Abstract
Mutation of the isocitrate dehydrogenase (IDH) gene is regarded a novel indicator for the prognosis of patients with glioma. However, the role of the IDH1 gene mutations in carcinogenesis and the mechanisms underlying their function in glioblastoma multiforme (GBM) remain unknown. The present study aimed to determine whether the association of RLIP76 with the different IDH1 mutational status could serve as a putative biomarker for improving disease prognosis. Quantitative PCR, western blotting and immunohistochemical staining assays were used to investigate the expression levels of RLIP76 in 124 patients with GBM with different IDH1 mutational status. In addition, the association between RLIP76 expression, IDH1 mutational status and clinicopathological characteristics was investigated. The effects of RLIP76 expression and IDH1 mutational status on cell proliferation, cell apoptosis, and cell signaling were examined by Cell Counting Kit-8, flow cytometry and western blot assays, respectively. The data demonstrated that IDH1 wild-type (IDH1Wt) patients with low RLIP76 expression exhibited improved overall and progression-free survival. This effect was not observed in patients with IDH1 mutant (IDH1Mut) GBM. In vitro assays demonstrated that knockdown of IDH1 or overexpression of the IDH1 R132H mutation suppressed cell proliferation and promoted cell apoptosis in U87 glioma cells. Mechanistic studies further indicated that although the IDH1 R132H mutant phenotype exhibited similar antitumor effects on GBM cells as those observed with the IDH1 knockdown, it acted via a different mechanism with regard to the regulation of the apoptosis signaling pathway. IDH1 R132H mutant cells promoted p53-induced apoptosis, while the IDH1 knockdown inhibited the RLIP76-dependent apoptotic pathway in glioma cells. The findings of the present study provided insight to the contribution of IDH1 mutation in the development of GBM and indicated that RLIP76 may be considered as a prognostic biomarker of IDH1Wt GBM.
Keywords: isocitrate dehydrogenase 1, ralA binding protein 1, glioblastoma, apoptosis, prognosis
Introduction
Gliomas, which originate from glial cells, are the most frequent primary malignant tumors of the central nervous system (CNS) in humans (1). Glioblastoma (GBM) is the most aggressive form of human astrocytoma with a median survival of only 14–15 months (2). It has been reported that isocitrate dehydrogenase (IDH) 1 or 2 gene mutations are commonly found in human gliomas (3). In light of the 2016 update of the World Health Organization (WHO) CNS tumor classification, IDH mutations have been considered novel indicators in predicting the outcome of glioma patients (4). The IDH1 R132 mutation accounts for >80% of all IDH mutations (5). Recent studies have confirmed that wild-type IDH1 (IDH1Wt) and IDH1 mutant (IDH1Mut) gliomas are biologically different tumor types (6). An increasing number of studies demonstrate that patients with IDH1Mut GBM exhibit a better prognosis compared with patients with IDH1Wt GBM (7,8). However, a higher proportion of distant relapses were noted in IDH1Mut compared with IDH1Wt glioma patients. Therefore, there is an urgent need to understand the molecular mechanisms underlying glioma tumorigenesis in patients with different IDH1 mutational status. This can aid the investigation and development of novel therapeutic strategies.
It has been shown that a hypoxic microenvironment contributes to glioma growth by inducing the glioma stem cell phenotype (9). Recent evidence suggests that IDH1 has a critical role in regulating the concentration of several antioxidants, such as α-ketoglutarate (α-KG), nicotinamide adenine dinucleotide phosphate (NADPH) and glutathione (GSH) (10). Redox imbalance can result in the inactivation of significant cell cycle and cell signaling regulators, such as the p53, activator protein 1 and nuclear factor erythroid 2-related factor 2 transcription factors (11,12). However, adverse effects were noted following treatment of glioma patients who were stratified according to their IDH1 mutational status and tumor grade. Therefore, it is reasonable to suspect that the different IDH1 mutational status may participate in the development of GBM under hypoxic conditions, although this hypothesis has not been clarified to date.
RLIP76, which is also known as ralA binding protein 1 (RalBP1), is associated with oxidative stress-induced cell apoptosis by affecting the intracellular levels of GSH (13,14). It has been demonstrated that downregulation of RLIP76 in tumor cells can reverse various tumor biological processes, such as proliferation, apoptosis and differentiation (15–17). It is important to note that the expression levels of RLIP76 are closely associated with the degree of malignancy in gliomas (18–20). It is also noteworthy that RLIP76 can serve as an oncogene in glioma via its interaction with important signaling pathways of tumorigenesis, such as the Rac1, AKT and JNK pathways (18,20). However, the contribution of the dysregulated RLIP76 expression in association with the different IDH1 mutational status to GBM tumorigenesis remains unclear.
The present study investigated the expression levels of RLIP76 in 124 GBM tissues (98 IDH1Wt and 26 IDH1Mut) using reverse transcription-quantitative PCR (RT-qPCR), western blot and immunohistochemical assays. The prognostic value of RLIP76 expression and IDH1 mutational status for GBM was investigated. The potential roles of RLIP76 on tumor proliferation and apoptosis in IDH1Wt or IDH1Mut GBM cells were also explored in a separate set of in vitro experiments.
Materials and methods
Tissue samples
The present study was granted approval by the Specialty Committee on Ethics of Biomedicine Research at the Tongji University. Written informed consent was obtained from all patients. The selection criteria were previously described (18). Briefly, the selection criteria were as follows: i) The subject had a diagnosis of primary GBM and no history of other tumors; ii) the subject had complete clinical data, including age, sex, clinical manifestations, mean tumor diameter (defined as the geometric mean of the three diameters by MRI scan), extent of resection and adjuvant therapy; and iii) the subject underwent evaluation by enhanced head MRI scans for tumor relapse or progression after surgery at least once every six months. A total of 124 patients who received glioma-resection surgery were recruited at the Department of Neurosurgery, Shanghai Tongji Hospital of Tongji University and of Changzheng Hospital, the Second Military Medical University. The subjects were recruited from July 2016 to December 2018. The follow-up was carried out by telephone or mail every 6 months and the survival time was evaluated until March 2019. GBM was diagnosed according to the 2016 WHO classification of tumors of the CNS by two independent experienced pathologists. The clinical characteristics of the patients with GBM are listed in Table I.
Table I.
Demographic and clinicopathologic characteristics of patients with glioblastoma multiforme.
| RLIP76 expression | |||
|---|---|---|---|
| Characteristic | Number of patients (%) | Low | High |
| Number of patients | 124 | 62 | 62 |
| Age, years | |||
| <60 | 88 (70.97) | 40 | 48 |
| ≥60 | 36 (29.03) | 22 | 14 |
| Sex | |||
| Male | 71 (57.26) | 35 | 36 |
| Female | 53 (42.74) | 27 | 26 |
| Mean tumor diameter, cm | |||
| <5 | 43 (34.69) | 24 | 19 |
| ≥5 | 81 (65.32) | 38 | 43 |
| IDH1 status | |||
| Wild-type | 98 (79.03) | 42 | 56 |
| Mutant | 26 (20.97) | 20 | 6 |
| Resection degree | |||
| Gross total resection | 96 (77.42) | 49 | 47 |
| Sub-total resection | 24 (19.35) | 11 | 13 |
| Partial resection | 4 (3.26) | 2 | 2 |
| Biopsy | 0 (0) | 0 | 0 |
| Survival status | |||
| Alive | 25 (20.16) | 21 | 4 |
| Dead | 99 (79.84) | 41 | 58 |
| Recurrence | |||
| No | 18 (14.52) | 15 | 3 |
| Yes | 106 (85.48) | 47 | 59 |
RLIP76, ralA binding protein 1; IDH1, isocitrate dehydrogenase 1.
Cell culture
The U87 cell line (glioblastoma of unknown origin) was purchased from the American Type Culture Collection (cat. no. HTB-14™). The cells were grown in DMEM (Sigma-Aldrich; Merck KGaA) supplemented with 8% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), penicillin G (100 U/ml) and streptomycin (100 g/ml) and maintained at 37°C in humidified air with 5% CO2. The cells were incubated in anoxic and/or hypoxic (3–5% O2) environments. Hypoxic conditions were obtained by replacing oxygen with N2, using a Heracell 150i CO2 incubator (Thermo Fisher Scientific, Inc.).
Bioinformatic analysis
To examine the expression of RLIP76 in GBM, a LinkedOmics analysis was performed of The Cancer Genome Atlas (TCGA) GBM databases according to their IDH1 status. LinkedOmics (http://www.linkedomics.org) is a publicly-available portal comprising multi-omics data from all 32 types of cancer in the TCGA database.
To examine the hypoxia-induced gene expression profiles of IDH1wt and IDH1Mut glioma stem cell lines, a previously published microarray study was downloaded from the Gene Expression Omnibus (GEO) database (accession no. GSE118683) (21). Differentially expressed genes (DEGs) were identified using the edgeR package (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) in R software; P<0.05 and fold change >2 were considered as statistically significant. The expressions of all DEGs were presented in the heatmap.
The DAVID bioinformatics resources 6.8 (https://david.ncifcrf.gov) were used to perform gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses on the differentially expressed genes (22,23). Following the analyses for significance and false discovery rate (FDR), the GO terms were selected from the significantly enriched gene sets (P<0.05 and FDR <0.05).
Transfection and stable clone selection
The IDH1-Flag and IDH1 R132H-Flag plasmids were constructed into the pCMV-Tag2B vector as previously described (24). Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection according to the manufacturer's instructions. The efficacy of the transfection was tested by examining the expression of the Flag protein using western blotting. IDH1 siRNA (cat. no. sc-60829) and siRNA reagent system (cat. no. sc-45064), and RLIP76 siRNA (cat. no. sc-36376) and control siRNA (cat. no. sc-37007), were purchased from Santa Cruz Biotechnology, Inc., and all transfections were conducted according to the manufacturer's instructions. On the 3rd day following transfection, protein expression was examined by western blotting in order to evaluate the knockdown efficacy.
RNA isolation and RT-qPCR
Total RNA, including miRNA, was extracted from cells/tissues with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using the ReverTra Ace qPCR RT kit (FSQ-101; Toyobo Life Science), according to the manufacturer's protocol. cDNA was amplified using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen; Thermo Fisher Scientific, Inc.) on a 7500 Fast Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR conditions were: 10 min at 95°C, 1 min at 55°C, followed by 40 cycles of 15 sec at 95°C and 30 sec at 55°C. Relative fold expression of the target gene was normalized to β-actin and calculated according to the 2−∆∆Cq method (25). The sequences for the forward and reverse primers of RLIP76 and β-actin were previously described (18).
Apoptosis assay
Cell apoptosis was assessed using the ApoScreen Annexin V Apoptosis Kit (Bender MedSystems, GmbH). After washing twice with PBS, cells were collected and stained with FITC-Annexin V and propidium iodide, as per the manufacturer's instructions. All cells were analyzed by flow cytometry (FACScan; BD Biosciences). Data analyses were performed using CellQuest software Version 5.0 (BD Biosciences). Experiments were conducted in triplicates.
Western blot analysis
Western blot analyses were performed as described previously (26). The primary antibodies used were the following: Anti-Flag antibody (cat. no. LT0420; LifeTein, LLC), IDH1 (Cell Signaling Technology, Inc.; cat. no. 8137; 1:500), RLIP76 (Abcam; cat. no. ab56815; 1:1,000), caspase-9 (Santa Cruz Biotechnology, Inc.; cat. no. sc-17784; 1:500), Bcl-2 (Santa Cruz Biotechnology, Inc.; cat. no. sc-509; 1:500), p53 (Abcam; cat. no. ab-32389; 1:1,000) and β-actin (Abcam; cat. no. ab-8227; 1:1,000). Western blot analysis was quantified by normalizing the signal intensity of each sample to that of β-actin.
Cell proliferation assay
Cell viability was assessed by the Cell Counting Kit-8 (CCK-8) assay. Briefly, all cells were seeded in collagen-coated 96-well plates at a density of 5×103 cells/ml and incubated with 10 µl CCK-8 solution for 4 h at 37°C. The optical density of each sample was recorded with a microplate reader (Bio-Rad Laboratories, Inc.) at 450 nm. The assays were performed in triplicate.
Immunohistochemical assays
The immunohistochemical assay (IHC) was conducted as previously described (18). The RLIP76 monoclonal antibody (Abcam; cat. no. ab-133549) was used at a dilution of 1:1,000. The assessment of RLIP76 expression was conducted as described previously (18). Briefly, RLIP76 expression was divided into ‘high’ (++, final score 4–6 and +++, final score >6) vs. ‘low’ (+, final score 1–3 and -, final score 0).
Quantification of GSH
GSH was quantified using a GSH-Glo™ Glutathione Assay kit (cat. no. V6912; Promega Corporation), according to the manufacturers' instructions, following incubation of 1×104 cells/well in 96-well plates for 1 h at 37°C with 5% CO2 in serum-free HBSS with added Ca2+ and Mg2+.
Statistical analysis
The experimental data were presented as mean ± standard deviation. Statistical analysis was performed by the Student's t-test. Kaplan-Meier survival analysis was used to compare the overall survival (OS) in glioma patients. Univariate survival analysis was carried out by the Kaplan-Meier method and analyzed via the log-rank test to assess differences in survival between the groups. The Cox proportional hazard model for multivariate survival analysis was used to assess predictors of survival. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of RLIP76 in patients with IDH1Wt and IDH1Mut GBM
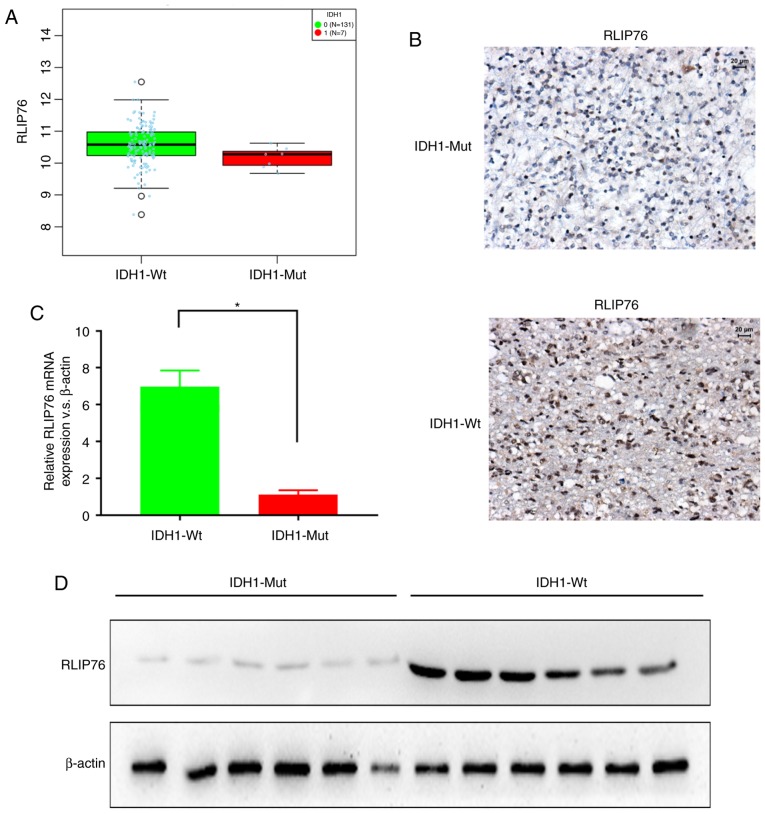
The expression levels of RLIP76 in GBM were investigated from the TCGA database using LinkedOmics (http://www.linkedomics.org). RLIP76 mRNA expression levels were significantly higher in the IDH1Wt (n=131) group compared with the IDH1Mut (n=7) group (Fig. 1A; P=0.0406). Subsequently, the protein expression levels of RLIP76 were investigated in 124 human GBM specimens by IHC staining. The GBM tissues were divided into IDH1Mut (n=26) and IDH1Wt (n=98) groups, according to their WHO grading. The IHC results revealed that the IDH1Wt group exhibited higher immunoreactivity for RLIP76 in their GBM tissues compared with the IDH1Mut group (Fig. 1B). By RT-qPCR analysis, it was also revealed that the IDH1Wt group exhibited significantly higher RLIP76 mRNA expression levels compared with the IDH1Mut group (Fig. 1C). Similar differences were also observed by western blotting, with regard to the RLIP76 protein expression levels between the two glioma groups (Fig. 1D). These results indicated that the expression levels of RLIP76 were significantly upregulated in IDH1Wt gliomas compared with the IDH1Mut gliomas.
Figure 1.
Expression levels of RLIP76 in patients with IDH1Wt and IDH1Mut GBM. (A) LinkedOmics analysis of The Cancer Genome Atlas database revealed that RLIP76 is overexpressed in IDH1Wt GBM tissues compared with IDH1Mut GBM tissues. P=0.0406. (B) Representative images of immunohistochemical staining for RLIP76 in IDH1Wt and IDH1Mut GBM tissues. (C) mRNA expression levels of RLIP76 measured in IDH1Wt and IDH1Mut GBM tissues by reverse transcription-quantitative PCR. *P<0.05. (D) Protein expression levels of RLIP76 measured in IDH1Wt and IDH1Mut GBM tissues by western blot analysis. RLIP76, ralA binding protein 1; IDH1, IDH1, isocitrate dehydrogenase 1; Wt, wild-type; Mut, mutant; GBM, glioblastoma multiforme.
Prognostic value of RLIP76 in IDH1Wt GBM
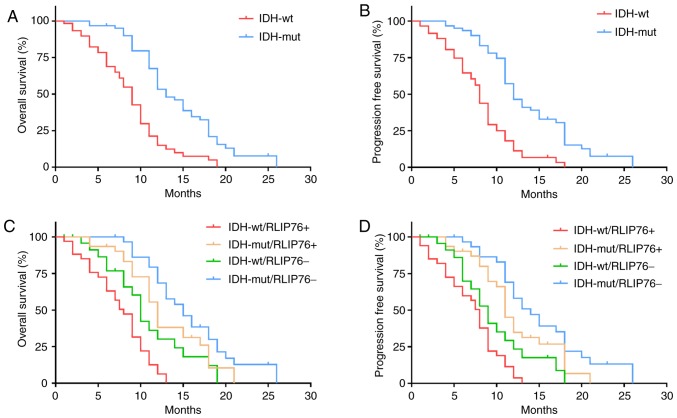
To further explore the prognostic value of RLIP76 in GBM tissues with different IDH1 mutational status, the patients (n=124) were divided into the IDH1Mut (n=26) and IDH1Wt (n=98) groups. The results confirmed that IDH1Wt was significantly associated with poor OS (P=0.0241; Fig. 2A) and progression-free survival (PFS) in patients with GBM (P=0.0178; Fig. 2B). Kaplan-Meier analysis further revealed that increased RLIP76 expression levels were associated with poor OS and PFS in the IDH1Wt group (P=0.0297 and P=0.0374, respectively), while no significant difference was noted with regard to the prognostic value of RLIP76 in the IDH1Mut group (P=0.162 and P=0.219, respectively; Fig. 2C and D). These results suggested that RLIP76 may exert a specific role in IDH1Wt GBM that is different from that noted in IDH1Mut GBM.
Figure 2.
Prognostic value of RLIP76 in IDH1Wt GBM. (A) Kaplan-Meier curves for overall survival and (B) progression-free survival according to IDH1 status in patient with GBM. (C) Kaplan-Meier curves for overall survival and (D) progression-free survival after stratification of the patients with IDH1Wt and IDH1Mut GBM based on tumor RLIP76 expression levels. IDH1, IDH1, isocitrate dehydrogenase 1; GBM, glioblastoma multiforme; Wt, wild-type; Mut, mutant; RLIP76, ralA binding protein 1.
Univariate survival analysis indicated that the expression levels of RLIP76 were a prognostic factor for OS and for PFS in patients with IDH1Wt glioma (Table II). Multivariate analysis further confirmed that high expression levels of RLIP76 at diagnosis were a critical and independent prognostic factor for OS and PFS in patients with IDH1Wt glioma (Table III).
Table II.
Univariate analysis of factors associated with survival of patients with IDH1 wild-type glioblastoma multiforme.
| A, Overall survival | |||
|---|---|---|---|
| Variable | HR | 95% CI | P-value |
| Sex (male vs. female) | 0.754 | 0.435–1.269 | 0.526 |
| Age (≥60 vs. <60) | 1.857 | 1.113–2.648 | 0.038 |
| RLIP76 (high vs. low) | 1.986 | 1.045–2.976 | 0.008 |
| MTD (≥5 cm vs. <5 cm) | 0.776 | 0.418–1.397 | 0.716 |
| Resection degree | 0.039 | ||
| Total vs. partly | 0.324 | 0.098–0.912 | 0.023 |
| Subtotal vs. partly | 0.275 | 0.078–0.936 | 0.032 |
| B, Progression-free survival | |||
| Variable | HR | 95% CI | P-value |
| Sex (male vs. female) | 0.684 | 0.468–1.078 | 0.493 |
| Age (≥60 vs. <60) | 1.568 | 1.213–2.336 | 0.042 |
| RLIP76 (high vs. low) | 1.756 | 1.129–2.743 | 0.008 |
| MTD (≥5 cm vs. <5 cm) | 0.721 | 0.356–1.463 | 0.841 |
| Resection degree | 0.047 | ||
| Total vs. partly | 0.411 | 0.0618–0.985 | 0.031 |
| Subtotal vs. partly | 0.178 | 0.043–0.574 | 0.013 |
Univariate analysis, Cox proportional hazards regression model (n=98). Significant P-values are denoted in bold font. IDH1, isocitrate dehydrogenase 1; RLIP76, ralA binding protein 1; MTD, mean tumor diameter; HR, Hazard ratio; CI, confidence interval.
Table III.
Multivariate analysis of factors associated with survival of patients with IDH1 wild-type glioblastoma multiforme.
| A, Overall survival | |||||
|---|---|---|---|---|---|
| Variable | Median survival (months, 95% CI) | HR | 95% CI | P-value | |
| RLIP76 (high vs. low) | 11 (6.124–13.142) | 16 (8.187–23.879) | 1.872 | 1.104–3.267 | 0.012 |
| Resection degree | NA | NA | NA | NA | 0.047 |
| Total vs. partial | 13 (11.256–14.784) | 4 (0–6.998) | 0.242 | 0.053–0.816 | 0.019 |
| Subtotal vs. partial | 16 (9.872–20.121) | 4 (0–6.998) | 0.195 | 0.049–0.788 | 0.018 |
| Age (≥60 vs. <60 years) | 8 (3.477–12.236) | 15 (10.121–19.689) | NA | NA | 0.154 |
| B, Progression-free survival | |||||
| Variable | Median survival (months, 95% CI) | HR | 95% CI | P-value | |
| RLIP76 (high vs. low) | 7 (6.124–13.142) | 12 (8.187–23.879) | 1.579 | 1.212–3.978 | 0.021 |
| Resection degree | NA | NA | NA | NA | 0.034 |
| Total vs. partial | 11 (8.117–13.168) | 3 (0–5.168) | 0.172 | 0.036–0.775 | 0.026 |
| Subtotal vs. partial | 12 (7.981–16.336) | 3 (0–5.099) | 0.217 | 0.057–0.819 | 0.031 |
| Age (≥60 vs. <60 years) | 7 (3.117–10.148) | 10 (8.564–16.375) | NA | NA | 0.182 |
Multivariate analysis, Cox proportional hazards regression model (stepwise backward method; n=98). Variables were adopted for their prognostic significance by univariate analysis. Significant P-values are denoted in bold font. IDH1, isocitrate dehydrogenase 1; RLIP76, ralA binding protein 1; HR, hazard ratio; CI, confidence interval.
Microarray-based GO analysis and pathway analysis
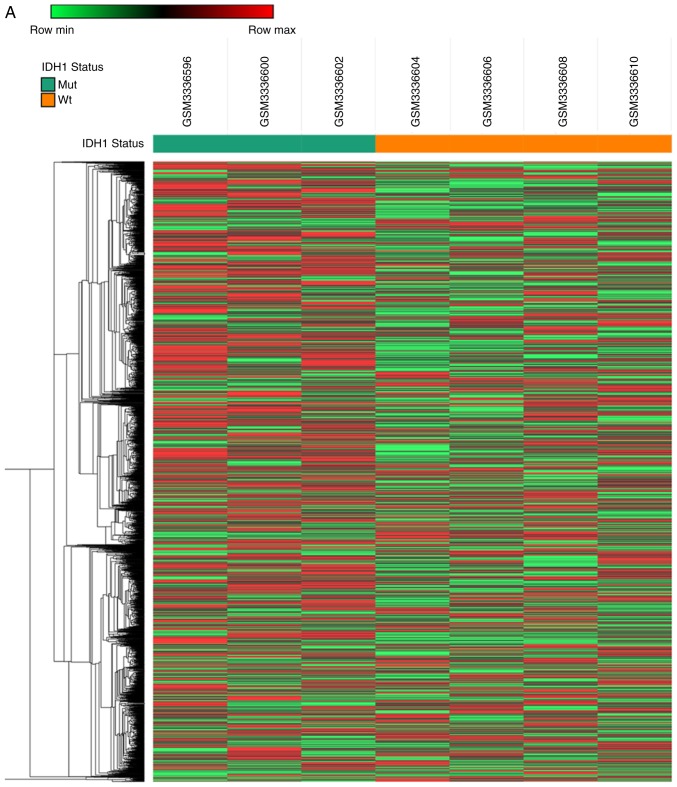
To identify novel oncogenic mRNAs in GBM tissues with different IDH1 mutational status, the present study analyzed a previously published microarray study of hypoxia-treated IDH1wt and IDH1Mut glioma stem cell lines (full data available at GEO, accession no. GSE118683) (21). The present analysis identified 2,928 mRNAs that were upregulated in IDH1Wt glioma stem cells compared with IDH1Mut glioma stem cells (fold change >2 and P<0.05; Fig. 3A) under hypoxia conditions. In addition, 1,263 mRNAs were downregulated in IDH1Wt glioma stem cells compared with IDH1Mut glioma stem cells (fold change >2 and P<0.05; Fig. 3A).
Figure 3.
Gene expression analysis of IDH1Wt and IDH1Mut glioma cells under hypoxia. A previously published database (GSE118683) reporting the profiling of glioma cells grown in hypoxic vs. normoxic conditions in vitro was used. (A) Heatmap of differentially expressed genes in IDH1Wt and IDH1Mut glioma cells in hypoxic conditions. (B) GO term classification of differentially expressed genes. Count represents the number of genes annotated by gene ontology database to each of the GO terms. (C) KEGG pathway analysis of differentially expressed genes. IDH1, IDH1, isocitrate dehydrogenase 1; Wt, wild-type; Mut, mutant; GO, gene ontology; KEGG; Kyoto Encyclopedia of Genes and Genomes; BP, biological process; MF, molecular function; CC, cellular component.
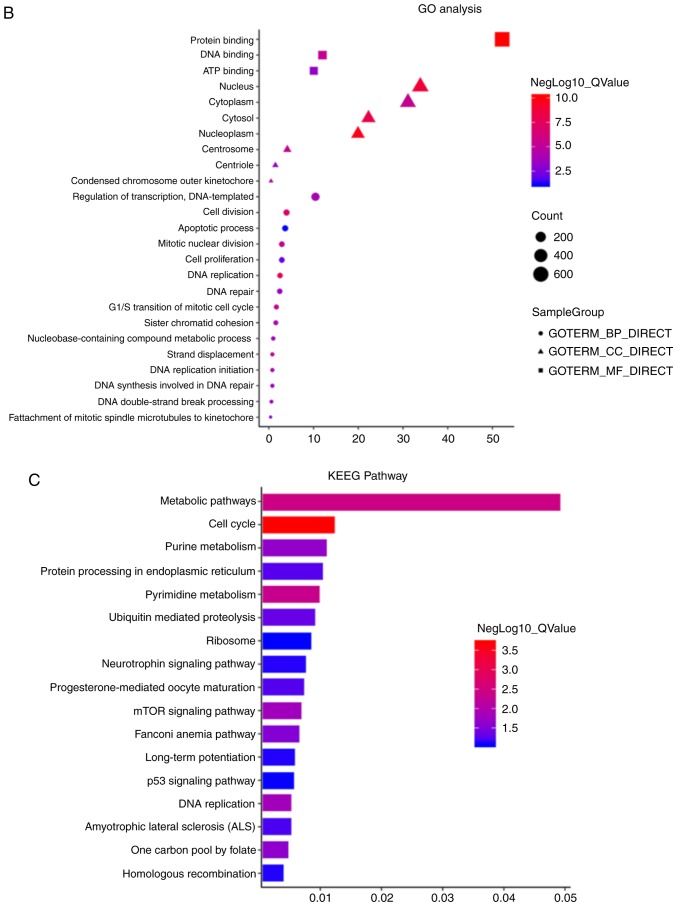
GO analysis on the targeted genes was conducted using DAVID 6.8 (https://david.ncifcrf.gov). Based on GO analysis, ~1,234 differentially expressed genes (|fold change|>4 and P<0.05) were classified (Fig. 3B). GO analysis revealed that specific biological processes were enriched, including ‘DNA replication’, ‘cell division’, ‘cell proliferation’ and the ‘apoptotic process’. In addition to the biological processes, the differentially expressed genes were also enriched in the GO terms associated with ‘cellular component’ and ‘molecular function’, such as ‘protein binding’, ‘DNA binding’, ‘ATP binding’ and ‘nucleoplasm’ (Fig. 3B). KEGG pathway enrichment analyses were also performed. The differentially expressed genes were significantly and predominantly associated with the ‘metabolic pathway’, a significant process in the progression of tumor proliferation and apoptosis. Other enriched pathways involved the ‘cell cycle’, ‘purine metabolism’, the mTOR and p53 signaling pathways, the ‘long-term potentiation’ and the ‘pyrimidine metabolism’ (Fig. 3C).
Overexpression of IDH1 R132H mutant, but not IDH1Wt, inhibits cell growth and increases cell apoptosis via p53-mediated apoptosis in a hypoxic microenvironment
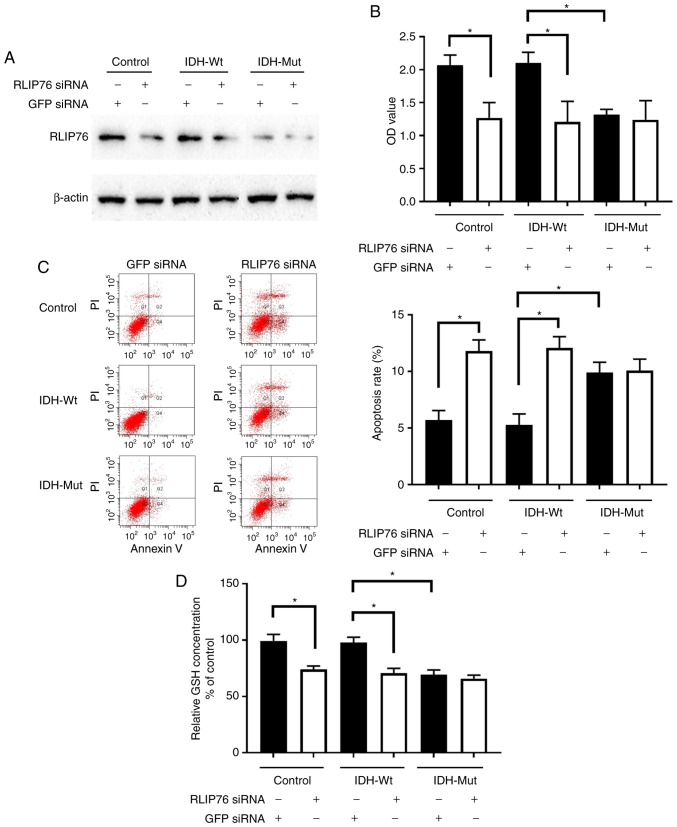
U87 cells that overexpressed either empty vector or pCMVtag-2B containing IDH1Wt or IDH1Mut were used to investigate the effects of IDH1Wt and IDH1Mut proteins on glioma cell growth and apoptosis under hypoxia. U87 cells with stable overexpression of IDH1 and IDH1 R132H mutant proteins were successfully established (Fig. 4).
Figure 4.
Overexpression of IDH1 constructs in U87 cells. Protein expression levels of IDH1 were detected by western blotting using an anti-Flag antibody in U87 empty vector control stable cells, IDH1 Wt-expressing stable cells and IDH1 R132H Mut-expressing stable cells. IDH1, IDH1, isocitrate dehydrogenase 1; Wt, wild-type; Mut, mutant.
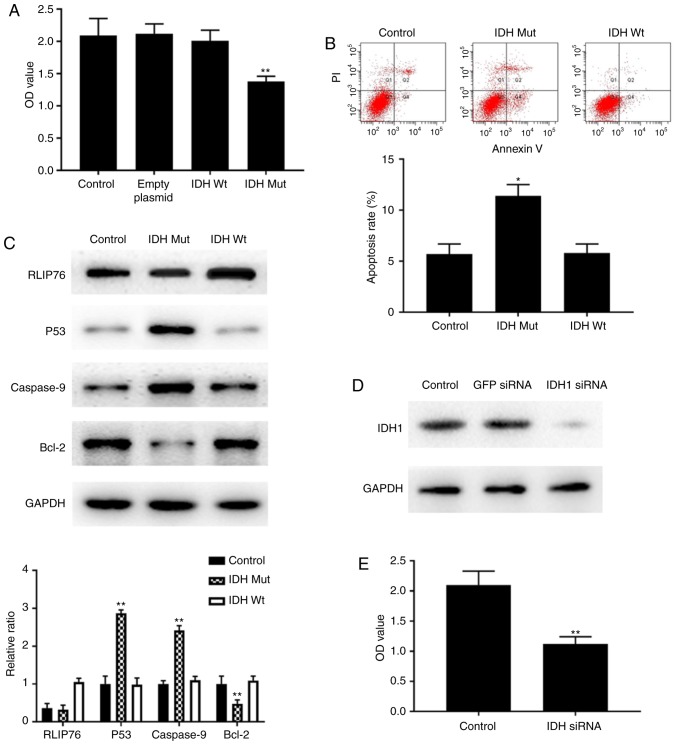
Subsequently, the contribution of the IDH1 mutational status to the growth of U87 cells under hypoxia was explored by the CCK-8 assay. Overexpression of the IDH1 R132H mutant protein significantly suppressed cell proliferation in U87 cells compared with cells transfected with empty plasmid or control parental cells (Fig. 5A). By contrast, overexpression of the IDH1Wt protein did not influence cell proliferation compared with cells transfected with empty plasmid or control parental cells (Fig. 5A). In addition, overexpression of the IDH1Mut protein resulted in enhanced apoptosis of U87 cells (Fig. 5B). No differences were noted with regard to cell apoptosis in IDH1Wt cells compared with the parental control cells (Fig. 5B). These findings suggested that overexpression of IDH1Mut, but not IDH1Wt, suppressed cell proliferation and promoted cell apoptosis in GBM cells under hypoxic conditions.
Figure 5.
Effects of IDH1 overexpression on the proliferation and apoptosis of U87 cells. (A) Cell proliferation was measured by CCK-8 assay at 72 h. (B) Apoptosis rates were measured by flow cytometry. (C) Representative images and quantification from western blot analysis for RLIP76, p53, Caspase-9 and Bcl-2 protein expression levels. (D) Western blotting results confirming successful knockdown of IDH1 in U87 cells by siRNA transfection. (E) Proliferation of control cells and cells transfected with IDH1 siRNA was measured by CCK-8 assay at 72 h. (F) Apoptosis rates of control cells and IDH1 siRNA-transfected cells were measured by flow cytometry. (G) Representative images and quantification from western blot analysis for RLIP76, p53, Caspase-9 and Bcl-2 protein expression levels in control cells and IDH1 siRNA-transfected cells. *P<0.05 and **P<0.01 vs. control. IDH1, IDH1, isocitrate dehydrogenase 1; CCK-8, Cell Counting Kit-8; RLIP76, ralA binding protein 1; siRNA, small interfering RNA; Wt, wild-type; Mut, mutant; OD, optical density; GFP, green fluorescent protein; PI, propidium iodide.
To further explore the mechanism by which the IDH1 R132H mutant protein mediated cell growth inhibition and apoptosis, the expression levels of RLIP76, p53, caspase-9 and Bcl-2 proteins were detected by western blotting. The bioinformatic analysis of the GSE118683 database performed in the present study indicated that the expression levels of the aforementioned genes were significantly altered between IDH1Mut and IDH1Wt GBM cells under hypoxic conditions (data not shown). Western blot analysis confirmed that the IDH1 R132H mutant-overexpressing cells exhibited higher protein expression levels of p53 and caspase-9, and lower protein expression levels of Bcl-2 and RLIP76, compared with the IDH1Wt-overexpressing and the control cells (Fig. 5C). Notably, no difference was observed in the protein expression levels of RLIP76 between the control and IDH1Mut groups. These results suggested that the antitumor effects of the IDH1 R132H mutant proteins on GBM were associated with an induction of the p53-dependent apoptotic pathway under hypoxia.
Knockdown of IDH1 inhibits cell growth and enhances cell apoptosis via induction of the RLIP76-dependent apoptotic pathway under hypoxia
U87 cells were transfected with IDH1 siRNA and western blotting results demonstrated that the protein expression levels of IDH1 were successfully reduced (Fig. 5D). Following IDH1 siRNA transfection, U87 cell proliferation was significantly suppressed under hypoxic conditions compared with that of the control cells, as demonstrated by the CCK-8 assay (Fig. 5E). In addition, flow cytometric analysis indicated that the apoptosis rate was significantly increased in U87 cells following IDH1 silencing compared with that of the control cells (Fig. 5F). These results indicated that IDH1 functioned as a tumor oncogene in U87 cells under hypoxic conditions.
Since RLIP76 has a critical role in the prognosis of IDH1Wt GBM (Fig. 2C), the expression levels of RLIP76, p53, caspase-9 and Bcl-2 were measured in order to investigate their ability to regulate tumor progression under hypoxia. Knockdown of IDH1 resulted in a significant decrease in RLIP76 expression levels without affecting p53 expression in U87 cells (Fig. 5G). This result was not noted in the tissues containing the IDH1 R132H mutant phenotype (Fig. 5C). The data demonstrated that although the IDH1 R132H mutant phenotype exerted similar antitumor effects on GBM with those noted in the presence of IDH1 knockdown, it promoted apoptosis under hypoxia via a distinct mechanism of action. The IDH1 R132H mutant protein promoted p53-induced apoptosis, whereas IDH1 knockdown inhibited the RLIP76-dependent apoptotic pathway. This may partially explain why RLIP76 was a better prognostic indicator in IDH1Wt GBM compared with IDH1Mut GBM.
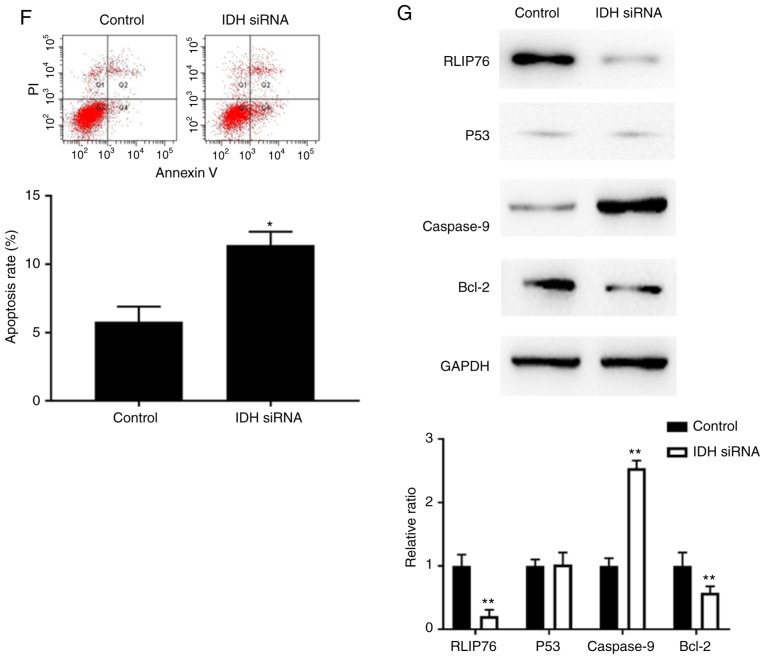
Knockdown of RLIP76 inhibits cell proliferation and decreases GSH levels in IDH1Wt, but not in IDH1Mut, glioma cells under hypoxia
Transfection of parental control, IDH1Wt or IDH1Mut U87 cells with RLIP76 siRNA significantly suppressed RLIP76 protein expression (Fig. 6A). RLIP76 knockdown significantly inhibited cell growth and promoted the induction of apoptosis in IDH1Wt U87 cells, although it did not affect the proliferation and apoptosis of IDH1Mut U87 cells under hypoxia (Fig. 6B and C). The results suggested that RLIP76 expression contributed to the malignant progression of IDH1Wt glioma cells under hypoxic conditions.
Figure 6.
Effects of RLIP76 knockdown on proliferation and apoptosis of U87 cells overexpressing Wt or Mut IDH1. Control cells and cells stably overexpressing Wt or R132H Mut IDH1 were transfected with either a control siRNA or a RLIP76-specific siRNA for 72 h. (A) Western blotting results confirming successful knockdown of RLIP76 after siRNA transfection. (B) Cell proliferation was measured by Cell Counting Kit-8 assay. (C) Apoptosis rates were measured by flow cytometry. Representative plots and quantification is shown. (D) GSH levels were measured using a commercial glutathione assay kit. *P<0.05, with comparisons indicated by brackets. RLIP76, ralA binding protein 1; IDH1, IDH1, isocitrate dehydrogenase 1; Wt, wild-type; Mut, mutant; siRNA, small interfering RNA; GSH, glutathione; GFP, green fluorescent protein; OD, optical density; PI, propidium iodide.
The present bioinformatic analysis revealed that the differential gene expression noted in IDH1Mut glioma cells was mainly associated with the metabolic pathway (Fig. 3). In addition, a previous study has reported that RLIP76 has a critical role in the regulation of the metabolic pathway required for GSH detoxification (27). In light of the aforementioned findings, the expression levels of GSH were investigated. GSH is considered the main antioxidative defense mechanism that prevents intracellular damage under hypoxia. IDH1Mut cells exhibited a dramatic decrease in GSH levels compared with those of the control and IDH1Wt cells under hypoxia. In addition, knockdown of RLIP76 in IDH1Wt glioma cells significantly decreased GSH levels. However, GSH levels were not significantly different in IDH1Mut cells following transfection with either control or RLIP76 siRNA. These results suggested that RLIP76 may influence the metabolic pathway of IDH1Wt glioma cells by regulating GSH levels in the hypoxic microenvironment.
Discussion
GBM represents one of the most malignant forms of astrocytoma in humans. It has been reported that IDH1Wt glioma patients exhibit poorer prognosis and lower functional connectivity compared with IDH1Mut glioma patients (28). The present study confirmed that the IDH1Wt phenotype was a prognostic factor for poor disease outcome in patients with GBM. Previous studies have demonstrated that IDH1 mutations result in depleted levels of crucial antioxidant molecules, such as GSH, NADPH and α-KG. GSH is regarded as the major intracellular free radical scavenger and the elimination of GSH-conjugates (GS-E) is critical for cell survival, since the accumulation of GS-E results in cell toxicity. RLIP76 has demonstrated dinitrophenyl-S-glutathione conjugate-dependent ATPase (DNP-SG ATPase) activity, which accounts for up to 80% of the GS-E efflux and is the major member of the cell detoxification system. It has been shown that the GS-E detoxification process is reversible. The present study demonstrated that the expression levels of RLIP76 in the IDH1Mut specimens were significantly lower compared with those in the IDH1Wt glioma specimens. The present results indicated that IDH mutation may inhibit RLIP76 expression levels. However, the precise mechanism of the regulation of RLIP76 expression by the IDH mutation remains unclear.
The traditional WHO classification has been revised in 2016 and includes novel molecular markers in addition to histological evaluation. IDH1 mutation is one of the most robust markers used in glioma patients. IDH1 is frequently mutated in low grade glioma and secondary GBM. However, these IDH1 mutations are rare in primary GBM (29). IDH1-driven metabolic reprogramming has been regarded as a critical progress for retaining the glioma stem cell compartment (24). Diminished IDH1 activity results in exhaustion of reduced glutathione and stimulates the production of reactive oxygen species (ROS) (24). ROS-induced lipid peroxidation that occurs in plasma and mitochondrial membranes is the major stimulus for the production of 4-hydroxynonenal (4-HNE) (30). It is believed that the metabolic pathway for 4-HNE detoxification is activated by RLIP76. This pathway may represent an ideal gene-targeting strategy for malignant tumor treatment (31). Therefore, it is reasonable to speculate that RLIP76 regulates numerous cellular signaling pathways, notably the IDH1-induced metabolic pathway, under hypoxic conditions, and by doing so it may ultimately promote tumor development. Bioinformatic analysis in the present study demonstrated that the metabolic pathway was significantly altered between IDH1wt GBM and IDH1Mut GBM tissues under hypoxia. In addition, the current data revealed that high levels of RLIP76 expression were associated with lower OS and PFS in patients with IDH1Wt GBM. RLIP76 expression is not an ideal marker for prognosis prediction in the IDH1Mut group. This may be due to the considerable differences between the metabolic adaptation in IDH1Wt and IDH1Mut GBM, which are driven by IDH1 activity (3). However, these issues need to be further clarified in future studies.
The fast and incurable tumor recurrence is more common in IDH1Wt GBM subjects and requires effective treatment approaches (32). However, the markers available to predict prognosis of patients with IDH1Wt GBM are insufficient. Several studies point to the application of RLIP76 as a prognostic marker of various malignant cancers, such as melanoma, kidney, colon and prostate cancer (16,33,34). In addition, it was previously demonstrated that RLIP76 expression exhibited elevated levels in high-grade gliomas (18). This finding led to the current investigation that RLIP76 could be used as a prognostic marker for GBM with a different IDH1 mutational status on the basis of the 2016 WHO classification. In the present study, statistical analysis indicated that RLIP76 was a critical factor for OS and PFS in the IDH1Wt, but not in the IDH1Mut group. The findings suggested that RLIP76 expression could be employed to predict the clinical outcome of IDH1Wt GBM.
RLIP76 functions as an oncogene by increasing the expression of the Rac1/JNK pathway proteins and by activating the PI3K/AKT pathway in glioma (18,20). Notably, RLIP76 regulates apoptosis independent of the p53 status in malignant glioma and neuroblastoma (18,35). The present study demonstrated that the decrease in IDH1 activity, either by siRNA transfection or by upregulation of the IDH1 R132H mutant protein, could suppress cell growth and enhance cell apoptosis in a hypoxic microenvironment. To identify the molecular mechanisms of this process in U87 cells, the expression levels of Bcl-2, caspase-9, p53 and RLIP76 proteins were investigated and the results indicated that the IDH1 R132H mutant protein promoted p53-induced apoptosis, while the IDH1Wt protein suppressed cell apoptosis via the RLIP76-dependent pathway. Notably, knockdown of IDH1 significantly inhibited RLIP76 expression, without affecting p53 expression. It is possible that RLIP76 had a critical role in IDH1Wt-mediated apoptosis, whereas p53 status was highly associated with IDH1 mutation-induced apoptosis under hypoxic conditions. Mounting evidence indicates that IDH1Wt and IDH1Mut gliomas are biologically different tumor types (36,37). In agreement with this hypothesis, the results of the present study provided experimental evidence that IDH1Wt and IDH1Mut regulated the induction of GBM apoptosis via distinct biological mechanisms. These data suggested that agents that specifically target RLIP76 in IDH1Wt, but not in IDH1Mut cell types, may be promising therapeutic agents for GBM treatment.
Taken collectively, the present study demonstrated that RLIP76 may be an ideal prognostic biomarker of IDH1Wt GBM. In addition, the present findings improved our understanding on the fundamental function of IDH1 mutation in glioma. RLIP76 may be used to further stratify patients with IDH1Wt glioma into high and low risk subjects, in order to optimize their treatment.
Acknowledgments
Not applicable.
Funding
This work was supported by the National Science Foundation of China (grant no. 81802489), the Fundamental Research Funds for the Central Universities (grant no. 22120180353), the Shanghai Natural Science Foundation (grant nos. 18ZR1434500, 19ZR1448900 and 18411962500) and the Scientific Research Initial Funding of Shanghai Tongji Hospital (grant no. RCQD1704).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QW and LZ performed the experiments. QW and YC wrote the manuscript. JG, HC, CZ collected and interpreted the data and performed the bioinformatics analysis. JQ and CL provided technical assistance. QW, JQ and CL designed the study and revised the manuscript. All authors read and approved the final manuscript..
Ethics approval and consent to participate
The present study was granted approval by the Specialty Committee on Ethics of Biomedicine Research at the Tongji University. Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 2.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaminska B, Czapski B, Guzik R, Król SK, Gielniewski B. Consequences of IDH1/2 mutations in gliomas and an assessment of inhibitors targeting mutated IDH proteins. Molecules. 2019;24(pii):E968. doi: 10.3390/molecules24050968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stancheva G, Goranova T, Laleva M, Kamenova M, Mitkova A, Velinov N, Poptodorov G, Mitev V, Kaneva R, Gabrovsky N. IDH1/IDH2 but not TP53 mutations predict prognosis in Bulgarian glioblastoma patients. Biomed Res Int. 2014;2014:654727. doi: 10.1155/2014/654727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31:272–281. doi: 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akyerli CB, Yüksel S, Can Ö, Erson-Omay EZ, Oktay Y, Coşgun E, Ülgen E, Erdemgil Y, Sav A, von Deimling A, et al. Use of telomerase promoter mutations to mark specific molecular subsets with reciprocal clinical behavior in IDH mutant and IDH wild-type diffuse gliomas. J Neurosurg. 2018;128:1102–1114. doi: 10.3171/2016.11.JNS16973. [DOI] [PubMed] [Google Scholar]
- 7.Hata N, Hatae R, Yoshimoto K, Murata H, Kuga D, Akagi Y, Sangatsuda Y, Suzuki SO, Iwaki T, Mizoguchi M, Iihara K. Insular primary glioblastomas with IDH mutations: Clinical and biological specificities. Neuropathology. 2017;37:200–206. doi: 10.1111/neup.12362. [DOI] [PubMed] [Google Scholar]
- 8.Labussiere M, Boisselier B, Mokhtari K, Di Stefano AL, Rahimian A, Rossetto M, Ciccarino P, Saulnier O, Paterra R, Marie Y, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83:1200–1206. doi: 10.1212/WNL.0000000000000814. [DOI] [PubMed] [Google Scholar]
- 9.Chen JE, Lumibao J, Blazek A, Gaskins HR, Harley B. Hypoxia activates enhanced invasive potential and endogenous hyaluronic acid production by glioblastoma cells. Biomater Sci. 2018;6:854–862. doi: 10.1039/C7BM01195D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi N, Das GM. Metabolic reprogramming in breast cancer and its therapeutic implications. Cells. 2019;8(pii):E89. doi: 10.3390/cells8020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox AG, Tsomides A, Kim AJ, Saunders D, Hwang KL, Evason KJ, Heidel J, Brown KK, Yuan M, Lien EC, et al. Selenoprotein H is an essential regulator of redox homeostasis that cooperates with p53 in development and tumorigenesis. Proc Natl Acad Sci USA. 2016;113:E5562–E5571. doi: 10.1073/pnas.1600204113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McArdle A, Pollock N, Staunton CA, Jackson MJ. Aberrant redox signalling and stress response in age-related muscle decline: Role in inter- and intra-cellular signalling. Free Radic Biol Med. 2019;132:50–57. doi: 10.1016/j.freeradbiomed.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhal SS, Singh SP, Singhal P, Horne D, Singhal J, Awasthi S. Antioxidant role of glutathione S-transferases: 4-Hydroxynonenal, a key molecule in stress-mediated signaling. Toxicol Appl Pharmacol. 2015;289:361–370. doi: 10.1016/j.taap.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singhal SS, Yadav S, Roth C, Singhal J. RLIP76: A novel glutathione-conjugate and multi-drug transporter. Biochem Pharmacol. 2009;77:761–769. doi: 10.1016/j.bcp.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagaprashantha LD, Singhal J, Li H, Warden C, Liu X, Horne D, Awasthi S, Salgia R, Singhal SS. 2′-Hydroxyflavanone effectively targets RLIP76-mediated drug transport and regulates critical signaling networks in breast cancer. Oncotarget. 2018;9:18053–18068. doi: 10.18632/oncotarget.24720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N, Du CH. RLIP76 silencing inhibits cell proliferation and invasion in melanoma cell line A375. Eur Rev Med Pharmacol Sci. 2017;21:2054–2060. [PubMed] [Google Scholar]
- 17.Wang W, Liu J, Qi J, Zhang J, Zhu Q, Qin C. RLIP76 increases apoptosis through Akt/mTOR signaling pathway in gastric cancer. Oncol Rep. 2016;36:2216–2224. doi: 10.3892/or.2016.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Wang JY, Zhang XP, Lv ZW, Fu D, Lu YC, Hu GH, Luo C, Chen JX. RLIP76 is overexpressed in human glioblastomas and is required for proliferation, tumorigenesis and suppression of apoptosis. Carcinogenesis. 2013;34:916–926. doi: 10.1093/carcin/bgs401. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Qian J, Wang J, Luo C, Chen J, Hu G, Lu Y. Knockdown of RLIP76 expression by RNA interference inhibits invasion, induces cell cycle arrest, and increases chemosensitivity to the anticancer drug temozolomide in glioma cells. J Neurooncol. 2013;112:73–82. doi: 10.1007/s11060-013-1045-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C, Cai Z, Liang Q, Wang Q, Lu Y, Hu L, Hu G. RLIP76 depletion enhances autophagic flux in U251 cells. Cell Mol Neurobiol. 2017;37:555–562. doi: 10.1007/s10571-016-0410-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dao Trong P, Rosch S, Mairbaurl H, Pusch S, Unterberg A, Herold-Mende C, Warta R. Identification of a prognostic hypoxia-associated gene set in IDH-mutant glioma. Int J Mol Sci. 2018;19(pii):E2903. doi: 10.3390/ijms19102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Calvert AE, Chalastanis A, Wu Y, Hurley LA, Kouri FM, Bi Y, Kachman M, May JL, Bartom E, Hua Y, et al. Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep. 2017;19:1858–1873. doi: 10.1016/j.celrep.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Wang Q, Cui Y, Liu ZY, Zhao W, Wang CL, Dong Y, Hou L, Hu G, Luo C, et al. Knockdown of cyclin D1 inhibits proliferation, induces apoptosis, and attenuates the invasive capacity of human glioblastoma cells. J Neurooncol. 2012;106:473–484. doi: 10.1007/s11060-011-0692-4. [DOI] [PubMed] [Google Scholar]
- 27.Sharma R, Singhal SS, Cheng J, Yang Y, Sharma A, Zimniak P, Awasthi S, Awasthi YC. RLIP76 is the major ATP-dependent transporter of glutathione-conjugates and doxorubicin in human erythrocytes. Arch Biochem Biophys. 2001;391:171–179. doi: 10.1006/abbi.2001.2395. [DOI] [PubMed] [Google Scholar]
- 28.Derks J, Kulik S, Wesseling P, Numan T, Hillebrand A, van Dellen E, de Witt Hamer PC, Geurts JJG, Reijneveld JC, Stam CJ, et al. Understanding cognitive functioning in glioma patients: The relevance of IDH-mutation status and functional connectivity. Brain Behav. 2019;9:e01204. doi: 10.1002/brb3.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013;125:621–636. doi: 10.1007/s00401-013-1106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: Focusing on mitochondria. Redox Biol. 2015;4:193–199. doi: 10.1016/j.redox.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasparovic AC, Milkovic L, Sunjic SB, Zarkovic N. Cancer growth regulation by 4-hydroxynonenal. Free Radic Biol Med. 2017;111:226–234. doi: 10.1016/j.freeradbiomed.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 32.Wang PF, Song HW, Cai HQ, Kong LW, Yao K, Jiang T, Li SW, Yan CX. Preoperative inflammation markers and IDH mutation status predict glioblastoma patient survival. Oncotarget. 2017;8:50117–50123. doi: 10.18632/oncotarget.15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Song X, Gong W, Zhu Z, Liu X, Hou Q, Sun Y, Chai J, Zou L, Guan J. RLIP76 blockade by siRNA inhibits proliferation, enhances apoptosis, and suppresses invasion in HT29 colon cancer cells. Cell Biochem Biophys. 2015;71:579–585. doi: 10.1007/s12013-014-0237-5. [DOI] [PubMed] [Google Scholar]
- 34.Singhal SS, Singhal J, Yadav S, Sahu M, Awasthi YC, Awasthi S. RLIP76: A target for kidney cancer therapy. Cancer Res. 2009;69:4244–4251. doi: 10.1158/0008-5472.CAN-08-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singhal SS, Nagaprashantha L, Singhal P, Singhal S, Singhal J, Awasthi S, Horne D. RLIP76 inhibition: A promising developmental therapy for neuroblastoma. Pharm Res. 2017;34:1673–1682. doi: 10.1007/s11095-017-2154-y. [DOI] [PubMed] [Google Scholar]
- 36.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker SJ, Metallo CM. Metabolic consequences of oncogenic IDH mutations. Pharmacol Ther. 2015;152:54–62. doi: 10.1016/j.pharmthera.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.