Abstract
Long non-coding RNAs (lncRNAs) are considered to be important regulators in breast cancer. In the present study, the potential mechanisms and functional roles of lncRNA PSMG3-antisense (AS)1 were investigated in vivo and in vitro. The relative expression levels of lncRNA PSMG3-AS1 and microRNA (miR)-143-3p were determined using reverse-transcription quantitative PCR. The protein expression levels of collagen type 1 alpha 1 (COL1A1) and proliferating cell nuclear antigen (PCNA) were obtained using western blot analysis. Bioinformatics analysis was used to identify the relationship between PSMG3-AS1, miR-143-3p and COL1A1. Colony forming and Cell Counting Kit-8 assays were used to detect cell proliferation. Transwell and wound-healing assays were used to determine cell migration. The results of the present study demonstrated that PSMG3-AS1 expression was increased in breast cancer tumor tissues and cell lines, and that of miR-143-3p was decreased. Knockdown of PSMG3-AS1 increased the level of miR-143-3p expression, which led to the mitigation of proliferation and migration capacity in breast carcinoma cells. Additionally, PSMG3-AS1 knockdown was demonstrated to reduce the mRNA and protein expression levels of COL1A1. miR-143-3p mimic transfection reduced proliferation and migration in MDA-MB-231 and MCF-7 cell lines. Furthermore, miR-143-3p inhibition significantly increased the proliferation and migration of breast cancer cells compared with the negative control group. The mRNA and protein expression levels of PCNA were reduced in the MCF-7 cell line when transfected with miR-143-3p mimics and si-PSMG3-AS1. However, PCNA expression was increased in cells transfected with a miR-143-3p inhibitor. In conclusion, the results of the present study identified a novel lncRNA PSMG3-AS1, which serves as a sponge for miR-143-3p in the pathogenesis of breast cancer. PSMG3-AS1 may be used as a potential therapeutic target gene in breast cancer treatment.
Keywords: breast cancer, PSMG3-AS1, miR-143-3p, COL1A1, proliferation, migration
Introduction
Breast cancer is recognized as the most common malignancy in women, with high incidence and mortality rates around the world (1,2). A study indicated that in 2018, 2.1 million individuals would be diagnosed with breast cancer, accounting for ~25% of all cancer diagnoses in women (3). Breast cancer is the most frequently diagnosed cancer in the majority of countries (154 out of 185) and is the leading cause of cancer-associated mortality in >100 locations (3). Breast carcinoma ranks second and fourth for incidence and mortality worldwide, respectively, and has escalated as an emerging epidemic. Therefore, determining the pathogenesis of breast cancer and developing more effective therapeutic strategies for this disease is crucial.
Recent research has documented that non-coding RNAs, which serve key roles in the pervasive transcription of the mammalian genome, constitute the majority of the transcribed genome (4,5). It has also been previously reported that ~2% of the human genome is associated with protein coding, while ~70–90% is transcribed into non-coding RNAs (6). MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are two significant forms of non-coding RNAs (7,8). lncRNAs are characterized as having >200 nucleotides, and serve an essential role in a variety of biological processes, including cellular differentiation, metabolism, migration and apoptosis (9). Previous research has demonstrated that lncRNAs act as tumor promoters in breast cancer. For example, lncROR can serve as an oncogenic factor in breast cancer cells, leading to poor prognostic outcome (10). EZR-antisense (AS)1 was revealed to be overexpressed in breast cancer tissues and may serve as a prognostic marker, leading to the promotion of migration, proliferation and invasion of breast cancer cells (11). lncRNA BLAT1 was revealed to be increased in basal-like breast cancer through epigenetic modifications (12). miRNAs are highly conserved small non-coding RNAs, consisting of 18–24 nucleotides, that can inhibit translation or induce mRNA degradation by binding to the 3′-untranslated regions (3′-UTRs) of target genes (13). Previous research has indicated that miR-143-3p can inhibit the proliferation, migration and invasion of breast cancer cells (1,14).
Previous research has proposed the competing endogenous RNAs (ceRNAs) hypothesis. This hypothesis states that endogenous RNAs, including mRNAs, pseudogenes and lncRNAs, include the miRNA recognition elements (MREs) that competitively combine with common miRNAs and reduce the inhibition of miRNAs on target genes, sequentially, increasing the expression of target genes and regulating cell activities (15). An increasing number of studies have demonstrated that lncRNAs can serve as ceRNAs to regulate mRNAs by sponging target miRNAs. Some studies have indicated that lncRNA-SNHG16 may act as a ceRNA through sponging miR-216-5p competitively to regulate the target gene zinc finger E-box binding homeobox 1 in cervical cancer cell lines (16). LncRNA-SNHG16 has been indicated to increase the expression of the gene PRMT5 by serving as an endogenous sponge of miR-4518 in glioma cell lines (17). The HIF-2α/MALAT1/miR-216b axis has been demonstrated to regulate the multi-drug resistance of HCC cells by modulating autophagy (18).
The present study aimed to identify a signal axis for the lncRNA-miRNA-mRNA interaction in breast carcinoma. The results identified six lncRNAs to be associated with breast cancer, including PSMG3-AS1, through preliminary bioinformatics analysis. The present study revealed that PSMG3-AS1 was increased in breast carcinoma cell lines and tissues. Furthermore, PSMG3-AS1 regulated the expression of COL1A1 by sponging miR-143-3p to affect breast carcinoma development. As a consequence, a PSMG3-AS1/miR-143-3p/COL1A1 regulatory axis was identified in the present study, which served a role in the pathogenic mechanism of breast cancer, and PSMG3-AS1 may act as a potential therapeutic target gene in breast carcinoma.
Materials and methods
Breast cancer tissue samples
The protocol of the present study conformed to the ethical guidelines of the 1975 Declaration of Helsinki and its later amendments. The present study was approved by the Ethics Committee of the Fifth Affiliated Hospital of Harbin Medical University. All participants signed informed consent, and patients provided informed consent for undergoing the procedures and for having data collected and analyzed for research purposes. A total of 33 patients with breast cancer were selected between October 2016 and August 2018. Patients were aged 26–80 years, with an average age of 56.42±11.12 years. The breast cancer tissue and para-carcinoma tissue specimens were collected at the Fifth Affiliated Hospital of Harbin Medical University and maintained at −80°C for further use.
Cell lines
The normal breast epithelial cells MCF-10A and the breast cancer cell lines MDA-MB-468, MDA-MB-231 and MCF-7 were obtained from the American Type Culture Collection. MCF-10A cells were cultured in MEGM (Cobioer Biosciences) with 100 ng/ml cholera toxin. MDA-MB-468, MDA-MB-231 and MCF-7 cells were cultured in DMEM (Sangon Biotech Co., Ltd.) supplemented with 10% fetal bovine serum (Sangon Biotech Co., Ltd.) and 1% antibiotics (penicillin 100 U/ml; streptomycin 100 µg/ml; Sangon Biotech Co., Ltd.). All cell lines were cultured in an incubator at 37°C with 5% CO2.
RNA extraction, cDNA synthesis and reverse-transcription quantitative (RT-q) PCR
Total RNA was extracted from the tissue samples or cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.), and the RNA concentration and purity were determined using BioSpec-nano (Beijing Daojin Medical Appliance Co., Ltd.). The cDNA of total RNA was synthesized using the ReverTra Ace® qPCR RT kit (cat. no. FSQ-101; Toyobo Life Science) and the cDNA of miR-143-3p was synthesized using the Reverse transcription kit (Suzhou GenePharma Co., Ltd.). RT-qPCR for lncRNA PSMG3-AS1, COL1A1 and PCNA was performed using the THUNDERBIRD SYBR qPCR Mix (cat. no. QPS-201; Toyobo Life Science), and the reaction procedure was 95°C for 1 min, followed by 40 cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 1 min. miR-143-3p RT-qPCR was performed using the miR-143-3p Hairpin-it™ Real-Time PCR kit, and the reaction procedure was 95°C for 3 min, followed by 40 cycles at 95°C for 12 sec and 60°C for 40 sec. RT-qPCR was performed using a StepOnePlus RT-qPCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) and all procedures were carried out in accordance with the manufacturer's protocol. Relative expression levels were determined using the 2−ΔΔCq (19) method and normalized to β-Actin and U6. The primer sequences used were as follows. PSMG3-AS1 forward, 5′-GAAGCAGAACCAACGCACAG-3′ and reverse, 5′-GCATAATCCAATCCCTCAAGAA-3′; ACTB forward, 5′-CCTGGCACCCAGCACAAT-3′ and reverse, 5′-GGGCCGGACTCGTCATAC-3′; miR-143-3p forward, 5′-CTGGCGTTGAGATGAAGCAC-3′ and reverse, 5′-CAGAGCAGGGTCCGAGGTA-3′; U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse, 5′-TTCACGAATTTGCGTGTCATC-3′; COL1A1 forward, 5′-GCGTGTACCCCACTCAGC-3′ and reverse, 5′-CCGAACCAGACATGCCTC-3′.
Cell transfection
miR-143-3p mimics and negative control (NC), miR-143-3p inhibitor and NC and si-PSMG-AS1 and NC, were obtained from Suzhou GenePharma Co., Ltd. The sequences were as follows: miR-143-3p mimics forward, 5′-UGAGAUGAAGCACUGUAGCUC-3′ and reverse, 5′-GCUACAGUGCUUCAUCUCAUU-3′); mimics-NC forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′); miR-143-3p inhibitor forward, 5′-GAGCUACAGUGCUUCAUCUCA-3′) and inhibitor-NC reverse, 5′-CAGUACUUUUGUGUAGUACAA-3′); si-PSMG3-AS1 forward, 5′-GGACGUCUCCCAUUCUGAATT-3′ and reverse, 5′-UUCAGAAUGGGAGACGUCCTT-3′); si-PSMG3-NC forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. Prior to transfection, 5×105 cells/well were seeded into six-well plates. The RNA oligo was transfected in MDA-MB-231, MDA-MB-468 and MCF-7 cells using Lipofectamine 2000® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
Cell proliferation assay
The Cell Counting Kit (CCK)-8 assay was performed to calculate the proliferation of the breast cancer cell lines. Cells with different processing methods in different experimental groups were digested and seeded in 96-well plates following transfection. Cells were then cultured for 24, 48 and 72 h, respectively. At the end of the experiment, 10 µl CCK-8 solution (Sangon Biotech Co., Ltd.) was added into each well and the cells were incubated at 37°C in a humidified incubator with 5% CO2 for 2 h. The absorbance of each well was measured at 450 nm using a microplate reader (Sunrise; Tecan Group Ltd.). The result of relative cell proliferation was obtained by dividing the value measured at each time-point by the value at 0 h and setting the measured value at 0 h as 1 (20).
For the colony formation assay, 400 cells/well were plated and after 7 days of culture, the cells were washed with PBS, fixed with methanol and treated with 0.2% crystal violet, respectively. The colonies containing >50 cells were counted manually and images were obtained concurrently.
Cell migration assay
Wound-healing and Transwell assays were used to evaluate migration of RNA oligo-treated cell lines. For the wound healing assay, 200 µl pipette tips were used to create scratches when cells were 90% confluent. Images were obtained at 0 and 24 h, at the same position under a microscope (Olympus Corp.) at an ×100 magnification, to compare cell migration. For the Transwell assay, the Transwell cups were placed in 24-well plates (Corning, Inc.). Following treatment with RNA oligo, 5×104 cells/well were placed in the upper chamber with 200 µl DMEM (Sangon Biotech Co., Ltd.). The lower chamber was filled with 600 µl of 10% FBS DMEM. The cells were then cultured for 24 h. The outer surface was subsequently washed three times with PBS, fixed with methanol for 20 min and stained with 0.2% crystal violet for 15 min at room temperature. The images were obtained immediately after drying. The number of migrated cells were counted in five randomly microscopic fields of view (Olympus Corp.) at an ×400 magnification.
Western blot analysis
Total protein was isolated in cell lysis buffer for western blot analysis and immunoprecipitation in breast cancer cells (Beyotime Institute of Biotechnology). The proteins were quantified using an Enhanced BCA protein Assay kit (Beyotime Institute of Biotechnology) and equal amounts of protein lysates were subjected to 10% SDS-PAGE and transferred to a 0.45 µm PVDF membrane (Beyotime Institute of Biotechnology). The membranes were then blocked with blocking buffer and incubated at room temperature for 1 h. Membranes were subsequently incubated with primary antibodies at 4°C overnight. Incubation with secondary antibodies was performed at room temperature for 1 h. The primary antibodies used were as follows: COL1A1/Collagen I rabbit monoclonal antibody (1:750; cat. no. AF1840, rabbit anti-human), β-actin mouse monoclonal antibody (1:1,000; cat. no. AF0003, mouse anti-human), PCNA rabbit monoclonal antibody (1:1,000; cat. no. AF1363, rabbit anti-human). The secondary antibodies used were as follows: HRP-labeled goat anti-rabbit IgG (1:1,000; cat. no. A0208; goat anti-rabbit), HRP-labeled goat anti-mouse IgG (1:2,000; cat. no. A0216; goat anti-mouse). All antibodies were purchased from Beyotime Institute of Biotechnology. The signals were detected using BeyoECL Plus (Beyotime Institute of Biotechnology). Finally, all proteins were exposed to X-ray film and the protein bands were analyzed using ImageJ software (version 1.48; NIH; National Institutes of Health, Bethesda, MD, USA).
Bioinformatics analysis
miRNA-mRNA interactions and lncRNA-miRNA intersections were downloaded from starBase v2.0 (http://starbase.sysu.edu.cn/starbase2/index.php). mRNA and lncRNA expression profiles GSE47860 of breast cancer were downloaded [currently the chip platform suitable for this analysis is (huex-1_0-st) Affymetrix Human Exon 1.0 ST Array], and highly competitive lncRNA regulated subnetworks were constructed. Potential target genes of miR-143-3p were predicted using TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://www.mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php) and DIANA TOOLS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/page&view=software).
Statistical analysis
SPSS 18.0 software (SPSS, Inc.) was used for statistical analyses. All data are presented as the mean ± SEM from >two independent experiments. Differences between groups were analyzed using a Student's t-test (two groups) and a χ2 test was used to determine the correlation between the expression of PSMG3-AS1 and clinicopathological features. P<0.05 was considered to indicate a statistically significant result.
Results
PSMG3-AS1 is upregulated in breast cancer tissues and cell lines
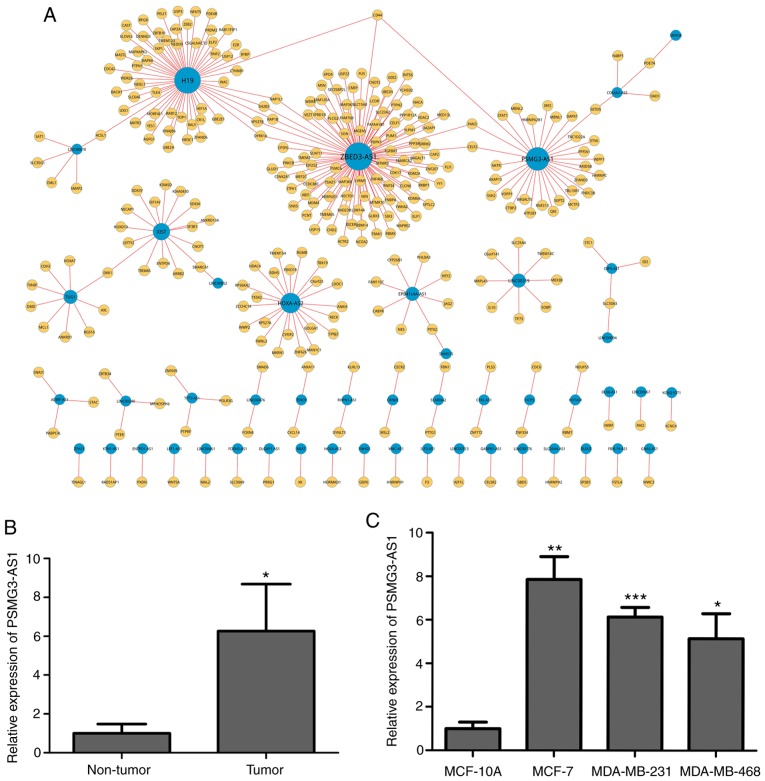
Through preliminary bioinformatics analysis six lncRNAs were revealed to be associated with breast cancer, including PSMG3-AS1 (Fig. 1A). To examine PSMG3-AS1 expression in breast cancer tissues and cell lines, a total of 33 pairs of breast cancer tissues (Tumor) and para-carcinoma tissues (Non-tumor), and three breast cancer cell lines (MDA-MB-468, MDA-MB-231 and MCF-7) were used to detect PSMG3-AS1 expression. The results indicated that the expression of PSMG3-AS1 in cancer tissues was significantly higher compared with non-tumor tissues (P<0.05; Fig. 1B). Similarly, PSMG3-AS1 was increased in MDA-MB-468, MDA-MB-231 and MCF-7 cell lines compared with non-metastatic human mammary epithelial cell line MCF-10A (P<0.05; Fig. 1C). Additionally, the present study investigated the association between the expression of PSMG3-AS1 and the clinicopathological characteristics of 33 patients with breast cancer (Table I). The results demonstrated that PSMG3-AS1 served a critical role in the development of breast cancer.
Figure 1.
PSMG3-AS1 is significantly increased in breast carcinoma tissues and cell lines. (A) LncRNA screening was closely associated with breast cancer. (B) RT-qPCR was performed to examine the expression of PSMG3-AS1 in tumor tissues and adjacent cancerous tissues. (C) The expression of PSMG3-AS1 in three breast cancer cell lines (MDA-MB-468, MDA-MB-231 and MCF-7) and a normal breast epithelial cell line MCF-10A was assessed using RT-qPCR. Error bars represented the mean ± SEM acquired from three independent experiments. *P<0.05, **P<0.01, ***P<0.001. AS, antisense; RT-qPCR, reverse transcription-quantitative PCR; lncRNA, long non-coding RNA.
Table I.
Relationship of PSMG3-AS1 expression with clinic pathological features in breast cancer patients.
| Variable | ||||||
|---|---|---|---|---|---|---|
| Age (years) | Tumor size | WHO grade | ||||
| Expression of lnc-PSMG3-AS1 | ≤60 | >60 | ≤5 cm | >5 cm | I–II | III |
| Low (%) | 13 (81.25) | 3 (18. 75) | 13 (81.25) | 3 (18.75) | 11 (68.75) | 5 (31.25) |
| High (%) | 10 (58.82) | 7 (41.18) | 15 (88.24) | 2 (11.76) | 14 (82.35) | 3 (17.65) |
| P-value | 0.259 | 0.656 | 0.438 | |||
Silencing of PSMG3-AS1 attenuates the progression of breast cancer cells
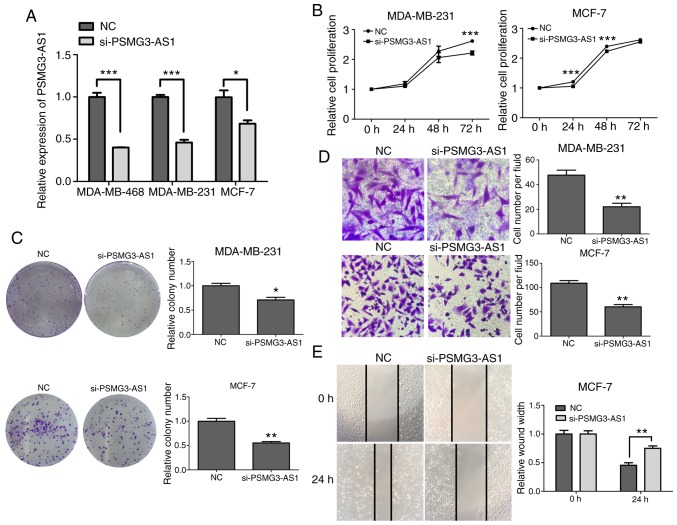
PSMG3-AS1 was silenced in MDA-MB-468, MDA-MB-231 and MCF-7 cell lines. RT-qPCR was performed to examine the efficiency of PSMG3-AS1 small interfering (si)RNAs. As presented in Fig. 2, the expression of PSMG3-AS1 was decreased in the PSMG3-AS1 siRNA-transfected groups (P<0.05; Fig. 2A). The results of the CCK-8 and colony forming assays indicated that silencing of PSMG3-AS1 inhibited the proliferation of MDA-MB-231 and MCF-7 breast cancer cells (P<0.05; Fig 2B and C). Furthermore, the results of the Transwell assay demonstrated that the migration of MDA-MB-231 and MCF-7 breast cancer cells were significantly decreased after PSMG3-AS1 knockdown (P<0.05; Fig. 2D), and wound-healing assay demonstrated that the migration of MCF-7 breast cancer cells was significantly reduced after PSMG3-AS1 knockdown (P<0.05; Fig. 2E).
Figure 2.
Silencing of PSMG3-AS1 suppresses proliferation and migration in breast cancer cells. (A) MDA-MB-468, MDA-MB-231 and MCF-7 breast carcinoma cell lines were transfected with si-PSMG3-AS1 and RT-qPCR was used to examine the transfection efficiency. (B and C) Colony formation and CCK-8 assays were performed to detect the proliferative ability of breast cancer cells incubated with si-PSMG3-AS1. (D and E) Transwell and wound healing assays were used to examine the migratory ability of cells treated with si-PSMG3-AS1. Error bars represented the mean ± SEM acquired from three independent experiments. *P<0.05, **P<0.01, ***P<0.001. AS, antisense; si, small interfering; RT-qPCR, reverse transcription-quantitative PCR; CCK-8, Cell Counting Kit-8.
miR-143-3p is a target of PSMG3-AS1 in breast cancer
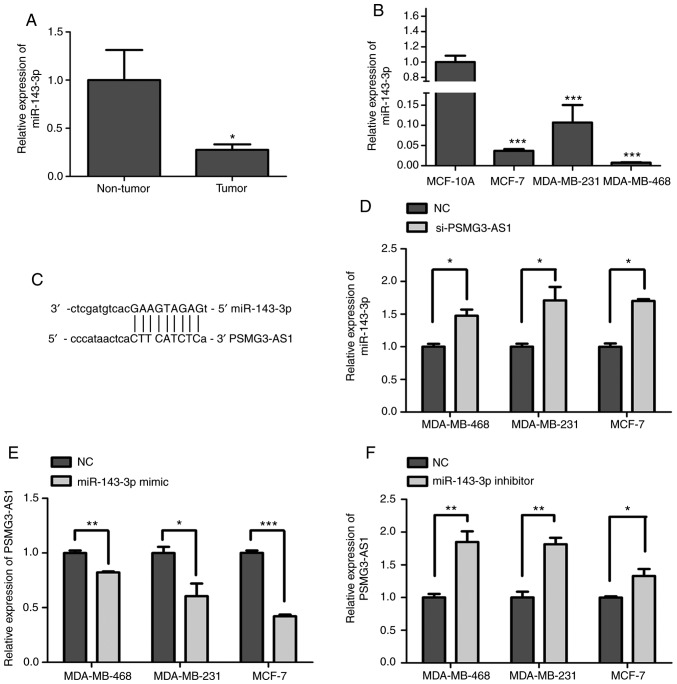
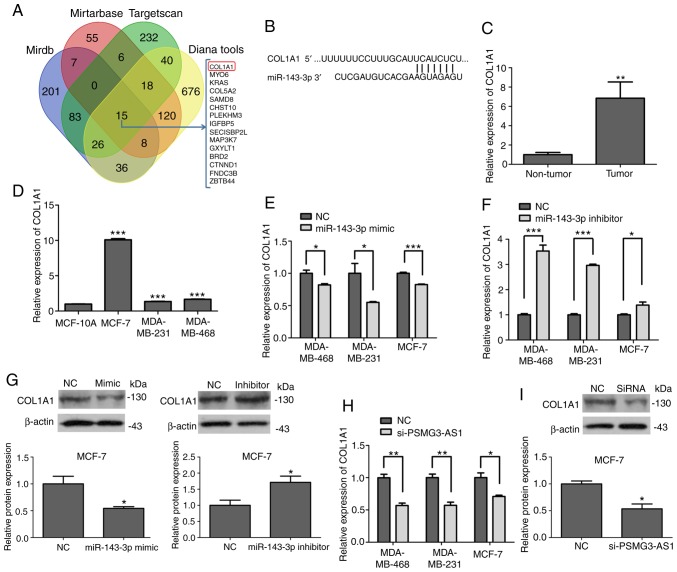
Increasing evidence has indicated that lncRNAs may function as ceRNAs or a molecular sponge in the regulation of a variety of miRNA biological functions (21). The present study identified three miRNAs, which could bind with PSMG3-AS1, from starBase v 2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php), including miR-143-3p, which has been reported to be associated with breast cancer (1,14). In the present study, the expression of miR-143-3p was downregulated in tumor tissues compared with non-tumor tissues (P<0.05; Fig. 3A). miR-143-3p was also revealed to be reduced in MDA-MB-468, MDA-MB-231 and MCF-7 breast cancer cell lines compared with MCF-10A (P<0.05; Fig. 3B).
Figure 3.
miR-143-3p is identified as a target gene of PSMG3-AS1. (A) RT-qPCR was performed to detect the expression of miR-143-3p in tumor and non-tumor tissues. (B) The expression of miR-143-3p in three breast cancer cell lines (MDA-MB-468, MDA-MB-231 and MCF-7) and a normal breast epithelial cell line MCF-10A was assessed using RT-qPCR. (C) Bioinformatics analysis indicating the interaction of PSMG3-AS1 with miR-143-3p. (D) Relative expression of miR-143-3p in si-NC and si-PSMG3-AS1 in three breast cancer cell lines. (E and F) Relative expression of PSMG3-AS1 in three breast cancer cell lines transfected with miR-143-3p mimics, mimics-NC, miR-143-3p inhibitor and inhibitor-NC. Error bars represented the mean ± SEM acquired from three independent experiments. *P<0.05, **P<0.01, ***P<0.001. miR, microRNA; AS, antisense; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering; NC, negative control.
Whether lncRNA PSMG3-AS1 could bind with miR-143-3p was also assessed in the present study (Fig. 3C). PSMG3-AS1 was silenced in breast cancer cell lines MDA-MB-468, MDA-MB-231 and MCF-7, and the RT-qPCR results revealed that miR-143-3p was significantly upregulated compared with the NC group (P<0.05; Fig. 3D). The overexpression of miR-143-3p downregulated the level of PSMG3-AS1, and with the inhibition of miR-143-3p, the opposite results were obtained (P<0.05; Fig. 3E and F). Based on these results, PSMG3-AS1 may act as a ceRNA to bind with miR-143-3p.
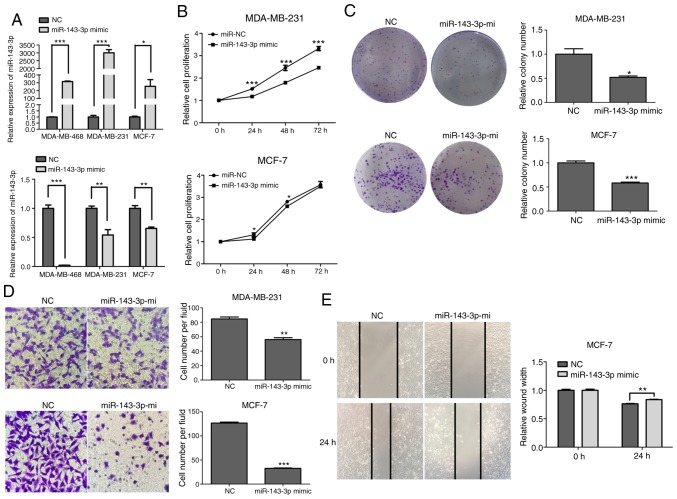
Overexpression or inhibition of miR-143-3p may influence progression of breast cancer cells
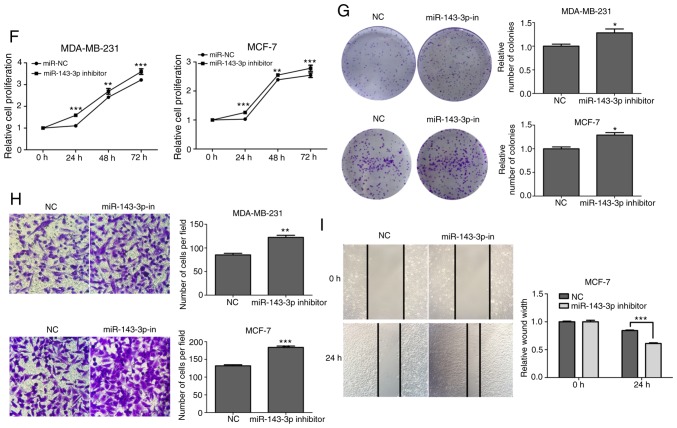
miR-143-3p mimics, mimics-NC, miR-143-3p inhibitor and inhibitor-NC were transfected into breast cancer cell lines. RT-qPCR was performed to examine transfection efficiency (Fig. 4A). CCK-8 and colony forming assays were used to assess cell proliferation, and Transwell and wound-healing assays were used to assess cell migration. The results demonstrated that the overexpression of miR-143-3p reduced the proliferative ability of MDA-MB-231 and MCF-7 cells (Fig. 4B and C). Additionally, Transwell assay revealed that the migratory capacities of MDA-MB-231 and MCF-7 cells were decreased (Fig. 4D), and wound-healing assay revealed that the migratory capacity of MCF-7 cells was reduced (Fig. 4E) with overexpression of miR-143-3p. In contrast, inhibiting miR-143-3p significantly increased the proliferation of MDA-MB-231 and MCF-7 cell lines compared with the inhibitor-NC group (Fig. 4F and G), and Transwell assay revealed that the migratory capacities of MDA-MB-231 and MCF-7 cells were increased (Fig. 4H) and wound-healing assay revealed that the migratory capacity of MCF-7 cells was enhanced (Fig. 4I). These results were consistent with previous research which determined this in other tumor types (22).
Figure 4.
Effect of miR-143-3p expression on cell proliferation and migration in breast cancer cells. (A) Relative expression of miR-143-3p in breast cancer cell lines MDA-MB-468, MDA-MB-231 and MCF-7 transfected with miR-143-3p mimics, mimics-NC, miR-143-3p inhibitor or inhibitor-NC. (B and C) Colony formation and CCK-8 assays were performed to detect the proliferative ability of breast cancer cells transfected with miR-143-3p mimics or mimics-NC. (D and E) Transwell and wound healing assays were performed to detect the migratory ability of cells transfected with miR-143-3p mimics and mimics-NC. (F and G) Colony formation and CCK-8 assays were used to examine the proliferative ability of breast cancer cells transfected with miR-143-3p inhibitor or inhibitor-NC. (H and I) Transwell and wound healing assays were performed to detect the migration of cells treated with miR-143-3p inhibitor or inhibitor-NC. Error bars represented the mean ± SEM acquired from three independent experiments. *P<0.05, **P<0.01, ***P<0.001. miR, microRNA; NC, negative control; CCK-8, Cell Counting Kit-8.
miR-143-3p may serve a role in regulating its target gene COL1A1
Using bioinformatic analysis, a total of 15 candidates were identified as common target genes for miR-143-3p (Fig. 5A). Among these genes, COL1A1 was increased in a number of malignant tumors and had been reported to contribute to tumor cell amplification and migration (23). The binding sites between miR-143-3p and COL1A1 were searched using bioinformatics analysis (Fig. 5B). The mRNA expression of COL1A1 in patient tumor tissues and breast cancer cell lines was increased, and these results were in accordance with those for PSMG3-AS1 (Fig. 5C and D). The present study indicated that the overexpression of miR-143-3p decreased the mRNA expression of COL1A1 in three breast cancer cell lines (Fig. 5E). However, the inhibition of miR-143-3p expression increased the mRNA of COL1A1 (Fig. 5F). Furthermore, the COL1A1 protein expression level in MCF-7 cells, which were treated with miR-143-3p mimics and inhibitor, was consistent with the COL1A1 mRNA expression level (Fig. 5G). In the present study, silencing of PSMG3-AS1 in breast cancer cells was revealed to reduce COL1A1 mRNA and protein expression (Fig. 5H and I). Therefore, the present study demonstrated that PSMG3-AS1 could regulate COL1A1 by binding with miR-143-3p.
Figure 5.
COL1A1 is identified as a target gene of miR-143-3p. (A) A Venn diagram presenting 4 software databases which predicted miRNA targets and identified 15 candidate genes that may interact with miR-143-3p. (B) A putative miR-143-3p site exists in the 3′-UTR of the COL1A1 mRNA. (C) The relative mRNA expression of COL1A1 in breast cancer tissues and adjacent cancerous tissues. (D) The relative mRNA expression of COL1A1 in three breast cancer cell lines (MDA-MB-468, MDA-MB-231 and MCF-7) and a normal breast epithelial cell line MCF-10A. (E and F) mRNA expression of COL1A1 in MDA-MB-468, MDA-MB-231 and MCF-7 cell lines transfected with miR-143-3p mimics, mimics-NC, miR-143-3p inhibitor or inhibitor-NC. (G) The COL1A1 protein expression in MCF-7 cells treated with miR-143-3p mimics and inhibitor. (H and I) Silencing of PSMG3-AS1 in MCF-7 cells reduced the mRNA and protein expression of COL1A1. *P<0.05, **P<0.01, ***P<0.001. COL1A1, collagen type 1 alpha 1; miR, microRNA; UTR, untranslated region; NC, negative control; si, small interfering; AS, antisense.
The expression of the cell proliferation marker PCNA is consistent with that of PSMG3-AS1
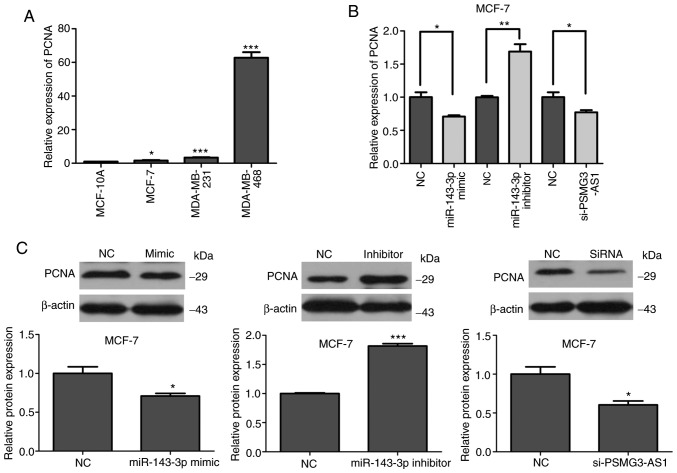
It is well established that PCNA is a cell proliferation-associated protein and manifests its expression during the cell division of normal and neoplastic cells (24). The cell proliferation marker PCNA expression is associated with the genesis and development of a variety of cancer types, including breast cancer (25) and gastric cancer (26). In the present study, PCNA mRNA expression was upregulated in three breast cancer cell lines compared with MCF-10A cells (Fig. 6A). This result was consistent with the results of PSMG3-AS1. Furthermore, the results also indicated that in the MCF-7 breast cancer cell line, the mRNA and protein expression of PCNA was consistent with PSMG3-AS1 after transfection with PSMG3-AS1 siRNAs, miR-143-3p mimics and miR-143-3p inhibitor (Fig. 6B and C). Therefore, it can be concluded that lncRNA PSMG3-AS1 may serve as a tumor promoter in breast cancer, and may combine with miR-143-3p to increase the expression of COL1A1.
Figure 6.
PCNA protein and mRNA levels in different transfection groups. (A) The relative mRNA expression of PCNA was upregulated in three breast cancer cell lines compared with MCF-10A cells. (B and C) The mRNA and protein expression of PCNA were decreased in MCF-7 breast cancer cell lines by transfection with miR-143-3p mimics and si-PSMG3-AS1, while they were increased in the cells by transfection with inhibitor-miR-143-3p. *P<0.05, **P<0.01, ***P<0.001. PCNA, proliferating cell nuclear antigen; miR, microRNA; si, small interfering; AS, antisense.
Discussion
Breast cancer is a type of malignant tumor, and is the most common cancer among women (2). Currently, combined with early diagnoses, radical surgery with adjuvant therapy could decrease breast cancer-associated mortality rates (27). However, breast cancer is still the most common gynecologic cancer. Multiple studies have demonstrated that lncRNAs are deregulated in a variety of human cancer types, including breast cancer (28,29), bladder cancer (30,31), glioma (17,32), lung cancer (33) and hepatocellular cancer (18,34). Recently, studies have indicated that lncRNAs serve a role in a variety of biological processes, including in chromatin modification, transcriptional regulation, genomic imprinting and the regulation of protein function (9,35). Studies have explored the abnormal expression of lncRNAs and their association with tumor initiation, growth and metastasis in tumor cells. Zhao et al (36) indicated that SNHG16 knockdown inhibited metastasis, proliferation and vasoformation of HemECs. Katsushima et al (37) revealed that the inhibition of TUG1 by siRNA reduced cell proliferation in glioma stem cells. Zhou et al (38) indicated that lncRNA H19 may serve an essential role in tumor metastasis, and that lncRNA H19 expression was correlated with metastasis in human breast cancer. Due to this, it can be concluded that lncRNAs may be used as diagnostic markers of metastatic tumors, and exhibit great clinical importance.
In the present study, a total of six lncRNAs (H19, XIST, TUG1, HOXA-AS2, ZBED3-AS1 and PSMG3-AS1) were screened using bioinformatics analysis. To date, little research has been performed assessing the role of lncRNAs in tumors, especially regarding the mechanism of PSMG3-AS1 in breast cancer. Therefore, the present study focused on determining PSMG3-AS1 expression. PSMG3-AS1 was demonstrated to be upregulated in breast cancer tumor tissues and breast cancer cell lines, suggesting it may serve an important role in the progression of breast cancer. The results indicated that knockdown of PSMG3-AS1 inhibited breast cancer cell proliferation and migration. CCK-8 and colony forming assays revealed that cell proliferation ability was reduced, and both Transwell and wound-healing assays indicated that migratory capacities were decreased. The mRNA and protein expression of cell proliferation marker PCNA were consistent with PSMG3-AS1. In summary, PSMG3-AS1 may serve a role in promoting tumor formation in breast cancer.
Bioinformatics analysis indicated that the miR-143-3p was a direct target gene of PSMG3-AS1, and previous research had indicated that miR-143-3p inhibited the progression of breast cancer cells and osteosarcoma cells (1,22). In the present study, miR-143-3p expression was reduced in breast cancer tissues and cell lines. Silencing of PSMG3-AS1 in breast cancer cells increased the expression level of miR-143-3p, and overexpression of miR-143-3p decreased the expression levels of PSMG3-AS1 and inhibited breast cancer cell proliferation and migration. Additionally, the opposite results were obtained after inhibition of miR-143-3p. Recent studies have demonstrated that COL1A1 promotes the metastasis of breast cancer and is a potential therapeutic target gene (39). The results of the present study revealed that COL1A1 is the target gene of miR-143-3p, which was identified using bioinformatics analysis. Overexpression of miR-143-3p may downregulate the mRNA and protein expression of COL1A1, and inhibition of miR-143-3p may induce the opposite experimental results.
The results of the present study indicated that lncRNA (PSMG3-AS1)-miRNA (miR-143-3p)-mRNA (COL1A1) may act as a signal axis in breast cancer. One of the main effects of lncRNAs is to act as a sponge for miRNAs, and to reduce the expression and activity of miRNAs (16). miRNAs serve a regulatory role by directly binding to target genes (40). The present study indicated that PSMG3-AS1 regulated the target gene COL1A1 of miR-143-3p by sponging miR-143-3p (Fig. 7). This correlation between PSMG3-AS1 and miR-143-3p highlighted the lncRNA-based mechanisms underlying diverse aspects of tumorigenesis. The present study is helpful in providing a novel therapeutic strategy for use in the treatment of breast cancer.
Figure 7.
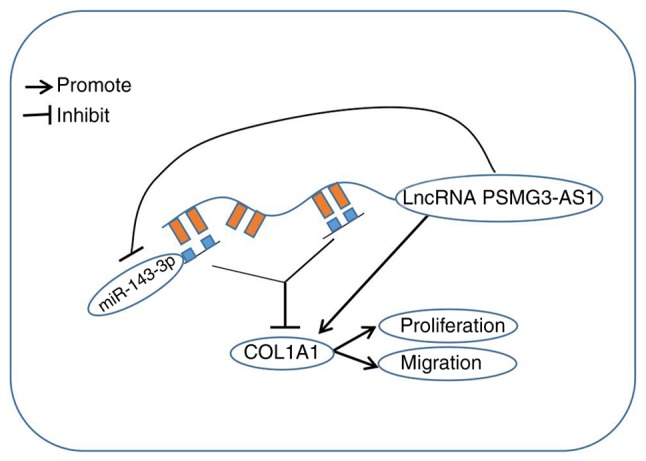
Schematic diagram of the PSMG3-AS1-miR-143-3p-COL1A1 regulatory axis. PSMG3-AS1 could promote the proliferation and migration of breast cancer cells by binding miR-143-3p and resulting in increased COL1A1 expression. AS, antisense; miR, microRNA; COL1A1, collagen type 1 alpha 1.
In summary, a PSMG3-AS1-miR-143-3p-COL1A1 regulatory axis in breast cancer was indicated in the present study. PSMG3-AS1 served as an oncogenic lncRNA that facilitated the genesis and development of breast cancer, by functioning as a ceRNA, which regulated the expression of COL1A1, and also through directly sponging miR-143-3p. In the present study, PSMG3-AS1 was indicated to be a new potential therapeutic target in breast carcinoma treatment. In vivo studies have revealed the important role of lncRNAs and miRNAs in tumors. For example, H19 was revealed to be associated with tumor development, progression, metastasis and drug resistance (41), and silencing of MALAT1 suppressed the proliferation of GSCs and in vivo tumor growth by upregulating miR-129 and inhibiting SOX2 (42).
However, there are some limitations in the present study: First, the sample size was only 33 cases, which was relatively small. Second, breast cancer cell lines and tissue samples from patients with breast cancer were assessed, however, the present research lacks analysis between expression and clinical course including genetic backgrounds, hormone receptor expression, chemo-sensitivity, local/distant relapse and prognosis. In Fig. 5D although the mRNA expression of COL1A1 in breast cancer cell lines was revealed to be increased compared with MCF-10A cells, its expression was higher in MCF-7 cells compared to the other 2 cell lines, which reflects the cell-line specific differences. In Fig. 3B, the level of miR-143-3p was reduced in all 3 cell lines however the reduction was less in MDA-MB-231 cells. It was speculated that different genetic backgrounds and receptor expression of these different cell lines may play a role. Third, experimental in vivo data were not collected in the present study. Therefore, future studies should increase the sample size and validate the function of the PSMG3-AS1-miR-143-3p-COL1A1 signaling axis in vivo. Selective inhibition and overexpression of PSMG3-AS1 and miR-143-3p should be performed to further study the effects of these two genes on the occurrence and development of tumors, hormone receptor expression, chemo-sensitivity, local/distant relapse and prognosis in vivo, and further assessment of the function of PSMG3-AS1 derived from different cell lines with a variety of genetic background is still required to a provide strong experimental basis for clinical practice.
Acknowledgements
Not applicable.
Funding
The present study was supported, in part, by the Fifth Affiliated Hospital of Harbin Medical University, Health Commission of Heilongjiang Province (grant no. 2017-183), the National Natural Science Foundation of China (grant no. 81503069), the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (grant no. UNPYSCT-2016040), the Natural Science Foundation of Heilongjiang Province (grant no. QC2016112), the Traditional Chinese Medicine Administration of Heilongjiang Province (grant no. IQG-050) and the Fundamental Research Funds for the Central Universities (grant no. 21619347).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YC performed all of the experiments. DY and YF designed the study and wrote the manuscript. GZ, QZ, YC and YB collected the tissue samples and participated in the statistical analysis and helped to draft the manuscript. ZY, GC, XP and FG participated in RNA extraction. JW and YB performed the western blot assay. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that accuracy or integrity of any parts of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Fifth Affiliated Hospital of Harbin Medical University and all the participants signed informed consent. All patients provided informed consent to undergo the procedures and for having their data collected and analyzed for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Xia C, Yang Y, Kong F, Kong Q, Shan C. MiR-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie. 2018;147:98–104. doi: 10.1016/j.biochi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Donepudi MS, Kondapalli K, Amos SJ, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10:506–511. doi: 10.4103/0973-1482.137927. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:E1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 6.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 7.Greco S, Gorospe M, Martelli F. Noncoding RNA in age-related cardiovascular diseases. J Mol Cell Cardiol. 2015;83:142–155. doi: 10.1016/j.yjmcc.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Hou L, Tu J, Cheng F, Yang H, Yu F, Wang M, Liu J, Fan J, Zhou G. Long noncoding RNA ROR promotes breast cancer by regulating the TGF-β pathway. Cancer Cell Int. 2018;18:142. doi: 10.1186/s12935-018-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai Y, Zhou X, Huang L, Wan Y, Li X, Wang Y. Long noncoding RNA EZR-AS1 promotes tumor growth and metastasis by modulating Wnt/β-catenin pathway in breast cancer. Exp Ther Med. 2018;16:2235–2242. doi: 10.3892/etm.2018.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han YJ, Boatman SM, Zhang J, Du XC, Yeh AC, Zheng Y, Mueller J, Olopade OI. LncRNA BLAT1 is upregulated in basal-like breast cancer through epigenetic modifications. Sci Rep. 2018;8:15572. doi: 10.1038/s41598-018-33629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y, Xu X, Wang T, Li Y, You W, Fu J, Liu Y, Jin S, Ji Q, Zhao W, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8:e2928. doi: 10.1038/cddis.2017.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, Hu J, Song H, Xu H, Wu C, Zhao B, Xie D, Wu T, Zhao J, Fang L. MiR-143-3p targeting LIM domain kinase 1 suppresses the progression of triple-negative breast cancer cells. Am J Transl Res. 2017;9:2276–2285. [PMC free article] [PubMed] [Google Scholar]
- 15.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. 2018;637:1–8. doi: 10.1016/j.abb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen JJ, Chen WJ, Sun WY, Liu XM, Chen JB. LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem. 2018;45:1975–1985. doi: 10.1159/000487974. [DOI] [PubMed] [Google Scholar]
- 18.Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478:1067–1073. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene exp ression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016;24:2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guil S, Esteller M. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem Sci. 2015;40:248–256. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Dai G, Yu L, Hu Q, Chen J, Guo W. MiR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci Rep. 2018;8:606. doi: 10.1038/s41598-017-18739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Fang C, Wang Y, Zhang J, Yu J, Zhang Y, Wang X, Zhong J. COL1A1: A potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. Int J Oncol. 2018;53:1869–1880. doi: 10.3892/ijo.2018.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurikova M, Danihel L, Polak S, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Guo JL, Gu SQ, Li Y, Zhang XY. Evaluation of clinical significance of endoglin expression during breast cancer and its correlation with ER and PCNA. Eur Rev Med Pharmacol Sci. 2017;21:5402–5407. doi: 10.26355/eurrev_201712_13927. [DOI] [PubMed] [Google Scholar]
- 26.Hu L, Li HL, Li WF, Chen JM, Yang JT, Gu JJ, Xin L. Clinical significance of expression of proliferating cell nuclear antigen and E-cadherin in gastric carcinoma. World J Gastroenterol. 2017;23:3721–3729. doi: 10.3748/wjg.v23.i20.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed M, Rubio IT, Klaase JM, Douek M. Surgical treatment of nonpalpable primary invasive and in situ breast cancer. Nat Rev Clin Oncol. 2015;12:645–663. doi: 10.1038/nrclinonc.2015.161. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Park HY, Kim WW, Lee SJ, Jeong JH, Kang SH, Jung JH, Chae YS. Biological function of long noncoding RNA snaR in HER2-positive breast cancer cells. Tumour Biol. 2017;39:1010428317707374. doi: 10.1177/1010428317707374. [DOI] [PubMed] [Google Scholar]
- 29.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Peter S, Borkowska E, Drayton RM, Rakhit CP, Noon A, Chen W, Catto JW. Identification of differentially expressed long noncoding RNAs in bladder cancer. Clin Cancer Res. 2014;20:5311–5321. doi: 10.1158/1078-0432.CCR-14-0706. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Yuan X, Yan D, Li D, Guan F, Dong Y, Wang H, Liu X, Yang B. Long Non-Coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell Physiol Biochem. 2017;42:1192–1201. doi: 10.1159/000478917. [DOI] [PubMed] [Google Scholar]
- 33.Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, Zhang W, Deng H, Zhou M, Peng S, et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–737. doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, Yang H, Deng Z, Su Y, Fang Q, Yin Z. HOX antisense lincRNA HOXA-AS2 promotes tumorigenesis of hepatocellular carcinoma. Cell Physiol Biochem. 2016;40:287–296. doi: 10.1159/000452545. [DOI] [PubMed] [Google Scholar]
- 35.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Fu H, Zhang S, Sun S, Liu Y. LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. 2018;29:10. doi: 10.1002/cam4.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et al. Targeting the notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, Guan GH, Liu Q, Qian YH, Xie D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:483. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du CW, Zhang GJ. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25:211–223. [PubMed] [Google Scholar]
- 40.Kobayashi M, Saito A, Tanaka Y, Michishita M, Kobayashi M, Irimajiri M, Kaneda T, Ochiai K, Bonkobara M, Takahashi K, et al. MicroRNA expression profiling in canine prostate cancer. J Vet Med Sci. 2017;79:719–725. doi: 10.1292/jvms.16-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng ZG, Xu H, Suo SS, Xu XL, Ni MW, Gu LH, Chen W, Wang LY, Zhao Y, Tian B, Hua YJ. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep. 2016;6:26093. doi: 10.1038/srep26093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong Z, Wang L, Wang Q. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J Cell Mol Med. 2018;29:10. doi: 10.1111/jcmm.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.