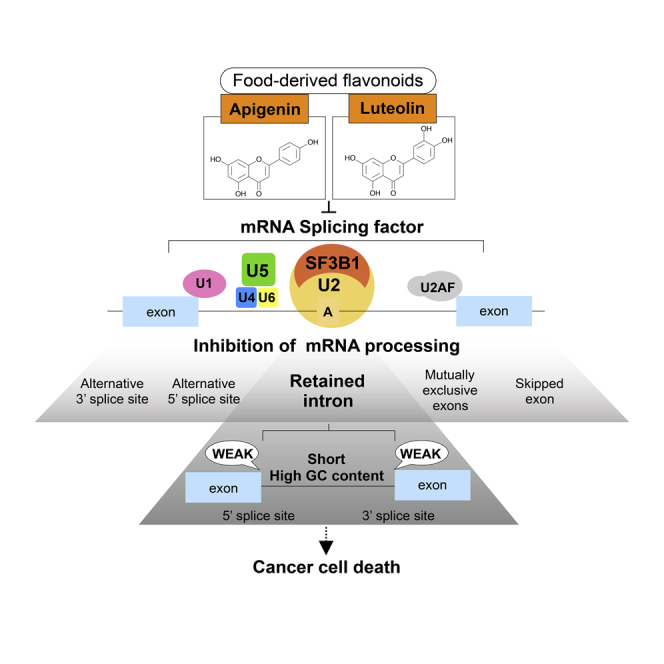
Summary
Cancer cells often exhibit extreme sensitivity to splicing inhibitors. We identified food-derived flavonoids, apigenin and luteolin, as compounds that modulate mRNA splicing at the genome-wide level, followed by proliferation inhibition. They bind to mRNA splicing-related proteins to induce a widespread change of splicing patterns in treated cells. Their inhibitory activity on splicing is relatively moderate, and introns with weak splice sites tend to be sensitive to them. Such introns remain unspliced, and the resulting intron-containing mRNAs are retained in the nucleus, resulting in the nuclear accumulation of poly(A)+ RNAs in these flavonoid-treated cells. Tumorigenic cells are more susceptible to these flavonoids than nontumorigenic cells, both for the nuclear poly(A)+ RNA-accumulating phenotype and cell viability. This study illustrates the possible mechanism of these flavonoids to suppress tumor progression in vivo that were demonstrated by previous studies and provides the potential of daily intake of moderate splicing inhibitors to prevent cancer development.
Subject Areas: Molecular Biology
Graphical Abstract
Highlights
-
•
Food-derived compounds, apigenin and luteolin, modulate mRNA splicing
-
•
The treatment of these flavonoids causes numerous alternative splicing events
-
•
Splicing of introns with weak splice sites tend to be inhibited by these flavonoids
-
•
Tumorigenic cells are more sensitive to these flavonoids than non-tumorigenic cells
Molecular Biology
Introduction
Messenger RNA processing (5′ capping, splicing, and 3′ end processing) is a fundamental step in eukaryotic gene expression to sustain various cell functions, such as proliferation, survival, and differentiation (Maniatis and Reed, 2002, Millevoi and Vagner, 2009, Orphanides and Reinberg, 2002). Among them, mRNA splicing is a crucial process through which noncoding introns are removed from pre-mRNA by the spliceosome, composed of five snRNPs (small nuclear ribonucleoproteins: U1, U2, U4, U5, and U6 snRNPs) and an assembly of accessory proteins (Hoskins and Moore, 2012, Wahl and Lührmann, 2015). This machinery recognizes short consensus sequences in the pre-mRNA, at the 5′ splice site, the branchpoint sequence (BPS), and the 3′ splice site. Alternative splicing plays an important role in conferring remarkable variety on the eukaryotic transcriptome through the differential usage of splice sites (Kim et al., 2014, Pan et al., 2008, Wang et al., 2008). It has been suggested that abnormalities in mRNA splicing lead to many pathological conditions, such as cancer onset and progression (Scotti and Swanson, 2016, Yoshida et al., 2011). For this reason, the inhibition of inadequate splicing is attracting attention as an important therapeutic strategy (Daguenet et al., 2015, Hahn et al., 2015, Maguire et al., 2015, Nlend Nlend et al., 2010, Salton and Misteli, 2016).
Various studies to detect compounds with inhibitory activity on splicing have been performed, and these successfully identified several microorganism-derived compounds with activities that modulate mRNA splicing (Effenberger et al., 2017). For example, spliceostatin A, pladienolide B, GEX1A, and H3B-8800 inhibit pre-mRNA splicing by disturbing spliceosome complex formation via binding to a component of the SF3B complex, one of the constituents of U2 snRNP (Hasegawa et al., 2011, Kaida et al., 2007, Kotake et al., 2007, Seiler et al., 2018).
Regarding food-derived compounds, epigallocatechin gallate, apigenin, and resveratrol were first identified by their inhibitory effects on the growth of cancer cells and later revealed to induce the alternative splicing of several mRNAs (Arango et al., 2013, Markus et al., 2011, Sakla and Lorson, 2008). However, only a few reports to date have been published on food-derived compounds with the ability to modulate mRNA processing. Because various previous studies have focused on compounds with strong inhibitory effects on splicing, the compounds with relatively moderate activity, such as food-derived compounds, have been overlooked.
In our established method, a relatively moderate inhibitory effect on mRNA processing can be captured as bulk poly(A)+ RNA accumulation in the nucleus (Fujita et al., 2012, Fujiwara et al., 2010). Utilizing our method, we previously screened food-derived compounds and identified mRNA processing inhibition activities in the soybean-derived isoflavonoid fraction and performed further analysis that elucidated that this activity was mainly exerted by compounds with a flavone skeleton (Kurata et al., 2016, Kurata et al., 2017).
In this study, we assessed the activity of 21 compounds with a flavone skeleton to inhibit mRNA processing and found that two of these flavonoids with highly related structures, namely, apigenin and luteolin, exhibited the strongest activity. LC-MS/MS analysis of proteins bound to these compounds suggested that their cellular targets are mRNA splicing-related proteins. RNA-seq data revealed that many alternative splicing events were induced upon treatment with these flavonoids, strongly suggesting the effect of these compounds on mRNA splicing. Further extensive analysis of the sequence features of target introns indicated that these flavonoids tend to influence the splicing of introns with weak splice sites.
This study provides an effective approach to identify compounds exerting moderate inhibitory activity against mRNA processing, with a particular emphasis on the usefulness of global analysis to clarify the mechanisms underlying the perturbations induced by these compounds in mRNA processing. Using the method described in this study, previously overlooked chemical libraries, such as those of food-derived compounds, are worth revisiting as potential sources of compounds with moderate activity of modulating mRNA processing.
Results
Apigenin and Luteolin Exhibit Inhibitory Activity against mRNA Processing
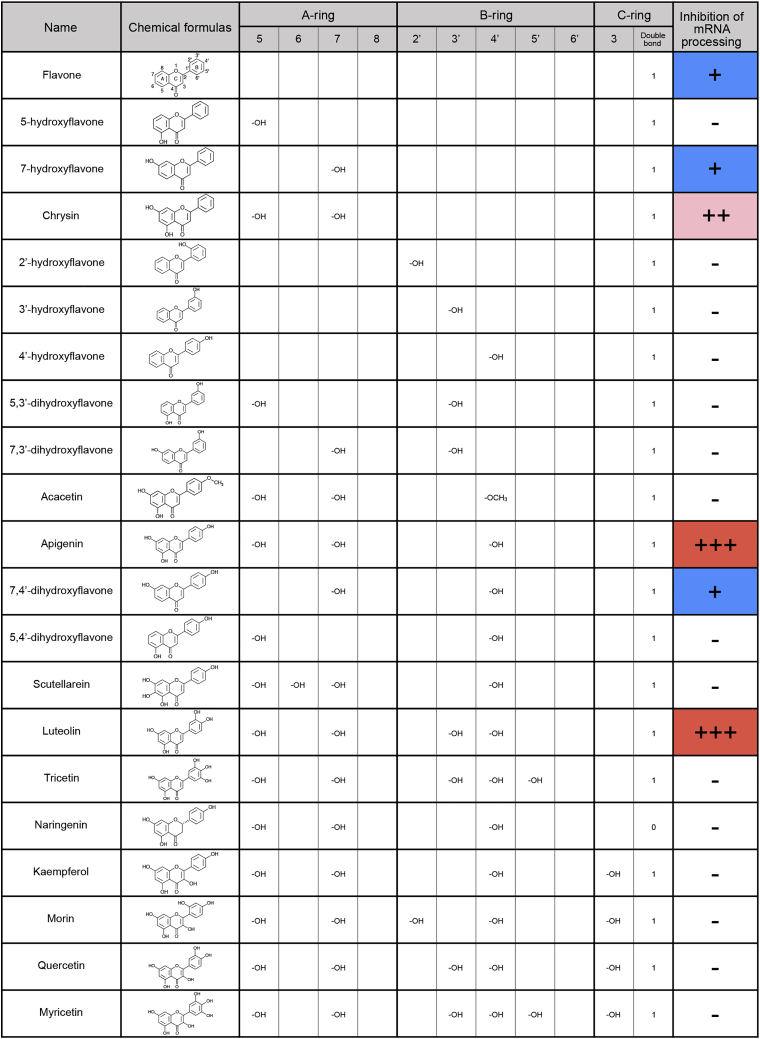
Previously, we identified several flavonoids that have inhibitory activity against mRNA processing (Kurata et al., 2016, Kurata et al., 2017). In this study, we explored the structure-activity relationship of compounds with a flavone skeleton by RNA-FISH (Figures 1 and S1). GEX1A, a well-known splicing inhibitor, was used as a positive control. The structure of flavones contains a skeleton of diphenylpropane, namely, two benzene rings (A-ring and B-ring) linked via a closed pyran ring (C-ring) with a benzenic A-ring. In addition, they have a double bond between the 2 and 3 positions and a ketone at the 4 position of the C-ring. Among the 21 compounds tested, apigenin and luteolin exhibited the most intense and dose-dependent phenotype of accumulating poly(A)+ RNA in the nucleus (Figures 2A–2C). These compounds harbor OH groups at the 5 and 7 positions of the A-ring, an OH group at the 4′ position of the B-ring, and a double bond in the C-ring as common structures, whereas an OH group at the 6 position of the A-ring, or the 2′ or 5′ position of the B-ring, appears to eliminate the flavonoids' activity on mRNA processing (Figures 1 and S1B). In apigenin- and luteolin-treated cells, poly(A)+ RNAs mainly co-localized with SC35, a nuclear speckle marker protein, indicating the accumulation of RNAs residing within the nuclear speckles (Figure 2D). This phenomenon was also observed in cells treated with GEX1A.
Figure 1.
Activity of Compounds with Flavone Skeleton Examined for mRNA Processing Inhibition
See also Figure S1.
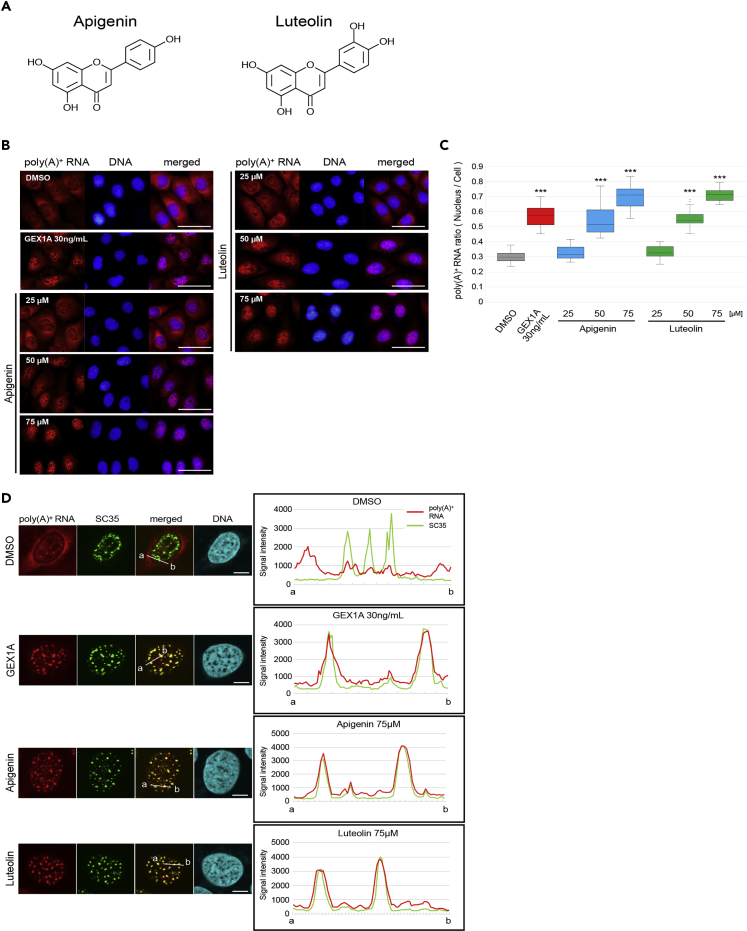
Figure 2.
Inhibitory Activity against mRNA Processing of Apigenin and Luteolin
(A) Chemical structures of apigenin and luteolin.
(B) RNA-FISH was performed to determine the localization of bulk poly(A)+ RNA. U2OS cells were treated with the indicated concentrations of each compound for 24 h GEX1A (30 ng/mL) was used as a positive control. The bulk poly(A)+ RNA was visualized by Alexa Fluor 594-labeled oligo-dT45 probe. The nuclei were visualized with DAPI. Scale bar, 50 μm.
(C) The ratio of the nuclear distribution of mRNA was analyzed. The signal intensities of the whole cell and the nucleus were quantified using ImageJ (n = 35). Boxes show median (center line) and upper and lower quartiles. Whiskers show the lowest and highest values. Statistical analysis was performed using one-way ANOVA followed by Dunnett's test. ***p < 0.001.
(D) The localization of bulk poly(A)+ RNA and nuclear speckles. Poly(A)+ RNA (red), speckles (green), and chromosomal DNA (blue) were visualized in U2OS cells for co-localization analysis. Cells were treated with each indicated compound for 24 h. Scale bar, 10 μm. In right panels, signal intensities of poly(A)+ RNA and SC35 were plotted between a and b lines in the left panels. Poly(A)+ RNA and SC35 signal are shown with red and green lines, respectively.
See also Figure S2.
mRNA Splicing Is a Candidate for the Cellular Target of Apigenin and Luteolin
The observed nuclear poly(A)+ RNA accumulation in apigenin- and luteolin-treated cells may reflect the disruption of an mRNA export pathway (Lee et al., 2008). To examine this possibility, we assessed the subcellular localization of transiently expressed β-globin mRNA in control cells and apigenin- or luteolin-treated cells. Expressed reporter β-globin mRNA was visualized using Alexa Fluor 546-labeled β-globin FISH probe (Figures 3A and 3B). Even in cells exhibiting a prominent phenotype of poly(A)+ RNA accumulation in the nucleus induced by treatment with these flavonoids, β-globin mRNA was exported to the cytoplasm as efficiently as in control cells, indicating that the general mRNA export machinery is not disturbed in these cells. In addition, the β-globin gene used in this assay contains introns with constitutive splice sites, suggesting that this reporter mRNA properly undergoes splicing. Indeed, when we assessed the splicing of this reporter mRNA, no obvious inhibition caused by these flavonoids was observed (Figure 3C). On the other hand, it has been reported that apigenin and luteolin treatment alters the splicing pattern of Caspase-9 and c-FLIP into an apoptosis-inducible form, thus inhibiting the proliferation of cancer cells (Arango et al., 2013). Those previously described changes in splicing pattern, namely, the decreases in both Caspase-9b, a variant with skipping of exons 3–6, and c-FLIPs, a variant with exon 7 instead of 8, were also induced by apigenin and luteolin treatment in our experimental conditions, implying that these flavonoids have some effect on splicing (Figure 3D). Supporting the idea that apigenin and luteolin affect splicing rather than export, the apigenin- and luteolin-derived nuclear speckle-associated poly(A)+ RNA accumulation pattern was similar to that observed in cells depleted of SF3B1, a component of U2 snRNP (Figure S2A), and was distinguishable from the uniform poly(A)+ RNA accumulation throughout the nucleus observed in cells depleted of TAP (also known as NXF1), an mRNA exporter (Figure S2B) (Wang et al., 2018). The subnuclear localization of accumulated poly(A)+ RNA observed in apigenin- and luteolin-treated cells was also reminiscent of a phenotype exhibited by cells treated with spliceostatin A, a well-known splicing inhibitor, also implicating the mRNA splicing process as a cellular target of these flavonoids (Figure 2D) (Carvalho et al., 2017, Kaida et al., 2007).
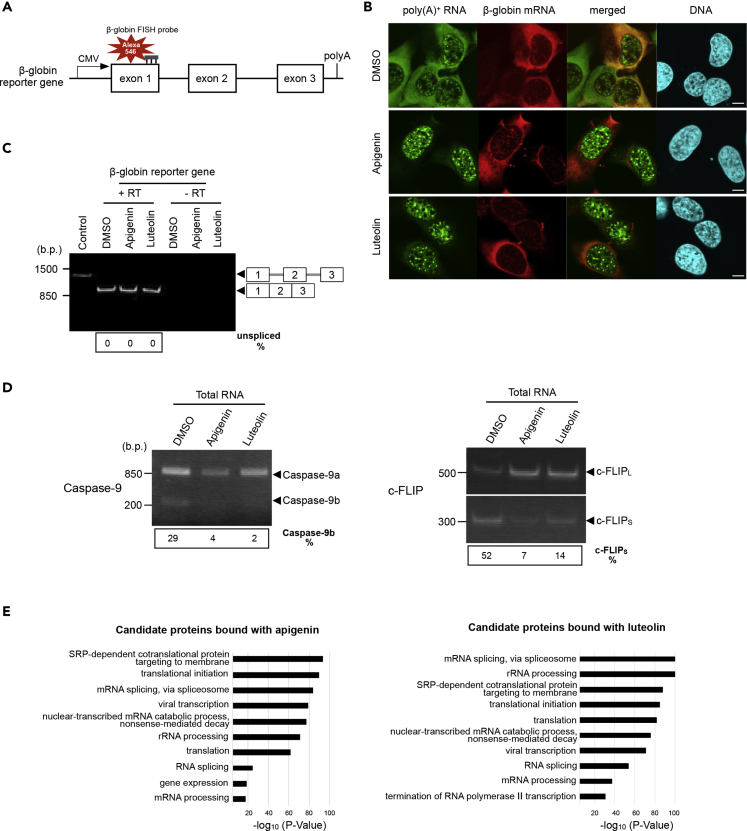
Figure 3.
The Effect on Splicing of Some Genes and Identification of Apigenin- and Luteolin-Targeted Proteins
(A) Schematic of β-globin construct containing the CMV promoter and BGH poly(A) site is presented. Horizontal lines indicate introns, and boxes indicate exons.
(B) The localization of β-globin mRNA was observed. Bulk poly(A)+ RNA (green), β-globin mRNA (red), and chromosomal DNA (blue) were visualized in U2OS cells. Scale bar, 10 μm.
(C) The splicing pattern of β-globin gene was observed by RT-PCR using total RNA. The digit panel below the photo shows the percentage of unspliced mRNA band intensity and representative results of triplicate experiments. DNA size in base pairs (b.p.) is indicated on the left side. Control: marker for unspliced β-globin mRNA. RT: reverse transcription.
(D) The change of splicing pattern of Caspase-9 and c-FLIP. RT-PCR was performed using total RNA. DNA size in base pairs (b.p.) is indicated on the right side. The digit panels below the photo show the percentage of the indicated isoform and representative results of triplicate experiments.
(E) Gene ontology analysis of proteins bound with apigenin or luteolin is shown. Proteins with protein score >0 in Tables S1 and S2 were uploaded to the DAVID database for GO analysis.
See also Figures S2–S4.
To explore the way in which these flavonoids influence poly(A)+ RNA metabolism in human cells, proteins bound to apigenin and luteolin were analyzed. Flavonoid was fixed on magnetic FG beads with an epoxy linker and was incubated with nuclear extract from HEK293 cells. Purified proteins were analyzed by LC-MS/MS (Figures 3E, S3A, and S3B; Tables S1 and S2). The interaction between some of the LC-MS/MS-detected proteins and either apigenin or luteolin was confirmed by the western blotting using specific antibodies against these proteins (Figure S3C). Gene ontology analysis demonstrated that many of the detected proteins have mRNA splicing-related functions (Figure 3E). Further detailed classification of the splicing-related proteins according to a previous report indicated that these compounds most intensely interact with U2 and U5 snRNP, components of the spliceosome (Figure S3B) (Hegele et al., 2012). HNRNPA2B1, a protein that has been reported to interact with apigenin (Arango et al., 2013), was also detected to a lesser extent than U2 and U5 snRNP components. To obtain insights into the interaction of apigenin and luteolin with their potential targets, docking studies were performed using the crystal structure of human SF3B1, a component of the SF3B subcomplex in the U2 snRNP, in a complex with E7107, a pladienolide derivative analog (PDB code: 5ZYA) (Finci et al., 2018). Our docking algorithm predicted that apigenin and luteolin are likely to bind to SF3B1 near the E7107-binding site (Figure S4A). Both flavonoids interact with several hydrophobic residues such as Leu1066, Val1114, Val1110, and Lys1067, which are also key residues for the hydrophobic interactions between E7107 and SF3B1 (Figure S4B). In addition, the overexpression of SF3B1 attenuated the nuclear accumulation of poly(A)+ RNA induced by the apigenin- and luteolin-treatment (Figure S4C). These results provide evidence that SF3B1 is one of the cellular targets for apigenin and luteolin and plays a crucial role in the modulation of mRNA splicing by these flavonoids.
Taking these findings together, apigenin and luteolin bind to spliceosome components, and in the cells treated with these compounds, changes in the splicing of at least some mRNAs can be effectively induced, whereas the splicing of other mRNAs, such as the β-globin mRNA reporter, is less influenced. These findings imply that apigenin and luteolin affect the splicing of a subset of introns, rather than generally inhibiting splicing reactions.
Apigenin and Luteolin Treatment Causes Numerous Alternative Splicing Events
To elucidate the effect of these flavonoids on mRNA splicing, we compared the poly(A)+ RNA profile in flavonoid-treated cells with that in control cells using RNA-seq. Poly(A)+ RNA was purified from total RNAs derived from either control cells or flavonoid-treated cells. RNA-seq reads were mapped with STAR (Dobin et al., 2013) against the hg38 human genome and to the obtained mapping data; we applied rMATS, a bioinformatic tool, to detect alternative splicing events (Figure 4A) (Shen et al., 2014). We detected a wide variety of alternative splicing events induced by treatment with either flavonoid (Figures 4B and 4C). Among the altered splicing patterns, retained intron (RI) was the most frequently detected, followed by skipped exon (SE). Since apigenin and luteolin have similar structures and binding proteins, we hypothesized that they commonly regulate mRNA splicing. To investigate the regulatory mechanism involved, we focused on their common events (Figures 4D and S5). Specifically, we performed gene ontology analysis on the events overlapping between apigenin- and luteolin-treated cells (Figures 4E and S5), indicating that the mRNAs with the retention of introns due to these flavonoids are particularly associated with RNA processing, mRNA metabolic process, and mRNA splicing. RT-PCR using cDNA generated from total RNA successfully confirmed some of the apigenin- and luteolin-derived RIs and another alternative splicing event identified in our rMATS analysis, whereas neither tricetin nor naringenin induced these changes. This demonstrated the significance of the specific structure within these flavonoids for their ability to exert such activity (Figures 4F and S6). Previous studies suggested that unspliced mRNAs are inefficiently exported to the cytoplasm, leading to poly(A)+ RNA accumulation in nuclear speckles (Johnson et al., 2000, Kaida et al., 2007). Based on these findings, we hypothesized that a phenotype derived from apigenin and luteolin treatment, namely, the nuclear speckle-associated poly(A)+ RNA accumulation, reflects nuclear-retained mRNAs with introns. Therefore, we focused on the RIs for further investigation.
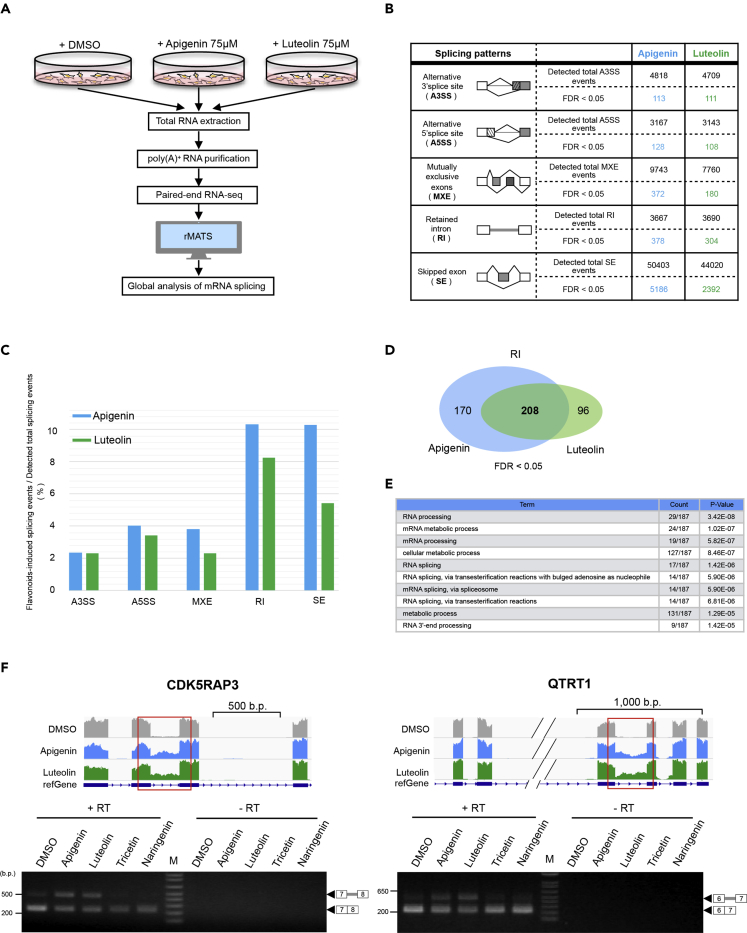
Figure 4.
Global Analysis of mRNA Splicing Affected by Apigenin and Luteolin
(A) Alternative splicing analysis scheme. Cells were treated with DMSO, apigenin (75 μM), or luteolin (75 μM) for 24 h; then, total RNA was extracted from the cells. Next, poly(A)+ RNA was purified and fragmented for RNA sequencing. Alternative splicing analysis was performed using rMATS.
(B) Alternative splicing analysis revealed five alternative splicing patterns that were output by rMATS. Upper row indicates the numbers of total splicing events detected in each alternative splicing pattern; bottom row indicates the numbers of statistically significant (false discovery rate: FDR <0.05) splicing pattern changes upon apigenin (blue) or luteolin treatment (green).
(C) Frequency of splicing pattern changes by apigenin or luteolin relative to the total splicing events is calculated using the data in (B).
(D) Venn diagram shows the numbers of apigenin- or luteolin-induced retained intron events.
(E) Gene ontology analysis of apigenin- and luteolin-induced retained intron events. Common target genes of apigenin and luteolin (the number shown in bold in Figure 4D) were uploaded to the DAVID database for GO analysis. Apigenin- and luteolin-targeted genes in U2OS cells were enriched for several GO terms categorized in “Biological Process.” Ten GO terms are listed in order of their p values. The “Count” column denominator indicates the number of genes assigned a DAVID ID among the uploaded target genes of apigenin and luteolin in retained intron events. The numerator indicates the number of genes associated with each GO term.
(F) Validation of retained introns regulated by apigenin and luteolin. In the upper panels, IGV snapshots of retained introns induced by apigenin (blue) and luteolin (green) are shown. Gene structure is depicted at the bottom of each snapshot, with horizontal lines indicating introns and boxes indicating exons. Red square lines surrounding introns indicate regions affected by apigenin and luteolin. Lower panels: RT-PCR was performed using total RNA samples to detect apigenin- or luteolin-induced retained introns. Representative results of triplicate experiments are shown. DNA size in base pairs (b.p.) is indicated on the left side. RT: reverse transcription. M: DNA size marker.
See also Figures S5 and S6.
Apigenin- and Luteolin-Induced Alternative Splicing Leads to a Change in Gene Expression
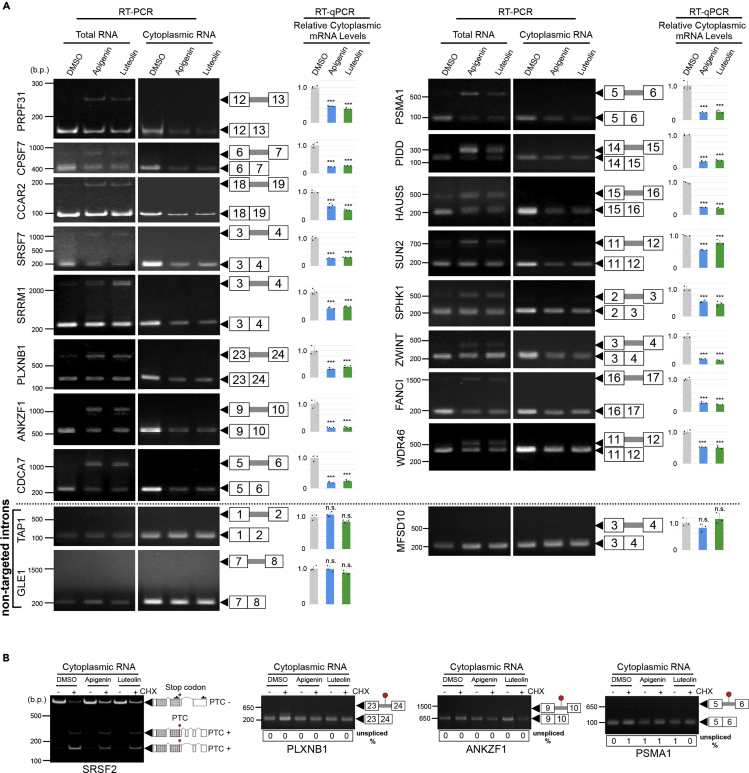
Taking into account the possibility that mRNAs with RIs are less effectively exported to the cytoplasm, it was assumed that the expression of genes with apigenin- and luteolin-sensitive introns decreases upon treatment with these flavonoids. To test this hypothesis, we fractionated the cytoplasmic RNA (Figure S7A). Our RT-PCR analysis clearly detected RIs caused by the treatment with these flavonoids in the whole-cell fractions, but not in the cytoplasmic fractions, indicating that mRNAs harboring those introns are not exported to and/or eliminated in the cytoplasm (Figure 5A). A significant decrease in the expression of genes whose splicing is inhibited by apigenin and luteolin treatment was demonstrated for the matured mRNAs in the cytoplasm (Figure 5A), as well as at the protein level (ANKZF1, PSMA1, and ZWINT) (Figure S7B). In contrast, SEs triggered by apigenin and luteolin treatment were efficiently detected in either fraction (Figure S7C), indicating that these mRNAs associated with exon skipping are efficiently exported to the cytoplasm. Neither the splicing nor the cytoplasmic expression level of nontarget introns appeared to be affected following treatment with apigenin and luteolin.
Figure 5.
The Expression Level of Retained Intron-Containing mRNA in the Cytoplasm
(A) Detection of retained intron events in the cytoplasm after apigenin and luteolin treatment. RT-PCR was performed using total RNA and cytoplasmic RNA. DNA size in base pairs (b.p.) is indicated on the left side. Non-targeted introns that are not regulated by apigenin and luteolin are shown at the bottom of (A). Representative results of triplicate experiments are shown. To compare the mRNA expression in cytoplasmic fractions, RT-qPCR was performed using cytoplasmic RNA. Values represent the relative expression of the indicated mRNA normalized to TBP. Each value is mean ± SD of three independent experiments. Statistical analysis was performed using one-way ANOVA followed by Dunnett's test. ***p < 0.001, n.s.: not significant.
(B) The effect of nonsense-mediated decay (NMD) on mRNA with retained intron in the cytoplasmic fraction was examined. Cycloheximide (CHX) was used as an NMD inhibitor. U2OS cells were treated with CHX (50 μg/mL) in the presence of apigenin (75 μM) or luteolin (75 μM) for 24 h. RT-PCR products of variant transcripts were detected. The digit panels below the photo shows the percentage of unspliced mRNA band intensity and representative results of triplicate experiments. Blue circle indicates a reference stop codon, and red circle indicates a premature termination codon (PTC). PTC + means that the transcript variant has PTC. SRSF2, a well-known target of NMD, was used as a positive control (Feng et al., 2015). Dotted region in the SRSF2 figure indicates protein-coding sequence, and primer locations (black arrows) are marked on the SRSF2 gene.
See also Figure S7.
Intron retention often produces premature termination codon (PTC) that makes the associated coding mRNAs sensitive to nonsense-mediated mRNA decay (NMD) in the cytoplasm (Ge and Porse, 2014). Indeed, we found PTCs within apigenin- and luteolin-derived unspliced mRNAs (Figure 5B). The absence of these unspliced mRNAs in the cytoplasm might be explained by the intense susceptibility of these RNAs to NMD, besides the retention of RNAs within the nucleus. To discriminate between these two possibilities, an NMD assay was performed (Figure 5B). In the cytoplasmic fraction, a PTC-containing transcript variant of SRSF2, a well-known NMD target (Feng et al., 2015), was detected only in the presence of cycloheximide (CHX), an NMD inhibitor, demonstrating disturbed NMD under these conditions. In contrast, unspliced mRNAs caused by apigenin and luteolin were not detected irrespective of the integrity of NMD, indicating that apigenin- and luteolin-elicited unspliced mRNAs are eliminated from the cytoplasm mainly by their nuclear retention, rather than by the digestion in the cytoplasm.
Taking these findings together, as a consequence of various alternative splicing events, such as the nuclear retention of unspliced mRNAs and the expression of exon skipping-caused transcript variants, these flavonoids induce a change in the cytoplasmic transcriptome.
Apigenin and Luteolin Inhibit Splicing of Introns with Weak Splice Sites
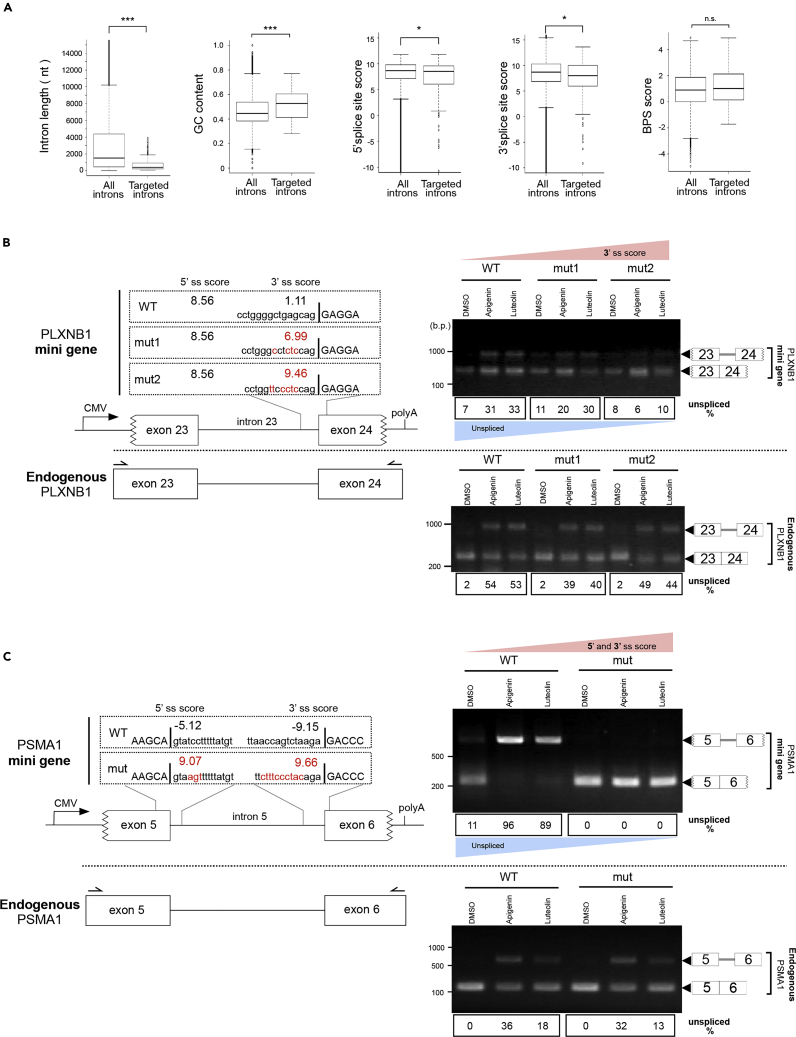
To characterize introns whose splicing is affected by apigenin and luteolin, we compared their intron length, GC content, splice site score, and BPS score with those of all of the introns annotated in the human genome (Figure 6A). Splice site score was calculated using MaxEntScan (Yeo and Burge, 2004). The BPS score was calculated using the SVM-BP program (Corvelo et al., 2010). The results suggested that RIs caused by apigenin and luteolin treatment are shorter, have weaker 5′ and 3′ splice sites, and show higher GC content, whereas the BPS scores are similar between these flavonoid-target and nontarget introns.
Figure 6.
Apigenin and Luteolin Induce Intron Retention with Weak Splice Sites
(A) The bioinformatic analysis on retained introns induced by apigenin and luteolin. All introns of human reference genes and retained introns induced by apigenin and luteolin were analyzed. Boxes show median (center line) and upper and lower quartile ranges. Whiskers show the lowest and highest values. Statistical analysis was performed using the two-sided Mann–Whitney U test. *p < 0.05, ***p < 0.001, n.s.: not significant.
(B and C) Upper left panels in (B and C) present schematic representations of mini gene constructs affected by apigenin and luteolin. The CMV promoter and BGH poly(A) sites are shown. Jagged line in the figure of the exon indicates that the edges of the exons have been partially deleted in order to distinguish endogenous mRNA from transgene-derived mRNA. The mutated nucleotides in each mutated-mini gene construct are shown in red, and the exon sequence is shown in upper case. Splice site score (ss score) was calculated using MaxEntScan. Lower left panels in (B and C) present schematic representations of the endogenous gene. The locations of the primers that amplify the endogenous target are marked in the schematic representation of the endogenous gene (black arrows). Right panels in (B and C) show the results of RT-PCR. DNA size in base pairs (b.p.) is indicated on the left side. The mRNAs derived from the transgene are shown in the upper right panels, and endogenous mRNAs are shown in the lower right panels. The digit panels below the photo show the percentage of unspliced mRNA band intensity and representative results of triplicate experiments.
See also Figure S8.
To confirm the significance of weak splice sites for the susceptibility to these flavonoids, sequences of 5′ and 3′ splice sites within introns of interest were manipulated to display a higher splice site score, and it was examined whether the splicing of these engineered introns is still sensitive to apigenin and luteolin. For this purpose, introns accompanied by portions of both neighboring exons were cloned into an expression vector, and these vectors were transiently transfected into U2OS cells, achieving the expression of these reporter RNAs under the control of the CMV promoter. We successfully obtained several mini genes, PLXNB1, PSMA1, and ANKZF1, which can efficiently reproduce apigenin- and luteolin-induced intron retention exhibited by the endogenous genes (Figure 5A). Mutating the splice sites of the introns in these mini genes to raise the splice sites with a higher score, namely, changes of 1.11→6.99 or 1.11→9.46 in the 3′ splice site score (3′ ss score) of the PLXNB1 mini gene, −5.12→9.07 in the 5′ splice site score (5′ ss score), −9.15→9.66 in the 3′ ss score in the PSMA1 mini gene, and −0.11→10.45 in the 5′ ss score in the ANKZF1 mini gene, almost abolished their responses to apigenin and luteolin (Figures 6B, 6C, and S8). In addition, the PLXNB1 mini gene with its 3′ ss mutated to achieve a moderate score of 6.99 still exhibited subtle susceptibility to these flavonoids. The efficiencies of either apigenin or luteolin treatment in these experiments were confirmed by RT-PCR analysis of the endogenous mRNAs from the same samples transiently expressing each reporter mini gene (Figures 6B, 6C, and S8). And as mentioned above, the β-globin reporter indicated that its transcript escaped from flavonoid-induced splicing inhibition (Figures 3B and 3C). These findings suggest that weak splice sites are crucial for an intron's susceptibility to apigenin and luteolin.
Apigenin- and Luteolin-Derived Unspliced mRNAs Accumulate at Nuclear Speckles
Next, we assessed the subcellular localization of these mini gene-derived mRNAs in the presence or absence of apigenin and luteolin to examine whether the nuclear accumulation of bulk poly(A)+ RNAs observed in the flavonoid-treated cells contains the flavonoid-caused unspliced mRNAs. To visualize these reporter mRNAs, a sequence complementary to the β-globin FISH probe used as shown in Figure 3A was inserted into either the PLXNB1 or PSMA1 mini gene. RT-PCR analysis demonstrated that β-globin sequence-fused mini genes maintained their response to the flavonoids (Figure 7A). The results also showed that mutating the splice sites to have a higher score abolished their response to the flavonoids, as observed with native sequence reporters (Figures 6B, 6C, and 7A). In either apigenin-/luteolin-treated or untreated cells, these mini genes were transiently expressed and visualized by FISH using β-globin probe (Figure 7B). Bulk poly(A)+ RNA was simultaneously visualized in the same cells to verify the effect of the treatment with flavonoids. In DMSO-treated cells, expressed reporter mRNAs were efficiently exported to the cytoplasm. In apigenin- and luteolin-treated cells, WT reporter mRNAs were diminished from the cytoplasm and accumulated in the nucleus, whereas apigenin- and luteolin-insensitive mutated mRNAs were efficiently exported to the cytoplasm under these conditions. Notably, in the flavonoid-treated nucleus, WT mRNA reporters co-localized with bulk poly(A)+ RNAs, indicating that flavonoid-caused unspliced reporter mRNAs accumulate at the nuclear speckles.
Figure 7.
Localization of mRNAs with Retained Intron Induced by Apigenin and Luteolin
(A) Schematic of mini gene constructs with β-globin FISH probe tag. The CMV promoter and BGH poly(A) sites are shown. DNA size in base pairs (b.p.) is indicated on the left side. The digit panels below the photo show the percentage of unspliced mRNA band intensity and representative results of triplicate experiments.
(B) The localization of target mRNA was observed. Bulk poly(A)+ RNA (green), target mRNA (red), and chromosomal DNA (blue) were visualized in U2OS cells. Scale bar, 10 μm.
Tumorigenic Cells Exhibit More Prominent Sensitivity to Apigenin and Luteolin than Non-tumorigenic Cells
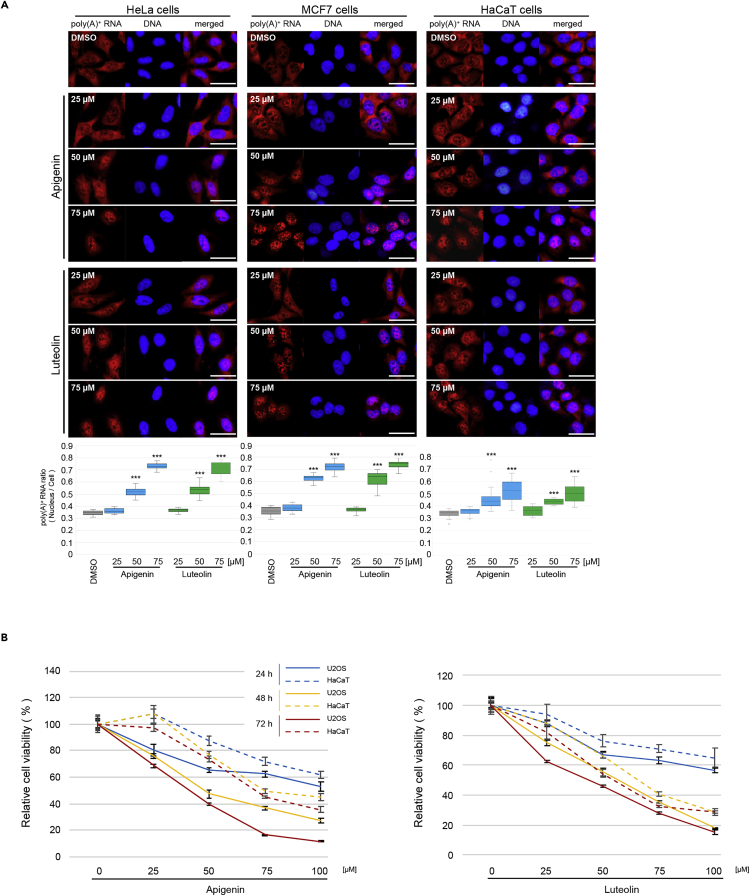
It has been shown that cancer cells are more sensitive to reagents inhibiting splicing (see Discussion) (Dvinge et al., 2016). We thus assessed whether this is also the case with apigenin and luteolin by comparing their effects on U2OS, HeLa, and MCF7 tumorigenic cell lines with those on non-tumorigenic HaCaT cells. When added at 75 μM, apigenin and luteolin caused prominent nuclear poly(A)+ RNA accumulation in all of the tumorigenic cell lines tested, whereas the nuclear poly(A)+ RNA accumulation in HaCaT cells under the same conditions was moderate (Figure 8A). In addition, tumorigenic cells exhibited a steeper dose response than HaCaT cells.
Figure 8.
Inhibition of mRNA Processing and Proliferation of Tumorigenic Cells by Apigenin and Luteolin
(A) Bulk poly(A)+ RNA (red) and chromosomal DNA (blue) were visualized in HeLa, MCF7, and HaCaT cells. Cells were treated with the indicated concentrations of each compound for 24 h. Scale bar, 50 μm. In lower panels, the signal intensities of bulk poly(A)+ RNA in the whole cell and in the nucleus were quantified using ImageJ (n = 35). Boxes show median (center line) and upper and lower quartiles. Whiskers show the lowest and highest values. Statistical analysis was performed using one-way ANOVA followed by Dunnett's test. ***p < 0.001.
(B) Proliferation of U2OS and HaCaT cells treated with the indicated concentrations of each compound for 24, 48, or 72 h. Cell viability was determined by MTT assay. Cell viability in comparison with that of DMSO-treated cells, which was set to 100%, is reported as mean ± SD of six independent experiments.
See also Figure S9.
We also examined the effect of these compounds on the cell viability (Figures 8B and S9). These flavonoids significantly inhibited the proliferation of U2OS cells in a dose- and time-dependent manner. HaCaT cells also exhibited a dose- and time-dependent response to these flavonoids but to a lesser extent than U2OS cells at all doses and time points. Other non-tumorigenic cells, WI-38 and TIG-1, also gave similar results with HaCaT cells. Taken together, apigenin and luteolin seem to be more effective for tumorigenic cells than for non-tumorigenic cells (Figure S9).
Discussion
Food not only provides nutrition, taste, and texture but also regulates biological function. The daily intake of bioactive food compounds is expected to be effective for the prevention of chronic diseases. For example, polyphenols and carotenoids have antioxidant activity and function in preventing lifestyle-related diseases (Pandey and Rizvi, 2009, Reinisalo et al., 2015, Young and Lowe, 2001). Flavonoids, which are abundant in fruit and vegetables, have been reported to reduce the risk of lifestyle-related diseases and carcinogenesis (Bo et al., 2016, López-lázaro, 2009). Despite the broad bioactivity of flavonoids, few studies on their regulatory activity against mRNA processing, especially splicing, have been reported. We previously searched for compounds derived from food that have the ability to inhibit mRNA processing and identified such effects in compounds with a flavone skeleton (Kurata et al., 2016, Kurata et al., 2017).
In this study, we have shown that, among the flavonoids, apigenin and luteolin have remarkable activity of inhibiting mRNA processing. Several lines of evidence indicate that this activity is derived from the direct inhibition of splicing. Our rMATS analysis demonstrated that all types of alternative splicing events are provoked by treatment with apigenin and luteolin, and the retention of introns is the event having the highest frequency. It was previously shown that splicing inhibition by either spliceostatin A treatment or the knockdown of a component of U2 snRNP or U5 snRNP, such as SF3B1 or PRPF8, causes the retention of numerous introns in mRNAs coding RNA processing factors (Wickramasinghe et al., 2015, Yoshimoto et al., 2016), implying that these mRNAs are highly sensitive to splicing inhibition. Such enrichment of RNA processing factors, especially of mRNA splicing, was also observed in the genes that we identified to harbor introns whose retention was induced by apigenin and luteolin. It has been reported that unspliced mRNAs resulting from the global inhibition of splicing by spliceostatin A treatment manifest as a robust accumulation of bulk poly(A)+ RNAs at the nuclear speckles (Carvalho et al., 2017, Kaida et al., 2007). Similarly, we observed that apigenin and luteolin treatment induced the accumulation of bulk poly(A)+ RNA, as well as the retention of intron-containing transcripts from target mini genes, in the nuclear speckles, implying that these flavonoids affect a broad range of mRNA splicing events. Our results from LC-MS/MS, docking studies and SF3B1 overexpression analysis simply suggest that these flavonoids interact with U2 and U5 snRNP, possibly through the interaction with SF3B1, to modulate mRNA splicing, although we cannot exclude other possible splicing regulatory mechanisms by these flavonoids, such as SUMOylation of spliceosomal components previously reported for an apigenin- and luteolin-related biflavonoid, hinokiflavone (Pawellek et al., 2017). Together, these findings suggest that apigenin and luteolin associate with spliceosomal components to directly prevent the function of spliceosomes, thus affecting alternative splicing at the genome-wide level.
Our rMATS analysis did not detect previously demonstrated alternative splicing events induced by apigenin and luteolin treatment, such as the change of splicing pattern of Caspase-9 and c-FLIP mRNA (Arango et al., 2013), although it was efficiently recapitulated by RT-PCR analysis, indicating that these flavonoids actually affect a broader range of splicing events beyond our criteria of rMATS analysis. Such events may include the very subtle splicing perturbation that, if it occurs to numerous targets, can contribute to the flavonoid-induced nuclear bulk poly(A)+ RNA accumulation.
Our data also illustrated a mechanism by which these flavonoids inhibit splicing. The bioinformatic analysis raises the possibility that weak splice sites are a general feature for the retention of introns caused by apigenin and luteolin. Supporting this hypothesis, manipulating the splice site score in the target introns abolished their sensitivity to these flavonoids and led them to be spliced out. Therefore, apigenin and luteolin presumably perturb the association of splicing-related factors with the weak splice sites to inhibit splicing.
We observed more sensitivity of tumorigenic cells to apigenin and luteolin than that observed for non-tumorigenic cells, both for the bulk mRNA processing and cell viability. The exact relationship between splicing and cancer progression is still unclear. However, splicing abnormality has been found to be involved in the onset and progression of various cancers (Scotti and Swanson, 2016, Sveen et al., 2016), and it has been suggested that cancers harboring mutations in splicing factors are highly susceptible to splicing inhibition (Lee et al., 2016). For example, acute myeloid leukemia cells with SF3B1 mutation exhibit reduced viability in the presence of a splicing inhibitor (Seiler et al., 2018). High sensitivity to splicing inhibition has also been demonstrated for cancers caused by c-Myc oncogene overexpression (Hsu et al., 2015). In this case, it has been suggested that massive amounts of c-Myc-driven pre-mRNA sequester a substantial population of spliceosomes to weaken the splicing activity available for endogenous pre-mRNAs, thus conferring on cells similar sensitivity for splicing inhibition as observed in cancer cells resulting from mutations in splicing factors. Hence, splicing inhibitors are attractive candidates for anti-cancer drugs. Indeed, splicing inhibitors such as H3B-8800 (Seiler et al., 2018) are under clinical trials for anti-cancer therapy. Because the concentrations at which apigenin and luteolin can effectively exert their splicing-inhibiting activities are much higher than those of previously identified compounds, such as spliceostatin A and H3B-8800, the employment of these flavonoids as anti-cancer drugs may not be so promising. Nonetheless, further evaluation of these flavonoids as anti-cancer drugs should be conducted. On the other hand, it has been reported that xenograft progression of human tumors was repressed by the oral administration of apigenin or luteolin into nude mice (Li et al., 2018, Shukla et al., 2015, Xu et al., 2018) and that the daily intake of bulk flavonoids reduced carcinogenesis in humans (Bo et al., 2016, López-lázaro, 2009). These findings suggest a beneficial effect of these compounds on preventing cancer development when they are consumed in food on a daily basis for a considerable period.
Many screening systems to identify anti-cancer compounds are highly focused on finding compounds with strong activity. Therefore, compounds with moderate activity have been overlooked. Our study provides an effective strategy to discover compounds harboring moderate inhibitory activity on mRNA processing and to elucidate the mechanism underlying the disruption of mRNA processing induced by candidate compounds.
Limitations of the Study
We have revealed the underlying mechanism that food-derived compounds apigenin and luteolin cause intron retention. Other than intron retention, we also observed various alternative splicing events induced by these flavonoids and whether an analogous molecular mechanism contributes to these events remains to be assessed.
A hypersensitivity of cancer cells to the splicing inhibition has been shown in many studies, implying our observed in vitro inhibition of cancer cell proliferation by apigenin and luteolin, as well as the previously demonstrated suppression of human tumor xenograft progression by apigenin-treatment in a mouse model, is derived from the inhibited splicing. However, a further investigation should be required to elucidate a direct relationship between these flavonoids' inhibitory activity on splicing and their exhibited tumor growth suppressive effect.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Robin Reed for the gifts of antibodies against SF3B1 and U2AF65 and Mr. Yuzo Watanabe for help with the LC-MS/MS analysis. We also thank Dr. Michael Antoniou for providing the β-globin reporter and Mr. Soichi Nishimura for technical assistance. Moreover, we are grateful to Kyowa Hakko Kirin Co., Ltd., for providing GEX1A. This work was supported in part by “Grants-in-Aid” from JSPS KAKENHI (Grant Numbers 26292053, 17K19232, 19K22280, and 19H02884 to S.M, 19K15807 to K.F, and 15K11299 to Y.S.). This work was also supported in part by “Grants-in-Aid” from The Tojuro Iijima Foundation for Food Science and Technology, The Public Foundation of Elizabeth Arnold-Fuji, The Kieikai Research Foundation, Fuji Foundation for Protein Research, The Skylark Food Science Institute, and The Kyoto University Foundation to S.M. Moreover, this work was supported in part by “Grants-in-Aid” from The Sasakawa Scientific Research Grant from The Japan Science Society to M.K. This work was also supported by the Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture. The authors would like to thank Enago (www.enago.com) and Ms. Nozomi Ojima for the English language review.
Author Contributions
M.K., Y.S., and S.M. conceived the study; M.K. and Y.Y. conducted biological experiments; H.K. and N.T. performed RNA-seq; M.K., N.F., K.F., and S.S. performed bioinformatic analysis; Y.M. performed docking studies; M.K. and S.S. performed statistical analysis; and M.K., N.F., K.F., and S.M. analyzed the results and wrote the paper. All authors reviewed the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: December 20, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.11.033.
Data and Code Availability
The RNA-seq data were submitted to Gene Expression Omnibus (accession number: GSE128097). Original imaging data have been deposited to Mendeley Data and are available at https://doi.org/10.17632/t6r9jftdnr.1.
Supplemental Information
References
- Arango D., Morohashi K., Yilmaz A., Kuramochi K., Parihar A., Brahimaj B., Grotewold E., Doseff A.I. Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc. Natl. Acad. Sci. U S A. 2013;110:E2153–E2162. doi: 10.1073/pnas.1303726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo Y., Sun J., Wang M., Ding J., Lu Q., Yuan L. Dietary flavonoid intake and the risk of digestive tract cancers: a systematic review and meta-analysis. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep24836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Martins S., Rino J., Marinho S., Carmo-Fonseca M. Pharmacological inhibition of the spliceosome subunit SF3b triggers exon junction complex-independent nonsense-mediated decay. J. Cell Sci. 2017;130:1519–1531. doi: 10.1242/jcs.202200. [DOI] [PubMed] [Google Scholar]
- Corvelo A., Hallegger M., Smith C.W.J., Eyras E. Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 2010;6:e1001016. doi: 10.1371/journal.pcbi.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daguenet E., Dujardin G., Valcarcel J. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16:1640–1655. doi: 10.15252/embr.201541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger K.A., Urabe V.K., Jurica M.S. Modulating splicing with small molecular inhibitors of the spliceosome. Wiley Interdiscip. Rev. RNA. 2017;8:e1381. doi: 10.1002/wrna.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Su R.C., Zou L., Triggs-Raine B., Huang S., Xie J. Increase of a group of PTC+ transcripts by curcumin through inhibition of the NMD pathway. Biochim. Biophys. Acta. 2015;1849:1104–1115. doi: 10.1016/j.bbagrm.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Finci L.I., Zhang X., Huang X., Zhou Q., Tsai J., Teng T., Agrawal A., Chan B., Irwin S., Karr C. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 2018;32:309–320. doi: 10.1101/gad.311043.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Okamura M., Nishimoto S., Kurihara T., Momma K., Miyamae Y., Kambe T., Nagao M., Narita H., Masuda S. Establishment of a monitoring system to detect inhibition of mRNA processing. Biosci. Biotechnol. Biochem. 2012;76:1248–1251. doi: 10.1271/bbb.120226. [DOI] [PubMed] [Google Scholar]
- Fujiwara N., Yoshikawa M., Yamazaki T., Kambe T., Nagao M., Masuda S. A screening method tuned for mRNA processing factors in human cells by evaluation of the luciferase reporter activity and the subcellular distribution of bulk poly(A)+ RNA. Biosci. Biotechnol. Biochem. 2010;74:1512–1516. doi: 10.1271/bbb.100363. [DOI] [PubMed] [Google Scholar]
- Ge Y., Porse B.T. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays. 2014;36:236–243. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- Hahn C.N., Venugopal P., Scott H.S., Hiwase D.K. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol. Rev. 2015;263:257–278. doi: 10.1111/imr.12241. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Miura T., Kuzuya K., Inoue A., Ki S.W., Horinouchi S., Yoshida T., Kunoh T., Koseki K., Mino K. Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS Chem. Biol. 2011;6:229–233. doi: 10.1021/cb100248e. [DOI] [PubMed] [Google Scholar]
- Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C.L., Pena V., Lührmann R., Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Hoskins A.A., Moore M.J. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem. Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T.Y.T., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., Echeverria G.V., Sun T., Kurley S.J., Tyagi S. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Primorac D., McKinstry M., McNeil J., Rowe D., Lawrence J.B. Tracking COL1A1 RNA in osteogenesis imperfecta: splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 2000;150:417–431. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Kurata M., Murata Y., Momma K., Fouad Ali Mursi I., Takahashi M., Miyamae Y., Kambe T., Nagao M., Narita H., Shibuya Y. The isoflavone fraction from soybean presents mRNA maturation inhibition activity. Biosci. Biotechnol. Biochem. 2016;81:551–554. doi: 10.1080/09168451.2016.1249451. [DOI] [PubMed] [Google Scholar]
- Kurata M., Morimoto M., Kawamura Y., Fouad Ali Mursi I., Momma K., Takahashi M., Miyamae Y., Kambe T., Nagao M., Narita H. Inhibition of mRNA maturation by compounds which have a flavonoid skeleton. Biochem. Mol. Biol. 2017;2:46–53. [Google Scholar]
- Lee C.S., Dias A.P., Jedrychowski M., Patel A.H., Hsu J.L., Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C.W., Dvinge H., Kim E., Cho H., Micol J.B., Chung Y.R., Durham B.H., Yoshimi A., Kim Y.J., Thomas M. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 2016;22:672–692. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Chen L., Li H. The dietary compound luteolin inhibits pancreatic cancer growth by targeting BCL-2. Food Funct. 2018;9:3018–3027. doi: 10.1039/c8fo00033f. [DOI] [PubMed] [Google Scholar]
- López-lázaro M. Distribution and biological activities of the flavonoid luteolin. Mini Rev. Med. Chem. 2009;9:31–59. doi: 10.2174/138955709787001712. [DOI] [PubMed] [Google Scholar]
- Maguire S.L., Leonidou A., Wai P., Marchiò C., Ng C.K., Sapino A., Salomon A.V., Reis-Filho J.S., Weigelt B., Natrajan R.C. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J. Pathol. 2015;235:571–580. doi: 10.1002/path.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Markus M.A., Marques F.Z., Morris B.J. Resveratrol, by modulating RNA processing factor levels, can influence the alternative splicing of Pre-mRNAs. PLoS One. 2011;6:e28926. doi: 10.1371/journal.pone.0028926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S., Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3’ end processing regulation. Nucleic Acids Res. 2009;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nlend Nlend R., Meyer K., Schümperli D. Repair of pre-mRNA splicing: prospects for a therapy for spinal muscular atrophy. RNA Biol. 2010;7:430–440. doi: 10.4161/rna.7.4.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pandey K.B., Rizvi S.I. Plant polyphenols as dietary antioxidants in human health and disease. Oxid. Med. Cell. Longev. 2009;2:270–278. doi: 10.4161/oxim.2.5.9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawellek A., Ryder U., Tammsalu T., King L.J., Kreinin H., Ly T., Hay R.T., Hartley R.C., Lamond A.I. Characterisation of the biflavonoid hinokiflavone as a pre-mRNA splicing modulator that inhibits SENP. Elife. 2017;6:e27402. doi: 10.7554/eLife.27402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisalo M., Kårlund A., Koskela A., Kaarniranta K., Karjalainen R.O. Polyphenol stilbenes: molecular mechanisms of defence against oxidative stress and aging- related diseases. Oxid. Med. Cell. Longev. 2015;2015:1–24. doi: 10.1155/2015/340520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakla M.S., Lorson C.L. Induction of full-length survival motor neuron by polyphenol botanical compounds. Hum. Genet. 2008;122:635–643. doi: 10.1007/s00439-007-0441-0. [DOI] [PubMed] [Google Scholar]
- Salton M., Misteli T. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol. Med. 2016;22:28–37. doi: 10.1016/j.molmed.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M., Yoshimi A., Darman R., Chan B., Keaney G., Thomas M., Agrawal A.A., Caleb B., Csibi A., Sean E. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 2018;24:497–504. doi: 10.1038/nm.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U S A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Kanwal R., Shankar E., Datt M., Chance M.R., Fu P., MacLennan G.T., Gupta S. Apigenin blocks IKKα activation and suppresses prostate cancer progression. Oncotarget. 2015;6:31216–31232. doi: 10.18632/oncotarget.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A., Kilpinen S., Ruusulehto A., Lothe R.A., Skotheim R.I. Aberrant RNA splicing in cancer; Expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35:2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- Wahl M.C., Lührmann R. SnapShot: spliceosome dynamics III. Cell. 2015;162:690. doi: 10.1016/j.cell.2015.07.033. [DOI] [PubMed] [Google Scholar]
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang L., Wang J., Chen S., Shi M., Cheng H. Intronless mRNAs transit through nuclear speckles to gain export competence. J. Cell Biol. 2018;217:3912–3929. doi: 10.1083/jcb.201801184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Gonzàlez-Porta M., Perera D., Bartolozzi A.R., Sibley C.R., Hallegger M., Ule J., Marioni J.C., Venkitaraman A.R. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5’ splice site strength. Genome Biol. 2015;16:1–21. doi: 10.1186/s13059-015-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhang Y., Tian K., Chen X., Zhang R., Mu X., Wu Y., Wang D., Wang S., Liu F. Apigenin suppresses PD-L1 expression in melanoma and host dendritic cells to elicit synergistic therapeutic effects. J. Exp. Clin. Cancer Res. 2018;37:1–15. doi: 10.1186/s13046-018-0929-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R., Kaida D., Furuno M., Maxwell B.A., Noma S., Suzuki H., Kawamura Y., Hayashizaki Y., Mayeda A., Yoshida M. Global analysis of pre-mRNA subcellular localization following splicing inhibition by spliceostatin A. RNA. 2016;23:47–57. doi: 10.1261/rna.058065.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.J., Lowe G.M. Antioxidant and prooxidant properties of carotenoids. Arch. Biochem. Biophys. 2001;385:20–27. doi: 10.1006/abbi.2000.2149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data were submitted to Gene Expression Omnibus (accession number: GSE128097). Original imaging data have been deposited to Mendeley Data and are available at https://doi.org/10.17632/t6r9jftdnr.1.