and Shorebird Communities’, The ISME Journal, 13: 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. (2010) ‘Comparative Analysis of Six Genome Sequences of Three Novel Picornaviruses, Turdiviruses 1, 2 and 3, in Dead Wild Birds, and Proposal of Two Novel Genera, Orthoturdivirus and Paraturdivirus, in the Family Picornaviridae’, Journal of General Virology, 91: 2433–48. [DOI] [PubMed] [Google Scholar]
- Yinda C. K. et al. (2019) ‘Penguin Megrivirus, a Novel Picornavirus from an Adelie Penguin (Pygoscelis adeliae)’, Archives of Virology, 164: 2887. [DOI] [PubMed] [Google Scholar]
- Zell R. et al. (2017) ‘ICTV Virus Taxonomy Profile: Picornaviridae’, Journal of General Virology, 98: 2421–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R. (2018) ‘Picornaviridae-the ever-growing Virus Family’, Archives of Virology, 163: 299–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence reads generated in this project are available at the Short Read Archive (SRA) under accessions SAMN12324479–SAMN12324487 (BioProject ID: PRJNA555609) and the consensus virus genome sequence was deposited in GenBank (accession number: MH255796).