Abstract
The G-quadruplexes that form in the HIV-1 RNA genome hinder progression of reverse transcriptase in vitro, but not in infected cells. We investigated the possibility that the HIV-1 nucleocapsid protein NCp7, which remains associated with the viral RNA during reverse transcription, modulated HIV-1 RNA G-quadruplex stability. By electrophoresis, circular dichroism, mass spectrometry, and reverse transcriptase stop assays, we demonstrated that NCp7 binds and unfolds the HIV-1 RNA G-quadruplexes and promotes DNA/RNA duplex formation, allowing reverse transcription to proceed. The G-quadruplex ligand BRACO-19 was able to partially counteract this effect. These results indicate NCp7 as the first known viral protein able to unfold RNA G-quadruplexes, and they explain how the extra-stable HIV-1 RNA G-quadruplexes are processed; they also point out that the reverse transcription process is hindered by G-quadruplex ligands at both reverse transcriptase and NCp7 level. This information can lead to the development of more effective anti-HIV-1 drugs with a new mechanism of action.
Keywords: G-quadruplexes, HIV-1 nucleocapsid protein, NCp7, reverse transcription, BRACO-19
The G-quadruplexes (G4s) are noncanonical secondary structures formed by single-stranded guanine (G)-rich DNA or RNA.1,2 Four Gs bind through Hoogsteen hydrogen bonds to form G-quartets, planar square structures that stack upon each other to form the G4, which in turn is significantly stabilized by monovalent cations.3 In the human genome, many G4s have been characterized as regulatory elements of critical cellular processes, including inhibition of telomerase activity, modulation of oncogene expression via transcriptional suppression, polymerase stalling during replication, and translational regulation in the untranslated regions (UTRs), where RNA G4s support both positive and negative control of mRNA translation.4−7 G4 folding has been shown to be triggered or hindered by cellular proteins.8,9 Nucleolin is an example of G4-binding protein, first identified as the binding and stabilizing partner of the G4-structured transcriptionally inactive form of the c-myc promoter10 and later demonstrated to bind G4s in viruses,11−14 with a preference for long-looped G4s.15 Various families of helicases have been shown to unfold G4 structures,16,17 thus allowing polymerase progression that otherwise would be impaired.
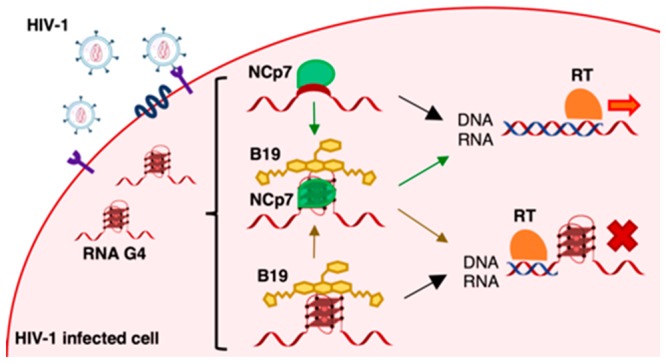
The human immunodeficiency virus type 1 (HIV-1) is a retrovirus with two identical copies of RNA genome encapsidated in the viral particle together with the enzyme machinery necessary for viral replication. Right after reverse transcription, which takes place in the cytoplasm of the host cell, the newly synthesized dsDNA genome is transported in the nucleus and integrated into the host genome thanks to the two long terminal repeat (LTR) flanking regions.18,19 In the proviral DNA, the 5′-LTR serves as promoter of all HIV-1 genes and is segmented into U3, R, and U5 regions.20,21 The U3 region is also present at the 3′-end of the viral RNA genome. We have previously demonstrated formation of multiple G4 structures in the U3 region of both the proviral DNA and RNA genome.22,23 G4-ligands with both low and high selectivity for viral G4s displayed antiviral activity ascribable to the interaction with the viral G4s.23,24 The antiviral activity of the G4 ligand BRACO-19 (B19) was linked to inhibition of both post- and preintegration steps of the viral cycle.23 At the preintegration steps, B19 hindered progression of reverse transcriptase (RT) on the RNA template, therefore inhibiting HIV-1 DNA synthesis and thus viral production. In infected cells, HIV-1 RT is assisted by the viral nucleocapsid protein (NCp7).25 HIV-1 NCp7 is a small 55-amino acid basic protein with two CCHC zinc-finger domains, produced as part of the Gag polyprotein.26 During reverse transcription, NCp7 assists RT by solving HIV-1 RNA genome secondary structures to ensure enzyme progression and complete synthesis of the viral DNA.27−29 NCp7 was shown to be able to recognize DNA G4s, in particular the intermolecular G4 that forms in the HIV-1 DNA flap, an intermediate and ultimately removed sequence of the reverse transcription process,30 and to unfold an intramolecular synthetic DNA G4.31 In addition, NCp7 was able to induce surface-attached oligonucleotides to fold into tetramolecular G4.32 Because the G4 structures that form in the U3 region of the HIV-1 RNA genome have been reported to be extremely stable,23 we here investigated the possibility that NCp7 was also involved in the unfolding of these structures and tested if G4 ligands could inhibit this process. We demonstrated that NCp7 is indeed able to bind and unfold the HIV-1 RNA G4s and stimulate RNA/DNA hybrid formation, thus greatly enhancing the processivity of RT and allowing synthesis of the proviral DNA genome. The presence of the G4-ligand B19 was able to partially counteract these effects.
Results
NCp7 Binds and Unfolds the RNA G4s in the U3 Region of the HIV-1 RNA Genome
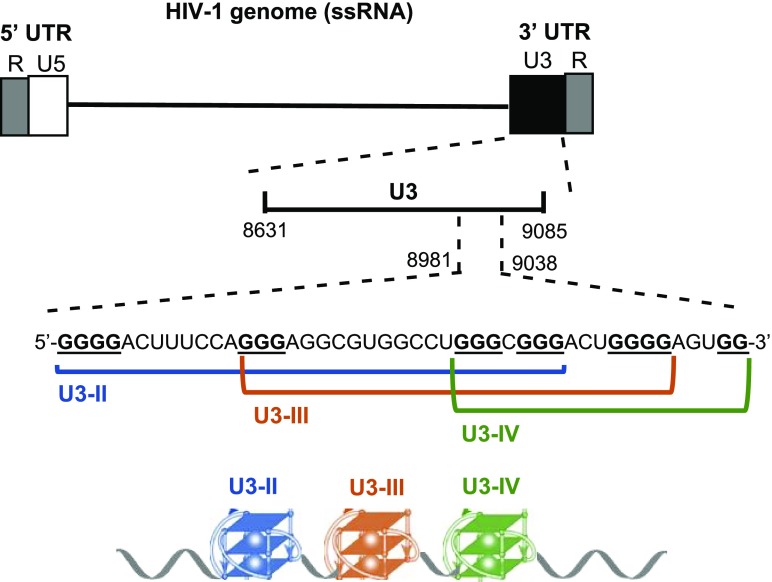
We have previously shown that two RNA G-quadruplexes (G4s), i.e., U3-III and U3-IV, form in the G-rich U3 region of the HIV-1 RNA genome (Figure 1).22 Because these are extremely stable in physiological conditions (i.e., Tm = 82.1 and 71.2 °C for U3-III and U3-IV, respectively, in 100 mM K+), we envisaged the presence of a protein with G4-unfolding activity that would allow progression of the reverse transcriptase (RT) in infected cells. Since the viral nucleocapsid protein (NCp7) associates with RT during reverse transcription,25,29 we explored the possibility that NCp7 itself was able to unfold the U3 G4s.
Figure 1.
Schematic representation of the G4 forming region in the HIV-1 RNA genome. Position of U3 region within the viral genome, along with the G-rich sequence and the tracts that form the three possible G4 structures (U3-II, U3-III, and U30IV) are shown.
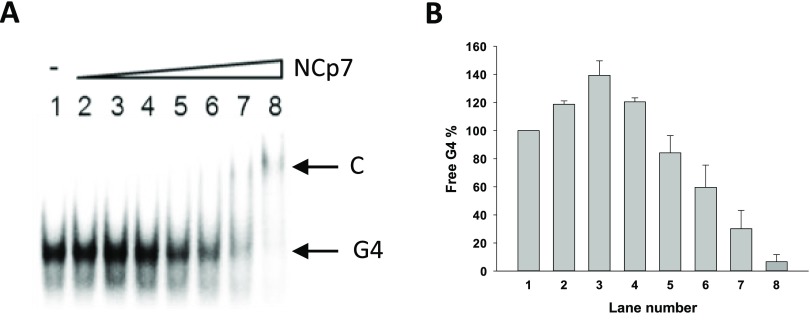
We first assessed the ability of NCp7 to bind the G4-folded U3 sequences. To this purpose, the RNA U3-III+IV oligonucleotide, folded into G4 in the presence of increasing amounts of NCp7, was analyzed by electrophoretic mobility shift assay (EMSA) (Figure 2A).
Figure 2.
EMSA-mediated assessment of the binding of NCp7 toward the HIV-1 U3-III+IV RNA G4. (A) Increasing concentrations of NCp7 (7.5, 15, 37.5, 75, 112.5, 150, and 300 nM) were incubated with RNA U3-III+IV G4 folded in 50 mM KCl buffer. C indicates protein/oligonucleotide complex. (B) Quantification of the free G4 band shown in panel (A) (average of two experiments; bars represent standard deviation).
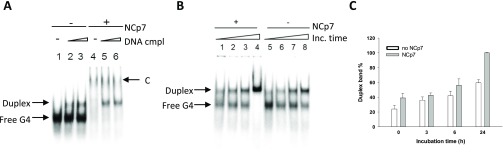
Initial increase of the free G4 band (Figure 2B) was ascribed to the aggregation properties of NCp7, mediated by unspecific electrostatic binding, too weak to make the protein/RNA complex visible in these conditions. Alternatively, different RNA conformations unable to enter the gel and/or species below detection in the gel may be unfolded by NCp7 to yield the most stable G4 structure. At the highest protein concentration, we observed a decrease in the free G4 band. Complexes with NCp7 were reported to aggregate/precipitate,30 and thus, the disappearance of the free G4 starting from NCp7 75 nM was deemed indicative of the binding. At NCp7 300 nM (Figure 2A, lane 8), a faint slower migrating band became visible, alongside the almost complete disappearance of the free G4, which indicated binding saturation (Figure 2A). We deemed the slower migrating band attributable to the G4-NCp7 complex, which was visible because, besides electrostatic interactions, it involves also higher energy bonds, such as hydrogen, hydrophobic, and stacking interactions. Since during reverse transcription a DNA/RNA intermediate forms, we next evaluated the binding properties of NCp7 to the G4-forming U3-III+IV sequence in the presence of the complementary DNA oligonucleotide (Figure 3A). Addition of the cold complementary DNA strand (i.e., the unlabeled DNA strand complementary to the U3-III+IV RNA sequence) to the labeled and G4-folded RNA sequence induced formation of a hybrid RNA-DNA duplex, which had a slower migration rate and thus could be separated from the single-stranded G4-folded species on a native polyacrylamide gel (Figure 3A, lanes 1–3). In the presence of NCp7, the RNA/DNA duplex competed for the formation of the RNA G4–NCp7 complex (Figure 3A, lanes 4–6). The free G4 species completely disappeared upon G4–NCp7 complex formation, while the amount of the free duplex was not perturbed. These data indicate the binding of NCp7 to the G4-folded RNA vs the DNA/RNA duplex. To form the duplex, the thermodynamically stable RNA G4 has to be unfolded prior to base pairing with its complementary strand. We thus analyzed duplex formation kinetics: the amount of duplex increased over time and reached 60% at 24 h (Figure 3C). Addition of NCp7 highly increased the amount of the duplex species, especially at 24 h when the G4 folded oligonucleotide completely converted to the duplex form (Figure 3C). These results indicate that when NCp7 binds to the HIV-1 RNA G4 structures, it stimulates their unfolding in the presence of the complementary strand.
Figure 3.
EMSA-mediated assessment of the unfolding properties of NCp7 toward the HIV-1 U3-III+IV RNA G4. (A) NCp7 binding to the G4 in the presence of the cold complementary DNA strand. RNA U3-III+IV G4 was annealed in 50 mM KCl buffer and incubated with 0.5 (lanes 2 and 5) and equimolar (lanes 3 and 6) ratios of complementary DNA for 1 h and then NCp7 was added at a final concentration of 300 nM and incubated for 10 min at 37 °C. C indicates protein/oligonucleotide complex. (B) NCp7 binding to the G4 in the presence of the cold complementary DNA strand at increasing time. RNA G4 was folded in 50 mM KCl buffer and incubated with a 2-fold excess of cold complementary DNA strand for 0, 3, 6, and 24 h in the presence (lanes 1–4) and absence (lanes 4–8) of 300 nM NCp7. Duplex and free G4 species are indicated by arrows. (C) Quantification of the duplex band shown in panel (B) (average of two experiments; bars represent standard deviation) in the presence and absence of NCp7.
To confirm this observation, NCp7 activity was investigated by circular dichroism (CD). The G4-folded U3-III+IV RNA was incubated in the presence/absence of NCp7 and analyzed by CD (Figures 4, S1, and S2). Upon addition of NCp7, the molar ellipticity of U3-III+IV G4 drastically decreased, while the CD spectrum maintained the G4 signature, with a maximum at 265 nm and a minimum at 238 nm. Usually, low molar ellipticity indicates low stability of the tested G4s. To check the actual stability of U3-III+IV G4 in the presence/absence of NCp7, CD spectra were recorded at increasing temperature (Figure 4). The melting temperature (Tm) calculated according to the van ’t Hoff equation applied to the molar ellipticity signal at 265 nm vs the temperature was 68.3 and 44.8 °C in the absence and presence of NCp7, respectively, indicating the effective unfolding of the RNA G4 mediated by NCp7. When the unfolded U3-III+IV G4 was reannealed by steadily decreasing the temperature from 90 to 20 °C, in the absence of NCp7 the Tm maintained its value of 68.3 °C, whereas in the presence of the protein the oligonucleotide was unable to regain the folded G4 structure (Figure 4), indicating that NCp7 was able to maintain its unfolding properties in this condition. We next investigated if the G4-ligand B19, which has been shown to stabilize the HIV-1 U3 G4s,23 could inhibit the G4-unfolding activity of NCp7. We incubated U3-III+IV G4 with B19 before addition and further incubation with NCp7 (Figure 4). B19 increased the G4 Tm up to >90 °C; in the presence of NCp7, this was reduced to 67.8 °C (Figure 4), indicating that B19 was able to in part suppress NCp7 unfolding activity.
Figure 4.
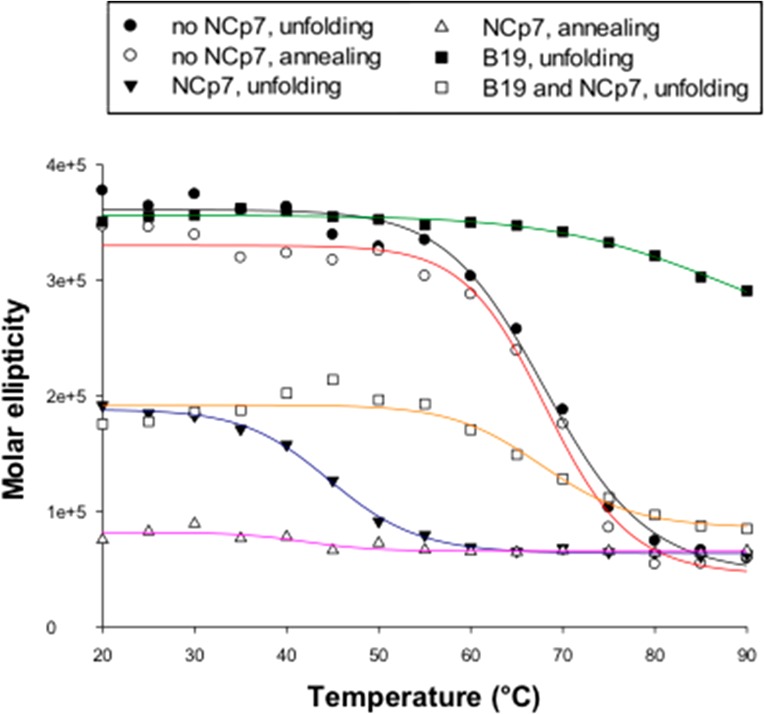
CD analysis of NCp7-mediated U3-III+IV RNA G4 unfolding. CD melting and annealing curves of the G4-folded RNA U3-III+IV in the presence/absence of 10-fold excess of NCp7 in 50 mM K+ and CD melting curves of the G4-folded RNA U3-III+IV in the presence of the G4-ligand B19 (8 μM) and in the presence/absence of NCp7 (10×). The molar ellipticity at the peak wavelength (265 nm) is shown as a function of the temperature.
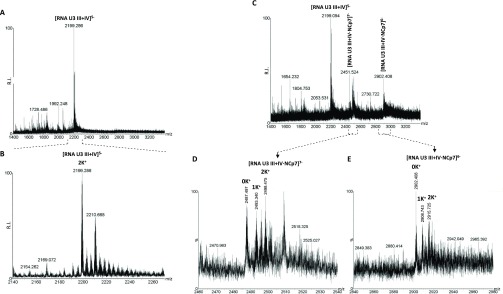
NCp7 unfolding properties were further assessed by electrospray ionization (ESI) mass spectrometry (MS) (Figure 5), a powerful technique to investigate both G4 structures and G4/small molecules binding.33 The number of the coordinated K+ ions is diagnostic of the number of G-quartets involved in the G4 structure, and therefore of the G4 folded conformation.
Figure 5.
ESI-MS analysis of NCp7 binding to and unfolding of U3-III+IV RNA G4. ESI-MS spectra of U3-III+IV RNA G4 in the absence or presence of 1 equiv of NCp7 in 0.8 mM KCl, 120 mM TMAA adjusted to pH 7.4 with TEA. (A) ESI-MS spectra showing the entire sample peak charge distribution of the U3-III+IV RNA G4 in the absence of NCp7. (B) Zoomed-in spectrum corresponding to the U3-III+IV G4 with a 5– charge state. (C) ESI-MS spectra of the U3-III+IV G4 in the presence of 1 equiv of NCp7. (D) Zoomed-in spectrum corresponding to the U3-III+IV G4 complexed with NCp7 with a 7– charge state. (E) Zoomed-in spectrum corresponding to the oligo-protein complex with a 6– charge state.
The MS spectrum of the RNA U3-III+IV G4 presented a peak corresponding to the oligonucleotide coordinated to two K+ ions, which indicated the expected three-layered U3-III+IV G4 in this condition (Figure 5A, B). In the presence of NCp7, two additional peaks corresponding to the oligo-protein complex appeared (Figure 5C). In the complex, the species with 0 and 1 K+ ions were prevalent with respect to the 2 K+ ion species (Figures 5D, E), indicating the unfolded state of the oligonucleotide in the presence and in complex with NCp7, and thus confirming the unfolding activity of NCp7 toward the HIV-1 RNA G4s.
NCp7-Mediated Unfolding of the HIV-1 RNA G4s Promotes Reverse Transcriptase Processivity
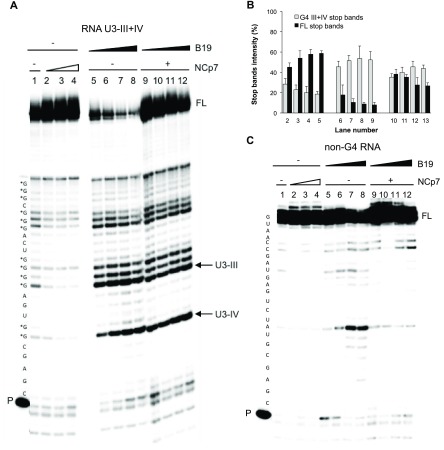
To investigate whether NCp7 unfolding properties could abolish the previously observed HIV-1 RNA G4-mediated RT stalling,22 we performed the RT stop assay in the presence of increasing concentrations of NCp7. The U3-III+IV sequence in the presence of K+ induced RT pausing at all G-tracts involved in formation of the overlapping G4s (i.e., U3-III and U3-IV, Figure 6A, lane 1). Upon addition of NCp7, the stop sites decreased and the full-length RT product increased (Figure 6A, lanes 2–4, and Figure 6B). When the RNA U3-III+IV G4 template was treated with increasing concentrations of B19, the stop sites significantly increased at the expense of the full-length product (Figure 6A, lanes 5–8, and Figure 6B). When NCp7 was added to the B19-treated samples, the full-length product was restored, while the stop sites were mainly maintained (Figure 6A, lanes 9–12, and Figure 6B), with a visible B19 concentration-dependent effect on both the full-length product and stop sites (Figure 6A, lanes 9–12, and Figure 6B). We next tested a control G-rich sequence unable to fold into G4 (Figure 6C). Minor pausing sites were observed (lane 1) likely due to transient base pairing within short RNA tracts. Addition of NCp7 also in this case was able to solve most of these pausing sites (lanes 2–4). In the presence of B19, one stop site paralleled by reduction of the full-length products was visible (lanes 5–8): this behavior is compatible with the reported not absolute G4 specificity of B19, especially at high concentrations.34 Incubation with NCp7 fully released the pausing sites (lanes 9–12), indicating that, in the absence of a G4 structure, unspecific binding of B19 to the RNA is not able to inhibit NCp7 and that B19 does not inhibit the protein activity per se. The full-length product in the presence of NCp7 was more abundant than that in the absence of the protein (lanes 9–12 vs lanes 2–4), confirming the previously reported NCp7 higher polymerization rates in crowding conditions.42 Altogether these data indicate that NCp7 unfolds G4 structures that form in the HIV-1 RNA genome, favoring the proceeding of reverse transcription. In the absence of G4 stabilizing compounds, NCp7 is able to destabilize the structures to allow complete synthesis of the DNA, while the G4-ligand B19 can in part counteract this effect.
Figure 6.
RT stop assay on the RNA U3-III+IV sequence. (A) The U3-II+IV G4 forming template was treated with increasing concentrations of NCp7 (350 nM, 700 nM, and 1.4 μM, lanes 2–4) prior to elongation. When folded, samples were stabilized by B19 (500 nM, 1 μM, 2 μM, 4 μM, lanes 5–8), elongation was performed in the absence (lanes 5–8)/presence (lanes 9–12) of NCp7 at 1.4 μM concentration. Stop bands corresponding to the U3-IV and U3-III G4 species in the RNA U3-III+IV template are indicated by arrows. P and FL indicate primer and full-length product bands, respectively. G bases potentially involved in G4 formation are labeled with asterisks. (B) Stop band intensity quantification relative to the RT stop assay represented in panel (A) (average of two technical replicates, bands represent standard deviation). (C) A control template unable to fold into G4 was treated with increasing concentrations of NCp7 (350 nM, 700 nM, and 1.4 μM, lanes 2–4) prior to elongation. The template was also treated with B19 (500 nM, 1 μM, 2 μM, and 4 μM, lanes 5–8), and elongation was performed in the absence (lanes 5–8)/presence (lanes 9–12) of NCp7 at 1.4 μM concentration. P and FL indicate primer and full-length product bands, respectively.
Discussion
We have previously observed that the U3 region of the HIV-1 genome can fold into extremely stable G4 structures that inhibit RT progression in vitro.22 In contrast to DNA G-rich regions that may form G4s only when dissociated from their complementary sequence, for example, temporarily by DNA- and RNA-polymerase processing during replication and transcription,35 the HIV-1 RNA genome is single-stranded and therefore folded into secondary structures most of the time. We reasoned that the very stable RNA G4 structures would be deleterious for viral survival and therefore viral/cellular proteins need to be present to solve them. While a cellular protein, i.e., hnRNP A2/B1, has been reported to unfold the HIV-1 DNA G4s,36 we hypothesized that, in this case, a viral protein could solve RNA G4s to allow the correct completion of the RT process: in fact, reverse transcription has recently being reported to initiate within the capsid of the mature virus, where cellular proteins do not have access.37
We thus focused on NCp7, a small retroviral protein generated by proteolytic cleavage of the Gag precursor: several hundred molecules of NCp7 coat and protect the HIV-1 dimeric RNA genome in the virion and later assist various steps of the HIV-1 replication cycle, including reverse transcription, genome dimerization, and selective genome packaging.26,28,38−42 NCp7 displays nucleic acid chaperone activity that, during reverse transcription, facilitates the rearrangement of nucleic acids into their most thermodynamically stable structures.27 NCp7 has been reported to bind DNA G4s: it was able to unfold a short and synthetic monomeric DNA G431 and to assemble tetramolecular G4 structures.32 This latter activity probably resulted from the nucleic acids aggregation properties of NCp7. However, NCp7 ability to bind and process RNA G4s has never been presented so far.
Using different and complementary techniques, we proved here that NCp7 was able to bind and unfold the U3 G4s in vitro. The presence of NCp7 stimulated production of full-length amplification products by RT, as assessed in the RT stop assay. In addition, we proved that NCp7 preferentially binds the G4 sequence vs its duplex counterpart. NCp7 has been reported to preferentially bind single-stranded nucleic acid regions.38 Here we take this concept further and demonstrate that NCp7 binds to conformationally structured single-stranded regions, such as G4s, and unfolds them. In our case, this activity resulted in the increased formation of duplex RNA/DNA hybrid, a structure that is thermodynamically more stable than the G4, as demonstrated by competition experiments in EMSA. A similar activity has been reported for another single-stranded structured RNA in HIV-1, the TAR hairpin, which gets unfolded by NCp7 to favor annealing to the complementary sequence and thus formation of the double-stranded molecule.43−45 In the case of TAR RNA, exposed G bases are the sites preferentially bound by NCp7.45 This evidence supports the unfolding activity observed on the HIV-1 RNA U3 G4s, where exposed G bases are present in the G4 loops. In addition, in the G4 conformation, Gs base-pair through the Hoogsteen-type hydrogen bonds that are less thermodynamically stable than the Watson and Crick ones and may thus be recognition sites for NCp7 as well.46 These results indicate that NCp7 is indeed the protein able to solve the stable RNA G4s, thus allowing viral reverse transcription to occur in vivo. This is the first time a viral protein is reported to unfold RNA G4s. So far, only one other viral protein, i.e., EBNA 1 of the Epstein–Barr virus, has been shown to bind to folded RNA G4s to promote viral DNA replication.47
The initial stability of the secondary structure processed by NCp7 dictates the efficiency of the unfolding activity. In fact, as previously reported for the annealing of TAR to its complementary strand, NCp7 destabilizes the less stable complementary TAR hairpin at a higher rate.44 Therefore, from a therapeutic point of view, increasing the stability of the HIV-1 RNA structures could be a valuable strategy to decrease the unfolding capacity of NCp7 and thus further inhibit RT progression.48 We have previously shown that the U3 G4s could be stabilized by the G4-ligand B19, which was able to inhibit RT progression at the sites of G formation. This activity resulted in inhibition of the viral life cycle at the preintegration step,23 which we proposed due exclusively to inhibition of RT progression at the G4 site. We proved here by CD and RT stop assay, that B19 is also able to counteract the unfolding activity of NCp7.
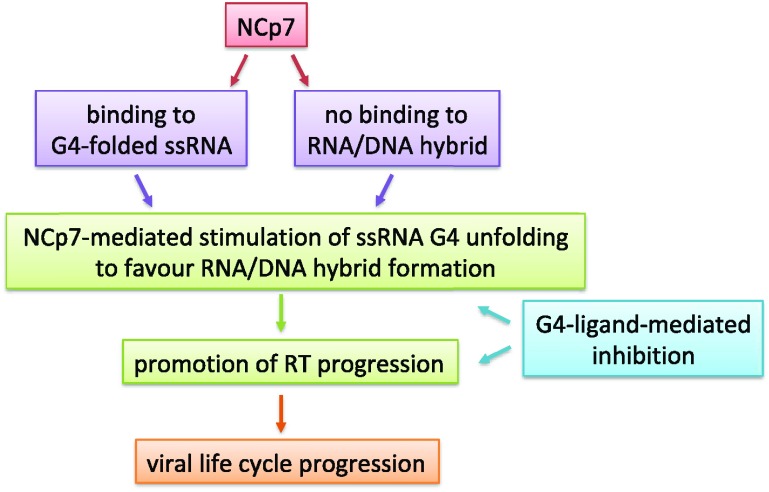
Therefore, our data indicate that G4 ligands possess a dual activity at the U3 RNA level (Figure 7): on one hand they sterically hinder RT processing of the structured RNA template; on the other, they inhibit the chaperone activity of NCp7, which in turn assists RT activity. Thus, both activities contribute to the final effect of inhibition of the reverse transcription process and thus of the viral life cycle. Our data also point out the strength of NCp7 as a chaperone, as this protein is able to process extremely stable structures, both naturally occurring and further stabilized by small molecules. Therefore, to take advantage of the dual inhibition at the HIV-1 RNA level, we envisage the need of G4 ligands able to potently stabilize the U3 region. In addition, since G4s are also largely present in the cell genome and their stabilization may not be beneficial to noninfected cells, G4 ligands that are also selective for the HIV-1 G4s are highly wished for.
Figure 7.
Conceptual diagram of the present work. ssRNA stands for single-stranded RNA.
Conclusions
Inhibition of NCp7 is an additional and previously unknown activity of G4 ligands. G4 ligands with improved U3 G4 stabilizing activity will likely allow researchers to exploit inhibition of both NCp7 and RT to the fullest extent, and they may lead to the development of anti-HIV-1 drugs with new targets and mechanism of action.
Methods
HIV-1 Recombinant Nucleocapsid Protein
The full-length recombinant nucleocapsid protein (NCp7) was prepared as previously reported.49 The stock solution was stored in aliquots at −80 °C until use. For each analysis, the lowest possible amount of protein was used.
Oligonucleotides and Compound
Synthetic oligonucleotides were purchased from Sigma-Aldrich (Milan, Italy). Table 1 provides specific information about oligos sequences and applications. The G4 ligand BRACO-19 (B19) was obtained from Endotherm (Saarbruecken, Germany).
Table 1. Oligonucleotides Used for This Study.
| application | name | sequence 5′-3′ |
|---|---|---|
| EMSA, CD, MS analysis | RNA U3-III+IV | GGGAGGCGUGGCCUGGGCGGGACUGGGGAGUGG |
| RT stop assay | RNA U3-III+IV RT | GGGAGGCGUGGCCUGGGCGGGACUGGGGAGUGGCGAGCCCUCAGAUCCUGCAUAUAAGCA |
| non-G4 RNA | GUAACCGAUGAGUCUAUGCGAGCCCUCAGAUCCU GCAUAUAAGCA | |
| Primer RT | TGCTTATATGCAGGATCTGAGG |
Electrophoretic Mobility Shift Assay (EMSA)
RNA oligonucleotides, labeled with [γ-32P-ATP] using T4 polynucleotide kinase at 37 °C for 30 min, were annealed by heating at 95 °C for 5 min in lithium cacodylate (10 mM, pH 7.4) and KCl (50 mM) buffer and gradually cooled to room temperature. The annealed oligonucleotides at 15 nM final concentration were added to 20 μL of binding reaction (8% glycerol, 30 mM Tris-HCl, 15 mM MgCl2, 50 μM ZnCl2) containing appropriate concentrations of NCp7. For EMSA unfolding assays, labeled RNA oligonucleotides were annealed to form G4s, and cold DNA complementary oligonucleotides were added to the binding reactions at equimolar or 2-fold excess strand ratio. Binding reactions were incubated for the indicated time at 37 °C in the presence of appropriate protein concentrations. Mixtures (80% of the sample) were then loaded on a 12% polyacrylamide native gel and run at 4 °C for 90 min at 90 V. Gels were dried, exposed overnight, and visualized by phosphorimaging (Typhoon FLA 9000, GE Healthcare).
Circular Dichroism (CD) Analysis
For CD analysis, the RNA oligonucleotides were diluted to 2 μM concentration in 10 mM lithium cacodylate buffer (pH 7.4) supplemented with 50 mM KCl. Samples were annealed by heating at 95 °C for 5 min and gradually cooled to room temperature to allow G4 formation. When the unfolding properties of NCp7 were analyzed, it was added to the samples at 10-fold NCp7/oligonucleotide ratio and incubated for 3 h before CD analysis. Where specified, B19 was added at 8 μM concentration 4 h after the annealing step, and the samples were placed at 4 °C for 24 h to permit G4 stabilization. CD spectra were recorded on a Chirascan-Plus (Applied Photophysics, Leatherhead, UK) instrument equipped with a Peltier temperature controller using a quartz cell of 5 mm optical path length and an instrument scanning speed of 50 nm/min over a wavelength range of 230–320 nm. The reported spectrum of each sample represents the average of 2 scans, and it is baseline corrected for signal contributions due to the buffer. Observed ellipticities were converted to mean residue ellipticity (θ) = deg × cm2 × dmol–1(mol ellip). Unfolding spectra were recorded over a temperature range of 20–90 °C, while 90–20 °C was used for annealing experiments, with a temperature increase/decrease rate of 1 °C/min. Tm values were calculated according to the van ’t Hoff equation, applied for a two-state transition from a folded to unfolded state, assuming that the heat capacity of the folded and unfolded states are equal.
Mass Spectrometry (MS) Analysis
The RNA oligonucleotides were diluted to 5 μM concentration in a final buffer composition consisting of 0.8 mM KCl, 120 mM trimethylammonium acetate (TMAA) adjusted from pH ∼ 7 to 7.4 with triethylamine (TEA). Samples were annealed by heating at 95 °C for 5 min, gradually cooled to room temperature and incubated overnight at 4 °C. Where appropriate, NCp7 was added to the sample at a 1:1 protein/oligonucleotide ratio: the high sensitivity of MS allowed the use of a lower amount of protein compared to the CD analysis. At the time of analysis, a volume of 5 μL of each sample was typically scanned by direct infusion using electrospray ionization (ESI) on a Xevo G2-XS QTOF mass spectrometer (Waters, Manchester, UK). The ESI source settings were as follows: electrospray capillary voltage 1.8 kV; source and desolvation temperatures 45 and 65 °C, respectively; sampling cone voltage 65 V. All these parameters ensured minimal fragmentation of the DNA complexes. The instrument was calibrated using a 2 mg/mL solution of sodium iodide in 50% of isopropanol (IPA). Additionally, the use of the internal standard LockSpray (a solution of leu-enkephalin 1 μg/mL in acetonitrile/water (50:50, v/v) containing 0.1% of formic acid) provided a typical <5 ppm mass accuracy. This high-resolution system allowed us to visualize the isotopic pattern, identify the charge state, and therefore unambiguously calculate the neutral mass of the detected species.
Reverse Transcriptase (RT) Stop Assay
DNA primer was 5′-labeled with [γ-32P-ATP] using T4 polynucleotide kinase at 37 °C for 30 min. The labeled primer (70 nM) was annealed to the RNA U3-III+IV in the presence of 50 mM KCl. The primer extension reaction was performed by recombinant HIV-1 Reverse Transcriptase (1 U/reaction; Calbiochem) in the provided buffer (50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2) at 44 °C for 1 h. Where specified, samples were incubated overnight with increasing concentrations of B19 (500 nM to 4 μM) at room temperature before primer extension. When NCp7 was used, appropriate concentrations (350 nM to 1.5 uM) of it were added immediately before the elongation reaction. Reaction products were treated with NaOH (2 N) at 95 °C for 3 min to permit the alkaline hydrolysis of RNA, and the pH was adjusted with HCl (2 N) to neutrality. Samples were ethanol precipitated, and extension products were separated on 16% denaturating gel and visualized by phosphorimaging (Typhon FLA9000; GE Healthcare).
Acknowledgments
We are thankful to D. Fabris (University of Albany, USA) for helpful discussions. This work was supported by grants to S.N.R. from the European Research Council (ERC Consolidator grant number 615879) and the Bill and Melinda Gates Foundation (grant numbers OPP1035881, OPP1097238).
Glossary
Abbreviations
- NCp7
nucleocapsid protein
- G4
G-quadruplex
- UTRs
untranslated regions
- LTR
long terminal repeat
- RT
reverse transcriptase
- B19
BRACO-19
- EMSA
electrophoretic mobility shift assay
- CD
circular dichroism
- MS
mass spectrometry
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.9b00272.
CD spectra and CD melting/annealing analysis of NCp7-mediated unfolding of U3 III+IV RNA G4 (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Lipps H. J.; Rhodes D. (2009) G-Quadruplex Structures: In Vivo Evidence and Function. Trends Cell Biol. 19 (8), 414–422. 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Kwok C. K.; Merrick C. J. (2017) G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 35 (10), 997. 10.1016/j.tibtech.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Campbell N. H.; Neidle S. (2012) G-Quadruplexes and Metal Ions. Met. Ions Life Sci. 10, 119–134. 10.1007/978-94-007-2172-2_4. [DOI] [PubMed] [Google Scholar]
- Métifiot M.; Amrane S.; Litvak S.; Andreola M.-L. (2014) G-Quadruplexes in Viruses: Function and Potential Therapeutic Applications. Nucleic Acids Res. 42 (20), 12352–12366. 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S.; Moine H.; Vagner S. (2012) G-Quadruplexes in RNA Biology. Wiley Interdiscip. Rev. RNA 3 (4), 495–507. 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- Bugaut A.; Balasubramanian S. (2012) 5′-UTR RNA G-Quadruplexes: Translation Regulation and Targeting. Nucleic Acids Res. 40 (11), 4727–4741. 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay M. M.; Lyons S. M.; Ivanov P. (2017) RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 429 (14), 2127–2147. 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brázda V.; Hároníková L.; Liao J.; Fojta M. (2014) DNA and RNA Quadruplex-Binding Proteins. Int. J. Mol. Sci. 15 (10), 17493–17517. 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. K.; Tawani A.; Mishra A.; Kumar A. (2016) G4IPDB: A Database for G-Quadruplex Structure Forming Nucleic Acid Interacting Proteins. Sci. Rep. 6 (1), 38144. 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González V.; Guo K.; Hurley L.; Sun D. (2009) Identification and Characterization of Nucleolin as a C-Myc G-Quadruplex-Binding Protein. J. Biol. Chem. 284 (35), 23622–23635. 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian W.-X.; Xie Y.; Wang X.-N.; Xu G.-H.; Fu B.-S.; Li S.; Long G.; Zhou X.; Zhang X.-L. (2019) Binding of Cellular Nucleolin with the Viral Core RNA G-Quadruplex Structure Suppresses HCV Replication. Nucleic Acids Res. 47 (1), 56–68. 10.1093/nar/gky1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista M. J.; Martins R. P.; Billant O.; Contesse M.-A.; Findakly S.; Pochard P.; Daskalogianni C.; Beauvineau C.; Guetta C.; Jamin C.; Teulade-Fichou M.-P.; Fåhraeus R.; Voisset C.; Blondel M. (2017) Nucleolin Directly Mediates Epstein-Barr Virus Immune Evasion through Binding to G-Quadruplexes of EBNA1MRNA. Nat. Commun. 8, 16043. 10.1038/ncomms16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista M. J.; Martins R. P.; Angrand G.; Quillévéré A.; Daskalogianni C.; Voisset C.; Teulade-Fichou M.-P.; Fåhraeus R.; Blondel M. (2017) A Yeast Model for the Mechanism of the Epstein-Barr Virus Immune Evasion Identifies a New Therapeutic Target to Interfere with the Virus Stealthiness. Microb. cell (Graz, Austria) 4 (9), 305–307. 10.15698/mic2017.09.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni E.; Frasson I.; Scalabrin M.; Perrone R.; Butovskaya E.; Nadai M.; Palù G.; Fabris D.; Richter S. N. (2015) Nucleolin Stabilizes G-Quadruplex Structures Folded by the LTR Promoter and Silences HIV-1 Viral Transcription. Nucleic Acids Res. 43 (18), 8884–8897. 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago S.; Tosoni E.; Nadai M.; Palumbo M.; Richter S. N. (2017) The Cellular Protein Nucleolin Preferentially Binds Long-Looped G-Quadruplex Nucleic Acids. Biochim. Biophys. Acta, Gen. Subj. 1861 (5 Pt B), 1371–1381. 10.1016/j.bbagen.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd A. K.; Bell M. R.; Raney K. D. (2018) Pif1 Helicase Unfolding of G-Quadruplex DNA Is Highly Dependent on Sequence and Reaction Conditions. J. Biol. Chem. 293, 17792. 10.1074/jbc.RA118.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. C.; Tippana R.; Demeshkina N. A.; Murat P.; Balasubramanian S.; Myong S.; Ferré-D’Amaré A. R. (2018) Structural Basis of G-Quadruplex Unfolding by the DEAH/RHA Helicase DHX36. Nature 558 (7710), 465–469. 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T.; Nakai M.; Ikuta K. (1998) The Life-Cycle of Human Immunodeficiency Virus Type 1. Micron 29 (2–3), 123–138. 10.1016/S0968-4328(98)00002-X. [DOI] [PubMed] [Google Scholar]
- Engelman A.; Cherepanov P. (2012) The Structural Biology of HIV-1: Mechanistic and Therapeutic Insights. Nat. Rev. Microbiol. 10 (4), 279–290. 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L. A.; Bentley K.; Peeters A.; Churchill M. J.; Deacon N. J. (2000) A Compilation of Cellular Transcription Factor Interactions with the HIV-1 LTR Promoter. Nucleic Acids Res. 28 (3), 663–668. 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbondji-Wonje C.; Dong M.; Wang X.; Zhao J.; Ragupathy V.; Sanchez A. M.; Denny T. N.; Hewlett I. (2018) Distinctive Variation in the U3R Region of the 5′ Long Terminal Repeat from Diverse HIV-1 Strains. PLoS One 13 (4), e0195661 10.1371/journal.pone.0195661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Nadai M.; Frasson I.; Poe J. A.; Butovskaya E.; Smithgall T. E.; Palumbo M.; Palù G.; Richter S. N. (2013) A Dynamic G-Quadruplex Region Regulates the HIV-1 Long Terminal Repeat Promoter. J. Med. Chem. 56 (16), 6521–6530. 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Butovskaya E.; Daelemans D.; Palu G.; Pannecouque C.; Richter S. N. (2014) Anti-HIV-1 Activity of the G-Quadruplex Ligand BRACO-19. J. Antimicrob. Chemother. 69 (12), 3248–3258. 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- Perrone R.; Doria F.; Butovskaya E.; Frasson I.; Botti S.; Scalabrin M.; Lago S.; Grande V.; Nadai M.; Freccero M.; Richter S. N. (2015) Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 58 (24), 9639–9652. 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druillennec S.; Caneparo A.; de Rocquigny H.; Roques B. P. (1999) Evidence of Interactions between the Nucleocapsid Protein NCp7 and the Reverse Transcriptase of HIV-1. J. Biol. Chem. 274 (16), 11283–11288. 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- Darlix J.-L.; Godet J.; Ivanyi-Nagy R.; Fossé P.; Mauffret O.; Mély Y. (2011) Flexible Nature and Specific Functions of the HIV-1 Nucleocapsid Protein. J. Mol. Biol. 410 (4), 565–581. 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Williams M. C.; Gorelick R. J.; Musier-Forsyth K. (2002) Specific Zinc-Finger Architecture Required for HIV-1 Nucleocapsid Protein’s Nucleic Acid Chaperone Function. Proc. Natl. Acad. Sci. U. S. A. 99 (13), 8614–8619. 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. G.; Mitra M.; Mascarenhas A.; Musier-Forsyth K. (2010) Role of HIV-1 Nucleocapsid Protein in HIV-1 Reverse Transcription. RNA Biol. 7 (6), 754–774. 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y.; Rosen A. E.; Guo J.; Musier-Forsyth K.; Levin J. G. (2003) Efficient Initiation of HIV-1 Reverse Transcription in Vitro: Requirement for RNA Sequences Downstream of the Primer Binding Site Abrogated by Nucleocapsid Protein-Dependent Primer-Template Interactions. J. Biol. Chem. 278 (16), 14185–14195. 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- Lyonnais S.; Gorelick R. J.; Mergny J.; Le Cam E.; Mirambeau G. (2003) G-Quartets Direct Assembly of HIV-1 Nucleocapsid Protein along Single-Stranded DNA. Nucleic Acids Res. 31 (19), 5754–5763. 10.1093/nar/gkg716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankia B. I.; Barany G.; Musier-Forsyth K. (2005) Unfolding of DNA Quadruplexes Induced by HIV-1 Nucleocapsid Protein. Nucleic Acids Res. 33 (14), 4395–4403. 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran A.; Endo M.; Hidaka K.; Tran P. L. T.; Mergny J.-L.; Gorelick R. J.; Sugiyama H. (2013) HIV-1 Nucleocapsid Proteins as Molecular Chaperones for Tetramolecular Antiparallel G-Quadruplex Formation. J. Am. Chem. Soc. 135 (49), 18575–18585. 10.1021/ja409085j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand A.; Gabelica V. (2014) Native Electrospray Mass Spectrometry of DNA G-Quadruplexes in Potassium Solution. J. Am. Soc. Mass Spectrom. 25 (7), 1146–1154. 10.1007/s13361-014-0890-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M.; Harrison R. J.; Romagnoli B.; Tanious F. A.; Gowan S. H.; Reszka A. P.; Wilson W. D.; Kelland L. R.; Neidle S. (2001) Structure-Based Design of Selective and Potent G Quadruplex-Mediated Telomerase Inhibitors. Proc. Natl. Acad. Sci. U. S. A. 98 (9), 4844–4849. 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Liu H.-H.; Zheng K.-W.; Hao Y.-H.; Tan Z. (2013) DNA G-Quadruplex Formation in Response to Remote Downstream Transcription Activity: Long-Range Sensing and Signal Transducing in DNA Double Helix. Nucleic Acids Res. 41 (14), 7144–7152. 10.1093/nar/gkt443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalabrin M.; Frasson I.; Ruggiero E.; Perrone R.; Tosoni E.; Lago S.; Tassinari M.; Palù G.; Richter S. N. (2017) The Cellular Protein HnRNP A2/B1 Enhances HIV-1 Transcription by Unfolding LTR Promoter G-Quadruplexes. Sci. Rep. 7, 45244. 10.1038/srep45244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankovic S.; Varadarajan J.; Ramalho R.; Aiken C.; Rousso I. (2017) Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J. Virol. 91 (12), e00289-17. 10.1128/JVI.00289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. G.; Guo J.; Rouzina I.; Musier-Forsyth K. (2005) Nucleic Acid Chaperone Activity of HIV-1 Nucleocapsid Protein: Critical Role in Reverse Transcription and Molecular Mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80, 217–286. 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Darlix J.; Garrido J. L.; Morellet N.; Mély Y.; Rocquigny H. de. (2007) Properties, Functions, and Drug Targeting of the Multifunctional Nucleocapsid Protein of the Human Immunodeficiency Virus. Adv. Pharmacol. 55, 299–346. 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- Darlix J.-L.; de Rocquigny H.; Mauffret O.; Mély Y. (2014) Retrospective on the All-in-One Retroviral Nucleocapsid Protein. Virus Res. 193, 2–15. 10.1016/j.virusres.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman D.; Goldschmidt V.; Barraud P.; Marquet R.; Paillart J.-C.; Tisné C. (2012) Initiation of HIV-1 Reverse Transcription and Functional Role of Nucleocapsid-Mediated TRNA/Viral Genome Interactions. Virus Res. 169 (2), 324–339. 10.1016/j.virusres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Bampi C.; Jacquenet S.; Lener D.; Décimo D.; Darlix J.-L. (2004) The Chaperoning and Assistance Roles of the HIV-1 Nucleocapsid Protein in Proviral DNA Synthesis and Maintenance. Int. J. Biochem. Cell Biol. 36 (9), 1668–1686. 10.1016/j.biocel.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Kanevsky I.; Chaminade F.; Chen Y.; Godet J.; René B.; Darlix J.-L.; Mély Y.; Mauffret O.; Fossé P. (2011) Structural Determinants of TAR RNA-DNA Annealing in the Absence and Presence of HIV-1 Nucleocapsid Protein. Nucleic Acids Res. 39 (18), 8148–8162. 10.1093/nar/gkr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S.; Stoylov S.; Piémont E.; Ficheux D.; Roques B. P.; Darlix J. L.; Mély Y. (2002) HIV-1 Nucleocapsid Protein Activates Transient Melting of Least Stable Parts of the Secondary Structure of TAR and Its Complementary Sequence. J. Mol. Biol. 317 (3), 385–399. 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- McCauley M. J.; Rouzina I.; Manthei K. A.; Gorelick R. J.; Musier-Forsyth K.; Williams M. C. (2015) Targeted Binding of Nucleocapsid Protein Transforms the Folding Landscape of HIV-1 TAR RNA. Proc. Natl. Acad. Sci. U. S. A. 112 (44), 13555–13560. 10.1073/pnas.1510100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum G. E.; Park Y. W.; Singleton S. F.; Dervan P. B.; Breslauer K. J. (1990) Thermodynamic Characterization of the Stability and the Melting Behavior of a DNA Triplex: A Spectroscopic and Calorimetric Study. Proc. Natl. Acad. Sci. U. S. A. 87 (23), 9436–9440. 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norseen J.; Johnson F. B.; Lieberman P. M. (2009) Role for G-Quadruplex RNA Binding by Epstein-Barr Virus Nuclear Antigen 1 in DNA Replication and Metaphase Chromosome Attachment. J. Virol. 83 (20), 10336. 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosic A.; Saccone I.; Carraro C.; Kenderdine T.; Gamba E.; Caliendo G.; Corvino A.; Di Vaio P.; Fiorino F.; Magli E.; Perissutti E.; Santagada V.; Severino B.; Spada V.; Fabris D.; Frecentese F.; Gatto B. (2018) Non-Natural Linker Configuration in 2,6-Dipeptidyl-Anthraquinones Enhances the Inhibition of TAR RNA Binding/Annealing Activities by HIV-1 NC and Tat Proteins. Bioconjugate Chem. 29 (7), 2195–2207. 10.1021/acs.bioconjchem.8b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan N.; Fabris D. (2003) Direct Mass Spectrometric Determination of the Stoichiometry and Binding Affinity of the Complexes between Nucleocapsid Protein and RNA Stem-Loop Hairpins of the HIV-1 Ψ-Recognition Element. Biochemistry 42 (36), 10736–10745. 10.1021/bi0348922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.