Abstract
In response to large-scale radiological incidents, rapid, accurate, and early triage biodosimeters are urgently required. In this study, we investigated candidate radiation-responsive biomarkers using proteomics approaches in mouse models. A total of 452 dysregulated proteins were identified in the serum samples of mice exposed to 0, 2, 5.5, 7, and 8 Gy at 6, 24, and 72 hours postirradiation. Ninety-eight proteins, including 46 at 6 hours, 36 at 24 hours, and 36 at 72 hours, were identified as radiation-responsive proteins (RRPs). Gene Ontology analysis showed the RRPs were involved in proteolysis, extracellular space, hydrolase activity, and carbohydrate binding. Kyoto Encyclopedia of Genes and Genome enrichment showed the RRPs were regulated in “the pentose phosphate pathway,” “the proteasome,” and “AGE-RAGE signaling in diabetic complications.” There were 3 proteins changed and overlapped at all the 3 time points, 8 proteins changed at 6 and 24 hours, 4 proteins changed at 24 and 72hours, and 2 proteins changed at both 6 and 72 hours. Of these proteins, ORM2, HP, SAA1, SAA2, MBL2, COL1A1, and APCS were identified as candidate biomarkers for biodosimeter-based diagnosis through Pearson correlation analysis.
Keywords: proteomics, biomarkers, serum, inflammatory, radiation
Introduction
In large-scale nuclear and radiation accidents such as the Chernobyl disaster on April 26, 1986, and the Fukushima Daiichi reactor incident on March 11, 2011, it is crucial that exposed individuals are distinguished from nonexposed groups to predict received radiation doses that can guide timely and effective medical treatment.1-3 In real-life situations, the radiation dose is variable and accurate dose estimations with appropriate radiation biodosimeters are required. Techniques such as lymphocyte reduction kinetics analysis and γ-H2AX foci assessments have been used for dose estimations and can guide patient management and triage when used in conjunction with other clinical and laboratory evidence. However, both methods are generally unsuitable for the assessment of partial-body exposure, which is common in radiation accidents,4,5 and both are time-consuming.6,7 The rapid immunodetection of radiation-responsive biomarkers would represent an alternative diagnostic tool for the mass screening of exposed individuals.
Flt3 ligand, C-reactive protein, and interleukin-6 are expressed in both mouse and NHP serum/plasma and have been proposed as early indicators of dose estimations and radiation-induced injury.8,9 More recently, proteomics-based approaches have been used to identify novel biomarkers for radiation biodosimeters.10 Robert et al showed how 5 proteins (AMY, FLT3L, MCP1, AACT, and NGAL) could discriminate radiation exposure in nonhuman primates receiving 1 to 10 Gy, 7 days postirradiation.11 Younghyun et al showed that the combined expression of FDXR, DDB2, and ACTN1 could accurately predict the received dose through humanized mice models irradiated with 0 to 2 Gy after 3 days.12 Due to differences in experimental animal models, the utility of radiation-responsive proteins (RRPs) as radiation biomarkers remains at the discovery stage and more robust validation studies are required.
In this study, to fully characterize the range of RRPs for dose estimations, we used proteomics to screen distinct biomarkers in mice exposed to different doses of radiation over a range of time points. Functional and pathway enrichment analyses were implemented for the RRPs to understand radiation pathogenesis. Based on the identified correlations between protein expression and irradiation doses at different time points, 7 proteins were selected as candidate biomarkers for radiation biodosimeters.
Materials and Methods
Mice, Total Body Irradiation, and Serum Collection
C57BL/6J male mice (6-8 weeks old) were obtained from and raised in the Academy of Military Medical Sciences (Beijing, China). Mice were irradiated in 0, 2, 5.5, 7, and 8 Gy using 60Co source γ-ray at a dose rate of 101.90 cGy/min. At different time points (6, 24, and 72 hours), blood was collected from the orbital plexus and mice were killed. Serum was isolated by centrifugation at 3000 rpm/min for 5 minutes at 4°C and stored at −80°C. Five mice were included in each dose group (excluding mice irradiated in 0 Gy at 6 and 72 hours, n = 4 per group) for biological replication of proteomic analysis, with 73 animals in total. And we established another group of mouse exposed to 0, 2, 5.5, 7, and 8 Gy, 8 mice were included in each dose group, to assess survival, body weight, and blood counts. Animal care and handling were performed in accordance with the “Guide for the Care and Use of Laboratory Animal of AMMS in China” and all animal experiments were approved by the Animal Care and Use Committee of the Beijing Institute of Radiation Medicine (Beijing, China).
Mass Spectrometry
High performance liquid chromatography-mass spectrometer (HPLC-MS)/MS analysis was performed on a Q-Exactive HF Mass Spectrometer (ThermoScientific, Waltham, MA) equipped with a nanoelectrospray ionization source and an EASY-nLC 1000 system (ThermoScientific, Waltham, MA). Samples were dissolved in formic acid (FA; 0.1%) and separated on a capillary column (150 µm id × 120 mm) packed with C18 (3 µm, 100 Å) at a flow rate of 600 nL/min. Mobile phase A (99.9% water/0.1% FA) and mobile phase B (99.9% acetonitrile/0.1% FA) were employed. Elution gradients were from 6% to 32% mobile phase B for 78 minutes. Data acquisition was performed using the data-dependent mode. Q-Exactive HF was run under the positive mode. MS1 survey scans were implemented with a mass range of 300 to 2000 at a resolution of 120 000 at 200 m/z. The automatic gain control (AGC) was set as 3e6 with a maximum injection time (MIT) of 80 milliseconds in the MS1 survey. The 20 most intense ions were subjected to fragmentation in the MS2 and were analyzed. The AGC was set at 5e4 and the MIT was set at 80 milliseconds for each M2 scan. A dynamic range of 12 seconds was established to avoid repeated detection of the same ion peaks. Raw data files of the peptides from the Q-Exactive HF MS were searched against the UniProt database (Release 2014-09, 140 916 entries) using MaxQuant software (version 1.5.3.8).
Statistical and Bioinformatics Analysis
A 2-sample Student t-test (2-tailed) was performed to compare protein levels between the groups. A total of 98 RRPs were identified with the cutoff criteria of the P values ≤.05 and |log2 fold-change (FC)|>1 in the highest dose of 8 Gy compared to 0 Gy at 6, 24, and 72 hours. Gene Ontology analysis according to DAVID (https://david.ncifcrf.gov) was applied for the identification and annotation of the 98 RRPs in the 3 categories of biological processes (BPs), cellular components (CCs), and molecular functions (MFs). KOBAS version 3.0 (http://kobas.cbi.pku.edu.cn/index.php) was used for the Kyoto Encyclopedia of Genes and Genome (KEGG) signaling pathway of the 98 proteins. To assess the correlation between protein expression and irradiated doses at all time points, protein intensities were log10 transformed for all 98 RRPs. Pearson correlation analysis was performed and values of P <.05 were considered statistically significant. Student t tests, hierarchical clustering analysis, and Pearson correlation analysis were performed using OmicShare (http://www.omicshare.com/tools).
Results
Protein Expression in C57BL/6J Mice After Ionizing Radiation Exposure
A total of 452 proteins were identified across the samples (Supplementary Table 1). To establish the mouse model in identical radiation conditions, mice were exposed to 0, 2, 5.5, 7, and 8 Gy and survival, body weight, and blood counts were assessed. Our published data13 show that mice treated with 8 Gy die after 16 days, while only 4 of the 8 mice survive for 30 days following 7 Gy treatment. Mice treated with 0, 2, and 5.5 Gy all survived. We, therefore, investigated the RRPs in mice treated with 8 Gy at 6, 24, and 72 hours in which FCs ≥2 and P values ≤.05 were considered candidate proteins. A total of 98 RRPs were identified, including 46 at 6 hours, 36 at 24 hours, and 36 at 72 hours postradiation (Supplementary Table 2). Hierarchical clustering analysis was performed to cluster the 98 RRPs at each time point. Figure 1 shows the heat maps of normalized protein abundance and protein clustering. Figure 2A shows that at after 6 hours of radiation treatment, 46 RRPs clustered in 0, 2, 5.5, and 7 Gy treatment groups. After 24 hours of radiation, 36 proteins were differentially expressed between 0 and 2, 5.5, 7, 8 Gy in which 2 and 5.5 Gy groups and 7 and 8 Gy groups clustered together. At 72 hours postradiation, 0 and 2 Gy groups and 5.5, 7, and 8 Gy groups showed clustering.
Figure 1.
Heat map showing normalized protein abundance, protein clustering, and irradiation doses (0, 2, 5.5, 7, and 8 Gy) of 98 proteins at (A) 6 hours, (B) 24 hours, and (C) 72 hours.
Figure 2.
Gene Ontology terms related to biological processes, cellular components, and molecular functions at (A) 6 hours, (B) 24 hours, and (C) 72 hours postradiation.
Gene Ontology Analysis of the RRPs
We performed GO analysis of (1) BPs, (2) CCs, and (3) MFs to discern the functional process and BP influenced by the RRPs (Supplementary Table 3). Figure 2A shows that after 6 hours radiation, RRPs mapping to proteolysis (n = 6) was the most abundant GO terms. Considering CCs, RRPs in the extracellular space (n = 28) were the most abundant. In the MFs category, the RRPs were mostly assigned to carbohydrate binding (n = 6). Figure 2B shows that after 24 hours of radiation in the BPs category, RRPs mapping to proteolysis (n = 7) was the most abundant GO terms. For CCs, RRPs in extracellular exosomes (n = 23) were the most abundant. Regarding MFs, the RRPs were mostly assigned to hydrolase activity (n = 8). Figure 2C shows that after 72 hours of radiation in the BPs category, RRPs involved in proteolysis (n = 6) were the most abundant. Considering CCs, proteins mapping to the extracellular space (n = 18) was the most abundant. Regarding MFs, RRPs were mostly assigned to hydrolase activity (n = 7).
Kyoto Encyclopedia of Genes and Genome Enrichment Analysis of the RRPs
To identify the latent involvement of the RRPs at different time points postradiation, KEGG enrichment was performed. At 6 hours postradiation, 46 RRPs were identified and found to regulate “the pentose phosphate pathway,” “the biosynthesis of amino acids,” and “carbon metabolism.” At 24 hours postradiation, the regulated pathways of the 36 RRPs were “the proteasome,” “complement and coagulation cascades,” and “extracellular matrix (ECM)–receptor interactions.” At 72 hours postradiation, 36 RRPs were identified and found to regulate “AGE-RAGE signaling in diabetic complications,” “ECM–receptor interactions,” and “leukocyte transendothelial migration” (Figure 3).
Figure 3.
Kyoto Encyclopedia of Genes and Genome signaling analysis of radiation-responsive proteins at (A) 6 hours, (B) 24 hours, and (C) 72 hours postradiation.
Selection of Candidate Protein Markers for Radiation Biodosimeters
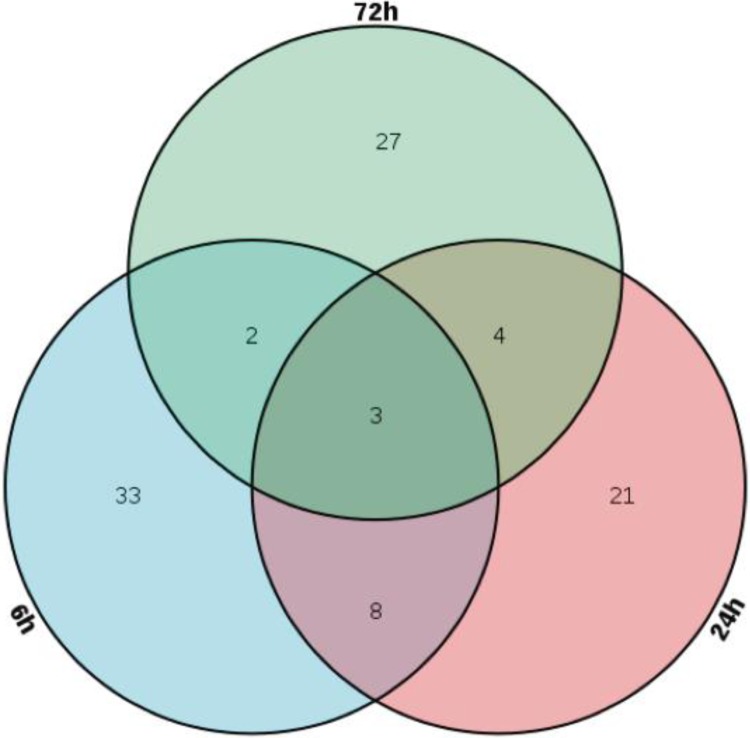
The overlapping distribution of the 98 RRPs at 6, 24, and 72 hours is shown in Figure 4. There were 33 RRPs in the 8 Gy group at 6 hours, 24 proteins in the 8 Gy group at 24 hours, and 27 proteins in the 8 Gy group at 72 hours. ORM2, HP, and APCS were common at all time points. Table 1 lists the overlapping FCs at each time point. Eight proteins (TKT, PGAM1, SAA1, SPP1, LCP1, SAA2, APOH, and PF4) showed significant changes at 6 and 24 hours after 8 Gy irradiation. Four proteins (MBL2, BPIFA2, COL1A2, and COL1A1) changed at both 24 and 72 hours after 8 Gy irradiation. Only 2 proteins (Ltf and Sell) significantly changed at both 6 and 72 hours after 8 Gy irradiation. SAA1 and SAA2 showed the largest changes (∼1000-fold). To assess dose-dependent responses, the correlation between protein expression and irradiated dose at all time points was evaluated by Pearson correlation analysis. Proteins with high correlations (correlation coefficient ≥0.85; P value ≤ .05) are indicated in Table 2. Figure 5 shows 7 candidate proteins (ORM2, HP, SAA1, SAA2, MBL2, COL1A1, and APCS) that were selected due to their change in abundance at more than 1 time point and relatively low variability between samples. Not all the 7 candidate proteins could meet the criteria at all the 3 time points postradiation. Therefore, ORM2 and HP were the only 2 proteins in good correlation with irradiated dose at all the 3 time points. Then, SAA1 and SAA2 were selected as candidate protein markers for radiation biodosimeters at just 6 and 24 hours postradiation. Finally, at 24 and 72 hours postradiation, MBL2, COL1A1, and APCS were in fine correlation with irradiated dose.
Figure 4.
Venn diagram of all 98 radiation-responsive proteins at 6, 24, and 72 hours postradiation.
Table 1.
Fold-Changes of Candidate Radiation-Responsive Proteins at Overlapping Time Points.
| Gene Name | 6 Hours | 24 Hours | 72 Hours | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 Gy | 5.5 Gy | 7 Gy | 8 Gy | 2 Gy | 5.5 Gy | 7 Gy | 8 Gy | 2 Gy | 5.5 Gy | 7 Gy | 8 Gy | |
| ORM2 | 2.35 | 3.37 | 3.32 | 8.46 | 9.23 | 9.09 | 15.02 | 13.08 | 5.78 | 22.05 | 18.74 | 23.44 |
| HP | 2.34 | 8.29 | 7.92 | 14.84 | 3.36 | 7.30 | 7.62 | 9.62 | 70.69 | 327.48 | 231.09 | 264.50 |
| APCS | 0.75 | 1.72 | 1.51 | 2.59 | 4.39 | 6.61 | 9.44 | 10.29 | 1.00 | 6.89 | 4.07 | 4.15 |
| SAA1 | 25.41 | 57.44 | 89.41 | 267.96 | 494.95 | 3314.42 | 5152.64 | 8774.95 | ||||
| SAA2 | 7.44 | 36.73 | 504.29 | 1758.73 | 20.72 | 502.73 | 655.40 | 1068.04 | ||||
| TKT | 9.65 | 41.57 | 18.20 | 70.56 | 1.67 | 2.20 | 3.59 | 4.64 | ||||
| LCP1 | 1.82 | 5.94 | 6.63 | 22.77 | 1.90 | 2.49 | 2.18 | 3.03 | ||||
| APOH | 23.66 | 5.90 | 9.30 | 13.19 | 1.57 | 2.08 | 3.07 | 5.19 | ||||
| PGAM1 | 2.43 | 1.51 | 1.86 | 3.17 | 2.11 | 3.05 | 4.87 | 3.17 | ||||
| SPP1 | 1.16 | 0.57 | 0.41 | 0.09 | 0.15 | 1.15 | 1.29 | 2.63 | ||||
| PF4 | 2.57 | 1.97 | 3.41 | 3.72 | 1.53 | 1.15 | 2.02 | 2.06 | ||||
| BPIFA2 | 11.07 | 5.79 | 6.80 | 10.03 | 22.47 | 22.42 | 94.85 | 93.50 | ||||
| MBL2 | 1.70 | 2.14 | 2.45 | 2.16 | 1.92 | 3.66 | 3.47 | 4.61 | ||||
| COL1A2 | 2.34 | 3.50 | 3.30 | 4.67 | 3.74 | 7.23 | 5.19 | 7.13 | ||||
| COL1A1 | 1.68 | 2.16 | 2.21 | 2.32 | 2.72 | 4.13 | 4.35 | 4.89 | ||||
| LTF | 2.47 | 6.13 | 4.77 | 7.91 | 1.45 | 0.10 | 0.10 | 0.12 | ||||
| SELL | 1.70 | 2.52 | 2.37 | 2.22 | 0.37 | 0.04 | 0.01 | 0.04 | ||||
Table 2.
Correlation Between Protein Expression and Irradiated Doses at Different Time Points Postradiation.
| Gene Name | 6 Hours | 24 Hours | 72 Hours | |||
|---|---|---|---|---|---|---|
| Correlation Coefficient | P Value | Correlation Coefficient | P Value | Correlation Coefficient | P Value | |
| ORM2 | 0.925 | .024 | 0.939 | .018 | 0.963 | .008 |
| HP | 0.980 | .003 | 0.948 | .014 | 0.915 | .029 |
| SAA1 | 0.996 | .000 | 0.964 | .008 | ||
| SAA2 | 0.943 | .016 | 0.981 | .003 | ||
| APOH | 0.886 | .045 | 0.979 | .004 | ||
| APCS | 0.945 | .015 | 0.900 | .038 | ||
| COL1A1 | 0.933 | .021 | 0.921 | .026 | ||
| MBL2 | 0.922 | .026 | 0.968 | .007 | ||
| LTF | 0.950 | .013 | −0.928 | .023 | ||
| SELL | 0.898 | .039 | −0.942 | .017 | ||
| ACTC1 | 0.980 | .003 | ||||
| LCP1 | 0.962 | .009 | ||||
| CTSD | 0.944 | .016 | ||||
| RBP4 | 0.918 | .028 | ||||
| IGHV1-75 | 0.896 | .040 | ||||
| ACTB | 0.892 | .042 | ||||
| PPBP | −0.956 | .011 | ||||
| PROZ | −0.959 | .010 | ||||
| SPP1 | −0.988 | .002 | ||||
| FLT4 | −0.995 | .000 | ||||
| PLA2G7 | 0.981 | .003 | ||||
| MMRN1 | 0.970 | .006 | ||||
| TRF | 0.960 | .009 | ||||
| LRG1 | 0.956 | .011 | ||||
| ITIH4 | 0.953 | .012 | ||||
| PSMA1 | 0.948 | .014 | ||||
| SERPING1 | 0.930 | .022 | ||||
| PSMA3 | 0.913 | .030 | ||||
| TKT | 0.912 | .031 | ||||
| COL1A2 | 0.886 | .045 | ||||
| CPB2 | 0.885 | .046 | ||||
| MMP2 | 0.995 | .000 | ||||
| IGHV10-1 | 0.977 | .004 | ||||
| C4BPA | 0.963 | .009 | ||||
| TNC | 0.954 | .012 | ||||
| IGHV1-9 | 0.936 | .019 | ||||
| IGKV4-71 | 0.886 | .045 | ||||
| FGB | 0.882 | .048 | ||||
| MUP3 | −0.960 | .009 | ||||
| CES2A | −0.973 | .005 | ||||
Figure 5.
Dose–response relationship of the 7 candidate proteins biomarkers (ORM2, HP, SAA1, SAA2, MBL2, COL1A1, and APCS) at 6, 24, and 72 hours after radiation. The error bar in curve was calculated from data of protein abundance transformed by log10 from mouse in each dose group. Data indicate mean ± standard deviation.
Discussion
A radiation biodosimeter was used to triage individuals exposed to ionizing radiation, which could impact the capacity to provide timely and effective medical treatment following large-scale nuclear and radiation incidents.14 The principle of radiation biodosimeter is to utilize changes induced in individual by ionizing radiation as a quantitative measure of the amount of radiation energy observed.15 In recent years, several biodosimeters including lymphocyte reduction kinetics analysis and γ-H2AX foci assessments have been used to distinguish nonexposed groups and to predict the received radiation dose.6,7 However, lymphocyte reduction kinetics are generally unsuitable for the assessment of partial-body exposure, which is common in radiation incidents. γ-H2AX foci analysis is also time-consuming and requires technical expertise. An increasing number of studies have employed MS-based proteomics to detect radiation biomarkers in whole blood serum/plasma samples. In this study, we used proteomics to identify novel radiation biomarkers in the serum of γ-irradiated female C57BL/6J mice. At 6, 24, and 72 hours postirradiation (0, 2, 5.5, 7, and 8 Gy), 452 biomarkers were identified. These RRPs were present in the 8 Gy group at each time point, with FCs ≥2 and P values ≤ .05 compared to the 0 Gy group. We identified 98 candidate RRPs, including 46 at 6 hours, 36 at 24 hours, and 36 at 72 hours postirradiation. TKT, PGAM1, SAA1, SPP1, LCP1, SAA2, APOH, and PF4 showed significant changes at both 6 and 24 hours after 8 Gy irradiation. MBL2, BPIFA2, COL1A2, and COL1A1 showed significant changes at both 24 and 72 hours following 8 Gy irradiation. Only Ltf and Sell significantly changed at both 6 and 72 hours following 8 Gy irradiation. To select candidate biomarkers, Pearson correlation analysis was performed for all 98 RRPs. Those with a high correlation (correlation coefficient ≥0.85; P value ≤ .05) are given in Table 2.
ORM2, HP, SAA1, SAA2, MBL2, COL1A1, and APCS were selected for radiation biodosimeters based on their overlap at several time points and consistency between donors. Most of these proteins regulate inflammatory responses. ORM2 (orosomucoid-2) and α1-acid glycoprotein are acute-phase proteins that are synthesized by the liver and secreted into the plasma.16 The concentration of ORM2 in mouse plasma is approximately 0.2 to 0.4 mg/mL, which increases 10- to 200-fold within 24 hours of disease stimuli, including acute inflammation,17 physical trauma,18 bacterial infections,18 cholangiocarcinoma,19 chronic kidney disease,20 asthma,21 and colorectal cancer.22 Studies on ORM2 in response to radiation are sparse. Sharma et al analyzed mouse urine samples by LC-MS/MS and two-dimensional electrophoresis and showed that α1-acid glycoproteins increased ≥5-fold within 24 hours of total body irradiation (TBI).23 Haptoglobin is an acute-phase glycoprotein generated by the liver in response to inflammation.24 Serum haptoglobin levels significantly increase in various tumors, including breast carcinoma,25 oral squamous cell carcinoma,26 ovarian cancer,27 lung cancer,28 and colorectal cancer.29 A number of studies have shown the effects of radiation on the levels of haptoglobin. Plasma haptoglobin values for 12 Gy partially irradiated rats indicate that from 1 to 101 days postirradiation, the average plasma haptoglobin levels significantly increase in irradiated rats compared to control groups.30 Magic et al reported that plasma haptoglobin levels increase in rats after 4 to 12 Gy irradiation early after exposure. Later, time points were not investigated.31 Through proteomic approaches, the haptoglobin levels of bone marrow tissue in 4 Gy irradiated mice were shown to be upregulated 24 hours after exposure.32 Serum amyloid A1 (SAA1) and serum amyloid A2 (SAA2) are synthesized and secreted by the liver and are collectively referred to as acute-phase protein serum amyloid A, as both increase during acute-phase inflammatory events including acute bacterial infection, trauma, inflammatory and autoimmune disease, and neoplasia.33 Plasma SAA1 levels in C57BL/6J mice treated with ≤6 Gy were elevated on the first 2 days and showed maximal expression on day 1.34 The utility of SAA1 as a radiation biomarker was studied by Kim et al,35 who indicated that plasma SAA1 levels increase in a dose-dependent manner during the first 2 days of 0.1 to 6 Gy irradiation exposure in female C57BL/6J mice. At doses of 1 to 8 Gy, serum SAA levels at 24 hours could accurately differentiate TBI mice into control/1 Gy and ≥2 Gy groups in blind studies.36
Serum amyloid P component (APCS) is a plasma glycoprotein that is synthesized in the liver and increases approximately 10-fold after 24 hours of endotoxin treatment.37 APCS is a biomarker for mild cognitive impairment, Alzheimer disease,38 and ankylosing spondylitis,39 but its response to ionizing radiation has not been reported. Mannose-binding protein (MBL) plays a key role in the innate immune response to pathogen infections.40 MBL2 (mannose-binding protein C) is a genetic variant41 that regulates pulmonary severity in cystic fibrosis42 and is a risk factor for immune-mediated complications in chronic granulomatous disease.43 MBL2 was overexpressed in the small intestine of mice at 24 hours and 3.5 days after TBI by laser capture microdissection.44 During the early stages of radiation, inflammatory reactions are a common pathophysiological response.45,46 Collagen type I α1 (COL1A1) is an important protein of bone, skin, tendons, and blood vessel walls that regulates tumor cell adhesion, gap junctions, and the ECM.47 Previous studies indicate that COL1A1 is overexpressed in a series of cancers, including gastric and breast cancer,48 and is a biomarker for the prognosis of hepatocellular carcinoma and gastric cancer.49 Radiation-induced tissue fibrosis occurs due to the excessive synthesis of collagen.49 Combing the identified RRPs with known radiation-induced genes may increase the accuracy of diagnostic predictions.
We note some limitations to this study. Disparity exists between humans and mice, and although mammalian and human irradiation models are rare, primates can be used to verify the utility of the biomarkers. Future studies should assess the dose–response relationships of the RRPs over days to weeks. Then, the 7 candidate biomarkers we identified were not able to be special radiation biodosimeter by reading-related literature. To this end, studies should be designed to sustain and validate the candidate biomarkers, permitting the development of biodosimeters for rapid, accurate, and early triage in large-scale radiological incidents.
Conclusions
Based on LC-MS/MS-based label-free quantitative analysis, 452 proteins were identified in the serum of mice at 6, 24, and 72 hours posttreatment with 0, 2, 5.5, 7, and 8 Gy. A total of 98 proteins, including 46 at 6 hours, 36 at 24 hours, and 36 at 72 hours, were identified as RRPs. Through Pearson correlation analysis, ORM2, HP, SAA1, SAA2, MBL2, COL1A1, and APCS were shown to be candidate biomarkers for radiation biodosimeters. Further studies to validate the dose–response relationships of these biomarkers up to a week after exposure in nonhuman primates should now be performed to validate their utility as rapid, accurate, and early triage biodosimeters for large-scale radiological incidents.
Supplemental Material
Supplemental Material, S1-452_proteins_expression_profiling for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Supplemental Material
Supplemental Material, S2-98_radiation-responsive_proteins for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Supplemental Material
Supplemental Material, S3-GO_analysis_of_98_radiation-responsive_proteins for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Supplemental Material
Supplemental Material, S4-KEGG_analysis_of_98_radiation-responsive_proteins for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Footnotes
Authors’ Note: Jinfeng Huang and Qi Wang contributed equally to this study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Major Project: BWS18J008, AWS14C014, BWS14J052, and 16CXZ027.
ORCID iD: Meijuan Zhou  https://orcid.org/0000-0001-6822-9774
https://orcid.org/0000-0001-6822-9774
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Chin FKC. Scenario of a dirty bomb in an urban environment and acute management of radiation poisoning and injuries. Singapore Med J. 2007;48(10):950–957. [PubMed] [Google Scholar]
- 2. Coeytaux K, Bey E, Christensen D, Glassman ES, Murdock B, Doucet C. Reported radiation overexposure accidents worldwide, 1980–2013: a systematic review. PLoS One. 2015;10(3):e0118709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pernot E, Hall J, Baatout S, et al. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat Res. 2012;751(2):258–286. doi:10.1016/j.mrrev.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 4. Goans RE, Holloway EC, Berger ME, Ricks RC. Early dose assessment following severe radiation accidents. Health Phys. 1997;72(4):513–518. [DOI] [PubMed] [Google Scholar]
- 5. Berger ME, Christensen DM, Lowry PC, Jones OW, Wiley AL. Medical management of radiation injuries: current approaches. Occupat Med. 2006;56(3):162–172. [DOI] [PubMed] [Google Scholar]
- 6. Sandrine R-L, Tania M, Pascale V, et al. Quantification of gamma-H2AX foci in human lymphocytes: a method for biological dosimetry after ionizing radiation exposure. Radiat Res. 2010;174(2):185–194. [DOI] [PubMed] [Google Scholar]
- 7. Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327(1-2):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blakely WF, Bolduc DL, Debad J. et al. Use of proteomic and hematology biomarkers for prediction of hematopoietic acute radiation syndrome severity in baboon radiation models. Health Phys. 2018;115(1):29–36. [DOI] [PubMed] [Google Scholar]
- 9. Ossetrova NI, Sandgren DJ, Blakely WF. Protein biomarkers for enhancement of radiation dose and injury assessment in nonhuman primate total-body irradiation model. Radiat Prot Dosimetry. 2014;159(1-4):61–76. [DOI] [PubMed] [Google Scholar]
- 10. Fenech M. Current status, new frontiers and challenges in radiation biodosimetry using cytogenetic, transcriptomic and proteomic technologies. Radiat Measure. 2011;46(9):737–741. [Google Scholar]
- 11. Balog RP, Chang P, Javitz HS, et al. Development of a point-of-care radiation biodosimeter: studies using novel protein biomarker panels in non-human primates. Int J Radiat Biol. 2018. [DOI] [PubMed] [Google Scholar]
- 12. Lee Y, Pujol Canadell M, Shuryak I, et al. Candidate protein markers for radiation biodosimetry in the hematopoietically humanized mouse model. Sci Rep. 2018;8(1):13557 doi:10.1038/s41598-018-31740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li L, Xiao R, Wang Q, et al. SERS detection of radiation injury biomarkers in mouse serum. RSC Adv. 2018;8(10):5119–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flood AB, Boyle HK, Du G, et al. Advances in a framework to compare biodosimetry methods for triage in large-scale radiation events. Radiat Protect Dosimetry. 2014;159(1-4):77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swartz HM, Williams BB, Flood AB. Overview of the principles and practice of biodosimetry. Radiat Environ Biophys. 2014;53(2):221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ceciliani F, Pocacqua V. The acute phase protein alpha1-acid glycoprotein: a model for altered glycosylation during diseases. Curr Protein Pept Sci. 2007;8(1):91–108. [DOI] [PubMed] [Google Scholar]
- 17. Magid E, Guldager H, Hesse D, Christiansen MS. Monitoring urinary orosomucoid in acute inflammation: observations on urinary excretion of orosomucoid, albumin, alpha1-microglobulin, and IgG. Clin Chem. 2005;51(11):2052–2058. doi:10.1373/clinchem.2005.055442. [DOI] [PubMed] [Google Scholar]
- 18. Petersen HH, Nielsen JP, Heegaard PM. Application of acute phase protein measurements in veterinary clinical chemistry. Vet Res. 2004;35(2):163–187. doi:10.1051/vetres:2004002. [DOI] [PubMed] [Google Scholar]
- 19. Rucksaken R, Charoensuk L, Pinlaor P, Pairojkul C, Khuntikeo N, Pinlaor S. Plasma orosomucoid 2 as a potential risk marker of cholangiocarcinoma. Cancer Biomar. 2017;18(1):27–34. doi:10.3233/cbm-160670. [DOI] [PubMed] [Google Scholar]
- 20. Romao JE, Jr, Haiashi AR, Elias RM, et al. Positive acute-phase inflammatory markers in different stages of chronic kidney disease. Am J Nephrol. 2006;26(1):59–66. doi:10.1159/000091806. [DOI] [PubMed] [Google Scholar]
- 21. Van Den Heuvel MM, Poland DC, De Graaff CS, et al. The degree of branching of the glycans of alpha(1)-acid glycoprotein in asthma. A correlation with lung function and inflammatory parameters. Am J Respir Crit Care Med. 2000;161(6):1972–1978. doi:10.1164/ajrccm.161.6.9812022. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X, Xiao Z, Liu X, et al. The potential role of ORM2 in the development of colorectal cancer. PLoS One. 2012;7(2):e31868 doi:10.1371/journal.pone.0031868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharma M, Halligan BD, Wakim BT, Savin VJ, Moulder JE. The urine proteome as a biomarker of radiation injury: submitted to proteomics clinical applications special issue: “Renal and Urinary Proteomics (Thongboonkerd)”. Proteomics Clin Appl. 2010;2(7-8):1065–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sadrzadeh SM, Bozorgmehr J. Haptoglobin phenotypes in health and disorders. Am J Clin Pathol. 2004;121:S97–S104. doi:10.1309/8glx5798y5xhq0vw. [DOI] [PubMed] [Google Scholar]
- 25. Hamrita B, Chahed K, Trimeche M, et al. Proteomics-based identification of α1-antitrypsin and haptoglobin precursors as novel serum markers in infiltrating ductal breast carcinomas. Clin Chim Acta. 2009;404(2):111–118. [DOI] [PubMed] [Google Scholar]
- 26. Lai CH, Chang N-W, Lin C-F, et al. Proteomics-based identification of haptoglobin as a novel plasma biomarker in oral squamous cell carcinoma. Clin Chim Acta. 2010;411(13-14):984–991. [DOI] [PubMed] [Google Scholar]
- 27. Mandato VD, Magnani E, Abrate M, et al. Haptoglobin phenotype and epithelial ovarian cancer. Anticancer Res. 2012;32(10):4353–4358. [PubMed] [Google Scholar]
- 28. Park J, Yang JS, Jung G, et al. Subunit-specific mass spectrometry method identifies haptoglobin subunit alpha as a diagnostic marker in non-small cell lung cancer. J Proteomics. 2013;94(20):302–310. [DOI] [PubMed] [Google Scholar]
- 29. Sun L, Hu S, Yu L, et al. Serum haptoglobin as a novel molecular biomarker predicting colorectal cancer hepatic metastasis. Int J Cancer. 2016;138(11):2724–2731. [DOI] [PubMed] [Google Scholar]
- 30. Boittin FX, Denis J, Mayol JF, et al. The extent of irradiation-induced long-term visceral organ damage depends on cranial/brain exposure. PLoS One. 2015;10(4):e0122900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Widlak P, Pietrowska M, Rutkowski T, et al. Radiation-related changes in serum proteome profiles detected by mass spectrometry in blood of patients treated with radiotherapy due to larynx cancer. J Radiat Res. 2011;81(2):S161–S162. [DOI] [PubMed] [Google Scholar]
- 32. Chen C, Lorimore SA, Evans CA, Whetton AD, Wright EG. A proteomic analysis of murine bone marrow and its response to ionizing radiation. Proteomics. 2010;5(16):4254–4263. [DOI] [PubMed] [Google Scholar]
- 33. De Buck M, Gouwy M, Wang JM, et al. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr Med Chem. 2016;23(17):1725–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiolog Rev. 1995;59(1):143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim D, Marchetti F, Chen Z, et al. Nanosensor dosimetry of mouse blood proteins after exposure to ionizing radiation. Sci Rep. 2013;3:2234 doi:10.1038/srep02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ao X, Hanggi P, Schmid G. In-phase and anti-phase synchronization in noisy Hodgkin-Huxley neurons. Math Biosci. 2013;245(1):49–55. doi:10.1016/j.mbs.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 37. Taktak Y, Stenning B. Solid phase enzyme immunoassays for the quantification of serum amyloid P (SAP) and complement component 3 (C3) proteins in acute-phase mouse sera. Horm Metab Res. 1992;24(08):371–374. [DOI] [PubMed] [Google Scholar]
- 38. Verwey NA, Schuitemaker A, Flier WMVD, et al. Serum amyloid P component as a biomarker in mild cognitive impairment and Alzheimer’s disease. Dement Geriatr Cogn Disord. 2008;26(6):522–527. [DOI] [PubMed] [Google Scholar]
- 39. Fischer R, Trudgian DC, Wright C, Thomas G, Bradbury LA. Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Mol Cell Proteomics. 2012;11(2): M111.013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Epstein J, Eichbaum Q, Sheriff S, Ezekowitz RAB. The collectins in innate immunity. Curr Opin Immunol. 1996;8(1):29–35. [DOI] [PubMed] [Google Scholar]
- 41. Madsen HO, Garred P, Thiel S, Kurtzhals JAL, Svejgaard A. Interplay between promoter and structural gene variants control basal level of mannan-binding protein. J Immunol. 1995;155(6):3013–3020. [PubMed] [Google Scholar]
- 42. Alton EW, Turner GDH, Klein N, Davies MJ, Neth O. Differential binding of mannose-binding lectin to respiratory pathogens in cystic fibrosis. Lancet. 2000;355(9218):1885–1886. [DOI] [PubMed] [Google Scholar]
- 43. Foster CB, Lehrnbecher T, Mol F, Steinberg SM, Chanock SJ. Host defense molecule polymorphisms influence the risk for immune-mediated complications in chronic granulomatous disease. J Clin Investig. 1998;102(12):2146–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng J, Garg S, Wang J, Loose DS, Hauer-Jensen M. Laser capture microdissected mucosa versus whole tissue specimens for assessment of radiation-induced dynamic molecular and pathway changes in the small intestine. PLoS One. 2013;8(1):e53711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Der A, Monti P, Lebaron-Jacobs L, Marquette C, Gourmelon P. Characterization of the acute inflammatory response after irradiation in mice and its regulation by interleukin 4 (Il4). Radiat Res. 2001;155(6):858–865. [DOI] [PubMed] [Google Scholar]
- 46. Gordon LE, Ruml D, Hahne HJ, Miller CP. Studies on susceptibility to infection following ionizing radiation: IV. The pathogenesis of the endogenous bacteremias in mice. J Exper Med. 1955;102(4):413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ao X, Li Y, Wang F, et al. The Sulfolobus initiator element is an important contributor to promoter strength. J Bacteriol. 2013;195(22):5216–5222. doi:10.1128/JB.00768-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol. 2016;14(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, S1-452_proteins_expression_profiling for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Supplemental Material, S2-98_radiation-responsive_proteins for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Supplemental Material, S3-GO_analysis_of_98_radiation-responsive_proteins for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response
Supplemental Material, S4-KEGG_analysis_of_98_radiation-responsive_proteins for Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation by Jinfeng Huang, Qi Wang, Yingchun Hu, Zhenhua Qi, Zhongwu Lin, Wantao Ying and Meijuan Zhou in Dose-Response