Abstract
Prenylated flavonoids possess a wide variety of biological activities, including estrogenic, antioxidant, antimicrobial, and anticancer activities. Hence, they have potential applications in food products, medicines, or supplements with health-promoting activities. However, the low abundance of prenylated flavonoids in nature is limiting their exploitation. Therefore, we investigated the prospect of producing prenylated flavonoids in the yeast Saccharomyces cerevisiae. As a proof of concept, we focused on the production of the potent phytoestrogen 8-prenylnaringenin. Introduction of the flavonoid prenyltransferase SfFPT from Sophora flavescens in naringenin-producing yeast strains resulted in de novo production of 8-prenylnaringenin. We generated several strains with increased production of the intermediate precursor naringenin, which finally resulted in a production of 0.12 mg L–1 (0.35 μM) 8-prenylnaringenin under shake flask conditions. A number of bottlenecks in prenylated flavonoid production were identified and are discussed.
Keywords: metabolic engineering, Saccharomyces cerevisiae, de novo, prenylated flavonoids, naringenin, 8-prenylnaringenin
Introduction
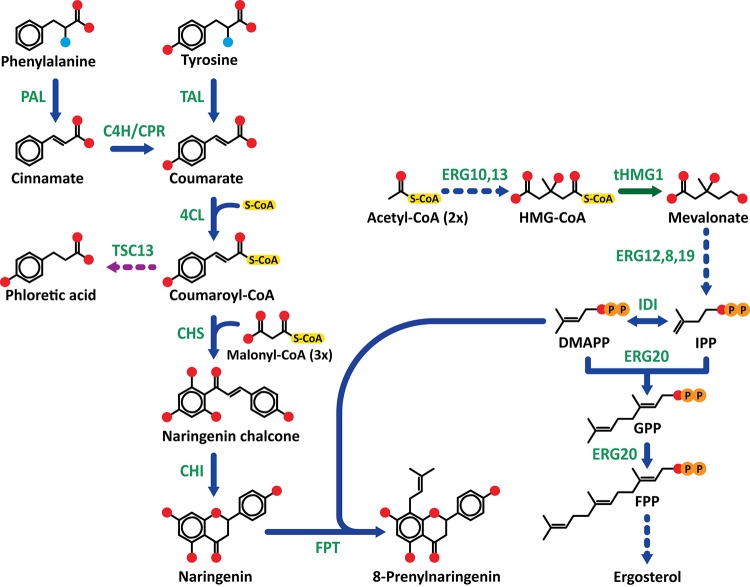
Prenylated flavonoids are a subclass of plant phenolics, which combine a flavonoid skeleton with a lipophilic prenyl side chain (see structures of naringenin and 8-prenylnaringenin in Figure 1). Unlike other flavonoids, they have a narrow distribution in plants, limited to only several plant families, including Leguminosae, Moraceae, and Cannabaceae.1,2 Prenylated flavonoids may act as phytoalexins and protect plants by their antimicrobial activity against pathogens.3 Plants that contain these compounds have frequently been applied as medicinal plants. Their pharmaceutical activities, including anticancer, anti-inflammatory, antimicrobial, and estrogenic biological activities, are often mediated by the prenylated flavonoids.4−6 It is hypothesized that the prenyl side chain improves the membrane permeability of prenylated flavonoids, which consequently confers enhanced biological activities.1,7−10 As a result of their health benefits, prenylated flavonoids are currently investigated as potential biopharmaceuticals and functional foods. Last but not least, prenylated (iso)flavonoids have also received interest because of their promising antimicrobial activity,11−14 suggesting that they might also be used as food preservatives or antibiotics in clinical applications.
Figure 1.
Representation of the 8-prenylnaringenin and isoprenoid biosynthesis pathways. Six A. thaliana genes were overexpressed: PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; CPR, cytochrome P450 reductase; 4CL, 4-coumarate-CoA ligase; CHS, chalcone synthase; and CHI, chalcone isomerase; one gene from S. flavescens: FPT, flavonoid prenyltransferase; one gene from R. capsulatus: TAL, tyrosine ammonia lyase; and one truncated gene from S. cerevisiae: tHMG1, truncated 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase. All enzyme names are in green. Dark blue arrows indicate the 8-prenylnaringenin and isoprenoid biosynthesis pathways. The purple dashed arrow indicates a route to the side-product phloretic acid, which is produced by the catalytic activity of endogenous TSC13 (a double-bond reductase). The green arrow indicates a truncated and deregulated copy of the rate-limiting enzyme HMG-CoA reductase (tHMG1).
A prenylated flavonoid with a very potent phytoestrogen activity is 8-prenylnaringenin, which is produced in several plant species, including Sophora flavescens and Humulus lupulus (hops).15,16 As a result of its estrogenic activity, 8-prenylnaringenin may have an application in treating the adverse symptoms of menopause, such as hot flashes and increased risk of osteoporosis, that result from changes in hormonal levels in women.17−19 Prenylated flavonoids are complex molecules, and to produce them by chemical synthesis is often challenging.20 In plants, they are present at low levels: for example, 8-prenylnaringenin is present at ±50 ppm in hop tissues.21 Clearly, large-scale purification of 8-prenylnaringenin from plants for use as a phytoestrogen supplement would likely not be technically and economically feasible. As an alternative, microbial production systems may be used for production of plant compounds. Microbial production systems for plant compounds offer the advantage of a predictable yield, price, and quality, and such systems have been implemented at commercial scale for an increasing number of plant molecules, including pharmaceuticals, antioxidants, and fragrance molecules.22,23
Flavonoids have been successfully produced in microorganisms by the expression of enzymes from the plant biosynthetic pathways in the microorganism.24,25 In plants, the flavonoid biosynthetic pathway starts from l-phenylalanine (Figure 1). By action of three enzymes, phenylalanine is converted to coumaroyl-CoA, which is subsequently condensed with three molecules of malonyl-CoA to naringenin chalcone. Naringenin chalcone is then isomerized to naringenin, which can be further converted to other flavonoids by a variety of other enzymes. Attempts to produce naringenin in the yeast Saccharomyces cerevisiae included deregulation of aromatic amino acid synthesis and introduction of phenylalanine ammonia lyase (PAL), tyrosine ammonia lyase (TAL), cinnamate 4-hydroxylase (C4H), cytochrome P450 reductase (CPR), 4-coumaric acid-CoA ligase (4CL), chalcone synthase (CHS), and chalcone isomerase (CHI) from Arabidopsis thaliana, which resulted in 400 μM naringenin in the culture medium, starting from glucose.26 Similar studies have been performed in bacteria, including Escherichia coli and Corynebacterium glutamicum.27,28
To produce prenylated flavonoids, a prenyltransferase would need to be added to the microorganism. Flavonoid prenylation in plants is mediated by flavonoid-specific prenyltransferases. These enzymes are membrane-bound proteins, comprising a number of transmembrane helices, related to the ubiA protein involved in ubiquinone biosynthesis.29 In plants, prenyltransferases are often localized to plastids.30,31 Dimethylallyl diphosphate (DMAPP) is the main prenyl donor accepted by these enzymes, although some enzymes also accept longer prenyl donors, such as geranyl diphosphate (GPP) and farnesyl diphosphate (FPP). A well-characterized example of a flavonoid prenyltransferase is SfFPT from S. flavescens, which was shown to display a high specific activity for naringenin and DMAPP as substrates.32
Whereas naringenin production has been demonstrated in both eukaryotic and prokaryotic microorganisms, yeast may provide a favorable chassis for producing 8-prenylnaringenin, because it is capable of de novo production of naringenin and a number of prenyltransferases have been successfully expressed in S. cerevisiae.32−34 Another issue of importance is the availability of prenyl donors, such as DMAPP, which is often tightly regulated in microorganisms. In yeast, prenyl diphosphates function as intermediates in the ergosterol biosynthetic pathway.35 Availability of longer chain prenyl diphosphates, such as FPP, has been enhanced in yeast, mainly for the production of sesquiterpenoids, such as artemisinin.36 However, it is as yet not clear whether such engineering also results in a pool of available DMAPP in yeast for flavonoid prenylation. Therefore, engineering production of 8-prenylnaringenin in yeast may pose additional challenges, because alongside the availability of phenylalanine, also the availability of prenyl donors may need to be engineered. In this paper, we investigate S. cerevisiae as a production host for plant-derived prenylated flavonoids. As a proof of principle, we produced the potent phytoestrogen 8-prenylnaringenin from glucose.
Materials and Methods
Strains and Maintenance
The S. cerevisiae strains used in this work are listed in Table 1. In general, yeast strains were grown and maintained on synthetic medium (6.8 g/L yeast nitrogen base without amino acids) with 20 g/L glucose (SMG medium) and appropriate growth factors to supplement the specific auxotrophic requirements of the strains (30 mg/L uracil, 125 mg/L histidine, 50 mg/L tryptophan, and 200 mg/L leucine). Some strains were grown in the above medium supplemented with 250 μM naringenin (SMNar medium), corresponding to de novo naringenin titers achieved in shake flasks.26E. coli DH5α electrocompetent cells were used for bacterial transformations. For plasmid propagation, E. coli DH5α cells were cultured in lysogeny broth (LB) medium supplemented with ampicillin (100 mg/L) at 37 °C with 250 rpm agitation. Optical density was measured at 600 nm using an Ultraspec 10 cell density meter (Amersham Biosciences). Glycerol stocks were prepared by adding a final concentration of 20% (v/v) glycerol to the culture, and 1.6 mL aliquots were stored at −80 °C.
Table 1. S. cerevisiae Strains Used in This Study.
| name | relevant genotype | contains plasmid | origin |
|---|---|---|---|
| IMK393 | MATalpha ura3-52 his3-Δ1 leu2-3,112 trp1-289 MAL2-8cSUC2 | (26) | |
| Δaro3::loxP ARO4G226S pdc6Δ::loxP pdc5Δ::loxP aro10Δ::loxP | |||
| PATW066 | IMK393 | (39) | |
| X-2::TEF1P-At4CL3-TEF1T TPI1P-AtCHS3-ADH1T TDH3P-AtCHI1-CYC1T | |||
| XII-2::TDH3P-AtPAL1-CYC1T TPI1P-coC4H-ADH1T PGI1P-coCPR1-PGI1T TEF1P-coCHS3-TEF1T | |||
| PATW083 | PATW066 | p414-TEF1p-Cas9-CYC1t | this study |
| PATW088 | PATW066 tsc13Δ::TSC13P-coMdECR-TSC13T | this study | |
| PATW089 | PATW088 | p414-TEF1p-Cas9-CYC1t | this study |
| PATW103 | PATW088 spr1Δ::TDH3P-coTAL1-CYC1T | this study | |
| PATW104 | PATW103 | p414-TEF1p-Cas9-CYC1t | this study |
| PPF3 | IMK393 | pMEN2 | this study |
| PPF4 | PATW066 | pMEN2 | this study |
| PPF5 | PATW088 | pMEN2 | this study |
| PPF6 | PATW103 | pMEN2 | this study |
Molecular Biology Techniques
All primers were supplied by Integrated DNA Technologies and are listed in Table 2. DNA amplification was performed by polymerase chain reaction (PCR) using Q5 High-Fidelity DNA polymerase (New England Biolabs), and PCR conditions were adapted to the instructions of the manufacturer. Plasmids were isolated from E. coli using the NucleoSpin Plasmid EasyPure kit (Macherey-Nagel). Restriction enzymes were obtained from New England Biolabs. DNA fragments were separated in 1% (w/v) agarose gel. DNA fragments were purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). DNA concentrations were measured with a NanoDrop spectrophotometer. Yeast transformations were performed with the lithium acetate method.37 Yeast plasmid and genomic DNA was extracted as previously described.38
Table 2. Primers Used in This Study.
| name | sequence | description |
|---|---|---|
| Assembly of pMEN2 | ||
| MH5 | CAGAAATGACTGTTTTATTGGTTAAAACCATAAAACTTAGATTAGATTGCTATGCTTTC | fragment GAPP-TEF1P |
| MH6 | AGAAAGCATAGCAATCTAATCTAAGTTTTATGGTTTTAACCAATAAAACAGTCATTTCTG | fragment tHMG1 |
| MH14 | GACAAGTTCTTGAAAACAAGAATCTTTTTATTGTCTTAGGATTTAATGCAGGTGACG | fragment tHMG1 |
| MH15 | GTCCGTCACCTGCATTAAATCCTAAGACAATAAAAAGATTCTTGTTTTCAAGAACTTGTC | fragment TEF1T-AmpR-2 μm |
| MH16 | AAAACACCAGAACTTAGTTTCGACGGATTCATGGGTTCTATGTTGTTGGCTTC | fragment coSfFPT |
| MH17 | TGGGAAAGAAGCCAACAACATAGAACCCATGAATCCGTCGAAACTAAGTTCTG | fragment GAPP-TEF1P |
| MH18 | GACTACTTCTTGATCCCATTGTTCAGATAATCATGTAATTAGTTATGTCACGCTTAC | fragment 2 μm-HIS3-CYC1T |
| MH19 | AATGTAAGCGTGACATAACTAATTACATGATTATCTGAACAATGGGATCAAGAAGT | fragment coSfFPT |
| ML009 | TCGGTATAGAATATAATCGGGGATGCC | fragment TEF1T-AmpR-2 μm |
| ML010 | GCGTTTACTGATTACTAGCGAAGCTG | fragment 2 μm-HIS3-CYC1T |
| CP100 | CGGTCTTCAATTTCTCAAGTTTCAG | diagnostic pMEN2 |
| MH20 | GGGTCTCTAACTTGTGGTTCG | diagnostic pMEN2 |
| MH21 | GATGCTAATACAGGAGCTTCTGC | diagnostic pMEN2 |
| ML050 | GGGACCTAGACTTCAGGTTGTC | diagnostic pMEN2 |
| MM25 | CTCTTAGCGCAACTACAGAGAACAGG | diagnostic pMEN2 |
| MM28 | ACCAGCATTCACATACGATTGACG | diagnostic pMEN2 |
| TSC13::coMdECR Integration | ||
| CP182 | GCTATCTAGAAACCAATTGAGCTATTTGAGAGAGATACATATTTTGAATTTAATTTGAAAATGAAGGTTACTGTTGTTTC | coMdECR repair fragment |
| CP183 | CCACTTCGTGAAAGCTAATATCTCTTTACCTTGCATTTGGGCATGTTGCAAACAGGAGGATTACAAGAATGGTGGCAAG | coMdECR repair fragment |
| CP184 | TGAAAAGGGACTAAGAGCGTG | diagnostic TSC13_int |
| CP185 | GATGAAAGCACCGAAAGACC | diagnostic TSC13_int |
| CP186 | GACTTTGCCAGTTCAACCAGG | diagnostic TSC13_int |
| CP187 | TGCTACTACGCCACTTCGTG | diagnostic TSC13_int |
| SPR1::coTAL1 Integration | ||
| CP190 | ACACCTTCTTTATTCGAGACTTTCCGTACTAATCCGTACAACGATGACGGTATTCCTGTTTGTAAAACGACGGCCAGT | coTAL1 repair fragment |
| CP169 | CTTCAAAAGCAAATTTTTCAATCTTTCCATGTCAATAACTGGACCTAACGGTTCATTGAGGCAAATTAAAGCCTTCGAGC | coTAL1 repair fragment |
| CP162 | GGTGGGTGGCTAGTATTGGAG | diagnostic SPR1_int |
| CP163 | GATGGTCAATTATGACGCCATATTCG | diagnostic SPR1_int |
| CP174 | ATTAATGGAAGTTTTGAGTGGTCATG | diagnostic SPR1_int |
| CP175 | CGTCTTGTGCAGGATGATC | diagnostic SPR1_int |
Plasmid Construction
The episomal expression vector pMEN2 was assembled in vivo from five separate DNA fragments using 60 bp homologous recombination sequences. The S. flavescensSfFPT (coSfFPT; GenBank accession number AHA36633) gene was ordered as a yeast codon-optimized synthetic gBlock gene fragment from Integrated DNA Technologies. The truncated HMG1 gene fragment (tHMG1) was amplified from genomic DNA of S. cerevisiae IMK393. The fragments TEF1T-AmpR-2 μm, GAPP-TEF1P, and 2 μm-HIS3-CYC1T were amplified from plasmid pUDE188.26 Correct assembly of the plasmid was confirmed via restriction enzyme analysis and sequencing (Macrogen). All plasmids used in this study are listed in Table 3.
Table 3. Plasmids Used in This Study.
| name | relevant characteristics | origin |
|---|---|---|
| p414-TEF1p-Cas9-CYC1t | centromeric plasmid, AmpR, TRP1, TEF1p-Cas9-CYC1t | (56); Addgene 43802 |
| p426-SNR52p-gRNA.SPR1.Y-SUP4t | 2 μm ori, AmpR, URA3, gRNA-SPR1.Y | (39) |
| p426-SNR52p-gRNA.TSC13.Y-SUP4t | 2 μm ori, AmpR, URA3, gRNA-TSC13.Y | (39) |
| pMEN2 | 2 μm ori, AmpR, HIS3, TEF1P-tHMG1-TEF1T, GAP1P-coSfFPT-CYC1T | this study |
| pUDE188 | template for TEF1T-AmpR-2 μm, GAPP-TEF1P, 2 μm-HIS3-CYC1T | (26) |
| pUDI069 | template for coTAL1 | (26) |
Strain Construction
Integration of gene fragments and knockout of genes was obtained using CRISPR-Cas9, as previously described.39S. cerevisiae strain PATW066 was transformed with p414-TEF1p-Cas9-CYC1t, yielding strain PATW083. The native open reading frame of TSC13 was replaced by its gene orthologue from Malus × domestica (MdECR; GenBank accession number XP_008382818), as described previously.39 Strain PATW083 was transformed with the gRNA.TSC13 plasmid and the coMdECR integration fragment. Correct integration of coMdECR was verified by colony PCR and sequencing. After gRNA.TSC13 and Cas9 plasmid removal, this resulted in strains PATW088 and PATW089 (Table 1).
The integration of Rhodobacter capsulatuscoTAL1 (RcTAL1; GenBank accession number WP_013066811) was combined with a SPR1 knockout, as described previously.39 Strain PATW089 was transformed with the gRNA.SPR1 plasmid and the coTAL1 integration fragment. Correct integration was verified by colony PCR and sequencing. After gRNA and Cas9 plasmid removal, this resulted in strains PATW103 and PATW104 (Table 1).
Strains IMK393, PATW066, PATW088, and PATW103 were transformed with construct pMEN2, resulting in strains PPF3, PPF4, PPF5, and PPF6, respectively (Table 1).
Sample Preparation and Analytical Methods
Chemicals and Standards
Ethyl acetate for extractions was purchased from Biosolve (Valkenswaard, Netherlands). Ultra-high-performance liquid chromatography–mass spectrometry (UHPLC–MS)-grade methanol (MeOH), acetonitrile (ACN) with 0.1% (v/v) formic acid (FA), and water with 0.1% (v/v) FA were purchased from Biosolve (Valkenswaard, Netherlands). Standards of naringenin (≥98%, w/w) and 8-prenylnaringenin (≥98%, w/w) were purchased from Cayman Chemical (Ann Arbor, MI, U.S.A.). A standard of 6-prenylnaringenin (≥98%, w/w) was purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.).
Production of 8-Prenylnaringenin in S. cerevisiae
Strains PATW066 and PPF4 were cultured in duplicate under shake flask conditions on SMG and SMNar medium for 120 h. Shake flask cultures were grown in 250 mL shake flasks with 50 mL of medium at 30 °C while shaking at 250 rpm. The shake flasks were inoculated to an OD600 of 0.2 with cells, resuspended in 2 mL of medium, and obtained from a preculture grown in similar conditions. For measurement of intra- and extracellular 8-prenylnaringenin, medium and biomass were separated by centrifugation. The medium was extracted twice with 10 mL of ethyl acetate using a separation funnel. The cell pellet was lyophilized and extracted twice with 10 mL of ethyl acetate. The soluble ethyl acetate portion of extracted medium or cell pellet was collected and dried under a stream of nitrogen. Dried compounds were dissolved in 0.5 mL of absolute ethanol (Merck, Darmstadt, Germany).
Composition analysis of the ethanol extracts was performed according to a similar method (i.e., same column, eluents, and mass spectrometer) as described for quantification (see the next section), with some adaptations. In short, 1 μL was injected on a Accela UHPLC system (Thermo Scientific, San Jose, CA, U.S.A.) equipped with a pump, autosampler, and photodiode array (PDA) detector coupled in-line to a Velos Pro mass spectrometer (Thermo Scientific). UHPLC separation was performed at 35 °C, and the flow rate was 300 μL min–1. The elution program was started by running isocratically at 5% B for 1.5 min, followed by 1.5–20 min linear gradient to 100% B, and 20–25 min isocratically at 100% B.
Mass spectrometric (MS) data were acquired in negative mode over the m/z range of 150–1500. The source conditions used were a capillary temperature of 400 °C, source heater temperature of 50 °C, source voltage of 3.5 kV, and S-lens radio frequency (RF) level of 61.36. Nitrogen was used as sheath gas (20 arbitrary units) and auxiliary gas (10 arbitrary units).
Standard solutions of naringenin, 8-prenylnaringenin (8-PN), and 6-prenynaringenin (6-PN) were used for screening and identification.
Effect of Enhanced Naringenin Biosynthesis on 8-Prenylnaringenin Production
Strains PPF3, PPF4, PPF5, and PPF6 were cultured in triplicate under shake flask conditions on SMG medium for 140 h. Shake flask cultures were grown in 250 mL shake flasks with 50 mL of medium at 30 °C while shaking at 250 rpm. The shake flasks were inoculated to an OD600 of 0.2 with cells, resuspended in 2 mL of medium, and obtained from a preculture grown in similar conditions. At the end of culturing, the whole culture was extracted with 10 mL of ethyl acetate using a separation funnel. The soluble ethyl acetate portion was collected, and 2 mL was dried by SpeedVac. Dried compounds were dissolved in 3 mL of methanol.
All dilutions were made in methanol. Naringenin and 8-prenylnaringenin were quantified using a Vanquish UHPLC system (Thermo Scientific) equipped with a pump, autosampler, and photodiode array detector. The flow rate was 400 μL min–1, of which two-thirds (266 μL min–1) was directed toward the mass spectrometer by a splitter behind the PDA detector. MS data were collected on a Velos Pro linear ion trap mass spectrometer (Thermo Scientific) equipped with a heated electrospray ionization (ESI) probe (Thermo Scientific).
For UHPLC separation, the preheater was set to 45 °C, the column compartment heater was set to 45 °C, and the post-column cooler was set to 40 °C. The column used was an Acquity UPLC BEH C18 (150 × 2.1 mm inner diameter, 1.7 μm) with a VanGuard guard column (5 × 2.1 mm inner diameter, 1.7 μm) of the same material (Waters, Milford, MA, U.S.A.). Eluents used were water (A) and ACN (B), both with 0.1% (v/v) FA. The elution program was started by running isocratically at 10% B for 1.09 min, followed by 1.09–7.45 min linear gradient to 80% B, 7.45–8.54 min linear gradient to 100% B, and 8.54–13.99 min isocratically at 100% B. The eluent was adjusted to its starting composition in 1.09 min, followed by equilibration for 5.41 min. Detection wavelengths for ultraviolet–visible (UV–vis) were set to the range of 190–680 nm, and data were recorded at 20 Hz.
Full MS data were collected in negative ionization mode over the m/z range of 200–800. Additionally, selected ion monitoring (SIM) was performed for naringenin (m/z 271) and 8-prenylnaringenin (m/z 339) with an isolation width of 1.0. The source conditions used were a capillary temperature of 254 °C, source heater temperature of 408 °C, source voltage of 2.5 kV, and S-lens RF level of 68.85. Nitrogen was used as sheath gas (50 arbitrary units), auxiliary gas (13 arbitrary units), and sweep gas (2 arbitrary units).
Data were processed using Xcalibur 4.1 (Thermo Scientific). Naringenin was quantified on the basis of UV absorbance at 280 nm with a calibration curve of the standard ranging from 0.1 to 50 μg mL–1 in MeOH (R2 = 0.9997). 8-Prenylnaringenin was quantified on the basis of SIM with a calibration curve of the standard ranging from 0.01 to 1.0 μg mL–1 in MeOH (R2 = 0.9996).
Results
Production of 8-Prenylnaringenin in S. cerevisiae
In previous work, the S. cerevisiae strain PATW066 was developed, which was capable of producing naringenin up to a concentration of 40 μM in the culture medium.39 PATW066 (aro3Δ, ARO4G226S, pdc6Δ, pdc5Δ, aro10Δ, atPAL1↑, coC4H↑, coCPR1↑, atCHI1↑, atCHS3↑, coCHS3↑, and at4CL3↑) has been engineered to overproduce aromatic amino acids, and all naringenin pathway genes have been integrated in its genome. Therefore, it provides a good platform for the current study. Two prenyltransferases, SfN8DT-1 and SfFPT from the plant S. flavescens, have been described that can prenylate naringenin specifically at the C-8 position.30,32 We selected the enzyme SfFPT as a prenyltransferase, because it was reported to have higher affinity and catalytic efficiency with the substrates naringenin and DMAPP than SfN8DT-1.32 Moreover, a truncated form of S. cerevisiae 3-methylglutaryl coenzyme A reductase (tHMG1) was used, to supply DMAPP. HMG1 is the key regulatory enzyme of the mevalonate pathway in yeast. Truncated versions of this enzyme, lacking 530 amino acids of the N terminus, have been shown to promote availability of prenyl building blocks for terpene biosynthesis in several host systems, including yeast.36 Overexpression of a cytosolic HMG-CoA reductase leads to squalene accumulation in yeast.40,41 We anticipated that it would also contribute to availability of DMAPP.
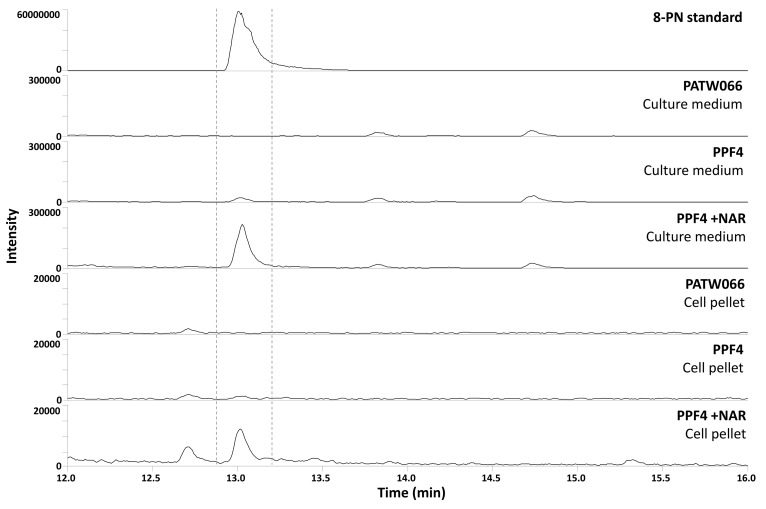
The episomal expression vector pMEN2, carrying the yeast codon-optimized version of S. flavescensSfFPT and a copy of tHMG1, was constructed. Plasmid pMEN2 was transformed into the naringenin-producing strain PATW066, resulting in strain PPF4. To investigate the formation of prenylated flavonoids, strains PATW066 and PPF4 were cultured in shake flasks on SMG and SMNar medium (supplemented with 250 μM naringenin) with glucose as the sole carbon source for 120 h. Supplementation with naringenin was performed to increase substrate availability for 8-prenylnaringenin production. At the end of 120 h, biomass was harvested. The culture media and yeast cell pellets were extracted and analyzed for 8-prenylnaringenin using UHPLC–MS (Figure 2 and Figure S1 of the Supporting Information). Production of 8-prenylnaringenin was detected in the culture medium of PPF4 cultures and not in the culture medium of control strains. Only trace amounts of 8-prenylnaringenin was detected in the pellet fraction of PPF4 cultures, suggesting that, like naringenin, the majority of 8-prenylnaringenin is exported; however, their export mechanism has not yet been elucidated. Other prenylated flavonoids, such as 6-prenylnaringenin, were not detected. This experiment showed that de novo 8-prenylnaringenin production is possible in S. cerevisiae. Interestingly, cultures of PPF4 that were supplemented with naringenin showed approximately 12-fold higher production of 8-prenylnaringenin (on the basis of peak areas), indicating that production of 8-prenylnaringenin is limited by availability of naringenin in strain PPF4.
Figure 2.
UHPLC–MS-extracted chromatograms of m/z 339 (negative mode) of culture media and cell pellets of S. cerevisiae strains PATW066 and PPF4. The strains were cultured in duplicate in shake flasks on SMG and SMNar medium (+NAR). At the end of 120 h cultivation, biomass was harvested. The culture media and cell pellets were both extracted with ethyl acetate. One representative of each duplicate shake flask culture is shown. 8-Prenylnaringenin (8-PN) standard was injected at 250 μM.
Effect of Enhanced Naringenin Biosynthesis on 8-Prenylnaringenin Production
As a next step, strain PATW066 was engineered for enhanced naringenin production, with the aim to improve de novo 8-prenylnaringenin production. Strain PATW066 not only showed naringenin production up to a concentration of 40 μM but also production of phloretic acid up to 160 μM.39 Recently, S. cerevisiaeTSC13, an essential endogenous double-bond reductase involved in fatty acid synthesis, was identified as the responsible enzyme for the formation of phloretic acid via the reduction of coumaroyl-CoA. When TSC13 was replaced by MdECR, an orthologue from apple (M. domesticus), phloretic acid production was eliminated and naringenin production improved.42 Therefore, the TSC13 coding sequence in strain PATW066 was replaced with a yeast codon-optimized version of MdECR (coMdECR), which resulted in strain PATW088 (aro3Δ, ARO4G226S, pdc6Δ, pdc5Δ, aro10Δ, tsc13Δ, atPAL1↑, coC4H↑, coCPR1↑, atCHI1↑, atCHS3↑, coCHS3↑, at4CL3↑, and coMdECR). In addition, the tyrosine ammonia lyase gene from R. capsulatus (coTAL1) was introduced in strain PATW088, with the aim to also tap from the yeast tyrosine pool. This resulted in strain PATW103 (aro3Δ, ARO4G226S, pdc6Δ, pdc5Δ, aro10Δ, tsc13Δ, spr1Δ, atPAL1↑, coC4H↑, coCPR1↑, atCHI1↑, atCHS3↑, coCHS3↑, at4CL3↑, coMdECR, and coTAL1↑). Plasmid pMEN2 was transformed to strains IMK393, PATW066, PATW08, and PATW103, resulting in strains PPF3, PPF4, PPF5, and PPF6, respectively. These strains were cultured under shake flask conditions using minimal medium and glucose as the sole carbon source for 140 h. At the end of cultivation, the total culture was extracted and extracts were analyzed for naringenin and 8-prenylnaringenin production using UHPLC–MS (Table 4). Naringenin production increased 5-fold from 18 mg L–1 (66 μM) by strain PATW066 to 100 mg L–1 (367 μM) by strain PATW103. The amount of 8-prenylnaringenin in the medium increased approximately 10-fold to a concentration of 0.12 mg L–1 (0.35 μM).
Table 4. Product Formation in S. cerevisiae Strains PPF3, PPF4, PPF5, and PPF6 in Shake Flask Culturesa.
| concentration (mg L–1 medium ± StDev) |
||
|---|---|---|
| sampleb | naringenin | 8-prenylnaringenin |
| strain PPF3 | ndc | nd |
| strain PPF4 | 18 ± 2 | 0.010 ± 0.004d |
| strain PPF5 | 95 ± 5 | 0.015 ± 0.0002d |
| strain PPF6 | 100 ± 8 | 0.119 ± 0.028 |
The strains were grown in shake flasks with 50 mL of SMG. The whole culture was extracted after 140 h of culturing at 30 °C. The metabolite concentrations of naringenin and 8-prenylnaringenin expressed in mg L–1 were measured by liquid chromatography–mass spectrometry (LC–MS). Data represent the average ± standard deviation (StDev) of independent biological triplicates.
Strain PPF3 was used as a negative control with only one biological replicate.
nd = not detected in LC–MS with SIM.
Quantification of 8-prenylnaringenin in these cases was based on a standard curve, which did not extend beyond 0.03 μg mL–1 medium.
Discussion
In this work, we describe de novo production of a prenylated flavonoid in S. cerevisiae, starting from glucose. Although yields of 8-prenylnaringenin are still low, this opens the opportunity to produce prenylated flavonoids in microbial systems, as an alternative to extraction from plants. In previous studies, the production of plant-derived prenylated flavonoids in microorganisms was only achieved via bioconversion of an intermediate that was supplied to the culture. For example, the β-bitter acid and desmethyl xanthohumol pathways from hops were recently reconstructed in yeast.33,43 In both studies, coumarate was added to the culture medium. Also, yeast expressing the SfN8DT-1 prenyltransferase was fed with naringenin in a biotransformation experiment for the production of 8-prenylnaringenin.34
Enhancing Naringenin Production Promotes the Formation of 8-Prenylnaringenin
In this study, we identified metabolic bottlenecks that limit the production of 8-prenylnaringenin in S. cerevisiae. Our strategy to increase yields was aimed at strengthening the supply of naringenin as a precursor. Previously, we observed improved anthocyanin production upon integration of coTAL1 and by preventing phloretic acid production through gene replacement of TSC13.39 Implementing these modifications in the naringenin-producing strain PATW066 indeed improved naringenin production 5-fold and resulted in approximately 10-fold higher yields of 8-prenylnaringenin (0.12 mg L–1). Still, only a small fraction of produced naringenin is prenylated. This suggests that other limitations still exist for prenylation of flavonoids. One likely limitation is the availability of the prenyl donor DMAPP. As a first strategy to boost the flow through the mevalonate pathway and improve DMAPP levels, we overexpressed tHMG1. In a recent study, a similar strategy was deployed for enhancing the production of prenylated β-carbolines, which derive from tryptophan.44 Interestingly, the overproduction of the prenyl donor DMAPP was more effective than overproduction of tryptophan.
Enhancing Prenyl Donor Availability To Improve 8-Prenylnaringenin Production
Downregulation of endogenous FPP synthase (ERG20) activity could be a second strategy to engineer the availability of DMAPP and increase 8-prenylnaringenin production in yeast. DMAPP and its isomer isopentenyl pyrophosphate (IPP) are converted to farnesyl pyrophosphate (FPP) by activity of the FPP synthase. In a previous study, a 44-fold increase in bioconversion of naringenin to 8-prenylnaringenin by SfN8DT-1 was observed for an engineered yeast strain (DD104).33 In this strain, the FPP synthase has been mutated (K197E), by which its activity was reduced, and the squalene synthase (ERG9) has been disrupted.45 However, the DD104 strain has major growth defects and needs supplementation of ergosterol to its medium. Possibly, to improve 8-prenylnaringenin production in our best naringenin-producing strain (PATW103), downregulating FPP synthase activity by introduction of mutations in the FPP synthase gene may increase availability of DMAPP, especially in combination with overexpression of tHMG1. Several mutations that downregulate FPP synthase activity have been described, such as K197S, F96W-N127W, and K254A.46−48 Targeted mutations in ERG20 may be introduced using CRISPR RNA-guided programmable deaminases (base editors).49 Alternatively, ergosterol biosynthesis can also be downregulated chemically, as was demonstrated for the production of lupulones in yeast.50 Several other strategies that could improve prenyl donor availability have been described, including overexpression of IDI1 (IPP isomerase)51 and disruption of the polyprenyl transferase COQ2 gene, which disables the use of prenyl diphosphates for ubiquinone synthesis.34
Subcellular Location of Enzymes May Affect 8-Prenylnaringenin Production
Another important consideration is the subcellular compartment in which the ectopic metabolic enzymes and intermediates are localized. Like other plant prenyltransferases, SfFPT prenyltransferase is predicted to be localized in the plant plastid. In the absence of a plastid organelle in yeast, it is probably localized to the outer membrane. On the other hand, sterol biosynthesis in yeast is known to largely take place in the cytosol and mitochondria and on the endoplasmatic reticulum (ER),52,53 and also naringenin seems to be produced largely in the cytosol and on the ER.54,55 Thus, it could make sense to relocate the prenyltransferase to the ER membrane, to bring it closer to its substrates.
In conclusion, we have successfully shown de novo production of 8-prenylnaringenin in S. cerevisiae. The engineered yeast generated in this study produced up to 0.12 mg L–1 (0.35 μM) 8-prenylnaringenin under shake flask conditions. At the same time, a number of bottlenecks were identified, in particular with regard to the efficiency of prenylation. Engineering these bottlenecks will require tuning the balance between ergosterol formation and availability of DMAPP as a donor for the prenyltransferase. Higher yields are expected from increasing the availability of the prenyl donor. The strains developed in this work represent an important platform for future development of economical production of 8-prenylnaringenin and potentially other relevant prenylated flavonoids, such as xanthohumol. The current platform can also be used to discover novel genes coding for flavonoid prenyltransferases. This study indicates the potential of engineered yeast for the production of plant prenylated flavonoids.
Glossary
Abbreviations Used
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
- GPP
geranyl diphosphate
- FPP
farnesyl diphosphate
- PAL
phenylalanine ammonia lyase
- TAL
tyrosine ammonia lyase
- C4H
cinnamate 4-hydroxylase
- CPR
cytochrome P450 reductase
- 4CL
4-coumaric acid-CoA ligase
- CHS
chalcone synthase
- CHI
chalcone isomerase
- tHMG1
truncated HMG-CoA reductase
- 8-PN
8-prenylnaringenin
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jafc.9b01367.
(A) UHPLC–MS extracted ion chromatograms of m/z 339 (negative mode) of the culture medium of S. cerevisiae strain PPF4 + NAR (solid line) and 8-prenylnaringenin standard (dashed line), (B) MS2 spectrum of m/z 339 of the 8-prenylnaringenin product in the culture medium of strain PPF4 + NAR, and (C) MS2 spectrum of m/z 339 in the 8-prenylnaringenin standard (Figure S1) (PDF)
Mark Levisson was funded by a NWO-ECHO grant (713.015.001) from the Netherlands Organisation for Scientific Research (NWO).
The authors declare no competing financial interest.
Supplementary Material
References
- Barron D.; Ibrahim R. K. Isoprenylated flavonoids—A survey. Phytochemistry 1996, 43 (5), 921–982. 10.1016/S0031-9422(96)00344-5. [DOI] [Google Scholar]
- Yazaki K.; Sasaki K.; Tsurumaru Y. Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 2009, 70 (15–16), 1739–1745. 10.1016/j.phytochem.2009.08.023. [DOI] [PubMed] [Google Scholar]
- Ahuja I.; Kissen R.; Bones A. M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17 (2), 73–90. 10.1016/j.tplants.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Botta B.; Vitali A.; Menendez P.; Misiti D.; Delle Monache G. Prenylated flavonoids: Pharmacology and biotechnology. Curr. Med. Chem. 2005, 12 (6), 713–739. 10.2174/0929867053202241. [DOI] [PubMed] [Google Scholar]
- Chen X.; Mukwaya E.; Wong M. S.; Zhang Y. A systematic review on biological activities of prenylated flavonoids. Pharm. Biol. 2014, 52 (5), 655–660. 10.3109/13880209.2013.853809. [DOI] [PubMed] [Google Scholar]
- Yang X.; Jiang Y.; Yang J.; He J.; Sun J.; Chen F.; Zhang M.; Yang B. Prenylated flavonoids, promising nutraceuticals with impressive biological activities. Trends Food Sci. Technol. 2015, 44 (1), 93–104. 10.1016/j.tifs.2015.03.007. [DOI] [Google Scholar]
- Barron D.; Balland C.; Possety F.; Ravanel P.; Desfougères A. Flavonoïdes prénylés et perméabilité membranaire. Acta Bot. Gallica 1996, 143 (6), 509–520. 10.1080/12538078.1996.10515348. [DOI] [Google Scholar]
- Simons R.; Gruppen H.; Bovee T. F. H.; Verbruggen M. A.; Vincken J. P. Prenylated isoflavonoids from plants as selective estrogen receptor modulators (phytoSERMs). Food Funct. 2012, 3 (8), 810–827. 10.1039/c2fo10290k. [DOI] [PubMed] [Google Scholar]
- van de Schans M. G. M.; Ritschel T.; Bovee T. F. H.; Sanders M. G.; de Waard P.; Gruppen H.; Vincken J. P. Involvement of a Hydrophobic Pocket and Helix 11 in Determining the Modes of Action of Prenylated Flavonoids and Isoflavonoids in the Human Estrogen Receptor. ChemBioChem 2015, 16 (18), 2668–2677. 10.1002/cbic.201500343. [DOI] [PubMed] [Google Scholar]
- van de Schans M. G. M.; Vincken J. P.; de Waard P.; Hamers A. R. M.; Bovee T. F. H.; Gruppen H. Glyceollins and dehydroglyceollins isolated from soybean act as SERMs and ER subtype-selective phytoestrogens. J. Steroid Biochem. Mol. Biol. 2016, 156, 53–63. 10.1016/j.jsbmb.2015.11.020. [DOI] [PubMed] [Google Scholar]
- Araya-Cloutier C.; den Besten H. M. W.; Aisyah S.; Gruppen H.; Vincken J. P. The position of prenylation of isoflavonoids and stilbenoids from legumes (Fabaceae) modulates the antimicrobial activity against Gram positive pathogens. Food Chem. 2017, 226, 193–201. 10.1016/j.foodchem.2017.01.026. [DOI] [PubMed] [Google Scholar]
- Araya-Cloutier C.; Vincken J. P.; van de Schans M. G. M.; Hageman J.; Schaftenaar G.; den Besten H. M. W.; Gruppen H. QSAR-based molecular signatures of prenylated (iso)flavonoids underlying antimicrobial potency against and membrane-disruption in Gram positive and Gram negative bacteria. Sci. Rep. 2018, 8 (1), 9267. 10.1038/s41598-018-27545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya-Cloutier C.; Vincken J. P.; van Ederen R.; den Besten H. M. W.; Gruppen H. Rapid membrane permeabilization of Listeria monocytogenes and Escherichia coli induced by antibacterial prenylated phenolic compounds from legumes. Food Chem. 2018, 240, 147–155. 10.1016/j.foodchem.2017.07.074. [DOI] [PubMed] [Google Scholar]
- de Bruijn W. J. C.; Araya-Cloutier C.; Bijlsma J.; de Swart A.; Sanders M. G.; de Waard P.; Gruppen H.; Vincken J. P. Antibacterial prenylated stilbenoids from peanut (Arachis hypogaea). Phytochem. Lett. 2018, 28, 13–18. 10.1016/j.phytol.2018.09.004. [DOI] [Google Scholar]
- Stevens J. F.; Ivancic M.; Hsu V. L.; Deinzer M. L. Prenylflavonoids from Humulus lupulus. Phytochemistry 1997, 44 (8), 1575–1585. 10.1016/S0031-9422(96)00744-3. [DOI] [Google Scholar]
- Zhao P.; Hamada C.; Inoue K.; Yamamoto H. Efficient production and capture of 8-prenylnaringenin and leachianone G—Biosynthetic intermediates of sophoraflavanone G—By the addition of cork tissue to cell suspension cultures of Sophora flavescens. Phytochemistry 2003, 62 (7), 1093–1099. 10.1016/S0031-9422(02)00671-4. [DOI] [PubMed] [Google Scholar]
- Christoffel J.; Rimoldi G.; Wuttke W. Effects of 8-prenylnaringenin on the hypothalamo-pituitary-uterine axis in rats after 3-month treatment. J. Endocrinol. 2006, 188 (3), 397–405. 10.1677/joe.1.06384. [DOI] [PubMed] [Google Scholar]
- Milligan S.; Kalita J.; Pocock V.; Heyerick A.; De Cooman L.; Rong H.; De Keukeleire D. Oestrogenic activity of the hop phyto-oestrogen, 8-prenylnaringenin. Reproduction 2002, 123 (2), 235–242. 10.1530/rep.0.1230235. [DOI] [PubMed] [Google Scholar]
- Stulikova K.; Karabin M.; Nespor J.; Dostalek P. Therapeutic Perspectives of 8-Prenylnaringenin, a Potent Phytoestrogen from Hops. Molecules 2018, 23 (3), 660. 10.3390/molecules23030660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khupse R. S.; Erhardt P. W. Total synthesis of xanthohumol. J. Nat. Prod. 2007, 70 (9), 1507–1509. 10.1021/np070158y. [DOI] [PubMed] [Google Scholar]
- Rong H.; Zhao Y.; Lazou K.; De Keukeleire D.; Milligan S. R.; Sandra P. Quantitation of 8-prenylnaringenin, a novel phytoestrogen in hops (Humulus lupulus L.), hop products, and beers, by benchtop HPLC-MS using electrospray ionization. Chromatographia 2000, 51 (9–10), 545–552. 10.1007/BF02490811. [DOI] [Google Scholar]
- Beekwilder J.; van Houwelingen A.; Cankar K.; van Dijk A. D. J.; de Jong R. M.; Stoopen G.; Bouwmeester H.; Achkar J.; Sonke T.; Bosch D. Valencene synthase from the heartwood of Nootka cypress (Callitropsis nootkatensis) for biotechnological production of valencene. Plant Biotechnol J. 2014, 12 (2), 174–182. 10.1111/pbi.12124. [DOI] [PubMed] [Google Scholar]
- Liu X.; Ding W.; Jiang H. Engineering microbial cell factories for the production of plant natural products: From design principles to industrial-scale production. Microb. Cell Fact. 2017, 16 (1), 125. 10.1186/s12934-017-0732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. P.; Parajuli P.; Koffas M. A. G.; Sohng J. K. Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 2016, 34 (5), 634–662. 10.1016/j.biotechadv.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Wang J.; Guleria S.; Koffas M. A. G.; Yan Y. J. Microbial production, of value-added nutraceuticals. Curr. Opin. Biotechnol. 2016, 37, 97–104. 10.1016/j.copbio.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Koopman F.; Beekwilder J.; Crimi B.; van Houwelingen A.; Hall R. D.; Bosch D.; van Maris A. J.; Pronk J. T.; Daran J.-M. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2012, 11 (1), 155. 10.1186/1475-2859-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milke L.; Ferreira P.; Kallscheuer N.; Braga A.; Vogt M.; Kappelmann J.; Oliveira J.; Silva A. R.; Rocha I.; Bott M.; Noack S.; Faria N.; Marienhagen J. Modulation of the central carbon metabolism of Corynebacterium glutamicum improves malonyl-CoA availability and increases plant polyphenol synthesis. Biotechnol. Bioeng. 2019, 10.1002/bit.26939. [DOI] [PubMed] [Google Scholar]
- Zhou S.; Lyu Y.; Li H.; Koffas M. A. G.; Zhou J. Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol. Bioeng. 2019, 10.1002/bit.26941. [DOI] [PubMed] [Google Scholar]
- Wang J.; Chu S.; Zhu Y.; Cheng H.; Yu D. Positive selection drives neofunctionalization of the UbiA prenyltransferase gene family. Plant Mol. Biol. 2015, 87 (4–5), 383–394. 10.1007/s11103-015-0285-2. [DOI] [PubMed] [Google Scholar]
- Sasaki K.; Mito K.; Ohara K.; Yamamoto H.; Yazaki K. Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol. 2008, 146 (3), 1075–1084. 10.1104/pp.107.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G.; Huhman D.; Lei Z.; Snyder J.; Sumner L. W.; Dixon R. A. Characterization of an Isoflavonoid-Specific Prenyltransferase from Lupinus albus. Plant Physiol. 2012, 159 (1), 70–80. 10.1104/pp.112.195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. D.; Liu X.; Zou J. H.; Yin Y. Z.; Ou B.; Li J. H.; Wang R. S.; Xie D.; Zhang P. C.; Dai J. G. Regio- and Stereospecific Prenylation of Flavonoids by Sophora flavescens Prenyltransferase. Adv. Synth. Catal. 2013, 355 (9), 1817–1828. 10.1002/adsc.201300196. [DOI] [Google Scholar]
- Li H.; Ban Z.; Qin H.; Ma L.; King A. J.; Wang G. A Heteromeric Membrane-Bound Prenyltransferase Complex from Hop Catalyzes Three Sequential Aromatic Prenylations in the Bitter Acid Pathway. Plant Physiol. 2015, 167 (3), 650–659. 10.1104/pp.114.253682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K.; Tsurumaru Y.; Yazaki K. Prenylation of Flavonoids by Biotransformation of Yeast Expressing Plant Membrane-Bound Prenyltransferase SfN8DT-1. Biosci., Biotechnol., Biochem. 2009, 73 (3), 759–761. 10.1271/bbb.80729. [DOI] [PubMed] [Google Scholar]
- Parks L. W.; Casey W. M. Physiological Implications of Sterol Biosynthesis in Yeast. Annu. Rev. Microbiol. 1995, 49, 95–116. 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- Ro D. K.; Paradise E. M.; Ouellet M.; Fisher K. J.; Newman K. L.; Ndungu J. M.; Ho K. A.; Eachus R. A.; Ham T. S.; Kirby J.; Chang M. C. Y.; Withers S. T.; Shiba Y.; Sarpong R.; Keasling J. D. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440 (7086), 940–943. 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Gietz R. D.; Woods R. A.. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology; Elsevier: Amsterdam, Netherlands, 2002; Vol. 350, pp 87–96, 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Lõoke M.; Kristjuhan K.; Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2011, 50 (5), 325. 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levisson M.; Patinios C.; Hein S.; de Groot P. A.; Daran J. M.; Hall R. D.; Martens S.; Beekwilder J. Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Fact. 2018, 17, 103. 10.1186/s12934-018-0951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald K. A. G.; Hampton R. Y.; Fritz I. B. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63 (9), 3341–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakowski T.; Stahl U.; Lang C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 1998, 49 (1), 66–71. 10.1007/s002530051138. [DOI] [PubMed] [Google Scholar]
- Lehka B. J.; Eichenberger M.; Bjørn-Yoshimoto W. E.; Vanegas K. G.; Buijs N.; Jensen N. B.; Dyekjær J. D.; Jenssen H.; Simon E.; Naesby M. Improving heterologous production of phenylpropanoids in Saccharomyces cerevisiae by tackling an unwanted side reaction of Tsc13, an endogenous double-bond reductase. FEMS Yeast Research 2017, 17 (1), fox004. 10.1093/femsyr/fox004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Z.; Qin H.; Mitchell A. J.; Liu B.; Zhang F.; Weng J. K.; Dixon R. A.; Wang G. Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (22), E5223–E5232. 10.1073/pnas.1802223115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhaus K.; Ludwig-Radtke L.; Xie X.; Li S. M. Manipulation of the Precursor Supply in Yeast Significantly Enhances the Accumulation of Prenylated β-Carbolines. ACS Synth. Biol. 2017, 6 (6), 1056–1064. 10.1021/acssynbio.6b00387. [DOI] [PubMed] [Google Scholar]
- Szkopinska A.; Grabinska K.; Delourme D.; Karst F.; Rytka J.; Palamarczyk G. Polyprenol formation in the yeast Saccharomyces cerevisiae: Effect of farnesyl diphosphate synthase overexpression. J. Lipid Res. 1997, 38 (5), 962–968. [PubMed] [Google Scholar]
- Fischer M. J. C.; Meyer S.; Claudel P.; Bergdoll M.; Karst F. Metabolic Engineering of Monoterpene Synthesis in Yeast. Biotechnol. Bioeng. 2011, 108 (8), 1883–1892. 10.1002/bit.23129. [DOI] [PubMed] [Google Scholar]
- Fischer M. J. C.; Meyer S.; Claudel P.; Bergdoll M.; Karst F. Identification of a Lysine Residue Important for the Catalytic Activity of Yeast Farnesyl Diphosphate Synthase. Protein J. 2011, 30 (5), 334–339. 10.1007/s10930-011-9336-y. [DOI] [PubMed] [Google Scholar]
- Ignea C.; Pontini M.; Maffei M. E.; Makris A. M.; Kampranis S. C. Engineering Monoterpene Production in Yeast Using a Synthetic Dominant Negative Geranyl Diphosphate Synthase. ACS Synth. Biol. 2014, 3 (5), 298–306. 10.1021/sb400115e. [DOI] [PubMed] [Google Scholar]
- Komor A. C.; Badran A. H.; Liu D. R. Editing the Genome Without Double-Stranded DNA Breaks. ACS Chem. Biol. 2018, 13 (2), 383–388. 10.1021/acschembio.7b00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.; Shen H.; Liu Y.; Wang Q.; Wang X.; Peng C.; Liu W.; Zhao Z. K. Enabling Heterologous Synthesis of Lupulones in the Yeast Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2019, 1–11. 10.1007/s12010-019-02957-8. [DOI] [PubMed] [Google Scholar]
- Liu J.; Zhang W.; Du G.; Chen J.; Zhou J. Overproduction of geraniol by enhanced precursor supply in Saccharomyces cerevisiae. J. Biotechnol. 2013, 168 (4), 446–51. 10.1016/j.jbiotec.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Koning A. J.; Roberts C. J.; Wright R. L. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol. Biol. Cell 1996, 7 (5), 769–789. 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T.; Hata S.; Taketani S.; Yabusaki Y.; Katsuki H. Subcellular-Localization of the Enzymes Involved in the Late Stage of Ergosterol Biosynthesis in Yeast. J. Biochem. 1981, 89 (5), 1391–1396. 10.1093/oxfordjournals.jbchem.a133330. [DOI] [PubMed] [Google Scholar]
- Hrazdina G.; Jensen R. A. Spatial-Organization of Enzymes in Plant Metabolic Pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 241–267. 10.1146/annurev.pp.43.060192.001325. [DOI] [Google Scholar]
- Saslowsky D.; Winkel-Shirley B. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 2001, 27 (1), 37–48. 10.1046/j.1365-313x.2001.01073.x. [DOI] [PubMed] [Google Scholar]
- DiCarlo J. E.; Norville J. E.; Mali P.; Rios X.; Aach J.; Church G. M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41 (7), 4336–4343. 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.