Abstract
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer-related deaths among cancer patients. Genes correlated with the progression and prognosis of HCC are critically needed to be identified. In the present study, 3 Gene Expression Omnibus (GEO) datasets (GSE46408, GSE65372 and GSE84402) were used to analyze the differentially expressed genes (DEGs) between HCC and non-tumor liver tissues. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted to clarify the functional roles of DEGs. A protein-protein interaction network was established to screen the hub genes associated with HCC. The prognostic values of hub genes in HCC patients were analyzed using The Cancer Genome Atlas (TCGA) database. The expression levels of hub genes were validated based on ONCOMINE, TCGA and Human Protein Atlas (HPA) databases. Notably, 56 upregulated and 33 downregulated DEGs were markedly enriched under various GO terms and four KEGG terms. Among these DEGs, 10 hub genes with high connectivity degree were identified, including cyclin B1, cyclin A2, cyclin B2, condensin complex subunit 3, PDZ binding kinase, nucleolar and spindle-associated protein 1, aurora kinase A, ZW10 interacting kinetochore protein, protein regulator of cytokinesis 1 and kinesin family member 4A. The upregulated expression levels of these hub genes in HCC tissues were further confirmed by ONCOMINE, TCGA, and HPA databases. Additionally, the increased mRNA expression of each hub gene was related to the unfavorable disease-free survival and overall survival of HCC patients. The present study identified ten genes associated with HCC, which may help to provide candidate targets for the diagnosis and treatment of HCC.
Keywords: hepatocellular carcinoma, differently expressed genes, bioinformatics analysis, hub gene, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most commonly diagnosed cancers and the fourth most common cause of cancer-related deaths in the world (1). More than 466,100 people are diagnosed with liver cancer, and approximately 422,100 individuals succumb to liver cancer annually in China (2). Although multimodal therapies have been used to treat HCC in the past several decades, the therapeutic outcomes of HCC are still unsatisfactory due to post-surgical recurrence and treatment resistance. Moreover, although numerous genes and signaling pathways participating in the initiation and evolution of HCC have been extensively discussed, the mechanisms underlying HCC development and progression remain unclear. Recently, microarray technology coupled with bioinformatics tools has been used to identify the novel genes related to cancer progression, diagnosis and prognosis. The major public databases such as The Cancer Genome Atlas (TCGA), ONCOMINE, and Gene Expression Omnibus (GEO) are powerful tools used to screen the differentially expressed genes (DEGs) generated from microarray data corresponding to the carcinogenesis and progression of HCC (3–7). These tools can assist in the comprehension of the mechanisms behind the occurrence and progression of HCC, and identify novel targets for the diagnosis and prognosis of HCC. Thus, bioinformatics analysis is a feasible and valuable method to screen DEGs from microarray data and identify the core genes related to HCC progression and prognosis.
In the present study, several mRNA microarray datasets (GSE46408, GSE65372, and GSE84402) were selected from the GEO database, in order to identify the genes correlated to HCC progression and prognosis. Using the online tool GEO2R, DEGs between HCC and non-cancerous liver tissues were obtained. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted to further provide an overview of the function of the screened DEGs. A protein-protein interaction network (PPIN) was constructed to determine the hub genes associated with HCC. Survival analyses of the screened hub genes were carried out using cBioPortal, Kaplan-Meier plotter and Gene Expression Profiling Interactive Analysis (GEPIA). The expression levels of the identified hub genes were validated based on GEPIA, ONCOMINE, UCSC Xena browser and Human Protein Atlas (HPA) online databases.
Materials and methods
Microarray data source
To obtain the mRNA expression datasets of HCC, the following keywords: ‘Hepatocellular carcinoma’ and ‘Homo sapiens’[porgn: txid9606]’, and ‘Expression profiling by array’ were searched against the GEO database. After a systematic review, three GSE profiles (GSE46408, GSE65372, and GSE84402) were selected and downloaded. GSE46408, GSE65372 and GSE84402 were based on GPL4133 (Agilent-014850 Whole Human Genome Microarray 4×44K G4112), GPL14951 (Illumina HumanHT-12 WG-DASL V4.0 R2 expression beadchip) and GPL570 [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array], respectively. The array data for GSE46408, GSE65372 and GSE84402 consisted of 6 HCC patients vs. 6 controls, 17 HCC patients vs. 15 controls and 14 HCC patients vs. 14 controls, respectively. All data were freely accessible, and the present study did not involve any human or animal experimentation.
DEG identification
GEO2R was adopted to identify the DEGs between HCC and non-cancerous liver tissues. The log-fold change (FC) in expression and adjusted P-values (adj. P) were determined. The adj. P using the Benjamini-Hochberg method with default values were applied to correct the potential false-positive results. Genes that met the specific cut-off criteria of adj. P<0.05 and |logFC|>1.0 were regarded as DEGs. The intersecting genes were examined using the Venn diagram web tool. Visual hierarchical cluster analysis was also conducted to display the volcano plot of DEGs.
GO annotation and KEGG pathway enrichment analyses of DEGs
To reveal the functions of DEGs, Enrichr database was used to conduct GO annotation and KEGG pathway enrichment analyses (8). The GO terms were comprised of the following three divisions: biological process (BP), cellular component (CC) and molecular function (MF). Adj. P<0.05 was regarded as statistically significant.
Construction of PPIN and screening of hub genes
Search Tool for the Retrieval of Interacting Genes (STRING) (9) is a database used for analyzing the functional protein association networks. The screened DEGs had previously been submitted to the STRING database. All PPI pairs with a combined score of >0.4 were extracted. High-degree nodes appear crucial for ensuring the stability of the overall network. The degree of all nodes was calculated by Cytoscape (v3.6.1) plugin cytoHubba (10). In this experiment, the genes with the top 10 highest degree values were considered as hub genes.
Validation of hub genes
To validate the mRNA expression level of the screened hub genes in HCC vs. non-tumor liver tissues, ONCOMINE microarray database (https://www.oncomine.org) was used (11). Gene rank is the median rank for one target gene over all analyses. The threshold was defined as P=0.05, a 2-fold change and top 10% gene rank. The hierarchical clustering analysis of hub genes in primary liver cancer (TCGA Liver Cancer, n=438) was performed by UCSC Xena browser. The GEPIA database, containing data from 9,736 tumors and 8,587 controls (12), was employed to visualize the mRNA expression of each hub gene in liver hepatocellular carcinoma (LIHC) and non-cancerous liver samples. The protein expression levels of the 10 hub genes in human normal and HCC tissues were determined using the Human Protein Atlas (HPA), a website that contains immunohistochemistry-based expression data for approximately 20 most common types of cancers, 12 individual tumors in each cancer type (13).
Genetic alterations of hub genes
The LIHC (TCGA, Provisional) dataset, including the data of 442 samples, was selected for the analyses of the genetic alterations in hub genes using cBioPortal. This portal allows for the visualization, analysis and downloading of a large-scale cancer genomic dataset (14). The genomic alterations included gene mutations, copy number variations (GISTIC), mRNA expression z-scores (RNA Seq V2 RSEM) with a z-score threshold of ±2.0 and protein expression z-scores. In accordance with the online instructions of cBioPortal, the analyses on disease-free survival (DFS), progression-free survival (PFS) and overall survival (OS) were carried out.
Survival analyses for hub genes
Kaplan-Meier plotter is widely applied to explore the roles of 54,675 genes in OS based on 10,461 tumor samples from GEO, European Genome-phenome Archive and TCGA datasets including 364 patients with liver cancer. The relationship between OS and hub genes expressed in patients with liver cancer was evaluated by the Kaplan-Meier survival analysis (15). Moreover, the association between DFS and the genes expressed in LIHC patients was determined using the online tool GEPIA. The lower and upper 50% of gene expression were set as the standard for analysis. In the present study, HCC patients were categorized into 2 groups based on the median expression values of hub genes. Log-rank test results with P<0.01 were regarded as statistically significant.
Results
DEG identification
Following GSE46408 dataset analysis, 1,417 DEGs were successfully identified, including 1,066 upregulated and 351 downregulated genes. For the GSE65372 dataset, 985 DEGs involving 399 upregulated and 586 downregulated genes were observed. For the GSE84402 dataset, 1,218 DEGs were identified, including 675 upregulated and 543 downregulated genes. Venn analysis was conducted to examine the intersection among the DEG profiles. Among them, 89 DEGs were identified from the three profile datasets. Notably, 33 DEGs were markedly downregulated (Fig. 1A), while 56 DEGs were significantly upregulated (Fig. 1B) in HCC tissues compared to non-cancerous liver tissues (Table I). These 89 DEGs in GSE84402 were plotted in Fig. 1C, where the red and green dots represented the up and downregulated genes, respectively.
Figure 1.

Identification of common DEGs from GSE46408, GSE65372 and GSE84402 datasets. Venn diagram of (A) downregulated and (B) upregulated DEGs based on the three GEO datasets. (C) Volcano plot of the 89 DEGs. Red, upregulation; green, downregulation. The intersecting areas represent the commonly altered DEGs. The t-test was used to analyze DEGs, with the cut-off criteria of |logFC|>1.0 and adj. P<0.05. GSE46408 (6 HCC patients vs. 6 controls), GSE65372 (17 HCC patients vs. 15 controls), GSE84402 (14 HCC patients vs. 14 controls); DEG, differentially expressed gene; GEO, Gene Expression Omnibus; logFC, log-fold change; HCC, hepatocellular carcinoma.
Table I.
The common DEGs of three gene expression profiles (adj. P-val. <0.05, |logFC|>1.0).
| Common DEGs | Gene symbol |
|---|---|
| Upregulated DEGs | E2F8; TKT; HSPB1; TPX2; IGF2BP3; CCNB1; KPNA2; SMYD3; EZH2; HIST1H3B; KIF18B; CDC25C; ENAH; AURKA; FAM189B; KIF4A; SQLE; CENPN; ZWINT; CCNA2; PBK; NUF2; PTTG1; CDCA5; CKS2; TRIM24; ECT2; ASPM; ATAD2; MND1; CCNB2; MCM7; PRC1; CDT1; CENPW; CEP55; MCM4; KIF15; DLGAP5; CKAP2L; NVL; FAM83D; CDCA2; HMMR; GPC3; CENPI; ORC6; CDKN2A; SGO2; CDKN3; NCAPG; NEK2; CENPM; CENPF; NUSAP1; ESM1 |
| Downregulated DEGs | CXCL14; CLEC1B; CRHBP; IGFALS; DBH; MT1M; ECM1; FCN2; ASPA; KBTBD11; CLEC4M; TMEM27; BMPER; GPM6A; ANGPTL1; DPT; SLC25A47; COLEC10; LYVE1; CXCL12; NTF3; ANGPTL6; HHIP; FCN3; CCBE1; MASP1; MARCO; LCAT; VIPR1; HAMP; PTH1R; ADGRG7; SYT9 |
DEGs, differentially expressed genes.
GO annotation and KEGG pathway enrichment analyses
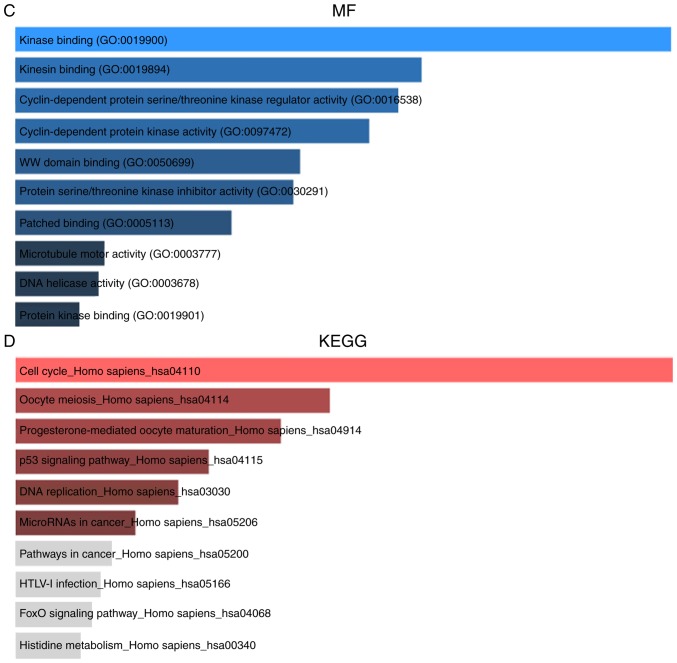
To obtain a deeper insight into the biological roles of these 89 DEGs, the Enrichr database (http://amp.pharm.mssm.edu/Enrichr/.) was employed to conduct GO annotation and KEGG pathway enrichment analyses. Fig. 2 lists the top 10 enriched GO terms and KEGG pathways. GO BP analysis revealed that these 89 DEGs were markedly enriched in the lectin pathway of complement activation, kinetochore organization, spindle microtubule attachment regulation to kinetochore and mitotic sister chromatid segregation (Fig. 2A). For GO CC analysis, the top four significantly enriched terms were microtubule cytoskeleton, mitotic spindle, chromosome centromeric region and spindle pole (Fig. 2B). The top four significantly enriched MF terms included kinase binding, kinesin binding, cyclin-dependent protein serine/threonine kinase regulator activity and cyclin-dependent protein kinase activity (Fig. 2C). In addition, the top four markedly enriched pathways for these 89 DEGs were cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation and p53 signaling pathway (Fig. 2D).
Figure 2.
GO annotation and KEGG pathway enrichment analysis of DEGs. The top 10 enriched GO (A) BP, (B) CC and (C) MF terms as well (D) KEGG pathways. GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; DEG, differentially expressed gene; BP, biological process; CC, cellular component; MF, molecular function.
PPIN construction and hub gene identification
The STRING database was adopted to determine the PPI pairs among the 89 DEGs. As revealed in Fig. 3A, 89 nodes (genes) and 715 edges (interactions) were established in the constructed PPIN (PPI enrichment P-value=1.0e-16). The top ten hub genes were identified based on their connectivity degree. The results revealed that cyclin B1 (CCNB1) was the most crucial gene with the highest connectivity degree=42, followed by cyclin A2 (CCNA2) at degree=41, cyclin B2 (CCNB2) at degree=40, condensin complex subunit 3 (NCAPG) at degree=40, PDZ binding kinase (PBK) at degree=39, nucleolar and spindle-associated protein 1 (NUSAP1) at degree=39, Aurora kinase A (AURKA) at degree=39, ZW10 interacting kinetochore protein (ZWINT) at degree=39, protein regulator of cytokinesis 1 (PRC1) at degree=38, and kinesin family member 4A (KIF4A) at degree=38 (Table II). The PPIN of the identified ten hub genes were also constructed, which indicated a strong interaction among each other (Fig. 3B). KEGG analysis revealed that the markedly enriched pathways for the 10 hub genes were progesterone-mediated oocyte maturation, the cell cycle, oocyte meiosis, cellular senescence, the p53 signaling pathway, the FOXO signaling pathway, human immunodeficiency virus 1 infection, and human T-cell leukemia virus 1 infection (Fig. 3C). The mRNA expression levels of these ten hub genes were markedly upregulated in HCC tissues.
Figure 3.
PPIN and hub gene identification. (A) PPIN was constructed by all the 89 DEGs using STRING database. (B) The top 10 hub genes in the PPIN were screened by Cytoscape (v3.6.1) plugin cytoHubba based on their connectivity degree. The 10 identified hub genes such as CCNB1, CCNA2, CCNB2, NCAPG, PBK, NUSAP1, AURKA, ZWINT, PRC1 and KIF4A are displayed from red (high degree value) to yellow (low degree value). (C) KEGG pathway enrichment analysis of the 10 hub genes. PPIN, protein-protein interaction network; DEG, differentially expressed gene; STRING, search tool for the retrieval of interacting genes; KEGG, Kyoto encyclopedia of genes and genomes.
Table II.
Top ten hub genes with higher degree of connectivity.
| Gene symbol | Gene description | Degree |
|---|---|---|
| CCNB1 | Cyclin B1 | 42 |
| CCNA2 | Cyclin A2 | 41 |
| CCNB2 | Cyclin B2 | 40 |
| NCAPG | Condensin complex subunit 3 | 40 |
| PBK | PDZ binding kinase | 39 |
| NUSAP1 | Nucleolar and spindle associated protein 1 | 39 |
| AURKA | Aurora kinase A | 39 |
| ZWINT | ZW10 interacting kinetochore protein | 39 |
| PRC1 | Protein regulator of cytokinesis 1 | 38 |
| KIF4A | Kinesin family member 4A | 38 |
Validation of mRNA expression of the top 10 hub genes in HCC
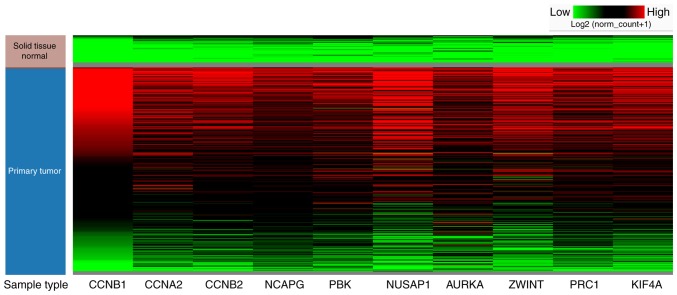
First, a meta-analysis on the mRNA expression levels of CCNB1, CCNA2, CCNB2, NCAPG, PBK, NUSAP1, AURKA, ZWINT, PRC1, and KIF4A between HCC and non-tumor liver tissues was performed using the ONCOMINE database. As demonstrated in Fig. 4, the mRNA expression levels of (Fig. 4A) CCNB1, (Fig. 4B) CCNA2, (Fig. 4C) CCNB2, (Fig. 4D) NCAPG, (Fig. 4E) PBK, (Fig. 4F) NUSAP1, (Fig. 4G) AURKA, (Fig. 4H) ZWINT, (Fig. 4I) PRC1, and (Fig. 4J) KIF4A were markedly upregulated in HCC tissues (P<0.05) compared to those in non-cancerous liver tissues. Furthermore, the median rank of NUSAP1 was the lowest (15) among the top 10 hub genes in HCC tissues (Fig. 4F). Hierarchical clustering analysis with UCSC Xena Browser also revealed that the mRNA expression levels of all the 10 hub genes were basically increased in primary hepatic cancer tissues compared to non-tumor tissue samples (Fig. 5). The results from the GEPIA database also revealed that the mRNA expression levels of all the 10 hub genes were significantly higher (P<0.01) in HCC tissues than those in normal liver tissues (Fig. 6). These findings were consistent with the obtained microarray data.
Figure 4.
Meta-analysis on the mRNA expression levels of (A) CCNB1, (B) CCNA2, (C) CCNB2, (D) NCAPG, (E) PBK, (F) NUSAP1, (G) AURKA, (H) ZWINT, (I) PRC1 and (J) KIF4A in HCC tissues vs. non-cancerous liver tissues using the five ONCOMINE datasets. The colored squares represent the median rank of these genes (vs. normal tissue) across the five datasets. The significance level for the median rank analysis was set at P<0.05. HCC, hepatocellular carcinoma.
Figure 5.
Hierarchical clustering analysis of the hub genes in HCC (n=371) and normal liver tissue (n=50) was conducted using the UCSC Xena browser. HCC, hepatocellular carcinoma.
Figure 6.
Validation of the mRNA expression levels of (A) CCNB1, (B) CCNA2, (C) CCNB2, (D) NCAPG, (E) PBK, (F) NUSAP1, (G) AURKA, (H) ZWINT, (I) PRC1, and (J) KIF4A in LIHC tissues and normal liver tissues using GEPIA. These ten box plots are based on 360 HCC samples (marked in red) and 160 normal samples (marked in gray). *P<0.01 was considered statistically significant. LIHC, liver hepatocellular carcinoma; HCC, hepatocellular carcinoma.
After examining the mRNA expression levels of the 10 hub genes in HCC, the protein expression levels of these hub genes in HCC were explored using the HPA database. Notably, the protein levels of (Fig. 7A) CCNB1, (Fig. 7B) CCNA2, (Fig. 7C) CCNB2, (Fig. 7D) NCAPG, (Fig. 7E) PBK, (Fig. 7F) NUSAP1, (Fig. 7G) AURKA and (Fig. 7I) PRC1 were not expressed in normal liver tissues, whereas medium and high expression levels of these genes were observed in liver cancer tissues (Fig. 7A-G and I). Moreover, the low protein expression levels of ZWINT and KIF4A were revealed in normal liver tissues, while medium protein expression levels of these genes were observed in liver cancer tissues (Fig. 7H and J). In summary, the present results indicated that the transcriptional and translational expression levels of the 10 hub genes were overexpressed in patients with HCC.
Figure 7.
Representative immunohistochemistry images of (A) CCNB1, (B) CCNA2, (C) CCNB2, (D) NCAPG, (E) PBK, (F) NUSAP1, (G) AURKA, (H) ZWINT, (I) PRC1, and (J) KIF4A in HCC and non-cancerous liver tissues derived from the HPA database. HCC, hepatocellular carcinoma; HPA, Human Protein Atlas.
Alteration in the frequency and prognostic values of hub genes
The frequencies of genetic alterations of the 10 hub genes in LIHC were evaluated using the cBioPortal database. Approximately 32.5% of LIHC clinical cases exhibited significant alterations in the 10 hub genes (Fig. 8A). The mRNA upregulation was one of the most important single factors for the altered 10 hub genes in 64 cases (17.78%) of LIHC. The mRNA expression (RNA Seq V2 RSEM) of the top 10 hub genes in LIHC was further analyzed. The results revealed that the percentage change in the mRNA expression levels of CCNB1, CCNA2, CCNB2, NCAPG, PBK, NUSAP1, AURKA, ZWINT, PRC1 and KIF4A in LIHC were 8, 1.9, 8, 5, 5, 8, 8, 7, 8 and 5%, respectively (Fig. 8B). Through the cBioportal database, the relationship between the changes in hub gene expression and LIHC prognosis was examined. Kaplan-Meier plots were used to compare DFS, PFS and OS in LIHC patients with or without alterations in the mRNA expression levels of the top 10 hub genes. As revealed in Fig. 8C, LIHC cases with altered hub gene expression exhibited significantly worse OS compared to those with unaltered hub gene expression (P=7.735e-3). Similarly, LIHC cases with altered hub gene expression displayed significantly worse DFS (P=0.0226) compared to those with unaltered hub gene expression (Fig. 8D).
Figure 8.
Alteration frequency and prognosis of the 10 hub genes. (A) The summary of the cancer types in the cBioPortal was used to calculate the percentages of LIHC cases with the 10 altered hub genes. (B) mRNA expression alterations (RNA Seq V2 RSEM) of the 10 hub genes in LIHC patients. (C) OS and (D) DFS/PFS of LIHC patients with altered (red) and unaltered (blue) mRNA expression of the 10 hub genes. LIHC, liver hepatocellular carcinoma; OS, overall survival; DFS/PFS, disease-free survival/progression-free survival.
Survival analysis of the hub genes in liver cancer
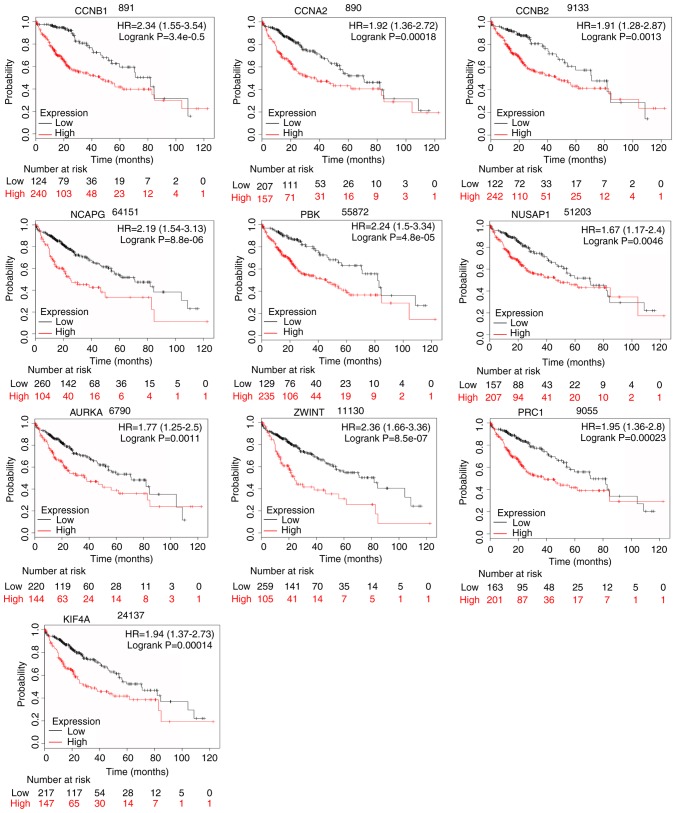
OS and DFS analyses of the 10 hub genes selected by PPI were further conducted by Kaplan-Meier plotter, bioinformatics analysis and the GEPIA database. As demonstrated in Fig. 9, the high expression levels of CCNB1, CCNA2, CCNB2, NCAPG, PBK, NUSAP1, AURKA, ZWINT, PRC1 and KIF4A in patients with liver cancer were associated with poor OS. The unfavorable DFS was also markedly observed in LIHC patients with increased expression levels of the top ten hub genes (Fig. 10).
Figure 9.
OS of the 10 hub genes overexpressed in patients with liver cancer was analyzed by Kaplan-Meier plotter. Data are presented as the hazard ratio with a 95% confidence interval. CCNB1, log-rank P=3.4e-05; CCNA2, log-rank P=0.00018; CCNB2, log-rank P=0.0013; NCAPG, log-rank P=8.8e-06; PBK, log-rank P=4.8e-05; NUSAP1, log-rank P=0.0046; AURKA, log-rank P=0.0011; ZWINT, log-rank P=8.5e-07; PRC1, log-rank P=0.00023; and KIF4A, log-rank P=0.00014. Log-rank P<0.01 was regarded as statistically significant. OS, overall survival.
Figure 10.
DFS of the 10 hub genes overexpressed in LIHC patients. Data are presented as the hazard ratio with a 95% confidence interval. CCNB1, log-rank P=2.8e-06; CCNA2, log-rank P=0.0037; CCNB2, log-rank P=0.0064; NCAPG, log-rank P=0.00246; PBK, log-rank P=0.006; NUSAP1, log-rank P=7e-04; AURKA, log-rank P=0.0012; ZWINT, log-rank P=7.8e-05; PRC1, log-rank P=0.00045; and KIF4A, log-rank P=0.0011. Log-rank P<0.01 was considered statistically significant. DFS, disease-free survival; LIHC, liver hepatocellular carcinoma.
Discussion
In the present experiments, bioinformatics analysis was performed to identify the potential key genes correlated with HCC. By comparing the three DEG profiles of HCC retrieved from the GEO database, 56 upregulated and 33 downregulated DEGs were successfully identified, respectively. Based on the degree of connectivity in PPIN, the top ten hub genes were ranked, including CCNB1, CCNA2, CCNB2, NCAPG, PBK, NUSAP1, AURKA, ZWINT, PRC1 and KIF4A. These identified hub genes were functioned as a group, and may play a crucial role in HCC.
KEGG enrichment analysis revealed that the cell cycle was the most significantly enriched pathway for these 89 DEGs. Cell-cycle deregulation is the major reason for the unlimited proliferation of cancer cells. Cyclins belong to a family of closely related proteins that drive the cell division cycle entry and progression, repair DNA damage, and control cell death by activating cyclin-dependent kinases (16). The up-regulation of cyclins causes cell-cycle deregulation and uncontrolled cell growth (17), indicating that cyclins play a vital role in the pathogenesis of cancer. CCNB1 (18,19), CCNA2 (20,21) and CCNB2 (22) are the founding members of the cyclin gene family, which regulate the proliferation, growth and apoptosis of cells, and have been associated with cancer progression and survival. CCNB1 (23,24), CCNA2 (20,25) and CCNB2 (22,26) have been identified in various types of tumors. Liu et al (3) revealed that CCNB1 and CCNB2 are highly expressed in HCC tissues compared to non-cancerous liver tissues. Based on the microarray studies of human liver tumors, CCNA2 was also overexpressed in human HCC tissues (27). The overexpression of CCNB1 (4,5,28,29) and CCNB2 (4) was correlated with poor OS and DFS in HCC patients by bioinformatics analysis. The results of the present study demonstrated that the upregulated levels of CCNB1 and CCNB2 significantly contributed to unfavorable OS and DFS in patients with HCC. In addition, it has been reported that CCNA2 is associated with a decrease in OS for patients with HCC, based on the survival and expression data from TCGA (30). In agreement, this study revealed that HCC patients with a low CCNA2 expression level exhibited longer OS and DFS compared to those with a high CCNA2 expression level.
NCAPG is the regulatory subunit of the condensin complex, and its related pathways are the cell cycle, mitotic and cell cycle chromosome condensation, which converts the interphase chromatin into mitotic-like condense chromosomes during mitosis and meiosis (31). Although the roles of NCAPG in cancers have not been studied extensively, it can potentially act as a novel oncogene for HCC progression. NCAPG knockdown suppresses the growth and proliferation of HCC cells (32,33), and inhibits the growth of HuH7 and HCCLM3 tumor xenografts (32). NCAPG was revealed to be overexpressed in HCC tissues, which contributed to the recurrence and OS of HCC patients (32,33). The overexpression of CCNB1 was revealed to exhibit a significant positive correlation with NCAPG overexpression in HCC patients (32). NCAPG has been identified as a hub gene in HCC in several studies by bioinformatics analysis (34–36). The findings of the present study demonstrated that the increased expression of NCAPG could confer a poor prognosis in HCC patients. Thus, NCAPG plays important roles in HCC progression, and serves as a novel therapeutic target for improving the treatment of HCC.
PBK is highly expressed in several cancers, including prostate cancer (37), breast cancer (38), gastric carcinoma (39), and lung cancer (40), and its overexpression may be correlated with tumor progression and poor prognosis. Although the clinical significance and biological role of PBK in HCC have not yet been extensively studied, PBK is suggested to function as an oncogene for HCC. First, Yue et al identified the upregulation of PBK as a potential prognostic biomarker for HCC by integrated GEO and TCGA datasets (41). Second, HCC patients with PBK overexpression were revealed to be more susceptible to increased tumor size, poor OS and DFS, occurrence of vascular invasion and incidence lymph node metastasis (42). Third, previous in vivo and in vitro data have demonstrated that PBK exerts an oncogenic role in HCC by activating the β-catenin signaling pathway (42). These results supported the present findings which revealed that the increased expression levels of PBK were markedly associated to poor OS and DFS in HCC patients.
NUSAP1 is a microtubule-associated protein that controls the cell cycle by promoting the aggregation of microtubules (43). Its expression levels are highly up-regulated in various tumor types, including prostate (44), pancreatic (45) and invasive breast cancers (46). In HCC patients, the expression of NUSAP1 was revealed to be upregulated, and its overexpression may serve as a prognostic factor (47). NUSAP1 knockdown could decrease the proliferation, migration and survival of tumor cells in a human liver cancer xenograft model (47). In this study, the present findings also revealed that the increased expression of NUSAP1 was correlated with poor OS and DFS in HCC patients.
AURKA is a mitotic serine/threonine kinase that is associated with the regulation of mitosis, cell division and cell cycle progression (48). The clinical role of AURKA in HCC has been studied extensively. AURKA overexpression has been detected in an HCC cell line (49) and HCC tissue samples (50,51). AURKA overexpression has been closely associated with the aggressive tumor characteristics (52), poor outcome (51) and chemoresistance (53) of HCC. The findings on AURKA gene polymorphisms have indicated that AURKA can act as a predictive biomarker for early-stage HCC (54). Another mechanistic study has revealed that AURKA can promote HCC metastasis by modulating epithelial-mesenchymal transition and cancer stem cell-like features (51). In HCC cells, AURKA at the transcriptional level was revealed to be regulated by c-Myc, which contributes to HCC progression (55). The inhibition of AURKA by alisertib (56), a compound that is currently being tested in phase II/III clinical trials of patients with hematological malignancies and solid tumors, can potentially reduce viability and induce apoptosis in HCC cells (49). Zhou et al revealed the overexpression of AURKA was negatively correlated with OS, based on the survival and expression data from TCGA (36). The present study also indicated that the increased expression levels of AURKA were associated with unfavorable OS and DFS in HCC patients.
ZWINT belongs to a component of a kinetochore complex, which can recruit ZW10 to kinetochores. The expression of ZWINT has been revealed to be downregulated in HCC (57). However, other findings have suggested that ZWINT mRNA and protein are overexpressed in HCC cell and tissue samples. The increased expression of ZWINT in HCC tissues was markedly correlated with tumor size and number, poor OS, and a great tendency for tumor recurrence (58). Furthermore, the overexpression of ZWINT increased the proliferation of HCC cells by modulating cell cycle-related proteins (58). Using bioinformatics analysis, the hub gene ZWINT was identified and higher expression of ZWINT in HCC predicted poor prognosis in several studies (34,35). The results of our studies also indicated that ZWINT could exert oncogenic effects rather than tumor inhibitory effects on HCC. However, these findings should be further investigated.
PRC1 is a microtubule-associated protein that is related to cell motion and microtubule dynamics. The expression level of PRC1 in HCC tissue samples has been revealed to be higher than that in non-tumor adjacent tissue samples (59). The increased expression of PRC1 in HCC patients was correlated with the decreased survival rates of HCC patients (59). PRC1 promoted early HCC recurrence by activating the Wnt/beta-catenin signaling pathway (60). Wang et al (61) reported that the high expression of PRC1 exacerbates chemoresistance in HCC cells. Li et al analyzed the HCC data from TCGA by weighted gene co-expression network analysis and identified PRC1 as a novel biomarker for HCC (34). The results of this study indicated that the increased expression of PRC1 was a prognostic indicator for DFS and OS in HCC patients.
KIF4A is a microtubule-based motor protein that regulates the segregation of chromosomes and organization of mitotic spindles during mitosis (62). KIF4A has been revealed to be highly expressed in HCC cells and tissues (63), and its overexpression in HCC patients indicates a poor prognosis (63,64). The findings on an HCC cell model overexpressing KIF4A revealed that KIF4A enhanced HCC cell survival and clonogenicity, by maintaining mitotic progression and protecting against cell death (63). KIF4A knockdown markedly decreased the proliferation and migration abilities of HCC-LM3 and PLC/PRF/5 (65). The hub gene KIF4A has been identified to be involved in the development of HCC by bioinformatics analysis (6). The findings of this study indicated that the upregulated expression of KIF4A could confer poor OS and DFS in HCC patients.
In summary, using three cohort profile datasets and integrated bioinformatics analysis, 10 HCC-associated hub genes were identified. The expression of the hub genes was revealed to be increased in HCC, and the overexpression level predicted poor prognosis. These results were consistent with previous studies (4–7,34–36,41). Further studies with larger sample sizes should be carried out to validate the present findings. Additionally, experimental evidence is warranted to investigate the functional roles of the identified genes in HCC. Collectively, it is our sincere hope that this present study will contribute to the discovery of novel diagnostic and prognostic biomarkers as well as therapeutic targets for HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the research funds from the National Natural Science Foundation of China (grant no. 81801559), the Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016259), the Suzhou Youth Science and Education Project (grant no. KJXW2016017).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
All authors contributed to the study concept and design, as well as the interpretation of the data. XS, RD, HG, MZ, WZ, CM and JM acquired and analyzed the data. XS, RD, CM and JM drafted the manuscript. CM and JM are responsible for the integrity of the work as a whole. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Yao X, Zhang D, Sheng J, Wen X, Wang Q, Chen G, Li Z, Du Z, Zhang X. Analysis of transcription factor-related regulatory networks based on bioinformatics analysis and validation in hepatocellular carcinoma. Biomed Res Int. 2018;2018:1431396. doi: 10.1155/2018/1431396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao X, Wang X, Zhang S. Bioinformatics identification of crucial genes and pathways associated with hepatocellular carcinoma. Biosci Rep. 2018;38 doi: 10.1042/BSR20181441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang L, Yang Z, Meng Z. Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted worse overall survival and disease-free survival in hepatocellular carcinoma patients. Biomed Res Int. 2018;2018:7897346. doi: 10.1155/2018/7897346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, Li L, Chen Y. The identification of core gene expression signature in hepatocellular carcinoma. Oxid Med Cell Longev. 2018;2018:3478305. doi: 10.1155/2018/3478305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Q, Sun Y, Zhou Q, He Q, Qian H. Identification of key genes and pathways by bioinformatics analysis with TCGA RNA sequencing data in hepatocellular carcinoma. Mol Clin Oncol. 2018;9:597–606. doi: 10.3892/mco.2018.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asplund A, Edqvist PH, Schwenk JM, Pontén F. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12:2067–2077. doi: 10.1002/pmic.201100504. [DOI] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Bergwelt-Baildon MS, Kondo E, Klein-González N, Wendtner CM. The cyclins: A family of widely expressed tumor antigens? Expert Rev Vaccines. 2011;10:389–395. doi: 10.1586/erv.10.170. [DOI] [PubMed] [Google Scholar]
- 18.Ding K, Li W, Zou Z, Zou X, Wang C. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses. 2014;83:359–364. doi: 10.1016/j.mehy.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Fang Y, Yu H, Liang X, Xu J, Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther. 2014;15:1268–1279. doi: 10.4161/cbt.29691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobashi Y, Shoji M, Jiang SX, Kobayashi M, Kawakubo Y, Kameya T. Active cyclin A-CDK2 complex, a possible critical factor for cell proliferation in human primary lung carcinomas. Am J Pathol. 1998;153:963–972. doi: 10.1016/S0002-9440(10)65638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volm M, Koomägi R, Mattern J, Stammler G. Cyclin A is associated with an unfavourable outcome in patients with non-small-cell lung carcinomas. Br J Cancer. 1997;75:1774–1778. doi: 10.1038/bjc.1997.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kühling H, Alm P, Olsson H, Fernö M, Baldetorp B, Parwaresch R, Rudolph P. Expression of cyclins E, A, and B, and prognosis in lymph node-negative breast cancer. J Pathol. 2003;199:424–431. doi: 10.1002/path.1322. [DOI] [PubMed] [Google Scholar]
- 23.Wang A, Yoshimi N, Ino N, Tanaka T, Mori H. Overexpression of cyclin B1 in human colorectal cancers. J Cancer Res Clin Oncol. 1997;123:124–127. doi: 10.1007/BF01269890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 25.Handa K, Yamakawa M, Takeda H, Kimura S, Takahashi T. Expression of cell cycle markers in colorectal carcinoma: Superiority of cyclin A as an indicator of poor prognosis. Int J Cancer. 1999;84:225–233. doi: 10.1002/(SICI)1097-0215(19990621)84:3<225::AID-IJC5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann HS, Hansen G, Burdach S, Bartling B, Silber RE, Simm A. Discrimination of human lung neoplasm from normal lung by two target genes. Am J Respir Crit Care Med. 2004;170:516–519. doi: 10.1164/rccm.200401-127OC. [DOI] [PubMed] [Google Scholar]
- 27.Andrisani OM, Studach L, Merle P. Gene signatures in hepatocellular carcinoma (HCC) Semin Cancer Biol. 2011;21:4–9. doi: 10.1016/j.semcancer.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin L, Chang C, Xu C. G2/M checkpoint plays a vital role at the early stage of HCC by analysis of key pathways and genes. Oncotarget. 2017;8:76305–76317. doi: 10.18632/oncotarget.19351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 2018;500:924–929. doi: 10.1016/j.bbrc.2018.04.201. [DOI] [PubMed] [Google Scholar]
- 30.Chen QF, Xia JG, Li W, Shen LJ, Huang T, Wu P. Examining the key genes and pathways in hepatocellular carcinoma development from hepatitis B virus-positive cirrhosis. Mol Med Rep. 2018;18:4940–4950. doi: 10.3892/mmr.2018.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen Y, Gutwein O, Garach-Jehoshua O, Bar-Haim A, Kornberg A. The proliferation arrest of primary tumor cells out-of-niche is associated with widespread downregulation of mitotic and transcriptional genes. Hematology. 2014;19:286–292. doi: 10.1179/1607845413Y.0000000125. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Su R, Shan C, Gao C, Wu P. Non-SMC condensin I complex, subunit G (NCAPG) is a novel mitotic gene required for hepatocellular cancer cell proliferation and migration. Oncol Res. 2018;26:269–276. doi: 10.3727/096504017X15075967560980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, Liang B, Liu H, Huang Y, Yin X, Zhou F, Yu X, Feng Q, Li E, Zou Z, Wu L. Overexpression of non-SMC condensin I complex subunit G serves as a promising prognostic marker and therapeutic target for hepatocellular carcinoma. Int J Mol Med. 2017;40:731–738. doi: 10.3892/ijmm.2017.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Pu K, Wu X. Identifying novel biomarkers in hepatocellular carcinoma by weighted gene co-expression network analysis. J Cell Biochem. 2019 Feb 11; doi: 10.1002/jcb.28420. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 35.Liu ZK, Zhang RY, Yong YL, Zhang ZY, Li C, Chen ZN, Bian H. Identification of crucial genes based on expression profiles of hepatocellular carcinomas by bioinformatics analysis. PeerJ. 2019;7:e7436. doi: 10.7717/peerj.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Du Y, Kong L, Zhang X, Chen Q. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther. 2018;11:1861–1869. doi: 10.2147/OTT.S156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, Zhang L, Shi C, Hu P, Yan W, Wang Z, Duan Q, Lu F, Qin L, Lu T, et al. TOPK is highly expressed in circulating tumor cells, enabling metastasis of prostate cancer. Oncotarget. 2015;6:12392–12404. doi: 10.18632/oncotarget.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JH, Lin ML, Nishidate T, Nakamura Y, Katagiri T. PDZ-binding kinase/T-LAK cell-originated protein kinase, a putative cancer/testis antigen with an oncogenic activity in breast cancer. Cancer Res. 2006;66:9186–9195. doi: 10.1158/0008-5472.CAN-06-1601. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Imamura T, Kiuchi J, Kosuga T, Konishi H, Shiozaki A, et al. Overexpression of PBK/TOPK relates to tumour malignant potential and poor outcome of gastric carcinoma. Br J Cancer. 2017;116:218–226. doi: 10.1038/bjc.2016.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei DC, Yeh YC, Hung JJ, Chou TY, Wu YC, Lu PJ, Cheng HC, Hsu YL, Kuo YL, Chen KY, Lai JM. Overexpression of T-LAK cell-originated protein kinase predicts poor prognosis in patients with stage I lung adenocarcinoma. Cancer Sci. 2012;103:731–738. doi: 10.1111/j.1349-7006.2011.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue C, Ren Y, Ge H, Liang C, Xu Y, Li G, Wu J. Comprehensive analysis of potential prognostic genes for the construction of a competing endogenous RNA regulatory network in hepatocellular carcinoma. Onco Targets Ther. 2019;12:561–576. doi: 10.2147/OTT.S188913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang YF, Pan YH, Cao Y, Fu J, Yang X, Zhang MF, Tian QH. PDZ binding kinase, regulated by FoxM1, enhances malignant phenotype via activation of β-catenin signaling in hepatocellular carcinoma. Oncotarget. 2017;8:47195–47205. doi: 10.18632/oncotarget.17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raemaekers T, Ribbeck K, Beaudouin J, Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg J, Carmeliet G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162:1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32:70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokkinakis DM, Liu X, Neuner RD. Modulation of cell cycle and gene expression in pancreatic tumor cell lines by methionine deprivation (methionine stress): Implications to the therapy of pancreatic adenocarcinoma. Mol Cancer Ther. 2005;4:1338–1348. doi: 10.1158/1535-7163.MCT-05-0141. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Pan Y, Fu H, Zhang J. Nucleolar and spindle associated protein 1 (NUSAP1) inhibits cell proliferation and enhances susceptibility to epirubicin in invasive breast cancer cells by regulating cyclin D kinase (CDK1) and DLGAP5 expression. Med Sci Monit. 2018;24:8553–8564. doi: 10.12659/MSM.910364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy S, Hooiveld GJ, Seehawer M, Caruso S, Heinzmann F, Schneider AT, Frank AK, Cardenas DV, Sonntag R, Luedde M, et al. microRNA 193a-5p regulates levels of nucleolar- and spindle-associated protein 1 to suppress hepatocarcinogenesis. Gastroenterology. 2018;155:1951.e26–1966.e26. doi: 10.1053/j.gastro.2018.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Xu W, Kang W, Wong SH, Wang M, Zhou Y, Fang X, Zhang X, Yang H, Wong CH, et al. Genomic analysis of liver cancer unveils novel driver genes and distinct prognostic features. Theranostics. 2018;8:1740–1751. doi: 10.7150/thno.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon EP, Freije CA, Farber BA, Lalazar G, Darcy DG, Honeyman JN, Chiaroni-Clarke R, Dill BD, Molina H, Bhanot UK, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci USA. 2015;112:E5916–E5925. doi: 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C, Song G, Xiang J, Zhang H, Zhao S, Zhan Y. AURKA promotes cancer metastasis by regulating epithelial-mesenchymal transition and cancer stem cell properties in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;486:514–520. doi: 10.1016/j.bbrc.2017.03.075. [DOI] [PubMed] [Google Scholar]
- 52.Jeng YM, Peng SY, Lin CY, Hsu HC. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clin Cancer Res. 2004;10:2065–2071. doi: 10.1158/1078-0432.CCR-1057-03. [DOI] [PubMed] [Google Scholar]
- 53.Zhang K, Chen J, Chen D, Huang J, Feng B, Han S, Chen Y, Song H, De W, Zhu Z, et al. Aurora-A promotes chemoresistance in hepatocelluar carcinoma by targeting NF-kappaB/microRNA-21/PTEN signaling pathway. Oncotarget. 2014;5:12916–12935. doi: 10.18632/oncotarget.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang B, Hsu CJ, Chou CH, Lee HL, Chiang WL, Su CM, Tsai HC, Yang SF, Tang CH. Variations in the AURKA gene: Biomarkers for the development and progression of hepatocellular carcinoma. Int J Med Sci. 2018;15:170–175. doi: 10.7150/ijms.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu L, Han H, Tian Y, Li W, Zhang J, Feng M, Li Y. Aurora kinase A mediates c-Myc's oncogenic effects in hepatocellular carcinoma. Mol Carcinog. 2015;54:1467–1479. doi: 10.1002/mc.22223. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Q, Luo M, Zhou C, Zhou Z, He Z, Yu X, Zhou S. A proteomics-based investigation on the anticancer activity of alisertib, an Aurora kinase A inhibitor, in hepatocellular carcinoma Hep3B cells. Am J Transl Res. 2017;9:3558–3572. [PMC free article] [PubMed] [Google Scholar]
- 57.Yang XY, Wu B, Ma SL, Yin L, Wu MC, Li AJ. Decreased expression of ZWINT is associated with poor prognosis in patients with HCC after surgery. Technol Cancer Res Treat. 2018;17:1533033818794190. doi: 10.1177/1533033818794190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ying H, Xu Z, Chen M, Zhou S, Liang X, Cai X. Overexpression of Zwint predicts poor prognosis and promotes the proliferation of hepatocellular carcinoma by regulating cell-cycle-related proteins. Onco Targets Ther. 2018;11:689–702. doi: 10.2147/OTT.S152138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Li Y, Meng L, Liu XY, Peng A, Chen Y, Liu C, Chen H, Sun S, Miao X, et al. Reducing protein regulator of cytokinesis 1 as a prospective therapy for hepatocellular carcinoma. Cell Death Dis. 2018;9:534. doi: 10.1038/s41419-018-0555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Shi F, Xing GH, Xie P, Zhao N, Yin YF, Sun SY, He J, Wang Y, Xuan SY. Protein regulator of cytokinesis PRC1 confers chemoresistance and predicts an unfavorable postoperative survival of hepatocellular carcinoma patients. J Cancer. 2017;8:801–808. doi: 10.7150/jca.17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu G, Chen PL. Structural requirements of chromokinesin Kif4A for its proper function in mitosis. Biochem Biophys Res Commun. 2008;372:454–458. doi: 10.1016/j.bbrc.2008.05.065. [DOI] [PubMed] [Google Scholar]
- 63.Huang Y, Wang H, Lian Y, Wu X, Zhou L, Wang J, Deng M, Huang Y. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis. 2018;9:141. doi: 10.1038/s41419-017-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Li S, Zhou S, Cao S, Lou Y, Shen H, Yin J, Li G. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther. 2017;13:651–659. doi: 10.4103/jcrt.JCRT_491_17. [DOI] [PubMed] [Google Scholar]
- 65.Hou G, Dong C, Dong Z, Liu G, Xu H, Chen L, Liu L, Wang H, Zhou W. Upregulate KIF4A enhances proliferation, invasion of hepatocellular carcinoma and indicates poor prognosis across human cancer types. Sci Rep. 2017;7:4148. doi: 10.1038/s41598-017-04176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.