Abstract
Several proteins in the iRhom family function as oncogenic regulators in certain cancers. However, the function of these proteins in cervical cancer (CC) is unknown. The relationship of iRhom1 and iRhom2 expression with the clinicopathological features and prognosis of patients with CC was investigated, and their possible molecular mechanisms were examined using in vitro experiments. The expression of iRhom1 and iRhom2 in CC samples of 83 patients was determined by immunohistochemistry (IHC), and the associations of their expression with the clinicopathological features of patients were determined. The relationship of iRhom1, iRhom2, and Ki-67 expression with survival rates was determined using Kaplan-Meier analysis and Cox regression analyses. HeLa cells were analyzed using MTT assays, cell cycle analysis, and apoptosis assays. The results revealed that CC tissues had higher levels of iRhom1 and iRhom2 than adjacent normal tissues. Increased expression of iRhom1, iRhom2, and K-i67 was significantly associated with tumor stage, size, and parametrium invasion. High expression of iRhom1, iRhom2 and Ki-67 was correlated with poor outcomes. Cancer stage and iRhom2 expression were independent prognostic indicators of CC. Knockdown of iRhom1 and iRhom2 in HeLa cells inhibited cell proliferation, promoted the G1 phase and relieved S-phase arrest, and induced apoptosis. Genomic microarray analysis indicated that iRhom2 knockdown altered several pathways with roles in oncogenesis, including the expression of five genes in the Wnt/β-catenin pathway. Western blotting in HeLa cells revealed that iRhom1 knockdown significantly suppressed the expression of β-catenin, Myc, p-EGFR and TGFBR2, and increased the expression of FAS; iRhom2 knockdown significantly suppressed the expression of β-catenin, GSK3β, p-EGFR and Myc. These results were consistent with the genomic microarray data. Collectively, the results indicated that iRhom1 and iRhom2 may function as oncogenes in CC and are potential therapeutic targets.
Keywords: iRhom1, iRhom2, uterine cervical cancer, prognostic biomarker, Wnt/β-catenin pathway, genomic microarray
Introduction
Cervical cancer (CC) is among the most common cancers in women worldwide. It mostly affects women in poor countries that lack public health infrastructures, CC screening, and vaccines against the human papillomavirus (HPV). In 2016, 511,000 women [95% confidence interval (CI), 414,000-542,000] developed CC, and 247,000 women (95% CI, 204,000-263,000) succumbed to CC worldwide (1). Radical surgery and radiotherapy are well-established and effective; however, tumors recur or progress in >30% of patients (2,3). The risk factors for CC are well known, but few targeted therapies are available. Thus, it is important to identify the genetic and molecular abnormalities underlying CC for the development of new and effective therapies.
Rhomboid genes were first described in Drosophila melanogaster, where they function as serine proteases and key regulators of epidermal growth factor receptor (EGFR) signaling (4). The inactive rhomboid protein 1 (iRhom1 or RHBDF1) is a member of the rhomboid family that lacks protease activity. This protein traverses the membrane 7 times, and has a long N-terminal cytoplasmic extension that comprises approximately half of the polypeptide sequence. This protein occurs in the endoplasmic reticulum and Golgi apparatus, rather than the plasma membrane. Several recent studies demonstrated that iRhom1 functions as an oncogene in head and neck neoplasms (5), breast (6) and colorectal cancer (7). Molecular studies reported that iRhom1 has a critical function in G-protein coupled receptor (GPCR) ligand-stimulated processes that activate EGFR ligands in head and neck squamous cancer cells (5) and breast cancer (6). Overexpression of iRhom1 was revealed to have a significantly positive correlation with poor overall survival (OS) in patients with colorectal cancer (7).
Inactive rhomboid protein 2 (iRhom2 or RHBDF2) is also a catalytically inactive member of the rhomboid family. This protein mediates the maturation of TNFα converting enzyme (TACE, also called ADAM17) and trafficking to the cell surface (8). iRhom2 functions as an essential endoplasmic reticulum-to-Golgi trafficking factor for metalloproteinase ADAM17 when macrophages secrete TNFα (9,10). We previously demonstrated that ADAM17 expression was an independent prognostic indicator for CC (11). In esophageal cancer, the hyperproliferative phenotype was revealed to be associated with altered iRhom2 levels and increased EGFR signaling (12,13). Identification of a novel p63-associated pathway indicated that therapeutic modulation of iRhom2 has potential for treatment of hyperproliferative skin disease and neoplasia (14). Blaydon et al recently reported that missense mutations in iRhom2 were responsible for the autosomal dominant condition tylosis with oesophageal cancer (TOC) in three families from the United Kingdom, United States and Germany (10). Cancer-associated fibroblasts (CAFs) promote tumorigenesis in several types of cancers. iRhom2 overexpression was revealed to occur in CAFs isolated from human diffuse-type gastric cancers (15). However, the levels and roles of iRhom1 and iRhom2 in the onset and progression of CC remain unknown.
In the present study, the expression of iRhom1 and iRhom2 in CC clinical samples was first assessed. Then, their associations with the clinicopathological features of the CC patients were determined and their prognostic value was assessed. Subsequently, HeLa cells were used to evaluate the effects of knockdown of iRhom1 and iRhom2 on cell proliferation, cell cycle distribution, and apoptosis. Finally, microarray analysis was used to identify the molecular mechanisms responsible for iRhom-mediated promotion of CC by examination of pathways that have critical roles in the development and progression of CC.
Materials and methods
Patients and tissue collection
The Ethics Committee of Fujian Provincial Cancer Hospital, which is affiliated with Fujian Medical University, provided approval of this study. Samples from 83 consecutive patients (age range, 32–80 years) with CC were collected for immunohistochemical (IHC) analysis from January 2010 to December 2012. Prior to surgery, none of the enrolled patients received chemotherapy or radiotherapy. Cancer classification followed the 2009 Federation International of Gynecology and Obstetrics (FIGO) protocol, and enrolled patients were followed-up until December 2017 or death. Eligibility was determined following hysterectomy (total, modified-radical, or radical), bilateral salpingo-oophorectomy, pelvic lymphadenectomy, or para-aortic lymphadenectomy. Surgical staging was performed no more than 8 weeks before initiation of radiotherapy. The included patients had hematological, liver, renal function, and other laboratory variables within normal ranges (creatinine clearance ≥40 ml/min, leucocytes ≥4.0×109/l, platelets ≥100×109/l, and hemoglobin ≥10 g/dl). Any patient with a secondary malignancy, a serious concomitant systemic disorder, or a psychiatric disease was deemed ineligible. For validation of each diagnosis, two independent pathologists evaluated the IHC results. After patients provided written informed consent, samples were used for analysis. Twenty fresh CC tissues specimens (11 from squamous cell carcinomas, 5 from adenocarcinomas, 1 from small-cell carcinoma, and 3 from adenosquamous carcinomas) and matching non-cancerous adjacent cervical tissue samples were used for immunoblotting.
Study endpoints
Progression-free survival (PFS, the duration from enrollment to disease progression or death) was the primary endpoint, and local-regional failure, distant failure, and OS were the secondary endpoints.
Reagents
The following antibodies were obtained from Abcam: iRhom1 (ID product code ab81342), iRhom2 (ID product code ab116139), Ki67 (ID product code ab92742), β-catenin (ID product code ab32572), Fas (ID product code ab82419), GSK3B (ID product code ab32391), Myc (ID product code ab32), TGFBR2 (ID product code ab78419), EGFR (ID product code ab52894), p-EGFR (ID product code ab40815), GADPH (ID product code ab8245), and actin (ID product code ab179467). Horseradish peroxidase (HRP)-labeled secondary antibodies (ID product code sc-2354 and sc-2357) were purchased from Santa Cruz Biotechnology, Inc.
Immunohistochemistry
IHC analyses employed formalin-fixed and paraffin-embedded 5 µm-thick sections. These sections were deparaffinized in xylene and rehydrated in a graded series of alcohol. For labeling, the avidin-biotin complex (ABC) method was used. For antigen retrieval, a steamer was used for 20 min in an EDTA buffer. Then, samples were treated with 3% hydrogen peroxide in methanol for 20 min to block endogenous peroxidase activity and the reaction was blocked for 1 h by incubation in a 5% solution of non-fat milk. Slides were then incubated with polyclonal rabbit antibodies against the iRhom1 (1:50 dilution), iRhom2 (1:50 dilution), or Ki67 (1:100 dilution) antibody at room temperature for 1 h. The secondary antibody (MaxVision HRP-polymer anti-rabbit; Fuzhou Maixin-Biotech Co., Ltd.) was added, and counterstaining was performed using Mayer's hematoxylin and diaminobenzidine. Tumor cells that had staining in the cytoplasm were considered iRhom1- or iRhom2-positive, and tumor cells that had staining in the nucleus were considered Ki67-positive.
For IHC analysis of iRhom1, iRhom2 and Ki-67, images of five typical visual fields of each section were analyzed using software (Image-Pro Plus 6.0; Media Cybernetics, Inc.), and the mean absorption was used to score expression. All sections were scored separately by two pathologists who were unaware of the clinical parameters of the corresponding patients. Stain intensity was scored as 0 (negative), 1 (weak), 2 (strong). The percentage of positively-stained area was 0 (fewer than 10% positive cells), 1 (10 to 25% positive cells), 2 (26 to 50% positive cells), and 3 (50% or more positive cells). The final score was determined from the average of products in stain intensity and percentage with staining that were immunoreactive on the cell membranes and/or cytoplasm by examination of 10 typical microscopic fields. The final IHC score was defined as 0 (−, negative expression), 1–2 (+, weak expression), 3–4 (++, medium expression), 5–6 (+++, strong expression).
Cell culture
HeLa cells (Chinese Academy of Sciences Cell Bank) were plated in 6-well plates (~2×106 cells/well), in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS; 1 ml/well). These cells were grown at 37°C with 5% CO2 and 95% air. After 24 h, cell attachment had occurred and cells were then incubated in SBF-free medium for 16 to 18 h.
Recombinant lentiviral vector construction and cell infection
Short hairpin RNAs (shRNAs) that targeted the iRhom1 gene (NM_022450.3) and iRhom2 gene (NM_024599.5) were designed, and lentiviruses were constructed to knockdown these genes. The shRNA targeting iRhom1 was GAGGCTGGCGGAAGCAGAA, the shRNA targeting iRhom2 was CGTGTCTGTGGTCTTTCAA, and the negative control (NC) sequence was TTCTCCGAACGTGTCACGT. Related stem-loop DNA oligonucleotides were prepared and inserted into a lentiviral vector (pGCSIL-GFP; Shanghai GeneChem Co., Ltd.). The Lentivector Expression System (Shanghai GeneChem Co., Ltd.) was used to produce lentiviruses expressing iRhom1 shRNA, iRhom2 shRNA, or NC shRNA.
Infection of HeLa cells with lentivirus
HeLa cells were grown in 12-well plates, then infected with a shRNA-expressing lentivirus, according to the desired multiplicity of infection (MOI). Cells were then observed using fluorescence microscopy (MicroPublisher 3.3RTV; Olympus Corp.) at 72 h. Cells were harvested at 120 h to determine knockdown efficiency using immunoblotting.
Western blotting
Samples were initially added to a radio-immunoprecipitation buffer (Beyotime Institute of Biotechnology) with 100 nM phenylmethylsulfonyl fluoride (PMSF). After assessment of protein concentrations with a bicinchoninic acid kit (Pierce; Thermo Fisher Scientific, Inc.), proteins (50 µg per lane) were separated by electrophoresis on 12% SDS-PAGE gels, and then transferred to PVDF membranes (EMD Millipore). These membranes were incubated with the primary antibody such as iRhom1 (1:50 dilution), iRhom2 (1:50 dilution), β-catenin (1:1,000 dilution), Fas (1:150 dilution), GSK3B (1:100 dilution), Myc (1:1,000 dilution), TGFBR2 (1:100 dilution), EGFR (1:1000 dilution), p-EGFR (1:1,000 dilution), GADPH (1:1,500 dilution) and actin (1:1,500 dilution) in Tris-buffered saline with 0.05% Tween-20 and 5% non-fat dry milk. After incubation at room temperature for 2 h, the membranes were washed, and then incubated with the HRP-labeled secondary antibody (1:2,000 dilution) at room temperature for 1 h. Electrochemiluminescence (Elecsys 2010; Roche Diagnostics, Basel, Switzerland) was used for visualization of proteins. Image Lab version 3.0 software was used to perform the densitometric analysis of blots.
MTT assay
Transfected HeLa cells were added to a 96-well plate (1×104 cells/well) and incubated for 24, 48, 72 and 96 h. Then MTT (10 µl; 5 mg/ml) in PBS (pH 7.4, Sigma-Aldrich; Merck KGaA) was added to each well for 4 h. After removal of the supernatant, dimethyl sulfoxide (DMSO, 100 µl/well; Thermo Fisher Scientific, Inc.) was added and the absorbance was measured at 490 nm using a microplate reader. Each experiment was performed in triplicate, and each measurement was also performed in triplicate.
Cell cycle analysis
Approximately 1×106 HeLa cells were treated with trypsin, washed twice in PBS, and then incubated overnight in 10 ml of cold ethanol. Cells were then centrifuged with 1,000 × g and the ethanol was removed. They were then washed twice in PBS, added to 100 µl RNase (30 min at 37°C), and then centrifuged at 1,000 × g. The pellet was added to 400 µl propidium iodide, and allowed to stand in darkness for 30 min at 4°C. The BD FACSCalibur system (BD Biosciences) was then used for flow cytometry and analysis of cell cycle distribution.
Clone formation assay
For each group, ~1×104 HeLa cells were added to each well of a 6-well culture plate (3 wells/group). The cells were incubated for 14 days at 37°C, and the growth medium was replaced every 3 days. After 14 days, the cells were washed two times with PBS and 0.5% crystal violet was added for staining in 5 min under ambient temperature. Microscopy (light, 10×10) was used to determine the number of colonies that had at least 50 cells. The efficiency of plate clone formation was defined as 100% × (number of colonies/number of cells inoculated), and there were 3 replicates per experiment.
Cell apoptosis assay with Annexin V-APC single-color staining
After collection of HeLa cells (~2×105), allophycocyanin (APC)-labeled Annexin V (cat. no. 88-8007; eBioscience) were added for single-color staining to detect apoptotic (Annexin V positive) cells. A total of 1.0×106 cells were washed twice with pre-cooled PBS (pH 7.4), and incubated for 15 min in 100 µl staining buffer including 5 µl APC-labeled Annexin V. FACS analysis for Annexin V staining was subsequently performed by flow cytometry (FACSCalibur; BD Biosciences).
Microarray processing and analysis
Total RNA was extracted from HeLa cells that were infected with lentivirus expressing NC/shRNA (n=3) or iRhom2/shRNA (n=3) by use of the TRIzol reagent. RNA was assessed using the Thermo NanoDrop 2000 and Agilent 2100 Bioanalyzer. Microarray processing and measurement of gene expression profiles were performed using the Affymetrix human GeneChip primeview (Affymetrix Inc.; Thermo Fisher Scientific, Inc.), using the manufacturer's protocols. The GeneChip 3′ IVT Expression Kit was used to perform reverse transcription, dsDNA template transformation, in vitro transcription, and biotin labeling. The GeneChip Hybridization Wash and Stain Kit was then used to perform microarray hybridization, washing, and staining. Finally, the GeneChip Scanner 3000 was employed to perform scanning of the arrays and to produce the raw data.
Integrative analyses
The integrative analyses were generated using Ingenuity Pathway Analysis (IPA; Ingenuity Systems®; www.ingenuity.com). A file that had 596 gene IDs and mean log2(fold-change) data was uploaded to identify canonical pathways, upstream regulators, relationships with cellular functions and diseases, effects on regulators, and network analysis. A P-value and a Z-score were determined for each pathway/network so that random data had a low probability of producing significant predictions. The P-value (from a right-sided Fisher's exact test) considers the number of input genes and the number of molecules in the database that could be in networks. The Z-score indicates activation or inhibition of a pathway/network. The IPA database is based on experiments and results published in numerous professional journals. The final graphical representation of each pathway/network has nodes (genes) and edges (biological relationships between nodes), with unique symbols used for different classes of molecules.
Statistical analyses
The statistical analyses were performed using SPSS (version 19.0; IBM Corp.). The correlations in the expression of different markers were evaluated using Pearson's coefficient, and the Chi-square test was used to calculate the statistical significance of differences between groups. Kaplan-Meier survival curves were plotted, and the log-rank test was used to determine the significance of differences between these curves. A multivariate analysis (Cox's non-parametric model) was performed to assess the relationship of iRhom1, iRhom2 and Ki-67 expression on progression-free survival (PFS) and overall survival (OS). The results from western blotting experiments (means ± standard deviations) were obtained from at least three independent experiments. The statistical significance of differences was determined using Student's t-test and a one-way analysis of variance (ANOVA) with post hoc Student-Newman-Keuls test. For all data, a P-value <0.05 was considered to indicate a statistically significant difference.
Results
Expression of iRhom1, iRhom2 and Ki-67
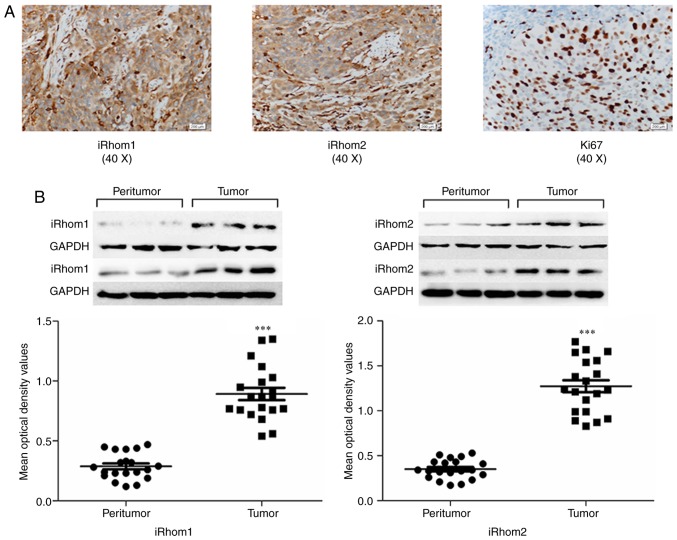
Tissue specimens from 83 CC patients were examined using IHC analysis for iRhom1, iRhom2 and Ki-67. iRhoms (−/+) and iRhoms (++/+++) were based on the average of products in stain intensity and percentage of positively-stained area. Pathological analysis indicated squamous cell carcinomas (n=44), adenocarcinomas (n=29), small-cell carcinomas (n=4), and adenosquamous carcinomas (n=6). iRhom1 and iRhom2 were present in the cytoplasm of tumor cells, and Ki67 was mainly in the nucleus, as indicated by dark yellow granules (Fig. 1A). There was high expression of iRhom1 in 55 out of the 83 cases (66.3%), of iRhom2 in 49 out of the 83 cases (59.1%), and of Ki-67 in 48 out of the 83 cases (57.8%) (Table I).
Figure 1.
Expression of iRhom1, iRhom2 and Ki-67 in cancerous tissues and adjacent normal tissues of patients with CC. (A) Representative photomicrographs (40X) of CC specimens stained with anti-iRhom1, anti-iRhom2 and anti-Ki67 antibodies. iRhom1 and iRhom2 were in the cytoplasm, and Ki-67 was mainly in the nucleus, as indicated by dark-yellow granules. (B) Quantitation of results, revealing increased levels of iRhom1 and iRhom2 in cancerous tissues relative to paired adjacent non-cancerous tissues (***P<0.001). iRhom1, inactive rhomboid protein 1; iRhom2, inactive rhomboid protein 2; CC, cervical cancer.
Table I.
Relationship of iRhom1, iRhom2, Ki-67 expression with clinicopathological features of patients with cervical cancer.
| Clinical features | n | iRhom1 (−/+) | iRhom1 (++- +++) | P-value | iRhom2 (−/+) | iRhom2 (++- +++) | P-value | Ki-67 (−/+) | Ki-67 (++- +++) | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| 83 | 28 | 55 | 34 | 49 | 35 | 48 | ||||
| Age, years | 0.176 | 0.479 | 0.146 | |||||||
| ≤45 | 40 | 16 | 24 | 17 | 23 | 14 | 26 | |||
| >45 | 43 | 12 | 31 | 17 | 26 | 21 | 22 | |||
| Clinical stage (FIGO) | 0.042a | 0.010a | <0.001a | |||||||
| I | 35 | 16 | 19 | 20 | 15 | 23 | 12 | |||
| II | 48 | 12 | 36 | 14 | 34 | 12 | 36 | |||
| Histology | 0.786 | 0.728 | 0.116 | |||||||
| Adenocarcinoma | 29 | 10 | 19 | 14 | 15 | 11 | 18 | |||
| Squamous carcinoma | 44 | 16 | 28 | 16 | 28 | 18 | 26 | |||
| Small cell carcinoma | 4 | 1 | 3 | 2 | 2 | 4 | 0 | |||
| Adenosquamous carcinoma | 6 | 1 | 5 | 2 | 4 | 2 | 4 | |||
| Lymph node metastasis | 0.081 | 0.288 | 0.002a | |||||||
| Positive | 21 | 4 | 17 | 7 | 14 | 3 | 18 | |||
| Negative | 62 | 24 | 38 | 27 | 35 | 32 | 30 | |||
| Differentiation | 0.287 | 0.507 | 0.472 | |||||||
| Well-differentiated | 10 | 4 | 6 | 5 | 5 | 6 | 4 | |||
| Moderately-differentiated | 56 | 21 | 35 | 24 | 32 | 22 | 34 | |||
| Poorly-differentiated | 17 | 3 | 14 | 5 | 12 | 7 | 10 | |||
| Tumor size (cm) | 0.004a | 0.403 | 0.069 | |||||||
| >4 | 17 | 1 | 16 | 6 | 11 | 4 | 13 | |||
| ≤4 | 66 | 27 | 39 | 28 | 38 | 31 | 35 | |||
| Parametrium invasion | 0.039a | 0.136 | 0.448 | |||||||
| Positive | 16 | 2 | 14 | 9 | 7 | 6 | 10 | |||
| Negative | 67 | 26 | 41 | 25 | 42 | 29 | 38 |
Bold values are statistically significant (P<0.05). iRhom1, inactive rhomboid protein 1; iRhom2, inactive rhomboid protein 2.
The possible function of iRhom1 and iRhom2 was investigated in CC tumorigenesis by performing western blotting of normal cervical tissues and adjacent fresh CC tissues. The results indicated that iRhom1 and iRhom2 had higher expression in CC tissues than adjacent noncancerous tissues (Fig. 1B, P<0.001).
Correlation of iRhom1, iRhom2 and Ki-67 with demographic and clinical factors
The correlations of expression of iRhom1, iRhom2 and Ki-67 with various clinical factors were examined (Table I). All three proteins had a significant association with FIGO stage (PiRhom1=0.042; PiRhom2=0.010; PKi-67<0.001). There was also a significant association between Ki-67 expression and lymph node metastasis (P=0.002), and significant associations of iRhom1 expression with parametrium invasion (P=0.039) and tumor size (P=0.04).
Correlation of iRhom1, iRhom2 and Ki-67 with clinical outcomes
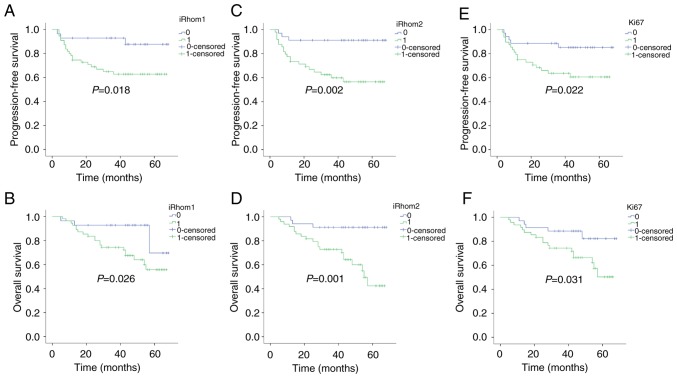
Kaplan-Meier analysis was also used to determine the independent prognostic value of iRhom1, iRhom2, and Ki-67 on PFS and OS (Fig. 2). iRhoms (−/+) represented weak expression and iRhoms (++/+++) represented strong expression. The results revealed that patients with strong expression of each protein had a significantly worse PFS than those with weak expression (PiRhom1=0.018; PiRhom2=0.002; PKi-67=0.022). In addition, patients with strong expression of each protein had significantly worse OS than those with weak expression (PiRhom1=0.026; PiRhom2=0.001; PKi-67=0.031).
Figure 2.
Kaplan-Meier analysis of PFS and OS with stratification by expression of iRhom1, iRhom2, and Ki-67 in patients with CC (n=83). (A and B) Strong expression (++/+++) of iRhom1 was associated with poor PFS (P=0.018) and poor OS (P=0.026). (C and D) Strong expression (++/+++) of iRhom2 was associated with poor PFS (P=0.002) and poor OS (P=0.001). (E and F) Patients with strong expression (++/+++) of Ki67 had poor PFS (P=0.022) and poor OS (P=0.031). PFS, progression-free survival; OS, overall survival; iRhom1, inactive rhomboid protein 1; iRhom2, inactive rhomboid protein 2; CC, cervical cancer.
Multivariate analysis of prognostic variables
multivariate Cox regression analysis was used to examine the relationship of OS with iRhom1, iRhom2, Ki-67, age, histology, stage, metastases to lymph nodes, and invasion of the parametrium (Table II). The results indicated that iRhom2 expression [hazard ratio (HR)=4.374; P=0.033; 95% confidence interval (CI)=1.130–16.937] and FIGO stage (HR=1.778; P=0.027; 95% CI=1.069–2.598) were statistically significant and independent prognostic factors (Table II). Multivariate analysis of PFS indicated that iRhom2 expression (HR=4.074; P=0.036; 95% CI=1.097–15.129) and FIGO stage (HR=1.892; P=0.014; 95% CI=1.137–3.147) were also statistically significant and independent prognostic factors (Table II).
Table II.
Multivariate analysis of prognostic factors for patients with cervical cancer (n=83).
| OS | |||
|---|---|---|---|
| Parameters | Hazard ratio | P-value | 95%CI |
| Age, years | 1.047 | 0.916 | 0.443–2.476 |
| ≤45 | |||
| >45 | |||
| Histology | 0.97 | 0.903 | 0.597–1.576 |
| Adenocarcinoma | |||
| Squamous carcinoma | |||
| Small cell carcinoma | |||
| Adenosquamous carcinoma | |||
| Clinical stage (FIGO) | 1.778 | 0.027a | 1.069–2.598 |
| I | |||
| II | |||
| Lymph node metastasis | 0.475 | 0.152 | 0.156–1.335 |
| Positive | |||
| Negative | |||
| Parametrium invasion | 1.767 | 0.342 | 0.546–5.714 |
| Positive | |||
| Negative | |||
| iRhom1 | 2.737 | 0.119 | 0.771–9.712 |
| iRhom2 | 4.374 | 0.033a | 1.130–16.937 |
| Ki-67 | 1.904 | 0.251 | 0.634–5.719 |
| PFS | |||
| Age, years | 1.2 | 0.68 | 0.506–2.846 |
| ≤45 | |||
| >45 | |||
| Histology | 0.986 | 0.956 | 0.597–1.630 |
| Adenocarcinoma | |||
| Squamous carcinoma | |||
| Small cell carcinoma | |||
| Adenosquamous carcinoma | |||
| Clinical stage (FIGO) | 1.892 | 0.014a | 1.137–3.147 |
| I | |||
| II | |||
| Lymph node metastasis | 0.403 | 0.106 | 0.134–1.213 |
| Positive | |||
| Negative | |||
| Parametrium invasion | 1.429 | 0.545 | 0.450–4.539 |
| Positive | |||
| Negative | |||
| iRhom1 | 3.037 | 0.085 | 0.858–10.753 |
| iRhom2 | 4.074 | 0.036a | 1.097–15.129 |
| Ki-67 | 1.876 | 0.272 | 0.611–5.759 |
Bold values are statistically significant (P<0.05). OS, overall survival; iRhom1, inactive rhomboid protein 1; iRhom2, inactive rhomboid protein 2; PFS, progression-free survival.
Oncogenic roles of iRhom1 and iRhom2 knockdown in HeLa cells
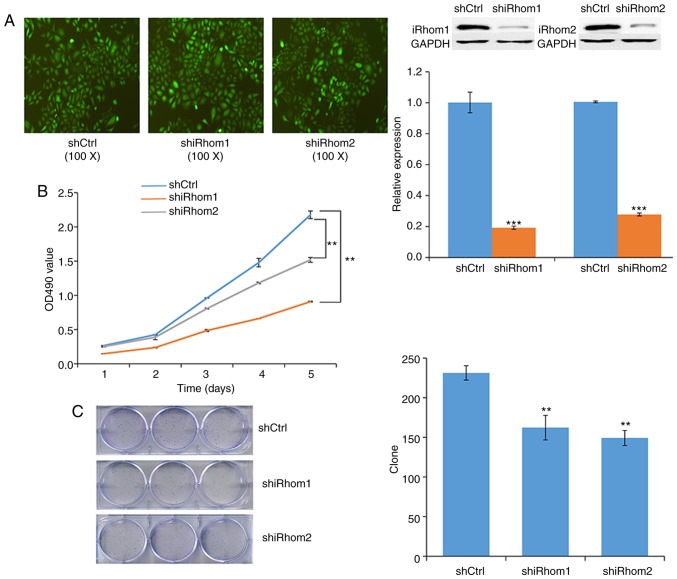
Next, transfected HeLa cells were used to examine the effect of knockdown of iRhom1 and iRhom2 on CC cell proliferation in vitro. The reduced expression of each protein in these cells confirmed the efficiency of silencing (Fig. 3A). In particular, each shRNA knocked down expression by >50%. The MTT assay indicated that cell viability in the controls (OD490 nm) was greater than in cells with knockdown of iRhom1 or iRhom2 from day 1 to day 5, and that iRhom2 knockdown had a stronger effect than iRhom1 knockdown (Fig. 3B). Moreover, knockdown of each gene significantly decreased the formation of cell colonies (Fig. 3C).
Figure 3.
Oncogenic roles of iRhom1 and iRhom2 knockdown on HeLa cells. (A) Expression of iRhom1 and iRhom2 (western blotting) in cells transfected with iRhom1 or iRhom2 shRNA for 72 h. Knockdown of iRhom1 suppressed its expression by 80.8% (***P<0.01) and knockdown of iRhom2 suppressed its expression by 72.2% (***P<0.01). (B) Changes over time in the viability (OD490 in MTT assay) of cells transfected with shCtrl, shRhom1, and shRhom2 (**P<0.05). (C) Colonies of cells formed following transfection with shCtrl, shRhom1, and shRhom2 (**P<0.05). iRhom1, inactive rhomboid protein 1; iRhom2, inactive rhomboid protein 2.
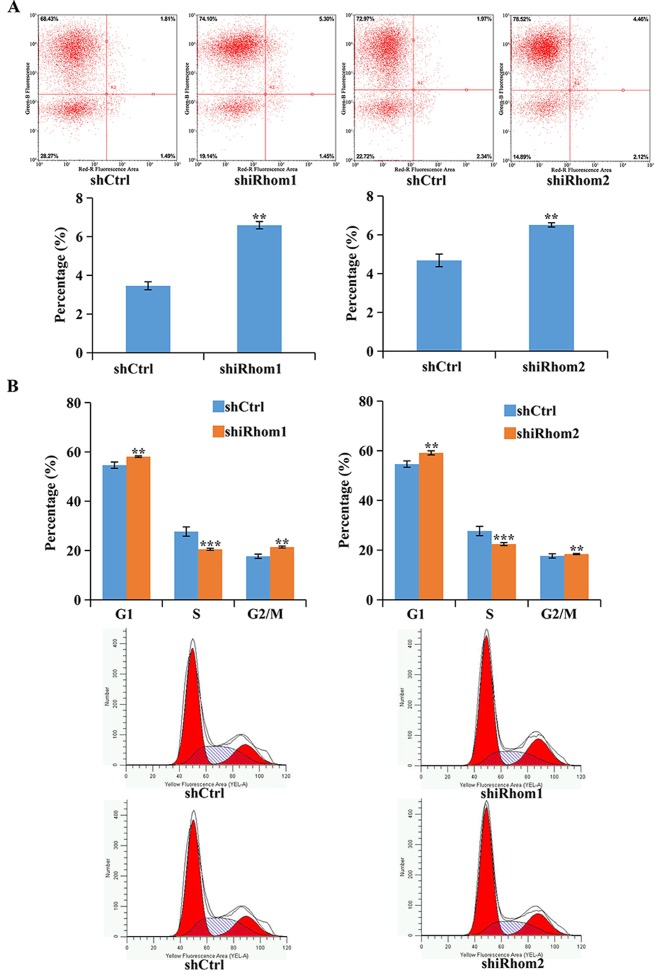
The effect of knockdown of iRhom1 and iRhom2 was also determined on apoptosis using Annexin V-APC staining and flow cytometry (Fig. 4A). In these assays, cells in the upper right quadrant (necrotic) and the lower right quadrant (apoptotic) were apoptotic (Annexin V positive). The results revealed that knockdown of iRhom1 or iRhom2 significantly increased the percentages of cells in apoptosis (Fig. 4A). Dysregulation of the cell cycle in tumors helps to sustain cell proliferation. Thus, flow cytometry was used to identify the effects of iRhom1 and iRhom2 knockdown on HeLa cell cycle progression (Fig. 4B). The results revealed that downregulation of iRhom1 or iRhom2 induced cell cycle arrest at the G1 phase, thus, there was a considerable decrease in the percentage of cells in the S phase and an increase in the percentage of cells in G1-phase relative to control cells (Fig. 4B). However, overexpression of iRhom1 and iRhom2 facilitated cell proliferation of cervical cancer.
Figure 4.
Effect of iRhom1 and iRhom2 knockdown on HeLa cells. (A) Effect of iRhom1 and iRhom2 knockdown on apoptosis was presented as flow cytometry, with Annexin V staining and fluorescence-activated cell sorting analysis. (B) Effect of iRhom1 and iRhom2 knockdown on cell cycle progression, based on flow cytometry. **P<0.05, ***P<0.01 relative to shCtrl (iRhom/NC). iRhom1, inactive rhomboid protein 1; iRhom2, inactive rhomboid protein 2.
Pathways and genes affected by iRhom2 knockdown
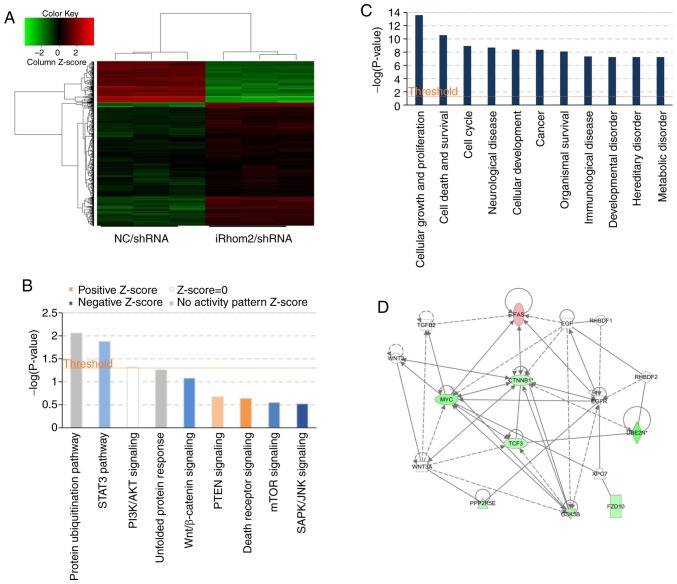
HeLa cells were used infected with iRhom2/shRNA or NC/shRNA and a microarray platform for genome-wide gene expression profiling (Fig. 5). The results revealed that iRhom2 knockdown led to significant differences in the expression of 596 genes [P<0.05 for absolute fold change (FC absolute)>1.5], with 147 upregulated genes and 449 downregulated genes (Fig. 5A). Use of the IPA system for analysis of the microarray data revealed that iRhom2 knockdown led to dysregulation of several cellular and molecular functions, including impairment of cell death and survival, cell growth and proliferation, cancer, neurological disease, immunological disease, metabolic disease and cell cycle progression (Fig. 5C). The ‘canonical pathway’ module was also used for further analysis of the microarray data and downstream pathways that function in carcinogenesis (STAT3, PI3K/AKT, Wnt/β-catenin, PTEN, mTOR, unfolded protein response signaling, Fig. 5B) were identified. This analysis considers a pathway to be inhibited when the IPA Z-score is negative. The results indicated significant inhibition of Wnt/β-catenin signaling, which had an IPA Z-score of −1.134 (Fig. 5D).
Figure 5.
Microarray and IPA analysis of HeLa cells following iRhom2 knockdown. (A) Heatmap of genes whose expression differed significantly when HeLa cells were transfected with iRhom2/shRNA vs. NC/shRNA. Genes are in rows and samples are in columns; red indicates upregulation and green indicates downregulation in cells expressing iRhom2/shRNA. (B) Enrichment of classical signaling pathways. Stimulated (orange) and inhibited (blue) canonical pathways (from the IPA database), are in descending order by the inverse of log(P). (C) Dysregulated cellular and molecular functions based on IPA analysis. The top ten disease types/functions are in descending order by the inverse of log(P). (D) Portion of the gene function network diagram, revealing major molecule-molecule interaction networks in known functional areas. Red indicates upregulation and green indicates downregulation, and brighter colors indicate stronger effects. Line indicates the interaction of two molecules, and full lines indicate stronger effects than imaginary lines. The arrows indicate the direction of action. IPA, Ingenuity Pathway Analysis; iRhom2, inactive rhomboid protein 2.
Effect of iRhom1 and iRhom2 knockdown on the Wnt/β-catenin pathway
The present results revealed that expression of iRhom1 and iRhom2 are linked to carcinogenesis and aggressiveness of CC cells in vitro, however, the mechanisms of this effect have not been systematically investigated. The results from the IPA system indicated that iRhom2 knockdown affected several proteins that function in Wnt/β-catenin signaling (Table III). This pathway has well established roles in the migration and invasion of tumor cells.
Table III.
Genes in the Wnt/β-catenin signaling pathway that are downregulated or upregulated following iRhom2 knockdown in HeLa cells (IPA system).
| Gene | Regulation | Absolute fold change |
|---|---|---|
| β-catenin | Down | 1.5029672 |
| GSK3B | Down | 2.0011744 |
| Myc | Down | 2.4327161 |
| TGFBR2 | Up | 1.55694 |
| FAS | Up | 1.7245258 |
iRhom2, inactive rhomboid protein 2; IPA, Ingenuity Pathway Analysis.
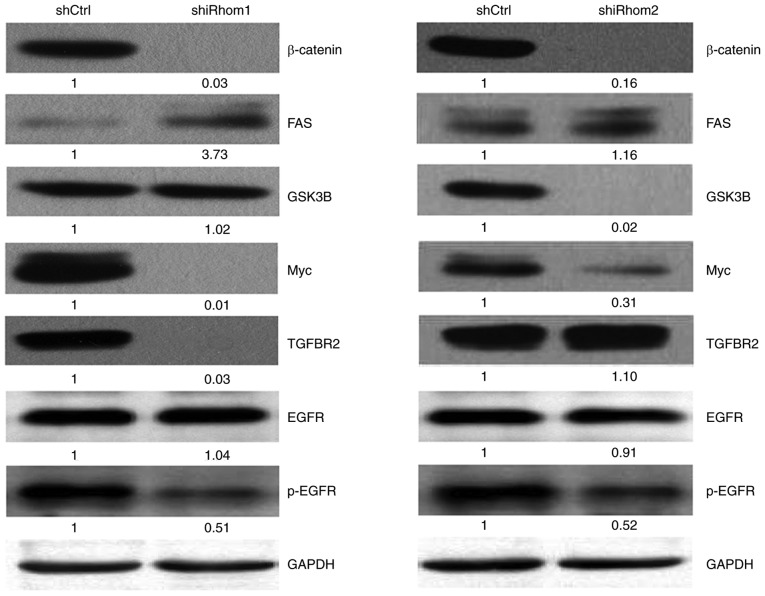
Thus, the effect of iRhom1 and iRhom2 knockdown on the Wnt/β-catenin signaling in HeLa cells was also determined by assessing the expression of several key proteins in this pathway (Fig. 6). The western blotting results indicated that iRhom1 silencing decreased the expression of β-catenin, Myc, p-EGFR and TGFBR2, and increased the expression of FAS (P<0.05 for each comparison); iRhom2 silencing significantly decreased the expression of β-catenin, GSK3β, p-EGFR and Myc (P<0.05 for each comparison).
Figure 6.
Western blotting of β-catenin, FAS, GSK3B, Myc, TGFBR2, EGFR and p-EGFR in HeLa cells transfected with shCtrl, shRhom1, or shRhom2. The number under each image is the grayscale value from quantitative analysis. EGFR, epidermal growth factor receptor.
Discussion
CC is a common gynecological cancer. An improved molecular understanding of its onset and pathogenesis is urgently required to develop better treatments. The impact of iRhom proteins on survival from malignant CC has not yet been fully clarified. To the best of our knowledge, no previous studies have examined the expression of iRhom proteins in CC, nor examined the prognostic value of iRhom1 and iRhom2 expression. The present western blot analyses of CC and adjacent non-malignant normal tissues revealed that iRhom1 and iRhom2 were upregulated in tumors. Additionally, the IHC results indicated positive associations of iRhom1, iRhom2, and Ki-67 expression with cancer stage, tumor size, parametrium invasion, and patient survival. Multivariate analysis indicated that cancer stage and iRhom2 expression were independent prognostic indicators for CC. Previous research reported that expression of iRhom1 was significantly elevated in clinical specimens of early-stage breast cancer (13). In addition, Yuan et al reported that iRhom1 expression was significantly correlated with tumor size, lymph-node invasion, TNM stage, and poor prognosis in colorectal cancer (CRC) (7), similar to our findings for CC. Another study reported that iRhom2 had low or undetectable expression in most ovarian tumors that were benign or had low malignant potential (LMP), and in a subset of malignant ovarian tumors (TOV800EPT, TOV881MT, TOV908DT, TOV974EPT, and TOV1007EPT) (16). These results indicated that expression of iRhom proteins in tumor cells may be cell-specific.
The present finding of positive correlations of iRhom1 and iRhom2 expression with several clinicopathological characteristics suggests that these proteins may have roles in the onset or progression of CC. Thus, to confirm the roles of iRhom1 and iRhom2 in CC, HeLa cells were used to evaluate the effects of iRhom1 and iRhom2 knockdown on cell proliferation, cell cycle progression, and apoptosis. The results indicated that knockdown of each mRNA inhibited cell proliferation and induced apoptosis, similar to their effects on CRC (16). To the best of our knowledge, the present study is the first to quantify the function of iRhom1 and iRhom2 knockdown on the cell cycle of cancer cells. Downregulation of each mRNA induced cell cycle arrest at the G1 phase, and there were therefore fewer cells in the S phase, and more cells in the G1 phase. These results demonstrated that iRhom1 and iRhom2 function as oncogenes in CC, and therefore may have potential as diagnostic and prognostic biomarkers and as therapeutic targets.
iRhoms1/2 regulated ADAM17-dependent EGFR signaling during mouse development, suggesting that iRhoms1/2 could emerge as novel targets for treatment of ADAM17/EGFR-dependent pathologies (17). Etheridge et al indicated that mutant iRhom2 proteins present in TOC patients provide a ‘gain-of-function’, leading to increased ADAM17-mediated shedding of EGF signaling molecules (including amphiregulin) from the cell surface (18). Overexpression of iRhom1 was revealed to coincide with increased secretion of TGF-α in HeLa cells (5). The Uev1A-Ubc13 complex with CHIP promoted K63-linked ubiquitination of iRhom2, increasing its effect on ADAM17 activation, and subsequently blocking TNFα-induced NF-κB signaling in HeLa cells (19). Our previous study reported that blockade of ADAM17 decreased the expression of EMMPRIN, p-EGFR, p-ERK, MMP-2, and MMP-9 proteins in SiHa and HeLa cells (11). In present study, it was demonstrated that p-EGFR was downregulated in HeLa cells with iRhom1 and iRhom2 knockdown, further indicating the activation of iRhoms1/2-ADAM17-EGFR signaling in cervical cancer. A previous study also demonstrated that iRhom2 was implicated in epithelial regeneration and cancer growth through constitutive activation of epidermal growth factor receptor (EGFR) signaling (20), indicating the positive link between iRhoms and EGFR signaling (21). Conversely, Vembar and Brodsky revealed that iRhom1 and iRhom2 bind to EGF ligands in the ER, downregulate EGFR signaling, and target these ligands for degradation (22). ADAM17 was revealed to bind to iRhom2, and activate Notch signaling in lung cancer stem cells (LCSCs). Moreover, iNOS was revealed to have an important role in hepatocellular carcinoma due to its effect on iRhom2/ADAM17 and Notch signaling (23). MET activation in keratinocytes enhanced Src activity and iRhom expression (particularly iRhom2), and this led to ADAM17 activation, possibly due to translocation and maturation of ADAM17 proteolytic activity in squamous tumors (24). We identified a critical role of iRhom proteins in CC development and progression based on studies of clinical samples and HeLa cells. However, the associated signaling mechanisms remain uncertain.
Genomic microarray analysis of HeLa cells in which iRhom2 was knocked down was also used. The results indicated this treatment significantly altered the expression of hundreds of genes. The IPA data identified multiple pathways that contribute to the onset and progression of cancer. For instance, knockdown of iRhom2 enriched the Wnt/β-catenin signaling pathway. This pathway has been revealed to play a critical role in cell proliferation, survival, differentiation, and epithelial-mesenchymal transition (EMT) in cancer cells (25–27). Studies of canonical Wnt signaling indicated that cytoplasmic stabilization and nuclear translocation of catenin β1 (CTNNB1), a regulator of transcription, has a key role (25). However, in the absence of Wnt ligands, the intracellular levels of β-catenin are low because of the constitutive activity the ‘β-catenin destruction complex’. This multiprotein complex consists of two tumor suppressors (APC and AXIN1), two serine/threonine kinases (CK1 and GSK3) a protein phosphatase (PP2A), and an E3-ubiquitin ligase (β-TrCP) (28). Wnt/β-catenin signaling plays an important role in several cancers, including CC (29), lung cancer (30), hepatocellular carcinoma (31), breast cancer (32) and leukemia (33). Although there have not been extensive studies of Wnt/β-catenin signaling in CC, there is some evidence of involvement. In particular, previous studies revealed that epigenetic silencing via hypermethylation of the promoters of SFRP1, SFRP2, and SFRP4 activated the Wnt pathway, and promoted cervical adenocarcinoma progression via activation of EMT (34,35). Another study reported that DKK1 was downregulated in CC, and this activated the β-catenin pathway (36). However, it remains unknown whether iRhom proteins directly or indirectly interact with the Wnt/β-catenin pathway in CC.
The IPA results revealed that iRhom2 knockdown in HeLa cells altered the expression of five mRNAs (β-catenin, FAS, Myc, TGFBR2 and GSK3β) that are involved in Wnt/β-catenin signaling. These results were verified by western blotting. Thus, the HeLa experiments demonstrated that iRhom1 silencing significantly decreased the expression of β-catenin, Myc and TGFBR2, and increased the expression of FAS. iRhom2 silencing significantly suppressed the expression of β-catenin, GSK3β, and Myc. A previous study of transgenic mice reported that upregulated β-catenin-dependent signaling accelerated carcinogenesis in HPV-mediated CC (37). There is also evidence that downregulation of a Wnt antagonist (Wnt inhibitory factor 1) reduced apoptosis and promoted growth, invasion, and angiogenesis of CC in vivo (38). DAX1 was revealed to be overexpressed in CC, and promoted cell growth and tumorigenicity by activating the Wnt/β-catenin pathway via GSK3β (39). A study on CRC reported that iRhom1 regulated the activity of APC and stimulated EMT and cell proliferation, in part via Wnt/β-catenin signaling (39). A study on breast cancer cells reported that translocation of accumulated β-catenin into the nucleus and formation of TCF/Lef/β-catenin complexes induced sequential expression of c-MYC, CCDN1, SNAIL1, and MMP2, and this increased cell proliferation, migration, and invasion (40). Our previous study of patients with CC indicated that ADAM17 appears to target matrix metalloproteinase MMP-2 and MMP-9 through the AREG/EMMPRIN and the EGFR-MEK-ERK pathway (11).
The HeLa experiments revealed that iRhom1 knockdown decreased the expression of TGFBR2, and increased the expression of FAS, and that iRhom2 knockdown suppressed the expression of GSK3β. The reason for the different effects of iRhom1 and iRhom2 knockdown on Wnt/β-catenin signaling is unknown. The present study is the first to report different functions of iRhom1 and iRhom2 in CC. The downregulation of iRhom proteins could inhibit proliferation, promote apoptosis maybe in part by altering Wnt/β-catenin signaling in CC. Our future studies will examine the nature of the interactions of iRhom proteins with these molecules and their effect on remodeling of the ECM.
In conclusion, it was revealed that expression of iRhom1 and iRhom2 were greater in CC tissues than adjacent normal tissues, and that upregulation of iRhom1 and iRhom2 was correlated with certain clinical characteristics and with poor OS. Knockdown of iRhom1 and iRhom2 in HeLa cells inhibited cell growth, disrupted the cell cycle, and promoted apotosis due to alterations in the expression of multiple genes and cancer-associated pathways. Collectively, the present results demonstrated that iRhom proteins act as oncogenes in CC, and therefore have potential use as diagnostic and prognostic biomarkers, as well as therapeutic targets.
Acknowledgements
The authors would like to thank the Department of Pathology of Fujian Cancer Hospital for their support of the present study.
Funding
The present study was supported by Grants-in-Aid for Scientific Research 2015J01379 from the Fujian Natural Science Foundation, Fujian Province Health Technology Project (No. 2015-ZQN-ZD-8), and the Science and Technology Program of Fujian Province, China, No. 2018Y2003).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
QX and CC provided substantial contributions to the conception and design of the work. BL and YL were responsible for the literature search. YL and CG conducted the clinical studies. DZ and YX progressed in the experimental studies. JL and LL provided the data acquisition and carried out the statistical analysis. QX drafted the work, edited the manuscript and revised it critically for important intellectual content. All authors critically revised and approved the final manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The Ethics Committee of Fujian Provincial Cancer Hospital which is affiliated with Fujian Medical University, provided approval of this study. Patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4:1553–1568. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SW, Chun M, Ryu HS, Chang SJ, Kong TW, Lee EJ, Lee YH, Oh YT. Salvage radiotherapy with or without concurrent chemotherapy for pelvic recurrence after hysterectomy alone for early-stage uterine cervical cancer. Strahlenther Onkol. 2017;193:534–542. doi: 10.1007/s00066-017-1122-0. [DOI] [PubMed] [Google Scholar]
- 3.Munro A, Codde J, Spilsbury K, Steel N, Stewart CJ, Salfinger SG, Tan J, Mohan GR, Leung Y, Semmens JB, et al. Risk of persistent and recurrent cervical neoplasia following incidentally detected adenocarcinoma in situ. Am J Obstet Gynecol. 2017;216:272.e1–272.e7. doi: 10.1016/j.ajog.2016.11.1044. [DOI] [PubMed] [Google Scholar]
- 4.Sturtevant MA, Roark M, Bier E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 1993;7:961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- 5.Zou H, Thomas SM, Yan ZW, Grandis JR, Vogt A, Li LY. Human rhomboid family-1 gene RHBDF1 participates in GPCR-mediated transactivation of EGFR growth signals in head and neck squamous cancer cells. FASEB J. 2009;23:425–432. doi: 10.1096/fj.08-112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Bai TR, Gao S, Zhou Z, Peng XM, Zhang LS, Dou DL, Zhang ZS, Li LY. Human rhomboid family-1 modulates clathrin coated vesicle-dependent pro-transforming growth factor α membrane trafficking to promote breast cancer progression. Ebiomedicine. 2018;36:229–240. doi: 10.1016/j.ebiom.2018.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan H, Wei R, Xiao Y, Song Y, Wang J, Yu H, Fang T, Xu W, Mao S. RHBDF1 regulates APC-mediated stimulation of the epithelial-to-mesenchymal transition and proliferation of colorectal cancer cells in part via the Wnt/β-catenin signalling pathway. Exp Cell Res. 2018;368:24–36. doi: 10.1016/j.yexcr.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaydon DC, Etheridge SL, Risk JM, Hennies HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr NK, et al. RHBDF2 mutations are associated with tylosis, a familial esophageal cancer syndrome. Am J Hum Genet. 2012;90:340–346. doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Q, Ying M, Chen G, Lin A, Xie Y, Ohara N, Zhou D. ADAM17 is associated with EMMPRIN and predicts poor prognosis in patients with uterine cervical carcinoma. Tumour Biol. 2014;35:7575–7586. doi: 10.1007/s13277-014-1990-1. [DOI] [PubMed] [Google Scholar]
- 12.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Z, Zou H, Tian F, Grandis JR, Mixson AJ, Lu PY, Li LY. Human rhomboid family-1 gene silencing causes apoptosis or autophagy to epithelial cancer cells and inhibits xenograft tumor growth. Mol Cancer Ther. 2008;7:1355–1364. doi: 10.1158/1535-7163.MCT-08-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arcidiacono P, Webb CM, Brooke MA, Zhou H, Delaney PJ, Ng KE, Blaydon DC, Tinker A, Kelsell DP, Chikh A. p63 is a key regulator of iRHOM2 signalling in the keratinocyte stress response. Nat Commun. 2018;9:1021. doi: 10.1038/s41467-018-03470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishimoto T, Miyake K, Nandi T, Yashiro M, Onishi N, Huang KK, Lin SJ, Kalpana R, Tay ST, Suzuki Y, et al. Activation of transforming growth factor beta 1 signaling in gastric cancer-associated fibroblasts increases their motility, via expression of rhomboid 5 homolog 2, and ability to induce invasiveness of gastric cancer cells. Gastroenterology. 2017;153:191–204.e16. doi: 10.1053/j.gastro.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 16.Wojnarowicz PM, Provencher DM, Mes-Masson AM, Tonin PN. Chromosome 17q25 genes, RHBDF2 and CYGB, in ovarian cancer. Int J Oncol. 2012;40:1865–1880. doi: 10.3892/ijo.2012.1371. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Maretzky T, Weskamp G, Monette S, Qing X, Issuree PD, Crawford HC, McIlwain DR, Mak TW, Salmon JE, Blobel CP. iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proc Natl Acad Sci USA. 2015;112:6080–6085. doi: 10.1073/pnas.1505649112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etheridge SL, Brooke MA, Kelsell DP, Blaydon DC. Rhomboid proteins: A role in keratinocyte proliferation and cancer. Tissue Cell Res. 2013;351:301–307. doi: 10.1007/s00441-012-1542-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Li Y, Yang X, Wang J, Wang R, Qian X, Zhang W, Xiao W. Uev1A-Ubc13 catalyzes K63-linked ubiquitination of RHBDF2 to promote TACE maturation. Cell Signal. 2018;42:155–164. doi: 10.1016/j.cellsig.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Hosur V, Johnson KR, Burzenski LM, Stearns TM, Maser RS, Shultz LD. Rhbdf2 mutations increase its protein stability and drive EGFR hyperactivation through enhanced secretion of amphiregulin. Proc Natl Acad Sci USA. 2014;111:E2200–E2209. doi: 10.1073/pnas.1323908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siggs OM, Grieve A, Xu H, Bambrough P, Christova Y, Freeman M. Genetic interaction implicates iRhom2 in the regulation of EGF receptor signalling in mice. Biol Open. 2014;3:1151–1157. doi: 10.1242/bio.201410116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, Deng M, Xiong S, Wang X, Zhang L, et al. iNOS promotes CD24+CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci USA. 2018;115:E10127–E10136. doi: 10.1073/pnas.1722100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cataisson C, Michalowski AM, Shibuya K, Ryscavage A, Klosterman M, Wright L, Dubois W, Liu F, Zhuang A, Rodrigues KB, et al. MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis. Sci Signal. 2016;9:ra62. doi: 10.1126/scisignal.aaf5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCrea PD, Gottardi CJ. Beyond β-catenin: Prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda T, Tsubamoto H, Inoue K, Sakata K, Shibahara H, Sonoda T. Itraconazole modulates hedgehog, WNT/β-catenin, as well as Akt signalling, and inhibits proliferation of cervical cancer cells. Anticancer Res. 2017;37:3521–3526. doi: 10.21873/anticanres.11720. [DOI] [PubMed] [Google Scholar]
- 30.Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, Reguart N, Rosell R, McCormick F, Jablons DM. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 31.Takigawa Y, Brown AM. Wnt signaling in liver cancer. Curr Drug Targets. 2008;9:1013–1024. doi: 10.2174/138945008786786127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911–2920. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, Kipps TJ, Corr M, Carson DA. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung MT, Lai HC, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Liu HS, Chu DW, Lin YW. SFRP1 and SFRP2 suppress the transformation and invasion abilities of cervical cancer cells through Wnt signal pathway. Gynecol Oncol. 2009;112:646–653. doi: 10.1016/j.ygyno.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Lin YW, Chung MT, Lai HC, De Yan M, Shih YL, Chang CC, Yu MH. Methylation analysis of SFRP genes family in cervical adenocarcinoma. J Cancer Res Clin Oncol. 2009;135:1665–1674. doi: 10.1007/s00432-009-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Yoon YS, Chung JH. Epigenetic silencing of the WNT antagonist DICKKOPF-1 in cervical cancer cell lines. Gynecol Oncol. 2008;109:270–274. doi: 10.1016/j.ygyno.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 37.Bulut G, Fallen S, Beauchamp EM, Drebing LE, Sun J, Berry DL, Kallakury B, Crum CP, Toretsky JA, Schlegel R, Üren A. Beta-catenin accelerates human papilloma virus type-16 mediated cervical carcinogenesis in transgenic mice. PLoS One. 2011;6:e27243. doi: 10.1371/journal.pone.0027243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandran I, Thavathiru E, Ramalingam S, Natarajan G, Mills WK, Benbrook DM, Zuna R, Lightfoot S, Reis A, Anant S, Queimado L. Wnt inhibitory factor 1 induces apoptosis and inhibits cervical cancer growth, invasion and angiogenesis in vivo. Oncogene. 2012;31:2725–2737. doi: 10.1038/onc.2011.455. [DOI] [PubMed] [Google Scholar]
- 39.Liu XF, Li XY, Zheng PS, Yang WT. DAX1 promotes cervical cancer cell growth and tumorigenicity through activation of Wnt/β-catenin pathway via GSK3β. Cell Death Dis. 2018;9:339. doi: 10.1038/s41419-018-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Yepez EA, Ayala-Sumuano JT, Lezama R, Meza I. A novel β-catenin signaling pathway activated by IL-1β leads to the onset of epithelial-mesenchymal transition in breast cancer cells. Cancer Lett. 2014;354:164–171. doi: 10.1016/j.canlet.2014.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.