Abstract
Retinitis pigmentosa (RP) is the most common form of inherited vision loss and is characterized by degeneration of retinal photoreceptor cells and the retinal pigment epithelium (RPE). Mutations in pre-mRNA processing factor 31 (PRPF31) cause dominant RP via haploinsufficiency with incomplete penetrance. There is good evidence that the diverse severity of this disease is a result of differing levels of expression of the wild-type allele among patients. Thus, we hypothesize that PRPF31-related RP will be amenable to treatment by adeno-associated virus (AAV)-mediated gene augmentation therapy. To test this hypothesis, we used induced pluripotent stem cells (iPSCs) with mutations in PRPF31 and differentiated them into RPE cells. The mutant PRPF31 iPSC-RPE cells recapitulate the cellular phenotype associated with the PRPF31 pathology, including defective cell structure, diminished phagocytic function, defects in ciliogenesis, and compromised barrier function. Treatment of the mutant PRPF31 iPSC-RPE cells with AAV-PRPF31 restored normal phagocytosis and cilia formation, and it partially restored structure and barrier function. These results suggest that AAV-based gene therapy targeting RPE cells holds therapeutic promise for patients with PRPF31-related RP.
Keywords: PRPF31, RPE, phagocytosis, haploinsuficiency, gene therapy, AAV, cilia, microvilli, iPSC-RPE
Graphical Abstract

Introduction
With a prevalence rate of approximately 1 in 3,500, retinitis pigmentosa (RP) is the most common form of inherited blindness, and it can be inherited in autosomal dominant, autosomal recessive, or X-linked patterns.1 The general pathology of RP is degeneration of the retinal pigment epithelium (RPE) and photoreceptor cells, sometimes leading to complete vision loss.2 The most common causes of autosomal dominant RP are mutations in rhodopsin, followed by mutations in precursor mRNA (pre-mRNA) processing factors PRPF3, PRPF4, PRPF6, PRPF8, PRPF31, and SNRNP200.3, 4, 5, 6, 7 Among these, mutations in PRPF31 are the most common and are estimated to account for approximately 10% of dominant RP.8 PRPF31 encodes a ubiquitously expressed splicing factor, which binds and stabilizes the tri-small nuclear ribonucleoprotein particle (snRNP) U4/U6-U5 ribonucleoprotein complex.9, 10, 11 It remains unclear why mutations in ubiquitously expressed splicing factors result in disease specific to the retina. Data obtained from studies of PRPF31-associated disease suggest that alterations in RNA splicing underlie these forms of inherited retinal degeneration (IRD).12
RP from mutations in PRPF31 stems from nonsense mutations, large-scale deletions, and premature stop codons affecting one allele.10 These mutations create null alleles and cause disease via haploinsufficiency. Complete loss of PRPF31 function results in embryonic lethality.10 Since mutations in PRPF31 cause disease via haploinsufficiency, it is a dominant disease that is a good candidate for treatment via gene augmentation therapy. Furthermore, evidence from studies of the reduced penetrance of disease observed in some families with PRPF31-associated retinal degeneration shows that increased expression of PRPF31 from the wild-type allele can reduce disease severity.13, 14, 15 For gene-based therapies, adeno-associated virus (AAV) vectors are at the forefront, since they are known to be non-pathogenic while simultaneously staying successful at penetrating cell membranes and mostly evading the immune system.16 Last year, the first US Food and Drug Administration (FDA)-approved gene therapy treatment for inherited retinal diseases was successfully performed in patients with mutations in the RPE-specific 65-kDa protein (RPE65) gene. Sub-retinal injection of the RPE65-expressing AAV vector restores normal function of this protein and leads to vision improvement.17 Stimulated by this initial success, clinical trials of AAV-mediated gene augmentation therapies are in progress for multiple genetic subtypes of IRD.18, 19, 20, 21, 22, 23
Among other functions, the RPE nourishes photoreceptor cells and phagocytoses shed photoreceptor outer segments (POSs).24 Mutations in PRPF31 primarily led to RPE degeneration in cellular and mouse models of PRPF31-linked RP.9,12,25 Specifically, RPE cells from Prpf31 mutant mice show progressive degeneration and a cell-autonomous phagocytic defect associated with decreased binding and internalization of POSs that eventually leads to photoreceptor loss.6 Since RPE can be derived from induced pluripotent stem cells (iPSCs), the RPE pathology associated with mutations in PRPF31 can be modeled using patient derived iPSC-RPE. Indeed, iPSC-RPE generated from patients with PRPF31-associated retinal degeneration show decreased phagocytosis and abnormal cilia growth.12 In the studies reported here, we used one of these patient-derived iPSC lines and a newly generated PRPF31+/− iPSC cell line to test the use of AAV-mediated gene augmentation therapy to treat PRPF31-associated retinal degeneration. Data obtained from these studies support the use of AAV-mediated gene augmentation therapy to treat PRPF31-associated disease.
Results
Generation of iPSC-RPE Cells PRPF31+/− via CRISPR-Cas9 Editing
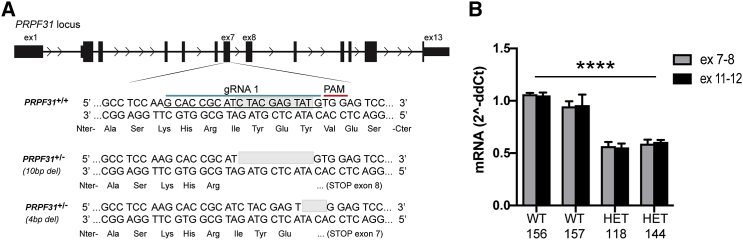
To test AAV-mediated gene augmentation therapy for PRPF31-associated IRD, we used iPSC-derived RPE cells from two sources. The first source is iPSC-derived RPE cells from a patient with an 11-bp deletion in exon 11 of PRPF31, causing RP.12 RPE cells derived from the same iPSC line in which the 11-bp deletion was corrected were used as controls.12 These PRPF31 mutant iPSC-derived RPE cells reproduce in vitro key features associated with PRPF31 pathology, such as defective splicing, decreased phagocytosis, and shorter cilia.12 The second source of iPSCs is wild-type IMR90 iPSCs into which we introduced a null allele of PRPF31 using CRISPR/Cas9-mediated genome editing. To accomplish this modification, we transfected wild-type iPSCs with the pSpCas9(BB)2A-EGFP (PX458) plasmid carrying the Cas9 nuclease and a guide RNA (gRNA) targeting exon 7 of PRPF31 (Figure 1). EGFP-positive cells were sorted and expanded to generate clonal cell lines. Screening of the clones via PCR and sequencing identified 18/255 clones with mutations in PRPF31 (8%). The most common indels found in these clones were 4-bp and 10-bp deletions in exon 7 of PRPF31, which resulted in frameshift mutations, causing premature stop codons in exons 7 and 8, respectively (Figure 1). Cells transfected with editing reagents, but that did not have a mutation in PRPF31, were expanded for use as controls. In clones harboring both the 4-bp and 10-bp deletions, the mRNA levels of PRPF31 were reduced to half compared to counterpart wild-type clones (Figure 1B; two-way ANOVA, p < 0.0001).
Figure 1.
CRISPR-Edited iPSC PRPF31+/−
(A) Schematic representation of the PRPF31 locus. A 20-bp nucleotide gRNA sequence (blue line) is followed by PAM (red line) designed to target exon 7. Bottom sequence shows the 10-bp deletion found in clone no. 144, which was used for differentiation into RPE. (B) mRNA levels of PRPF31 normalized to GAPDH measured in triplicate, expressed by CRISPR-edited iPSC PRPF31+/+ (wild-type [WT]) clones 156 and 157, and PRPF31+/− (heterozygous [HET]) mutant clones 118 (4-bp deletion) and 144 (10-bp deletion). The average expression of WT cells was used as a value of 1 for relative quantification (two-way ANOVA, ****p < 0.0001; data are represented as mean ± SD).
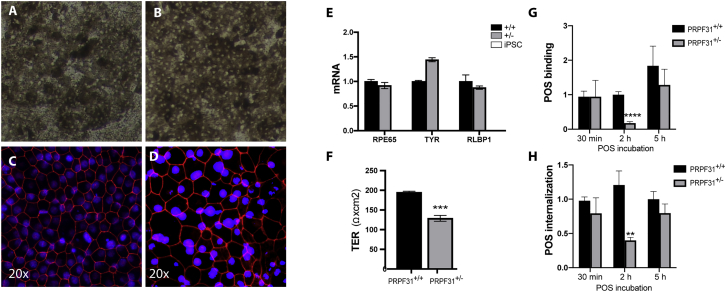
One wild-type clone (clone no. 157) and one clone harboring the 10-bp deletion in one allele of PRPF31 (clone no. 144) were chosen for further differentiation into RPE cells, according to a previously established protocol.26,27 At passage 2 (p2), iPSC-RPE cells on transwells displayed typical honeycomb morphology, pigmentation, and polarization (Figure 2). The RPE monolayer was formed as shown by the expression of the tight-junction protein ZO-1 (Figures 2C and 2D). Successful differentiation into RPE cells was determined through expression of the RPE markers RPE65, TYR (pigmentation enzyme), and RLBP1 (a visual cycle gene), which were not expressed in the iPSCs (Figure 2E). To be functional, the RPE monolayer needs to be highly polarized.24 One of the methods to assay RPE polarization is measuring the transepithelial electrical resistance (TER). Despite the normal expression of ZO-1, the engineered iPSC-RPE PRPF31+/− cells showed significantly lower TER than did the counterpart wild-type cells (t test, n = 4/genotype; p = 0.0009), corroborating results found in patient-derived iPSC-RPE cells (Figure 2F).12
Figure 2.
Characterization of the CRISPR-Edited iPSC-RPE Cell Monolayer
(A and B) Brightfield micrograph of mature iPSC-RPE PRPF31+/+ (A) and PRPF31+/− (B) cells on transwells. (C and D) Fluorescent micrographs of mature CRISPR-edited iPSC-RPE PRPF31+/+ (C) and PRPF31+/− (D) cells grown on transwells and immunostained with anti-ZO-1 antibody (red) and DAPI (blue). (E) RPE markers normalized to UBE2R2 expressed by mature iPSC-RPE cells wild-type PRPF31+/+ (+/+ black bars) and mutant PRPF31+/− (+/− gray bars) compared to iPSCs (white bars, no expression found) (n = 3 replicates/cell type; data are represented as mean ± SD). (F) Transepithelial electrical resistance (TER) of mature iPSC-RPE cells on transwells (four replicates/genotype; two-way ANOVA, ***p < 0.001; data are represented as mean ± SD). (G and H) Quantification of FITC-labeled POSs (G) bound and (H) internalized by CRISPR-edited iPSC-RPE PRPF31+/+ and PRPF31+/− cells at 30 min, 2 h, and 5 h (two-way ANOVA, **p < 0.01, ****p < 0.0001; data are represented as mean ± SD).
To test phagocytosis function of the PRPF31+/− iPSC-RPE, CRISPR-edited PRPF31+/+ and PRPF31+/− iPSC-RPE cells were incubated with POSs for 30 min, 2 h, and 5 h. Analyses showed significant defects in both POS binding (two-way ANOVA, p = 0.0001) and internalization (two-way ANOVA, p = 0.0027) of POSs in PRPF31+/− cells compared to wild-type cells at 2 h (Figures 2G and 2H). Differences in binding and internalization were not significant at 30 min and 5 h (Figures 2G and 2H). The first time point is too short and did not allow enough time for the POSs to bind the cells, while at 5 h of incubation both wild-type and mutant cells are saturated with POSs.
Optimization of AAV Vectors to Be Used for Gene Therapy in iPSC-RPE Cells
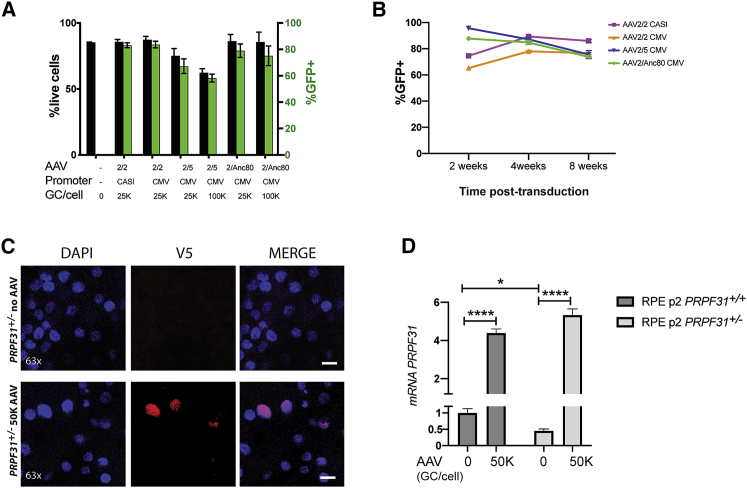
Since PRPF31-associated retinal degeneration results from haploinsufficiency, AAV-mediated gene augmentation therapy can theoretically be used to rescue the retinal degeneration phenotype. Using mature iPSC-RPE at p2 on transwells, we tested a panel of AAV with a variety of serotypes (AAV2/2, AAV2/5, and AAV2/Anc80) expressing EGFP under the control of two different promoters: the regular cytomegalovirus (CMV), and the synthetic CASI promoter, which contains a portion of CMV, a portion of the chicken β-actin promoter, and a portion of the UBC enhancer.28 In addition, different MOIs by AAV were tested at 25,000, 50,000 or 100,000 viral genome copies per cell (GC/cell). To determine the highest transduction efficiency and stability, EGFP-positive cells were counted at time points 72 h, 2 weeks, 4 weeks, and 8 weeks post-transduction using fluorescence-activated cell sorting (FACS) (Figure 3). At 72 h post-transduction, cell toxicity was highest when transduced with the AAV2/5 serotype (two-way ANOVA, n = 2 replicates/condition; p < 0.0001). Toxicity for each AAV tested was increased at the higher titer, except for cells transduced with AAV2/Anc80, which did not show significant differences between 25,000 and 100,000 GC/cell (Figure 3A). AAV 2/2 and AAV2/Anc80 serotypes showed similar levels of long-term transduction, with the CASI promoter expressing higher levels of GFP versus CMV (two-way ANOVA, n = 2 replicates/condition; p < 0.0001) (Figure 3B). For the construct expressing the PRPF31 gene, we chose the serotype AAV2/Anc80 with the CASI promoter, due to the lower cell toxicity at a higher titer, higher transduction expression of GFP, and high reported in vivo transduction efficiency.29,30 A detailed AAV-PRPF31 vector map is provided in Figure S1.
Figure 3.
Optimization of AAV Vectors
(A) Percentage of live cells (black bars) and GFP-positive cells (green bars) 72 h post-transduction with AAV measured by FACS (mean ± SEM). (B) Percentage of GFP-positive cells over the total population at 2, 4, and 8 weeks post-transduction with 25,000/cell (25K) AAV shows stable expression of GFP in iPSC-RPE cells over time. (C) RPE p2 transduced with 50,000 GC/cell (50K) AAV-PRPF31 and immunostained with anti-V5 (red) at 4 weeks post-transduction. AAV-derived PRPF31 WT protein localizes in the nuclei (blue) of the RPE cells. Scale bars, 50 μm. (D) mRNA levels of PRPF31 in iPSC-RPE PRPF31+/+ and PRPF31+/− cells before and after transduction with 50,000 (50K) GC/cell AAV. mRNA levels of PRPF31 normalized to GAPDH measured in triplicate are shown (n = 3 replicates/type; data are represented as mean ± SD; two-way ANOVA, *p < 0.05, ****p < 0.0001).
Functional Defects in Mutant iPSC-RPE PRPF31+/− Cells Can Be Rescued with AAV-Mediated Gene Augmentation
Phagocytic Defects in iPSC-RPE PRPF31+/− Can Be Rescued by AAV-Mediated Gene Augmentation
To test the use of gene augmentation therapy for PRPF31-associated disease, we treated patient-derived and CRISPR-edited iPSC-RPE PRPF31+/− cells and control cells with AAV-PRPF31.12 For these studies, differentiated iPSC-RPE cells at p2 were plated on Matrigel-coated transwells and cultured in X-VIVO 10 medium.27 After 4 weeks, cells were confluent and pigmented and were transduced with several doses of AAV2/Anc80 AAP.CASI.V5.PRPF31-mCHERRY.RBG (from now, AAV-PRPF31). Cells were cultured for an additional 4 weeks after AAV-PRPF31 treatment, and then PRPF31 expression and phagocytosis activity were evaluated. Transduced cells showed stable but weak expression of mCherry after fixation. Since mCherry is a C-terminal tag, the autocleavage rate is high and is detected in both nucleus and cytoplasm (Figure S2). Immunostaining with antibodies for the V5 N-terminal tag demonstrated stable transduction after 4 weeks and correct localization of the AAV-derived PRPF31 protein in the nucleus (Figure 3C).
To quantify the expression of AAV-derived PRPF31 versus endogenous PRPF31 by iPSC-RPE cells after treatment, mRNA levels were measured by qPCR and normalized to GAPDH. As shown in Figure 3D, endogenous mRNA levels of PRPF31 were reduced to half in iPSC-RPE PRPF31+/− cells compared to wild-type cells. After treatment, AAV-derived PRPF31 expression increased 4- to 5-fold compared to endogenous PRPF31 in both PRPF31+/+ and PRPF31+/− cells. No toxicity effects or morphological changes were observed in wild-type cells after treatment with AAV.
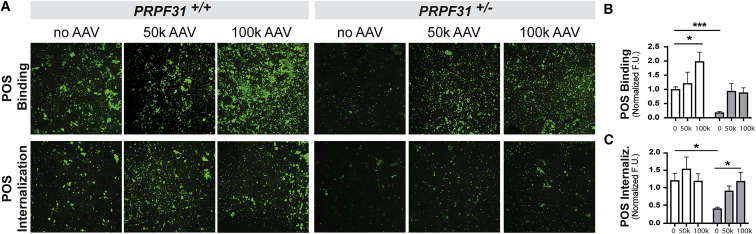
The phagocytic function of the AAV-PRPF31-treated cells was assayed by challenging the cells with bovine fluorescein isothiocyanate (FITC)-labeled POSs for 2 h. Treatment with AAV-PRPF31 resulted in rescue of binding and internalization efficiencies in iPSC-RPE PRPF31+/− cells (two-way ANOVA, p = 0.0322) (Figure 4).
Figure 4.
Phagocytosis Defect Is Rescued by AAV-PRPF31
(A–C) Representative confocal fluorescent images (×20 objective) and quantification (A) of POSs bound (B) and internalized (C) at 2 h show a significant phagocytosis defect in PRPF31 mutant cells PRPF31+/− (white bars) compared to wild-type PRPF31+/+ (gray bars). Phagocytic function is restored in mutant cells after gene therapy with AAV-PRPF31. AAV 50K, 50,000 GC/cell; AAV 100K, 100,000 GC/cell. (n = 2 replicates, 3 fields/replicate; data are represented as mean ± SEM; two-way ANOVA, *p < 0.05, ***p < 0.001).
Interestingly, the phagocytic activity was enhanced in wild-type iPSC-RPE PRPF31+/+ cells treated with AAV-PRPF31 (Figure 4). Specifically, the capacity of binding POSs increased by 2-fold when the cells were treated with 100,000 GC/cell (two-way ANOVA, p = 0.0293) (Figure 4B), while the amount of internalized POSs remained similar before and after AAV treatment (Figure 4C).
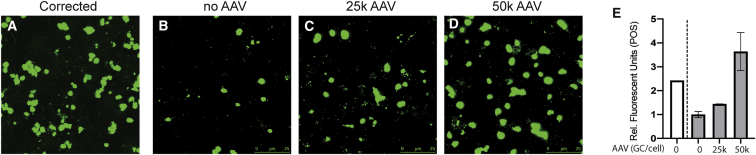
The effect of AAV-PRPF31 treatment on phagocytosis function was also evaluated in patient-derived iPSC-RPE PRPF31+/− cells and corrected counterparts. The results of these studies demonstrated that phagocytic activity was also restored in iPSC-RPE PRPF31+/− cells from a different genetic background in a dose-dependent manner (Figure 5).
Figure 5.
Patient-Derived iPSC-RPE PRPF31+/− Cells Show Dose-Dependent Rescued Phagocytic Function after Treatment with AAV-PRPF31
(A–D) Confocal fluorescent images show FITC-labeled POSs engulfed by the patient-derived iPSC-RPE p2 cells (corrected or mutant for PRPF31) on transwells treated with (A and B) no AAV, (C) 25,000 GC/cell AAV, or (D) 50,000 GC/cell AAV. Scale bars, 25 μm. (E) Fluorescence was quantified with ImageJ (n = 2/treatment; one-way ANOVA, p = 0.0655).
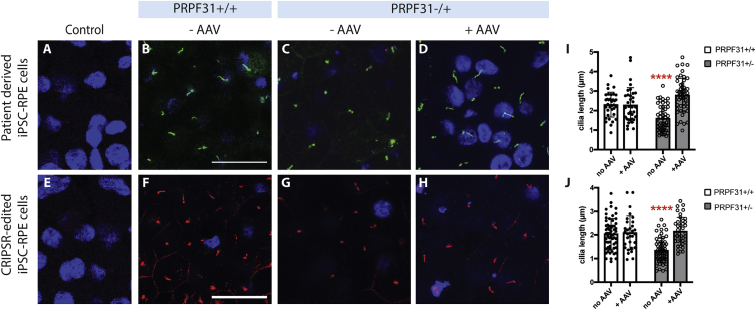
Defects in Cilia Growth in iPSC-RPE PRPF31+/− Can Be Rescued by AAV-Mediated Gene Augmentation
Among other defects, decreased cilia length was observed in patient-derived PRPF31+/− iPSC-RPE cells.12 Immunostaining with antibodies for the cilia-specific marker ARL13B showed similar results in CRISPR-edited iPSC-RPE PRPF31+/− cells grown on transwells for 8 weeks, compared to wild-type cells (Figure 6). To test the ability of gene augmentation to correct this cilia defect, patient-derived and CRISPR-edited iPSC-RPE PRPF31+/− cells were treated with AAV-PRPF31. Four weeks post-transduction with AAV-PRPF31, cilia length and incidence were restored to normal in both the patient-derived and CRISPR-edited iPSC-RPE PRPF31+/− cells (Figure 6) (two-way ANOVA, n = 50 cells/culture; p < 0.0001). No effect was observed in wild-type cells treated with the AAV-PRPF31 (Figure 6).
Figure 6.
Shorter Cilia Length in iPSC-RPE PRPF31+/− Cells Is Rescued by AAV-PRPF31
(A–H) Immunostaining for ARL13B shows shorter cilia in both patient-derived and CRISPR-edited iPSC-RPE PRPF31+/− cells compared to wild-type cells. The length of the cilia is restored after transduction with 50,000 GC/cell of AAV-PRPF31. Due to the physical distance with the cilia, nuclei are out of plane in some of the images. Immunostaining controls with secondary but no primary antibodies are shown in (A) and (E). Scale bars, 25 μm. (I and J) Cilia length measured with ImageJ demonstrates significantly shorter cilia in PRPF31+/− cells, which is rescued with AAV-based gene therapy (n = 50 cells/culture) (data are represented as mean ± SD; individual cell measurement values are shown as boxes [wild-type] or circles [PRPF31+/−]; two-way ANOVA, ****p < 0.0001).
Reduced Barrier Function in iPSC-RPE PRPF31+/− Cultures Is Partially Rescued by AAV-PRPF31
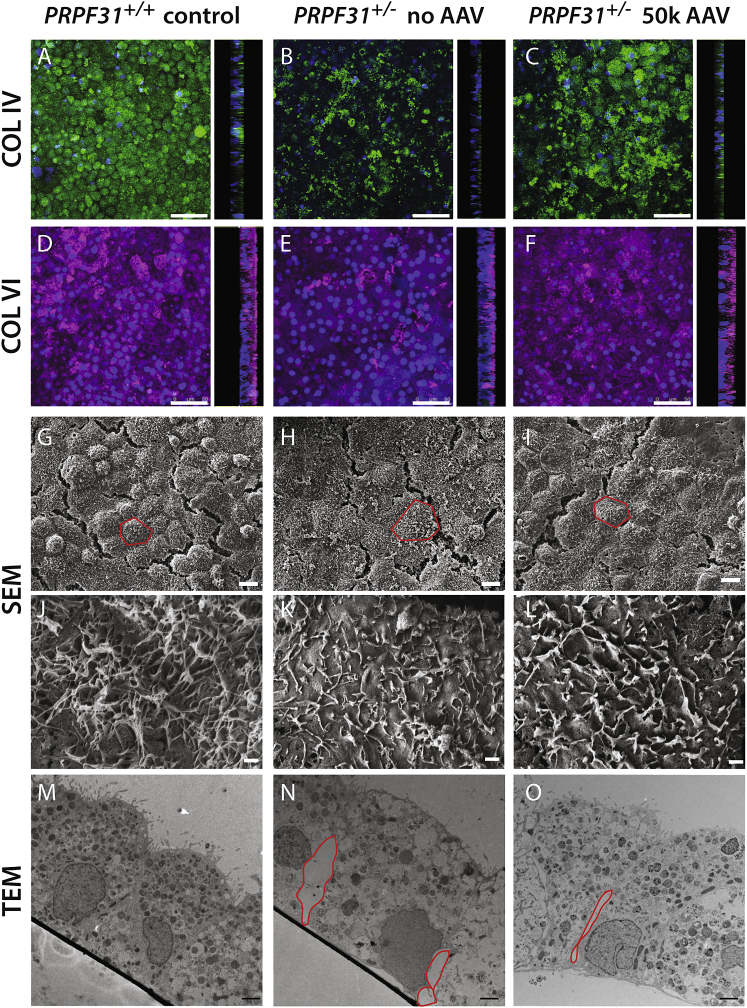
The barrier function of the iPSC-RPE PRPF31+/− cell monolayer was compromised, as indicated by lower levels of TER (Figure 2). To investigate the basis for this, we stained control and PRPF31+/− mutant iPSC-RPE with antibodies to collagen IV (COL IV) and VI (COL VI) to assess the polarity of the RPE monolayer and extracellular matrix elaboration. As shown in Figure 7, while the PRPF31+/− cells remain polarized and expressed COL IV on the basal membrane, the expression pattern was irregular, which indicates disruption of the basal lamina (Figures 7A and 7B). The basolateral secretion of COL VI was reduced in the mutant cells compared to wild-type cells, which denotes insufficient secretion of extracellular matrix by mutant cells (Figures 7D and 7E). Treatment of the iPSC-RPE PRPF31+/− cells with AAV-PRPF31 improved the expression pattern of COL IV and restored normal levels of COL VI production (Figures 7A–7C).
Figure 7.
Confocal and Electron Micrographs of iPSC-RPE Cell Monolayer after 8 Weeks on Transwells
(A–C) Confocal fluorescent images show the basal lamina of the RPE cells stained with antibodies for COL IV expressed at 8 weeks. A z stack was built from images taken every 0.13 μm with a confocal microscope. 90° projections show that COL IV is expressed on the basolateral membrane of the RPE cells. (D–F) Maximum intensity of COL VI expressed at 8 weeks. A z stack was built from images taken every 0.13 μm with a confocal microscope. 90° projections show that COL VI is deposited on the basal side of the RPE monolayer. (G–L) Flat view by SEM at low (G–I) and high (J–L) magnification of PRPF31+/+ and PRPF31+/− cell monolayers untreated or treated with AAV-PRPF31 (single cells delimited by red lines). (M–O) Section view by TEM of PRPF31+/+ and PRPF31+/− cell monolayers untreated or treated with AAV-PRPF31 (red lines frame gaps between cells). Scale bars: (A)–(F), 50 μm; (G)–(I), 10 μm; (J)–(L), 1 μm; (M)–(O), 2 μm.
Scanning electron microscopy images revealed the presence of flatter and harder-to-distinguish cells in PRPF31+/− cultures. The definition of the cells is partially restored following AAV treatment (Figures 7G–7I). At higher magnification, PRPF31+/− cells displayed fewer microvilli than controls with peculiarly coiled and flattened structure, which are also partially restored after treatment with AAV-PRPF31 (Figures 7J–7L).
Disruption of the RPE monolayer in PRPF31+/− cultures was confirmed by transmission electron microscopy (TEM), in which cross-section images showed gaps between adjacent cells at the basal side while apical tight junctions stayed intact (Figures 7M and 7N), explaining the normal pattern of ZO-1 exhibited by iPSC-RPE mutant cells (Figure 2). Additionally, stress vacuoles and shorter microvilli were observed in the mutant iPSC-PRPF31+/− cells. Treatment with AAV-PRPF31 results in fewer stress vacuoles and reduced gaps between the RPE cells (Figure 7O). Despite improved RPE cell morphology, TER values were not restored in iPSC-PRPF31+/− cultures after gene therapy treatment (Figure S2), possibly because basal gaps between cells, although smaller, were still present after treatment (Figure 7O).
Discussion
Autosomal dominant RP due to mutations in the PRPF31 gene is caused by haploinsufficiency,10,25 providing an opportunity for treatment with gene-augmentation therapy. In this study, we modeled the cellular phenotype of the disease to test AAV-based PRPF31 gene augmentation therapy. Mutant iPSC-RPE PRPF31+/− cells grown on transwells for 8 weeks recapitulated the cellular phenotype associated with mutations in PRPF31, including abnormal structure, decreased phagocytic activity, defective ciliogenesis, and compromised barrier function. AAV-mediated PRPF31 gene augmentation therapy restored normal phagocytosis function and cilia length and partially restored barrier function in patient-derived and genome-edited iPSC-RPE PRPF31+/− cells. To the best of our knowledge, this is the first proof of concept that AAV-based gene therapy has the potential to be used for treatment of PRPF31-associated RP.
The RPE monolayer is the primary cell type affected in PRPF31-related RP, and a number of cell and animal models have been developed that recapitulate the PRPF31-associated cellular phenotypes.6,9,12,25,31, 32, 33 Consistent with prior reports, both the patient-derived and genome-edited PRPF31 mutant iPSC-RPE cells used in the current study demonstrated defective phagocytosis and cilia formation.12 Our data suggest that mutations in PRPF31 affect both binding and internalization of POSs, with a larger impact in POS binding, analogous to the defect described in mutant mouse RPE Prpf31+/− cells.6 Phagocytosis is a critical support function in continuously renewing light-sensitive outer segment portions, which is necessary for vision.34 With diminished phagocytic activity, individual POS components are not degraded or recycled back to photoreceptors, eventually leading to photoreceptor loss.35
As expected by observations in RPE and optic cups derived from patients with mutations in PRPF31,12 iPSC-RPE PRPF31+/− cells displayed shorter cilia. The cilium is critical for the RPE cell development and maturation, and for correct polarization of the RPE monolayer, which supports the photoreceptor integrity.36 Defects in the cilia of RPE PRPF31+/− cells implies defects in maturation and polarization that compromise the function of the monolayer. A recent paper using ciliopathy patient-derived iPSC-RPE cells established the maturation of the RPE cilium as the primary cause for disease.36 Interestingly, the authors demonstrated that treatment with modulators of ciliogenesis can rescue the phagocytosis activity by the RPE cells.36 Based on those results, we think that the decreased phagocytosis activity observed in mutant iPSC-RPE PRPF31+/− cells may be secondary to the abnormal maturation of cilia and microvilli. In any case, the degeneration of the RPE occurs first, later leading to degeneration of the neural retina observed in patients.3,36 Thus, we postulate that the RPE is the best candidate tissue to be targeted by gene therapy.
In vivo, RPE microvilli interdigitate with photoreceptors, providing mechanical support and executing the diurnal phagocytic removal of shed POSs. Although degeneration in mutant mice is slow and photoreceptors do not show a phenotype associated with Prpf31 defect, mutations in this gene result in diminished adhesion between RPE microvilli and POSs, which delayed the phagocytic burst after light onset.6 Our cell-based model has allowed a better characterization of the microvilli structure in PRPF31 mutant cells, showing deficiencies in microvilli length and frequency, as well as a coiled morphology, which may reduce POS binding to the integrin receptors, thus delaying phagocytic function. Additional structural defects displayed by the iPSC-RPE PRPF31+/− cells were stress vacuoles and widened spaces between adjacent cells, which could be a consequence of high apoptotic rates derived from aberrations in gene splicing. Splicing defects due to mutations in PRPF31 have been specifically associated with functional and ultrastructural defects in the RPE.12 For instance, gaps between mutant cells could indicate reduced expression of focal adhesion genes, whereas diminished collagen may be associated with changes in regulators of extracellular matrix genes derived from mutations in PRPF31.12 Further transcriptome analyses are required to unravel the mechanisms underlying functional and structural defects in the RPE.
Patients with autosomal recessive IRD associated with mutations in RPE65 have been successfully treated and show improved visual function after sub-retinal injections with AAV-expressing wild-type RPE65.17 Furthermore, successful pre-clinical studies and clinical trials of gene therapies for other inherited retinal disorders are in progress.18, 19, 20, 21 Likewise, we think that the visual function in patients with RP caused by PRPF31 haploinsufficiency can be improved with AAV-mediated gene augmentation therapy. Based on this premise, we optimized an AAV vector that expresses wild-type PRPF31 under the CASI promoter, which showed no toxicity in RPE cells. Although our experiments were performed in vitro, it is hoped the development of this therapy can be advanced so that it can be used clinically. To support this goal, the AAV vector used for these studies was generated with the Anc80 serotype, in which modifications in the viral capsid makes it more potent to transduce retinal tissue in vivo, as proven in mice and non-human primate studies.29,30
The results presented here are a proof of concept that AAV-mediated PRPF31 expression can restore the phagocytic function and the cilia length of mutant RPE PRPF31+/− cells. Additionally, AAV-PRPF31 partially rescues the barrier function of the iPSC-RPE PRPF31+/− cell monolayer, which displays tighter cell junctions and healthier RPE cells after treatment. Interestingly, AAV-mediated PRPF31 expression also enhances phagocytic activity in wild-type RPE cells. Since AAV-based therapy is applied to pre-mature cells, this effect could imply an accelerated functional maturation of the RPE. Transcriptome analyses of these cells will contribute to understanding the mechanisms of RPE pathology in patients with PRPF31-linked RP.
It is also important to highlight that AAV-PRPF31 transduced the RPE cells with high efficiency, increasing 4- to 5-fold the endogenous levels of PRPF31 without any signs of toxicity, which supports its potential as a therapy for humans. However, based on the FACS analyses, there is a percentage of cells not targeted by the AAV, which will likely undergo apoptosis and prevent the TER values from being completely restored. These findings indicate that AAV-based gene therapy treatment may rescue the function of the RPE cells individually, while the global function of the RPE monolayer requires that most cells are targeted by the AAV. Further analyses are required to investigate whether earlier intervention with the AAV-PRPF31 vector may result in a total rescue of the RPE barrier function. Still, the rescue provided by AAV-derived PRPF31 may be limited to certain RPE functions, while others like the barrier function cannot be rescued. Nonetheless, partial restoration of the RPE function and structure may be enough to preserve vision in patients with PRPF31-linked RP. However, the PRPF31 pathology is complex in patients and has an impact on photoreceptor cells as well, which makes them additional potential targets for future gene therapy studies. Supplementary studies in vivo and using iPSC-derived optic cups may be warranted to identify the right dosage and cellular targets, as well as the safety of this treatment for human therapy.
Materials and Methods
Ethical Considerations
Patient-derived samples used were obtained by Dr. Majlinda Lako’s laboratory, with informed consent according to the protocols approved by Yorkshire and the Humber Research Ethics Committee (REC ref. no. 03/362). Further information on the patients and controls is provided in Buskin et al.12
Culture of iPSCs
IMR90 human iPSCs (hiPSCs) were purchased from WiCell (Madison, WI, USA) and cultured on growth factor-reduced (GFR) Matrigel basement membrane matrix (354230, Corning, Bedford, MA, USA)-coated plates in mTeSR1 (85850, STEMCELL Technologies, Cambridge, MA, USA). Patient-derived hiPSCs with a 1-bp deletion in PRPF31 and duplicate cells containing a CRISPR-Cas9 correction were generated in Dr. Majlinda Lako’s laboratory (Institute of Genetic Medicine, Newcastle University)12,37, 38, 39, 40 and cultured similarly to IMR90 hiPSCs.
gRNA Design and CRISPR/Cas9 Genome Editing
The single guide RNA (sgRNA) target sequence (5′-GCACCGCATCTACGAGTATG-3′) was generated using the tool http://www.e-crisp.org/E-CRISP/designcrispr.html, with a score of 91. The sgRNA was cloned into the vector pSpCas9(BB)-2A-GFP (PX458) (a gift from Feng Zhang, Addgene plasmid no. 48138).
Transfection of CRISPR-Edited IMR90 iPSCs
Cells were transfected using the Amaxa Nucleofector Kit V (Lonza, Morristown, NJ, USA) per the manufacturer’s instructions. Four micrograms of plasmid DNA was transfected per 10 million cells in a GFR-Matrigel-coated 10-cm dish. Cells were incubated undisturbed for 48 h in mTeSR1 with 1 μM selective ROCK inhibitor (Y27632, Tocris, Minneapolis, MN, USA) to increase cell survival rate. After 48 h, positively transfected cells were separated via FACS, due to transient GFP expression from the vector pSpCas9(BB)-2A-GFP (PX458). Positive clones were plated on GFR-Matrigel-coated 10-cm dishes in media composed of 50% fresh mTESR1 and 50% filtered conditioned media from confluent iPSC cultures until small colonies formed (roughly 8 days later). To prevent cross-contamination between clones, colonies were manually dissected and transferred to one colony per well in 96-well plates and expanded until 60%–70% confluent (roughly 8 days later). Once split, three replica plates were created: one to keep cells growing in an incubator, one to analyze the genotype, and one to freeze clones.41
Recent studies have raised the concern that Cas9 produces a p53-mediated DNA damage response in iPSCs, which reduces the editing efficiency.42,43 This finding suggests that defects in p53 improve the efficiency of genome editing in iPSCs, potentially leading to the generation of a biased cell population with defective p53.43 To determine whether this occurred during editing of PRPF31, PBS only, or 1 μg total of the p53 antagonist MDM2 (E3-204-050, R&D Systems, Minneapolis, MN, USA) in PBS, was co-transfected with the plasmid DNA. No differences were observed in editing efficiency between cultures treated with or without rhMDM2 (Figure S3).
Genotyping Edited Clones by Sanger Sequencing
DNA was extracted from the expanded cell clones using QuickExtract DNA extraction solution (QE0905T, Lucigen, Middleton, WI, USA). PCR fragments were amplified using primers 5′-GGACAAGTGCAAGAACAATGAGAACC-3′ (forward) and 5′-GGATGTAGCTTTCCCAAGGTCACAGTG-3′ (reverse). Deletions were undetectable via gel electrophoresis, so PCR products were analyzed by Sanger sequencing. Two mutant clones with 4-bp and 10-bp out-of-frame deletions and two wild-type clones with no detected abnormalities were expanded. Normal karyotypes of each iPSC line were validated with the hPSC genetic analysis kit (07550, STEMCELL Technologies, Cambridge, MA, USA).
Differentiation of iPSCs
Confluent cultures of iPSCs were differentiated using the 14-day direct differentiation protocol previously described.26,27 Briefly, a given combination of Noggin, Dkk-1, insulin growth factor 1, nicotinamide, activin A, basic fibroblast growth factor, SU-5402, and CHIR99021 produces directed differentiation of iPSCs into RPE cells that exhibit key characteristics of the RPE, including pigmentation, honeycomb morphology, and expression of RPE markers. Every 30 days, cells were enzymatically digested with TryPLE Express (12604013, Thermo Fisher, Waltham, MA, USA), strained through a 40-μm filter, and seeded at a density of 105 cells/cm2 onto Matrigel-coated plates in X-VIVO 10 media. After 60 days in culture, cells were seeded onto Matrigel-coated 6.5-mm polystyrene transwells with 0.4-μm pores (3470, Corning, Bedford, MA, USA) at a density of 20,0000 (low-density) or 200,000 (high-density) cells per transwell. The CRISPR-edited PRPF31 mutant and wild-type clones with better overall RPE-like morphology were chosen for the remainder of the protocol.
qPCR
RNA from patient-derived iPSCs and CRISPR-edited iPSCs was extracted at passage 2 using the RNeasy Mini Kit (74104, QIAGEN, Germantown, MD, USA). Next, cDNA was synthesized using random primers with the AffinityScript cDNA synthesis kit (600105, Agilent) and used to assay the expression of the RPE markers using the TaqMan probes described by Leach et al.27 using the Stratagene Mx3005P qPCR system (Agilent, Santa Clara, CA, USA). mRNA levels of PRPF31 were measured with SYBR Green using exon-exon primers: exon 7/8 (forward, 5′-AAGATCATGGGTGTGGCCG-3′, reverse, 5′-GTAGACGAGAAGCCCGACAG-3′) and exon 11/12 (forward, 5′-TCGGAGAGATCGAGGAGGAC-3′, reverse, 5′-CTGCCCGACTTGCCCAG-3′). Samples were run in triplicate using 10 ng of cDNA per well and normalized to housekeeping genes using the delta-CT method. Statistical tests were performed with GraphPad Prism 7.
Transduction of RPE Cells
All RPE p2 cells were matured for 4 weeks on transwells before transduction. AAV2/Anc80 AAP.CASI.V5.PRPF31-mCHERRY.RBG was suspended in X-VIVO (BE04-743Q, Lonza, Basel, Switzerland) with Normocin (ant-nr-1, InvivoGen, San Diego, CA, USA) to achieve an MOI of 25,000, 50,000, and 100,000 vector genomes (vg)/cell in a final volume of 50 μL for low-density cultures or 100 μL for high-density cultures. AAV was added to the iPSC-RPE cells on transwells and incubated overnight. Media were changed the next day and twice/week afterward. All RPE cells were cultured for 4 weeks before performing a phagocytosis assay.
POS Phagocytosis Assay
POSs were purchased from InVision BioResources (98740, Seattle, WA, USA) and labeled with FITC for 1 h at room temperature (F6434, Fisher, Waltham, MA, USA). POSs were resuspended in enough cell media to constitute 10 POSs per cell. The patient-derived and CRISPR-edited iPSC-RPE cells were incubated with FITC-POSs for 30 min, 2 h, or 5 h. At the end of the incubation period, FITC-POSs were aspirated and samples washed with PBS three times for 1 min each to stop phagocytosis. To quench FITC-POSs bound outside of the cell, half of the samples were incubated with 0.4% trypan blue in PBS for 10 min at room temperature. All samples challenged with FITC-POSs were fixed with ice-cold 100% methanol for 10 min and rehydrated with PBS. Samples without FITC-POSs were fixed with 4% paraformaldehyde (PFA) for 10 min, followed by 1% glutaraldehyde for 30 min, and left in PBS.
Immunocytochemistry
Transwells were cut from the chamber, cut in half with a sharp blade, and cells were permeabilized with 0.1% Triton X-100 for 15 min, then blocked with 0.1% BSA in PBS for 1 h.44 Primary antibodies were diluted in 0.1% BSA in PBS and incubated with cells overnight at 4°. Primary antibodies used were as follows: ZO-1 (1:100, 61-7300, Invitrogen, Carlsbad, CA, USA), V5 (1:50, R96025, Invitrogen, Carlsbad, CA, USA), ARL13B (1:100, 17711-1-AP, Proteintech, Rosemont, IL, USA), COL IV (1:100, AB6586, Abcam, Cambridge, MA, USA), and COL VI (1:100, AB6588, Abcam, Cambridge, MA, USA). Secondary antibody was diluted 1:500 in 0.1% BSA in PBS and incubated with cells for 2 h at room temperature. Secondary antibodies used were labeled with Alexa Fluor 488 (A11034, Thermo Fisher Scientific, Waltham, MA, USA), Alexa Fluor 555 (A21429, Thermo Fisher Scientific, Waltham, MA, USA), or Alexa Fluor 647 (A21235, Thermo Fisher Scientific, Waltham, MA, USA). Lastly, cells were incubated with DAPI, rinsed with PBS, and mounted with Fluoromount-G (17984-25, Electron Microscopy Sciences, Hatfield, PA, USA). Samples were imaged with a TCS SP5 II confocal laser scanning microscope (Leica, Allendale, NJ, USA) or fluorescence microscope (Eclipse T, Nikon, Melville, NY, USA).
Measurement of Cilia Length
Cilia lengths were measured with ImageJ using scale bars to set the pixel/μm ratio.
Quantification of the Fluorescent Signal
Images were converted to a binary format with ImageJ. The integrated intensity was measured.45,46
TEM
Samples for TEM were prepared as previously described.47 Briefly, transwells were fixed with 2.5% glutaraldehyde overnight at 4°C in 0.1 M cacodylate buffer. The transwell inserts containing cell monolayers were cut into smaller pieces and post-fixed in 1.0% osmium tetroxide in cacodylate buffer for 1 h at room temperature (RT), then rinsed in cacodylate buffer. Insert pieces were then dehydrated through a graded series of ethanol and placed pre-infiltrated overnight with propylene oxide and Eponate 1:1. Specimens were embedded in Eponate resin. 70-nm sections were cut using a Leica EM UC7 ultramicrotome, collected onto Formvar-coated grids, stained with uranyl acetate and Reynold’s lead citrate, and examined in a JEOL JEM 1011 transmission electron microscope at 80 kV.
Scanning Electron Microscopy
Transwell inserts containing exposed extracellular matrix after decellularization were fixed with 4% PFA for 10 min and 1% glutaraldehyde for 30 min at RT, followed by critical dehydration and chromium coating as previously described.46,47 Samples were imaged by a field emission scanning electron microscope (JEOL JSM 7401F).
Statistics
Results are expressed as mean ± SD, with p < 0.05 considered statistically significant. Differences between groups were compared using the Student’s t test or two-way ANOVA as appropriate using GraphPad Prism software.
Author Contributions
Conceptualization, E.B., R.B., E.A.P, and R.F.-G.; Methodology, E.B, A.B., M.L., and R.F.-G.; Writing – Review, & Editing, E.B., E.A.P, and R.F.-G.; Funding Acquisition, E.A.P.; Supervision, E.A.P., and R.F.-G.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank Dr. Michael Farkas and Maria Sousa for their preliminary studies, and Dr. Clegg and Leah Foltz for technical support with the RPE differentiation method. We thank the MEEI/SERI Gene Transfer Vector Core for viral vector production. We thank Diane Capen for excellent technical assistance with the TEM, supported by the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology, which is funded partially by an Inflammatory Bowel Disease grant DK043351 and Boston Area Diabetes and Endocrinology Research Center (BADERC) Award DK057521. This work was supported by NIH grant EY020902. The patient-derived cells were generated with the support of RP Fighting Blindness (GR595), Fight for Sight (1456/1457), and ERC (CoG_614620).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.10.014.
Supplemental Information
References
- 1.Nishiguchi K.M., Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS ONE. 2012;7:e41902. doi: 10.1371/journal.pone.0041902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berson E.L. Retinitis pigmentosa: the Friedenwald lecture. Invest. Ophthalmol. Vis. Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]
- 3.Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol. Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 4.Tanackovic G., Ransijn A., Thibault P., Abou Elela S., Klinck R., Berson E.L., Chabot B., Rivolta C. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 2011;20:2116–2130. doi: 10.1093/hmg/ddr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafler B.P., Comander J., Weigel DiFranco C., Place E.M., Pierce E.A. Course of ocular function in PRPF31 retinitis pigmentosa. Semin. Ophthalmol. 2016;31:49–52. doi: 10.3109/08820538.2015.1114856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas M.H., Lew D.S., Sousa M.E., Bujakowska K., Chatagnon J., Bhattacharya S.S., Pierce E.A., Nandrot E.F. Mutations in pre-mRNA processing factors 3, 8, and 31 cause dysfunction of the retinal pigment epithelium. Am. J. Pathol. 2014;184:2641–2652. doi: 10.1016/j.ajpath.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X., Liu Y., Sheng X., Tam P.O.S., Zhao K., Chen X., Rong W., Liu Y., Liu X., Pan X. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2014;23:2926–2939. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- 8.Daiger S.P., Sullivan L.S., Bowne S.J. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013;84:132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graziotto J.J., Farkas M.H., Bujakowska K., Deramaudt B.M., Zhang Q., Nandrot E.F., Inglehearn C.F., Bhattacharya S.S., Pierce E.A. Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:190–198. doi: 10.1167/iovs.10-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rio Frio T., Wade N.M., Ransijn A., Berson E.L., Beckmann J.S., Rivolta C. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J. Clin. Invest. 2008;118:1519–1531. doi: 10.1172/JCI34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venturini G., Rose A.M., Shah A.Z., Bhattacharya S.S., Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012;8:e1003040. doi: 10.1371/journal.pgen.1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buskin A., Zhu L., Chichagova V., Basu B., Mozaffari-Jovin S., Dolan D., Droop A., Collin J., Bronstein R., Mehrotra S. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 2018;9:4234. doi: 10.1038/s41467-018-06448-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkas M.H., Grant G.R., White J.A., Sousa M.E., Consugar M.B., Pierce E.A. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivolta C., McGee T.L., Rio Frio T., Jensen R.V., Berson E.L., Dryja T.P. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum. Mutat. 2006;27:644–653. doi: 10.1002/humu.20325. [DOI] [PubMed] [Google Scholar]
- 15.Vithana E.N., Abu-Safieh L., Pelosini L., Winchester E., Hornan D., Bird A.C., Hunt D.M., Bustin S.A., Bhattacharya S.S. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest. Ophthalmol. Vis. Sci. 2003;44:4204–4209. doi: 10.1167/iovs.03-0253. [DOI] [PubMed] [Google Scholar]
- 16.Gao G.-P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues G.A., Shalaev E., Karami T.K., Cunningham J., Slater N.K.H., Rivers H.M. Pharmaceutical development of AAV-based gene therapy products for the eye. Pharm. Res. 2018;36:29. doi: 10.1007/s11095-018-2554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banin E., Gootwine E., Obolensky A., Ezra-Elia R., Ejzenberg A., Zelinger L., Honig H., Rosov A., Yamin E., Sharon D. Gene augmentation therapy restores retinal function and visual behavior in a sheep model of CNGA3 achromatopsia. Mol. Ther. 2015;23:1423–1433. doi: 10.1038/mt.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komáromy A.M., Alexander J.J., Rowlan J.S., Garcia M.M., Chiodo V.A., Kaya A., Tanaka J.C., Acland G.M., Hauswirth W.W., Aguirre G.D. Gene therapy rescues cone function in congenital achromatopsia. Hum. Mol. Genet. 2010;19:2581–2593. doi: 10.1093/hmg/ddq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zallocchi M., Binley K., Lad Y., Ellis S., Widdowson P., Iqball S., Scripps V., Kelleher M., Loader J., Miskin J. EIAV-based retinal gene therapy in the shaker1 mouse model for usher syndrome type 1B: development of UshStat. PLoS ONE. 2014;9:e94272. doi: 10.1371/journal.pone.0094272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bainbridge J.W.B.B., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 23.Lam B.L., Davis J.L., Gregori N.Z., MacLaren R.E., Girach A., Verriotto J.D., Rodriguez B., Rosa P.R., Zhang X., Feuer W.J. Choroideremia gene therapy phase 2 clinical trial: 24-month results. Am. J. Ophthalmol. 2019;197:65–73. doi: 10.1016/j.ajo.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 25.Bujakowska K., Maubaret C., Chakarova C.F., Tanimoto N., Beck S.C., Fahl E., Humphries M.M., Kenna P.F., Makarov E., Makarova O. Study of gene-targeted mouse models of splicing factor gene Prpf31 implicated in human autosomal dominant retinitis pigmentosa (RP) Invest. Ophthalmol. Vis. Sci. 2009;50:5927–5933. doi: 10.1167/iovs.08-3275. [DOI] [PubMed] [Google Scholar]
- 26.Foltz L.P., Clegg D.O. Rapid, directed differentiation of retinal pigment epithelial cells from human embryonic or induced pluripotent stem cells. J. Vis. Exp. 2017;(128):e56274. doi: 10.3791/56274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leach L.L., Croze R.H., Hu Q., Nadar V.P., Clevenger T.N., Pennington B.O., Gamm D.M., Clegg D.O. Induced pluripotent stem cell-derived retinal pigmented epithelium: a comparative study between cell lines and differentiation methods. J. Ocul. Pharmacol. Ther. 2016;32:317–330. doi: 10.1089/jop.2016.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balazs A.B., Chen J., Hong C.M., Rao D.S., Yang L., Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2011;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho L.S., Xiao R., Wassmer S.J., Langsdorf A., Zinn E., Pacouret S., Shah S., Comander J.I., Kim L.A., Lim L., Vandenberghe L.H. Synthetic adeno-associated viral vector efficiently targets mouse and nonhuman primate retina in vivo. Hum. Gene Ther. 2018;29:771–784. doi: 10.1089/hum.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinn E., Pacouret S., Khaychuk V., Turunen H.T., Carvalho L.S., Andres-Mateos E., Shah S., Shelke R., Maurer A.C., Plovie E. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 2015;12:1056–1068. doi: 10.1016/j.celrep.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin J., Brocher J., Fischer U., Winkler C. Mutant Prpf31 causes pre-mRNA splicing defects and rod photoreceptor cell degeneration in a zebrafish model for Retinitis pigmentosa. Mol. Neurodegener. 2011;6:56. doi: 10.1186/1750-1326-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan L., Kawada M., Havlioglu N., Tang H., Wu J.Y. Mutations in PRPF31 inhibit pre-mRNA splicing of rhodopsin gene and cause apoptosis of retinal cells. J. Neurosci. 2005;25:748–757. doi: 10.1523/JNEUROSCI.2399-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant G.R., Farkas M.H., Pizarro A.D., Lahens N.F., Schug J., Brunk B.P., Stoeckert C.J., Hogenesch J.B., Pierce E.A. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young R.W. The renewal of photoreceptor cell outer segments. J. Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosch E., Horwitz J., Bok D. Phagocytosis of outer segments by retinal pigment epithelium: phagosome-lysosome interaction. J. Histochem. Cytochem. 1993;41:253–263. doi: 10.1177/41.2.8419462. [DOI] [PubMed] [Google Scholar]
- 36.May-Simera H.L., Wan Q., Jha B.S., Hartford J., Khristov V., Dejene R., Chang J., Patnaik S., Lu Q., Banerjee P. Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep. 2018;22:189–205. doi: 10.1016/j.celrep.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 38.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu K., Yu J., Suknuntha K., Tian S., Montgomery K., Choi K.D., Stewart R., Thomson J.A., Slukvin I.I. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 43.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Godino R., Bujakowska K.M., Pierce E.A. Changes in extracellular matrix cause RPE cells to make basal deposits and activate the alternative complement pathway. Hum. Mol. Genet. 2018;27:147–159. doi: 10.1093/hmg/ddx392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Godino R., Garland D.L., Pierce E.A. A local complement response by RPE causes early-stage macular degeneration. Hum. Mol. Genet. 2015;24:5555–5569. doi: 10.1093/hmg/ddv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez-Godino R., Garland D.L., Pierce E.A. Isolation, culture and characterization of primary mouse RPE cells. Nat. Protoc. 2016;11:1206–1218. doi: 10.1038/nprot.2016.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.