Summary
Light filtered through dense planting initiates the shade avoidance syndrome (SAS) in plants, which helps them compete against their neighbors. Quantitative trait loci (QTL)-based analysis identified the nighttime-expressed clock component ELF3 as a new player in the SAS, but its detailed mechanism is unclear. Here, we show that the circadian clock gates shade-induced gene expression and hypocotyl elongation at night. ELF3 is involved in nighttime suppression via interaction with and inactivation of PHYTOCHROME-INTERACTING FACTOR 7 (PIF7). Loss of function of ELF3 restores the shade induction, which is largely reduced in the absence of PIF7, indicating that ELF3 acts upstream of PIF7. Finally, we found that the repressive activity of ELF3 on the shade response is stronger under short days than under long days. Our results reveal that the interaction between ELF3 and PIF7 mediates the circadian gating of the SAS, which coordinates the daily control of physiological outputs.
Subject Areas: Biological Sciences, Molecular Mechanism of Gene Regulation, Plant Biology, Plant Genetics
Graphical Abstract
Highlights
-
•
ELF3 is involved in the inhibition of the shade response at night
-
•
ELF3 interacts with PIF7 and prevents PIF7 from binding DNA
-
•
ELF3 acts upstream of PIF7 in shade-induced growth
-
•
Repressive activity of ELF3 is stronger under SDs than under LDs
Biological Sciences; Molecular Mechanism of Gene Regulation; Plant Biology; Plant Genetics
Introduction
Because of the selective absorption of blue and red wavelengths by chlorophyll, shaded plants perceive reduced ratios of red (R, 660 nm) to far red (FR, 730 nm), which triggers the shade avoidance syndrome (SAS) (Casal, 2012). A classic phenotype of the SAS is manifested as reallocation of energy resources from storage organs to stem-like organs, such as the hypocotyl and petiole, for plants to outgrow their competitors (Ballare and Pierik, 2017).
Several microarray and RNA sequencing analyses have revealed transcriptional regulation after shade treatment (Ciolfi et al., 2013, Leivar et al., 2012, Li et al., 2012, Salter et al., 2003, Sellaro et al., 2017, Yang et al., 2018). Interestingly, the transcriptional regulation of PIL1 (a marker gene of the shade response) by a low R/FR is reported to be gated by the circadian clock (Salter et al., 2003). The circadian clock limits the timing of maximum responsiveness to shade light to specific times of the day. However, the mechanism of circadian clock involvement in the shade response is unclear.
PHYTOCHROME-INTERACTING FACTOR 7 (PIF7) is a major regulator of shade-induced gene expression and hypocotyl elongation (Li et al., 2012, Mizuno et al., 2015, Peng et al., 2018). In the shade, PIF7 is dephosphorylated when the Pfr form of phyB is photoconverted to the Pr form, and PIF7 then regulates cell elongation by enhancing the transcription of growth-promoting genes (Li et al., 2012). PIF7 also regulates the shade induction of PIL1; therefore, whether the circadian clock is involved in the gating of PIF7 function is of particular interest.
EARLY FLOWERING 3 (ELF3) was first identified in Arabidopsis as an early-flowering mutant that is insensitive to the photoperiod (Zagotta et al., 1996). ELF4 association with ELF3 directs LUX action in the circadian clock (Herrero and Davis, 2012, Herrero et al., 2012), and this complex is referred to as the evening complex (EC) (Bujdoso and Davis, 2013). ELF3 functions as a transcriptional regulator, repressing clock- and growth-associated transcription factors to regulate the circadian rhythm and hypocotyl elongation (Chow et al., 2012, Dixon et al., 2011, Helfer et al., 2011, Nusinow et al., 2011). Furthermore, natural variation has revealed that the intracellular distribution of ELF3 proteins is associated with specific functions in the circadian clock (Anwer et al., 2014, Anwer and Davis, 2013). The photoreceptor phyB physically interacts with ELF3 in the central oscillator to provide a direct light input to the clock (Kolmos et al., 2011, Oakenfull and Davis, 2017). ELF3 can prevent PIF4 from activating its transcriptional targets in an EC independent manner (Nieto et al., 2015) and also directly regulate PIF4 expression in thermoresponsive growth (Raschke et al., 2015). Recently, ELF3 was implicated as a regulator of the shade avoidance response based on quantitative trait loci (QTL) mapping analysis in the Bay x Sha recombinant inbred line population (Coluccio et al., 2011, Jimenez-Gomez et al., 2010). However, the detailed mechanism governing ELF3 involvement in shade avoidance response is unclear.
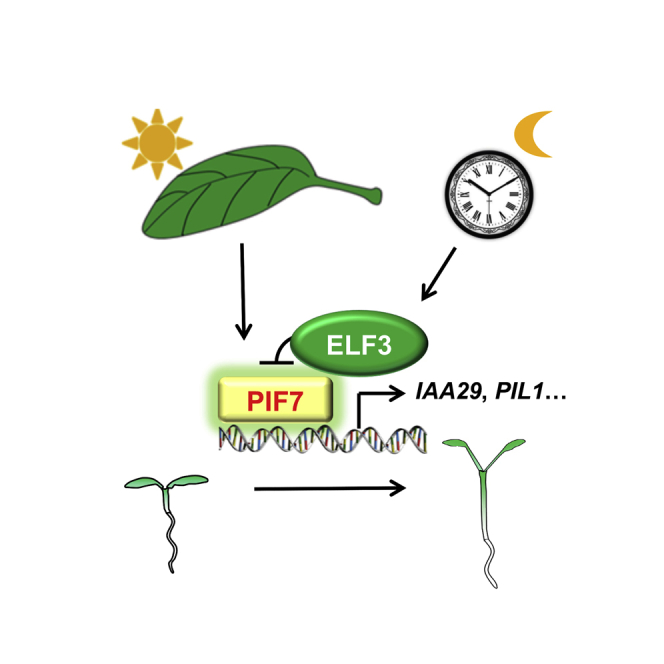
Here, we found that the time-specific inhibition of shade-induced gene expression and hypocotyl elongation appeared around CT16-CT20. ELF3 is responsible for this inhibition via interactions with PIF7 and represses PIF7's DNA-binding activity, antagonizing PIF7-induced gene expression and hypocotyl elongation. Our findings reveal that ELF3 affects the shade avoidance response with respect to circadian rhythm traits.
Results
Inhibition of the Shade Avoidance Response Occurs during the Night
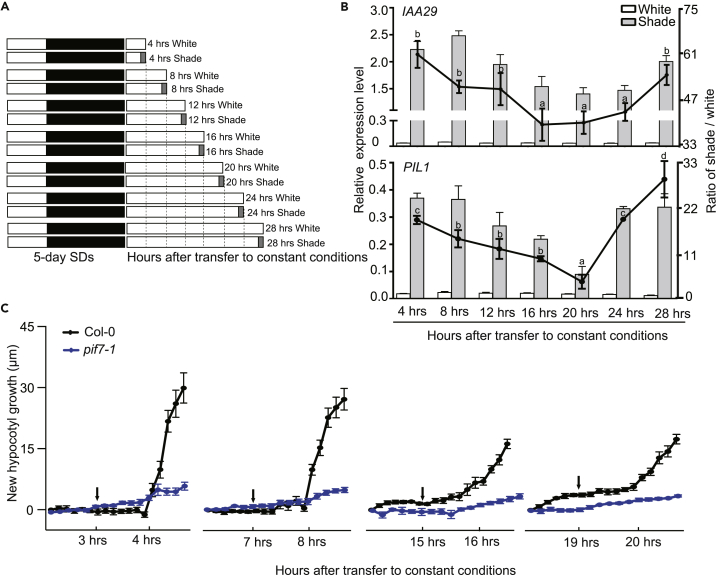
To determine whether shade-induced gene expression is gated by the circadian clock, wild-type seedlings were grown under short days (SDs) for 5 days and then transferred to continuous white light, under which the internal circadian clock of the plants was still functional. We considered these conditions to be the circadian conditions. Then, the seedlings were kept under white light or treated with shade light for 1 h every 4 h (Figure 1A), at which point the transcript levels of IAA29 and PIL1 were measured. As shown in Figure 1B, shade treatment induced the expression of these two genes from 4 to 8 h after transfer to constant conditions, whereas shade induction was inhibited from 16 to 20 h after transfer to constant conditions (Figure 1B, circadian conditions).
Figure 1.
Inhibition of the Shade Avoidance Response Occurs at Night
(A) Light treatment for the detection of shade-induced gene expression and hypocotyl growth. Wild-type seedlings were grown under short days (SDs) for 5 days and then transferred to continuous white light, after which the shade treatment was started or the seedlings were kept under continuous white light for 1 h every 4 h. The white, black, and gray colors represent white light, darkness, and shade, respectively.
(B) The expression levels of IAA29 and PIL1 when the seedlings were treated with shade or white light at different times of the day. The left y axis represents the relative expression level, and the right y axis represents the mean ratio of gene expression under shade and white light. The error bars indicate the SEMs of three independent studies. The lines marked with different letters denote significant differences (p < 0.05), calculated by Student's t test, in the mean ratio of gene expression.
(C) The effect of shifting the time of shade treatment on shade-induced hypocotyl growth. Col-0 and pif7-1 seedlings were grown under SD conditions for 5 days, transferred to continuous white light for 3 h/7 h/15 h/19 h and then treated with shade light for 2 h. The shade treatment times are indicated by arrows. The data are presented as the means with SEMs.
To assess whether shade-induced hypocotyl elongation is also gated by the circadian clock, we measured the new growth of hypocotyls during the first 2 h when SD-grown wild-type seedlings were treated with shade light at different times of the day using a seedling phenotyping platform (DynaPlant) (Figure 1C). We observed that the shade-induced growth response at 4 h was nearly identical to that at 8 h after transfer to constant conditions. However, shade-induced elongation was inhibited from 16 to 20 h after transfer to constant conditions (Figure 1C). Loss of function of PIF7 abolished the shade-induced hypocotyl elongation. Combined with shade-induced gene expression data, these data suggested that the inhibition of the shade avoidance response mainly occurs at night. As a master regulator for shade-induced hypocotyl elongation, PIF7 functions continuously.
Because IAA29 and PIL1 are targets of PIF7 and because PIF7 is a master regulator of shade-induced hypocotyl elongation, we were interested in whether the translational or transcript levels of PIF7 are gated by the circadian clock. However, shade light consistently dephosphorylates PIF7 regardless of the time (from 4 to 28 h after transfer to constant conditions, circadian conditions) (Figures S1A and S1B), indicating that the phosphorylation of PIF7 is regulated by light, but not the circadian clock. The phosphorylation and dephosphorylation of PIF7 consistently switched slowly following light conditions when the seedlings were kept under SD (light/dark cycle) conditions (diurnal conditions) (Figure S1C). Although the transcript level of PIF7 is lower at ZT12-ZT21 than that at ZT3-ZT6, the degree of oscillation is less than that of PIF4 (Figure S1D, diurnal conditions). Therefore, in addition to the weak transcriptional rhythm of PIF7, it is possible that a circadian component might be involved in the inhibition of shade induction at night.
ELF3 Is Involved in Shade-Induced Hypocotyl Elongation
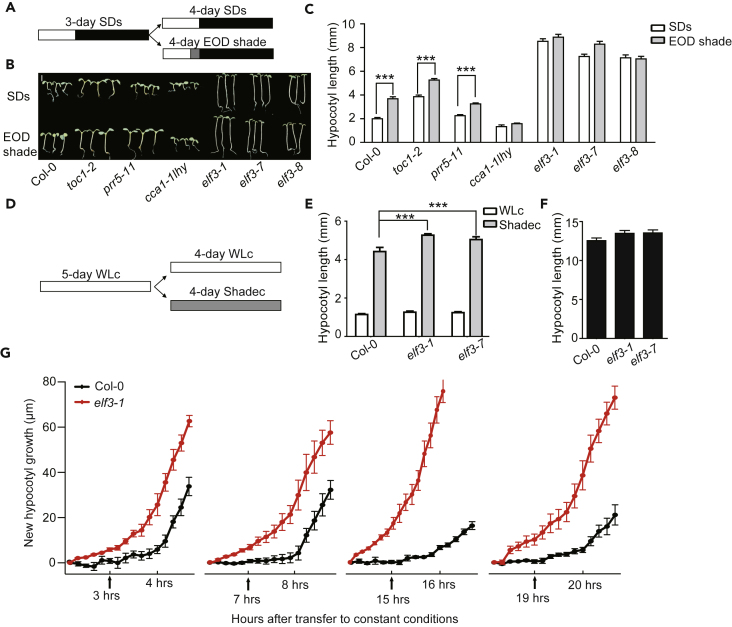
The circadian gating of the shade-induced increase in PIL1 and IAA29 transcript levels and hypocotyl elongation led us to investigate which circadian-related components are involved in this mechanism. We measured the hypocotyl lengths of SD-grown seedlings treated with end-of-day (EOD) shade in Col-0, toc1-2 (a mutant of TOC1 [Timing of CAB expression 1]), prr5-11 (a mutant of PRR5 [PSEUDO-RESPONSE REGULATOR 5]), cca1-1 lhy (a double mutant of CCA1 [Circadian Clock Associated 1] and LHY [Late elongated hypocotyl]), elf3-1, elf3-7, and elf3-8. Shade-induced hypocotyl elongation was manifested in Col-0, toc1-2, and prr5-11 but was diminished in cca1-1 lhy and the three elf3 mutants (Figures 2A–2C, diurnal conditions). Although TOC1 has been reported to interact with PIF3 to act as a gate of growth during predawn conditions (Soy et al., 2016) and interact with PIF4 to regulate the circadian gating of thermoresponsive growth (Zhu et al., 2016), toc1-2 displays a reduced but significant shade response. ELF3 and CCA1/LHY appear to be more important than TOC1 in the SAS. CCA1 has been reported to regulate hypocotyl elongation by modulating the transcription of DWARF4 (Zheng et al., 2018) and ELF3 (Lu et al., 2012, Reed et al., 2000). The maximal expression of ELF3 occurs around CT12-CT20 under SDs (Herrero et al., 2012, Liu et al., 2001, Nusinow et al., 2011), a time at which the repression of shade induction occurs. Therefore, ELF3 is a possible candidate to repress shade induction at night.
Figure 2.
ELF3 Might Be Involved in the Inhibition of Shade-Induced Gene Expression at Night
(A) Light treatment for the hypocotyl measurements of seedlings grown under SD conditions and SDs with end-of-day (EOD) shade conditions. Seedlings were grown for 3 days under SD conditions and either kept under SDs or treated with 2 h of shade at the end of each day for 4 days. The white, black, and gray colors represent white light, darkness, and shade, respectively.
(B) Images of the representative seedlings of Col-0 and circadian-related mutants (toc1-2, prr5-1, cca1-1 lhy, elf3-1, elf3-7 and elf3-8) under SDs or EOD shade treatment. Scale bar, 2 mm.
(C) Hypocotyl length of seedlings shown in (B). The error bars indicate SEMs of three independent measurements with at least 20 seedlings each. The asterisk indicates statistically significant differences between mean values according to Student's t test (***p < 0.001).
(D) Light treatment for the hypocotyl measurements of seedlings grown under continuous white light (WLc) or continuous shade (Shadec) conditions. Seedlings were grown for 5 days under WLc and either kept under WLc or transferred to Shadec conditions for 4 days. The white and gray colors represent white light and shade, respectively.
(E) Hypocotyl length of Col-0, elf3-1, and elf3-7 seedings grown under WLc or Shadec conditions. The data are presented as the means with SEMs; more than 20 seedlings were measured. The asterisk indicates statistically significant differences between mean values according to Student's t test (***p < 0.001).
(F) Hypocotyl length of Col-0, elf3-1, and elf3-7 seedlings grown in the dark. The data are presented as the means with SEMs; more than 20 seedlings were measured.
(G) The effect of shifting the time of shade treatment on the shade-induced hypocotyl growth of elf3-1. The seedlings were grown under SD conditions for 3 days and then transferred to continuous white light for 3 h/7 h/15 h/19 h and then treated with shade light for 2 h. The shade treatment times are indicated by arrows. The data are presented as the means with SEMs.
We measured the effects of shade on the transcriptional and translational levels of ELF3. As shown in Figures S2A and S2B (circadian conditions), there was no significant effect of shade treatment on the expression of ELF3 from 12 to 24 h after transfer to constant conditions. To measure the effects of shade on the translational level of ELF3, we obtained ELF3 antibodies (Ding et al., 2018). A western blot was performed to determine the specificity of the ELF3 antibodies, and the results are shown in Figure S2C. There was also no significant effect of shade treatment on the translational level of ELF3 at 16 or 20 h after transfer to constant conditions (Figure S2D).
We also measured the shade-induced hypocotyl elongation of elf3 mutants that were grown under continuous white light conditions and then transferred to shade conditions (Figures 2D and 2E). An enhanced shade response confirmed that ELF3 is a negative regulator of shade-induced hypocotyl elongation. Under dark conditions, elf3 mutants look like wild-type plants (Figure 2F). We then checked the new growth of hypocotyls during the first 2 h when SD-grown elf3-1 seedlings were treated with shade light at different times of the day. The results showed that the shade responses are almost the same at any time of day in elf3-1 (Figure 2G), indicating that ELF3 is involved in the inhibition of the shade response at night.
We took advantage of published RNA sequencing data of elf3-1 and checked the effect of ELF3 on the expression level of PIF7 targets (IAA29, PIL1, IAA19, YUCCA8, YUCCA9, IAA2, GH3.3, ATHB4, and HAT2) at different times under SDs (Ezer et al., 2017). The expression of all these genes dramatically increased at ZT12-ZT22 in elf3-1 (Figure S3A, diurnal conditions). The transcript levels of YUCCA8, IAA29, and PIL1 were confirmed by qRT-PCR (Figure S3B, diurnal conditions). We also obtained an ELF3ox overexpression line (Liu et al., 2001), which was confirmed by qRT-PCR (Figure S3C) and western blotting (Figure S3D). We then detected that the transcript levels of IAA29 and PIL1 were reduced in the ELF3ox line after shade treatment (Figure S3E), which suggests that ELF3 negatively moderates these genes' transcript levels.
ELF3 Physically Interacts with PIF7
It has been known that the circadian clock plays a pivotal role in the control of hypocotyl elongation by regulating the rhythmic expression of PIF4 and PIF5 (Nusinow et al., 2011). Therefore, we measured the transcript level of PIF7 in SD-grown Col-0 and elf3 at CT9-CT21. Compared with the expression of PIF4, the expression of PIF7 is not significantly altered in elf3 (Figure S4, diurnal conditions), indicating that there are other mechanisms of ELF3 in addition to the transcriptional regulation of PIF7.
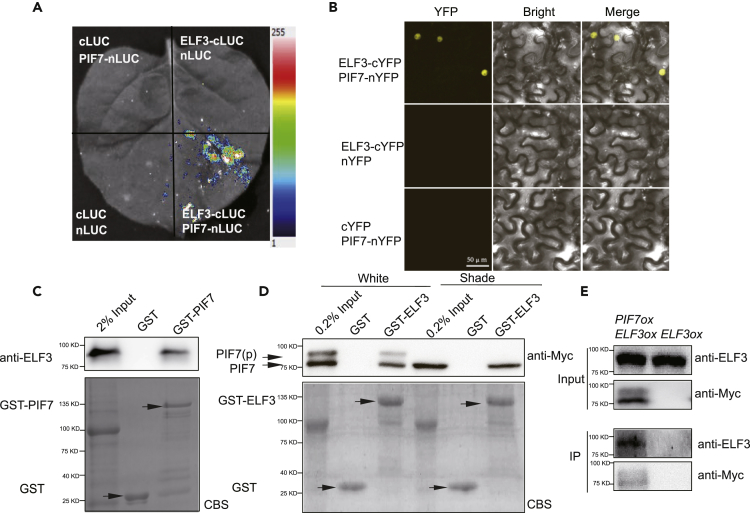
Recently, PIF7 was identified as a protein that is associated with ELF3 by affinity purification and mass spectrometry (AP-MS) (Huang et al., 2016). We examined the protein interaction by a luciferase complementation imaging (LCI) assay, which revealed that ELF3-cLUC could interact with nLUC-PIF7 when transiently expressed in Nicotiana benthamiana leaf cells (Figure 3A). A bimolecular fluorescence complementation assay also revealed that the interaction occurred in the nucleus (Figure 3B). In an in vitro pull-down experiment, ELF3 extracted from Col-0 seedlings directly bound with glutathione S-transferase (GST) fusion proteins of PIF7 (Figure 3C), and PIF7-Flash extracted from both white light-grown and shade-treated PIF7ox (35S:PIF7-Flash) seedlings (Li et al., 2012), could bind with the GST-fused ELF3 (Figure 3D). Coimmunoprecipitation experiments further confirmed that ELF3 precipitated with PIF7 (Figure 3E). These data indicate that ELF3 can physically interact with PIF7 both in vitro and in vivo.
Figure 3.
Interaction between PIF7 and ELF3
(A) Interactions between PIF7 and ELF3 were detected with a bimolecular fluorescence complementation assay based on firefly luciferase (LUC). The N- and C-terminal halves of LUC were fused to PIF7 and ELF3, respectively. Constructs were coexpressed in tobacco leaf cells. Luciferin was infiltrated before LUC activity was monitored.
(B) Bimolecular fluorescence complementation analysis of the interaction between PIF7 and ELF3 in tobacco leaf cells. The C-terminal half of YFP was fused to ELF3, and the N-terminal half of YFP was fused to PIF7. The constructs were cotransformed into tobacco leaf cells, and fluorescence images were obtained by confocal microscopy.
(C) The interaction between ELF3 extracted from plants and purified GST-PIF7 from E. coli in the GST pull-down assay. Top panel: the pull-down fractions and inputs were analyzed by western blots using anti-ELF3 antibodies. Bottom panel: Coomassie brilliant blue R250-stained (CBS) proteins on an SDS-PAGE gel are shown.
(D) The interaction between purified GST-fused ELF3 from E. coli and total protein extracts from plants overexpressing PIF7-Flash grown under white light conditions or treated with 1 h of shade, as indicated by GST pull-down assays. Top panel: the pull-down fractions and inputs were analyzed by western blots using anti-Myc antibodies. Bottom panel: Coomassie brilliant blue R250-stained (CBS) proteins on an SDS-PAGE gel are shown.
(E) Coimmunoprecipitation analysis of the interaction between PIF7 and ELF3. Anti-FLAG sepharose beads were used to precipitate PIF7-Flash from PIF7ox plants. Western blot was performed using anti-Myc and anti-ELF3 antibodies as indicated.
ELF3 Suppresses the Shade-Induced Expression of IAA29 and PIL1 by Preventing PIF7 from Binding DNA
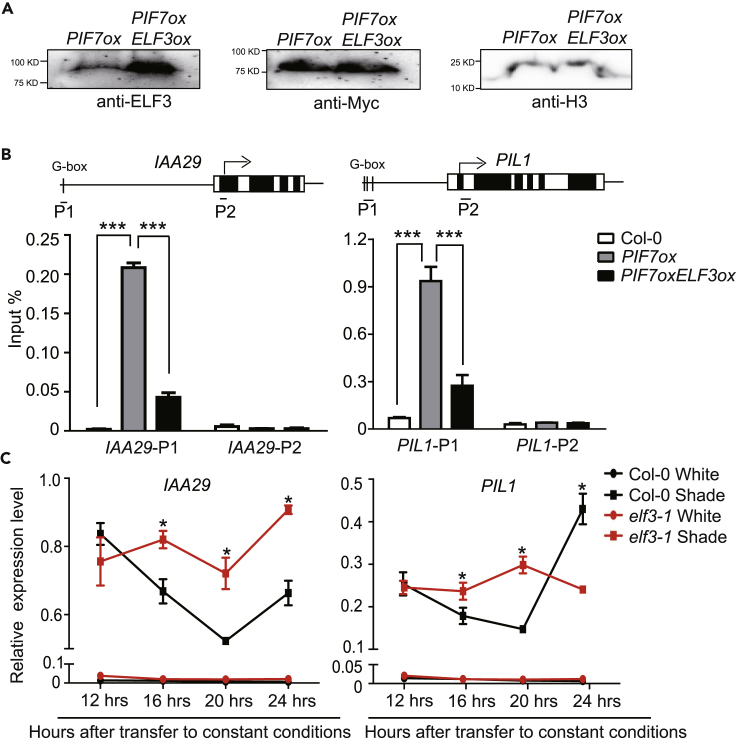
To explore the biological implications of this interaction, we generated PIF7oxELF3ox Arabidopsis lines, in which both the PIF7 and ELF3 genes were expressed under control of the 35S promoter. We then examined PIF7 and ELF3 protein levels in double-overexpression lines. As shown in Figure 4A, overexpression of ELF3 does not affect the protein level of PIF7 in continuous white light-grown seedlings. A chromatin immunoprecipitation-PCR assay that used primers that anneal to the G-box PIF7 recognition motifs and coding regions in IAA29 and PIL1 (Figures 4B and S5) was performed to detect the DNA-binding ability of PIF7 in both PIF7ox and PIF7oxELF3ox plants after shade treatment. The G-box regions were less enriched in the chromatin fractions from PIF7ox ELF3ox plants compared with PIF7ox plants (Figures 4B and S5). Moreover, no enrichment was detected with primers in the coding regions of IAA29 and PIL1. These results demonstrate that ELF3 suppresses PIF7 activity by a sequestration mechanism.
Figure 4.
ELF3 Suppresses IAA29 and PIL1 Expression by Inhibition of the DNA-Binding Activity of PIF7
(A) The PIF7-Flash and ELF3 protein levels in PIF7ox and PIF7oxELF3ox samples. Equal loading of samples is shown by anti-H3 antibodies.
(B) Chromatin immunoprecipitation-PCR analysis using anti-FLAG agarose at various chromatin regions of IAA29 and PIL1 chromatin in Col-0, PIF7ox, and PIF7oxELF3ox seedlings. Chromatin immunoprecipitation assays were performed via 4-day white light-grown seedlings treated with 1 h of shade. The top panels show a schematic of the gene structure of IAA29 and PIL1. The black boxes represent coding regions, and the white boxes represent untranslated regions. The G-box within the gene promoter is indicated. The bars labeled with numbers represent regions examined by PCR. The asterisk indicates statistically significant differences between mean values according to Student's t test (***p < 0.001).
(C) Relative expression of IAA29 and PIL1 in Col-0 and elf3-1 seedlings. Seedlings were grown under SD conditions for 5 days and then transferred to continuous white light, after which the shade treatment was started or the seedlings were kept under continuous white light for 1 h. The error bars indicate the SEMs of three independent experiments. The asterisk indicates statistically significant differences between mean values according to Student's t test (*p < 0.05).
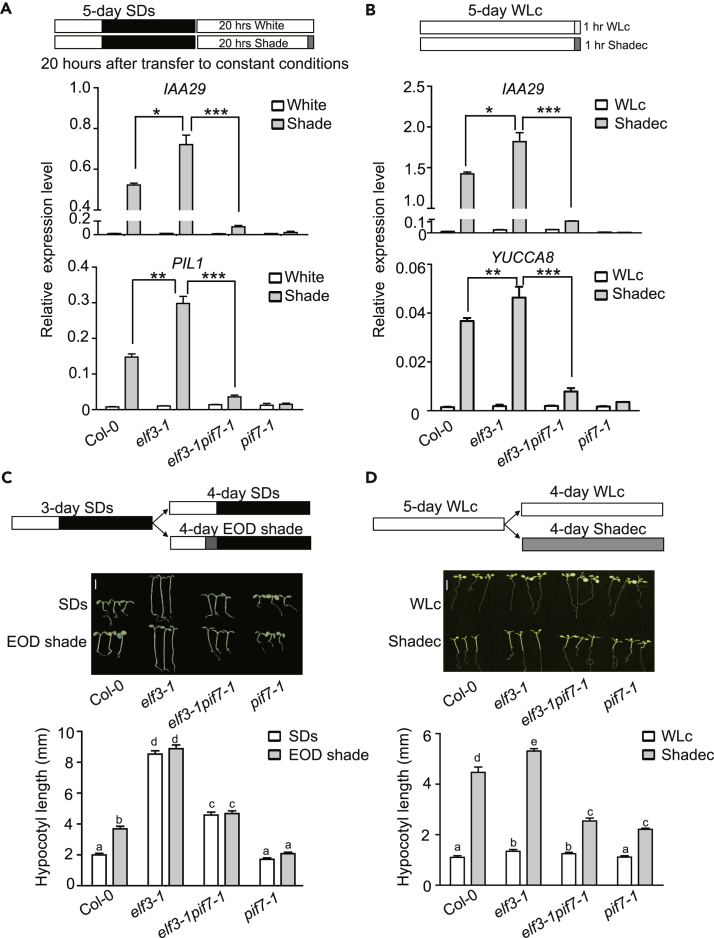
Then we would like to investigate whether loss of function of ELF3 could restore the downstream gene expression of PIF7 at night. The circadian clock regulates hypocotyl elongation by affecting cell elongation-related gene expression, and the expression of these genes is also modulated by light (Figure S3A). To exclude the effects of the circadian clock on the expression of these genes, we transferred the seedlings from diurnal conditions to continuous white light (Figures 1A and 4C, circadian conditions). Under these conditions, the expression of downstream genes such as IAA29 and PIL1 is strongly inhibited by white light (Figure 4C). As we expected, elf3 mutant restores the shade-induced expression of PIL1 and IAA29 from 16 to 20 h after transfer to constant conditions (Figure 4C, circadian conditions). These results suggest that ELF3 negatively regulates shade-induced gene expression by suppressing the DNA-binding activity of PIF7 during night in SD-grown seedlings.
ELF3 Acts Upstream of PIF7 in Shade-Induced Growth
To explore the genetic interactions between ELF3 and PIF7, we generated an elf3-1pif7-1 double mutant. The rescuing of the shade-induced expression of IAA29 and PIL1 at 16 h after transfer to constant conditions (Figure S6A, circadian conditions) and 20 h after transfer to constant conditions (Figure 5A, circadian conditions) by the loss of function of ELF3 was abolished again by a pif7 mutation, confirming that ELF3 acts upstream of PIF7 in shade-induced gene expression. We further tested the shade-induced expression of IAA29 and YUCCA8 in continuous white light-grown elf3-1 and elf3-1pif7-1 mutants. Similar expression patterns were found, which are shown in Figure 5B with Figure 5A, which suggests that the suppression of ELF3 on PIF7 activity also occurs under continuous white light conditions. Previous studies have shown that elf3 mutants are taller than phyB mutants in diurnal light/dark cycles but shorter than phyB mutants in cRL (Nieto et al., 2015), indicating that ELF3 plays a dominant role in the diurnal light/dark cycle, rather than in cRL. We investigated the hypocotyl length of seedlings grown under SD conditions or SDs with EOD shade treatments. The hypocotyl length of elf3-1pif7-1 double mutants is between that of elf3-1 and pif7-1 (Figure 5C), and the hypocotyl length of PIF7oxELF3ox is between that of PIF7ox and ELF3ox under SD conditions or EOD shade treatment (Figure S6B), which might result from redundant functions of PIF4 and PIF5 in circadian-regulated hypocotyl elongation (Hornitschek et al., 2009, Nieto et al., 2015, Nusinow et al., 2011). We further measured the hypocotyl length of seedlings that were grown under continuous white light (WLc) and then either kept in continuous white light or transferred to continuous shade conditions (Shadec), in which PIF7 is a dominant player. Under these conditions, the hypocotyl length of the elf3-1pif7-1 double mutant is more similar to that of pif7-1, which is consistent with the shade-induced gene expression results (Figure 5D), indicating the dominant action of PIF7. Together, these results suggested that ELF3 acts upstream of PIF7 in shade-induced gene expression and growth.
Figure 5.
ELF3 Acts Upstream of PIF7 in Shade-Induced Growth
(A) Relative expression of IAA29 and PIL1 in Col-0, elf3-1, elf3-1pif7-1 and pif7-1 seedlings at 20 h after transfer to constant conditions. The top panel represents the light treatment for the detection of shade-induced gene expression. Wild-type seedlings were grown under SD conditions for 5 days and transferred to continuous white light, after which the shade treatment was started or the seedlings were kept under continuous white light for 1 h. The white, black, and gray colors represent white light, darkness, and shade, respectively. The bottom panels represent the expression of IAA29 and PIL1. The error bars indicate the SEMs of three independent experiments. The asterisk indicates statistically significant differences between mean values according to Student's t test (*p < 0.05, **p < 0.01, ***p < 0.001).
(B) Relative expression of IAA29 and PIL1 in Col-0, elf3-1, elf3-1pif7-1, and pif7-1. The seedlings were grown under continuous white light (WLc) or transferred to continuous shade (Shadec) for 1 h. The error bars indicate the SEMs of three independent experiments. The asterisk indicates statistically significant differences between mean values according to Student's t test (*p < 0.05, **p < 0.01, ***p < 0.001).
(C) Hypocotyl phenotypes of Col-0, elf3-1, elf3-1pif7-1, and pif7-1 seedlings under SDs or end-of-day (EOD) shade treatment. The top panel represents the light treatment for the hypocotyl measurements of seedlings grown under SDs and EOD shade conditions. The seedlings were grown for 3 days under SD and either kept under SDs or treated for 2 h with shade at the end of each day for 4 days. The white, black, and gray colors represent white light, darkness and shade, respectively. The bottom panels represent the phenotypes of hypocotyl length. The data are presented as the means with SEMs; more than 20 seedlings were measured. The bars marked with different letters denote significant differences (p < 0.05), calculated by Student's t test. Scale bar, 2 mm.
(D) Hypocotyl length of Col-0, elf3-1, elf3-1pif7-1, and pif7-1 seedlings grown under WLc or Shadec conditions. The seedlings were germinated and grown for 5 days under continuous white light and either kept under white light or transferred to shade for 4 days. The data are presented as the means with SEMs; more than 20 seedlings were measured. The bars marked with different letters denote significant differences (p < 0.05), calculated by Student's t test. Scale bar, 2 mm.
Repression of ELF3 in the Shade Response Is Stronger under SDs than under LDs
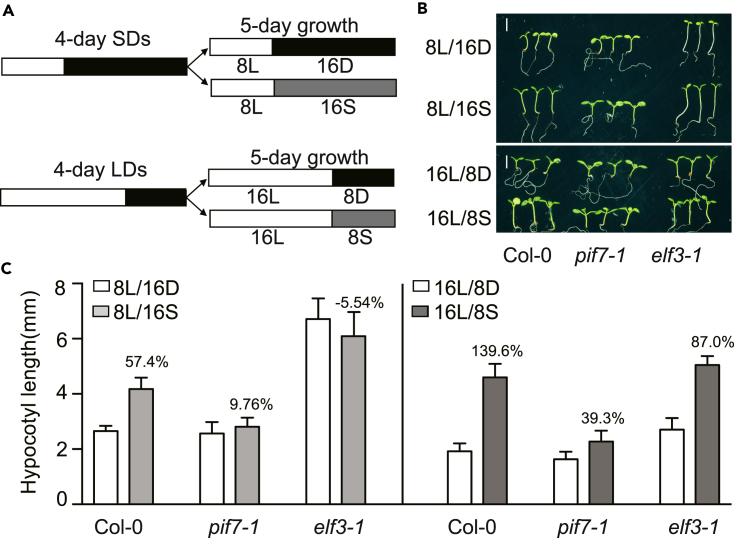
We found that ELF3 represses PIF7 binding activity at night. Therefore, we were interested in whether the effects of ELF3 are decreased under LD conditions with night-shade treatment compared with SDs with night-shade treatment (Figure 6A). The hypocotyl response to night shade was measured as the percent increase in hypocotyl length. As expected, the percent hypocotyl increase was 57.4% under SDs but 139.6% under LDs (Figures 6B and 6C). Although the shade duration is longer under SDs than under LDs, the stronger repression of ELF3 resulted in reduced shade response under SDs. These results suggest that PIF7 plays a major role in both conditions, whereas ELF3 is more important to night-shade treatment under SDs than under LDs.
Figure 6.
Repression of ELF3 in the Shade Response Is Stronger under SDs than under LDs
(A) Light treatment for the hypocotyl measurements of seedlings grown under SDs or LDs after night-shade treatment. The seedlings were germinated and grown for 4 days under SDs/LDs and either kept under SDs/LDs or transferred to shade for 5 days.
(B and C) Hypocotyl length of Col-0, pif7-1, and elf3-1 seedlings grown under different conditions. The data are presented as the means with SEMs; more than 20 seedlings were measured. The percent increase in response to shade is listed above the column. Scale bar, 2 mm.
Discussion
In the current study, we demonstrated that ELF3 negatively regulates shade-induced gene expression and hypocotyl elongation by suppressing the DNA-binding activity of PIF7 during ZT16-ZT20 both in SD-grown seedlings and in continuous white light-grown seedlings. Our data provide evidence that a core clock component directly connects to a transcription-centered signaling hub involved in various environmental sensory systems.
Similar to that which occurs with thermoresponsive growth (Zhu et al., 2016) and circadian-regulated growth, the circadian component is also involved in the inhibition of shade-induced growth at night (Figure 1). However, the detailed molecular mechanism is not the same. TOC1 has been reported to interact with PIF3 and repress PIF3 activity by preventing PIF3 from binding DNA (Soy et al., 2016). Moreover, TOC1 binds to PIF4 and suppresses thermomorphogenesis by directly repressing PIF4 activity without affecting its DNA-binding activity (Zhu et al., 2016). In our study, toc1-2 exhibited a reduced but significant shade response (Figures 2B and 2C). ELF3 is more important than TOC1 in the SAS. ELF3 can interact with PIF4 to inhibit the DNA-binding activity of PIF4 (Nieto et al., 2015). Here, it appears that a similar mechanism of ELF3 occurs with respect to PIF4 and PIF7. However, the PIF7 protein stability was not significantly affected by the overexpression of ELF3, whereas the protein stability of PIF4 and ELF3 influences each other. Although the PIF proteins belong to the same subfamily 15 of the Arabidopsis bHLH superfamily and similarly bind the Pfr form of phyB (Leivar and Quail, 2011), their regulatory mechanisms seem to be different in different physiological settings, indicative of additional layers of complexity for functional diversity. Owing to the importance of PIF7 in SAS, the biological significance of the PIF7-ELF3 interaction could be more relevant to shade response.
The circadian gating of shade-induced hypocotyl elongation has been previously published (Sellaro et al., 2012). In that study, the authors showed that afternoon shade events promote hypocotyl growth, whereas morning shade is ineffective. In their experiments, the seedlings returned to white light after a morning shade treatment, and darkness followed the afternoon shade treatment. The final phenotype was measured after 3 days. In the current study, we should emphasize that we did not check how the long-term phenotypes were affected by the timing of daily shade events. We investigated the short-term shade-induced gene expression and hypocotyl elongation and compared them between the daytime and nighttime (Figure 1). The window we focused on is the early stage of the shade response. Consistent with short-term changes in gene expression and hypocotyl elongation, there are related reports in which different shade starting points during the day result in the same growth rate pattern along short time scales via real-time monitoring methods (Cole et al., 2011, Gommers et al., 2017).
Our molecular and genetic tests present a compelling mechanistic model; however, it remains unclear why plants have evolved a mechanism for inhibiting shade responses at a time when shade signals are rare. The different shade responses under SDs and LDs are probably related to the various types of growth during the different seasons. However, thus far, more work needs to be done to understand the importance of the inhibition of ELF3 activity on PIF7 under shade.
Limitations of the Study
In this study, we revealed that, by repressing the activity of PIF7, ELF3 affects the expression of shade-induced genes (PIL1) involved in circadian rhythm traits. The regulatory circuit has both negative and positive gated points depending on the time of day a shade pulse has already occurred (Salter et al., 2003). However, the transcriptional regulation of PIF7 by PIL1 is still unknown. Therefore, ELF3 regulation of PIF7 target expression followed by ELF3-PIF7 direct protein interaction regulation creates a feedforward mechanism that would be interesting to investigate further. Additional studies are required to elucidate whether other circadian components such as CCA1/LHY are also involved in the circadian gating of the shade response.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Xingwang Deng (Peking University) for sharing the seeds of ELF3ox, elf3-1, elf3-7, and elf3-8 and Dr. Hongtao Liu (Shanghai Institutes for Biological Sciences-Institute of Plant Physiology and Ecology) for sharing the seeds of toc1-2, prr5-11, and cca1-1 lhy. This research was supported by the National Key R&D Program of China (2017YFA0503800).
Author Contributions
Y.J., C.Y., S.H., F.X., Y.X., and C.L. performed the experiments. L.L. conceived the project and wrote the paper.
Declaration of Interests
The authors declare that they have no competing or financial interests.
Published: December 20, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.11.029.
Supplemental Information
References
- Anwer M.U., Boikoglou E., Herrero E., Hallstein M., Davis A.M., James G.V., Nagy F., Davis S.J. Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. Elife. 2014;3:2206. doi: 10.7554/eLife.02206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwer M.U., Davis S.J. An overview of natural variation studies in the Arabidopsis thaliana circadian clock. Semin. Cell Dev. Biol. 2013;24:422–429. doi: 10.1016/j.semcdb.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Ballare C.L., Pierik R. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 2017;40:2530–2543. doi: 10.1111/pce.12914. [DOI] [PubMed] [Google Scholar]
- Bujdoso N., Davis S.J. Mathematical modeling of an oscillating gene circuit to unravel the circadian clock network of Arabidopsis thaliana. Front. Plant Sci. 2013;4:3. doi: 10.3389/fpls.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J. Shade avoidance. Arabidopsis Book. 2012;10:e0157. doi: 10.1199/tab.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B.Y., Helfer A., Nusinow D.A., Kay S.A. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 2012;7:170–173. doi: 10.4161/psb.18766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolfi A., Sessa G., Sassi M., Possenti M., Salvucci S., Carabelli M., Morelli G., Ruberti I. Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 2013;163:331–353. doi: 10.1104/pp.113.221549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole B., Kay S.A., Chory J. Automated analysis of hypocotyl growth dynamics during shade avoidance in Arabidopsis. Plant J. 2011;65:991–1000. doi: 10.1111/j.1365-313X.2010.04476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio M.P., Sanchez S.E., Kasulin L., Yanovsky M.J., Botto J.F. Genetic mapping of natural variation in a shade avoidance response: ELF3 is the candidate gene for a QTL in hypocotyl growth regulation. J. Exp. Bot. 2011;62:167–176. doi: 10.1093/jxb/erq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Wang S., Song Z.T., Jiang Y.P., Han J.J., Lu S.J., Li L., Liu J.X. Two B-box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep. 2018;25:1718–1728.e4. doi: 10.1016/j.celrep.2018.10.060. [DOI] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D., Jung J.H., Lan H., Biswas S., Gregoire L., Box M.S., Charoensawan V., Cortijo S., Lai X., Stockle D. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants. 2017;3:17087. doi: 10.1038/nplants.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers C.M.M., Keuskamp D.H., Buti S., van Veen H., Koevoets I.T., Reinen E., Voesenek L.A.C.J., Pierik R. Molecular profiles of contrasting shade response strategies in wild plants: differential control of immunity and shoot elongation. Plant Cell. 2017;29:331–344. doi: 10.1105/tpc.16.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., Davis S.J. Time for a nuclear meeting: protein trafficking and chromatin dynamics intersect in the plant circadian system. Mol. Plant. 2012;5:554–565. doi: 10.1093/mp/sss010. [DOI] [PubMed] [Google Scholar]
- Herrero E., Kolmos E., Bujdoso N., Yuan Y., Wang M.M., Berns M.C., Uhlworm H., Coupland G., Saini R., Jaskolski M. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M.J., Evans B.S., Briggs S.P., Hicks L.M., Kay S.A., Nusinow D.A. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics. 2016;15:201–217. doi: 10.1074/mcp.M115.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gomez J.M., Wallace A.D., Maloof J.N. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet. 2010;6:e1001100. doi: 10.1371/journal.pgen.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Herrero E., Bujdoso N., Millar A.J., Toth R., Gyula P., Nagy F., Davis S.J. A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell. 2011;23:3230–3246. doi: 10.1105/tpc.111.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24:1398–1419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ljung K., Breton G., Schmitz R.J., Pruneda-Paz J., Cowing-Zitron C., Cole B.J., Ivans L.J., Pedmale U.V., Jung H.S. Linking photoreceptor excitation to changes in plant architecture. Gene Dev. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wanger D.R. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Webb C.J., Knowles S.M., Kim S.H., Wang Z., Tobin E.M. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Oka H., Yoshimura F., Ishida K., Yamashino T. Insight into the mechanism of end-of-day far-red light (EODFR)-induced shade avoidance responses in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2015;79:1987–1994. doi: 10.1080/09168451.2015.1065171. [DOI] [PubMed] [Google Scholar]
- Nieto C., Lopez-Salmeron V., Daviere J.M., Prat S. ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr. Biol. 2015;25:187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farre E.M., Kay S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull R.J., Davis S.J. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 2017;40:2571–2585. doi: 10.1111/pce.13033. [DOI] [PubMed] [Google Scholar]
- Peng M., Li Z., Zhou N., Ma M., Jiang Y., Dong A., Shen W.H., Li L. Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol. 2018;176:1341–1351. doi: 10.1104/pp.17.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke A., Ibañez C., Ullrich K.K., Anwer M.U., Becker S., Glöckner A., Trenner J., Denk K., Saal B., Sun X. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 2015;15:197. doi: 10.1186/s12870-015-0566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Bastow R.M., Solomon K.S., Dowson-Day M.J., Elumalai R.P., Millar A.J. Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 2000;122:1149–1160. doi: 10.1104/pp.122.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- Sellaro R., Pacin M., Casal J.J. Diurnal dependence of growth responses to shade in Arabidopsis: role of hormone, clock, and light signaling. Mol. Plant. 2012;5:619–628. doi: 10.1093/mp/ssr122. [DOI] [PubMed] [Google Scholar]
- Sellaro R., Pacin M., Casal J.J. Meta-analysis of the transcriptome reveals a core set of shade-avoidance genes in Arabidopsis. Photochem. Photobiol. 2017;93:692–702. doi: 10.1111/php.12729. [DOI] [PubMed] [Google Scholar]
- Soy J., Leivar P., Gonzalez-Schain N., Martin G., Diaz C., Sentandreu M., Al-Sady B., Quail P.H., Monte E. Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U S A. 2016;113:4870–4875. doi: 10.1073/pnas.1603745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Xie F., Jiang Y., Li Z., Huang X., Li L. Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell. 2018;44:29–41.e4. doi: 10.1016/j.devcel.2017.11.017. [DOI] [PubMed] [Google Scholar]
- Zagotta M.T., Hicks K.A., Jacobs C.I., Young J.C., Hangarter R.P., Meeks-Wagner D.R. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- Zheng H., Zhang F., Wang S., Su Y., Ji X., Jiang P., Chen R., Hou S., Ding Y. MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell. 2018;30:67–82. doi: 10.1105/tpc.17.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Oh E., Wang T., Wang Z.Y. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 2016;7:13692. doi: 10.1038/ncomms13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.