Abstract
Emerging evidence has revealed that long noncoding RNAs (lncRNAs) play crucial roles in the development and progression of tumors. The present study aimed to examine the roles and illustrate the underlying mechanisms of lncRNA ferritin heavy chain 1 pseudogene 3 (FTH1P3) in cervical cancer. The expression of lncRNA FTH1P3 and microRNA-145 (miRNA-145 or miR-145) in human cervical cancer samples and cervical cancer cell lines was detected by qRT-PCR (reverse transcription-quantitative polymerase chain reaction). FTH1P3 overexpression, siRNA plasmid, hsa-miR-145 mimic or hsa-miR-145 inhibitor were transfected. The target of FTH1P3 was predicted by bioinformatics analysis and validated by luciferase assay. Statistical significance analysis was performed by SPSS software. The results revealed that FTH1P3 was significantly upregulated in cervical cancer tissues compared with normal tissues. Increased expression of FTH1P3 was revealed in human cervical cancer cell lines compared with cervical normal epithelial cells. Downregulation of FTH1P3 inhibited cell proliferation, invasion and migration, and promoted apoptosis in cervical cancer cells. miR-145 was predicted and validated as a direct target of FTH1P3. Moreover, FTH1P3 siRNA partially attenuated the effects of the miR-145 inhibitor on cell viability and mobility in cervical cancer cells. The present results demonstrated that lncRNA FTH1P3 functioned as a promoting factor in cervical cancer by targeting miR-145.
Keywords: ferritin heavy chain 1 pseudogene 3, miRNA-145, cervical cancer, proliferation, invasion, migration
Introduction
Cervical cancer ranks as the 4th most common female malignancy and the 2nd most common cancer in developing countries, which affects women globally (1–3). Surgery or a combination of chemotherapy and radiotherapy are the routine treatments for cervical cancer patients (4–7). However, the malignancy is largely incurable for the patient. The early stage of cervical cancer is predisposed to develop progressively into an advanced stage. Therefore, early detection and prognosis are crucial. It is critical to understand the molecular mechanisms of cervical cancer development and identify novel biomarkers for the early detection and treatment of cervical cancer.
Long non-coding RNAs (lncRNAs) emerged as important molecules involved in normal development and in tumorigenesis (8). Growing evidence has revealed that lncRNAs are involved in the aberrant pathological development of cervical cancer (9). For example, reduced lncRNA MEG3 (maternally expressed 3) is associated with increased cervical cancer cell proliferation (10). LncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) markedly increased after HPV infection in cervical cancer (11). FTH1P3 belongs to long non-coding RNAs, and is a member of the ferritin heavy chain (FHC) gene family located at chromosome 2p23.3 (12). FTH1P3 is widely expressed in different human cell lines and tissues and positively modulated during cell differentiation (12). Zhang et al revealed that FTH1P3 is a promoting factor in the growth and progression of OSCC by stimulating cell proliferation, migration, and invasion (13). However, the role of FTH1P3 in cervical cancer has not yet been elaborated.
MicroRNAs (miRNAs or miRs), ~18–25 nucleotides, are another component of the noncoding RNA family. They participate and regulate gene expression through a post-transcriptional pattern (14–16). miRNAs are aberrantly expressed in various types of malignancies and function either as oncogenes or as tumor suppressors (17–19). Accumulating evidence has demonstrated that miRNAs regulated various carcinogenesis processes, including cell maturation, cell proliferation, migration, invasion, autophagy, apoptosis, and metastasis (20–26). Therefore, miRNAs have a large potential to serve as promising markers in the diagnosis, prognosis, and personalized targeted therapies (22–26). miR-145 plays a tumor-suppressive role in several types of cancer, such as gastric (27), hepatic (28), breast (29), non-small cell lung (30), and cervical cancer (31). Zhou et al reported that miRNA-145 inhibited tumorigenesis and invasion of cervical cancer stem cells by inducing cancer stem cell (CSC) differentiation through downregulation of the stem cell transcription factors that maintain CSC pluripotency (32). Sathyanarayanan et al stated that miRNA-145 modulated epithelial-mesenchymal transition (EMT) and suppressed proliferation, migration, and invasion by targeting SIP1 in human cervical cancer cells (33).
However, the reason that miR-145 was downregulated in cervical cancer remains obscure. In our previous bioinformatics analysis (unpublished data), it was revealed that FTH1P3 was possibly regulating miR-145 by anti-sense sequence matching. Since miR-145 has a wide range of functions in a variety of tumors, the study aimed to determine whether FTH1P3 can target miR-145 in cervical cancer. In the present study, the functions and associations between FTH1P3 and miR-145 in cervical cells and tissues were demonstrated. The present findings may further provide a therapeutic target for the treatment and diagnosis of cervical cancer patients.
Materials and methods
Patients and materials
Fifty-two cervical cancer tissues (all females, 44–69 years, 55.7±5.5 years) were collected from patients who underwent surgery at our hospital from March 2015 to March 2017. The samples of patients with other major diseases were excluded. Normal cervix healthy tissues were used as a normal control. The dissected patient surgical specimen was immediately transferred to the surgical laboratory. Written consent was signed from each patient. The implemented protocol was approved by the Human Ethics Committee of Gansu Provincial Cancer Hospital. All the tissues were either fixed in 4% PFA (paraformaldehyde) or were snap-frozen in liquid nitrogen for later use. Cervical cancer patients (52) were divided into high- and low-expression groups according to the median values of FTH1P3 (fold change=4.32) and miR-145 (fold change=0.41) expression.
Cell culture
Human cervical cancer cell lines (SiHa, HeLa, CaSki, and C4-1) and normal cervical epithelial cells (Ect1/E6E7) were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM (Dulbecco's Modified Eagle's Medium) supplemented with 10% heated inactivated fetal bovine serum (Hi-FBS) and 100 U/ml penicillin-streptomycin (10,000 U/ml; all from Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C, and 5% CO2 in a humidified incubator.
Bioinformatics analysis
The binding candidates of FTH1P3 were predicted using miRcode software (http://www.mircode.org/). All parameters were default.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
Total RNA was extracted using TRIzol™ reagent (Thermo Fisher Scientific, Inc.). RNA concentrations were measured using an ND-1000 UV–Vis Spectrophotometer (Thermo Fisher Scientific, Inc.), and the quality was monitored by an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). qRT-PCR was carried out using a TaqMan miRNA Assay according to the manufacturer's protocol (Applied Biosystems; Thermo Fisher Scientific, Inc.). The amplification conditions were: 40 cycles of 15 sec at 95°C and 1 min at 60°C. The expression levels of miR-145 and FTH1P3 were normalized by U6.
Cell transfection
The hsa-miRNA-145 mimic/miRNA-145 scramble (negative-control for miRNA-145 mimic) and hsa-miRNA-145 inhibitor/miRNA-145 scramble (negative-control for miRNA-145 inhibitor) were designed and synthesized from Shanghai GeneChem Co., Ltd. The FTH1P3 siRNAs were synthesized by Invitrogen; Thermo Fisher Scientific, Inc. FTH1P3 siRNA (targeted region) sequences were: siRNA_1, CGCCUGUAAUCCCAGCUCUCA; siRNA_2, AUAAGCGUAACUUCCCUCAAA; siRNA_3, CGUAACUUCCCUCAAAGCAACAACC. pcDNA-3.1(+)-FTH1P3 (lncRNA-FTH1P3). The overexpression plasmid of FTH1P3 was purchased from Shanghai GeneChem Co., Ltd. A transfection assay was conducted using a Lipofectamine 3000 kit (Thermo Fisher Scientific, Inc.). The sequence of miRNA-145 mimic, miRNA-145 inhibitor or respective control were as follows: miRNA-145 mimic: 5′-UCCCUAAGGACCCUUUUGACCUG-3′ (sense) 3′-AGGGAUUCCUGGGAAAACUGGAC-5′ (antisense); miRNA-145 scramble (negative-control for miRNA-145 mimic): 5′-CUAUCCACCAGGUUGCUUUGACC-3′ (sense) 3′-GAUAGGUGGUCCAACGAAACUGG-5′ (antisense); miRNA-145 inhibitor: 5′-CAGGUCAAAAGGGUCCUUAGGGA-3′; miRNA-145 scramble (negative-control for miRNA-145 inhibitor): 5′-GUCCAGAGGAAACUUGUCGAAGG-3′.
CCK-8 analysis
Cell Counting Kit-8 (CCK-8) was used to determine the cell viability in cell proliferation and cytotoxicity. Briefly, human cervical cancer cells were seeded in a 96-well plate (5×103 cells/well) overnight. After cell confluence reached 70–80%, the miR-145 mimic or FTH1P3 siRNA was transfected into the cells using Lipofectamine 3000 kit (Thermo Fisher Scientific, Inc.). After 2 days, the CCK-8 reagent (5 mg/ml) was directly loaded into each well and incubated in the dark for 2 h at 37°C followed by measurement of the absorbance at OD 490 nm. Each measurement was repeated in triplicate.
Flow cytometric analysis
After transfection with the desired plasmid or negative control in 6-well plates, the cells were cultured for 48 h. Annexin V-FITC (3 µg/ml) and propidium iodide (5 µg/ml) were incubated with cells for 30 min at RT shielded from the light. Then the cells were collected, filtered and subjected to a BD FACSAria Fusion Cell Sorter (BD Biosciences) with Cytomics FC 500 with CXP Software (Beckman Coulter, Inc.). The data were quantified with FlowJo software v10.6.1 (FlowJo LLC).
Invasion and migration assays
For the invasion assay, 2×104 HeLa cells in FBS-free DMEM were placed in the upper chamber of an insert (8-µm pores; Merck KGaA). Matrigel (Merck KGaA) was employed to pre-coat the membrane of the Transwell chambers. The lower chambers were incubated in culture medium supplemented with 10% FBS for 24 h. Then, the HeLa cells on the upper surface were scraped and washed away. Subsequently, the cells on the lower surface were fixed and stained with Diff-Quik staining kit (Sysmex Corp.) at RT for 2 h. The invaded cells in the lower surface of membrane were also fixed and stained with Diff-Quik kit (Sysmex Corp.). The invaded cells were imaged and counted under a BX43 Upright Microscope (Olympus Corp.) at a magnification of ×200 from 10 random fields in each well. For the migration assay, 1.5×106 cells/well were seeded in 6-well plate for overnight culture until the cells reached an ~90% confluence. The scratch was generated using a 200-µl sterile pipet tip in the hood. After aspiration and washing, fresh complete medium was added. Then the cells were cultured for another 24 h. Cell migration was monitored under a BX43 Upright Microscope (Olympus Corporation) and images were captured at 0 and 24 h.
Luciferase reporter assays
The fragments of the 3′-UTR of FTH1P3 were amplified with Phusion® High-Fidelity DNA Polymerase (NEB, Inc.) from human genomic DNA. After gel extraction, the PCR product was cloned into the multiple cloning sites located downstream of the firefly luciferase coding region of the pGL3 Luciferase Reporter Vectors (Promega, USA). The recombinant vector was named pMIR-FTH1P3-wild-type. The mutations in the miR-145 binding sites were introduced using Site-Directed Mutagenesis (Thermo Fisher Scientific, Inc.). The mutation sequence was confirmed by sequencing (Shengong Bio Company). The resulting vector was designated as pMIR-BCYRN-mutant. pMIR-FTH1P3 or pMIR-FTH1P3-mut vector and miR-145 mimic or miR-145 inhibitor were co-transfected into a 24-well plate according to the Lipofectamine 3000 kit (Thermo Fisher Scientific, Inc.). After 36 h, the cells were harvested and lysed (lysis buffer; Promega Corporation). The luciferase reporter gene assay was detected by the GloMax® 20/20 Luminometer (Promega Corporation) according to the instructions.
Statistical analysis
Statistical analysis was performed using SPSS 13.0 (Pearson). All data were expressed as the mean ± standard deviation (SD). Correlation between FTH1P3 and miR-145 expression was determined using Spearman's correlation analysis. Differences were assessed by two-sample t-test or chi-square test or one-way ANOVA and Fisher's LSD post hoc tests. P<0.05 was considered to indicate a statistically significant difference.
Results
FTH1P3 expression is upregulated in cervical cancer tissues and cell lines
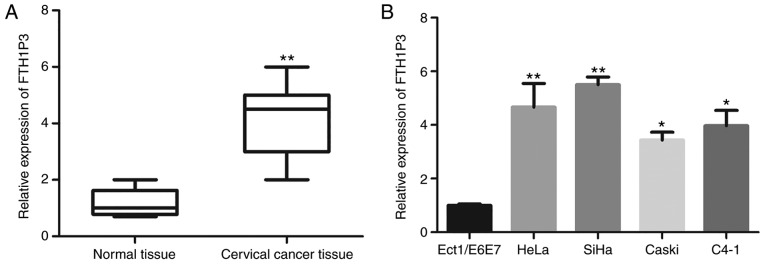
To investigate the expression of lncRNA FTH1P3 in cervical cancer tissues, total RNA was extracted from 52 cervical cancer tissues and paired adjacent non-cancerous normal tissues. qRT-PCR was carried out to detect FTH1P3 mRNA expression level. As revealed in Fig. 1A, FTH1P3 expression was significantly upregulated in cervical cancer tissues compared to paired adjacent non-cancerous normal tissues (P<0.01). Furthermore, the correlation between FTH1P3 expression and pathological parameters of cervical cancer patients were evaluated. The results of Table I revealed that FTH1P3 upregulation in cervical cancer tissues was significantly correlated with crucial clinicopathological factors, including tumor size (P=0.014), lymph node metastasis (P=0.005) and FIGO stage (P=0.002). However, there was no significant difference between FTH1P3 expression and other clinicopathological factors, such as age, menopause, depth of invasion and CEA level. Next, the expression of FTH1P3 in cervical cancer cell lines was further investigated. As revealed in Fig. 1B, it was determined that FTH1P3 expression was significantly increased in cervical cancer cell lines (HeLa, SiHa, Caski, and C4-1) compared to cervical normal epithelial cells (ECT1/E6E7). Notably, higher expression of FTH1P3 was observed in the HeLa and SiHa cell lines. Therefore, HeLa and SiHa cells were selected for further studies.
Figure 1.
FTH1P3 expression is upregulated in cervical cancer. (A) Relative expression of FTH1P3 in cervical cancer tissues and adjacent normal tissues was determined by qRT-PCR (**P<0.01 compared to normal tissues). (B) Expression of FTH1P3 in human cervical cancer cell lines (SiHa, HeLa, CaSki and C4-1) and cervical normal epithelial cells (Ect1/E6E7) was determined using qRT-PCR (*P<0.05 and **P<0.01 compared to Ect1/E6E7 cells). FTH1P3, ferritin heavy chain 1 pseudogene 3.
Table I.
Correlation analysis between FTH1P3 expression and clinicopathological index in cervical cancer patients.
| FTH1P3 | miR-145 | ||||||
|---|---|---|---|---|---|---|---|
| Parameters | n | Low (n) | High (n) | P-value | Low (n) | High (n) | P-value |
| Age (years) | |||||||
| <50 | 24 | 10 | 14 | 0.101 | 11 | 13 | 0.525 |
| ≥50 | 28 | 16 | 12 | 15 | 13 | ||
| Menopause | |||||||
| Yes | 22 | 10 | 12 | 0.438 | 12 | 10 | 0.438 |
| No | 30 | 16 | 14 | 14 | 16 | ||
| Tumor size (cm) | |||||||
| <4 | 18 | 12 | 6 | 0.014a | 7 | 11 | 0.035a |
| ≥4 | 34 | 14 | 20 | 19 | 15 | ||
| Depth of invasion | |||||||
| <2/3 | 21 | 12 | 9 | 0.085 | 11 | 10 | 0.621 |
| ≥2/3 | 31 | 14 | 17 | 15 | 16 | ||
| LNM stage | |||||||
| Negative | 35 | 21 | 14 | 0.005b | 15 | 20 | 0.011a |
| Positive | 17 | 5 | 12 | 11 | 6 | ||
| FIGO stage | |||||||
| I–II | 27 | 20 | 7 | 0.002b | 17 | 10 | 0.017a |
| III–IV | 25 | 6 | 19 | 9 | 16 | ||
| CEA | |||||||
| Negative | 33 | 18 | 15 | 0.083 | 16 | 17 | 0.662 |
| Positive | 19 | 8 | 11 | 10 | 9 | ||
P<0.05
P<0.01. FTH1P3, ferritin heavy chain 1 pseudogene 3.
FTH1P3 silencing inhibits cell viability and motility in cervical cancer cell lines
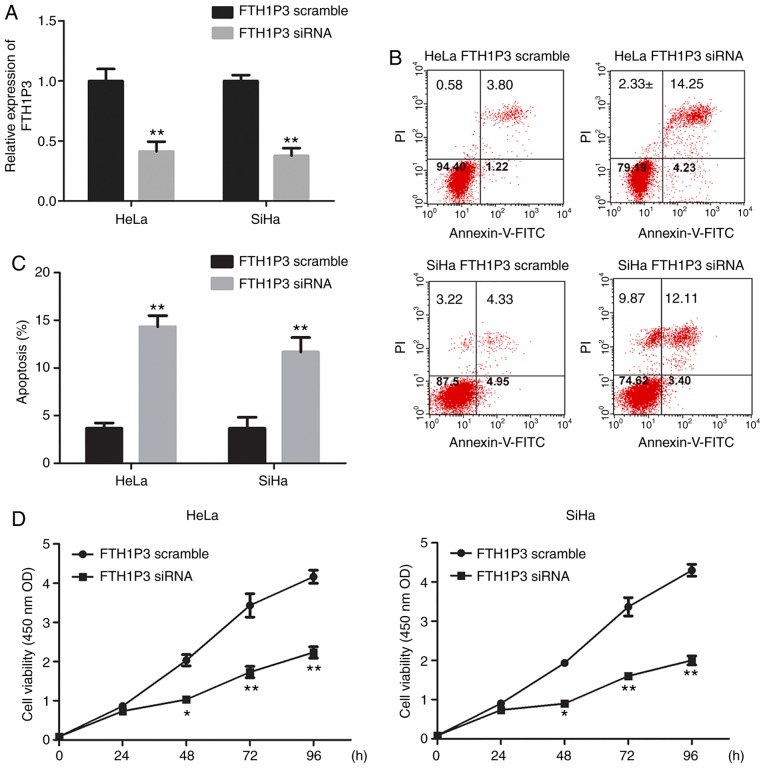
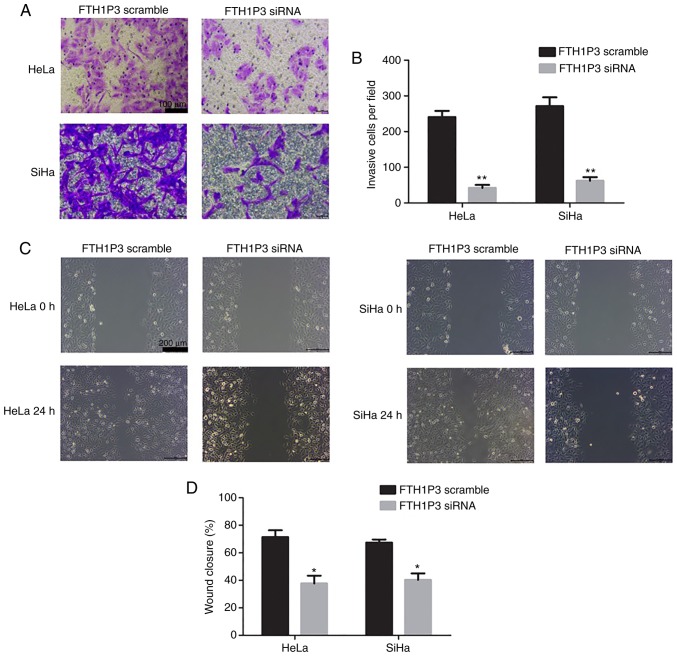
To determine the roles of FTH1P3 in cervical cancer, siRNA of FTH1P3 was constructed and transfected into HeLa and SiHa cells. As revealed in Fig. 2A, siRNA of FTH1P3 significantly suppressed the expression of FTH1P3 in both HeLa and SiHa cells (P<0.01). Furthermore, downregulation of FTH1P3 by siRNA induced significant apoptosis in HeLa and SiHa cells (Fig. 2B and C; P<0.01). CCK-8 assay also revealed that FTH1P3 downregulation significantly inhibited the proliferation of HeLa and SiHa cells (Fig. 2D; P<0.01). In addition, the effects of FTH1P3 on cell motility were examined by Transwell and wound-healing assays. The results revealed that the number of invasive cells was significantly reduced after transfection with FTH1P3 siRNA (P<0.01; Fig. 3A and B). Similarly, a significantly decreased closure rate of scratch wounds was observed in the FTH1P3 siRNA group compared with the siRNA scramble group in HeLa and SiHa cells (P<0.05; Fig. 3C and D).
Figure 2.
FTH1P3 silencing inhibits cell proliferation and promotes apoptosis in cervical cancer cells. HeLa and SiHa cells were transfected with FTH1P3 siRNA or siRNA scramble, respectively. (A) FTH1P3 expression in cervical cancer cells was assessed using qRT-PCR. (B and C) Cell apoptosis in cervical cancer cells was detected using flow cytometry. (D) Cell proliferation was monitored using CCK-8 assay. Data are presented as the means ± SD; three independent experiments were replicated. *P<0.05 and **P<0.01 compared to the FTH1P3 scramble group. FTH1P3, ferritin heavy chain 1 pseudogene 3.
Figure 3.
Downregulation of FTH1P3 inhibits the cell invasion and migration in cervical cancer cell lines. (A and B) The invasive capability of cervical cancer cells was detected by Transwell assay. (C and D) The migration of cervical cancer cells was measured by wound-healing assays. *P<0.05 and **P<0.01 compared to the FTH1P3 scramble group. FTH1P3, ferritin heavy chain 1 pseudogene 3.
FTH1P3 is identified as a potential target of miR-145
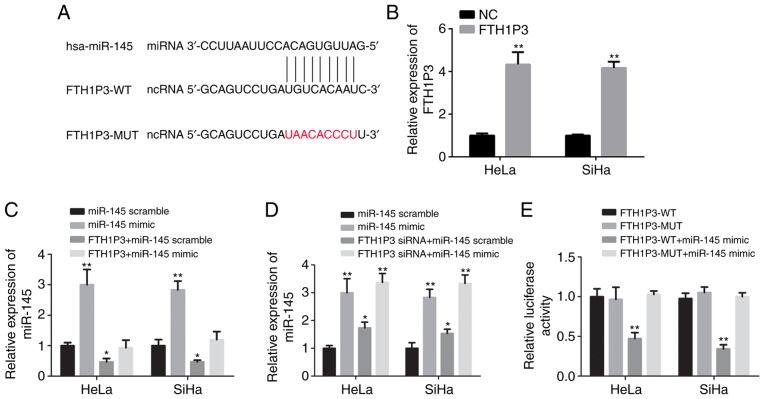
Bioinformatics analysis with miRcode software was performed and it was revealed that human miR-145 (hsa-miR-145) was a potential binding candidate of FTH1P3 (Fig. 4A). As revealed in Fig. 4B, FTH1P3 overexpression plasmid significantly increased the expression of FTH1P3 in both HeLa and SiHa cells compared with the negative control (P<0.01). After transfection of human miR-145 mimic, a significant increase of miR-145 expression in HeLa and SiHa cells was observed (Fig. 4C; P<0.01). The overexpression of FTH1P3 by plasmid transfection could significantly reduce the expression of miR-145, while miR-145 expression was inversely increased due to the downregulation of FTH1P3 (Fig. 4C and D; P<0.05). In addition, a luciferase reporter assay revealed that miR-145 mimic significantly suppressed the luciferase activities of FTH1P3-WT reporter vector. Conversely, after transfection with FTH1P3-MUT and the miRNA-145 mimic, the luciferase activities between these two cells were nearly comparable with that in the control cells (Fig. 4E; P<0.01). These data indicated that FTH1P3 was a potential target of miR-145.
Figure 4.
miR-145 is identified as a potential target of FTH1P3. (A) Alignment of potential FTH1P3 base pairing with miR-145 as identified by miRcode. (B) Cervical cancer cells were transfected with the overexpression plasmid of FTH1P3 [pcDNA-3.1(+)-FTH1P3] or negative control. Expression of lncRNA FTH1P3 was measured by qRT-PCR. (C and D) Cervical cancer cells were transfected with lncRNA FTH1P3 or siRNA FTH1P3 combined with miR-145 mimic or mimic control. Expression of miR-145 was assessed by qRT-PCR. (E) Wild-type (lncR-FTH1P3-WT) or mutant (lncR-FTH1P3-Mut) luciferase reporter and/or miR-145 mimic were co-transfected into cervical cancer cells to determine the luciferase activity. Data are presented as the means ± SD of three independent experiments. *P<0.05 and **P<0.01 compared with lncRNA-FTH1P3 WT group. MicroRNA-145, miR-145; FTH1P3, ferritin heavy chain 1 pseudogene 3.
miRNA-145 is downregulated in cervical cancer
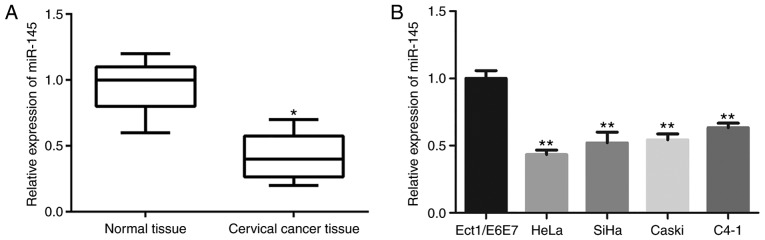
To investigate the role of miR-145 in cervical cancer progression, miR-145 expression in cervical cancer tissues and non-malignant tissues was detected by qRT-PCR. As revealed in Fig. 5A, miR-145 expression in cervical cancer tissues was significantly downregulated compared with the non-malignant tissues (P<0.01). Consistently, the expression of miR-145 in five assessed cervical cancer cell lines were all significantly lower than that in normal cervical epithelial cells (Ect1/E6E7) (P<0.01; Fig. 5B). Moreover, the downregulated expression of miR-145 demonstrated a significant association with tumor size (P=0.035), lymph node metastasis (P=0.011) and FIGO stage (P=0.017). Spearman's correlation analysis revealed that miR-145 expression was inversely associated with FTH1P3 expression in cervical cancer tissues (Table II, r=−0.265, P=0.002), indicating that upregulated FTH1P3 expression in cervical cancer was correlated with downregulated miR-145 expression.
Figure 5.
miR-145 expression is downregulated in cervical cancer. (A) miR-145 expression was determined by qRT-PCR in cervical cancer tissues and adjacent normal tissues (*P<0.05 compared to normal tissues). (B) Expression of miR-145 in human cervical cancer cell lines (SiHa, HeLa, CaSki and C4-1) and cervical normal epithelial cells (Ect1/E6E7) was determined using qRT-PCR (**P<0.01 compared to Ect1/E6E7 cells). MicroRNA-145, miR-145.
Table II.
Correlation between FTH1P3 expression and miR-145 expression in cervical cancer patients.
| miR-145 Expression | ||||||
|---|---|---|---|---|---|---|
| FTH1P3 expression | n | Low (n, %) | High (n, %) | rs | χ2 | P-value |
| Low (n, %) | 26 | 8 | 18 | −0.265 | 9.102 | 0.002 |
| High (n, %) | 26 | 18 | 8 | |||
FTH1P3, ferritin heavy chain 1 pseudogene 3.
Effects of miR-145 inhibitor on the cell proliferation, invasion, and migration in cervical cancer cells
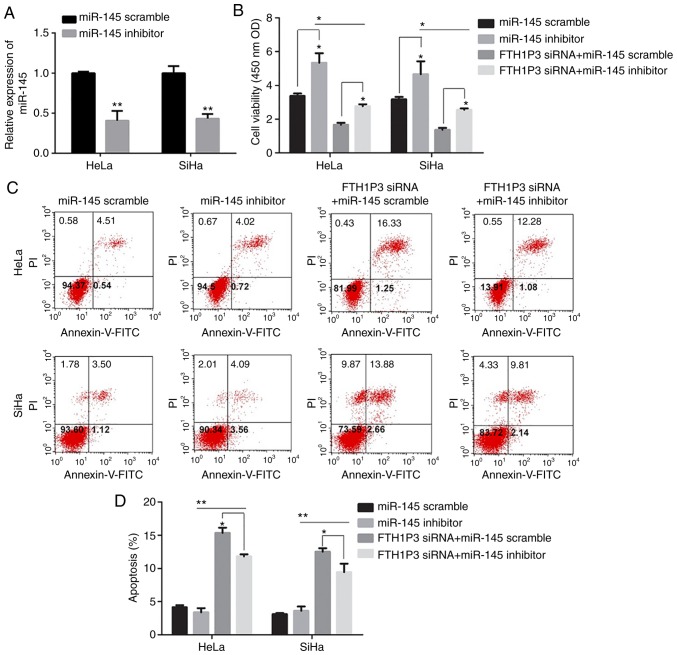
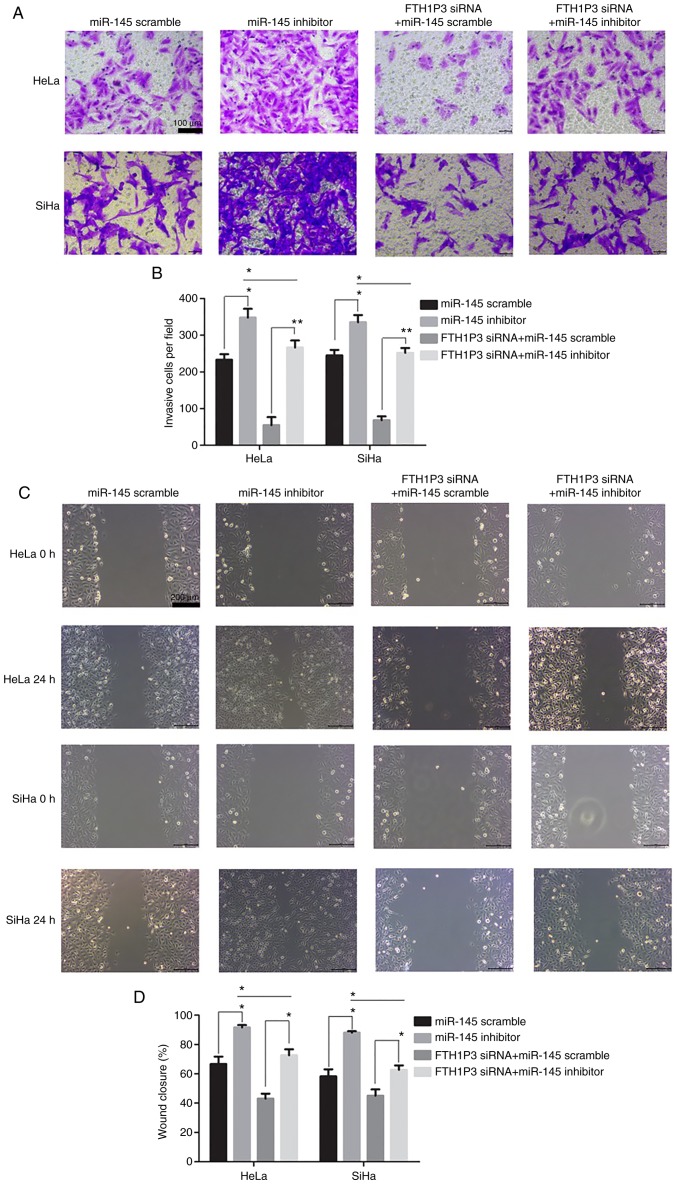
As illustrated in Fig. 6A, the transfection of miR-145 inhibitor caused a significant reduction of miR-145 in cervical cancer cells (P<0.01). Moreover, downregulation of FTH1P3 significantly inhibited the proliferation, invasion and migration abilities and promoted apoptosis in HeLa and SiHa cells (Figs. 6 and 7). Notably, the miRNA-145 inhibitor could enhance the cell proliferation, invasion and migration in cervical cancer cells compared to the RNA scramble group, while FTH1P3 siRNA revealed a significant bucking effect for miRNA-145 inhibitor in HeLa and SiHa cells compared to cells transfected with FTH1P3 siRNA+ RNA scramble group (Figs. 6 and 7).
Figure 6.
Effects of miR-145 inhibitor on cell proliferation and apoptosis in FTH1P3-siRNA-treated cervical cancer cells. (A) miR-145 expression was determined by qRT-PCR in HeLa and SiHa cells after transfection with miR-145 inhibitor. (B) Cell proliferation of HeLa and SiHa cells was detected using CCK-8 assay. (C and D) Cell apoptosis percentage was assessed using flow cytometry. *P<0.05 and **P<0.01 compared to the corresponding group. MicroRNA-145, miR-145; FTH1P3, ferritin heavy chain 1 pseudogene 3.
Figure 7.
Effects of miR-145 inhibitor on cell invasion and migration in FTH1P3-siRNA-treated cervical cancer cells. (A and B) Cell invasion was detected using Transwell assay. (C and D) Cell motility was detected using wound healing assay. *P<0.05 and **P<0.01 compared to the corresponding group. MicroRNA-145, miR-145; FTH1P3, ferritin heavy chain 1 pseudogene 3.
Discussion
The present study revealed that FTH1P3 was highly expressed in malignant cervical cancer tissues and cell lines. Downregulation of FTH1P3 significantly inhibited cell proliferation, invasion and migration, and promoted apoptosis in cervical cancer cells. The FTH1P3 and miR-145 axis was demonstrated, to have reciprocal repression functionally. These results demonstrated that lncRNA FTH1P3 functioned as a promoting factor in cervical cancer by targeting miR-145.
FTH1P3 expression was revealed to be enhanced in uveal melanoma cell lines and tissues, and FTH1P3 upregulation promoted cell proliferation, the cell cycle and migration in uveal melanoma by targeting miR-224-5p (34). FTH1P3 was abundantly expressed in paclitaxel-resistant breast cancer tissue and cells (35). FTH1P3 was revealed to play a role as a competing endogenous RNA (ceRNA) to sponge miR-206 and increase ABCB1 (ATP-binding cassette subfamily B member 1) protein expression (35). FTH1P3 was overexpressed in oral squamous cell carcinoma (OSCC) tissues (13). Overexpression of of FTH1P3 significantly promoted OSCC cell growth, while the cell growth was inhibited after knockout of FTH1P3 (13). In the present study, it was also revealed that FTH1P3 was highly expressed in malignant cervical cancer tissues and human cervical cancer cell lines (SiHa, HeLa, CaSki and C4-1) (Fig. 1). Collectively, these studies indicated that FTH1P3 may function as an oncogene and be favorable to tumorigenesis and progression of various cancers.
Emerging studies have indicated that miR-145 is implicated in a number of cancers, such as breast and cervical cancer, chondrosarcoma, colorectal and endometrial cancer, esophageal squamous cell carcinoma as well as gallbladder carcinoma (32,36,37), and its downregulation inhibited tumor proliferation, migration, invasion, metastasis, progression, and angiogenesis via distinctive signaling pathways. For instance, miR-145 downregulation inhibited invasion of bladder cancer cells by targeting PARK1 (38). Hsa-mir-145 downregulation was closely associated with aggressive progression and poor prognosis in human cervical cancer (31). miR-145 directly targeted p70S6K1 in cancer cells to inhibit colon cancer tumor growth and angiogenesis (39). miR-145 suppressed cell migration and invasion by targeting paxillin in human colorectal cancer cells (40). In endometrioid carcinomas, miR-145 and miR-143 were downregulated and associated with DNA methyltransferase 3B overexpression and worse prognosis (41). In the present study, miRNA-145 was predicted as a potential binding candidate of FTH1P33. Through in vitro transfection with lncRNA FTH1P3 or siRNA FTH1P3 combined with miR-145 mimic and mutant (lncRNA-FTH1P3-Mut) luciferase reporter, the association between FTH1P3 and miR-145 was further confirmed.
Moreover, it was revealed that miRNA-145 inhibitor could enhance cell proliferation, invasion, and migration in cervical cancer cells, while FTH1P3 siRNA exhibited a significant bucking effect for miRNA-145 inhibitor in HeLa and SiHa cells. FTH1P3 siRNA partially attenuated the effects of the miR-145 inhibitor on cell viability and mobility in cervical cancer cells, indicating that the FTH1P3-miR-145 axis functioned inversely in the cervical tumor.
It is worth noting that this study is limited by the small sample size. In addition, only in vitro cell experiments were performed. Future studies with a larger sample size and animal model experiments should be included to further confirm the conclusions.
In conclusion, the present study demonstrated the FTH1P3-miR-145 axis was involved in cell proliferation, invasion, migration, apoptosis, viability, and mobility in cervical cancer cells. These results may provide a potential therapeutic target and strategy to impede the progression of cervical cancer.
Acknowledgments
Not applicable.
Glossary
Abbreviations
- lncRNAs
long non-coding RNAs
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- FHC
ferritin heavy chain
- FTH1P3
ferritin heavy chain 1 pseudogene 3
- MEG3
maternally expressed 3
- miRNA
microRNA
- CSC
cancer stem cell
- EMT
epithelial-mesenchymal transition
- CCK-8
Cell Counting Kit-8
Funding
The present study was supported by Gansu Provincial Cancer Hospital.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
RL designed and conceived this study and performed the experiments and analysis of the data and wrote and supervised the manuscript. QWZ performed the research and manuscript revision. Both authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Gansu Provincial Cancer Hospital. All patients and healthy volunteers provided written informed consent prior to their inclusion in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zeferino LC, Derchain SF. Cervical cancer in the developing world. Best Pract Res Clin Obstet Gynaecol. 2006;20:339–354. doi: 10.1016/j.bpobgyn.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 2.Ohazurike E, Anorlu I, Okunade K, Okunowo A, Sajo A. Cervical cancer: An urgent need to scale up screening in the developing world. Int J Gynecol Cancer. 2017;27:842–842. [Google Scholar]
- 3.Nogueira-Rodrigues A, de Melo AC, Garces AHI, Paulino E, Alves FV, Vilaça Mdo N, Silva LG, Gonçalves CA, Fabrini JC, Carneiro AT, Thuler LC. Patterns of care and outcome of elderly women diagnosed with cervical cancer in the developing world. Int J Gynecol Cancer. 2016;26:1246–1251. doi: 10.1097/IGC.0000000000000756. [DOI] [PubMed] [Google Scholar]
- 4.Rao GG, Rogers P, Drake RD, Nguyen P, Coleman RL. Phase I clinical trial of weekly paclitaxel, weekly carboplatin, and concurrent radiotherapy for primary cervical cancer. Gynecol Oncol. 2005;96:168–172. doi: 10.1016/j.ygyno.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Falcetta FS, Medeiros LR, Edelweiss MI, Pohlmann PR, Stein AT, Rosa DD. Adjuvant platinum-based chemotherapy for early stage cervical cancer. Cochrane Database Syst Rev. 2016;11:CD005342. doi: 10.1002/14651858.CD005342.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosa DD, Medeiros LR, Edelweiss MI, Bozzetti MC, Pohlmann PR, Stein AT, Dickinson HO. Adjuvant platinum-based chemotherapy for early stage cervical cancer. Cochrane Database Syst Rev. 2009:CD005342. doi: 10.1002/14651858.CD005342.pub2. [DOI] [PubMed] [Google Scholar]
- 7.Rosa DD, Medeiros LR, Edelweiss MI, Pohlmann PR, Stein AT. Adjuvant platinum-based chemotherapy for early stage cervical cancer. Cochrane Database Syst Rev. 2012:CD005342. doi: 10.1002/14651858.CD005342.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin XJ, Chen XJ, Hu Y, Ying F, Zou R, Lin F, Shi Z, Zhu X, Yan X, Li S, Zhu H. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med. 2017;6:1409–1423. doi: 10.1002/cam4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–113. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, Zhou G, Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–2141. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Sanzo M, Aversa I, Santamaria G, Gagliardi M, Panebianco M, Biamonte F, Zolea F, Faniello MC, Cuda G, Costanzo F. FTH1P3, a Novel H-Ferritin pseudogene transcriptionally active, is ubiquitously expressed and regulated during cell differentiation. PLoS One. 2016;11:e0151359. doi: 10.1371/journal.pone.0151359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CZ. Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene. 2017;607:47–55. doi: 10.1016/j.gene.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Zovoilis A, Mungall AJ, Moore R, Varhol R, Chu A, Wong T, Marra M, Jones SJ. The expression level of small non-coding RNAs derived from the first exon of protein-coding genes is predictive of cancer status. EMBO Rep. 2014;15:402–410. doi: 10.1002/embr.201337950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhry MA. Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells. Int J Mol Sci. 2013;14:9099–9110. doi: 10.3390/ijms14059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol. 2012;24:333–340. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of MicroRNAs in human liver cancers. Semin Oncol. 2011;38:752–763. doi: 10.1053/j.seminoncol.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilmott JS, Zhang XD, Hersey P, Scolyer RA. The emerging important role of microRNAs in the pathogenesis, diagnosis and treatment of human cancers. Pathology. 2011;43:657–671. doi: 10.1097/PAT.0b013e32834a7358. [DOI] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A. From worms to humans: Understanding the role of microRNAs in cancer progression. Cancer Res. 2010;70:LB–353. [Google Scholar]
- 20.Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy- regulating microRNAs and cancer. Front Oncol. 2017;7:65. doi: 10.3389/fonc.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu R, Mo GQ, Duan Z, Huang M, Chang J, Li X, Liu P. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun Signal. 2014;12:45. doi: 10.1186/s12964-014-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad MK, Waseem M, Serajuddin M, Mahdi AA, Sankhwar SN, Mishra DP. MicroRNA: A new potential marker for prostate cancer. Cancer Med. 2018;7:17–17. [Google Scholar]
- 23.Hironaka-Mitsuhashi A, Matsuzaki J, Takahashi RU, Yoshida M, Nezu Y, Yamamoto Y, Shiino S, Kinoshita T, Ushijima T, Hiraoka N, et al. A tissue microRNA signature that predicts breast cancer recurrence in young women. PLoS One. 2017;12:e0187638. doi: 10.1371/journal.pone.0187638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldassari F, Zerbinati C, Galasso M, Corrà F, Minotti L, Agnoletto C, Previati M, Croce CM, Volinia S. Screen for MicroRNA and Drug interactions in breast cancer cell lines points to miR-126 as a modulator of CDK4/6 and PIK3CA inhibitors. Front Genet. 2018;9:174. doi: 10.3389/fgene.2018.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frixa T, Sacconi A, Cioce M, Roscilli G, Ferrara FF, Aurisicchio L, Pulito C, Telera S, Carosi M, Muti P, et al. MicroRNA-128-3p-mediated depletion of Drosha promotes lung cancer cell migration. Carcinogenesis. 2018;39:293–304. doi: 10.1093/carcin/bgx134. [DOI] [PubMed] [Google Scholar]
- 26.Heydari N, Nikbakhsh N, Sadeghi F, Farnoush N, Khafri S, Bastami M, Parsian H. Overexpression of serum MicroRNA-140-3p in premenopausal women with newly diagnosed breast cancer. Gene. 2018;655:25–29. doi: 10.1016/j.gene.2018.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Jiang SB, He XJ, Xia YJ, Hu WJ, Luo JG, Zhang J, Tao HQ. MicroRNA-145-5p inhibits gastric cancer invasiveness through targeting N-cadherin and ZEB2 to suppress epithelial-mesenchymal transition. Oncotargets Ther. 2016;9:2305–2315. doi: 10.2147/OTT.S101853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Noh JH, Chang YG, Kim MG, Jung KH, Kim JK, Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al. MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013;335:455–462. doi: 10.1016/j.canlet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Ding Y, Zhang C, Zhang J, Zhang N, Li T, Fang J, Zhang Y, Zuo F, Tao Z, Tang S, et al. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int J Oncol. 2017;50:1701–1710. doi: 10.3892/ijo.2017.3945. [DOI] [PubMed] [Google Scholar]
- 30.Wang M, Wang J, Deng J, Li X, Long W, Chang Y. MiR-145 acts as a metastasis suppressor by targeting metadherin in lung cancer. Med Oncol. 2015;32:344. doi: 10.1007/s12032-014-0344-6. [DOI] [PubMed] [Google Scholar]
- 31.Azizmohammadi S, Safari A, Azizmohammadi S, Kaghazian M, Sadrkhanlo M, Yahaghi E, Farshgar R, Seifoleslami M. Molecular identification of miR-145 and miR-9 expression level as prognostic biomarkers for early-stage cervical cancer detection. QJM. 2017;110:11–15. doi: 10.1093/qjmed/hcw101. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Yue Y, Wang R, Gong B, Duan Z. MicroRNA-145 inhibits tumorigenesis and invasion of cervical cancer stem cells. Int J Oncol. 2017;50:853–862. doi: 10.3892/ijo.2017.3857. [DOI] [PubMed] [Google Scholar]
- 33.Sathyanarayanan A, Chandrasekaran KS, Karunagaran D. microRNA-145 downregulates SIP1-expression but differentially regulates proliferation, migration, invasion and Wnt signaling in SW480 and SW620 cells. J Cell Biochem. 2018;119:2022–2035. doi: 10.1002/jcb.26365. [DOI] [PubMed] [Google Scholar]
- 34.Zheng X, Tang H, Zhao X, Sun Y, Jiang Y, Liu Y. Long non-coding RNA FTH1P3 facilitates uveal melanoma cell growth and invasion through miR-224-5p. PLoS One. 2017;12:e0184746. doi: 10.1371/journal.pone.0184746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Wang R, Zhang T, Yang Z, Jiang C, Seng J. Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J Cell Mol Med. 2018;22:4068–4075. doi: 10.1111/jcmm.13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Lu L, Feng B, Zhang K, Han S, Hou D, Chen L, Chu X, Wang R. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci Rep. 2017;7:4637. doi: 10.1038/s41598-017-04113-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Mak IW, Singh S, Turcotte R, Ghert M. The epigenetic regulation of SOX9 by miR-145 in human chondrosarcoma. J Cell Biochem. 2015;116:37–44. doi: 10.1002/jcb.24940. [DOI] [PubMed] [Google Scholar]
- 38.Kou B, Gao Y, Du C, Shi Q, Xu S, Wang CQ, Wang X, He D, Guo P. miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol Oncol. 2014;32:846–854. doi: 10.1016/j.urolonc.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761–774. doi: 10.1093/nar/gkr730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin J, Wang F, Jiang H, Xu J, Jiang Y, Wang Z. MicroRNA-145 suppresses cell migration and invasion by targeting paxillin in human colorectal cancer cells. Int J Clin Exp Patho. 2015;8:1328–1340. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Dong Y, Ti H, Zhao J, Wang Y, Li T, Zhang B. Down-regulation of miR-145 and miR-143 might be associated with DNA methyltransferase 3B overexpression and worse prognosis in endometrioid carcinomas. Hum Pathol. 2013;44:2571–2580. doi: 10.1016/j.humpath.2013.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.