Abstract
Lung cancer is one of the most common malignancies and the leading cause of cancer-associated mortality in Korea. A significant amount of effort has been put into the development of new and more effective treatments and biological markers for the prediction of therapeutic responses, which has led to the identification of various genetic changes in cancer, that are the so-called ‘growth drivers’ of carcinogenesis. Certain genetic alterations have become new treatment targets, and it has been suggested that different mutations are associated with different clinicopathological characteristics and prognosis. The present study aimed to evaluate the status of the key ‘driver’ mutation anaplastic lymphoma kinase (ALK) fusion in Korean patients with non-small cell lung cancer (NSCLC) and its association with clinicopathological characteristics, including the presence of other genetic mutations. The present study also compared different methods for ALK fusion detection, including fluorescence in situ hybridization (FISH), immunohistochemistry (IHC) and next-generation sequencing (NGS) to evaluate which method is the most effective. A total of 482 patients with NSCLC who underwent ALK FISH analysis were evaluated for clinicopathological features, such as age, sex, smoking history, tumor stage, histological subtype, immunohistochemical profile, including ALK and EGFR mutation statuses, and survival. Some ALK FISH-positive and -negative cancers were newly submitted to NGS analysis for DNA and RNA alterations. The ALK fusion-positive tumors were associated with a younger age, female patients, frequent nodal metastases, advanced stage and shorter survival. Comparing the results of ALK FISH, IHC and NGS analyses, it was concluded that in practice, ALK testing should better be diversified concerning FISH and IHC, and NGS analysis would be a good alternative to FISH, with an additional advantage of being able to concurrently detect different mutations.
Keywords: non-small cell lung cancer, ALK fusion, ALK FISH, ALK IHC, next-generation sequencing
Introduction
Lung cancer is one of the most common malignancies and is the leading cause of cancer-associated mortality worldwide (1). In Korea, the incidence of lung cancer has continuously increased and since 2005 it has been the most frequent cause of cancer-associated mortality in both men and women (2). Non-small cell lung cancer (NSCLC) accounts for ~80% of all lung cancer cases, and it includes adenocarcinoma, squamous cell carcinoma and large cell carcinoma. Despite the advancements in different treatment strategies, including surgery, chemotherapy, radiation and their combinations, the prognosis of NSCLC has not been markedly improved, with a 5-year overall survival rate <20% (3).
The identification of several genetic mutations, the so-called ‘growth drivers’ of NSCLC carcinogenesis, has led to the development of targeted cancer therapy and the search for biological markers for the prediction of therapy responses. Epidermal growth factor receptor (EGFR) mutation is a key mechanism of carcinogenesis in certain lung adenocarcinomas since it promotes the growth and division of tumor cells through sustained activation of a kinase receptor. Thus, inhibition of EGFR kinase activity with drugs, such as gefitinib is an example of effective targeted therapy (4–8). The KRAS gene mutation is considered crucial for the carcinogenesis of various cancer types, including NSCLC (9,10). KRAS can act in the downstream part of the cellular signaling pathways, which is activated by mutant EGFR. Therefore, increased KRAS activity in tumor cells due to mutations is a well-known predictor of therapeutic resistance to EGFR inhibitors; however, to the best of our knowledge, there are no treatments that directly target KRAS activation yet (11–13). In 2007, a fusion of echinoderm microtubule-associated protein-like 4 (EML4) gene and anaplastic lymphoma kinase (ALK) gene, EML4-ALK, was identified to be a strong carcinogenic factor leading to cell proliferation and cancer development by increasing the kinase activity of the ALK gene (14). The ALK gene fusion has >20 partners but is most frequently partnered with EML4 (15–17), and has been reported to occur in 2–10% of all lung cancer patients, primarily in adenocarcinomas (18–22). Although the EGFR mutation and ALK fusion may coexist in younger and non-smoking patients with adenocarcinomas, the simultaneous occurrence of EGFR mutation and ALK fusion has been reported to be very rare and considered to be virtually exclusive (23–25). Concerning the lung adenocarcinomas with EGFR mutation, numerous studies have reported the clinicopathological features in Korean patients (26–30); however, there has been a relatively small number of studies regarding lung cancer cases with ALK fusion in Korea (21,29–31), partly because the National Health Insurance Service (NHIS) in Korea did not start covering ALK inhibitor treatment for advanced ALK-positive lung cancer until 2015. The first aim of the present study was to evaluate the status of key driver mutations, particularly the ALK rearrangement in Korean patients with NSCLC and the associations with clinicopathological characteristics.
The current diagnostic methods for detection of ALK fusion include florescence in situ hybridization (FISH), immunohistochemistry (IHC), reverse transcription-quantitative PCR (RT-qPCR) and next-generation sequencing (NGS) analyses. Until recently the ALK FISH was the gold standard of diagnosis and the ALK IHC or NGS analyses had limited uses as screening or auxiliary tools. However, FISH has several well-known limitations. It is labor-intensive, time-consuming and operator-dependent in both preparation and interpretation processes (32). A number of studies have reported that ALK IHC produces almost 100% concordant results with ALK FISH, although there are always some discrepancies (22,33–35). After the anti-ALK (D5F3) CDx assay (Ventana®) was approved as a stand-alone ALK diagnostic test by the USA Food and Drug Administration, a large study in 2017 reported that dichotomous ALK IHC with D5F3 should be the standard diagnostic test to select patients with NSCLC who benefit from ALK inhibitor treatment, since it better predicted the tumor response rate and survival after crizotinib treatment compared with ALK FISH (36). NGS enables prompt detection of various genetic alterations, including ALK fusion, and is increasingly cost-effective; it is expected to overtake the existing ALK diagnostic tests. An evidence-based guideline for the molecular diagnosis and treatment of lung cancer, jointly reported by the International Association for the Study of Lung Cancer (IASLC), College of American Pathologists (CAP), and Association for American Molecular Pathology (AMP), recently reported that NGS panels are preferred over single gene tests to identify other treatment options beyond ALK, EGFR, and ROS1 inhibitors, emphasizing the importance of NGS for the detection of genetic alterations in lung cancer (37). In Korea, the NHIS recently began to contribute to the cost of NGS testing for cancer patients; however, it does not yet contribute to the costs of targeted drug treatments, including ALK inhibitors, according to the results of NGS analyses, partly due to the insufficient data on NGS results of Korean patients with cancer. Therefore, the second aim of the present study was to compare the different diagnostic tests for ALK fusion in Korean patients with lung cancer and to investigate the possibility of NGS as a new standard ALK diagnostic test.
Materials and methods
Case selection and clinical data collection
A total of 482 NSCLC specimens with ALK gene status evaluated by FISH were collected and stored in the Biobank of Korea University Guro Hospital between 2012 and 2018. The glass slides were reviewed for histological diagnosis and immunohistochemical features, including ALK (5A4; Novocastra), TTF-1 (8G7G3/1; Dako; Agilent Technologies, Inc.), and napsin A (polyclonal; Cell Marque). The formalin-fixed paraffin-embedded (FFPE) tissue blocks of 10 patients, stored for <3 years to minimize the degradation of DNA and RNA, were selected for NGS analysis, and consisted of five ALK FISH-positive and five ALK FISH-negative adenocarcinomas (38,39). The clinical information included age, sex, smoking history, cancer stage according to the 8th Edition of the American Joint Committee on Cancer staging manual, genetic mutation status and survival. All the glass slides, paraffin blocks and clinical information were provided by the Human Biobank of Korea University Guro Hospital, which collects patients' samples and medical information at the time of biopsies or surgical excisions of their tumors, with the written informed consent for future research use. The present study was conducted with the approval of the Institutional Review Board of Korea University Guro Hospital (approval no. 2018GR0357).
NGS analysis
HiSeq or MiSeq of Illumina, Inc. and Iron Torrent of Thermo Fisher Scientific, Inc. are the most widely used NGS platforms. A number of researchers prefer MiSeq of Illumina, Inc. due to its high diversity of gene selection and its ability to obtain large amounts of genetic information. However, when it is used with customized cancer panels, high quality DNA of ≥200–300 ng is required. In addition, the device is costly, operator-dependent and labor-intensive. Whereas, Ion Torrent, even though the selection range is limited, is cost-effective and can be used with low quality DNA if the target gene is spared. Sequencing with Ampliseq Cancer Hotspot Panel enables targeted sequencing from 10 ng of DNA; however, frequent errors and missing copy number variations (CNVs) are disadvantages (38,40). Therefore, selection of an appropriate platform depends on the purpose of the experiment, and the quantity and quality of the DNA extracted from the available samples. The present study selected the Miseq system (Illumina, Inc.) with customized cancer panels to obtain a large amount of genetic information.
For DNA analyses, the paraffin tissue blocks containing enough, viable tumor cells were selected and sectioned to a thickness of 10 µm. The DNA was extracted with Qiagen GeneRead DNA FFPE kit (Qiagen Sciences, Inc.). The sample purity was evaluated by the absorbance ratio at 260/280 and the samples with a ratio between 1.8 and 2.1 were considered appropriate for further analyses. The DNA integrity numbers (DINs) of samples were measured by Agilent 4200 TapeStation system (Agilent Technologies, Inc.) and those with a DIN >3 were selected for further analysis. The DNA library construction, target sequence hybridization and purification of captured sequences were conducted using xGen® Lockdown® probes and reagents (Integrated DNA Technologies, Inc.) and Dynabeads M-270 Streptavidin (Thermo Fisher Scientific, Inc), according to the manufacturers' protocols. The 80 different oncogenes included in the DNA hybridization probes are presented in Table SI. Following target hybridization and purification, the sequencing and analysis of captured DNA fragments was performed using Illumina MiSeq system with MiSeq Reagent kit v3 (Illumina, Inc.). Sample concentrations were measured using a Qubit® 3.3 Fluorometer (Thermo Fisher Scientific, Inc.) and Qubit™ dsDNA HS assay kit (Thermo Fisher Scientific, Inc.). The final concentrations for sequencing were adjusted to 10–12 pM with volumes between 350 and 420 µl.
The RNA analyses were conducted through similar processes to those of the DNA analysis, except for the early ribosomal RNA (rRNA) removal and complementary DNA (cDNA) library construction steps. The paraffin tissue blocks were sectioned to a thickness of 10 µm. Total RNA was extracted with Qiagen RNeasy FFPE kit (Qiagen Science, Inc.). The sample purity was evaluated by the absorbance ratio at 260/280 and those with a ratio between 1.8 and 2.1 were considered appropriate for further analyses. The DV200 of samples, the fraction of RNA >200 nucleotides, was measured with Agilent 4200 TapeStation system (Agilent Technologies, Inc.) and only those samples with a DV200>70% were selected for further analysis according to the manufacturer's guidelines. The rRNA was depleted from the total RNA samples using NEBNext® rRNA Depletion kit (New England Biolabs, Inc.), and then the RNA samples were amplified by RT-PCR to produce cDNA samples. Subsequently, the cDNA library construction, target sequence hybridization and purification were conducted using xGen® Lockdown® probes and reagents (Integrated DNA Technologies, Inc.) and Dynabeads M-270 Streptavidin (Thermo Fisher Scientific, Inc.), according to the manufacturers' protocols. The 30 different oncogenes included in the hybridization probes are presented in Table SII. The sequencing and analysis of captured cDNA fragments were performed using Illumina MiSeq system with MiSeq Reagent kit v3 (Illumina, Inc.), with the same sample concentrations as for the DNA analysis.
Statistical analysis
Fisher's exact test and Pearson's χ2 test were used for the statistical analyses. P<0.05 was considered to indicate a statistically significant difference. To compare survival data, Kaplan-Meier survival analysis and a log-rank test were performed. All statistical analyses were performed with SPSS version 18 (SPSS, Inc.).
Results
Clinicopathological features
Between 2012 and 2018, 482 patients underwent ALK FISH analysis with small biopsies or surgically excised specimens of primary or metastatic NSCLC. For all patients, sections from the FFPE tissue or cell blocks, but no cytological materials or body fluid samples, were used for FISH or other molecular studies. The majority of the patients had adenocarcinoma (n=451) and only a small number had other histological types, including adenosquamous carcinoma (n=7), squamous cell carcinoma (n=11), large cell carcinoma (n=8) and pleomorphic carcinoma (n=5). This was partly because the NHIS in Korea only covers the cost of ALK FISH analysis for patients with adenocarcinoma histology. The majority of the patients with other histological types had been suspected of having adenocarcinoma in small biopsies but confirmed as otherwise in excised specimens or with additional immunohistochemical studies. All 39 patients (8.1%) who were diagnosed as ALK FISH-positive had adenocarcinomas. The mean age of the ALK FISH-positive patients was 60.7 years (range, 34–80 years). This was significantly younger than the mean age of ALK FISH-negative patients with or without concurrent EGFR mutation (66.6 years; P<0.001). The male to female ratio in the ALK FISH-positive patients was 1.44 (23:16), while it was 1.79 (284:159) in the ALK-negative patients. The proportion of females was significantly higher in the ALK FISH-positive patients (P<0.001). The smoking history of the ALK FISH-positive and -negative patients demonstrated no statistically significant difference (P>0.5). Of the ALK FISH-positive patients, 21 were never-smokers, and the remaining 18 had a history of smoking, ranging between 6 and 200 pack years. However, when the EGFR mutation-positive patients were removed from the ALK FISH-negative group, and the ALK FISH-positive group was compared with the ALK FISH−/EGFR mutation-negative group, the proportion of never-smokers were significantly higher in ALK FISH-positive group (P<0.001) (Table I).
Table I.
Clinicopathological characteristics of patients according to the ALK fusion and EGFR mutation status.
| ALK FISH-positive (n=39) | ALK FISH-negative and EGFR mutation-negative (n=329) | ALK FISH-negative and EGFR mutation-positive (n=114) | ALK IHC-positive (n=74) | |
|---|---|---|---|---|
| Sex | ||||
| Male | 23 (59.0%) | 198 (60.2%) | 70 (61.4%) | 38 (51.4%) |
| Female | 16 (41.0%) | 131 (39.8%) | 44 (38.6%) | 36 (48.6%) |
| Smoking history | ||||
| Never | 21 (53.8%) | 141 (42.9%) | 72 (63.2%) | 39 (52.7%) |
| Previous or current | 18 (46.2%) | 188 (57.1%) | 42 (36.8%) | 35 (47.3%) |
| Mean age | 60.7 years | 66.5 years | 64.3 years | 63.7 years |
| Nodal metastasis | 31 (79.5%) | 223 (67.8%) | 72 (63.2%) | 58 (78.4%) |
| Tumor stage | ||||
| I | 6 (15.4%) | 83 (25.2%) | 37 (32.5%) | 7 (9.5%) |
| II | 3 (7.7%) | 30 (9.1%) | 15 (13.2%) | 10 (13.5%) |
| III | 9 (23.1%) | 59 (17.9%) | 21 (18.4%) | 25 (33.8%) |
| IV | 21 (53.8%) | 157 (47.7%) | 41 (36.0%) | 32 (43.2%) |
| 1-year mortality | 16 (41.0%) | 31 (41.9%) | ||
ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; FISH, fluorescence in situ hybridization.
At the time of diagnosis, the ALK FISH-positive tumors were significantly associated with more frequent nodal metastases (n=31; 79.5%) compared with ALK FISH-negative cases (n=295; 66.6%) (P<0.001). A total of 30 ALK FISH-positive patients (77.0%) had unresectable stage III tumors with mediastinal metastases or stage IV with distant tumor spread at the time of diagnosis, whereas among the ALK FISH−/EGFR mutation-negative (n=215; 65%) and ALK FISH-negative/EGFR mutation-positive patients (n=62; 54.4%) less proportions exhibited stage III and IV diseases (P<0.001); and only 6 ALK FISH-positive patients could undergo a lobectomy for stage I or II disease. This indicates that ALK FISH-positive cancer cases were more advanced compared with ALK FISH-negative cases at the time of diagnosis. In total, 16 ALK FISH-positive patients (41.0%) succumbed to the disease within 1 year of diagnosis, while 92 ALK FISH-negative patients (20.8%) succumbed to the disease in the same period (P<0.001) (Table I). All patients were treated with surgery, chemotherapy, radiation and targeted therapeutic agents according to the tumor stage and genetic mutation status. Of the 39 ALK FISH-positive patients, 12 were treated with ALK inhibitors. Among them, 7 patients received crizotinib as first-line therapy, whereas the others were treated with various chemotherapeutic agents and/or radiation before starting treatment with an ALK inhibitor. No patient could undergo surgical resection due to the advanced tumor stage. All 12 patients started ALK inhibitor treatment after 2015, and those who received first-line ALK inhibitor therapy started after 2017 according to the NHIS approval for payment. The duration of treatment ranged between 1 and 24 months. Overall, 1 patient had to stop crizotinib treatment after 1 month of administration due to severe hepatotoxicity. Another patient, who had been heavily treated with various chemotherapeutic regimens and radiation before ALK inhibitor treatment, demonstrated partial response to crizotinib and ceritinib for 13 months, then returned to chemotherapy due to disease progression, succumbed to the disease 3 months later. Considering that only a small number of patients had tried ALK inhibitor drugs, and an even lower number had received it as first-line treatment, it was considered that the influence of treatment with an ALK inhibitor on the survival of patients would not be substantial in the present study.
Of the 482 patients evaluated by ALK FISH, 313 were also evaluated by ALK (5A4) IHC staining. A total of 74 tumors were positively stained for ALK (23.6%), and among them 27 were also ALK FISH-positive. The concordance rate between ALK FISH and IHC in the present study was 81.2%. The clinicopathological features of ALK IHC-positive patients were similar to those of ALK FISH-positive patients. The mean age was 63.7 years. At the time of diagnosis, 58 patients had nodal metastases (78.4%) and 57 had stage III or IV diseases (77.0%). The number of patients who succumbed to the disease within 1 year of diagnosis was 31 (41.9%) (Table I).
Since all patients suspected of having lung adenocarcinoma were also evaluated for EGFR mutation status at the Korea University Guro Hospital, all 482 patients tested for ALK FISH were also evaluated for EGFR mutation by peptide nucleic acid (PNA) clamping real-time PCR method, and 114 patients were positive for EGFR mutation (23.7%). Among the 39 ALK FISH-positive patients, none were positive for EGFR mutation. The unexpectedly low EGFR-positive rate in the present study may be because the patients were first selected among those who underwent ALK FISH analysis and therefore, not all patients who underwent EGFR mutation tests were included. Therefore, the clinicopathological features according to the EGFR mutation status in the present study may also exhibit some discrepancies with the generally understood characteristics of EGFR mutation-positive lung cancer. Nevertheless, the comparisons of clinicopathological features among ALK FISH-positive, ALK FISH−/EGFR mutation-negative, and EGFR mutation-positive patients in the present study are summarized in Table I.
Histopathological features
Due to the aforementioned reason, adenocarcinoma (n=451) was the most frequent histological type of cancer included in the present study. The glass slides were reviewed for subtype identification of adenocarcinoma. Numerous patients only had biopsies and the histological subtypes of the entire tumor could not be evaluated. Nevertheless, a number of tumors demonstrated mixed patterns of more than two subtypes, even in small biopsy specimens. The ALK FISH-positive adenocarcinomas exhibited various histological subtypes, including solid (n=26), acinar (n=14), cribriform (n=5), micropapillary (n=5), papillary (n=4), mucinous (n=4), and enteric (n=1) types. The most frequently observed solid pattern was present in 66.7% of ALK FISH-positive lung adenocarcinomas and the proportion was significantly higher compared with that identified in ALK FISH-negative tumors (38.3%) irrespective of EGFR mutation status (P<0.0001). Notably, among the tumors with solid growth pattern, the proportions of ALK FISH-positive and EGFR mutation-positive cancers demonstrated no significant difference. The presence of the mucinous type was more frequent in ALK FISH-positive adenocarcinomas compared with ALK FISH-negative adenocarcinomas (P=0.0401). The cribriform pattern was significantly more frequent in ALK FISH-positive adenocarcinomas (14.3%) compared with in ALK FISH-negative tumors (5.4%; P<0.001). However, considering the EGFR mutation status, the difference was maintained only between ALK FISH-positive tumors and ALK FISH-negative/EGFR mutation-positive adenocarcinomas (P=0.0121), but not between ALK FISH-positive and ALK FISH-negative/EGFR mutation-negative cases (P=0.0804). The EGFR mutation also influenced the presence of acinar, papillary and lepidic subtypes, which were significantly more frequent in ALK FISH-negative/EGFR mutation-positive adenocarcinomas compared with in ALK FISH-positive or ALK FISH-negative/EGFR mutation-negative tumors. In addition, among the tumors exhibiting acinar, papillary, or lepidic growth, the presence of EGFR mutation was significantly more frequent than ALK fusion. The histological subtypes observed in adenocarcinomas according to the ALK and EGFR mutation status are summarized in Table II.
Table II.
Histopathologic subtypes of adenocarcinomas according to the ALK and EGFR mutation status.
| Histological | ALK FISH-positive adeno-carcinomas | ALK FISH-negative/EGFR mutation-negative adenocarcinomas | ALK FISH-negative/EGFR mutation-positive adenocarcinomas | Ratio of ALK FISH positive adenocarcinnomas according to the histological | Ratio of EGFR mutation-positive adenocarcinnomas according |
|---|---|---|---|---|---|
| Subtypes | (n=39) (%) | (n=298) (%) | (n=114) (%) | subtype (%) | to the histological subtype (%) |
| Solid | 26 (66.7) | 131 (44.0) | 27 (23.7) | 14.1 | 14.7 |
| Acinar | 14 (35.9) | 98 (32.9) | 72 (63.2) | 7.6 | 39.1 |
| Papillary | 4 (10.3) | 31 (10.4) | 26 (22.8) | 6.6 | 42.6 |
| Micro-papillary | 5 (14.3) | 57 (19.1) | 31 (27.2) | 5.4 | 33.3 |
| Cribriform | 5 (14.3) | 16 (5.4) | 2 (1.8) | 21.7 | 8.7 |
| Mucinous | 4 (10.3) | 25 (8.4) | 2 (1.8) | 12.9 | 6.5 |
| Enteric | 1 (2.6) | 3 (1.0) | 0 (0.0) | 25.0 | 0.0 |
| Lepidic | 0 (0.0) | 20 (6.7) | 15 (13.2) | 0.0 | 42.9 |
Numerous adenocarcinomas consisted of more than two types of histological growth pattern. In such cases, each histological type was counted separately. ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; FISH, fluorescence in situ hybridization.
Immunohistochemical features
The ALK FISH-positive adenocarcinomas were frequently positive for thyroid transcription factor 1 (TTF-1; n=34; 87.2%) and napsin A (n=29; 74.4%). TTF-1 expression was significantly more frequent in ALK FISH-positive tumors than in ALK FISH-negative/EGFR mutation-negative tumors (P=0.0361), but not compared with in ALK FISH-negative/EGFR-positive cases (P=0.0544). Between the ALK FISH-positive and negative tumors, napsin A expression demonstrated no statistically significant difference (P=0.0694); however, the ALK FISH-negative/EGFR mutation-positive adenocarcinomas expressed napsin A significantly more frequently than ALK FISH-positive adenocarcinomas (P=0.0046), although in both groups napsin A expression was very frequent (Table III).
Table III.
The immunohistochemical features of adenocarcinomas according to the ALK fusion and EGFR mutation status.
| ALK FISH-positive | ALK FISH-negative/EGFR mutation-negative | ALK FISH-negative/EGFR mutation-positive | |
|---|---|---|---|
| TTF-1 | 34/39 (87.2%) | 199/279 (71.3%) | 105/109 (96.3%) |
| Napsin | 29/39 (74.4%) | 154/231 (66.7%) | 86/93 (92.5%) |
The number of cases positively reactive for each antibody over the total number of stained cases was recorded.
NGS
The present study analyzed five ALK FISH-positive and five ALK FISH-negative adenocarcinomas with customized cancer DNA and RNA panels (Tables SI and SII). The DNA cancer panel consisted of 80 genes associated with single nucleotide variant (SNV), CNV, and insertion and deletion (INDEL) mutations, and the RNA panel included 30 genes associated with gene translocation and/or fusion. All samples for DNA analyses revealed >600× coverages, which were enough to detect variants of 5% frequency. All samples for RNA analyses revealed 1,000× coverages, enough for detection of gene fusions.
Among the patients with ALK FISH-positive adenocarcinomas, two cases of EML4-ALK translocation were detected, which were also positive in the ALK IHC analysis. However, three ALK FISH-positive tumors demonstrated no RNA fusion abnormalities. In total, 1 patient exhibited an EGFR L858R mutation, which was not detected in PNA clamping, and another had an ERBB2 exon 20 insertion (A775_G776 in YVMA) which had not been tested before. Among the ALK FISH-negative patients, none exhibited ALK fusion in NGS. Furthermore, 1 patient was identified to have TPM3-NTRK1 translocation, which had not been tested before. One EGFR exon 19 deletion which had also been detected in PNA clamping, one ERBB2 exon 20 insertion (A775_G776 in YVMA) and one KRAS G12D mutation were identified in 1 patient each. Since the KRAS or ERBB2 mutation test has not been allowed in patients with lung cancer by the NHIS in Korea, the majority of patients in the present study, including those with KRAS G12D or ERBB exon 20 insertion detected by NGS, had not been tested before. The comparison of the results of ALK FISH, ALK IHC, EGFR PNA clamping and NGS analysis are summarized in Table IV. The results of ALK FISH, ALK IHC and NGS demonstrated significant differences, and the EGFR PNA clamping and NGS DNA analysis also demonstrated some discordance. Considering the NGS results only, patients with ALK fusion-positive adenocarcinomas exhibited no mutations in other oncogenes such as EGFR, BRAF, and KRAS; however, those with no ALK alteration demonstrated frequent genetic mutations in EGFR, ERBB2 and KRAS, even with the small number of cases tested.
Table IV.
Comparison of the results of ALK FISH, ALK IHC, EGFR PNA clamping and NGS analysis.
| NGS analysis | |||||||
|---|---|---|---|---|---|---|---|
| Sex | Age | ALK FISH | ALK IHC | EGFR PNA clamping | RNA | DNA | |
| Case 1 | M | 63 | Positive (18/50; 36.0%) | Positive | Wild | EML4-ALK | None |
| Case 2 | F | 53 | Positive (8/50; 16.0%) | Negative | Wild | None | EGFR L858R |
| Case 3 | F | 53 | Positive (8/50; 16.0%) | Negative | Wild | None | ERBB2 Ins |
| Case 4 | F | 74 | Positive (11/50; 22.0%) | Positive | Wild | None | None |
| Case 5 | F | 44 | Positive (25/50; 50%) | Positive | Wild | EML4-ALK | None |
| Case 6 | F | 71 | Negative | Negative | 19Del | None | EGFR 19 Del |
| Case 7 | M | 53 | Negative | Negative | Wild | TPM3-NTRK1 | None |
| Case 8 | F | 49 | Negative | Negative | Wild | None | KRAS G12D |
| Case 9 | F | 53 | Negative | Negative | Wild | None | ERBB2 Ins |
| Case 10 | F | 73 | Negative | Positive | Wild | None | None |
In ALK FISH-positive cases, the number and percentage of tumor cells with split signals were presented. ALK FISH, ALK IHC, EGFR PNA clamping and NGS. ALK, anaplastic lymphoma kinase; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; EGFR, epidermal growth factor receptor; PNA, peptide nuclei acid; NGS, next-generation sequencing.
Survival analysis
The patients were categorized into the following four groups according to the ALK fusion detection methods and their results: i) Both ALK FISH- and ALK IHC-positive (n=27); ii) only ALK FISH-positive (n=12); iii) only ALK IHC-positive (n=47); and iv) both ALK FISH− and ALK IHC-negative (n=227). The mean survival time of the ALK FISH−/IHC-positive group was 40.42 months and that of the ALK FISH−/IHC-negative group was 56.96 months. The negative group survived significantly longer than the positive group (P<0.001). The mean survival time was 34.41 months in the ALK FISH-negative/IHC-positive group and 40.72 months in the ALK FISH-positive/IHC-negative group, indicating that the only ALK IHC-positive patients died significantly earlier than the only ALK FISH-positive patients (P<0.001; Table V and Fig. 1). The median follow-up time was 46.35 months.
Table V.
The mean survival months of patients according to the ALK fusion detection methods and their results.
| 95% confidence interval | ||||
|---|---|---|---|---|
| Patient group | Mean | Standard error | Lower bound | Upper bound |
| 1 | 30.172 | 3.713 | 22.896 | 37.449 |
| 2 | 40.718 | 3.446 | 33.963 | 47.473 |
| 3 | 34.412 | 2.020 | 30.452 | 38.372 |
| 4 | 56.962 | 1.270 | 54.472 | 59.451 |
The patients were categorized into four groups as follows: 1, ALK FISH/ALK IHC (+/+) (n=27); 2, ALK FISH/ALK IHC (+/-) (n=12); 3, ALK FISH/ALK IHC (−/+) (n=47); 4, Both ALK FISH/ALK IHC (−/-) (n=227). ALK, anaplastic lymphoma kinase; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry.
Figure 1.
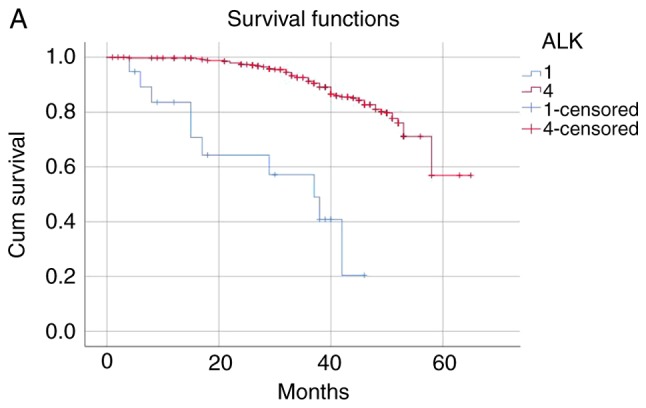
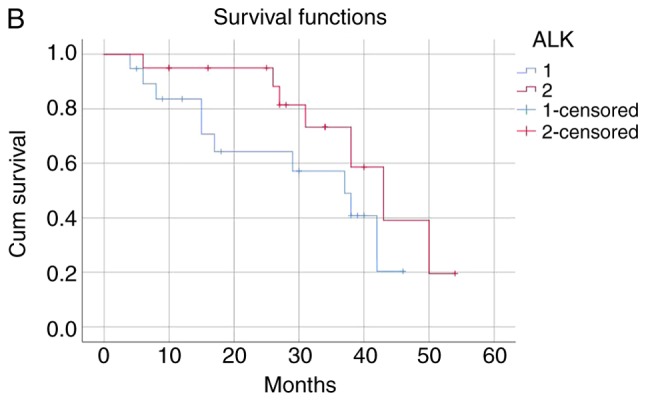
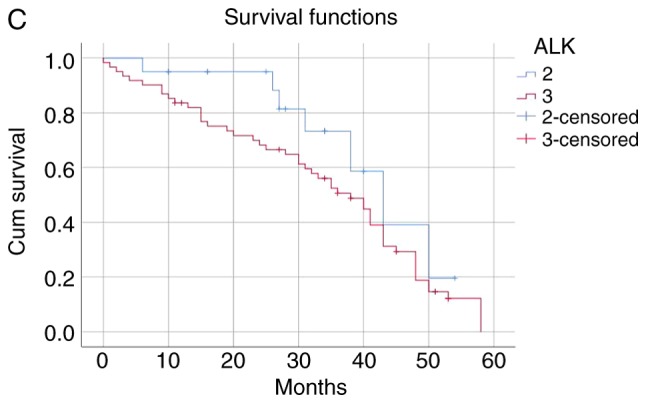
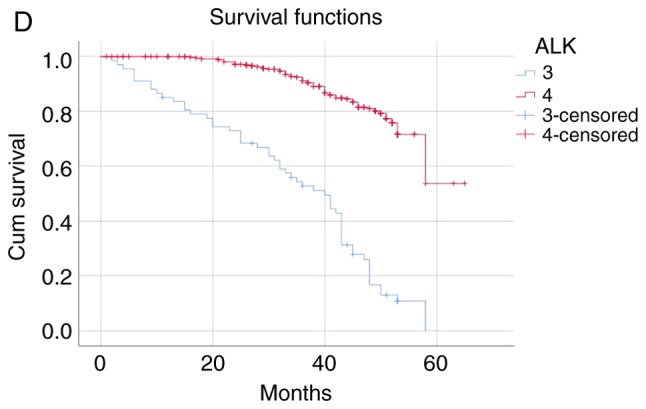
(A) The ALK FISH/ALK IHC (+/+) group was associated with worse survival than ALK FISH/ALK IHC (−/-) group (P<0.001). (B) ALK FISH/ALK IHC (+/+) group was associated with worse survival than ALK FISH/ALK IHC (+/-) group (P<0.001). (C) ALK FISH/ALK IHC (−/+) group was associated with worse survival than ALK FISH/ALK IHC (+/-) group (P<0.001). (D) ALK FISH (−/+) group was associated with worse survival than ALK FISH/IHC (−/-) group (P<0.001).
Discussion
In 2018, the World Conference on Lung Cancer reported the results of the first interim analysis from the ALTA-1L study, which compared the next-generation ALK inhibitor brigatinib and the traditional first-line ALK inhibitor drug crizotinib (41,42). ALTA-1L was a phase III, randomized, open-labeled, comparative, multicenter, international study with 275 participants who had ALK fusion-positive, locally advanced or metastatic NSCLC and had not been previously treated with an ALK inhibitor. The primary endpoint, progression-free survival (PFS), assessed by a double-blinded independent central review, was significantly longer among patients who received brigatinib than those who received crizotinib. The estimated 12-month PFS was 67% [95% confidence interval (CI), 56–75] in the brigatinib group and 43% (95% CI, 32–53) in the crizotinib group, and brigatinib was associated with a 51% lower risk of disease progression or mortality compared with crizotinib [(hazard ratio, 0.49; 95% CI, 0.33–0.74); P<0.001]. Currently, three ALK target therapeutic drugs have been approved in Korea, crizotinib (Xalkori®), alectinib (Alecensa®), and ceritinib (Zykadia®). Only crizotinib was approved as a first-line drug, while the other drugs are limited to second-line treatment. With the results of the ALTA-1L trial, there is an expected shift in generations among ALK target drugs in Korea. Investigations regarding the development of new drugs targeting ALK or other genetic alterations in cancer are proceeding rapidly and frequently as joint research with large pharmaceutical companies occurs. However, compared with the fast evolution of ALK-targeting treatment, the development of diagnostic methods of ALK aberration has been slow. Until recently, ALK FISH has been used as the gold standard for the detection of ALK fusion, and ALK IHC or NGS have only been used as screening tools or adjunctive diagnostic methods. Only recently has the Ventana® anti-ALK (D5F3) CDx assay been considered a more advanced diagnostic method compared with traditional ALK IHC, and it has been approved in Korea as an ALK-testing method for selecting patients eligible for crizotinib. In the case of NGS, although it is the newest technology, with continuous development of platforms and data analysis methods enabling rapid and simultaneous detection of various genetic alterations including ALK fusion, its use has been minimal due to the high cost and incomplete coverage by the NHIS in Korea.
Prompt and accurate diagnosis of genetic alterations including ALK fusion can provide patients with the administration of appropriate drugs and thereby improve disease prognosis. Therefore, the present study investigated clinical, histopathological and immunohistochemical features that may be associated with ALK alteration and searched for other genetic changes that may accompany ALK fusion in NSCLC. The present study also investigated whether NGS, the promising, new method, could replace the traditional method of ALK FISH for the detection of ALK-positive NSCLC. Both ALK fusion and EGFR mutation were more frequent in tumors in never-smokers. The ALK IHC- or ALK FISH-positive tumors were similarly associated with younger patient age, female patients, frequent nodal metastases and advanced stage (III or IV) at the time of diagnosis and higher 1-year mortality. The more advanced disease with frequent nodal and distant metastases at the time of diagnosis could explain the higher 1-year mortality and shorter survival in patients with ALK fusion-positive adenocarcinomas; however, definite evidence that ALK fusion was an independent poor prognostic factor was not identified in the present study. Several studies have attempted to clarify whether ALK fusion is an independent risk factor for poor prognosis in cancers; however, the results have been inconsistent thus far (18,43,44).
Although the positive rates of TTF-1 and napsin A immunohistochemical staining were significantly higher than those of other antibodies in ALK fusion-positive tumors, the relevance of these markers to ALK fusion could not be demonstrated since both TTF-1 and napsin A are well-known markers for lung adenocarcinoma and are also expressed in quite a high proportion of adenocarcinomas with no ALK fusion. However, in 2017, a study reported that the overall survival was significantly longer in stage IV patients with TTF-1-positive adenocarcinomas than in patients with TTF-1-negative tumors (18 vs. 9 months) (45). Considering that the patients with ALK fusion-positive cancers frequently had either advanced stage III or IV diseases, TTF-1 may act as a marker with prognostic value in these patients.
Although the NGS analysis in the present study revealed no concurrent mutations in EGFR, BRAF, and KRAS in the ALK fusion-positive adenocarcinomas, and historically ALK rearrangement is considered a virtually exclusive event with other driver mutations (23,25,44), several studies have revealed that more than two driver mutations can occur in a small portion of lung adenocarcinomas (24,29,46). In ALK fusion-negative tumors, different types of EGFR-activating mutations, ERBB2 insertion, KRAS mutation and TPM3-NTRK1 fusion were detected by NGS in one, two, one and one cases, respectively. It may have been due to the small number of cases submitted to the NGS analysis in the present study that no concurrent mutations were detected in ALK fusion-positive tumors. The NTRK1 gene, which encodes the high-affinity nerve growth factor receptor TRKA protein, has been known to fuse with various partners at low frequency and act as an oncogenic driver in different malignancies, including lung adenocarcinoma, colorectal adenocarcinoma, papillary thyroid carcinoma, neuroblastoma, prostate adenocarcinoma, and breast carcinoma (47–49). NPM3-NTRK1 is a type of oncogenic TRK1 fusion gene, which can be detected in various cancers, including lung adenocarcinoma, colorectal carcinoma and papillary thyroid carcinoma, and it is inhibited by TRK1 inhibitors, such as entrectinib and larotrectinib (50–54). The ERBB2 mutations, known as oncogenic drivers, are identified in 2–4% of NSCLCs, particularly adenocarcinomas of young and non-smoking women (55–57). Constitutive activation of ERBB2 gene by mutation causes overexpression of HER2 protein and subsequent activation of downstream PI3K/AKT and MEK/ERK signaling pathways, which leads to carcinogenesis (58). The ERBB exon 20 insertion identified in the present study is understood to be the most common type of ERBB2 mutation in lung cancer (57,59,60). Studies evaluating the possibility of HER2-targeted treatment for lung cancer have been conducted and it may be possible that HER2-targeted antibodies or tyrosine kinase inhibitors could be a novel therapeutic approach for lung cancer in the future (61–63). Simultaneous detection of multiple oncogenic alterations, such as TRK1 fusion or ERBB2 mutation, is one of the strongest advantages of NGS testing. No genetic changes had been previously detected in those tumors with NPM3-TRK rearrangement or ERBB2 mutation prior to NGS in the present study.
Concerning the EGFR mutation, the results of NGS demonstrated a 90% agreement with those of the PNA clamping method. However, three out of five ALK FISH-positive tumors were negative in the NGS RNA panel analysis. The diagnostic agreement rate between ALK FISH and NGS RNA panel analysis was only 70%, while the agreement rate between ALK IHC and NGS was 80% (Table IV). Although the number of patients submitted to NGS analysis was small, such discrepancies among the three detection methods for ALK fusion cast doubts on the reliability of various detection methods and the overwhelming excellence of any one approach. Investigating the three ALK FISH-positive but NGS-negative cases more closely, it was identified that the rearrangement-positive cells, i.e., with abnormal split signals under fluorescence microscopy, were 16% each in two cases and 22% in the other case, all falling within or close to the range of borderline positivity of 10 to 20% (Table IV), which has been known as the primary source of discrepancy between FISH and other modalities (64,65). The ALK IHC results in these patients were negative in two and positive in one. The FFPE blocks selected for NGS analysis were relatively fresh and contained enough viable tumor cells. The quantity and quality of the RNA samples were sufficient for NGS analyses. Therefore, in those two patients with ALK FISH-positive/ALK IHC-negative/NGS-negative tumors, it was assumed that the results of ALK FISH were false-positive. The borderline FISH positivity of 16% and the concordant negativity in ALK IHC and NGS also seemed supportive of this conclusion. These patients did not receive ALK inhibitor therapy. However, the third patient with ALK FISH-positive/ALK IHC-positive/NGS-negative disease started crizotinib treatment based on the FISH result and exhibited partial response and stable disease for 13 months prior to commencement of this study. Therefore, it was concluded that the NGS result was false-negative in this patient. The targeted RNA sequencing by NGS is known to be sensitive enough to detect gene fusions with FFPE tissue (39). However, various factors, including fixation time, specimen size during fixation, and storage temperature and duration, can influence the RNA quality from FFPE samples (39,66). Although the RNA sample of the third patient was extracted from the FFPE block stored at consistent room temperature for only 13 months and exhibited a DV200 value sufficiently high for Illumina sequencing, it still could have been degraded in the process of fixation and storage due to some unknown causes. The present study used stored FFPE tissue blocks that were not expected to be used for NGS analysis. However, if NGS is used routinely for the detection of genetic alterations and all specimens are processed from the fixation step to minimize the degradation of genetic material and submitted to NGS analysis without prolonged storage, it is expected that the test accuracy would be improved. Currently, considering the discrepancies among the results of ALK FISH, IHC and NGS, it can be concluded that no one detection method is completely reliable. Therefore, it would be reasonable to more frequently add other testing methods to ALK FISH for the accurate diagnosis of ALK fusion-positive lung cancer, even though the ALK FISH has been considered the standard diagnostic method for a long time.
The present study assumed that the ALK test results would be more reliable when two different modalities, for example FISH and IHC, agreed with each other. Thus, during Kaplan-Meier survival analysis, it was reasonably concluded that ALK fusion-positive lung cancer had a worse prognosis compared with ALK fusion-negative disease; the mean survival time was 30.17 months in patients with tumors positive for both ALK FISH and IHC and 56.96 months in those with tumors negative for both ALK FISH and IHC. However, in certain patients the results of ALK FISH and IHC were discordant, and the mean survival time was 40.72 months in ALK FISH-positive/IHC-negative patients and 34.41 in ALK FISH-negative/IHC-positive patients. The present study considered the possibility that this difference in the survival periods could reflect the accuracy of FISH and IHC tests, and the ALK IHC-positive group could include more ALK fusion-positive patients than the ALK FISH-positive group. However, also considering that the positive rate of ALK IHC was much higher in the present study than the previously determined ALK fusion-positive rate in lung cancer (19–24), the ALK FISH-negative/IHC-positive group may also include a number of false-positive cases. The shorter survival period could be due to other clinicopathological features, such as frequent nodal metastasis and advanced stage, which were frequent in this group. This high positive rate of ALK IHC was the result of staining with clone 5A4. It was hypothesized that the relatively weak stainability of this clone in NSCLC could have made it difficult to differentiate between weakly stained tumor cells and background staining, resulting in frequent false-positive cases (67).
Nonetheless, the present study suggests that ALK FISH and IHC should be used together for more accurate results, and NGS analysis with an advantage of simultaneous detection of different mutations could also be considered an alternative to ALK FISH as the diagnostic standard. In the present study, TPM3-NTRK1 fusion or ERBB2 mutation, which were not anticipated at the time of diagnosis, were first detected in NGS analysis with a rather high frequency even in the small number of cases submitted. Even if a patient is known to have TPM3-NTRK1 fusion, they would not be able to receive tyrosine kinase inhibitor treatment, such as entrectinib, in Korea due to the lack of drug approval and coverage by the NHIS. Likewise, patients with ERBB2 mutations would not have access to HER2-targeted drugs at the time of diagnosis. However, considering the speed of the discovery of new therapeutic targets and the development of appropriate drugs, additional NGS results, even with no proper treatment at the time of diagnosis, would be helpful for future treatment decisions while monitoring the progress of the patient.
The present study confirmed that ALK-rearranged lung adenocarcinomas have characteristic clinical, histological and immunohistochemical features. Rapid and accurate diagnosis of ALK rearrangement is closely associated with the treatment and prognosis of patients. The present results emphasized that in practice ALK testing should be diversified; ALK FISH and IHC should be used concurrently to complement each other, and NGS analysis could be a good alternative of FISH. These conclusions were in agreement with the new molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors, which was jointly reported by IASLC/AMP/CAP, in that the role of ALK IHC and/or NGS analysis could be expanded further in clinical practice (39).
Supplementary Material
Acknowledgements
We are sincerely grateful to Dr Ensel Oh for her technical advice on selecting an appropriate next-generation sequencing platform and troubleshooting during experiments.
Funding
Not applicable.
Availability of data and material
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
BKS designed the study and reviewed and analyzed the histological and immunohistochemical features of submitted cases. WCC reviewed and analyzed the clinical data of patients, conducted statistical analyses, and performed the experiments. HKK contributed to the analysis and interpretation of the experimental and clinical data. All authors drafted, reviewed, edited, read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This study was conducted with the approval of the Institutional Review Board of Korea University Guro Hospital (Approval no. 2018GR0357). The clinical information including age, sex, smoking history, cancer stage, genetic mutation status and survival was obtained through the Human Biobank of Korea University of Guro Hospital, without identifiable personal data, according to the approved protocol by the Institutional Review Board of Korea University Guro Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Information Center (NCIC), corp-author Main Cancer Mortality Fraction in 2017. https://cancer.go.kr/lay1/S1T645C647/contents.do [Google Scholar]
- 3.Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al., editors. National Cancer Institute; Bethesda, MD: 2017. SEER Cancer Statistics Review, 1975–2015. [Google Scholar]
- 4.Fukui T, Mitsudomi T. Mutations in the epidermal growth factor receptor gene and effects of EGFR-tyrosine kinase inhibitors on lung cancers. Gen Thorac Cardiovasc Surg. 2008;56:97–103. doi: 10.1007/s11748-007-0193-8. [DOI] [PubMed] [Google Scholar]
- 5.Tamura K, Okamoto I, Kashii T, Negoro S, Hirashima T, Kudoh S, Ichinose Y, Ebi N, Shibata K, Nishimura T, et al. Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: Results of the West Japan thoracic oncology group trial (WJTOG0403) Br J Cancer. 2008;98:907–914. doi: 10.1038/sj.bjc.6604249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DH, Han JY, Yu SY, Kim HY, Nam BH, Hong EK, Kim HT, Lee JS. The role of gefitinib treatment for Korean never-smokers with advanced or metastatic adenocarcinoma of the lung: A prospective study. J Thorac Oncol. 2006;1:965–971. doi: 10.1097/01243894-200611000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, et al. EGFR mutations in non-small-cell lung cancer: Analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 9.Bos JL. Ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 10.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 11.Zhao XD, Deng HB, Lu CL, Bao YX, Lu X, Deng LL. Association of EGFR and KRAS mutations with expression of p-AKT, DR5 and DcR1 in non-small cell lung cancer. Neoplasma. 2017;64:182–191. doi: 10.4149/neo_2017_203. [DOI] [PubMed] [Google Scholar]
- 12.Campos-Parra AD, Zuloaga C, Manriquez ME, Avilés A, Borbolla-Escoboza J, Cardona A, Meneses A, Arrieta O. KRAS mutation as the biomarker of response to chemotherapy and EGFR-TKIs in patients with advanced non-small cell lung cancer: Clues for its potential use in second-line therapy decision making. Am J Clin Oncol. 2015;38:33–40. doi: 10.1097/COC.0b013e318287bb23. [DOI] [PubMed] [Google Scholar]
- 13.Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, Ladanyi M, Riely GJ. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119:356–362. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 15.Hong M, Kim RN, Song JY, Choi SJ, Oh E, Lira ME, Mao M, Takeuchi K, Han J, Kim J, Choi YL. HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol. 2014;9:419–422. doi: 10.1097/JTO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 16.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 17.Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, Nakajima T, Mano H, Takeuchi K. KLC1-ALK: A novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One. 2012;7:e31323. doi: 10.1371/journal.pone.0031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng D, Wang R, Zhang Y, Pan Y, Cheng X, Cheng C, Zheng S, Li H, Gong R, Li Y, et al. Prevalence and clinicopathological characteristics of ALK fusion subtypes in lung adenocarcinomas from Chinese populations. J Cancer Res Clin Oncol. 2016;142:833–843. doi: 10.1007/s00432-015-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, Ouafik L, Besse B, Rouquette I, Westeel V, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French cooperative thoracic intergroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 20.Vidal J, Clave S, de Muga S, González I, Pijuan L, Gimeno J, Remón J, Reguart N, Viñolas N, Gironés R, et al. Assessment of ALK status by FISH on 1000 Spanish non-small cell lung cancer patients. J Thorac Oncol. 2014;9:1816–1820. doi: 10.1097/JTO.0000000000000361. [DOI] [PubMed] [Google Scholar]
- 21.Kim TJ, Park CK, Yeo CD, Park K, Rhee CK, Kim J, Kim SJ, Lee SH, Lee KY, Yoon HK. Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non-small-cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage. J Surg Oncol. 2014;110:245–251. doi: 10.1002/jso.23646. [DOI] [PubMed] [Google Scholar]
- 22.Ali G, Proietti A, Pelliccioni S, Niccoli C, Lupi C, Sensi E, Giannini R, Borrelli N, Menghi M, Chella A, et al. ALK rearrangement in a large series of consecutive non-small cell lung cancers: Comparison between a new immunohistochemical approach and fluorescence in situ hybridization for the screening of patients eligible for crizotinib treatment. Arch Pathol Lab Med. 2014;138:1449–1458. doi: 10.5858/arpa.2013-0388-OA. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhang S, Yang X, Yang J, Zhou Q, Yin L, An S, Lin J, Chen S, Xie Z, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu CW, Cai XY, Shao Y, Li Y, Shi MW, Zhang LY, Wang L, Zhang YP, Wang LP, Tian YW. A case of lung adenocarcinoma with a concurrent EGFR mutation and ALK rearrangement: A case report and literature review. Mol Med Rep. 2015;12:4370–4375. doi: 10.3892/mmr.2015.4001. [DOI] [PubMed] [Google Scholar]
- 25.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik Wong M, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 26.Kim HJ, Choi EY, Jin HJ, Shin KC. Relationship between EGFR mutations and clinicopathological features of lung adenocarcinomas diagnosed via small biopsies. Anticancer Res. 2014;34:3189–3195. [PubMed] [Google Scholar]
- 27.Lim JU, Yeo CD, Rhee CK, Kim YH, Park CK, Kim JS, Kim JW, Lee SH, Kim SJ, Yoon HK, et al. Chronic obstructive pulmonary disease-related non-small-cell lung cancer exhibits a low prevalence of EGFR and ALK Driver Mutations. PLoS One. 2015;10:e0142306. doi: 10.1371/journal.pone.0142306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae NC, Chae MH, Lee MH, Kim KM, Lee EB, Kim CH, Park TI, Han SB, Jheon S, Jung TH, Park JY. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet. 2007;173:107–113. doi: 10.1016/j.cancergencyto.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Lee T, Lee B, Choi YL, Han J, Ahn MJ, Um SW. Non-small cell lung cancer with concomitant EGFR, KRAS, and ALK mutation: Clinicopathologic features of 12 cases. J Pathol Transl Med. 2016;50:197–203. doi: 10.4132/jptm.2016.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha SY, Choi SJ, Cho JH, Choi HJ, Lee J, Jung K, Irwin D, Liu X, Lira ME, Mao M, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget. 2015;6:5465–5474. doi: 10.18632/oncotarget.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee B, Lee T, Lee SH, Choi YL, Han J. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget. 2016;7:23874–23884. doi: 10.18632/oncotarget.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thunnissen E, Bubendorf L, Dietel M, Elmberger G, Kerr K, Lopez-Rios F, Moch H, Olszewski W, Pauwels P, Penault-Llorca F, Rossi G. EML4-ALK testing in non-small cell carcinomas of the lung: A review with recommendations. Virchows Arch. 2012;461:245–257. doi: 10.1007/s00428-012-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yatabe Y. ALK FISH and IHC: You cannot have one without the other. J Thorac Oncol. 2015;10:548–550. doi: 10.1097/JTO.0000000000000461. [DOI] [PubMed] [Google Scholar]
- 34.Savic S, Diebold J, Zimmermann AK, Jochum W, Baschiera B, Grieshaber S, Tornillo L, Bisig B, Kerr K, Bubendorf L. Screening for ALK in non-small cell lung carcinomas: 5A4 and D5F3 antibodies perform equally well, but combined use with FISH is recommended. Lung Cancer. 2015;89:104–109. doi: 10.1016/j.lungcan.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Cabillic F, Gros A, Dugay F, Begueret H, Mesturoux L, Chiforeanu DC, Dufrenot L, Jauffret V, Dachary D, Corre R, et al. Parallel FISH and immunohistochemical studies of ALK status in 3244 non-small-cell lung cancers reveal major discordances. J Thorac Oncol. 2014;9:295–306. doi: 10.1097/JTO.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 36.van der Wekken AJ, Pelgrim R, 't Hart N, Werner N, Mastik MF, Hendriks L, van der Heijden EHFM, Looijen-Salamon M, de Langen AJ, Staal-van den Brekel J, et al. Dichotomous ALK-IHC is a better predictor for ALK inhibition outcome than traditional ALK-FISH in advanced non-small cell lung cancer. Clin Cancer Res. 2017;23:4251–4258. doi: 10.1158/1078-0432.CCR-16-1631. [DOI] [PubMed] [Google Scholar]
- 37.Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr K, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the college of American pathologists, the international association for the study of lung cancer, and the association for molecular pathology. J Mol Diagn. 2018;20:129–159. doi: 10.1016/j.jmoldx.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, Temple-Smolkin RL, Voelkerding KV, Nikiforova MN. Guidelines for validation of next-generation sequencing-based oncology panels: A joint consensus recommendation of the association for molecular pathology and college of American pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS One. 2007;2:e1261. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 41.Camidge R, Kim HR, Ahn M, Yang JC, Han J, Lee J, Hochmair M, Li JY, Chang G, Lee K, et al. PL02.03 brigatinib vs crizotinib in patients with ALK inhibitor-naive advanced ALK+ NSCLC: First report of a phase 3 trial (ALTA-1L) J Thorac Oncol. 2018;13:S184–S185. doi: 10.1016/j.jtho.2018.08.011. [DOI] [Google Scholar]
- 42.Camidge DR, Kim HR, Ahn MJ, Yang JC, Han JY, Lee JS, Hochmair MJ, Li JY, Chang GC, Lee KH, et al. Brigatinib versus Crizotinib in ALK-positive non-small-cell lung cancer. N Engl J Med. 2018;379:2027–2039. doi: 10.1056/NEJMoa1810171. [DOI] [PubMed] [Google Scholar]
- 43.Yang P, Kulig K, Boland JM, Erickson-Johnson MR, Oliveira AM, Wampfler J, Jatoi A, Deschamps C, Marks R, Fortner C, et al. Worse disease-free survival in never-smokers with ALK+ lung adenocarcinoma. J Thorac Oncol. 2012;7:90–97. doi: 10.1097/JTO.0b013e31823c5c32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu SG, Kuo YW, Chang YL, Shih JY, Chen YH, Tsai MF, Yu CJ, Yang CH, Yang PC. EML4-ALK translocation predicts better outcome in lung adenocarcinoma patients with wild-type EGFR. J Thorac Oncol. 2012;7:98–104. doi: 10.1097/JTO.0b013e3182370e30. [DOI] [PubMed] [Google Scholar]
- 45.Schilsky JB, Ni A, Ahn L, Datta S, Travis WD, Kris MG, Chaft JE, Rekhtman N, Hellmann MD. Prognostic impact of TTF-1 expression in patients with stage IV lung adenocarcinomas. Lung Cancer. 2017;108:205–211. doi: 10.1016/j.lungcan.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Won JK, Keam B, Koh J, Cho HJ, Jeon YK, Kim TM, Lee SH, Lee DS, Kim DW, Chung DH. Concomitant ALK translocation and EGFR mutation in lung cancer: A comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol. 2015;26:348–354. doi: 10.1093/annonc/mdu530. [DOI] [PubMed] [Google Scholar]
- 47.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019;32:147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 48.Ardini E, Bosotti R, Borgia AL, De Ponti C, Somaschini A, Cammarota R, Amboldi N, Raddrizzani L, Milani A, Magnaghi P, et al. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol Oncol. 2014;8:1495–1507. doi: 10.1016/j.molonc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, Mahale S, Davies KD, Aisner DL, Pilling AB, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Chi P. Basket trial of TRK inhibitors demonstrates efficacy in TRK fusion-positive cancers. J Hematol Oncol. 2018;11:78. doi: 10.1186/s13045-018-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolfo C, Raez L. New targets bring hope in squamous cell lung cancer: Neurotrophic tyrosine kinase gene fusions. Lab Invest. 2017;97:1268–1270. doi: 10.1038/labinvest.2017.91. [DOI] [PubMed] [Google Scholar]
- 53.Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1:e000023. doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farago AF, Le LP, Zheng Z, Muzikansky A, Drilon A, Patel M, Bauer TM, Liu SV, Ou SH, Jackman D, et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol. 2015;10:1670–1674. doi: 10.1097/01.JTO.0000473485.38553.f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z, Wu L, Cao J, Yang Z, Zhou C, Cao L, Wu H, Shen H, Jin M, Zhang Y, et al. Clinical characterization of ERBB2 exon 20 insertions and heterogeneity of outcomes responding to afatinib in Chinese lung cancer patients. Onco Targets Ther. 2018;11:7323–7331. doi: 10.2147/OTT.S173391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillai RN, Behera M, Berry LD, Rossi MR, Kris MG, Johnson BE, Bunn PA, Ramalingam SS, Khuri FR. HER2 mutations in lung adenocarcinomas: A report from the lung cancer mutation consortium. Cancer. 2017;123:4099–4105. doi: 10.1002/cncr.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonobe M, Manabe T, Wada H, Tanaka F. Lung adenocarcinoma harboring mutations in the ERBB2 kinase domain. J Mol Diagn. 2006;8:351–356. doi: 10.2353/jmoldx.2006.050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2009;27:5838–5847. doi: 10.1200/JCO.2009.22.1507. [DOI] [PubMed] [Google Scholar]
- 59.Lee JW, Soung YH, Kim SY, Nam SW, Park WS, Wang YP, Jo KH, Moon SW, Song SY, Lee JY, et al. ERBB2 kinase domain mutation in the lung squamous cell carcinoma. Cancer Lett. 2006;237:89–94. doi: 10.1016/j.canlet.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 60.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O'Meara S, Smith R, Parker A, et al. Lung cancer: Intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 61.Liu S, Li S, Hai J, Wang X, Chen T, Quinn MM, Gao P, Zhang Y, Ji H, Cross DAE, Wong KK. Targeting HER2 aberrations in non-small cell lung cancer with osimertinib. Clin Cancer Res. 2018;24:2594–2604. doi: 10.1158/1078-0432.CCR-17-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Langen AJ, Jebbink M, Hashemi SMS, Kuiper JL, de Bruin-Visser J, Monkhorst K, Thunnissen E, Smit EF. Trastuzumab and paclitaxel in patients with EGFR mutated NSCLC that express HER2 after progression on EGFR TKI treatment. Br J Cancer. 2018;119:558–564. doi: 10.1038/s41416-018-0194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mazieres J, Barlesi F, Filleron T, Besse B, Monnet I, Beau-Faller M, Peters S, Dansin E, Früh M, Pless M, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: Results from the European EUHER2 cohort. Ann Oncol. 2016;27:281–286. doi: 10.1093/annonc/mdv573. [DOI] [PubMed] [Google Scholar]
- 64.von Laffert M, Stenzinger A, Hummel M, Weichert W, Lenze D, Warth A, Penzel R, Herbst H, Kellner U, Jurmeister P, et al. ALK-FISH borderline cases in non-small cell lung cancer: Implications for diagnostics and clinical decision making. Lung Cancer. 2015;90:465–471. doi: 10.1016/j.lungcan.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 65.Ilie MI, Bence C, Hofman V, Long-Mira E, Butori C, Bouhlel L, Lalvée S, Mouroux J, Poudenx M, Otto J, et al. Discrepancies between FISH and immunohistochemistry for assessment of the ALK status are associated with ALK ‘borderline’-positive rearrangements or a high copy number: A potential major issue for anti-ALK therapeutic strategies. Ann Oncol. 2015;26:238–244. doi: 10.1093/annonc/mdu484. [DOI] [PubMed] [Google Scholar]
- 66.Heyer EE, Deveson IW, Wooi D, Selinger CI, Lyons RJ, Hayes VM, O'Toole SA, Ballinger ML, Gill D, Thomas DM, et al. Diagnosis of fusion genes using targeted RNA sequencing. Nat Commun. 2019;10:1388. doi: 10.1038/s41467-019-09374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ibrahim M, Parry S, Wilkinson D, Bilbe N, Allen D, Forrest S, Maxwell P, O'Grady A, Starczynski J, Tanier P, et al. ALK immunohistochemistry in NSCLC: Discordant staining can impact patient treatment regimen. J Thorac Oncol. 2016;11:2241–2247. doi: 10.1016/j.jtho.2016.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.


