Abstract
Monoclonal antibodies bind with high specificity to a wide range of diverse antigens, primarily mediated by their hypervariable complementarity determining regions (CDRs). The defined antigen binding loops are supported by the structurally conserved β-sandwich framework of the light chain (LC) and heavy chain (HC) variable regions. The LC genes are encoded by two separate loci, subdividing the entity of antibodies into kappa (LCκ) and lambda (LCλ) isotypes that exhibit distinct sequence and conformational preferences. In this work, a diverse set of techniques were employed including machine learning, force field analysis, statistical coupling analysis and mutual information analysis of a non-redundant antibody structure collection. Thereby, it was revealed how subtle changes between the structures of LCκ and LCλ isotypes increase the diversity of antibodies, extending the predetermined restrictions of the general antibody fold and expanding the diversity of antigen binding. Interestingly, it was found that the characteristic framework scaffolds of κ and λ are stabilized by diverse amino acid clusters that determine the interplay between the respective fold and the embedded CDR loops. In conclusion, this work reveals how antibodies use the remarkable plasticity of the beta-sandwich Ig fold to incorporate a large diversity of CDR loops.
Keywords: antibody, architecture, isotype
Introduction
The complexity of the antibody engineering process was dramatically increased by the stepwise development from murine monoclonal antibodies (mAbs), to chimeric mAbs with murine variable (V) and human constant (C) regions (Boulianne et al., 1984; Morrison et al., 1984; Neuberger et al., 1985; Sun et al., 1987), to humanized mAbs with mouse-derived complementarity determining regions (CDRs) fused on the β-sandwiches of the human V framework regions (FR) (Jones et al., 1986; Hale et al., 1988; Riechmann et al., 1988; Verhoeyen et al., 1988), to the state-of-the-art fully human mAbs (Bruggemann et al., 1989, 2015; McCafferty et al., 1990; Bruggemann and Neuberger, 1996; Lonberg, 2008). The genetically engineered mAbs share the characteristic composition of classical mAbs by comprising two identical heavy (HC) and two identical light chains (LC) (Fig. 1A) (Edelman, 1959; Alzari et al., 1988). The variable domain’s amino-terminal regions of the HC (VH) and LC (VL) each comprise three CDRs that mediate the antigen binding (Fig. 1B and C). Therefore, the chimeric and CDR-grafted antibodies retained the affinity of the original mouse antibodies for the antigen targets whilst comprising less sequence motifs that are recognized by the human immune system (Hwang and Foote, 2005).
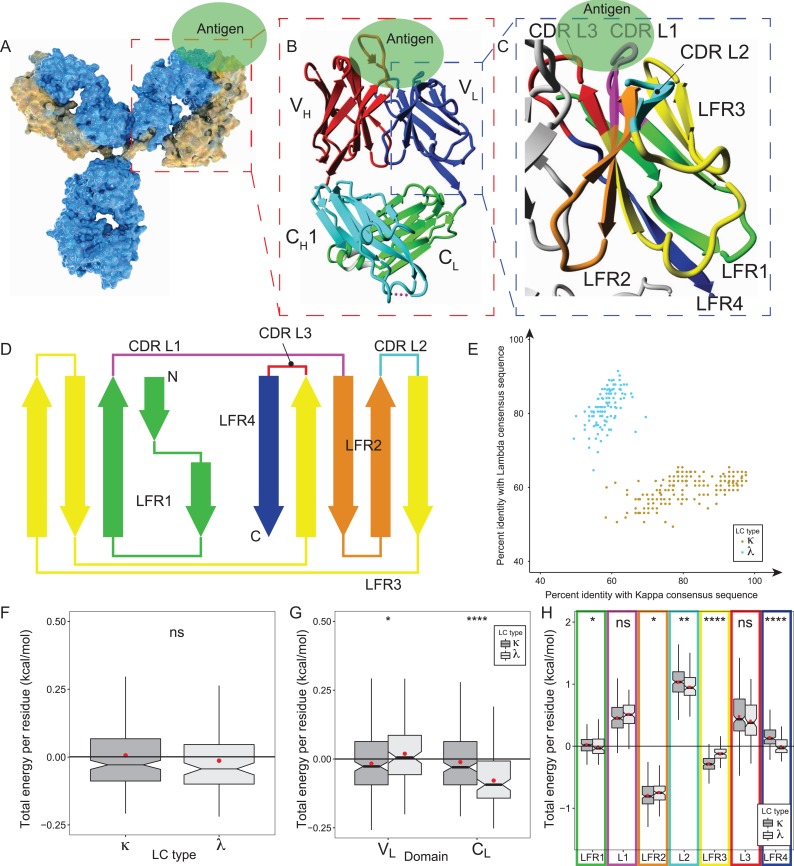
Fig. 1.
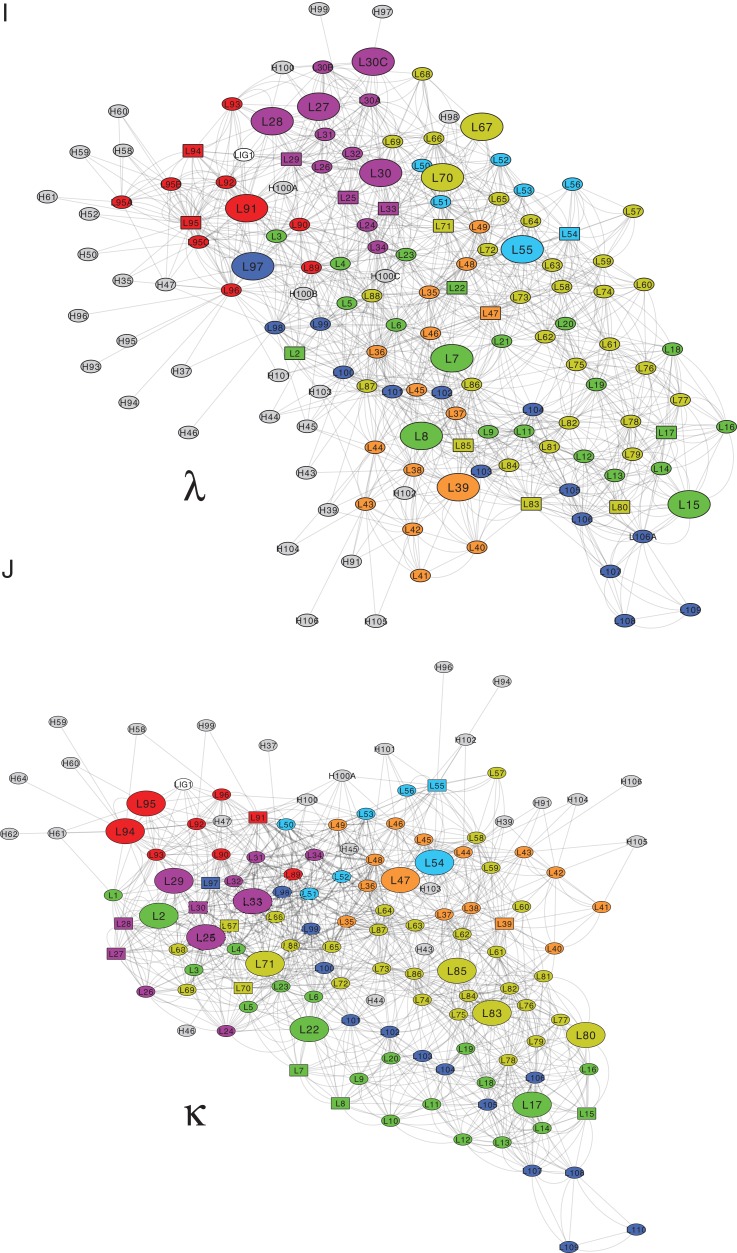
Global sequence-structure analysis of antibodies VL domains splitting into kappa and lambda isotypes. (A) Schematic representation of the surface of a full-length antibody that comprises two identical heavy (HC, blue) and light chains (LC, yellow), jointly forming the antigen (Ag, green) binding fragment (Fab, highlighted by red box). (B) The Fab fragment consists of HC’s constant CH1 (cyan) and variable VH domain (red), as well as LC’s constant CL (green) and variable VL domain (blue). Fab’s Ag affinity is cooperatively mediated by six complementarity-determining regions (CDRs) that are equally distributed over VH and VL (blue box). (C) VL contributes to the Ag binding via three CDR loops (CDR L1 in magenta, CDR L2 in cyan and CDR L3 in red, Chothia definition) that are structurally orientated by four neighboring framework regions (LFR1 green, LFR2 orange, LFR3 yellow, and LFR4 blue). (D) Scheme of the VL structure that comprises three CDR loops being embedded in four β-strand (arrows) rich LFRs. N, amino-terminal end; C, carboxy-terminal end. (E) A set of 333 non-redundant VL sequences was assigned to the lambda (λ, light gray) and kappa (κ; dark gray) subtype on the basis of their identity to the λ and κ consensus sequence. (F-H) Statistical analysis of the energy contributions to VL and CL structures. The boxplots incorporate the dataset’s entire value distribution (whiskers), 25th (lower limit of the box) and 75th percentile (upper limit), mean (red dot) as well as median (central line) with a 95% confidence interval (notches). Statistical significances obtained by a Wilcoxon rank-sum test are indicated by ns (P > 0.05), * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001) and **** (P ≤ 0.0001). (F) FoldX evaluations of the κ (dark gray) and λ (light gray) dataset provided the average residue contributions to the free energy of the folding stability of the entire LC (VL + CL). (G) The average free energies of the VL and CL fold are illustrated separately for the κ (dark gray) and λ (light gray) dataset. (H). The LFRs, and especially LFR2, contribute to the folding stability of VLκ (dark gray) and VLλ (light gray), while the CDRs (L1, L2, L3) destabilize both structures. (I & J) The cytoscape software (version 3.7.1. (Shannon et al., 2003)) generated 2D visualizations of the residue-residue interaction network that stabilizes the VLλ (I) and VLκ (J) folding. Each residue is shown as a node, and each edge represents a Van der Waals contact that was determined by FoldX. Representative protein structures of the VLκ and VLλ isotypes (pdb id 1L7I (Vajdos et al., 2002) and 6axk (Oyen et al., 2017) were used in consistence with Fig. 1E. In cytoscape, the network layout was set to perfuse force directed in order to ensure that residues buried in the structure are centered in the 2D representation. The color code corresponds to the regions of the domain as defined in Fig. 1D. Large nodes indicate residues that differentially stabilize the complementary isotypes as it was identified in Fig. 5B, and square-node representations of the corresponding residues in the contrary isotype simplify the cross-comparison. Residues that interact with the VH domain are shown in gray.
In particular, the modern-day engineering processes are more technically demanding and require the consideration of structural information on the mAb and the mAb-antigen complex to guide the engineering process and increase the probability of success (Haidar et al., 2012; Hanf et al., 2014). Strikingly, the identification of mutations that improve a given property (e.g. affinity) without comprising other properties (e.g. stability) challenges the engineering process of antibodies. The CDR grafting approach often requires back-mutation of key residues of the human FR to the amino acids of the original murine mAb to preserve the functional conformation of the CDRs and thus its high-affinity (Queen et al., 1989; Co and Queen, 1991; Chames et al., 2009). However, the complex interplay between residues of the FR and CDR is not completely understood so far, thereby complicating the rational design of mutations that increase the thermodynamic stability of the mAb structure and mediate the conformation as well as orientation of CDR loops.
In this context, it is interesting that approximately 90–95% of the human IgG1 sequence are conserved, whilst being assigned to the framework region (FR) and constant (C) domains (Harris et al., 2004). The constant FR regions of the LC (LFRs, Fig. 1C and D) have either of the two evolutionarily developed kappa (LCκ) or lambda (LCλ) sequences, coding for two distinct scaffold isotypes that are employed in grafting the antigen-binding loops (Titani et al., 1967). For reasons thus far unknown, the ratio of LCκ to LCλ varies considerably between species with an average ratio of 95:5 in mouse and 60:40 in human (Popov et al., 1999). The affinity for the antigen is specified by the remaining 5% of the variable sequence part that comprises the six CDRs. The sequence variability is initially obtained by somatic recombination of V(D)J regions, followed by somatic mutations to provide a broad diversity of CDR loops with varying physicochemical complementarities and predictable canonical structures that differ between LCκ and LCλ (Chothia and Lesk, 1987; North et al., 2011; Raghunathan et al., 2012; Nowak et al., 2016). Although the binding energy is predominantly determined by a limited number of critical interface residues, the antigen residues are stochastically mutated together with the non-binding residues of the CDRs (Clackson and Wells, 1995; Bogan and Thorn, 1998; Sheinerman et al., 2000). Interestingly, by preferring certain amino acids over others, each CDR has its own contact preferences that do not depend on the remaining five CDRs (Zhao and Li, 2010; Raghunathan et al., 2012; Kunik and Ofran, 2013).
For a long time, the antigen affinity was solely attributed to the residues of the CDRs that are oriented by the FR scaffold (Tramontano et al., 1990). However, over the past few years the importance of FR residues for antigen binding was emphasized (Sedrak et al., 2003; Haidar et al., 2012). Several studies confirmed that antigen binding of CDR grafted antibodies was successfully reobtained by back-mutation of FR residues to the original murine residues (Verhoeyen et al., 1988; Queen et al., 1989; Tramontano et al., 1990; Kettleborough et al., 1991; Carter et al., 1992; Foote and Winter, 1992; Xiang et al., 1995; Baca et al., 1997; Chiu et al., 2011; Rodriguez-Rodriguez et al., 2012). Reversely, in some cases the mAb fold was stabilized by mutations in the CDRs (Koenig et al., 2015, 2017). These findings underline the complex interplay between the binding loops and the FR scaffold. Up to now, it is still unclear why distinct structural motifs evolved in the human antibody repertoires of LCκ and LCλ, and whether the isotypes correlate with the antigen class and/or the antibody’s affinity, specificity and LFR structure (Knappik et al., 2000).
In first attempts, structure-based computational design methods were applied to a predetermined antibody scaffold to achieve new affinities and specificities (Liu et al., 2017). Low hit rates (Liu et al., 2017) and simplified binding systems (Chevalier et al., 2017) indicated that mAb engineering shows a growing need for increasing the probability of success by acquiring more knowledge on the relatively unknown interplay between antibody modules and residue interactions.
In this work, statistical evaluations of the abYsis antibody database (Swindells et al., 2017) provide deeper understanding of the complex structure-function relationship of single and clustered residues in LCκ and LCλ scaffolds. Defined networks of interacting residues that mediate the interplay between CDR’s distinct canonical structures and the surrounding LFR regions were identified for both isotypes. This work provides further knowledge on how the VLλ and VLκ structures evolutionarily adapted to fit to a broad variety of different CDR loop conformations. In summary, stabilizing networks of interacting residues increase the diversity of antibodies, extend the predetermined restrictions of the general antibody fold and expand the diversity of antigen binding.
Materials and methods
Database retrieval and analysis
The abYsis database (Swindells et al., 2017) was queried using the human sequences of the PDB as data source. Sequences with warnings and unclassified, unpaired, or unnumbered sequences were excluded, resulting in 1399 antibody structures. Duplicates were removed as well as antibodies that contained errors after downloading. Solely the structures that contained the Fab (300 and 500 amino acids) and provided a resolution lower than 2.8 Å were analyzable by FoldX. Redundancy removal was performed based on light chain sequence at a threshold of 98% sequence identity using the CD-hit algorithm (Li and Godzik, 2006; Fu et al., 2012). The final database contained 333 Fab structures that were renumbered via the Abnum script (Abhinandan and Martin, 2008) following the Chothia numbering scheme.
FoldX
FoldX version 3.0 Beta 6 (c) compilation for Linux was used for all analyses. Initially, the structures were repaired using the ‘RepairPDB’ command, and information of the residue level was obtained via the ‘SequenceDetail’ command (Schymkowitz et al., 2005). Interface analysis was performed by the ‘AnalyseComplex’ command. Intramolecular interface analysis (VL/CL) was obtained by splitting the molecule at the hinge region using python scripting and the ‘AnalyseComplex’ command. Residue level contributions to the interaction energy was obtained by removing the interaction partner (Antigen or HC), using the ‘SequenceDetail’ command, and subtracting the difference of contributions with and without interaction partner present.
Calculation of significance
Information on the residue level was analyzed using R-studio (RStudio Team, 2016) by running the ‘aggregate’ function, summing and averaging on the different levels of resolution. Significance levels and comparing means between distributions were determined using the Wilcoxon signed-rank test and ggpubr package (Alboukadel Kassambara, 2017). Sequence logos were calculated and plotted using the geom_logo command from ggseqlogo package (Wagih, 2017a). Box-plots were generated either by using the standard boxplot command or the geom_boxplot function from the ggplot2 package (Wickham, 2009).
Random Forest
R-studio was used to perform the RandomForest analyses (randomForest R-package version 4.6-14) (Breiman, 2001). The variable n-tree was set at 1000 and importance at TRUE. Only numerical variables were used in the analysis.
Statistical coupling analysis
The python implementation of Statistical Coupling Analysis was used with the default settings to generate the clusters of interacting amino acids (Rivoire et al., 2016).
Mutual information analysis
Mutual information between amino acid identities at Chothia positions and CDR canonical structures was calculated by converting one letter amino acid identities and canonical structures to arbitrarily chosen numbers. Subsequently, the mutual information was calculated using the ‘mutinformation’ command from the infotheo package (version 1.2.0 (Meyer, 2008)) with default settings. Chord diagrams were generated using the ‘chordDiagram’ function from the circlize package (version 0.4.4. (Gu et al., 2014)). A mutual information threshold of 0.5 was used for displaying the chords.
Structure viewer
All structural visualization was done using YASARA Structure (version 18.4.24 (Krieger and Vriend, 2014)). Images were generated using Ray-traced screenshots.
Availability of scripts and datasets
Datasets and scripts are available at Protein Engineering, Design and Selection online.
Results and discussion
In silico analysis of the abYsis database
The abYsis database is a web-based antibody repository for antibody sequence and structure-management, analysis, and prediction (Swindells et al., 2017). In this work, abYsis (Version 3.1.0. (Swindells et al., 2017)) was screened for crystal structures of human antibodies and fragments thereof that contained both, heavy (HC) and light chain (LC). The query resulted in 1399 structures of the antigen binding fragment (Fab) with HC and LC of similar size (Fig. 1B). In order to retain only high fidelity structural information and run reliable energy calculations with FoldX (Schymkowitz et al., 2005), the set was limited to crystal structures that exhibit a resolution better than 2.8 Å. To avoid a bias towards thoroughly investigated structures and close relatives thereof (e.g. point mutants), redundancy removal was performed at 98% sequence identity of the LC by performing the CD-Hit algorithm. This clustering approach selected representative entries in case of redundancy (Fu et al., 2012). The entire selection procedure resulted in a set of 333 non-redundant Fab structures that were assigned to 221 LCκ and to 112 LCλ structures (relative frequencies of 0.66 and 0.33, respectively, Supplementary Figure 1), reflecting the state-of-the-art diversity of publicly available Fab structures with sufficient resolution. A consensus sequence pursuant to the Chothia numbering system (Chothia and Lesk, 1987) was determined for LCκ and LCλ subgroups by identification of the amino acid with the highest propensity per position. The percent identity for both consensuses was then calculated for each sequence. Plotting the distances (i.e. identity) of all λ and κ sequences from the respective consensus visualized the clustering of both subgroups (Fig. 1E). In addition, the distribution of sequences to LCκ and LCλ isotypes also yielded a wide range of sequence diversity towards its consensus sequences. The sequence identity of both LC classes ranged from 70–95%, while the identity to the different groups accounted for 50–70%. As a result, the sequence identity of a small quantity of LCκ sequences to the LCκ consensus sequence equals that of the LCλ consensus sequence, underlining the difficulty in accurately assigning these ambiguous sequences. In these cases, the sequence was assigned to one of the isotypes via typical λ or κ key residues (Honegger and Pluckthun, 2001) such as position L10 (L, LC residue position derived from the sequence alignment (Chothia and Lesk, 1987)) that is present in κ but absent in the λ alignment, which in turn comprises a λ-typical position L106A that lacks in the κ alignment. More specifically, position L71 comprises conserved Phe/Tyr in κ, but Ala/Val/Arg in λ.
The destabilizing effect of incorporating CDRs is distributed differently in κ than in λ
Each Fab structure of the dataset was analyzed using the empirical FoldX force field (Schymkowitz et al., 2005), which was optimized to predict the effect of mutations on the thermodynamic stability of a protein. The resulting information of the in silico analysis were subdivided into that of the amino acid level, the region level (FR/CDR), the domain level (CL/VL), the chain level, as well as the full HC/LC complex level. These energies are predictions that computationally estimate various factors.
The statistical analysis of the structure dataset demonstrated that both architectural isotypes exhibit a comparable overall thermodynamic stability on the domain level (Fig. 1F). Therefore, the energies of the constituent atoms as calculated by FoldX were added to the residue and polypeptide chain level, thereby creating energy descriptors at the residue and the chain level. FoldX further allowed to characterize the LC-specific energy pattern of the thermodynamic stability of the VL and CL domains by unraveling the energy contributions e.g. electrostatics, solvation or Van der Waals packing of each residue (Table I). On average, the CL domain of LCλ is more stable than that of LCκ, however this trend is reversed for the VL domains (Fig. 1G). This suggests that the VL and CL regions are structurally independent of each other (i.e. stable by themselves) in the κ architecture, whilst in the λ architecture a highly stable CL domain seems to be needed to compensate for the rather less stable VL domain. In this view the CL of LCλ appears to be a stable platform that compensates through the VL-CL interface for the destabilizing effect of the CDRs, which was confirmed by a more detailed analysis below. Assigning the per-residue energy distributions to the specific VL regions clearly indicated that the scaffold (i.e. LFRs) stabilizes the overall structure, whilst the CDRs exert a destabilizing impact (Fig. 1H). Hence, the structurally destabilizing properties of CDRs are compensated by the stabilizing LFRs in both VL scaffolds. Despite this general trend, subtle but important differences were identified between both isotypes. Exemplarily, CDR L2 is on average more destabilizing in VLκ, which in return is compensated by the remarkably stable LFR3 found in the immediate proximity (Fig. 1H). In contrast, VLλ comprises a more stable LFR4 that forms the hinge region between the variable and constant domain. Again, the interface between the VL and the CL domain seems to compensate the incorporation of unstable CDRs in VL. To illustrate the specific integration of distinct structural elements in VLλ and VLκ, a two-dimensional representation of the residue-residue interaction networks that stabilize each domain was generated (Fig. 1I and J). To this end, the Van der Waals contacts were determined by FoldX from the representative protein structures of the VLκ and VLλ isotypes (pdb id 1L7I (Vajdos et al., 2002) and 6axk (Oyen et al., 2017)) and displayed as a network using Cytoscape (Shannon et al., 2003). While the networks of VLλ and VLκ show much similarity, such as an extensive network of residues interacting with the VH domain, there are also notable differences in the exact configuration of the network that are discussed below.
Table I.
Descriptions of FoldX variables.
| Variable | Definition | Unit |
|---|---|---|
| total.energy | The predicted overall stability | kcal/mol |
| Backbone.Hbond | The contribution of backbone H-bonds | kcal/mol |
| Sidechain.Hbond | The contribution of sidechain H-bonds | kcal/mol |
| Van.der.Waals | The contribution of VanderWaals forces | kcal/mol |
| Electrostatics | The contribution of electrostatic interactions | kcal/mol |
| Solvation.Polar | The penalty for burying polar residues | kcal/mol |
| Solvation.Hydrophobic | The contribution of hydrophobic groups | kcal/mol |
| Van.der.Waals.clashes | The penalty for VanderWaals’ clashes (interresidue) | kcal/mol |
| entropy.sidechain | The entropy cost of fixing the sidechain | kcal/mol |
| entropy.mainchain | The entropy cost of fixing the mainchain/backbone | kcal/mol |
| cis_bond | The penalty for having a cis peptide bond | kcal/mol |
| torsional.clash | The penalty for VanderWaals’ torsional clashes (intraresidue) | kcal/mol |
| backbone.clash | The penalty for VanderWaals’ backbone-backbone clashes | kcal/mol |
| helix.dipole | The contribution of the helix dipole (electrostatic) | kcal/mol |
| disulfide | The contribution of disulfide bonds | kcal/mol |
| electrostatic.kon | Th electrostatic interaction between molecules in the precomplex | kcal/mol |
| energy.Ionisation | The contribution of ionization energy | kcal/mol |
| sidechain.burial | Burial of the sidechain | fraction |
| mainchain.burial | Burial of the backbone | fraction |
| sidechain.Occ | Occupancy of the sidechain | fraction |
| mainchain.Occ | Occupancy of the backbone | fraction |
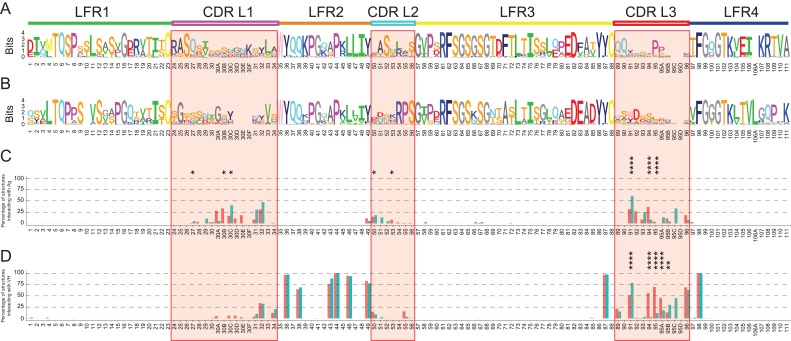
Next, differences between the amino acid sequences of VLκ and VLλ were investigated, and the frequency of amino acids at each position was visualized using sequence logos (Wagih, 2017a) (Fig. 2A and B). The size of the letter reflects the amino acid’s frequency in the alignment as calculated via Shannon’s information-entropy formalism (Shannon, 1948). Kolmogorov-Smirnov and Mann-Whitney tests confirmed that the entropy distributions of the positions did not significantly differ between VLκ and VLλ (Supplementary Figure 2), indicating that the quantity of conserved and variable positions was similar between both isotypes. The analysis further uncovered that the sequence variation is not restricted to the CDRs but extended to the LFRs (11 positions of VLκ and 14 of VLλ with an entropy of > 1.5 bits). The variability at distinct positions in the LFRs presumably counterbalances for effects arising from CDR’s sequence variability. Together with λ- and κ-specific and conserved amino acid positions, these data clearly point towards isotype-specific stabilizing networks of interacting amino acids.
Fig. 2.
Per-residue analysis of the isotype-specific sequence-structure relationship of VL. (A, B) “Sequence logo” (Wagih, 2017a) representation of the amino acid occurrence per Chothia positions in the LFR and CDR (red boxes) of VLκ (A) and VLλ (B) sequences. The stack height indicates the conservation of the position, whilst the character height reports the relative conservation of distinct amino acids. The theoretical maximum value for the sequence entropy of a protein alignment is 4.36 bits. 221 VLκ and 112 VLλ sequences were used for the alignment, significantly exceeding the critical quantity of 40 sequences allowing for accurate computation (Crooks et al., 2004). The percentage frequency of contribution to the interaction with antigen (C) and VH domain (D) was calculated per position for VLκ (red) and VLλ (blue). Residues were considered to be relevantly involved in binding if the FoldX force field calculated an interaction energy of at least −0.5 kcal/mol. CDRs are marked by red boxes, and significance levels of the differences > 0.2 kcal/mol between VLκ and VLλ are indicated (* for P ≤ 0.05, ** for P ≤ 0.01, *** for P ≤ 0.001, **** for P ≤ 0.0001).
Remarkably, several conserved positions comprise different amino acid identities in VLλ and VLκ (Table II). Exemplarily, VLκ incorporates at position L95 of CDR L3 a conserved proline that was previously identified to adopt the cis-conformation, which restricts the loop’s degrees of freedom (Chothia and Lesk, 1987; Al-Lazikani et al., 1997). Further striking variations include positions L7 (P in κ, S in λ), L11 (L in κ, V in λ), L71 (F in κ, A in λ), L105 (E in κ, T in λ), L106 (I in κ, V in λ), L109 (T in κ, P in λ) and L111 (P in κ, S in λ). In particular, the strong differential conservation of the structurally similar residues L and V at position 11 is notable. Interestingly, positions L105-L111 thereof are located in the C-terminal part of LFR4 that is known to affect the λ- and κ-specific elbow angle by forming the VL-CL interface (Stanfield et al., 2006). Consequently, these distinct amino acid identities at conserved positions highlight the architectural differences between λ and κ structures.
Table II.
Similarities and differences in the conservation of VLκ and VLλ as determined by the alignments.
| Definition | Chothia numbers |
|---|---|
| Conserved in both VLκ and VLλ, identical amino acids | L5, L6, L16, L23, L35, L36, L37, L38, L40, L41, L44, L46, L48, L49, L57, L61, L62, L63, L64, L67, L68, L73, L75, L82, L86, L87, L88, L98, L99, L101, L102, L103 |
| Conserved in both VLκ and VLλ, different amino acids | L7, L11, L71, L105, L106, L109 and L111 |
| Highly variable in both VLκ and VLλ | L3, L13, L17, L19, L22, L27, L28, L29, L30, L30B, L30C, L31, L32, L33, L34, L42, L43, L45, L50, L51, L53, L58, L60, L76, L77, L78, L79, L80, L89, L91, L92, L93, L94, L95A, L95B, L96, L97, L104 |
| Conserved in VLκ while variable in VLλ | L1, L2, L8, L18, L20, L24, L26, L39, L47, L52, L59, L66, L69, L70, L72, L81, L90, L95, L107, L108 |
| Conserved in VLλ while variable in VLκ | L4, L9, L12, L15, L21, L25, L30A, L54, L55, L56, L83, L92, L100 |
The isotypes share a common principle behind the antibody-antigen interaction
Interestingly, the C-terminal part of CDR L2 (L54-L56) is highly conserved in VLλ but variable in VLκ (t-test P-value = 0.03, Fig. 2A and B). The contrary sequence variability in this part of the CDR could emerge from differing antigen binding mechanisms between VLλ and VLκ. This binding hypothesis was verified by a more thorough analysis of 88 VLκ and 38 VLλ crystal structures being bound to a protein ligand. First, the total energy of the antibody-antigen interaction was unraveled into the contributions of the antibody’s heavy chain and λ, respectively, κ light chain (Supplementary Figure 3). As expected, the analysis confirmed the widespread theory (D’Angelo et al., 2018) that the heavy chain contributes on average more to the antigen binding than the light chain of both isotypes. Focusing on the light chain, the thermodynamic stability of VL with and without antigen bound was investigated for each residue’s contribution to the interaction energy with the ligand. Therefore, the frequency of contributing to the binding energy with more than 0.5 kcal/mol (typical FoldX-cutoff for interactions (Schymkowitz et al., 2005; Sanchez et al., 2008)) was calculated per position (Fig. 2C). The antibody interacts almost exclusively with the antigen via its CDRs, confirming the widely accepted theory that sequence variations in the LFRs accommodate sequence selection in the CDRs to mediate binding. Since in more than 50% of the cases only a single residue (L91) is critical for interacting with the antigen, the frequency plots further verified a high diversity in the residues that mediate the binding. By exhibiting interaction frequencies between 20% and 50%, positions L30A-L32 in CDR1 and L91-L94 in CDR3 were identified as the most frequently involved residues for antigen interaction. In contrast, residues of CDR L2, and, in particular, it’s highly conserved C-terminal part (L54-L56) are only marginally involved in antigen binding, indicating a rather LFR-like role. Neither of the residue position is involved in antigen binding in more than 50% of the VLλ or VLκ structures, which again confirmed that the CDRs of the light chain take a support role in both isotypes, while that of the heavy chain are most frequently involved in antigen recognition.
The VL–VH interaction occurs differently between the κ and λ isotypes
The energetic contributions of VL residues to the interaction with the VH domain were obtained for all 333 non-redundant Fab structures by removing VH and calculating the free energy differences. Amino acids that contribute with at least −0.5 kcal/mol (corresponding to the accepted uncertainty of FoldX (Schymkowitz et al., 2005)) to the free energy of the VL–VH complex were classified as key residues. The threshold allowed to identify conserved key residues that contribute significantly to the interaction with VH for the set of VL structures, and to calculate the percentage of key residues per position (Fig. 2D). There is a well-known network of conserved amino acid residues that mediates the interaction between VH and VL (Chothia et al., 1985), which were confirmed here for both isotypes by exhibiting a frequent contribution to the VH–VL interaction (>50% of the structures) and a high conservation (Fig. 2A and B) at positions L36 (Tyr), L38 (Gln), L43 (Ala/Ser), L44 (Pro), L46 (Leu), L49 (Tyr), L87 (Tyr) and L98 (Phe). Interestingly, the sidechain orientation of these conserved residues within the VL–VH interface is highly similar for both isotypes (Supplementary Figure 4A–D). The antigen-distant part of the VL–VH interface is capped by a hydrogen bonding network, connecting the backbone of a variable residue at position L42 with the conserved Gln at position L38 in LFR2 and the highly conserved Gln at position H39 of VH (HFR2) (Supplementary Figure 4D). Additionally, conserved residues form a hydrophobic core and interact via π-π stacking of two Tyrosine (H91 – HFR2 and a variable position in CDR H3) and two Tryptophan residues (H47 – HFR2 and H103 – HFR4).
While the majority of interactions between VL and VH are common for both subtypes, discrepancies are present. For example, the interaction of λ’s and κ’s VL regions with VH most significantly differs in CDR L3 (Fig. 2D). Strikingly, the amino acid composition and conformation of this region is well known to frequently contribute to the interaction with both, antigen and VH. CDR L3 exhibits on average a shorter and highly consistent length in VLκ if compared to the increased and more variable length of the loop in VLλ (Supplementary Figure 4E, Supplementary Table 1). The substantial sequence differences might be linked to isotype-specific canonical structures of CDR L3 (Chothia and Lesk, 1987; North et al., 2011; Nowak et al., 2016). In VLλ, the additionally introduced residues seem to shift the hotspot for the VH-interaction towards the C-terminus of CDR L3. For instance, CDR L3’s N-terminal position L91 interacts significantly more frequent with VH in VLλ than in VLκ. Interestingly, this position shows less Shannon entropy in VLλ (1.94) if compared to VLκ (3.00), indicating less variation in amino acid identity in VLκ, and is often an aromatic amino acid (Trp/Tyr) (Fig. 2A and B). Furthermore, positions L94, L95 and L95 A interact significantly more often with VH in VLκ than in VLλ. Taken together, this suggests that the amino acids at positions L91 and L95 C are more important interactors with both the VH as well as the antigen in VLλ than in VLκ.
VL–CL interactions modulate elbow-angle differences between κ and λ
In the next step, the free energy of inter-domain interactions between VH and VL (VL/VH), CL and CH (CL/CH) as well as VL and CL (VL/CL) were analyzed for λ and κ isotypes (Supplementary Figure 5). FoldX calculations yielded a stabilizing total interaction energy between CL and VLκ, however this interaction is significantly weaker in the VLλ/CL complex (Supplementary Figure 5A). The interaction network that stabilizes the VL/CL complex could be assigned to the extraordinary H-bonding of LCκ‘s backbone (Supplementary Figure 5B) and sidechain (Supplementary Figure 5C). This effect is decisively reinforced by LCκ’s significantly increased quantity of interacting residues with CL (Supplementary Figure 5D). Carefully inspecting representative structures of both isotypes enabled to pinpoint three highly conserved residue positions that formed discrete backbone H-bonding; L105 (Glu in κ and Thr in λ), L106 (Val in κ and Ile in λ) and L106A (missing in κ, Leu in λ) (Supplementary Figure 5E). Similarly, two positions responsible for the difference in sidechain H-bonding were identified; L105 (Glu in κ and Thr in λ) and L108 (Arg in κ and Gln in λ) (Supplementary Figure 5F). Moving to the structural level of a representative LCκ, a conserved Glutamine at CLκ’s position L165 was identified to form a conserved hydrogen bond with the backbone of residues L105-L106A (Supplementary Figure 5G). Furthermore, the highly conserved Arginine L108 forms a hydrogen bond bridge between the backbone of L109 and CLκ. Another hydrogen bond is formed between the Tyr at residue position 173 in CLκ and the negatively charged residue (Glu) at position L105 in VLκ. The absence of Gly at position L107 (exclusively present in VLλ) and a consistent hydrogen bonding pattern between VL and CL domains might restrain the flexibility of the hinge region in LCκ but. Moreover, a strong interaction energy between VLκ and CLκ is reached by the simultaneous formation of two hydrogens bonds via an Arg at position L108, whilst in λ only a singular and weaker hydrogen bond is formed via a conserved Glu at position L108 (Supplementary Figure 5G and H, respectively).
This analysis confirmed that in κ an extensive network of hydrogen bonds firmly staples the hinge region connecting VL and CL in place, thereby fixing the elbow angle between the domains. In contrast, λ exhibits significantly less interactions between VL and CL, thus creating a higher flexibility of the elbow angle (Stanfield et al., 2006).
Sequence selection in the CDRs is coupled to sequence variation in the entire VL domain
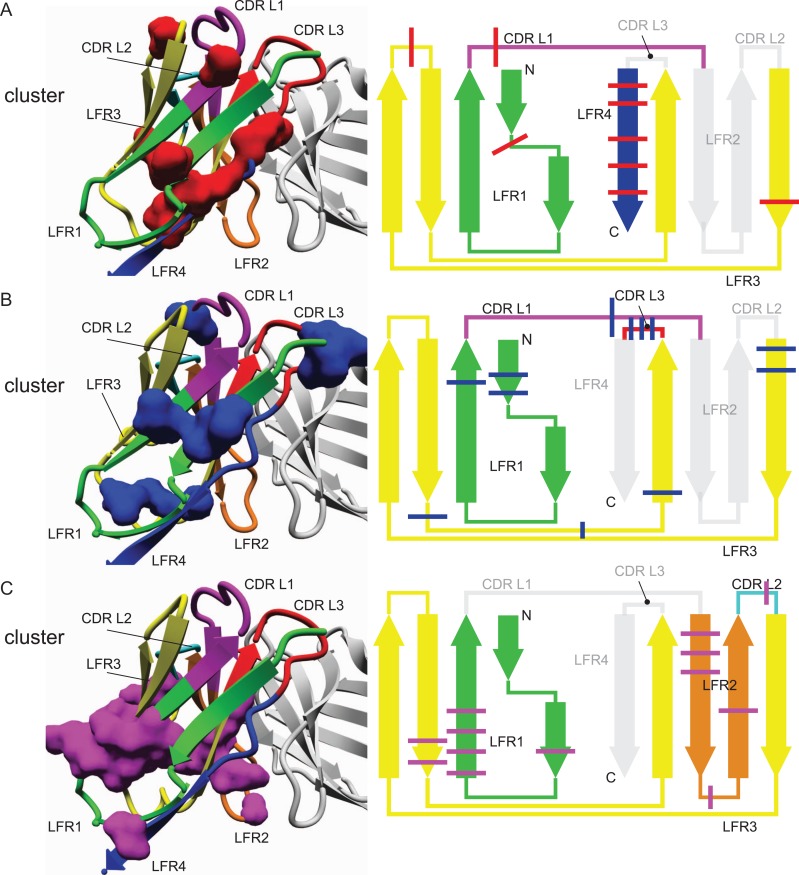
Besides characterizing individual contributions of single residues of VL to the binding of antigen and VH, the VL fold was further investigated for correlated networks of amino acids using statistical coupling analysis (SCA) (Halabi et al., 2009). The technique measures covariations between pairs of amino acids in a multiple sequence alignment. Thereby, high statistical coupling energies indicate evolutionary dependence between residue pairs. Ideally, clustering of statistical coupling energies yields networks of evolutionary dependent amino acids (Halabi et al., 2009), for which reason it is capable of revealing how sequence selection in one part of the antibody potentially affects the amino acid identity of further parts. While the technique itself is entirely unbiased, it depends critically on the number of sequences comprised within the alignment in order to detect meaningful coupling information (Zafra Ruano et al., 2016). However, given that our analysis is focused on the structure-sequence relationship, we limited the analysis to our non-redundant set of sequences for which reliable structural data was available, i.e. the analysis was performed on the entire multiple sequence alignment containing 333 VL sequences without subgrouping into λ and κ, which is on the low side for this method and hence only the most consistent results should be taken from this. Nevertheless, using typical settings of statistically significant coupling analysis (Lockless and Ranganathan, 1999), three conserved clusters of interacting amino acids were identified (Fig. 3 and Table III) that unequivocally connect sequence variations in the CDRs with different parts of the LFRs. For example, cluster 1 comprises statistically coupled amino acids of the framework regions LFR1 (L7), LFR3 (L62 and L67) and LFR4 (L98, L99, L101, L102 and L104) with a residue of CDR L1 (L25) (Fig. 3A). In other words, the selection outcome of position L25 at the N-terminal end of CDR L1 correlates with adaptive changes in the frame regions LFR1, LFR3, and most prominently in LFR4 that forms CDR L1’s opposite side of the domain. Intriguingly, position L25 is not frequently involved in binding (Fig. 2C), but seems to serve as a connection point between the loop to the rest of the structure. Notably, the coupled L7 (LFR1) is a conserved position that differs between the VL isotypes (Proline in κ, Serine in λ, Fig. 2A and B), suggesting that these different conserved residues help to accommodate a distinct selection of CDR sequences. The next identified cluster 2 shows statistical coupling between amino acid positions in the framework regions LFR1 (L5, L6 and L22) and LFR3 (L57, L58, L78, L82 and L84) with CDR L1 (L31) and CDR L3 (L94, L95 and L95B), thus connecting the rest of the fold with both CDRs that are strongly involved in interacting with the ligand (Fig. 3B). In particular, residues L31 and L94 are often critically involved in antigen binding (Fig. 2C), and their coupling to LFR residues provides direct evidence of the entire domain adjusting to selections at these critical positions for antigen binding. The role of L95 and L95B seems to be more adaptive as they are not as frequently involved in antigen binding (Fig. 2C). Additionally, cluster 3 comprised statistically coupled amino acid positions of the framework regions LFR1 (L12, L18, L19, L20 and L21), LFR2 (L36, L37, L38, L41 and L45) and LFR3 (L74 and L75) and a central position in CDR L2 (L54) (Fig. 3C). Again, the third cluster connects a sequence selection in one of the CDRs to positions distributed across the entire domain. Taken together, these three amino acid clusters provide clear evidence that CDRs and FRs cannot be studied individually. Since sequence variations in CDR positions impact throughout the entire antibody domain, adaptive changes may be required to accommodate the CDR sequence selection.
Fig. 3.
Statistical Coupling Analysis (SCA) of VL structures identified three conserved clusters of statistically coupled amino acids. Ranganathan’s SCA method determined three statistically coupled networks of amino acids (1, red; 2, blue; 3, magenta) in the VL domain. The clusters are shown on a representative VLκ structure (left, PDB: 1l7i (Vajdos et al., 2002)) and marked through bars on a topological scheme. The color-coding scheme for CDR and LFR is maintained from Fig. 1C and D.
Table III.
Ranganathan clusters
| Cluster # | Chothia # | Region |
|---|---|---|
| 1 | L7 | LFR1 |
| L25 | CDRL1 | |
| L62 | LFR3 | |
| L67 | LFR3 | |
| L98 | LFR4 | |
| L99 | LFR4 | |
| L101 | LFR4 | |
| L102 | LFR4 | |
| L104 | LFR4 | |
| 2 | L5 | LFR1 |
| L6 | LFR1 | |
| L22 | LFR1 | |
| L31 | CDRL1 | |
| L57 | LFR3 | |
| L58 | LFR3 | |
| L78 | LFR3 | |
| L82 | LFR3 | |
| L84 | LFR3 | |
| L94 | CDRL3 | |
| L95 | CDRL3 | |
| L95B | CDRL3 | |
| 3 | L12 | LFR1 |
| L18 | LFR1 | |
| L19 | LFR1 | |
| L20 | LFR1 | |
| L21 | LFR1 | |
| L36 | LFR2 | |
| L37 | LFR2 | |
| L38 | LFR2 | |
| L41 | LFR2 | |
| L45 | LFR2 | |
| L54 | CDRL2 | |
| L74 | LFR3 | |
| L75 | LFR3 |
Besides that, no specific differences in the network of co-evolving residues were found between VLκ and VLλ, in part due to this method’s large data requirements to detect subtle connections (Zafra Ruano et al., 2016). Additionally, the conservation analysis most readily identifies the commonalities in the entire family and is less sensitive for identifying differences in subgroups (Lockless and Ranganathan, 1999; Suel et al., 2003). Consequently, additional methods were applied to resolve potential differences between interacting residues in VLκ and VLλ.
Mining structural differences between λ and κ
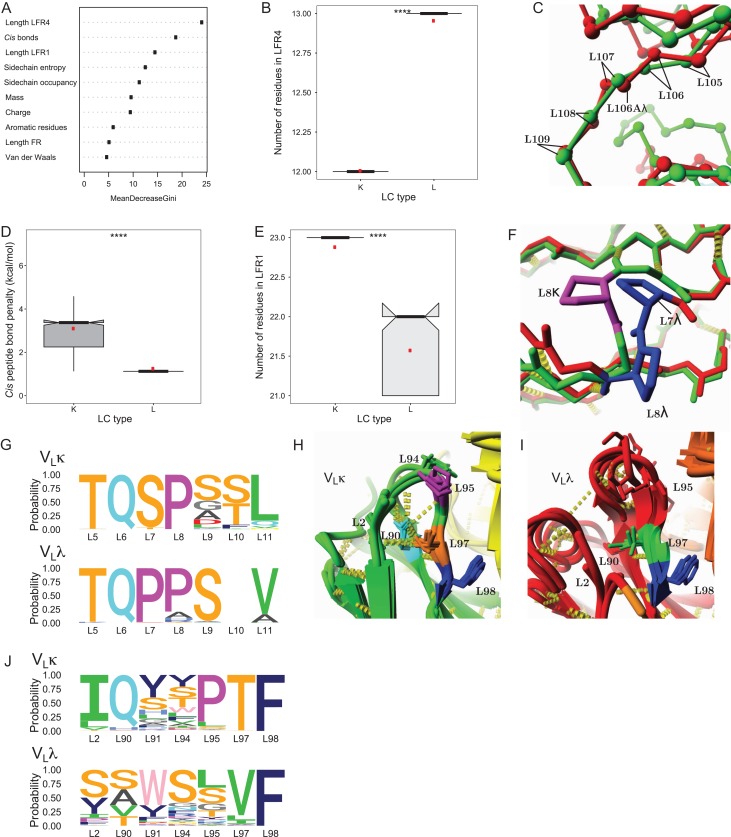
In order to elucidate structural factors that distinguish between VLκ and VLλ, a machine learning technique called Random Forest (Breiman, 2001) was used in order to identify the most convenient variables differentiating between both classes. According to this method, a large quantity of decision trees is generated that each splits instances of a dataset to their predefined classes (VLκ and VLλ), based on a subset of variables that are randomly selected from a predefined pool. Parameters describing the properties of λ and κ Fab structures were extracted from the FoldX analysis, supplemented with other structural descriptors and used as input variables (Supplementary Table 2). The ensemble of decision trees (the forest) was then used to rank the most important variables with respect to their ability to classify between kappa and lambda by determining the mean decrease gini, a factor that weighs the relative importance of each parameter in obtaining a reliable classification (Fig. 4A). The length of the LFR4 region was identified as the variable with the highest importance (Fig. 4B), referring to the fact that LFR4 comprises an additional residue (L106A) in VLλ but not in VLκ (Fig. 4C). The relatively small and flexible glycine is usually present as VLλ’s additional residue (Fig. 1J) in a stretch forming the VLλ-CLλ interface, which was previously hypothesized in literature (Stanfield et al., 2006) and confirmed above to allow λ light chains to adopt a more flexible switch region and larger elbow angles between VL and CL.
Fig. 4.
Structural differences between VLκ and VLλ were determined by RandomForest. (A) The most reliable classifiers for assigning the dataset of full light chain structures to the κ and λ isotypes are identified by the mean decrease gini (variables defined in Supplementary Table 1). (B) Unknown sequences can be reliably assigned to the κ and λ isotype according to the length of LFR4. (C) An overlay of representative Cα-traces displays how the exclusion (VLκ, green, 5ifa (Huang et al., 2004)) or incorporation (VLλ, red, 3ujj (Guan et al., 2013)) of L106A in LFR4 affects the structure. (D) VLλ structures exhibited a significantly lower energy penalty for peptide bonds in the cis-conformation if compared to VLκ structures. (E) The number of residues in LFR1 enables to distinguish between VLκ and VLλ sequences. (F) Representative structures of VLλ (3ujj (Gorny et al., 2011), red) and VLκ (5ifa (Jardine et al., 2016), green) illustrate structural differences in LFR1. VLκ structures frequently contain a cis-proline at position L8, stabilizing a fold that dramatically differs from VLλ structures that favor trans-prolines at positions L7 and L8. (G) ‘Sequence logo’ representation of the amino acids identities in LFR1 of VLκ (top) and VLλ (bottom). (H, I) Representative structures of VLκ (H, green, PDBs: 3drq (Julien et al., 2008), 4ygv (Schiele et al., 2015), 4jm4 (Kong et al., 2013) and 4zyk (Gilman et al., 2015)) and VLλ (I, red, PDBs: 4h8w (Acharya et al., 2014), 1aqk (Faber et al., 1998), 5d7s (Eylenstein et al., 2016), 5cck (Lee et al., 2015)) clearly differ in the conserved environment of residue L95. Yellow cylinders, hydrogen bonds; yellow and orange structure, heavy chain; cyan residue, Gln L90; magenta, Pro L95; orange, Thr L97; blue, Phe L98; green, Val L97. (J) ‘Sequence logo’ representation of the amino acids occurrence spatially neighboring residues of L95 in VLκ (top) and VLλ (bottom).
The penalty for peptide bonds in the cis-conformation i.e. cis-prolines was identified as the second most important variable to classify between κ and λ. Strikingly, VLλ comprises no cis-Prolines on average, and CLλ and CLκ both contain one cis-conformation (Fig. 4D). Contrarily, VLκ exhibits two peptide bonds in the cis-conformation in LFR1 (L8) and CDR L3 (L95) in the most cases (Fig. 4D) (Spada et al., 1998; Ewert et al., 2003). As a result, the N-terminal β-strand (LFR1) of VLκ is bent by a cis-proline at position L8, while in VLλ this kink in the β-strand results from two consecutive trans-prolines (Fig. 4F).
Correlating with the presence of cis-conformations in LFR1, the length of this region emerged as another important classification parameter between both isotypes. The LFR1 of VLκ exhibits exactly 23 amino acids, whereas the length of the region is restricted to 22 or 21 amino acids in VLλ (Fig. 4E). The cis-proline at position L8 allows for one extra residue to be placed in LFR1 of VLκ (Fig. 4F), where in VLλ two prolines at positions L7 and L8 form the bridge connecting the two halves of the beta-sandwich (Fig. 2A, B and F).
Being exclusively found in VLκ, the second cis-proline at residue L95 leads to a highly conserved structural feature in the middle of CDR L3 (Fig. 4H, J). Simultaneously, the consistent length of this loop clearly differentiates from the varying length of the corresponding CDR in VLλ lacking the cis-conformation (Supplementary Figure 4E). A clear pattern emerges from the structural environment of cis-Pro95 comprising a hydrogen bond between the conserved Thr97 and the frequently found (86%), β-branched Ile2 (Fig. 4H). As a consequence, the N-terminal region is fixed to the structure by a network of conserved hydrogen bonds. Interestingly, the VLλ structures relatively often lack the N-terminal positions L1 and L2, which might result from a low electron density in the X-ray diffraction pattern that is obtained during structure elucidation of this typically flexible region. This relates back to the hypothesis that the N-terminal end of LFR1 has a lower thermodynamic stability in VLλ, which was previously proposed to explain the susceptibility of human VLλ to amyloid formation and overrepresentation in light chain amyloidosis (Zhao et al., 2018).
Additionally distinguishing VLκ from VLλ, CDR L3 is fixed via hydrogen bonding between the backbone of position L93 and a conserved Gln90 (85%) that further hydrogen bridges with Thr97 (Fig. 4H). This considerably conserved network of hydrogen bonds consistently stabilizes the structural environment of the antigen-binding loop between VLκ chains. In contrast, VLλ exhibits a broad range of amino acids at position L90 (Ser: 46%, Ala: 21%, Thr: 17%, Val: 16%) as well as more unspecific interaction partners (Val at L97:76%), providing CDR L3 a greater level of variability to adopt versatile loop conformations and broad affinities to ligands. The isotype-specific structural features seem to be linked to the particular function of the respective CDR L3. While this region is on average only moderately involved in target binding in VLκ, the loop is critically important in VLλ with L91 significantly contributing to the antigen interaction (Fig. 2C).
λ and κ structures are stabilized by differential interaction networks
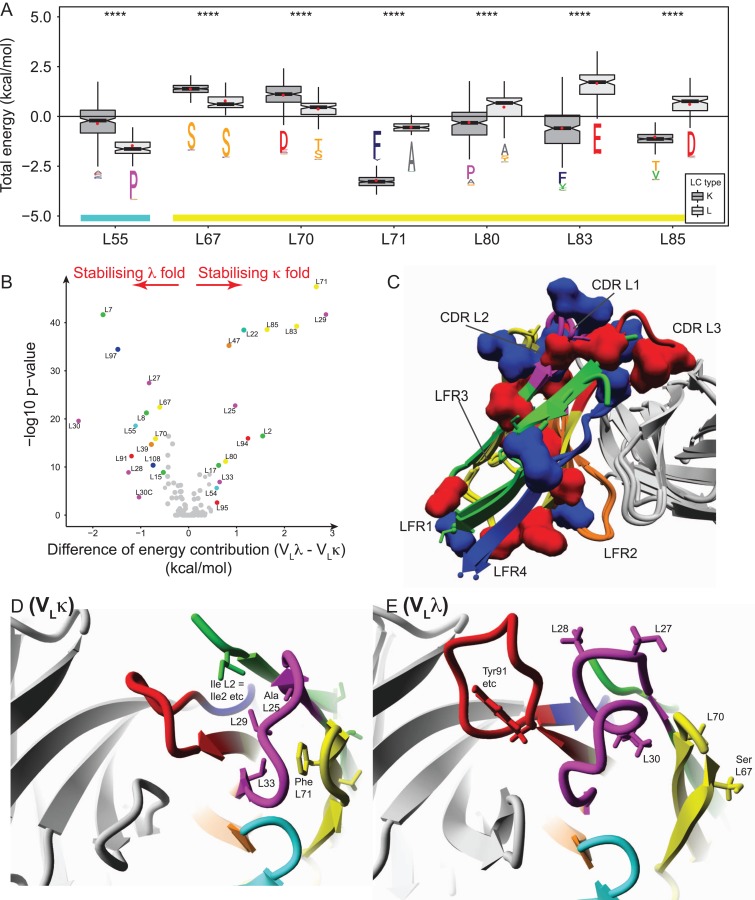
After determining differences between the sequence (Figs 1–3) and structural appearance (Figs 1, 2 and 4) of VLλ and VLκ, next the underlying scaffolds were investigated for stability differences. To this end, the contribution of each residue to the thermodynamic stability (ΔGcontrib) of the respective fold was calculated using FoldX (example data in Fig. 5A). To identify energetic differences between the scaffolds of both isotypes, the per-residue contribution to the thermodynamic stability of VLλ was subtracted by that of VLκ.
Fig. 5.
Key residues contribute significantly different to the folding stability of VLκ and VLλ structures. (A) Key residues that differed most clearly in their energy contribution to the folding stability of either VLκ or VLλ were identified. The Wilcoxon rank-sum test yielded a statistical significance of P ≤ 0.0001 (****) for key residues. The amino acid conservation in the ‘sequence logo’ format was extracted from Fig. 2A and B. In accordance to the schema in Fig. 1C, the color coding of the lower bar indicates the CDR L2 and LFR3. (B) The volcano plot visualizes the per-residue preference for stabilizing either the VLλ (negative energy values) or the VLκ fold (positive energy values) by plotting the difference of mean energy contribution to both structure types against the -log10 P-value (negative 10 base log of the P-value). Highly significant data points correspond to low P-values that appear at the top of the plot. The mean energy difference threshold was set at > 0.5 kcal/mol (corresponding to the accepted uncertainty of FoldX (Schymkowitz et al., 2005)) and the P-value threshold at P < 0.05 (corresponding to a -log10 P-value of < 1.30). The dots are color coded according to Fig. 1C and D. (C) The isotype-specific folds are stabilized by particular residue positions (Fig. 5B) that are highlighted on representative VLκ (red clusters, PDBid: 5ifa (Huang et al., 2004) and VLλ structures (blue clusters, PDBid: 3ujj (Guan et al., 2013). (D-E) Distinct clusters of stabilizing amino acids near the antigen binding site are shown for (D) VLκ (PDBid: 5ifa (Huang et al., 2004) and (E) VLλ (PDBid: 3ujj (Guan et al., 2013).
The statistical significance of energetic differences was calculated using the Wilcoxon signed rank test (Fig. 5A), and ΔΔGcontrib was plotted against the -log10 p-value resulting in a so-called volcano plot (Fig. 5B). Difference of at least ΔΔGcontrib = ±0.5 kcal/mol combined with a p-value <0.05 were assigned as significant. The method identified 28 positions that are evenly distributed throughout the domain, half of which contribute to the stability of the λ fold, while the remaining positions favor the κ scaffold. The identified residues were highlighted on a representative structure (Fig. 5C) and on a two-dimensional interaction network representation (Fig. 1I, J), both illustrating that the isotypes are stabilized by different networks of interacting residues. Positions that predominantly stabilize the VLλ structure (L7, L8, L15, L27, L28, L30, L30C, L39, L55, L67, L70, L91, L97 and L108) are widespread and differ in the conservation degree; L7, L8, L15 and L55 (all highly conserved Prolines, the last located in CDRL2), L27, L28, L30, L30C and L91 (variable positions in CDR L1 and CDR L3), L39 (variable position in LFR2), L67, L70, L97 and L108 (relatively conserved positions in LFR3 and LFR4). Similarly, the remaining positions that contribute to the stability of VLκ (L2, L17, L22, L25, L29, L33, L47, L54, L71, L80, L83, L85, L94, L95) are distributed throughout the domain while offering a broad range of conservation scores. L2, L17 and L22 (relatively highly conserved positions in LFR1), L25, L29 and L33 (relatively highly conserved positions in CDR L1), L47 (completely conserved position in LFR2), L54 (relatively highly conserved position in CDR L2), L71 (completely conserved position in LFR3), L80, L83 and L85 (variable positions in LFR3), L94 (highly variable position in CDR L3) and L95 (highly conserved proline in CDR L3).
In the following, the domain’s antigen binding site is exemplarily studied in more detail to illustrate isotype-specific interaction networks. In VLκ (Fig. 5D), the cluster of interacting residues connects CDR L1 (Ala/Ser at L25, an aliphatic residue at L29 and Leu33) with LFR1 (Ile2) and LFR3 (Phe71) (Fig. 5B). VLλ is stabilized by a different network in which several variable positions of CDR L1 (L27, L28, L30) interact with LFR3 (Thr/Ser at L70, Ser67) (Fig. 5E). Furthermore, the cluster comprises a frequently found Tyr/Trp at position L91 that was shown to mediate antigen and VH interaction. Other parts of the domain reveal further differences in stabilizing clusters, thus showing that despite the high similarity in overall structure, VLλ and VLκ are stabilized in a fundamentally different manner.
λ and κ frameworks adapt to the conformation of the CDRs
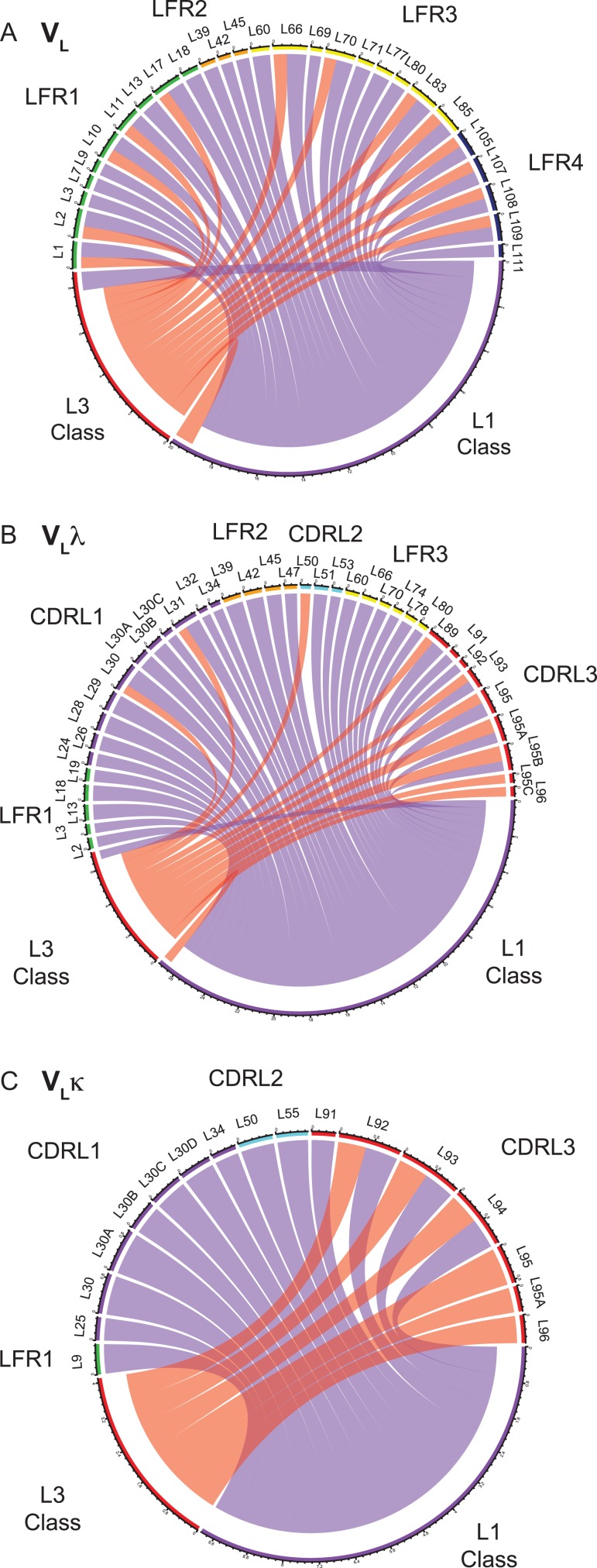
To elucidate how the selection of certain CDR loop conformations requires adaptations in the framework region, a variation of the Shannon information analysis was performed (Lenaerts et al., 2008, 2009a,b; Zafra Ruano et al., 2016). State-of-the-art CDR databases were used that classify canonical structures according to their length (Dunbrack (North et al., 2011; Adolf-Bryfogle et al., 2015)) and overall structural similarity (Deane (Nowak et al., 2016)) (Table IV). For the entirety of VL structures, the mutual information network between framework’s residue identities and both CDR loop libraries (Fig. 6A,Supplementary Figure 6A) were visualized as Chord diagrams, expressing the quantity of mutual information between residue positions (nodes) via the thickness of edges. Thereby, the connection between CDR conformations and sequence patterns of VLλ and VLκ, and thus adaptations in the LFR accommodating for CDR canonical classes was unraveled. An extensive list of couplings between residue positions of the scaffold and loop conformations of CDR L1 and L3 was identified. CDR L2 did not show any mutual information with framework reisdues, which is likely due to its limited number of canonical structures (Table IV, (North et al., 2011; Nowak et al., 2016)). Given that loop conformations of λ and κ are assigned to different canonical categories, the coupling is directly related to isotype-specific residue positions that were already determined by sequence alignments (Fig. 2A, B). Interestingly, the mutual information between framework positions and canonical structures of CDR L1 seems to exceed that of CDR L3 (Fig. 6A, quantity of edges), suggesting a particular dependence of CDR L1’s canonical structures on framework identities.
Table IV.
Counts of canonical structures for Dunbrack and Deane definitions.
| Dunbrack | Deane | |||||
|---|---|---|---|---|---|---|
| CDR | Canonical | K count | L count | Canonical | K count | L count |
| L1 | L1-8-* | 0 | 2 | L1-10,11,12-A | 52 | 0 |
| L1-9-* | 4 | 0 | L1-11-A | 0 | 10 | |
| L1-10-1 | 4 | 0 | L1-11-B | 0 | 4 | |
| L1-11-1 | 103 | 0 | L1-12-A | 2 | 0 | |
| L1-11-2 | 11 | 1 | L1-12-B | 3 | 0 | |
| L1-11-3 | 2 | 36 | L1-12-D | 1 | 0 | |
| L1-12-1 | 13 | 0 | L1-13-A | 0 | 1 | |
| L1-12-2 | 10 | 0 | L1-13,14-A | 0 | 26 | |
| L1-12-3 | 0 | 1 | L1-17-A | 1 | 0 | |
| L1-13-1 | 0 | 28 | Unclustered | 4 | 5 | |
| L1-13-2 | 1 | 3 | ||||
| L1-14-2 | 0 | 24 | ||||
| L1-14-cis9-* | 0 | 1 | ||||
| L1-15-1 | 9 | 0 | ||||
| L1-15-2 | 1 | 0 | ||||
| L1-16-1 | 24 | 0 | ||||
| L1-16-cis9-* | 1 | 0 | ||||
| L1-17-1 | 9 | 0 | ||||
| <NA> | 29 | 16 | <NA> | 158 | 66 | |
| L2 | L2-6-* | 0 | 1 | L2-7-A | 63 | 44 |
| L2-8-1 | 182 | 77 | L2-7-B | 2 | 0 | |
| L2-8-2 | 5 | 10 | ||||
| L2-8-3 | 2 | 0 | ||||
| L2-8-4 | 1 | 5 | ||||
| L2-8-5 | 1 | 1 | ||||
| L2-8-cis3-* | 2 | 1 | ||||
| L2-12-2 | 0 | 1 | ||||
| <NA> | 28 | 16 | <NA> | 156 | 68 | |
| L3 | L3-5-* | 10 | 1 | L3-5-A | 4 | 1 |
| L3-6-cis4-* | 0 | 1 | L3-7-A | 1 | 0 | |
| L3-7-1 | 1 | 0 | L3-8-A | 2 | 0 | |
| L3-8-1 | 9 | 0 | L3-9-A | 0 | 3 | |
| L3-8-2 | 4 | 0 | L3-9-B | 0 | 1 | |
| L3-8-cis6-1 | 1 | 0 | L3-9,10-A | 51 | 0 | |
| L3-9-1 | 4 | 12 | L3-10-A | 0 | 3 | |
| L3-9-2 | 12 | 2 | L3-10-C | 1 | 2 | |
| L3-9-cis5,7-* | 1 | 0 | L3-10-D | 0 | 1 | |
| L3-9-cis7-1 | 116 | 0 | L3-10,11-A | 0 | 15 | |
| L3-9-cis7-2 | 1 | 0 | L3-12-A | 0 | 1 | |
| L3-9-cis7-3 | 4 | 0 | L3-13-A | 0 | 2 | |
| L3-10-1 | 5 | 27 | Unclustered | 10 | 15 | |
| L3-10-cis5,8-* | 1 | 0 | ||||
| L3-10-cis6-* | 0 | 2 | ||||
| L3-10-cis7-* | 3 | 0 | ||||
| L3-10-cis7,8-1 | 6 | 0 | ||||
| L3-10-cis8-1 | 1 | 0 | ||||
| L3-11-1 | 8 | 37 | ||||
| L3-11-cis7-1 | 5 | 0 | ||||
| L3-11-cis8-* | 1 | 1 | ||||
| L3-12-1 | 1 | 11 | ||||
| L3-12-cis8-* | 0 | 1 | ||||
| L3-13-1 | 0 | 2 | ||||
| <NA> | 27 | 15 | <NA> | 152 | 68 | |
Fig. 6.
Chord diagrams of mutual information between canonical structures and framework residues. (A, B, C) Mutual information of Dunbrack’s canonical structures (North et al., 2011) and framework residues of (A) VL, (B) VLλ and (C) VLκ was calculated, and values > 0.5 are shown. The color of the chords corresponds with CDR’s canonical group (salmon: CDR L3, purple: CDR L1), and chord width correlates with the extent of mutual information (in bits). The coloring scheme of Chothia positions reflects their topological region similar to that used in Figs 1–3.
To study VLλ- and VLκ-specific sequence adaptions to its distinct CDR loop classes, equivalent chord diagrams were constructed isotype-specifically applying Dunbrack’s (Fig. 6B and C) and Deane’s (Supplementary Figure 6B and C) CDR loop classes. VLλ comprises an intricate network of connected amino acid residues, for which reason many positions throughout the domain accommodate the canonical structures of CDR L1 and CDR L3 (Fig. 6B). The conformational cluster of CDR L1 is connected to positions of the surrounding VLλ scaffold except region LFR4. This connection is used as an example to validate the mutual information method below.
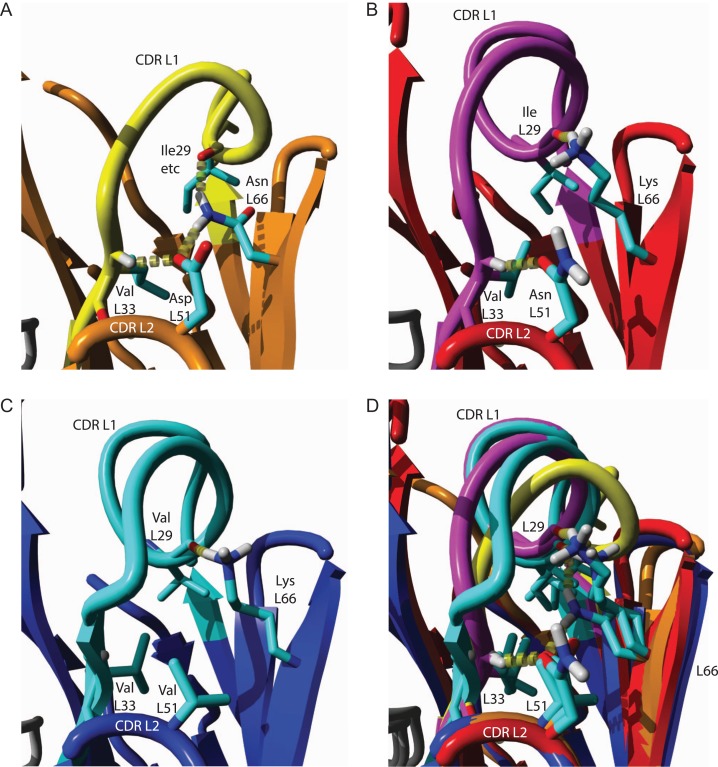
The canonical structures of VLλ’s CDR L1 favor three representative Dunbrack classes, i.e. L1-11-3, L1-13-1 and L1-14-2 (Table IV). Residues of CDR L2 (L51) and LFR3 (L66) clearly correlate with that of the surrounding CDR L1 (L29) and its C-terminal end (L33) by forming an interaction network (Fig. 7). Canonical structure L1-11-3 (Fig. 7A) is uniquely stabilized by a cross-linked network of hydrogen bonds involving the class-specific side chains of AspL51 and AsnL66 as well as the backbone at position L33 and L29(L29···AsnL66···AspL51···L33). Two of these positions (AsnL51, LysL66) are consistently modified in sequence of the second canonical structure L1-13-1, which dramatically affects the conformation of CDR L1 (Fig. 7B). The C-terminal end of the loop is fixed via hydrogen bonding of the altered AsnL51 side chain and the backbone of L33, while the middle of the loop is stabilized by a hydrogen bond between the varied LysL66 side chain and the L29 backbone. Similarly, the third canonical structure L1-14-2 is oriented by the unchanged LysL66 side chain and backbone of L29, but hydrogen binding between CDR L1 and L2 is prevented by a class-specific ValL51 (Fig. 7C). The rather nonspecific, hydrophobic interactions of ValL51 presumably permit a more flexible loop orientation of CDR L1. An overlay of these three canonical classes (Fig. 7D) indicates that the lacking hydrogen bond of L51 relaxes the backbone of CDR L1-14-2 into an extended beta-strand. In contrast, the loop conformation of L1-11-3 is precisely positioned by a well-defined hydrogen bond network between L33, L51, L66 and L28. The sequence variability of CDR L2 in VLλ seems to contribute less to antigen binding (Fig. 2C) rather than to support the remaining hypervariable regions to form distinct canonical structures. The herein identified unique interaction networks between framework residues and canonical CDR structures strongly supports the hypothesis stating that the conformation of CDRs of VLλ depend stronger on framework positions than in the case of VLκ. For grafting CDRs on VLλ scaffolds, this finding underlines an increased importance of considering key residues of the scaffold and verifying the loop orientation of its CDR L1, L2 and L3. VLκ exhibits less couplings between loop conformations and LRF scaffold, suggesting that loop grafting could potentially be less complicated (Fig. 6C).
Fig. 7.
Networks of interacting amino acids stabilize distinct canonical structures of CDR L1 in VLλ. (A) Dunbrack’s CDR cluster L1-11-3 is favored by a hydrogen bond network between Asp L51 of CDR L2 and Asn L66 of LFR3, cooperatively coordinating the C-terminal end of CDR L1 (L33) and the center of CDR L1 (L29). PDB 4m5y was representatively used to visualize the structure (Hong et al., 2013). (B) Canonical class L1-13-1 is stabilized by hydrogen bonding between Asn L51 of CDR L2 and Val L33 of CDR L1, and between Lys L66 of LFR3 and Ile L29 of CDR L1. The structure of PDB 3n9g was exemplary used for visualization (Kaufmann et al., 2010). (C) The canonical structure L1-14-2 is favored by a hydrogen bond between Val L29 of CDR L1 and Lys L66 of LFR3, whilst L33 and L51 do not interact as it was shown for PDB 3kdm (Niemi et al., 2011). (D) An overlay of (A-C) highlights the structural differences between the canonical classes L1-11-3, L1-13-1 and L1-14-2. The consistent hydrogen bonds between the sidechain of L66 and the central part CDR L1, and between the sidechain of L51 of CDR L2 and L33 of the C-terminus of CDR L1 define the Dunbrack canonical classes of CDR L1 in VLλ.
Conclusion
The role of loops, particularly longer ones, in protein structures is often underappreciated as the focus goes to the regular hydrogen bonded elements of (secondary and tertiary) structure and the hydrophobic core, with the loops required to connect more interesting structure parts (Pei et al., 1997; Fiser et al., 2000). This is particularly pronounced in the concept of CDR loop grafting in the field of antibody engineering. However, 20–30% of protein structures are composed of elements that can be classified as loop structures while playing an integral and essential role in the folding and stability of the protein structure (Vanhee et al., 2011). Moreover, loop structures can act as tensioned springs that propagates strain throughout the surrounding structure, a mechanistic concept that seems to occur typically around active sites (Rousseau et al., 2001; Gutteridge and Thornton, 2004).
The field of antibody engineering has gradually incorporated a more integrative view, in which the CDRs are fitted into the framework regions rather than being grafted on top of it. In the current study, advanced statistical analysis of protein structures was employed to comprehensively map the network of interactions that connect the CDRs to the rest of the VL domain. A special focus was further set on the structural difference between VLλ and VLκ isotypes to highlight the specific structural adaptations through the domains. The view that emerges is that there are residues distributed over the VL domain whose amino acid identity are adapted to the structure of the CDRs. The analysis forms the starting point to design a back-mutation algorithm that identifies and modifies problematic positions to optimize the placement of defined loop configurations on scaffold regions. The applied methods are in principle amenable to analyze any protein domain, but the availability of structural information with sufficient resolution and diversity are a prerequisite. Taking the latter into consideration, the analysis of novel developments such as more complex antibody formats are currently out of scope.
The structural differences between VLλ and VLκ reveal how the VL domain adapts to entirely different loop classes. Most notably, solely VLλ‘s CDR L1 contains a helical segment sittings tightly in the beta-sandwich of the Ig fold, partly distorting the inter-sheet interaction of the sandwich. Another important difference to VLκ is found in the linker region between VL and VC, which forms less interactions and a more flexible elbow angle conformation in VLλ. The structural differences are apparent in both, the conservation pattern of each isotype, and the network of interactions stabilizing each structure. It is hence consistent that grafting of loops on a VL domain will require isotype-specific adaptations of its key residues which identities associate with its CDR loop classes.
The analytical framework of this work provided new insight into the VL architecture, but have not yet been turned into a protocol for antibody framework engineering in response to loop grafting. A method still to be developed could predict the most appropriate amino acid choices at crucial framework positions under consideration of its CDR loop classes. Although machine learning algorithms appear to be ideally suited for this task at first sight, the available set of diverse and high-quality structural data is currently too limited to allow the training of a sufficiently accurate method. Besides that, in the most cases accurate loop class assignments are precluded by solely knowing the CDR loop sequences rather than their exact structures. The absence of accurate structural models further limits the potential application of force field calculations to identify optimal amino acid identities at critical positions. As a consequence of the mentioned restrictions, the best current approach may be based on selectively varying amino acids on critical positions in the VL domain followed by experimentally selecting the best variants with regard to its biophysical properties and antigen affinity. The fact that several of the aspects that stood out form our analysis have been incorporated in existing antibody design workflows (Lapidoth et al., 2015; Adolf-Bryfogle et al., 2018) further underlines the validity of our findings.
Supplementary Material
Funding
The VIB Switch Laboratory was supported by grants from VIB, University of Leuven, the Funds for Scientific Research Flanders (FWO), the Flanders Institute for Science and Technology (IWT) and the Federal Office for Scientific Affairs of Belgium (Belspo), IUAP P7/16 and by the European Research Council under the European Union’s Horizon 2020 Framework Programme, ERC Grant agreement 647458 (MANGO) to JS. RVDK was supported by Boehringer Ingelheim Pharma GmbH & Co and VIB.
Author contributions
JS, FR, JB, MB and PG designed the project. RVDK, JS and FR carried out the bioinformatics analysis. All authors contributed to data interpretation. RVDK, JB, JS and FR wrote the paper with input from all other authors.
Competing financial interests
MB, SK, AK, JB, PG were employees of Boehringer Ingelheim Pharma GmbH & Co. KG during the course of the project.
References
- Abhinandan K.R. and Martin A.C.R. (2008) Mol. Immunol., 45, 3832–3839. [DOI] [PubMed] [Google Scholar]
- Acharya P., Tolbert W.D., Gohain N. et al. (2014) J. Virol., 88, 12895–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolf-Bryfogle J., Kalyuzhniy O., Kubitz M., Weitzner B.D., Hu X., Adachi Y., Schief W.R. and Dunbrack R.L. Jr. (2018) PLoS Comput. Biol., 14, e1006112 10.1371/journal.pcbi.1006112. First published on 2018/04/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolf-Bryfogle J., Xu Q., North B., Lehmann A. and Dunbrack R.L. Jr. (2015) Nucleic Acids Res., 43, D432–D438. 10.1093/nar/gku1106. First published on 2014/11/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Lazikani B., Lesk A.M. and Chothia C. (1997) J. Mol. Biol., 273, 927–948. 10.1006/jmbi.1997.1354. First published on 1998/02/12. [DOI] [PubMed] [Google Scholar]
- Alzari P.M., Lascombe M.B. and Poljak R.J. (1988) Annu. Rev. Immunol., 6, 555–580. 10.1146/annurev.iy.06.040188.003011. First published on 1988/01/01. [DOI] [PubMed] [Google Scholar]
- Baca M., Presta L.G., O’Connor S.J. and Wells J.A. (1997) J. Biol. Chem., 272, 10678–10684. First published on 1997/04/18. [DOI] [PubMed] [Google Scholar]
- Bogan A.A. and Thorn K.S. (1998) J. Mol. Biol., 280, 1–9. [DOI] [PubMed] [Google Scholar]
- Boulianne G.L., Hozumi N. and Shulman M.J. (1984) Nature, 312, 643–646. First published on 1984/12/13. [DOI] [PubMed] [Google Scholar]
- Breiman L. (2001), Machine Learning. unknown.
- Bruggemann M., Caskey H.M., Teale C., Waldmann H., Williams G.T., Surani M.A. and Neuberger M.S. (1989) Proc. Natl Acad. Sci. USA., 86, 6709–6713. First published on 1989/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann M. and Neuberger M.S. (1996) Immunol. Today, 17, 391–397. First published on 1996/08/01. [DOI] [PubMed] [Google Scholar]
- Bruggemann M., Osborn M.J., Ma B., Hayre J., Avis S., Lundstrom B. and Buelow R. (2015) Arch. Immunol. Ther. Exp. (Warsz), 63, 101–108. 10.1007/s00005-014-0322-x. First published on 2014/12/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Presta L., Gorman C.M. et al. (1992) Proc. Natl Acad. Sci. USA., 89, 4285–4289. First published on 1992/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P., Van Regenmortel M., Weiss E. and Baty D. (2009) Br. J. Pharmacol., 157, 220–233. 10.1111/j.1476-5381.2009.00190.x. First published on 2009/05/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A., Silva D.A., Rocklin G.J. et al. (2017) Nature, 550, 74–79. 10.1038/nature23912. First published on 2017/09/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W.C., Lai Y.P. and Chou M.Y. (2011) PLoS One, 6, e16373 10.1371/journal.pone.0016373. First published on 2011/02/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. and Lesk A.M. (1987) J. Mol. Biol., 196, 901–917. [DOI] [PubMed] [Google Scholar]
- Chothia C., Novotny J., Bruccoleri R. and Karplus M. (1985) J. Mol. Biol., 186, 651–663. First published on 1985/12/05. [DOI] [PubMed] [Google Scholar]
- Clackson T. and Wells J.A. (1995) Science, 267, 383–386. [DOI] [PubMed] [Google Scholar]
- Co M.S. and Queen C. (1991) Nature, 351, 501–502. 10.1038/351501a0. First published on 1991/06/06. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M. and Brenner S.E. (2004) Genome Res., 14, 1188–1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo S., Ferrara F., Naranjo L., Erasmus M.F., Hraber P. and Bradbury A.R.M. (2018) Front. Immunol., 9, 395 10.3389/fimmu.2018.00395. First published on 2018/03/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman G.M. (1959) J. Am. Chem. Soc., 81, 3155–3156. 10.1021/ja01521a071. [DOI] [Google Scholar]
- Ewert S., Huber T., Honegger A. and Plückthun A. (2003) J. Mol. Biol., 325, 531–553. [DOI] [PubMed] [Google Scholar]
- Eylenstein R., Weinfurtner D., Härtle S., Strohner R., Böttcher J., Augustin M., Ostendorp R. and Steidl S. (2016) MAbs, 8, 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber C., Shan L., Fan Z., Guddat L.W., Furebring C., Ohlin M., Borrebaeck C.A. and Edmundson A.B. (1998) Immunotechnology, 18, 253–270. [DOI] [PubMed] [Google Scholar]
- Fiser A., Do R.K. and Sali A. (2000) Protein Sci., 9, 1753–1773. 10.1110/ps.9.9.1753. First published on 2000/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote J. and Winter G. (1992) J. Mol. Biol., 224, 487–499. First published on 1992/03/20. [DOI] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S. and Li W. (2012) Bioinformatics, 28, 3150–3152. 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M.S.A., Moin S.M., Mas V. et al. (2015) PLoS Pathog., 17, e1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny M.K., Sampson J., Li H. et al. (2011) PLoS One, 6, e27780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Gu L., Eils R., Schlesner M. and Brors B. (2014) Bioinformatics, 30, 2811–2812. 10.1093/bioinformatics/btu393. First published on 2014/06/16. [DOI] [PubMed] [Google Scholar]
- Guan Y., Pazgier M., Sajadi M.M. et al. (2013) Proc. Natl Acad. Sci. USA, 110, E69–E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge A. and Thornton J. (2004) FEBS Lett., 567, 67–73. 10.1016/j.febslet.2004.03.067. First published on 2004/05/29. [DOI] [PubMed] [Google Scholar]
- Haidar J.N., Yuan Q.A., Zeng L., Snavely M., Luna X., Zhang H., Zhu W., Ludwig D.L. and Zhu Z. (2012) Proteins, 80, 896–912. 10.1002/prot.23246. First published on 2011/12/20. [DOI] [PubMed] [Google Scholar]
- Halabi N., Rivoire O., Leibler S. and Ranganathan R. (2009) Cell, 138, 774–786. 10.1016/j.cell.2009.07.038. First published on 2009/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Dyer M.J., Clark M.R., Phillips J.M., Marcus R., Riechmann L., Winter G. and Waldmann H. (1988) Lancet, 2, 1394–1399. First published on 1988/12/17. [DOI] [PubMed] [Google Scholar]
- Hanf K.J., Arndt J.W., Chen L.L. et al. (2014) Methods, 65, 68–76. 10.1016/j.ymeth.2013.06.024. First published on 2013/07/03. [DOI] [PubMed] [Google Scholar]
- Harris R.J., Shire S.J. and Winter C. (2004) Drug Develop. Res., 61, 137–154. 10.1002/ddr.10344. [DOI] [Google Scholar]
- Honegger A. and Pluckthun A. (2001) J. Mol. Biol., 309, 657–670. 10.1006/jmbi.2001.4662. [DOI] [PubMed] [Google Scholar]
- Hong M., Lee P.S., Hoffman R.M.B. et al. (2013) J. Virol., 87, 12471–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-c., Venturi M., Majeed S. et al. (2004) Proc. Natl Acad. Sci. USA, 101, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.Y. and Foote J. (2005) Methods, 36, 3–10. 10.1016/j.ymeth.2005.01.001. First published on 2005/04/26. [DOI] [PubMed] [Google Scholar]
- Jardine J.G., Kulp D.W., Havenar-Daughton C. et al. (2016) Science, 351, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.T., Dear P.H., Foote J., Neuberger M.S. and Winter G. (1986) Nature, 321, 522–525. 10.1038/321522a0. First published on 1986/05/04. [DOI] [PubMed] [Google Scholar]
- Julien J.-P., Bryson S., Nieva J.L. and Pai E.F. (2008) J. Mol. Biol., 384, 377–392. [DOI] [PubMed] [Google Scholar]
- Kassambara A. (2017).
- Kaufmann B., Vogt M.R., Goudsmit J., Holdaway H.A., Aksyuk A.A., Chipman P.R., Kuhn R.J., Diamond M.S. and Rossmann M.G. (2010) Proc. Natl Acad. Sc.i USA., 107, 18950–18955. 10.1073/pnas.1011036107. First published on 2010/10/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough C.A., Saldanha J., Heath V.J., Morrison C.J. and Bendig M.M. (1991) Protein Eng., 4, 773–783. First published on 1991/10/01. [DOI] [PubMed] [Google Scholar]
- Knappik A., Ge L., Honegger A. et al. (2000) J. Mol. Biol., 296, 57–86. 10.1006/jmbi.1999.3444. First published on 2000/02/05. [DOI] [PubMed] [Google Scholar]
- Koenig P., Lee C.V., Sanowar S., Wu P., Stinson J., Harris S.F. and Fuh G. (2015) J. Biol. Chem., 290, 21773–21786. 10.1074/jbc.M115.662783. First published on 2015/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P., Lee C.V., Walters B.T., Janakiraman V., Stinson J., Patapoff T.W. and Fuh G. (2017) Proc. Natl Acad. Sci. USA., 114, E486–E495. 10.1073/pnas.1613231114. First published on 2017/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Lee J.H., Doores K.J. et al. (2013) Nat. Struct. Mol. Biol., 20, 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E. and Vriend G. (2014) Bioinformatics, 30, 2981–2982. 10.1093/bioinformatics/btu426. First published on 2014/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunik V. and Ofran Y. (2013) Protein Eng. Des. Sel., 26, 599–609. 10.1093/protein/gzt027. First published on 2013/06/12. [DOI] [PubMed] [Google Scholar]
- Lapidoth G.D., Baran D., Pszolla G.M., Norn C., Alon A., Tyka M.D. and Fleishman S.J. (2015) Proteins, 83, 1385–1406. 10.1002/prot.24779. First published on 2015/02/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Leaman D.P., Kim A.S. et al. (2015) Nat. Commun., 6, 8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts T., Ferkinghoff-Borg J., Schymkowitz J. and Rousseau F. (2009. a) BMC Syst. Biol., 3, 9 10.1186/1752-0509-3-9. First published on 2009/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts T., Ferkinghoff-Borg J., Stricher F., Serrano L., Schymkowitz J.W. and Rousseau F. (2008) BMC Struct. Biol., 8, 43 10.1186/1472-6807-8-43. First published on 2008/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts T., Schymkowitz J. and Rousseau F. (2009. b) Curr. Protein Pept. Sci., 10, 133–145. First published on 2009/04/10. [DOI] [PubMed] [Google Scholar]
- Li W. and Godzik A. (2006) Bioinformatics, 22, 1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu X., Taylor R.D., Griffin L., Coker S.F., Adams R., Ceska T., Shi J., Lawson A.D. and Baker T. (2017) Sci. Rep., 7, 41306 10.1038/srep41306. First published on 2017/01/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S.W. and Ranganathan R. (1999) Science, 286, 295–299. [DOI] [PubMed] [Google Scholar]
- Lonberg N. (2008) Handb. Exp. Pharmacol., 181, 69–97. 10.1007/978-3-540-73259-4_4. First published on 2007/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J., Griffiths A.D., Winter G. and Chiswell D.J. (1990) Nature, 348, 552–554. 10.1038/348552a0. First published on 1990/12/06. [DOI] [PubMed] [Google Scholar]
- Meyer P.E. (2008), PhD thesis of the Universite Libre de Bruxelles.
- Morrison S.L., Johnson M.J., Herzenberg L.A. and Oi V.T. (1984) Proc. Natl. Acad. Sci. USA., 81, 6851–6855. First published on 1984/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M.S., Williams G.T., Mitchell E.B., Jouhal S.S., Flanagan J.G. and Rabbitts T.H. (1985) Nature, 314, 268–270. First published on 1985/03/21. [DOI] [PubMed] [Google Scholar]
- Niemi M.H., Takkinen K., Amundsen L.K., Soderlund H., Rouvinen J. and Hoyhtya M. (2011) J. Mol. Recognit., 24, 209–219. 10.1002/jmr.1039. First published on 2011/03/02. [DOI] [PubMed] [Google Scholar]
- North B., Lehmann A. and Dunbrack R.L. Jr. (2011) J. Mol. Biol., 406, 228–256. 10.1016/j.jmb.2010.10.030. First published on 2010/11/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak J., Baker T., Georges G., Kelm S., Klostermann S., Shi J., Sridharan S. and Deane C.M. (2016) MAbs, 8, 751–760. 10.1080/19420862.2016.1158370. First published on 2016/03/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyen D., Torres J.L., Wille-Reece U. et al. (2017) Proc. Natl. Acad. Sci. USA., 114, E10438–E10445. 10.1073/pnas.1715812114. First published on 2017/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X.Y., Holliger P., Murzin A.G. and Williams R.L. (1997) Proc. Natl. Acad. Sci. USA., 94, 9637–9642. First published on 1997/09/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov A.V., Zou X., Xian J., Nicholson I.C. and Brüggemann M. (1999) J. Exp. Med., 189, 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Schneider W.P., Selick H.E. et al. (1989) Proc. Natl. Acad. Sci. USA., 86, 10029–10033. First published on 1989/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan G., Smart J., Williams J. and Almagro J.C. (2012) J. Mol. Recognit., 25, 103–113. 10.1002/jmr.2158. First published on 2012/03/13. [DOI] [PubMed] [Google Scholar]
- Riechmann L., Clark M., Waldmann H. and Winter G. (1988) Nature, 332, 323–327. 10.1038/332323a0. First published on 1988/03/24. [DOI] [PubMed] [Google Scholar]
- Rivoire O., Reynolds K.A. and Ranganathan R. (2016) PLoS Comput. Biol., 12, e1004817 10.1371/journal.pcbi.1004817. First published on 2016/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez E.R., Ledezma-Candanoza L.M., Contreras-Ferrat L.G., Olamendi-Portugal T., Possani L.D., Becerril B. and Riano-Umbarila L. (2012) J. Mol. Biol., 423, 337–350. 10.1016/j.jmb.2012.07.007. First published on 2012/07/28. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz J.W.H., Wilkinson H.R. and Itzhaki L.S. (2001) Proc. Natl. Acad. Sci. U. S. A., 98, 5596–5601. 10.1073/pnas.101542098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2016), RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. http://www.rstudio.com/.
- Sanchez I.E., Beltrao P., Stricher F., Schymkowitz J., Ferkinghoff-Borg J., Rousseau F. and Serrano L. (2008) PLoS Comput. Biol., 4, e1000052 10.1371/journal.pcbi.1000052. First published on 2008/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiele F., van Ryn J., Litzenburger T., Ritter M., Seeliger D. and Nar H. (2015) MAbs, 7, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F. and Serrano L. (2005) Nucleic Acids Res., 33, W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedrak P., Hsu K. and Mohan C. (2003) Mol. Immunol., 40, 491–499. First published on 2003/10/18. [DOI] [PubMed] [Google Scholar]
- Shannon C.E. (1948) Bell Syst. Tech. J., 27, 379–423. [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B. and Ideker T. (2003) Genome Res., 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman F.B., Norel R. and Honig B. (2000) Curr. Opin. Struct. Biol., 10, 153–159. First published on 2000/04/08. [DOI] [PubMed] [Google Scholar]
- Spada S., Honegger A. and Pluckthun A. (1998) J. Mol. Biol., 283, 395–407. 10.1006/jmbi.1998.2068. First published on 1998/10/14. [DOI] [PubMed] [Google Scholar]
- Stanfield R.L., Zemla A., Wilson I.A. and Rupp B. (2006) J. Mol. Biol., 357, 1566–1574. [DOI] [PubMed] [Google Scholar]
- Suel G.M., Lockless S.W., Wall M.A. and Ranganathan R. (2003) Nat. Struct. Biol., 10, 59–69. [DOI] [PubMed] [Google Scholar]
- Sun L.K., Curtis P., Rakowicz-Szulczynska E., Ghrayeb J., Chang N., Morrison S.L. and Koprowski H. (1987) Proc. Natl. Acad. Sci. USA., 84, 214–218. First published on 1987/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindells M.B., Porter C.T., Couch M., Hurst J., Abhinandan K.R., Nielsen J.H., Macindoe G., Hetherington J. and Martin A.C.R. (2017) J. Mol. Biol., 429, 356–364. [DOI] [PubMed] [Google Scholar]
- Titani K., Wikler M. and Putnam F.W. (1967) Science, 155, 828–835. First published on 1967/02/17. [DOI] [PubMed] [Google Scholar]
- Tramontano A., Chothia C. and Lesk A.M. (1990) J. Mol. Biol., 215, 175–182. 10.1016/S0022-2836(05)80102-0. First published on 1990/09/05. [DOI] [PubMed] [Google Scholar]
- Vajdos F.F., Adams C.W., Breece T.N., Presta L.G., de Vos A.M. and Sidhu S.S. (2002) J. Mol. Biol., 320, 415–428. 10.1016/S0022-2836(02)00264-4. First published on 2002/06/25. [DOI] [PubMed] [Google Scholar]
- Vanhee P., Verschueren E., Baeten L., Stricher F., Serrano L., Rousseau F. and Schymkowitz J. (2011) Nucleic Acids Res, 39, D435–D442. 10.1093/nar/gkq972. First published on 2010/10/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeyen M., Milstein C. and Winter G. (1988) Science, 239, 1534–1536. First published on 1988/03/25. [DOI] [PubMed] [Google Scholar]
- Wagih O. (2017. a) Bioinformatics, 33, 3645–3647. 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009) Elegant Graphics for Data Analysis. Springer-Verlag, Springer, New York, 260. [Google Scholar]
- Xiang J., Sha Y., Jia Z., Prasad L. and Delbaere L.T. (1995) J. Mol. Biol., 253, 385–390. 10.1006/jmbi.1995.0560. First published on 1995/10/27. [DOI] [PubMed] [Google Scholar]
- Zafra Ruano A., Cilia E., Couceiro J.R., Ruiz Sanz J., Schymkowitz J., Rousseau F., Luque I. and Lenaerts T. (2016) PLoS Comput. Biol., 12, e1004938 10.1371/journal.pcbi.1004938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. and Li J. (2010) BMC Struct. Biol., 10, S6 10.1186/1472-6807-10-S1-S6. First published on 2010/05/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang B., Zhu J., Nussinov R. and Ma B. (2018) Biochim. Biophys. Acta, 1864, 2294–2303. 10.1016/j.bbadis.2017.12.009. First published on 2017/12/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.