Abstract
The combination of chemotherapeutic modalities may be more effective in treating gastric cancer compared with any modality alone. Previous studies have demonstrated that the combination of anticancer bioactive peptides (ACBP) and oxaliplatin (OXA) significantly inhibited the growth of the gastric cancer cell line MKN-45, promoted the apoptosis of MKN-45 cells, and caused an irreversible arrest of the MKN-45 cell cycle in the G2/M phase. In the present study, an isobaric tag for relative and absolute quantitation (iTRAQ)-based quantitative proteomics technique was used to determine the effect of ACBP-OXA treatment on the proteomics profile of MKN-45 cells. Notably, a total of 6,210 proteins were detected. Proteins with a >1.2-fold change in expression (either up- or downregulation) and P<0.05 were considered to be differentially expressed. A total of 256 differentially expressed proteins were identified through alignments with different groups. Compared with the control group, MKN-45 cells treated with ACBP, OXA and ACBP-OXA exhibited 17 (10 up- and 7 downregulated), 111 (27 up- and 84 downregulated) and 128 (53 up- and 75 downregulated) differentially expressed proteins, respectively. Of the 256 differentially expressed proteins, 6 (TPX2, NUSAP1, TOP2A, YAP, MKi-67 and GPC4) were verified by the parallel reaction monitoring method, which revealed that TPX2, NUSAP1, TOP2A, YAP, MKi-67 and GPC4 expression decreased with ACBP-OXA treatment. The cellular localization, functional annotation and biological pathways of differentially expressed proteins were examined by Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis. The results indicated that ACBP-OXA may act through the ribosome or the AMP-activated protein kinase (AMPK) signaling pathway, and the AMPK signaling pathway may be an important mediator of the inhibitory effects of ACBP-OXA on MKN-45 gastric cancer cells. In summary, iTRAQ-based proteomics analysis of the effect of ACBP-OXA on MKN-45 cells may guide future therapeutic strategies for gastric cancer. In addition, the present study may help provide new insights into the therapeutic role of combined ACBP and OXA in gastric cancer.
Keywords: gastric cancer, anticancer bioactive peptides, oxaliplatin, isobaric tag for relative and absolute quantitation, proteomics analysis
Introduction
Gastric cancer is the fourth most common type of cancer and the second leading cause of cancer-related mortality worldwide (1). The etiology of gastric cancer is complex and its clinical symptoms are atypical (2,3). Due to the lack of early diagnosis, the majority of patients with gastric cancer are at advanced stage at the time of treatment and have missed the opportunity to undergo curative surgery. In addition, these patients are at high risk for local recurrence and distant metastases, and have worse prognosis and survival. Therefore, gastric cancer is associated with significant disease burden (4–6).
At present, the majority of patients with advanced and metastatic gastric cancer receive chemotherapy. Oxaliplatin (OXA) is a third-generation platinum drug that inhibits DNA replication and transcription. This drug has been widely used to treat malignant tumors of the gastrointestinal tract (7). However, the use of OXA as a chemotherapeutic drug is associated with certain disadvantages. Long-term treatment can reduce the initial therapeutic effect of OXA by increasing the risk of adverse effects and the occurrence of multidrug resistance (8). Therefore, identifying a new combination therapy for gastric cancer has become a research hotspot.
Anticancer bioactive peptides (ACBP) are novel antitumor agents isolated from goat liver that have been found to contain a mixture of peptides with molecular weights of ~8 kDa, including ubiquitin proteases and fatty acid-binding proteins. ACBP do not interfere with normal physiological functions and enzymatic reactions in vivo. Previous studies have demonstrated that ACBP effectively inhibit tumor cell proliferation in the stomach, nasopharynx and gallbladder (9–11).
The combination of ACBP with low-dose cisplatin can achieve the same therapeutic effect as continuous high-dose cisplatin treatment, which effectively reduces the dosage of cisplatin and the possibility of drug resistance (12). Moreover, the combination of ACBP with OXA inhibits proliferation, induces apoptosis, and causes an irreversible arrest of MKN-45 cells in the G2/M phase of the cell cycle. In addition, ACBP-OXA significantly improves the survival rate and inhibits the tumor formation ability in vivo (13,14). Therefore, ACBP-OXA may be used as a new strategy for gastric cancer treatment (13). However, the mechanisms underlying the therapeutic effect of ACBP-OXA in gastric cancer have yet to be fully elucidated.
In the era of post-genomics, proteins, as participants in life activities and executants of biological functions, have been widely investigated. High-throughput proteomics technologies may lead to more accurate identification of diagnostic and prognostic biomarkers by comprehensively analyzing the differential expression levels, interactions and post-translational modifications of proteins. Isobaric tag for relative and absolute quantitation (iTRAQ), as the latest high-throughput proteomics technique, may be useful for screening and identifying drug-targeting proteins in cancer cells (15–17).
MKN-45 is a tumorigenic human gastric cancer cell line that is resistant to chemotherapy and radiotherapy and exhibits stem-cell characteristics due to its self-renewal and proliferation abilities (18). In the present study, iTRAQ technology was used to perform a comprehensive proteomics analysis of MKN-45 cells treated with a combination of ACBP and OXA. In addition, bioinformatics and functional analyses, such as Gene Ontology (GO) annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, cluster analysis and protein-protein interaction (PPI) network analysis, were used to analyze the proteomics data. Furthermore, the proteomics results were verified by parallel reaction monitoring (PRM) of selected target proteins. The results of the present study may provide a basis for further research on the role of ACBP-OXA in the treatment of gastric cancer and introduce a basis for the clinical application of combined ACBP-OXA therapy in gastric cancer.
Materials and methods
Cell culture
The human gastric cancer cell line MKN-45 was purchased from the Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Sciences, Peking Union Medical College. Cell culture was performed at the Clinical Medical Research Center of the Inner Mongolia Medical University. MKN-45 cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences) and 1% penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) and maintained in a humidified CO2 incubator at 37°C. MKN-45 is a poorly differentiated human gastric adenocarcinoma cell line, and 90% of MKN-45 cells exhibit stem cell characteristics (19).
Extraction and purification of bioactive peptides
Extraction and purification of bioactive peptides were performed as previously reported (20). Additionally, the optimal concentration of 20 µg/ml bioactive peptides was determined and selected for the treatment of MKN-45 cells (10,11).
Cell treatment
OXA was purchased from Jiangsu Aosaikang Pharmaceutical Co., Ltd. and dissolved in DMSO as a stock solution. The yield of cultured MKN-45 cells in the laboratory was 1×106 cells/ml. After being cultured for 24 h, 20 µg/ml of induced ACBP, 15 µg/ml OXA, and a combination of 10 µg/ml induced ACBP and 7.5 µg/ml OXA were added to the cell culture medium. The negative control group was treated with phosphate-buffered saline (PBS). PBS is a phosphate buffer, which acts as a dissolving protective agent, does not affect cell growth and causes no damage to cells (11,21,22). After 36 h of incubation with all three treatments, the cells were collected for further analysis (13,20,23). All experiments were performed in triplicate.
Protein extraction
Following addition of an appropriate amount of SDT-lysis buffer, the cells were ultrasonicated at 80 W for 10 repeated cycles, which included sonication for 10 sec, pausing for 15 sec, and boiling for 15 min. The supernatants of the cell lysates were collected after centrifugation at 14,000 × g for 40 min. Proteins were quantified by the bicinchoninic acid assay. The samples were transferred to a dispensing pack and stored at −80°C. Three biological replicates were performed for each group (24).
SDS-PAGE
Protein samples (20 µg) were mixed with 5X loading buffer and boiled for 5 min. Subsequently, SDS-PAGE was conducted on a 12.5% (v/w) polyacrylamide gel. Three biological replicates were performed for each group.
Filter-aided sample preparation
Protein sample solution (30 µl) was mixed with DTT to a final concentration of 100 mM and then boiled for 5 min. After cooling to room temperature, 200 µl of UA buffer was added and the mixture was transferred to a 10-kDa ultrafiltration centrifuge tube. Following centrifugation at 14,000 × g for 15 min at 37°C, the filtrate was discarded. This step was repeated once. The tube was supplemented with 100 µl IAA buffer (100 mM IAA in UA), followed by shaking at 4,000 × g for 1 min at 37°C. Subsequently, the mixture was incubated in the dark for 30 min at room temperature, and then centrifuged at 14,000 × g for 15 min. Following addition of 100 µl UA buffer, centrifugation was performed at 14,000 × g for 15 min. This step was repeated twice. Next, the tube was loaded with 100 µl of 10-fold diluted dissolution buffer and centrifuged at 14,000 × g for 15 min. This step was repeated twice. After loading with 40 µl trypsin buffer (4 µg trypsin in 40 µl dissolution buffer), the tube was shaken at 4,000 × g for 1 min and incubated for 16–18 h at 37°C. Then, the tube was substituted with a new collecting tube, which was centrifuged at 14,000 × g for 15 min. Following addition of 40 µl of 10-fold diluted dissolution buffer, the filtrate was collected after centrifugation at 14,000 × g for 15 min. The peptides were desalted with a C18 cartridge and redissolved with 40 µl dissolution buffer after lyophilization. Finally, the peptide samples were quantified by measuring absorbance at 280 nm (OD280) (24).
iTRAQ labeling
Peptides (~100 µg) in each group were labeled with the iTRAQ Labeling Kit (AB SCIEX Co.) according to the manufacturer's instructions. Three biological replicates were performed for each group.
Strong cation-exchange chromatography fractionation
The labeled peptides of each group were mixed and fractionated using an AKTA Purifier 100 (GE Healthcare). Buffer A (pH 3.0) containing 10 mM KH2PO4 and 25% ACN was used as the mobile phase. Buffer B (pH 3.0) containing 10 mM KH2PO4, 500 mM KCl and 25% CAN was used as the eluent. Subsequently, the column was equilibrated with buffer A. The peptide samples were then separated by the column at a flow rate of 1 ml/min. The linear gradient of buffer B was from 0 to 8% in 22 min, 8 to 52% in 25 min and 52 to 100% in 3 min, maintained at 100% for 8 min, and then reset to 0%. The elution profile was monitored by UV absorbance at 214 nm. Finally, the fractions were collected every 1 min and desalted using a C18 cartridge.
High-performance liquid chromatography (HPLC)
All samples were separated using an HPLC system Easy nLC at a nanoliter flow rate. Buffer A was a 0.1% formic acid-water solution, whereas buffer B was a 0.1% formic acid-84% acetonitrile-water solution. The column was equilibrated with 95% buffer A. Samples loaded in the autosampler were transferred onto the loading column (Acclaim PepMap100, 100 µm ×2 cm, nanoViper C18; Thermo Fisher Scientific, Inc.) and then separated by an analytical column (EASY-Column, 10 cm, ID 75 µm, 3 µm, C18-A2; Thermo Fisher Scientific, Inc.) at a flow rate of 300 nl/min. The linear gradient of buffer B was from 0 to 35% in 50 min, 35 to 100% in 5 min, and maintained at 100% for 5 min.
Mass spectrometry (MS) identification
HPLC-fractionated samples were subjected to MS using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.). The parameters used in MS were as follows: Detection mode, positive ion; analysis time, 60 min; scanning range of parent ion, 300–1,800 m/z; MS1 resolution, 70,000 at 200 m/z; AGC target value, 3.0×10−6; first-order maximum IT, 10 msec; dynamic exclusion, 10 msec; The mass-to-charge ratios of peptides and their fragments were recorded. Ten fragmentographies were acquired from MS2 scans. MS2 activation type, higher energy collisional dissociation (HCD); isolation window, 2 m/z; resolution, 17,500 at 200 m/z; microscans, 1; second-order maximum IT, 60 msec; normalized collision energy, 30 eV; and underfill ratio, 0.1%.
Proteomics data analysis
The raw MS data were extracted from RAW files. Mascot version 2.2 (Matrix Science) and Proteome Discoverer version 1.4 (Thermo Electron) were used for molecular identification and quantitative analysis. The MS data were analyzed with the UniProt protein database. Carbamidomethylation of cysteines, iTRAQ labeling at the N-term and lysine side-chain amino groups were set as the fixed modifications, while the oxidation of methionine and iTRAQ4plex (Y) was set as a variable modification. The false discovery rate for each peptide was adjusted to 1%, and the minimum peptide length was specified to 6. In addition, the enzyme specificity was set to trypsin, and up to two missed cleavages were allowed. Mass tolerance was set as 20 ppm for precursor ions and 0.1 Da for fragment ions.
Functional GO annotation
GO annotation of the target proteins was conducted by Blast2GO (25), which consisted of four steps: i) Sequence alignment (BLAST), ii) GO entry extraction mapping, iii) GO annotation, and iv) data augmentation. First, the target proteins were aligned against the specific protein sequence database using the localized sequence alignment tool NCBI BLAST+ (ncbi-blast-2.2.28+-win32.exe), and the first 10 aligned sequences with an E-value ≤1e-3 were retained for subsequent analysis. Next, Blast2GO Command Line was used to extract the GO entries related to the target protein sets and the aligned proteins or homologous proteins with high sequence identity (database version: go_201608.obo, www.geneontology.org). During the annotation process, Blast2GO Command Line annotated the target proteins with the GO entries extracted from the mapping process based on the sequence similarity between the targeted and aligned proteins, the reliability of GO entry sources, and the structure of the GO-directed acyclic graph. Following annotation, the conserved motifs that matched with target proteins were searched against the EBI database using InterPro Scan to improve the annotation efficiency. Functional information related to the motifs was obtained and annotated to the target protein sequences. ANNEX was used to enhance the annotation information, and a link between different GO categories was established to improve the accuracy of GO annotations.
KEGG pathway annotation
KO (KEGG Orthology) in the KEGG database is a classification system for genes and their products. Orthologous genes with similar functions are grouped with their products on the same pathway, and a KO (or K) tag was assigned for their interaction. KO classification was performed on the target protein sequences by BLAST against the KEGG GENES database using KEGG Automatic Annotation Server software. Additionally, the details on the pathways associated with target protein sequences were obtained according to the KO classification.
Cluster analysis of protein sequences
For clustering analysis, the quantitative information of the target protein set was first normalized to the [-1, 1] interval. Next, Cluster 3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) was used to classify the two dimensions of the sample and protein expression (distance algorithm, Euclidean; connection, Average linkage) simultaneously. Finally, a hierarchical clustering heat map was constructed using Java TreeView software, version 3.0 (http://jtreeview.sourceforge.net).
Network analysis of protein-protein interactions (PPI)
Gene symbols obtained from the database of target protein sequences were used to investigate the direct and indirect interactions between target proteins and experimental evidence via the IntAct database (http://www.ebi.ac.uk/intact/main.xhtml). CytoScape version 3.2.1 (http://www.cytoscape.org/) and String database (https://string-db.org/) were used to generate the PPI network and analyze the network topologies.
PRM acquisition
To verify the protein expression levels obtained by iTRAQ analysis, the expression levels of selected proteins were quantified by LC-PRM-MS analysis (26). Briefly, peptides were prepared according to the iTRAQ reagents protocol. An AQUA stable isotope peptide was spiked in each sample as an internal standard reference. For desalting purposes, tryptic peptides were loaded into C18 stage tips on an Easy nLC-1200 system prior to reversed-phase chromatography. A LC gradient of acetonitrile ranging from 5 to 35% in 45 min was used. PRM analysis was performed using a Q-Exactive Plus mass spectrometer. The optimization of collision energy, charge state and retention times for the most significantly regulated peptides was conducted by unique peptides with the highest intensity and confidence. A full MS scan was carried out in a positive ion mode mass spectrometer with 70,000 resolution (at 200 m/z), an AGC target value of 3.0×10−6, and a maximum ion injection time of 250 msec. Then, 20 PRM scans were performed at 35,000 resolution (at 200 m/z), an AGC target value of 3.0×10−6 and a maximum injection time of 200 msec. The targeted peptides were isolated using a 2 Thomson (Th) window. Peptide fragmentation was induced by higher-energy collisional dissociation (HCD) at a normalized collision energy of 27. The raw proteomics data were analyzed with Skyline software, version 19.1 (MacCoss Lab, University of Washington) (27), where the signal intensities for the identified peptide sequences were relatively quantified and normalized with a reference standard.
PRM screening
To validate the results of MS, 6 differentially expressed proteins (TPX2, NUSAP1, TOP2A, YAP, MKi-67 and GPC4) were selected for PRM analysis. The criteria for the validation of proteomics data were as follows: i) Potential biological functions and significant differential expression; ii) the number of peptide fragments detected by LC-MS/MS was >1; and iii) novel oncoproteins that were decreasingly expressed in MKN-45 cells after treatment with ACBP-OXA compared with ACBP or OXA treatment alone.
Statistical analysis
Student's t-test was used to analyze the statistical software SPSS (version 22, IBM Corp.). Data are expressed as means ± standard deviation of three independent biological replicates. A P-value of <0.05 was considered to indicate statistically significant differences (Tables II, III, SII and SIII).
Table II.
Optimized differentially expressed proteins between groups.
| Protein ID | Protein | Fold change (ACBP-OXA/C) | P-value |
|---|---|---|---|
| Q8IZZ8 | Antithrombin (fragment) | 1.33 | 0.0012 |
| B7Z8Q4 | Hemopexin | 2.54 | 0.00126 |
| D6RBJ7 | Vitamin D-binding protein | 2.81 | 0.0016 |
| Q96RG4 | Insulin receptor substrate 2 insertion mutant (fragment) | 1.3 | 0.0018 |
| E5RJK7 | LYR motif-containing protein 2 | 1.23 | 0.0045 |
| A8MW49 | Fatty acid-binding protein | 4.8 | 0.0053 |
| Q99988 | Growth/differentiation factor 15 | 1.42 | 0.0054 |
| Q9BWT3 | Poly(A) polymerase gamma | 1.32 | 0.0055 |
| A0A087WVA8 | Testis-expressed sequence 2 protein | 1.39 | 0.0056 |
| Q8TB52 | F-box only protein 30 | 1.33 | 0.0066 |
| Q7Z6E9 | E3 ubiquitin-protein ligase RBBP6 | 1.21 | 0.0066 |
| Q9P1Y5 | Calmodulin-regulated spectrin-associated protein 3 | 1.24 | 0.008 |
| B3KRB7 | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | 1.45 | 0.008 |
| Q9ULR3 | Protein phosphatase 1H | 1.29 | 0.009 |
| B4DQK1 | Autophagy-related protein 7 | 1.22 | 0.0179 |
| Q5T985 | Inter-alpha-trypsin inhibitor heavy chain H2 | 1.74 | 0.02 |
| O60701 | UDP-glucose 6-dehydrogenase | 1.21 | 0.02 |
| Q5T440 | Putative transferase CAF17 | 1.27 | 0.022 |
| Q53RD8 | Putative uncharacterized protein LOC84524 (fragment) | 1.37 | 0.023 |
| Q5T123 | SH3 domain-binding glutamic acid-rich-like protein 3 | 1.26 | 0.023 |
| Q15125 | 3-beta-hydroxysteroid-Delta(8), Delta(7)-isomerase | 1.42 | 0.024 |
| I3L1D4 | RNA-binding protein fox-1 homolog 1 (fragment) | 1.23 | 0.025 |
| B4DMX4 | Alpha-fetoprotein | 2.03 | 0.025 |
| P48506 | Glutamate-cysteine ligase catalytic subunit | 1.25 | 0.026 |
| P01023 | Alpha-2-macroglobulin | 2.3 | 0.027 |
| A0A024R172 | Leukotriene B4 12-hydroxydehydrogenas | 1.22 | 0.028 |
| B4DTK6 | RNA polymerase I-specific transcription initiation factor RRN3 | 1.24 | 0.028 |
| I3L2L5 | Mapk-regulated corepressor-interacting protein 1 | 1.27 | 0.028 |
| F6KPG5 | Albumin (fragment) | 2.94 | 0.029 |
| B3KM35 | Beta-1,4-galactosyltransferase 4 | 1.27 | 0.031 |
| B4E1V0 | Lactotransferrin | 1.68 | 0.034 |
| B7Z8R6 | AMBP protein | 2.11 | 0.039 |
| B7Z2S5 | Thioredoxin reductase 1 | 1.22 | 0.038 |
| P07477 | Trypsin-1 | 1.24 | 0.039 |
| Q5HYD9 | Putative uncharacterized protein DKFZp686M0619 (fragment) | 1.2 | 0.041 |
| Q9BW34 | EEF1D protein (fragment) | 1.28 | 0.041 |
| O15173 | Membrane-associated progesterone receptor component 2 | 1.21 | 0.04 |
| H7C5E8 | Serotransferrin (fragment) | 2.02 | 0.042 |
| P09669 | Cytochrome c oxidase subunit 6C | 1.22 | 0.044 |
| Q71UM5 | 40S ribosomal protein S27-like | 1.79 | 0.046 |
| P46013 | Proliferation marker protein Ki-67 | 0.076 | 0.0008 |
| Q9H3K6 | BolA-like protein 2 | 0.79 | 0.0009 |
| P16402 | Histone H1.3 | 0.809 | 0.0016 |
| B4DMI9 | Discs large homolog 7 | 0.8 | 0.0048 |
| D3YTB1 | 60S ribosomal protein L32 (fragment) | 0.8 | 0.005 |
| H3BQH3 | Kelch domain-containing protein 4 (fragment) | 0.744 | 0.0079 |
| B3KMT5 | SGT1 protein | 0.81 | 0.009 |
| P63218 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) | 0.8 | 0.009 |
| Q9Y3U8 | 60S ribosomal protein L36 | 0.8 | 0.009 |
| P11388 | DNA topoisomerase 2-alpha | 0.78 | 0.114 |
| P0CJ79 | Zinc finger protein 888 | 0.77 | 0.012 |
| B3KQT6 | Tetraspanin-13 | 0.81 | 0.012 |
| B2RA70 | Tyrosine-protein kinase | 0.82 | 0.015 |
| Q5T7U1 | General transcription factor 3C polypeptide 5 | 0.81 | 0.015 |
| A8YQF4 | MHC class I antigen (fragment) | 0.71 | 0.017 |
| Q15397 | Pumilio homolog 3 | 0.799 | 0.018 |
| P50914 | 60S ribosomal protein L14 | 0.82 | 0.019 |
| G5E9A6 | Ubiquitin carboxyl-terminal hydrolase 11 | 0.74 | 0.02 |
| P39023 | 60S ribosomal protein L3 | 0.83 | 0.002 |
| P31350 | Ribonucleoside-diphosphate reductase subunit M2 | 0.8 | 0.02 |
| M0R0F0 | 40S ribosomal protein S5 (fragment) | 0.82 | 0.025 |
| Q6N075 | Molybdate-anion transporter | 0.82 | 0.026 |
| Q8TDD1 | ATP-dependent RNA helicase DDX54 | 0.74 | 0.031 |
| O60287 | Nucleolar pre-ribosomal-associated protein 1 | 0.81 | 0.031 |
| C9K025 | 60S ribosomal protein L35a (fragment) | 0.82 | 0.031 |
| Q9ULW0 | Targeting protein for Xklp2 | 0.79 | 0.032 |
| A0A0A0MRW6 | Nucleolar protein 6 | 0.79 | 0.033 |
| Q92876 | Kallikrein-6 | 0.77 | 0.033 |
| A8K800 | Homo sapiens brix domain containing 1 | 0.79 | 0.038 |
| P60604 | Ubiquitin-conjugating enzyme E2 | 0.77 | 0.038 |
| A8K7A2 | Cell division cycle associated 8 | 0.81 | 0.044 |
| S4R456 | 40S ribosomal protein S15 (fragment) | 0.78 | 0.04 |
| A8K4B4 | Homo sapiens nucleolar and spindle associated protein 1 | 0.8 | 0.04 |
| O75487 | Glypican-4 | 0.76 | 0.04 |
| A0A087WXM6 | 60S ribosomal protein L17 (fragment) | 0.81 | 0.04 |
| Q8TDN6 | Ribosome biogenesis protein BRX1 homolog | 0.73 | 0.04 |
| Q96HP0 | Dedicator of cytokinesis protein 6 | 0.73 | 0.04 |
The proteins listed above did not exhibit differences in expression between the ACBP- and OXA-treated groups. ACBP, anticancer bioactive peptides; OXA, oxaliplatin; C, control.
Table III.
Target protein expression quantity analysis.
| Protein name | C_mean | ACBP_mean | ACBP-OXA_mean | OXA _mean | Ratio_ACBP/C | Ratio_ACBP-OXA/C | Ratio_OXA/C | TTEST_ACBP/C | TTEST_ACBP-OXA/C | TTEST_OXA/C |
|---|---|---|---|---|---|---|---|---|---|---|
| TPX2 | 0.0488 | 0.0992 | 0.0251 | 0.0922 | 2.03 | 0.52 | 1.89 | 0.00679 | 0.09427 | 0.16063 |
| NUSAP1 | 0.0335 | 0.1656 | 0.0205 | 0.0661 | 4.95 | 0.61 | 1.97 | 0.00040 | 0.31784 | 0.20261 |
| TOP2A | 0.1362 | 0.2487 | 0.1210 | 0.1476 | 1.83 | 0.89 | 1.08 | 0.04032 | 0.76786 | 0.84153 |
| YAP | 0.0637 | 0.1024 | 0.0590 | 0.1546 | 1.61 | 0.93 | 2.43 | 0.13889 | 0.87597 | 0.19902 |
| MKi-67 | 0.0356 | 0.0950 | 0.0289 | 0.0456 | 2.66 | 0.81 | 1.28 | 0.00272 | 0.56360 | 0.57830 |
| GPC4 | 0.4369 | 0.6705 | 0.4233 | 0.4868 | 1.53 | 0.97 | 1.11 | 0.15360 | 0.94407 | 0.82274 |
Results
LC-MS/MS
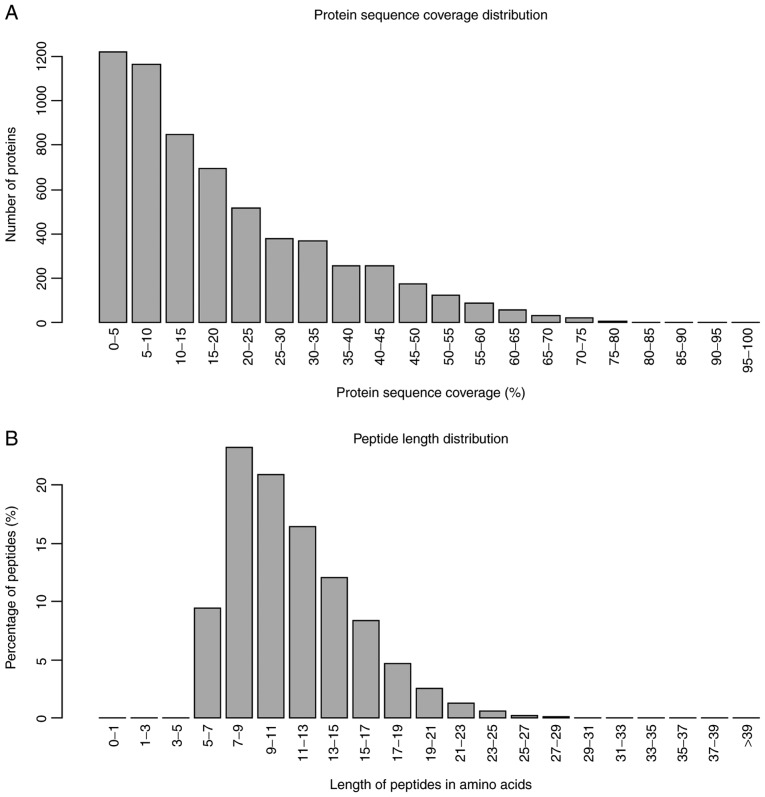
In the present study, the iTRAQ technique was applied to analyze the proteomics profile of MKN-45 cells treated with a combination of ACBP and OXA. The entire experimental procedure is illustrated in Fig. 1. A total of 6,210 proteins were detected (Table SI). Quality control of protein data revealed that the molecular mass of proteins fell in the range of 5–100 kDa (Fig. 2A), and the majority of the peptides were ~7–15 amino acids in length (Fig. 2B), which appeared to be similar to the known properties of tryptic peptides.
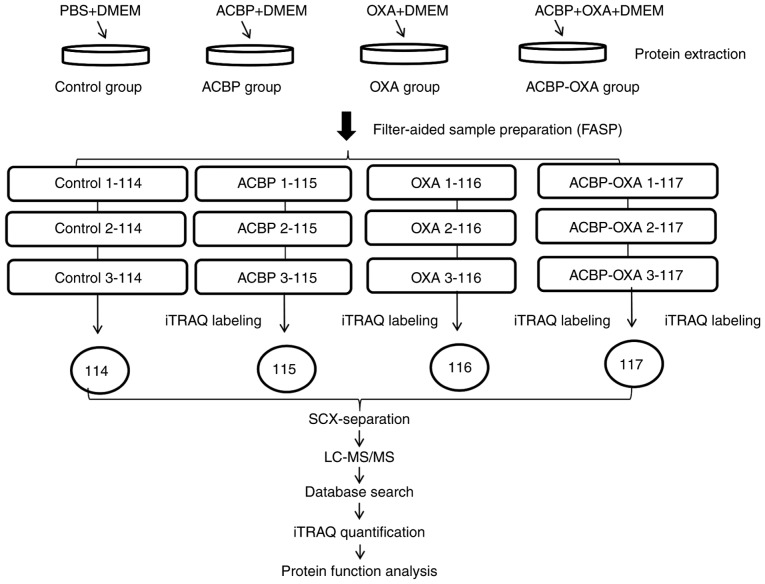
Figure 1.
Experimental process. Experimental design for the quantitative proteomics analysis, the experiment was divided into four groups (control, ACBP, OXA, ACBP-OXA), and proteins were extracted from three independent biological replicates per treatment. Extracted proteins were prepared via FASP and labeled with iTRAQ reagents. The labeled peptides were separated by SCX chromatography, and fractions were analyzed by reversed-phase LC-MS/MS. All data were analyzed by bioinformatics tools from different aspects. ACBP, anticancer bioactive peptides; OXA, oxaliplatin; SCX, strong cation exchange; LC, liquid chromatography; MS, mass spectrometry; FASP, filter-aided sample preparation.
Figure 2.
Quality control validation of protein data. (A) Protein mass distribution of all identified proteins. (B) Protein length distribution of all identified peptides.
Compared with the control group, MKN-45 cells treated with ACBP, OXA and ACBP-OXA exhibited 17 (10 up- and 7 downregulated), 111 (27 up- and 84 downregulated) and 128 (53 up- and 75 downregulated) differentially expressed proteins, respectively (Tables I and SII). Proteins with a >1.2-fold change in expression (either up- or downregulation) and P<0.05 were considered to be differentially expressed. The protein expression remained unchanged in the ACBP- or OXA-treated cells by the best screening criteria for the differentially expressed proteins (>1.2-fold change in expression and P<0.05). However, in ACBP-OXA-treated cells, the protein expression changed or changed in the opposite direction compared with the ACBP- or OXA-treated groups. According to the criteria mentioned above, a total of 77 differentially expressed proteins were subjected to further screening (Table II). Of these 77 proteins, 40 were significantly upregulated and 37 were significantly downregulated. Specifically, the top 5 differentially upregulated proteins were hemopexin, vitamin D-binding protein, fatty acid-binding protein, α-fetoprotein and α-2-macroglobulin, whereas the top 5 differentially downregulated proteins were ribosome biogenesis protein BRX1, dedicator of cytokinesis protein 6, targeting protein for Xklp2, DNA topoisomerase 2-α and proliferation marker protein Ki-67.
Table I.
Protein quantification in MKN45 cells treated by ACBP alone, OXA alone, and combined ACBP-OXA.
| Comparison between groups | Upregulation | Downregulation | Protein count |
|---|---|---|---|
| OXA vs. C | 27 | 84 | 111 |
| ACBP vs. C | 10 | 7 | 17 |
| ACBP-OXA vs. C | 53 | 75 | 128 |
ACBP, anticancer bioactive peptides; OXA, oxaliplatin; C, control.
PRM analysis of the target peptides
To improve the accuracy of protein identification, the PRM mode was first used to specifically monitor the peptide sequences of 6 target proteins in mixed samples. The mixed samples were repeatedly tested 3 times using LC-PRM/MS, and the PRM data were analyzed by Skyline software, version 19.1 (MacCoss Lab, University of Washington). The results demonstrated that the 6 target proteins had credible peptides. The corresponding peptides were selected for PRM quantitative analysis. The 3-time repeated tests of LC-PRM/MS stably detected 10 candidate peptides of the 6 target proteins, with a relative standard deviation of ~12%. These results support that PRM is an accurate method for the quantification analysis of peptides (Table SIII). Three daughter ions of the most abundant and consecutive peptides were selected for different analyses, such as quantitative analysis at the protein and peptide levels, data calibration and biostatistical analysis (Table SIV). First, the peak area of the daughter ion was integrated to obtain the original peak area of peptides. Second, the peak area of the internal standard peptide was labeled with heavy isotopes for correction purposes. The relative expression levels of each peptide in all samples were then measured. Finally, the mean relative expression level of the target peptide was calculated and statistically analyzed (Table SV).
Quantitative expression analysis of the target proteins
Based on the corresponding peptide fragments, the differences in the relative expression level of target proteins were further calculated in different samples. In other words, the mean ratios of multiple peptides were calculated (Table III). The LC-PRM/MS results revealed that quantitative information of target peptide fragments was obtained for all samples. Subsequently, the target proteins and peptide fragments were subjected to relative quantitative analysis by adding heavy isotope-labeled peptide fragments. The results demonstrated that differential protein expression was observed among the 6 target proteins. Notably, the protein expression levels were significantly decreased in MKN-45 cells treated with ACBP-OXA, but not cells treated with ACBP or OXA alone.
Bioinformatics analysis
All differentially expressed proteins detected by MS were further analyzed using bioinformatics methods.
Clustering analysis
Hierarchical clustering results are presented as heat maps, where the red color indicates upregulation and the green color downregulation. All samples were classified into four categories: C1-C3, ACBP1-ACBP3, OXA1-OXA3 and ACBP-OXA1-ACBP-OXA3. It was observed that the proteins may be divided into two categories via vertical comparison. The differential changes in protein expression between ACBP-OXA-treated MKN-45 cells and the control group are shown in Fig. 3. In addition, the differentially expressed proteins were significantly altered in ACBP-treated and OXA-treated MKN-45 cells (Figs. S1 and S2, respectively) compared with the control group, suggesting the rationality of differential expression patterns of selected target proteins. Therefore, clustering analysis may be used to assess the reasonability of screening differentially expressed proteins, e.g., whether changes in the expression levels of these target proteins can indicate the therapeutic effect of biological agents on cancer cells.
Figure 3.
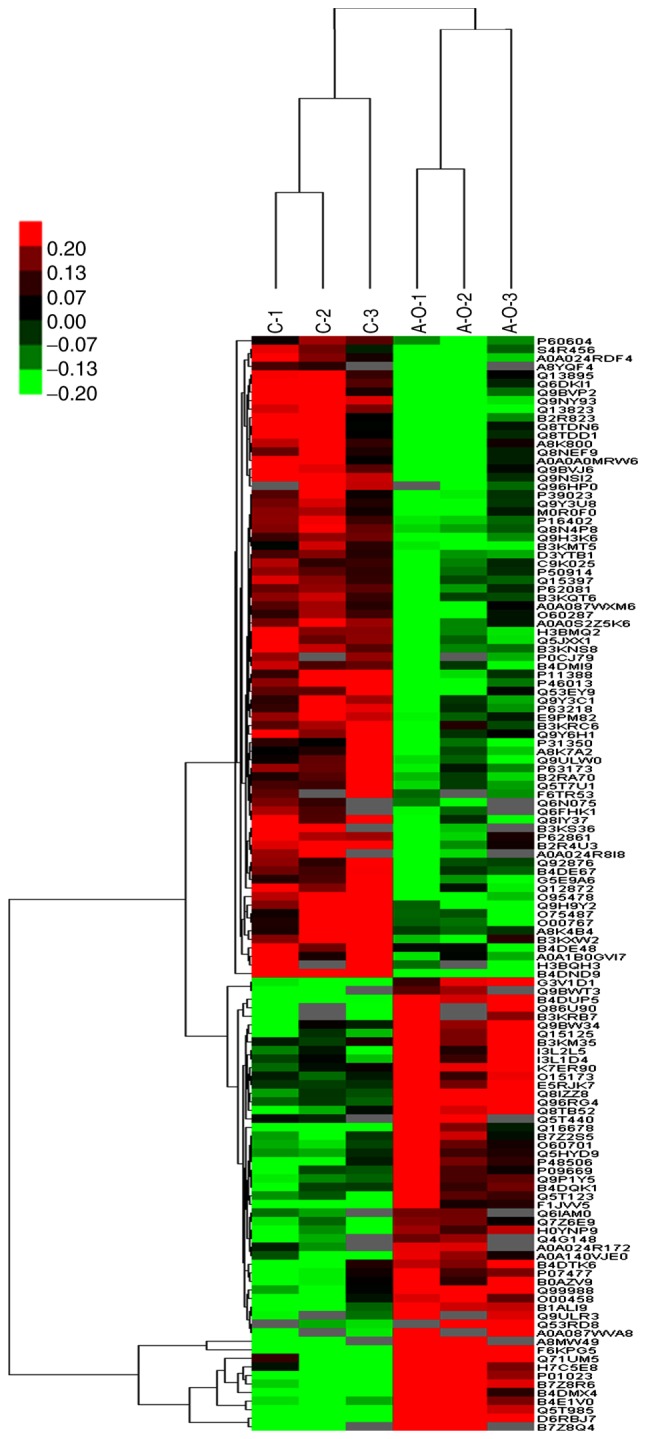
Cluster analysis of differential level proteins between ACBP-OXA and the control. The colors indicate the differential protein levels, which increase successively from green to dark red. Increased levels of proteins are indicated in red, and decreased levels are marked in green. ACBP, anticancer bioactive peptides; OXA, oxaliplatin.
GO function annotation and analysis
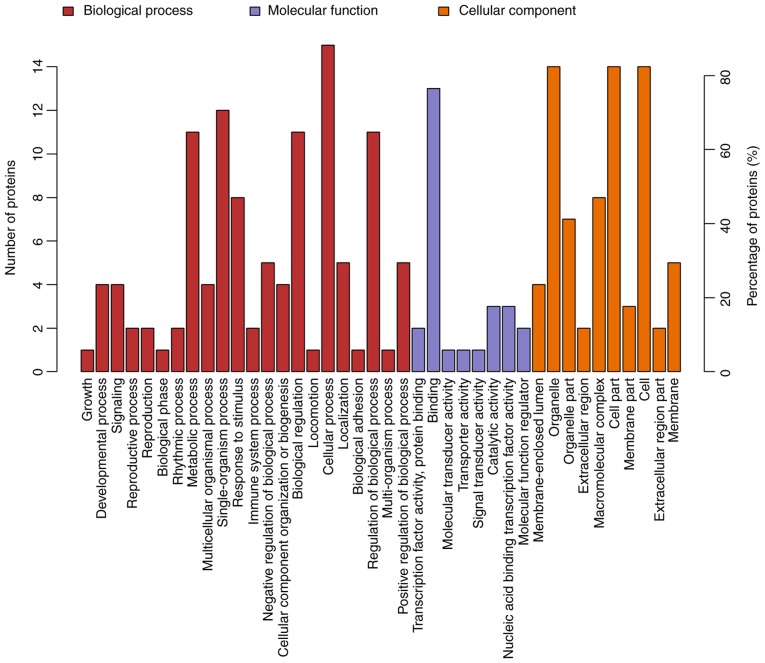
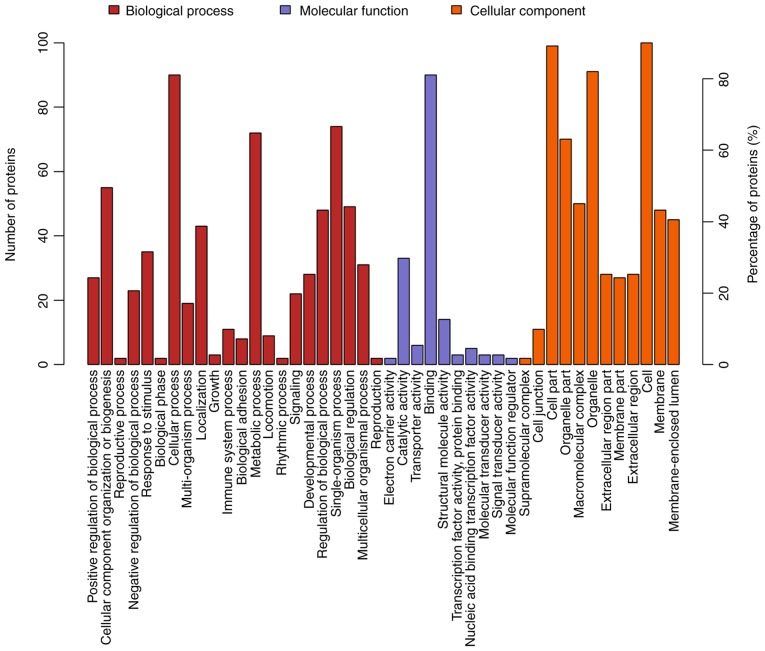
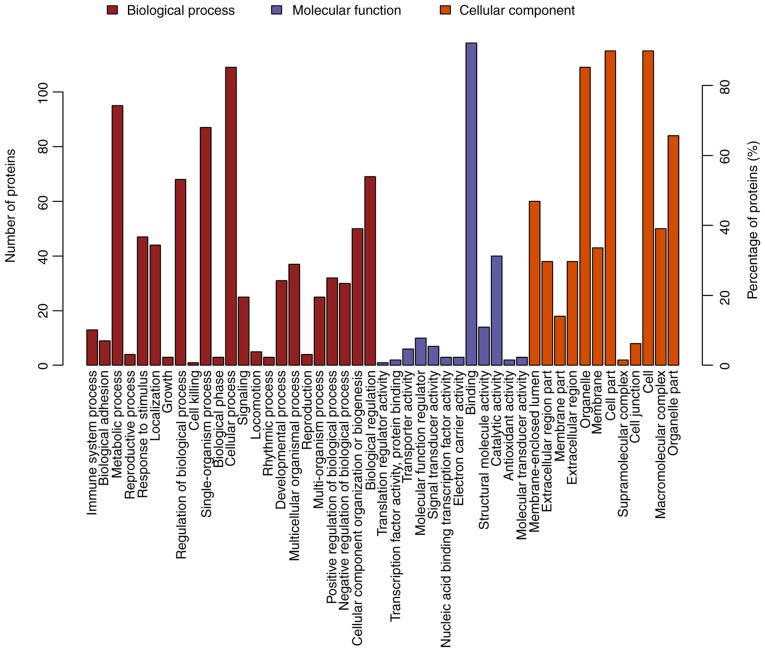
GO is a functional classification system that provides a set of dynamically updated standardized vocabulary to describe the properties of genes and gene products based on three different perspectives: i) The involved biological process, ii) molecular function and iii) cellular component. Differentially expressed proteins in ACBP- (n=17), OXA- (n=111) and ACBP-OXA-treated cells (n=128) exhibited a total of 734, 2,295 and 2,402 functional annotations, respectively (Tables SVI–SVIII). Similarly, the protein functions predicted from secondary GO enrichment analysis were divided into three categories: i) Molecular function, ii) cellular component and iii) the involved biological process. Each protein contained at least one functional GO annotation. Furthermore, GO functional annotations of differentially expressed proteins were compared between the four groups. Compared to the control group, 17 differentially expressed proteins in the ACBP group tended to be located at the macromolecular complex and were closely associated with catalytic activity, protein-binding and nucleic acid-binding transcription factor activity. These proteins were involved in cellular processes, metabolic processes and responses to stimuli (Fig. 4). The 111 differentially expressed proteins of the OXA group were found in the nucleus and cytosolic part and were associated with structural molecule activity. These proteins participated in diverse biological processes, such as metabolic processes, cellular component organization or biogenesis and localization (Fig. 5). The 128 differentially expressed proteins in the ACBP-OXA group were located in the nucleolus, membrane-enclosed lumen and organelle part and were involved in catalytic activity and structural molecule activity, which can affect signaling, cellular component organization or biogenesis and negative regulation of biological processes, indicating that the combination of two drugs exerts complementary and synergistic therapeutic effects on gastric cancer cells (Fig. 6).
Figure 4.
GO analyses of protein functions in ACBP-treated MKN-45 cells. GO functional annotations of 17 differentially expressed proteins in the ACBP-treated group compared with the control group. The 17 differentially expressed proteins were classified into biological processes, molecular functions and cellular components; GO, Gene Ontology; ACBP, anticancer bioactive peptides.
Figure 5.
GO analyses of protein functions in OXA-treated MKN-45 cells. GO functional annotations of 111 differentially expressed proteins in the OXA-treated group compared with the control group. The 111 differentially expressed proteins were classified into biological processes, molecular functions and cellular components. GO, Gene Ontology; OXA, oxaliplatin.
Figure 6.
GO analyses of protein functions in ACBP-OXA-treated MKN-45 cells. GO functional annotations of 128 differentially expressed proteins in the ACBP-OXA-treated group compared with the control group. The 128 differentially expressed proteins were classified into biological processes, molecular functions and cellular components. GO, Gene Ontology; ACBP, anticancer bioactive peptides; OXA, oxaliplatin.
KEGG pathway analysis
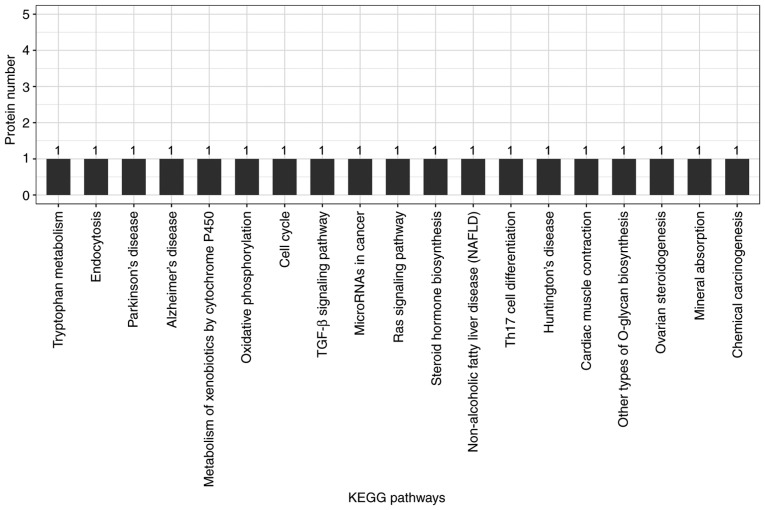
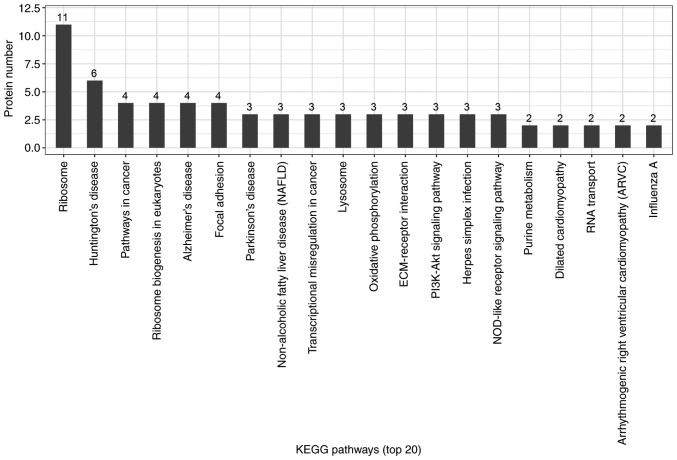
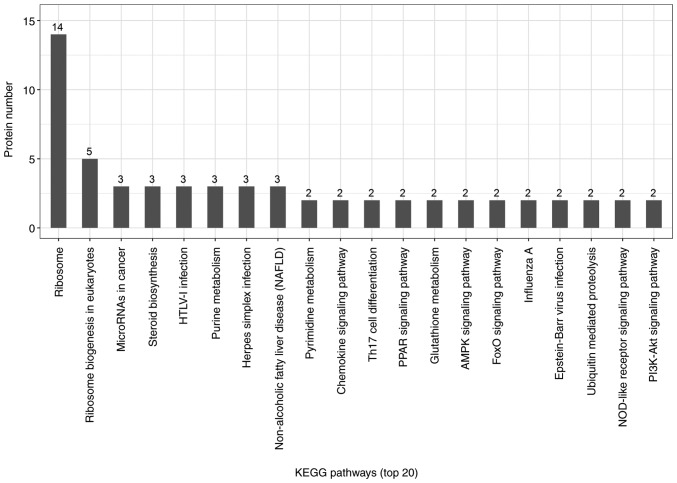
Candidate proteins regulate complex pathological and physiological processes through interaction and intercoordination with other proteins. KEGG pathway analysis was conducted to identify the key signaling pathways and related regulatory processes underlying the therapeutic effects of ACBP, OXA and ACBP-OXA (Tables SIX–XI). The identified signaling pathways in the ACBP group were associated with glycosylation biosynthesis, steroid hormone synthesis, mineral element absorption and cell cycle, among others (Fig. 7). KEGG signaling pathways in the OXA group were associated with cancer-related signaling pathways, adhesions, transcriptional error regulation in cancer, RNA transcription, ECM-receptor activation and PI3-AKT signaling, among others (Fig. 8). The KEGG signaling pathway in the ACBP-OXA group was associated with ribosomes, cancer-related signaling pathways, chemokines, PPAR and AMPK, among others (Fig. 9).
Figure 7.
KEGG analyses of protein functions in ACBP-treated MKN-45 cells. KEGG database pathway annotation was performed on 17 differentially expressed proteins in the ACBP-treated group compared with the control group. KEGG, Kyoto Encyclopedia of Genes and Genomes; ACBP, anticancer bioactive peptides.
Figure 8.
KEGG analyses of protein functions in OXA-treated MKN-45 cells. KEGG database pathway annotation was performed on 111 differentially expressed proteins in the OXA-treated group compared with the control group. KEGG, Kyoto Encyclopedia of Genes and Genomes; OXA, oxaliplatin.
Figure 9.
KEGG analyses of protein functions in ACBP-OXA-treated MKN-45 cells. KEGG database pathway annotation was performed on 128 differentially expressed proteins in the ACBP-OXA-treated group compared with the control group. KEGG, Kyoto Encyclopedia of Genes and Genomes; ACBP, anticancer bioactive peptides; OXA, oxaliplatin.
PPI network analysis
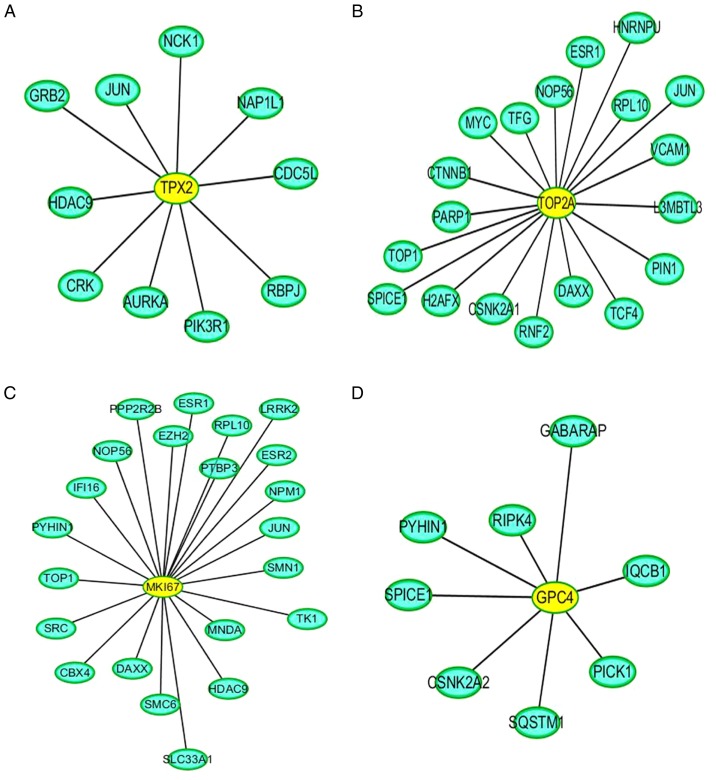
Direct interaction patterns between differently expressed proteins can be beneficial for extracting important information on target proteins. As shown in Fig. 10, the yellow color indicates the differentially expressed target proteins, and the blue color represents the proteins that interacted with these differentially expressed target proteins. In addition, PPI analysis revealed the connectivity degree of the protein interactions. A high connectivity degree may be more indicative of a protein complex. Through intergroup comparison between the ACBP-OXA and control groups (Fig. S3), it was demonstrated that TPX2 (Fig. 10A), TOP2A (Fig. 10B), MKi-67 (Fig. 10C) and GPC4 (Fig. 10D) exhibited a high degree of connectivity, which was located at the center of the network.
Figure 10.
ACBP-OXA-treated MKN-45 cells PPI analysis. There were four important node proteins among the ACBP-OXA groups: (A) TPX2, (B) TOP2A, (C) MKi-67 and (D) GPC4 exhibited the highest connectivity degree. ACBP, anticancer bioactive peptides; OXA, oxaliplatin; TPX2, targeting protein for xenopus kinesin-like protein 2; TOP2A, DNA topoisomerase 2-α; GPC4, glypican-4.
Discussion
The conventional single-agent chemotherapeutic approach may not be effective in treating gastric cancer (28,29). Combination therapy has obvious advantages in terms of enhancing the efficacy of chemotherapeutic drugs, minimizing multidrug resistance in cancer cells and preventing the potential adverse effects resulting from overdose and long-term use of a single drug. In recent years, exploring new combination therapies for treating gastric cancer has become a focus of research (30).
OXA is a third-generation platinum-based chemotherapy drug that has been widely used for gastrointestinal malignancies. However, its long-term use may lead to multidrug resistance and irreversible alterations in cancer cells, thus reducing the efficacy of the treatment (31). Thus, current clinical research has been focusing on finding novel anticancer drugs with higher efficiency, lower toxicity and improved targeting ability. Previous studies have reported that ACBP combined with chemotherapy drugs (e.g., cisplatin and OXA) can inhibit the proliferation of MKN-45 cells, promote apoptosis and induce G2/M phase arrest (13,20). However, the mechanisms underlying the inhibitory effects of ACBP and chemotherapeutic drugs on cell growth remain largely unclear. Several studies have reported that disordered protein expression is commonly found in various types of cancer, including gastric cancer. Hence, there is an urgent need to identify novel diagnostic biomarkers and new therapeutic targets for the treatment of gastric cancer.
In the post-genomic era, proteins have been proposed as the main regulators of biological functions. A high-throughput screening of the proteome expression patterns in cells, tissues and organs can further improve the reliability of disease diagnosis and prognosis prediction and accurately reflect the actual changes in the body compared to candidate protein expression. Therefore, to identify the key regulators and underlying mechanisms of action of combined ACBP and OXA in the inhibition of gastric cancer cell proliferation, iTRAQ technology was used to detect the proteomics profile of MKN-45 cells treated with ACBP and/or OXA.
The present study systematically identified and analyzed the differential proteome expression of the gastric cancer cell in response to combined treatment with ACBP and OXA. The ACBP-OXA, OXA and ACBP treatment groups exhibited 128, 111 and 17 differentially expressed proteins, respectively, compared with the control group. The protein expression patterns analyzed by PRM were consistent with the iTRAQ proteomics data, indicating that the proteomics results of iTRAQ were reliable and may be used for further analysis. Moreover, our findings demonstrated that the number of differentially expressed proteins in the combination therapy group was higher compared with that in either single-drug therapy group. Thus, these differentially expressed proteins may be used as important biomarkers for evaluating the therapeutic effect of ACBP-OXA on gastric cancer, and guide future strategies for treating gastric cancer. The present study revealed the important molecular mechanism underlying the role of combined ACBP and OXA in the treatment of gastric cancer.
Through GO annotation and KEGG pathway analysis, the specific regulatory mechanisms and signal transduction pathways during the process of ACBP-OXA treatment were identified, providing new insights into the development of gastric cancer and suggesting potential therapeutic strategies. GO functional annotation revealed a total of 128 differentially expressed proteins in the ACBP-OXA treatment group compared with the control group. These differentially expressed proteins are mainly found in the nucleus and the membrane-enclosed lumen, and can influence signaling, cellular component organization or biogenesis and negative regulation of biological processes. In addition, the results indicate that these two drugs exert complementary and synergistic effects, and their combination affects biological processes such as growth inhibition and metabolic processes. In addition, KEGG pathway analysis revealed that the signaling pathways in the ACBP-OXA treatment group were enriched in ribosomes, cancer-associated signaling pathways, chemokines, PPAR and AMPK, among others. Enriched signaling pathways are involved in the growth and metabolism of gastric cancer cells. Ribosomes are involved in protein translation, which is the key to regulating intracellular protein synthesis. KEGG analysis revealed that a total of 19 differentially expressed proteins in the ACBP-OXA treatment group regulate ribosome-related pathways, and may regulate intracellular protein synthesis, protein localization and protein transport in gastric cells. Furthermore, the ACBP treatment group exhibited significant differences in the regulatory mechanisms of gastric cancer cells compared with the OXA treatment group. The enrichment results of the AMPK metabolic pathway were consistent with those of lncRNA in MKN-45 cells treated with ACBP-OXA (20), suggesting a close relationship between the two. The AMPK signaling pathway mainly regulates cell metabolism and plays a key role in the regulation of cell energy homeostasis. ACBP-OXA may regulate the homeostasis and energy metabolism of gastric cancer cells by regulating the AMPK signaling pathway. Therefore, ACBP-OXA holds great promise for treating gastric cancer and its actions may be mediated via the AMPK signaling pathway. Furthermore, the cell cycle process was enriched in ACBP-treated MKN-45 cells, which is consistent with our previous report on cell cycle arrest in cancer cells treated with ACBP. The growth inhibition of gastric cancer cells was further supported by the findings that ACBP-OXA regulates cellular metabolism by modulating the AMPK signaling pathway. Taken together, the identified differentially expressed proteins exhibited a similar expression trend with the proteomics expression patterns validated through PRM analysis. Of note, the proteins selected from the ACBP-OXA group exhibited lower expression levels compared with the ACBP, OXA and control groups.
The present study demonstrated that downregulation of the targeting protein for xenopus kinesin-like protein 2 (TPX2) may be an important factor for promoting cell cycle arrest and apoptosis in gastric cancer cells after treatment with ACBP-OXA. TPX2 is a novel oncogene found in several types of cancer, and its overexpression is strongly associated with poor prognosis. TPX2 recruits other mitosis-related factors to activate spindle assembly and maintain the structural stability of the spindle (32–34). Thus, TPX2 appears to be a key mediator of cell mitosis. The binding of TPX2 to Aurora A can trigger the conformation change of Aurora A kinase, in which the N-terminus of TPX2 binds to Aurora A to activate and stabilize its kinase activities. In addition, TPX2 can prevent the premature degradation of Aurora A kinase and promote the local connection of Aurora A kinase with microtubules. The induction of Aurora A kinase activity may interfere with the DNA damage detection sites in the G2/M phase of the cell cycle, and the loss of genetic integrity may promote tumor cell proliferation and accelerate cancer progression (35–38). These observations are consistent with our findings that ACBP-OXA significantly induces G2/M phase arrest in MKN-45 cells. Therefore, it may be inferred that TPX2 is among the research priorities for combination therapy.
In addition, we found that the expression levels of the NUSAP1, TOP2A, YAP, MKi-67 and GPC4 proteins were significantly reduced in the ACBP-OXA group compared with the control group.
Nucleolar spindle-associated protein 1 (NuSAP1) is a microtubule-binding protein that ensures normal cell division and plays an important role in spindle assembly. In recent years, it has been found that NuSAP1 is overexpressed in several cancers and is significantly associated with tumor invasiveness (39–41). DNA topoisomerase 2-α (TOP2A) protein expression is closely associated with the proliferation rate of tumor cells. It has been considered a predictive biomarker for cancer development and represents a major target for chemotherapeutic drugs (42,43). TOP2A plays key roles in chromosome segregation, pairing, concentration, structure formation and alteration of DNA supercoiled structure (44).
Yes-associated protein 1 (YAP), a member of the SCr kinase family, is highly expressed in tumor cells and appears to be a promising chemotherapeutic target (45). Studies have shown that YAP protein overexpression promotes the proliferation and metastasis of tumor cells. Thus, it is of great clinical significance in the early diagnosis of cancer (46–48). The proliferation marker protein Ki-67 (MKi-67) antigen has been widely used as a cell proliferation protein biomarker, which is specifically expressed in the nucleus during the cell cycle (G1, S, G2 and M phases), but not in the resting (G0) phase (49). MKi-67 is the only protein whose expression pattern is closely associated with cell proliferation and the cell-division cycle, and is considered the best marker for discriminating proliferating, quiescent and apoptotic cell populations. An increase in the MKi-67 proliferation index is often associated with clinical deterioration in cancer patients. Moreover, it has been reported to be of value in predicting the survival of cancer patients and tumor recurrence (50–53). Glypican-4 (GPC4) is a member of the heparin proteoglycan family that plays a key role in regulating cell proliferation and differentiation. In addition, GPC4 has been shown to regulate cell migration (54). A previous study reported that a GPC4 gene polymorphism (rs1048369) is closely associated with the development of gastric cancer (55).
Notably, TPX2, TOP2A, MKi-67 and GPC4 are collectively found centrally in the PPI network. Therefore, our data indicate that these differentially expressed proteins can provide important proteomics information regarding the mechanisms of action of combination therapy in treating gastric cancer. The iTRAQ technique was applied to analyze the proteomics profile of MKN-45 cells treated with ACBP and OXA, identify the specific target proteins, and determine the effect of ACBP-OXA inhibition on MKN-45 gastric cancer cells. Taken together, these results may provide new insights into the therapeutic role of combined ACBP and OXA in gastric cancer.
In conclusion, the iTRAQ-based proteomics data and PRM analyses presented herein may help elucidate the proteomics profile of MKN-45 cells treated with ACBP and OXA. These data may also improve our understanding of the molecular mechanisms involved in these processes. Six differentially expressed proteins (i.e., TPX2, NUSAP1, TOP2A, YAP, MKi-67 and GPC4) were found to be significantly decreased in MKN-45 cells treated with ACBP-OXA. KEGG indicated that the AMPK signaling pathway may be one of the important ways through which ACBP-OXA inhibits MKN-45gastric cancer cells. PPI analyses indicated that TPX2, TOP2A, MKi-67 and GPC4 exhibited a high degree of connectivity, which was located at the center of the network. These differentially expressed proteins may be the key to the inhibitory effect of ACBP-OXA on MKN-45 cells, and indicate the potential molecular mechanisms underlying this effect. The limitation of the present study was the screening of differentially expressed proteins by in vitro experiments. In future studies, in vivo and in vitro experiments must be combined to further analyze the role and mechanism of ACBP-OXA from different perspectives and in a complementary manner.
Supplementary Material
Acknowledgements
The authors would like to thank Shanghai Applied Protein Technology Co., Ltd. for technical support of proteomics.
Glossary
Abbreviations
- iTRAQ
isobaric tag for relative and absolute quantitation
- ACBP
anticancer bioactive peptides
- OXA
oxaliplatin
- C
control group
- PRM
parallel reaction monitoring
- PPI
protein-protein interaction
- GO
Gene Ontology
- TPX2
targeting protein for xenopus kinesin-like protein 2
- NUSAP1
nucleolar spindle-associated protein 1
- TOP2A
DNA topoisomerase 2-α
- YAP
Yes-associated protein
- GPC4
glypican-4
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MS
mass spectrometry
- SCX
strong cation exchange
- HPLC
high-performance liquid chromatography
- BP
biological process
- MF
molecular function
- CC
cellular component
- PBS
phosphate-buffered saline
- HCD
higher-energy collisional dissociation
- BLAST
basic local alignment search tool
Funding
The present study was supported by grants from the National Natural Science Foundation of China (nos. 81660468 and 81960560) and the Autonomous Region Science and Technology Innovation Fund (nos. 1639005 and ×201002).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
XS conceived and designed the experiments. YX performed the experiments and analyzed the data. XL performed the language editing and data statistical analysis. All authors have read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
References
- 1.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki S, Gotoda T, Hatta W, Oyama T, Kawata N, Takahashi A, Yoshifuku Y, Hoteya S, Nakagawa M, Hirano M, et al. Survival benefit of additional surgery after non-curative endoscopic submucosal dissection for early gastric cancer: A propensity score matching analysis. Ann Surg Oncol. 2017;24:3353–3360. doi: 10.1245/s10434-017-6039-4. [DOI] [PubMed] [Google Scholar]
- 5.Ohnita K, Isomoto H, Shikuwa S, Yajima H, Minami H, Matsushima K, Akazawa Y, Yamaguchi N, Fukuda E, Nishiyama H, et al. Early and long-term outcomes of endoscopic submucosal dissection for early gastric cancer in a large patient series. Exp Ther Med. 2014;7:594–598. doi: 10.3892/etm.2014.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Caro S, Tao H, Grillo A, Franceschi F, Elia C, Zocco MA, Gasbarrini G, Sepulveda AR, Gasbarrini A. Bacillus clausii effect on gene expression pattern in small bowel mucosa using DNA microarray analysis. Eur J Gastroenterol Hepatol. 2005;17:951–960. doi: 10.1097/00042737-200509000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Ray B, Gupta B, Mehrotra R. Binding of platinum derivative, oxaliplatin to deoxyribonucleic acid: Structural insight into antitumor action. J Biomol Struct Dyn. 2019;37:3838–3847. doi: 10.1080/07391102.2018.1531059. [DOI] [PubMed] [Google Scholar]
- 8.Drott J, Starkhammar H, Kjellgren K, Berterö C. Neurotoxic side effects early in the oxaliplatin treatment period in patients with colorectal cancer. Oncol Nurs Forum. 2018;45:690–697. doi: 10.1188/18.ONF.690-697. [DOI] [PubMed] [Google Scholar]
- 9.Su LY, Xin HY, Liu YL, Zhang JL, Xin HW, Su XL. Anticancer bioactive peptide (ACBP) inhibits gastric cancer cells by upregulating growth arrest and DNA damage-inducible gene 45A (GADD45A) Tumour Biol. 2014;35:10051–10056. doi: 10.1007/s13277-014-2272-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhao YY, Peng SD, Su XL. Effects of anti-cancer bioactive peptide on cell cycle in human nasopharyngeal carcinoma strain CNE. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2006;41:607–611. (In Chinese) [PubMed] [Google Scholar]
- 11.Xing Z, Yu L, Li X, Su X. Anticancer bioactive peptide-3 inhibits human gastric cancer growth by targeting miR-338-5p. Cell Biosci. 2016;6:53. doi: 10.1186/s13578-016-0112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X, Dong C, Zhang J, Su L, Wang X, Cui H, Chen Z. Combination therapy of anti-cancer bioactive peptide with Cisplatin decreases chemotherapy dosing and toxicity to improve the quality of life in xenograft nude mice bearing human gastric cancer. Cell Biosci. 2014;4:7. doi: 10.1186/2045-3701-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Wu H, Ouyang X, Zhang B, Su X. New bioactive peptide reduces the toxicity of chemotherapy drugs and increases drug sensitivity. Oncol Rep. 2017;38:129–140. doi: 10.3892/or.2017.5674. [DOI] [PubMed] [Google Scholar]
- 14.Su L, Xu G, Shen J, Tuo Y, Zhang X, Jia S, Chen Z, Su X. Anticancer bioactive peptide suppresses human gastric cancer growth through modulation of apoptosis and the cell cycle. Oncol Rep. 2010;23:3–9. [PubMed] [Google Scholar]
- 15.Moulder R, Bhosale SD, Goodlett DR, Lahesmaa R. Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrom Rev. 2018;37:583–606. doi: 10.1002/mas.21550. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Dai Y, Xiong J, Zhu S, Zhao M, Ding S, Li J. iTRAQ-based differential proteomics analysis of the brains in a rat model of delayedcarbon monoxide encephalopathy. Brain Res Bull. 2018;137:329–337. doi: 10.1016/j.brainresbull.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Wang Z, Zhao Z, Cui Y. iTRAQ-based proteome profiling of hyposaline responses in zygotes of the Pacific oyster Crassostrea gigas. Comp Biochem Physiol Part D Genomics Proteomics. 2019;30:14–24. doi: 10.1016/j.cbd.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, Gordon SA, Shimada Y, Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Zhu S, Zhao M, Zhao P, Zhao H, Deng J, Li J. Identification of plasma biomarkers for diffuse axonal injury in rats by iTRAQ-coupled LC-MS/MS and bioinformatics analysis. Brain Res Bull. 2018;142:224–232. doi: 10.1016/j.brainresbull.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Han W, Xiao R, Zhang C, Suyila Q, Li X, Su X. Selecting lncRNAs in gastric cancer cells for directed therapy with bioactive peptides and chemotherapy drugs. Oncotarget. 2017;8:86082–86097. doi: 10.18632/oncotarget.20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhusal P, Rahiri JL, Sua B, McDonald JE, Bansal M, Hanning S, Sharma M, Chandramouli K, Harrison J, Procter G, et al. Comparing human peritoneal fluid and phosphate-buffered saline for drug delivery: Do we need bio-relevant media? Drug Deliv Transl Res. 2018;8:708–718. doi: 10.1007/s13346-018-0513-9. [DOI] [PubMed] [Google Scholar]
- 22.Thorat AA, Suryanarayanan R. Characterization of phosphate buffered saline (PBS) in frozen State and after Freeze-drying. Pharm Res. 2019;36:98. doi: 10.1007/s11095-019-2619-2. [DOI] [PubMed] [Google Scholar]
- 23.Yu L, Yang L, An W, Su X. Anticancer bioactive peptide-3 inhibits human gastric cancer growth by suppressing gastric cancer stem cells. J Cell Biochem. 2014;115:697–711. doi: 10.1002/jcb.24711. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 25.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito Y, Yoshikawa T, Fujiwara M, Kojima H, Matsui T, Mochizuki Y, Cho H, Aoyama T, Ito S, Misawa K, et al. Quality of life and nutritional consequences after aboral pouch reconstruction following total gastrectomy for gastric cancer: Randomized controlled trial CCG1101. Gastric Cancer. 2016;19:977–985. doi: 10.1007/s10120-015-0529-5. [DOI] [PubMed] [Google Scholar]
- 29.Kalfusova A, Hilska I, Krskova L, Kalinova M, Linke Z, Kodet R. Gastrointestinal stromal tumors-quantitative detection of the Ki-67, TPX2, TOP2A, and hTERT telomerase subunit mRNA levels to determine proliferation activity and a potential for aggressive biological behavior. Neoplasma. 2016;63:484–492. doi: 10.4149/320_150714N390. [DOI] [PubMed] [Google Scholar]
- 30.Villanueva MT. Combination therapy: Update on gastric cancer in East Asia. Nat Rev Clin Oncol. 2011;8:690. doi: 10.1038/nrclinonc.2011.171. [DOI] [PubMed] [Google Scholar]
- 31.Berretta M, Taibi R, Bearz A, La Mura N, Berretta S, Tirelli U, Frustaci S. Dysphonia as an unusual toxic event of oxaliplatin-based chemotherapy. J Chemother. 2004;16:595–598. doi: 10.1179/joc.2004.16.6.595. [DOI] [PubMed] [Google Scholar]
- 32.Wadsworth P. Tpx2. Curr Biol. 2015;25:R1156–R1158. doi: 10.1016/j.cub.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumayer G, Belzil C, Gruss OJ, Nguyen MD. TPX2: Of spindle assembly, DNA damage response, and cancer. Cell Mol Life Sci. 2014;71:3027–3047. doi: 10.1007/s00018-014-1582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfaro-Aco R, Petry S. How TPX2 helps microtubules branch out. Cell Cycle. 2017;16:1560–1561. doi: 10.1080/15384101.2017.1348080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SY, Kim EY, Kim KH, Lee KA. Bcl2l10, a new Tpx2 binding partner, is a master regulator of Aurora kinase A in mouse oocytes. Cell Cycle. 2016;15:3296–3305. doi: 10.1080/15384101.2016.1243630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrido G, Vernos I. Non-centrosomal TPX2-dependent regulation of the Aurora A Kinase: Functional implications for healthy and pathological cell division. Front Oncol. 2016;6:88. doi: 10.3389/fonc.2016.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grover A, Singh R, Shandilya A, Priyandoko D, Agrawal V, Bisaria VS, Wadhwa R, Kaul SC, Sundar D. Ashwagandha derived withanone targets TPX2-Aurora A complex: Computational and experimental evidence to its anticancer activity. PLoS One. 2012;7:e30890. doi: 10.1371/journal.pone.0030890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pascreau G, Eckerdt F, Lewellyn AL, Prigent C, Maller JL. Phosphorylation of p53 is regulated by TPX2-Aurora A in xenopus oocytes. J Biol Chem. 2009;284:5497–5505. doi: 10.1074/jbc.M805959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Guan C, Lu C, Liu Y, Ni R, Xiao M, Bian Z. High NUSAP1 expression predicts poor prognosis in colon cancer. Pathol Res Pract. 2018;214:968–973. doi: 10.1016/j.prp.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Gordon CA, Gong X, Ganesh D, Brooks JD. NUSAP1 promotes invasion and metastasis of prostate cancer. Oncotarget. 2017;8:29935–29950. doi: 10.18632/oncotarget.15604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han G, Wei Z, Cui H, Zhang W, Wei X, Lu Z, Bai X. NUSAP1 gene silencing inhibits cell proliferation, migration and invasion through inhibiting DNMT1 gene expression in human colorectal cancer. Exp Cell Res. 2018;367:216–221. doi: 10.1016/j.yexcr.2018.03.039. [DOI] [PubMed] [Google Scholar]
- 42.Ye M, He Z, Dai W, Li Z, Chen X, Liu J. A TOP2A-derived cancer panel drives cancer progression in papillary renal cell carcinoma. Oncol Lett. 2018;16:4169–4178. doi: 10.3892/ol.2018.9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engstrom MJ, Ytterhus B, Vatten LJ, Opdahl S, Bofin AM. TOP2A gene copy number change in breast cancer. J Clin Pathol. 2014;67:420–425. doi: 10.1136/jclinpath-2013-202052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Resende MF, Vieira S, Chinen LT, Chiappelli F, da Fonseca FP, Guimarães GC, Soares FA, Neves I, Pagotty S, Pellionisz PA, et al. Prognostication of prostate cancer based on TOP2A protein and gene assessment: TOP2A in prostate cancer. J Transl Med. 2013;11:36. doi: 10.1186/1479-5876-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Yang YC, Zhu JS, Zhou Z, Chen WX. Clinicopathologic characteristics of YES-associated protein 1 overexpression and its relationship to tumor biomarkers in gastric cancer. Int J Immunopathol Pharmacol. 2012;25:977–987. doi: 10.1177/039463201202500415. [DOI] [PubMed] [Google Scholar]
- 46.Chang HL, Chen HA, Bamodu OA, Lee KF, Tzeng YM, Lee WH, Tsai JT. Ovatodiolide suppresses yes-associated protein 1-modulated cancer stem cell phenotypes in highly malignant hepatocellular carcinoma and sensitizes cancer cells to chemotherapy in vitro. Toxicol In Vitro. 2018;51:74–82. doi: 10.1016/j.tiv.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, So KK, Wu K, Fan D, Yu J, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17:2130–2139. doi: 10.1158/1078-0432.CCR-10-2467. [DOI] [PubMed] [Google Scholar]
- 48.Lee SE, Lee JU, Lee MH, Ryu MJ, Kim SJ, Kim YK, Choi MJ, Kim KS, Kim JM, Kim JW, et al. RAF kinase inhibitor-independent constitutive activation of Yes-associated protein 1 promotes tumor progression in thyroid cancer. Oncogenesis. 2013;2:e55. doi: 10.1038/oncsis.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li HH, Qi LN, Ma L, Chen ZS, Xiang BD, Li LQ. Effect of KI-67 positive cellular index on prognosis after hepatectomy in Barcelona Clinic Liver Cancer stage A and B hepatocellular carcinoma with microvascular invasion. Onco Targets Ther. 2018;11:4747–4754. doi: 10.2147/OTT.S165244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshikawa K, Shimada M, Higashijima J, Nakao T, Nishi M, Takasu C, Kashihara H, Eto S, Bando Y. Ki-67 and survivin as predictive factors for rectal cancer treated with preoperative chemoradiotherapy. Anticancer Res. 2018;38:1735–1739. doi: 10.21873/anticanres.12409. [DOI] [PubMed] [Google Scholar]
- 51.Warli SM, Kadar DD, Siregar GP. Ki-67 expression as a predictive factor of muscle invasion in bladder cancer. Open Access Maced J Med Sci. 2018;6:260–262. doi: 10.3889/oamjms.2018.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belinsky I, Murchison AP, Evans JJ, Andrews DW, Farrell CJ, Casey JP, Curtis MT, Nowak Choi KA, Werner-Wasik M, Bilyk JR. Spheno-orbital meningiomas: An analysis based on World health organization classification and Ki-67 proliferative index. Ophthalmic Plast Reconstr Surg. 2018;34:143–150. doi: 10.1097/IOP.0000000000000904. [DOI] [PubMed] [Google Scholar]
- 53.Ishibashi N, Nishimaki H, Maebayashi T, Hata M, Adachi K, Sakurai K, Masuda S, Okada M. Changes in the Ki-67 labeling index between primary breast cancer and metachronous metastatic axillary lymph node: A retrospective observational study. Thorac Cancer. 2019;10:96–102. doi: 10.1111/1759-7714.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dono R. Glypican 4 down-regulation in pluripotent stem cells as a potential strategy to improve differentiation and to impair tumorigenicity of cell transplants. Neural Regen Res. 2015;10:1576–1577. doi: 10.4103/1673-5374.165274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao D, Liu S, Sun L, Zhao Z, Liu S, Kuang X, Shu J, Luo B. Glypican-4 gene polymorphism (rs1048369) and susceptibility to Epstein-Barr virus-associated and -negative gastric carcinoma. Virus Res. 2016;220:52–56. doi: 10.1016/j.virusres.2016.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.