Abstract
Objective
Obesity is the result of positive energy balance. It can be caused by excessive energy consumption but also by decreased energy dissipation, which occurs under several conditions including when the development or activation of brown adipose tissue (BAT) is impaired. Here we evaluated whether iRhom2, the essential cofactor for the Tumour Necrosis Factor (TNF) sheddase ADAM17/TACE, plays a role in the pathophysiology of metabolic syndrome.
Methods
We challenged WT versus iRhom2 KO mice to positive energy balance by chronic exposure to a high fat diet and then compared their metabolic phenotypes. We also carried out ex vivo assays with primary and immortalized mouse brown adipocytes to establish the autonomy of the effect of loss of iRhom2 on thermogenesis and respiration.
Results
Deletion of iRhom2 protected mice from weight gain, dyslipidemia, adipose tissue inflammation, and hepatic steatosis and improved insulin sensitivity when challenged by a high fat diet. Crucially, the loss of iRhom2 promotes thermogenesis via BAT activation and beige adipocyte recruitment, enabling iRhom2 KO mice to dissipate excess energy more efficiently than WT animals. This effect on enhanced thermogenesis is cell-autonomous in brown adipocytes as iRhom2 KOs exhibit elevated UCP1 levels and increased mitochondrial proton leak.
Conclusion
Our data suggest that iRhom2 is a negative regulator of thermogenesis and plays a role in the control of adipose tissue homeostasis during metabolic disease.
Keywords: iRhom2, ADAM17/TACE, Obesity, Insulin resistance, NAFLD, BAT, Browning, Thermogenesis, UCP1
Highlights
-
•
Deletion of iRhom2 protects mice from metabolic syndrome.
-
•
In obesity, iRhom2 deletion increases energy expenditure, thermogenesis and white adipose tissue beiging.
-
•
iRhom2 deletion enhances thermogenesis in naïve brown adipocytes.
1. Introduction
Mammals, including humans, have a homeostatic circuitry that balances food intake with energy expenditure. However, positive energy balance, caused by the availability of energy-rich food, combined with sedentary lifestyles, blunts this mechanism, resulting in obesity [1]. A variety of other factors can promote obesity, or exacerbate the effects of positive energy balance, including genetic susceptibility, endocrine disorders, and the microbiota [2]. Obesity drives important chronic metabolic diseases, collectively termed metabolic syndrome, comprising dyslipidemia, insulin resistance/type 2 diabetes, cardiovascular disease, and non-alcoholic fatty liver disease (NAFLD) [3]. These complications are a consequence of dealing with surplus energy, resulting in fat accumulation in non-adipose tissues, including muscle and liver, leading to lipotoxicity-induced insulin resistance in a range of organs [4,5].
Adipose tissues play a central role in metabolic homeostasis and metabolic disease [6] and are classified into two major types: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT stores energy within lipid droplets (LD) [7] and, upon metabolic demand, hydrolyzes triglycerides into glycerol and free fatty acids (FFA), to fuel the functions of peripheral organs. WAT also plays an important endocrine role during health and disease by secreting a range of signaling proteins collectively called adipokines [6,8,9].
In rodents, BAT serves a fundamentally different purpose: non-shivering thermogenesis. FFA activate uncoupling protein 1 (UCP1), a channel that diverts protons produced by oxidative phosphorylation; the resultant proton leakage into the mitochondrial matrix drives heat production [10]. Substantial evidence from mouse models indicates that BAT can counteract adiposity, insulin resistance, and hyperlipidemia [11]. Consequently, BAT biology and strategies to encourage browning of white adipose tissue are the subject of intense focus [12,13]. An additional category of thermogenic adipocytes called “Beige” (or “brite”) adipocytes express UCP1, but develop within the WAT [14]. These cells are recruited during prolonged cold exposure, exercise, or adrenergic activation, as well as by a range of other stimuli—a process termed “browning” or “beiging” [[15], [16], [17], [18]].
Lipid overloading within adipocytes has profound pathological consequences, such as triggering the stress-induced secretion of pro-inflammatory adipokines and cytokines including tumor necrosis factor (TNF), the first cytokine implicated as a link between inflammation and obesity [19]. Soluble TNF, generated by cleavage of its transmembrane form by the cell surface protease ADAM17 (a disintegrin and metalloprotease-17), also called TACE (TNF-α converting enzyme [20] plays an important role in the pathophysiology of metabolic disease. Crucially, the trafficking of ADAM17 within the secretory pathway, maturation from its pro-enzyme form [[21], [22], [23]], stimulation of its proteolytic activity on the cell surface [24,25], and control of its substrate specificity [26], all require a cofactor called iRhom2. iRhom2 is a polytopic membrane protein that is highly expressed in immune cells, particularly macrophages, where it plays an essential role in ADAM17 regulation [21,22]. Hence, the release of TNF from myeloid cells requires iRhom2 [21,22].
TNF regulates the pathobiology of metabolic syndrome in several important ways [27]. It is responsible for macrophage recruitment to adipose tissue and the establishment of metainflammation—the chronic low grade inflammatory state that is established in adipose tissue and other metabolic organs during obesity [28,29]. Metainflammation has important pleiotropic effects on metabolic disease, both within adipose tissue and systemically [19]. One of TNF's major pathological impacts is triggering insulin resistance in skeletal muscle and adipose tissue by promoting serine phosphorylation of insulin receptor substrate 1 (IRS-1) [30], which impairs the capacity of this adaptor protein to mediate insulin receptor signaling. TNF also enhances lipolysis in adipocytes, increases circulating FFA, and promotes leptin release from adipocytes, both of which can contribute to insulin resistance [31]. The role of TNF in the regulation of the BAT thermogenic capacity remains debated [27].
Mice null for TNF or its receptors are protected from obesity-associated insulin resistance but not from an increase in fat mass [32]. By contrast, ADAM17 null mice, also protected from insulin resistance, exhibit a hypermetabolic phenotype and resist weight gain in models of obesity [33,34]. These differences in the metabolic phenotype of TNF and ADAM17 null mice are likely associated with the fact that ADAM17 cleaves multiple cellular substrates in addition to TNF [35].
iRhom2 shares redundant functions in ADAM17 regulation with its paralog iRhom1. Consequently, the phenotype of iRhom2 KO mice is much milder than ADAM17 KO mice, which die perinatally with severe abnormalities in the eyes, skin, hair, and lung [36]. By contrast, iRhom2 KO mice appear normal but are protected from a range of conditions, most of which involve inflammation, including sepsis [21,22], arthritis [37], and lung injury [38]. However, whether global deletion of iRhom2 impacts energy balance and/or adipose tissue function is currently unknown.
In this work, we evaluated the contribution of iRhom2 to metabolic syndrome. Intriguingly, we find that iRhom2 KO mice are protected from a range of deleterious metabolic phenotypes including obesity, adipose tissue inflammation, hepatic steatosis, and insulin resistance. We also show that iRhom2 is expressed in adipocytes, and its expression levels are increased in brown adipocytes from obese animals. Strikingly, loss of iRhom2 enhances the capacity of the animals to dissipate excess energy via increased adaptive thermogenesis in BAT and through increased beiging of WAT. Consistent with this, iRhom2-null primary adipocytes exhibit elevated levels of UCP1 and increased mitochondrial proton leak, offering a potential mechanistic explanation for the observed phenotypes. Overall, our data reveal a fundamental organismal role for iRhom2 as a negative regulator of thermogenesis.
2. Results
2.1. iRhom2 is expressed in adipose tissues and its expression is elevated specifically in BAT
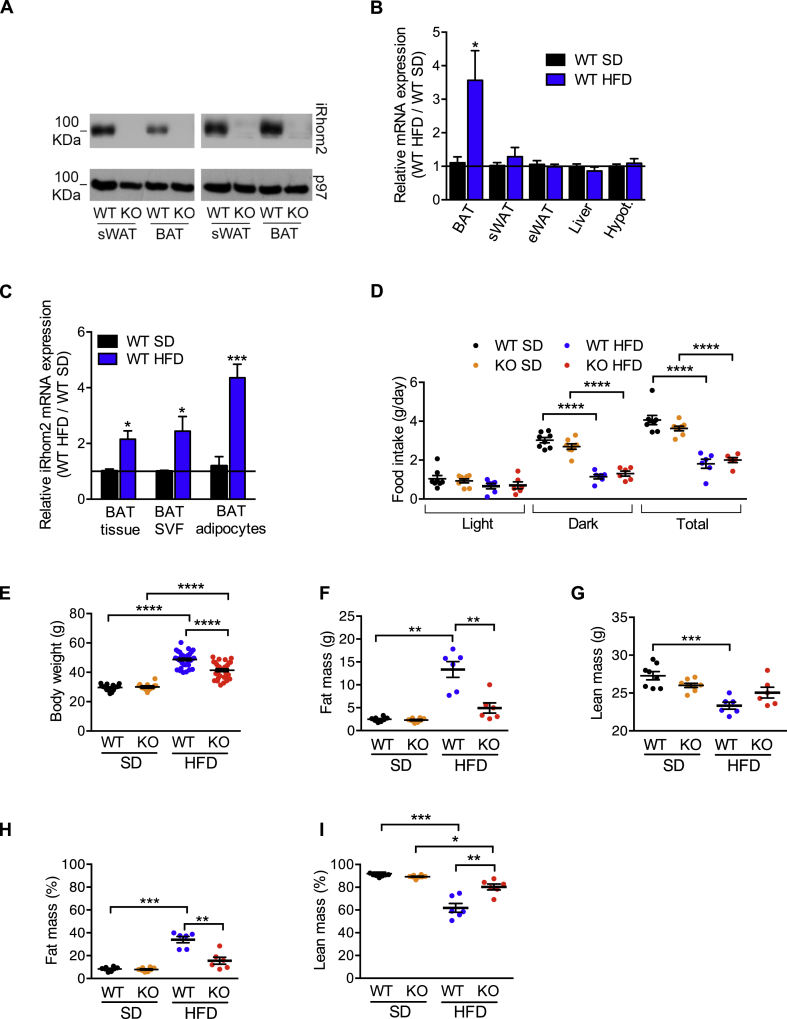
To examine objectively the expression pattern of iRhom2 in vivo, we generated a mouse strain in which the endogenous iRhom2 gene was modified to incorporate an in-frame N-terminal FLAG tag. Strikingly, western blots on lysates from tissues isolated from iRhom2-FLAG mice revealed that iRhom2 was significantly expressed in a range of metabolic organs including adipose tissues, pancreas, liver, and muscle (Fig. S1A). As iRhom2 is expressed in several major adipose depots, we focused in more detail on these organs. Notably, iRhom2 was expressed in primary adipocytes differentiated from progenitors from the stromal vascular fraction (SVF) from subcutaneous WAT (sWAT) or BAT (Figure 1A). Intriguingly, iRhom2 mRNA was upregulated in BAT and brown adipocytes biochemically isolated from mice chronically exposed to a high fat diet (HFD) for 30 weeks, a model for obesity/metabolic syndrome (Figure 1B,C). We confirmed that iRhom2 was also upregulated at the protein level in BAT from animals on HFD (Fig. S1B). In an in vitro inflammatory environment, iRhom2 mRNA was also modestly upregulated in primary WT brown adipocytes differentiated in vitro from SVF treated with TNF (Fig. S1C).
Figure 1.
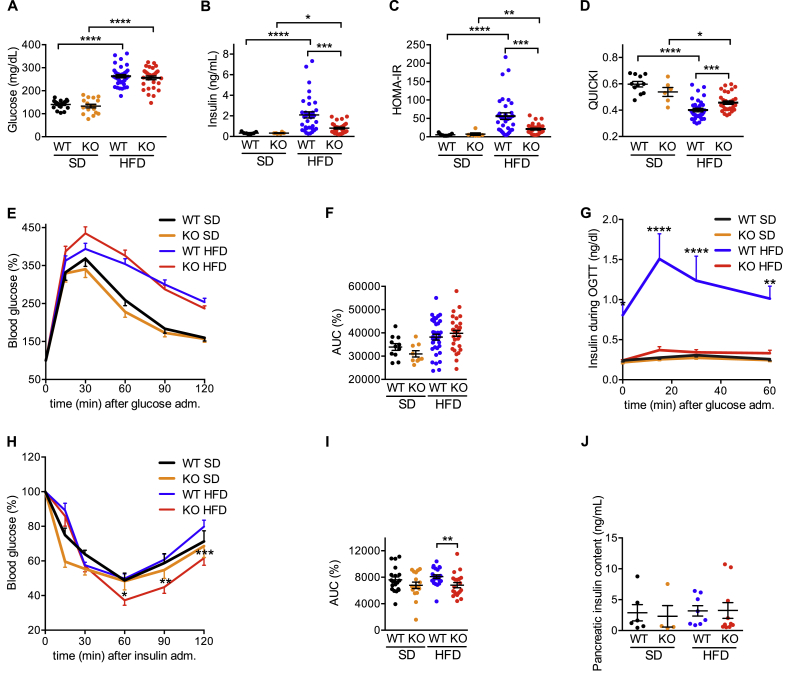
iRhom2 expression is increased in brown adipose tissue in HFD-induced obesity and iRhom2 KO mice are less prone to this disease. A Protein expression of iRhom2 in subcutaneous and brown adipocytes differentiated in vitro from the stromal vascular fraction of 4–5 pooled WT and iRhom2 KO mice. As a loading control, we measured p97 protein levels. Two independent experiments. B RT-PCR analysis of Rhbdf2 expression in brown adipose tissue (BAT), subcutaneous (sWAT), and epididymal (eWAT) white adipose tissues, liver, and hypothalamus from WT mice fed with HFD for 30 weeks compared to WT fed with SD for the same extent. Two experiments with 3–4 mice per group. C RT-PCR analysis of Rhbdf2 expression in mature brown adipocytes, stromal vascular fraction (SVF), and whole BAT tissue from HFD-fed WT mice compared to SD-fed ones. Two independent experiments with 3 replicates pooled from 2 mice each. D Food intake of WT and iRhom2 KO fed with SD or HFD for 17 weeks. One experiment, with 6–8 mice per group. E Body weight of iRhom2 KO and WT mice after 30 weeks of SD or HFD exposure. Three independent HFD-induced obesity experiments, with 12 WT and 11 KO in two, and 11 WT and 9 KO mice in the third were performed. The analysis of the SD-fed mice was performed on 3 groups of 4–7 mice per genotype. F–I Absolute values of fat (F) and lean (G) mass, and percentage of fat (H) and lean (I) mass relative to body weight of the mice described above. One experiment, with 6 and 8 mice per genotype fed with SD and HFD for 17 weeks, respectively. The legend colour coding of panel D applies also to panels E–I. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, **** represents p < 0.0001.
2.2. iRhom2 KO mice are protected from HFD-induced obesity, dyslipidemia, and hepatosteatosis
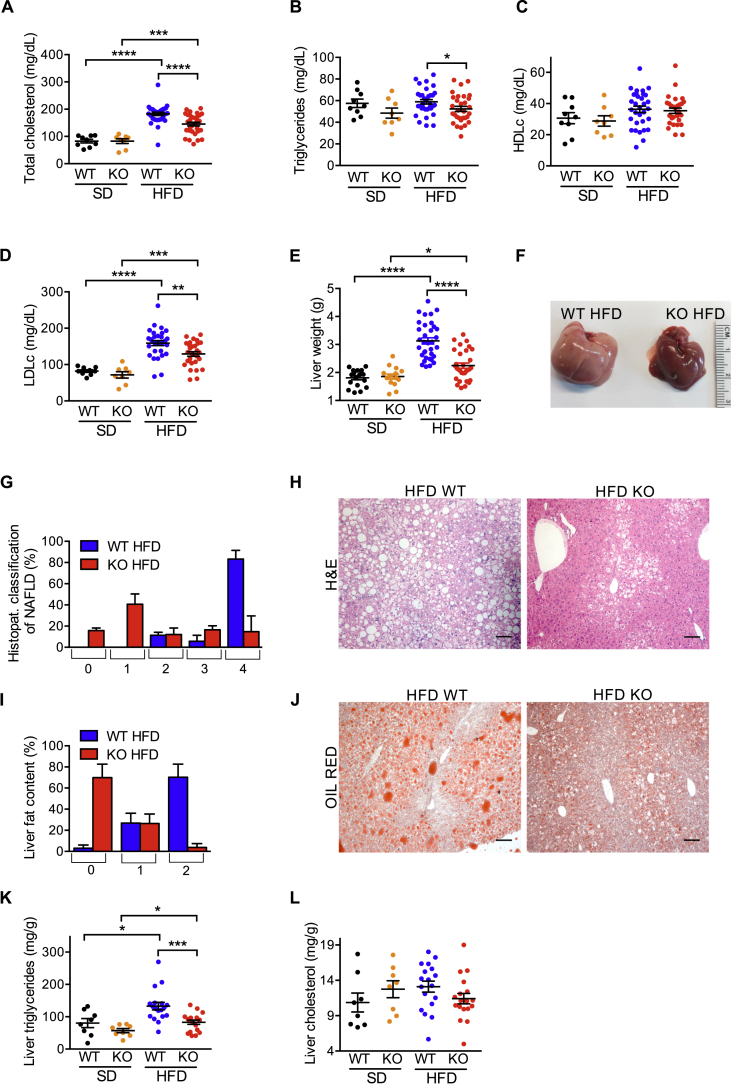
The upregulation of iRhom2 levels in BAT during obesity and the fact that iRhom2 plays a central role in ADAM17 regulation and TNF release [21,22] led us to determine whether the ablation of iRhom2 had an impact on the development of HFD-induced metabolic syndrome. Notably, after challenging WT versus iRhom2 KO mice with HFD for ∼30 weeks, we found that, in spite of food intake being equivalent between genotypes (Figure 1D), iRhom2 KOs were protected from HFD-induced obesity (Figure 1E,F,H, Fig. S1D–F). Therefore, compared to WTs, HFD-fed iRhom2 KO mice had reduced body weight (BW) (Figure 1E, Fig. S1D), adiposity (Figs. S1E–F), and absolute and relative fat mass (Figure 1F,H). iRhom2 KOs were also protected from the decrease in lean mass observed in the WT mice on HFD compared to the respective SD-fed controls (Figure 1G,I). As predicted by the decreased fat mass of iRhom2 KO mice, we observed a significant protection from HFD-induced dyslipidemia (Figure 2A–D). The most pronounced reduction was observed in fasting cholesterolemia (Figure 2A), which was associated with decreased levels of serum low density lipoproteins (LDL) (Figure 2D).
Figure 2.
iRhom2 KO mice are protected from dyslipidemia and NAFLD. A-D Colorimetric analysis of total cholesterol (A), triglycerides (B), HDL (C), LDL (D) in the serum of iRhom2 KO and WT mice fed with SD and HFD for 22 weeks. E Liver weight of iRhom2 KO and WT mice fed with SD and HFD for 30 weeks. Three independent HFD-induced obesity experiments, with 9–12 WT and 8–11 KO mice. The analysis of the SD-fed mice was performed on 2–3 groups of 3–7 mice per genotype. F–H Liver photographs (F), histopathological classification of NAFLD (scores ranging from no alterations (0) to a severe NAFLD phenotype (4) (G), and representative photographs of liver H-E staining (100x magnification) (H) of iRhom2 KO and WT mice described above fed with HFD for 30 weeks. I-J Liver fat content histopathological classification (scores ranging from within normal limits (0) to severe fat accumulation (2) (I), and representative photographs of liver Oil red staining (100x magnification) (J) of the mice described above fed with HFD. K-L Colorimetric analysis of liver triglycerides (K) and cholesterol (L) concentration in the animals described above fed with SD and HFD for 30 weeks. Two independent experiments with n = 6 or 12 HFD-fed mice and n = 2 or 6 SD-fed mice per genotype. Scale bar = 100 μm. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, **** represents p < 0.0001.
Lipid accumulation in non-adipose organs is a common hallmark of the pathophysiology of obesity [39]. As shown in Figure 2E, iRhom2 KO mice were protected from HFD-induced increased liver weight. This was associated with a striking protection from NAFLD-associated hepatic histopathological alterations and lipid accumulation in iRhom2 KO mice (Figure 2F–J). We confirmed that the reduced hepatic steatosis in iRhom2 KO mice correlated with reduced triglyceride deposition rather than cholesterol (Figure 2K,L). By contrast, the mild lipid accumulation observed in skeletal muscle on animals maintained on HFD was similar between WT and iRhom2 KOs (Fig. S2A and B).
2.3. iRhom2 KO mice are protected from HFD-induced adipose depot expansion
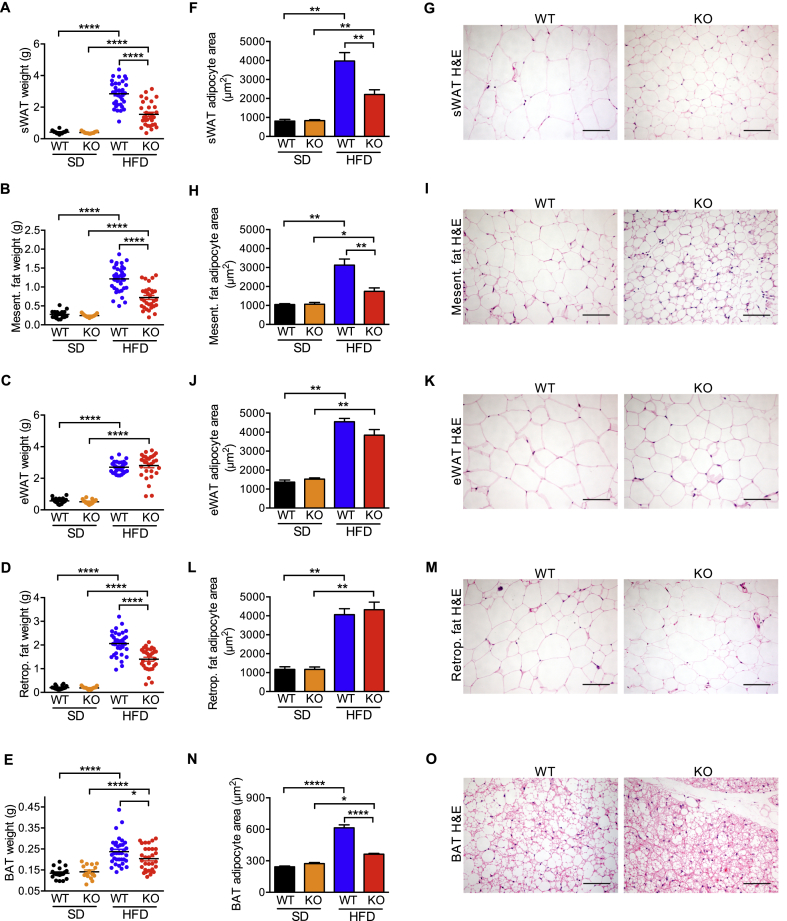
As expected from their overall reduced body weight on HFD (Figure 1E; Fig. S1D–F), iRhom2 KO mice presented with a significantly reduced adipose tissue weight in all anatomical regions except eWAT, where the effect was less marked (Figure 3A–E; Fig. S3A). Strikingly, on HFD we observed a significant reduction in adipocyte area in subcutaneous and mesenteric adipose tissues from iRhom2 KO mice, indicating a protection from adipocyte hypertrophy (Figure 3F–I, Fig. S3B and C). Nevertheless, there was no alteration of the adipocyte area in the epididymal and retroperitoneal adipose tissues (Fig. 3J-M, Fig. S3D and E).
Figure 3.
iRhom2 KO mice fed with HFD have reduced adipose tissue depots and smaller adipocytes. A-O Weight (A-E), adipocyte area (F, H, J, L, N), and representative photographs of H-E staining (200x magnification) (G, I, K, M, O) of subcutaneous (sWAT) (A, F, G), mesenteric (B, H, I), epididymal (eWAT) (C, J, K), retroperitoneal (D, L, M) white adipose tissues, and brown adipose tissue (BAT) (E, N, O) from iRhom2 KO and WT mice fed with SD and HFD for 30 weeks (photographs only from HFD-fed mice samples). Three independent HFD-induced obesity experiments, with 11–12 WT and 9–11 KO mice. The analysis of the SD-fed mice was performed on 3 groups of 4–7 mice per genotype. The average WAT adipocyte size [average cross-sectional area per cell (μm2)] was determined using a macro on H-E staining of adipose tissue from 3 HFD-fed animals and 1–3 SD-fed mice per genotype per experiment (3 experiments). The average BAT adipocyte size was calculated as described previously [83] in adipose tissue samples from the SD and HFD-fed animals described above. Scale bar = 100 μm. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, **** represents p < 0.0001.
Obesity-associated lipid loading causes brown adipocytes to lose their characteristic multilocular appearance, becoming morphologically more similar to white adipocytes [19,40,41]. Notably, in mice on HFD, the adipocytes from WT BAT developed large unilocular lipid droplets more characteristic of white adipocytes, whereas in iRhom2 KO BAT adipocytes were protected from this phenomenon, remaining multilocular and smaller (Figure 3N–O; Fig. S3F).
2.4. HFD-fed iRhom2 KO mice have reduced systemic and adipose tissue inflammation
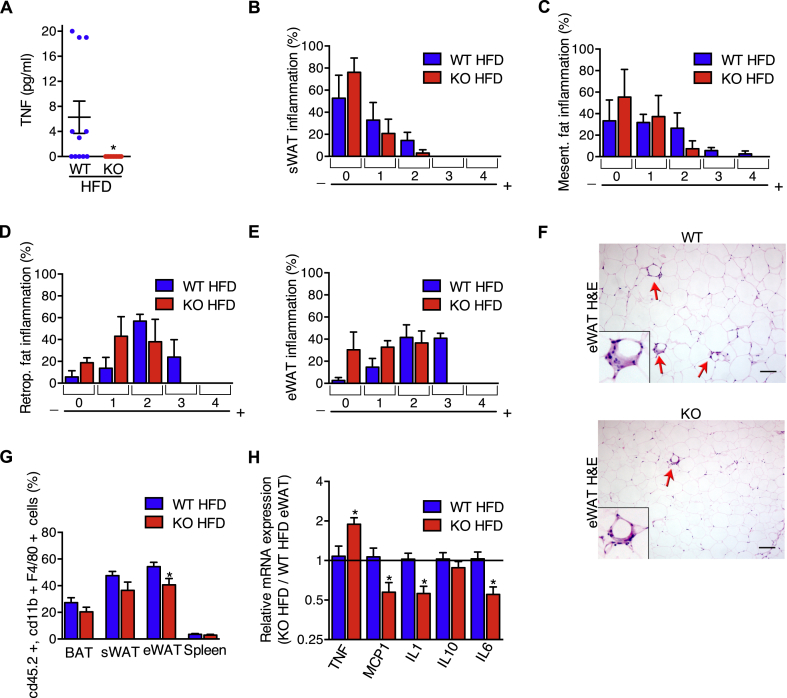
When the limits of adipocyte plasticity are reached during obesity, adipocyte stress triggers the secretion of adipokines, inflammatory cell recruitment, and the release of inflammatory cytokines, such as TNF. Consistent with the requirement for iRhom2 in TNF shedding in other models [21,22,37,42,43], circulating levels of soluble TNF in iRhom2 KOs were undetectable, compared to WT controls, in serum from animals subjected to the HFD protocol (Figure 4A). To determine the impact of loss of iRhom2 on adipose tissue inflammation, we used histopathology to score inflammatory cell infiltration. Overall, we observed a mild protection from immune cell infiltration into adipose tissues in iRhom2 KOs (Figure 4B–F), which was most obvious in epididymal adipose tissue (Figure 4E,F), where, during obesity, adipocyte death and inflammation are normally more pronounced compared to subcutaneous adipose tissue [44]. We stained the macrophages isolated from the adipose tissues, compared to the spleen of HFD-fed iRhom2 KO and WT mice, confirming that there was a significant reduction in the relative proportion of macrophages found in the epididymal WAT of iRhom2 KO mice (Figure 4G; gating strategy indicated in Fig. S4). Notably, this correlated with a decrease in inflammatory cytokine/chemokine mRNA expression in iRhom2 KO epididymal adipose tissue (Figure 4H). An exception was the increased TNF mRNA levels in iRhom2 KOs, which we speculate to be an attempted compensatory mechanism to mitigate the loss of TNF shedding, as observed previously in ADAM17 tissue-specific KO mice [45]. Hence, iRhom2 contributes to the development of an inflamed environment within adipose tissues.
Figure 4.
iRhom2 KO mice fed with HFD have no circulating TNF and reduced fat steatitis. A TNF concentration in the serum of iRhom2 KO and WT mice fed with HFD for 30 weeks. One experiment with 12 WT and 11 KO mice. B-E Histopathological classification of steatitis (scores 0–4) of iRhom2 KO and WT HFD-fed mice subcutaneous (sWAT) (B), mesenteric (C), retroperitoneal (D), and epididymal (eWAT) (E) adipose tissues. Steatitis was scored using the following classification: (0) no alterations, (1) rare macrophage infiltration, (2) mild multifocal macrophage infiltration, (3) moderate multifocal macrophage infiltration, (4) moderate multifocal macrophage infiltration with steatonecrosis. Three independent HFD-induced obesity experiments with 11–12 WT and 9–11 KO mice. F Representative photographs of HFD-fed KO and WT eWAT steatitis (indicated by red arrows) in the H-E staining (100x magnification), with an amplification of an area of steatitis. Scale bar = 100 μm. G FACS analysis showing the proportion of macrophages in the BAT, sWAT, eWAT, and spleen of IRhom2 KO and WT mice fed with HFD for 30 weeks. Three independent experiments in pools of 5–6 mice tissues from each genotype. H RT-PCR analysis of Tnf, Mcp1, Il1b, Il10, and Il6 expression in HFD-fed iRhom2 KO mice eWAT relatively to WT samples. Two independent analysis of 2–3 samples from each genotype. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, **** represents p < 0.0001.
2.5. iRhom2 KO mice develop less obesity-related insulin resistance
Next, we evaluated if the reduced inflammation caused by loss of iRhom2 could delay the development of insulin resistance. Although WT and iRhom2 KO mice had similarly raised fasting glycemia on HFD (Figure 5A), the KOs exhibited significantly reduced fasting insulinemia when compared to WT (Figure 5B). This equated with a reduced homeostatic model assessment of insulin resistance index (HOMA-IR) (Figure 5C) and increased quantitative insulin sensitivity check index (QUICKI) (Figure 5D) in HFD-fed iRhom2 KO mice. These data imply that on HFD the onset of insulin resistance in WT mice necessitates elevated insulin secretion to achieve equivalent glucose uptake to that reached by iRhom2 KOs, which retain insulin sensitivity. Consistent with this interpretation, although both genotypes cleared glucose similarly in oral glucose tolerance tests (Figure 5E,F), the HFD-fed WT mice that exhibited substantially raised basal insulin levels, also elevated insulin secretion significantly to achieve the same kinetics of glucose clearance (Figure 5G). By contrast, iRhom2 KO mice maintained insulinemia equivalent to animals on standard diet, confirming that lack of iRhom2 protects against the development of diabetes (Figure 5G) and predicts that iRhom2 KO mice are more insulin sensitive than their WT counterparts, which was confirmed in insulin tolerance tests (Figure 5H,I). Importantly, the reduced insulinemia in iRhom2 KOs was not a consequence of impaired insulin biogenesis, since the pancreatic insulin content was equivalent among all groups (Figure 5J). Together these data emphasize that loss of iRhom2 protects from the onset of insulin resistance.
Figure 5.
iRhom2 KO mice are less prone to HFD-induced insulin resistance. A-D Fasting glycemia (A) serum insulin concentration (B) homeostatic model assessment of insulin resistance index (HOMA-IR), (C) and quantitative insulin sensitivity check index (QUICKI) (D) in iRhom2 KO and WT fed with SD and HFD for 20–22 weeks. For the glucose analysis 3 experiments were performed in 11–12 WT versus 9–11 KO mice fed with HFD and 6–7 WT versus 5–6 KO mice fed with SD. For the insulin, QUICKI and HOMA-IR analysis, 2 experiments were performed in 11–22 WT versus 9–20 KO mice fed with HFD, and 3–4 WT versus 2–4 KO mice fed with SD. E, G, H Blood glucose (E) and serum insulin concentration (G) during OGTT and blood glucose level during ITT (H) of iRhom2 WT and KO mice fed with SD and HFD for 26–28 weeks. F, I AUC of glucose levels during the OGTT (F) and ITT (I). Three or two independent experiments were performed for OGTT and ITT, respectively. In each, 9–12 HFD-fed mice and 5–7 SD-fed controls were used for all the analysis. Insulin concentration during OGTT was evaluated in 1–4 SD and HFD-fed mice (total of 7 mice per group) from each experiment. J Pancreatic insulin content. Two independent experiments using 3–5 WT versus 3–7 KO mice fed with HFD and 1–5 WT versus 1–3 KO mice fed with SD for 30 weeks. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, **** represents p < 0.0001.
2.6. iRhom2 KO exhibit increased energy expenditure
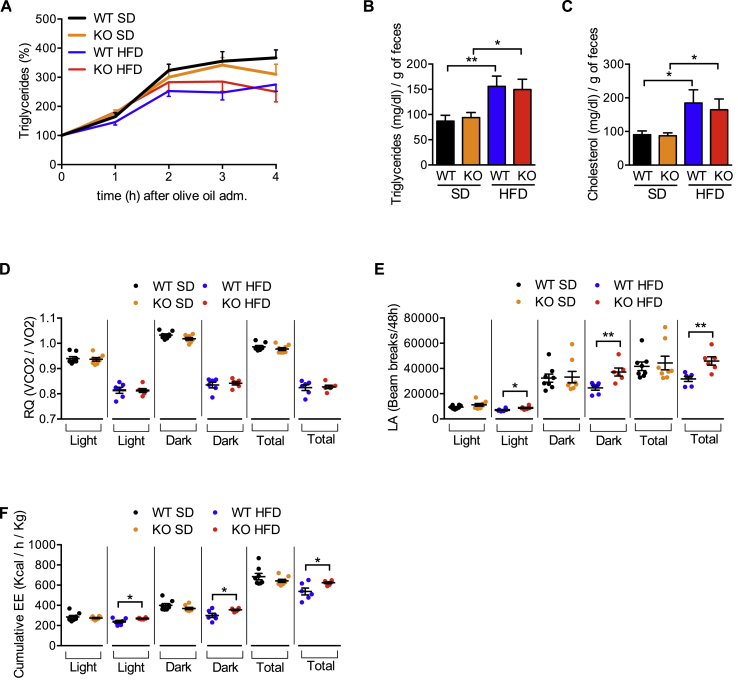
To investigate the basis of the pronounced protection from metabolic disease observed in iRhom2 KO mice, we analyzed WT versus iRhom2 KO mice fed with SD or HFD, by indirect calorimetry. As the protection from HFD-induced obesity observed in iRhom2 KOs (Figure 1E,F,H, Fig. S1D–F) was not caused by differences in food consumption (Figure 1D), one alternative basis for the improved metabolic phenotypes of iRhom2 KO mice is a reduced capacity to harvest dietary lipids from food. However, iRhom2 KO mice were indistinguishable from WT controls in their capacity to absorb triglycerides upon oral gavage (Figure 6A), and neither the concentration of triglycerides (Figure 6B) or cholesterol (Figure 6C) excreted in feces nor the overall mass of feces excreted (data not shown) was significantly different. Next, we analyzed the energy consumption of iRhom2 KOs using indirect calorimetry. Notably the respiratory quotient (RQ) (vCO2/vO2), was unaltered in iRhom2 KOs, indicating equivalent fat versus carbohydrate oxidation between both genotypes (Figure 6D). Hence, energy intake, usage and disposal, was normal in iRhom2 KO mice.
Figure 6.
HFD-fed iRhom2 KO mice have no differences in energy intake but increased energy expenditure. A Blood triglyceride content during lipid tolerance test of iRhom2 WT and KO mice fed with SD and HFD for 26 weeks. B–C Triglycerides (B) and cholesterol (C) content in one gram of dried feces of the mice described above. D-F Respiratory quotient (RQ) (vCO2/vO2) (D), locomotor activity (LA) (E), and cumulative energy expenditure (EE) (F) of iRhom2 KO and WT mice fed with SD and HFD for 17 weeks. Results are representative of two independent experiments, with 7–8 mice per group in A. Four experiments were performed in B–C, each with 2–6 (SD) or 8–9 (HFD) pooled mice per genotype, fed on SD or HFD for 24–25 weeks respectively. Data shown in panels D–F are derived from one experiment, with a total of 8 and 6 mice for SD and HFD, respectively. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, **** represents p < 0.0001.
If energy intake is unaltered, iRhom2 KOs must expend the energy obtained from HFD more efficiently. A common obesity-associated phenotype is a reduction in physical activity in the obese state. Notably, consistent with iRhom2 KOs on HFD being leaner and metabolically healthier than WT mice, iRhom2 KOs maintained a locomotor activity (LA) on HFD that was broadly similar to that observed with WT and iRhom2 KO animals on SD (Figure 6E). This difference correlated with significantly higher energy expenditure (EE) in iRhom2 KOs than WT animals on HFD (Figure 6F, Fig. S5A and C). By contrast, there were no significant differences in EE when both genotypes were maintained on SD (Figure 6F, Fig. S5B and D). Taken together, our data show that iRhom2 KO animals can better mitigate the excess energy intake when subjected to HFD.
2.7. iRhom2 KO mice dissipate energy through increased BAT thermogenesis and beiging of WAT
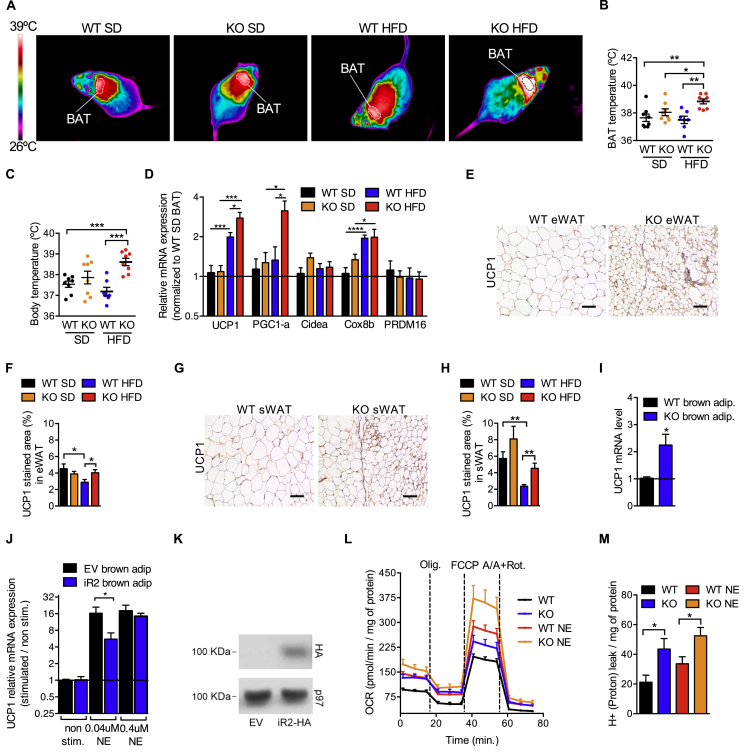
We looked for differences to explain the significantly increased energy expenditure of iRhom2 KOs on HFD. As BAT is the major organ responsible for energy expenditure [7,46] and iRhom2 levels are elevated in BAT and brown adipocytes from obese mice (Figure 1B,C), we next examined in detail whether the loss of iRhom2 had an impact on BAT physiology. Strikingly, thermal imaging experiments revealed that on HFD, iRhom2 KO animals exhibited a significantly elevated temperature (Figure 7A), with a pronounced temperature difference within the BAT itself (Figure 7B). This effect was physiologically relevant since it translated into a significantly elevated overall body temperature in iRhom2 KO mice on HFD (Figure 7C). Interestingly, and consistent with these observations, the BAT of iRhom2 KOs had significantly increased levels of the key thermogenic genes UCP1 and PGC1α (Peroxisome proliferator-activated receptor gamma coactivator 1-alpha) following a chronic (Figure 7D) or 2 week exposure to HFD (Fig. S6A), providing a molecular basis for the increased thermogenesis.
Figure 7.
HFD-fed iRhom2 KO mice have increased thermogenesis and browning of the white adipose tissue. A-C Thermal images (A), and BAT (B) or body (C) temperature of iRhom2 KO and WT mice fed with SD and HFD for 19 weeks. One experiment with 8 mice per group. D RT-PCR analysis of UCP1, PGC1α, Cidea, PRDM16, and Cox8b expression in SD-fed iRhom2 KO and HFD-fed WT and iRhom2 KO mice BAT samples compared to WT SD-fed control samples. Two experiments with 3–4 replicates in each. E, G Representative photographs of eWAT (E) and sWAT (G) UCP1 immunohistochemistry derived from iRhom2 KO and WT mice fed with HFD for 20 weeks. Scale bar = 100 μm. F, H Graphics showing the UCP1 percentage of area stained in the eWAT (F) and sWAT (H) of the mice described above. One experiment with 8 mice per group (with 2 photographs analyzed per mouse). I RT-PCR analysis of UCP1 in brown adipocytes differentiated in vitro from the stromal vascular fraction of 4–5 pooled WT or iRhom2 KO mice. Three independent experiments. J RT-PCR analysis of UCP1 in immortalized WT brown preadipocytes transduced with empty vector or iRhom2-HA, differentiated in vitro and stimulated with norepinephrine for 6 h. Two independent experiments. K HA expression in differentiated immortalized WT brown preadipocytes transduced with retrovirus containing iRhom2 cDNA fused to C-terminal HA tag (iR2-HA). As a negative control, we used the same cells transduced with retrovirus containing the empty vector (EV), and as a loading control we measured p97 protein level. Two independent experiments. L Mitochondrial oxygen consumption rate (OCR) of brown adipocytes differentiated in vitro from the stromal vascular fraction of 4–5 pooled WT and iRhom2 KO mice and stimulated or not with norepinephrine. The results were normalized to the protein content. Two experiments with one or two independent samples per genotype, respectively. M Mitochondrial proton leak of the cells described in L normalized to the protein content. Error bars represent SEM; * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001.
In agreement with the premise that loss of iRhom2 expression enhances the thermogenic capacity of BAT, we observed that iRhom2 mRNA levels were highest in mice maintained under thermoneutrality, reduced under conditions where thermogenesis was triggered by sub-thermoneutral temperatures, and increased upon acclimation from sub-thermoneutral to thermoneutral conditions (Fig. S6B).
These data are consistent with the premise that endogenous iRhom2 acts as a brake on thermogenesis and predict that loss of iRhom2 should lead to the beiging of adipose tissue under sub-thermoneutral conditions. Notably, the visceral (eWAT) and subcutaneous (sWAT) adipose tissues from iRhom2 KO mice on HFD exhibited increased UCP1 protein levels compared to WT mice (Figure 7E–H). Hence, loss of iRhom2 enhances the thermogenic capacity of BAT and promotes WAT beiging in response to positive energy intake.
2.8. Increased BAT thermogenesis is a cell-intrinsic feature of iRhom2 KO brown adipocytes
As our experiments were based on global iRhom2 KO mice, we wanted to address whether the increased thermogenic capacity in iRhom2 KO BAT reflected a cell-autonomous phenotype (i.e., within brown adipocytes themselves). Consistent with this premise, UCP1 mRNA levels were elevated in primary brown adipocytes differentiated in vitro from SVF from iRhom2 KO on SD compared to WT controls (Figure 7I), whereas overexpressing iRhom2 in immortalized brown adipocytes reduced the stimulation of UCP1 levels in response to norepinephrine (Figure 7J,K). These observations suggest that the circuitry through which iRhom2 controls UCP1 levels can be found within adipocytes themselves.
To obtain additional insights, we studied the oxygen consumption rate (OCR) of naïve WT versus iRhom2 KO brown adipocytes as a readout for mitochondrial function (Fig. 7L). Notably, the major phenotypic difference observed was an enhanced proton leak in iRhom2 KO brown adipocytes (Figure 7M; Fig. S6C–H). This is consistent with elevated UCP1 expression/function in iRhom2 KO tissues. Taken together, our data demonstrate that loss of iRhom2 enhances thermogenesis, implying that iRhom2 acts as an endogenous brake on thermogenesis.
3. Discussion
Our work identifies that lack of iRhom2 protects against a range of metabolic complications associated with diet-induced obesity, including adipose tissue expansion, hepatosteatosis, and insulin resistance. As anticipated, loss of iRhom2, probably through impaired ADAM17-dependent TNF shedding, results in a less inflamed adipose tissue environment—a feature known to drive many of the metabolic phenotypes observed in our study [19].
While we do not rule out a role for iRhom2 in myeloid cells in driving inflammation to contribute to the metabolic phenotypes observed, our work reveals an unanticipated intimate connection between iRhom2 and BAT homeostasis, identifying iRhom2 as an inhibitor of thermogenesis. Our data suggest that iRhom2 regulates a cell-autonomous circuitry that represses thermogenesis; isolated brown adipocytes from iRhom2 KO mice exhibit elevated UCP1 levels and correspondingly have enhanced mitochondrial proton leak. Additionally, the levels of iRhom2 correlate with ambient temperature: iRhom2 mRNA levels are highest in animals with the lowest requirement for thermogenesis, and vice versa. Finally, the levels of iRhom2 are elevated in brown adipocytes from obese animals. Together our data suggest that modulation of iRhom2 levels acts as a rheostat for the control of thermogenesis.
Speculatively, this suggests a potential mechanism whereby iRhom2 KOs are protected from metabolic syndrome; on HFD, the increased thermogenic capacity of BAT allows animals to dissipate surplus energy more efficiently, avoiding excess weight gain. Moreover, we observe that loss of iRhom2 triggers the beiging of WAT on HFD, providing an additional basis for enhanced thermogenesis. Future studies using tissue-specific ablation of iRhom2 are required to address this hypothesis directly and to delineate the sequence of phenotypic events that occur following the loss of iRhom2.
iRhom2 is required for the proteolytic activity of the cell surface protease ADAM17 [21,22], and our work identifies metabolic phenotypes common between iRhom2 and ADAM17 mutant mice [33,34,47]. For instance, the inducible systemic KO of ADAM17 [34] or global inactivation of a single ADAM17 allele [47] protects mice from obesity, hepatosteatosis, and insulin resistance.
In another study, global ADAM17 KO mice that escape the lethality associated with this strain [36] and survive to adulthood have increased UCP1 expression and exhibit a hypermetabolic phenotype on standard chow [33]. By contrast, iRhom2 KOs only exhibit significantly increased energy expenditure when challenged by HFD. Several potential factors could account for these phenotypic differences, most obviously the functional redundancy between iRhom1 and iRhom2 in ADAM17 regulation, with iRhom2 having a more restricted expression pattern than ADAM17 [48]. Hence, although the fur of iRhom2 KO mice appears normal [49], ADAM17 KOs have hair follicle density defects [33] that could potentially enhance heat loss, increasing thermogenic demand at sub-thermoneutral temperatures. However, mechanisms other than heat loss must contribute to the ADAM17 KO phenotype since the mice exhibit an increased oxygen consumption rate when maintained under conditions close to thermoneutrality [33]. The authors also found decreased corticotropin-releasing hormone (Crh) mRNA expression in the hypothalamic paraventricular nucleus of ADAM17 KOs [33]. Future studies involving tissue-specific deletion of iRhom2 or ADAM17 are required to delineate the precise mechanistic role and site(s) of action of iRhom2/ADAM17 in metabolic regulation, and to separate cause from effect.
Defective cleavage of TNF by ADAM17 is an obvious possibility to account for some of the phenotypes observed in our studies, including the development of adipose tissue inflammation and systemic insulin resistance [32]. Interestingly, blockade of TNF signaling has been reported to improve the thermogenic capacity of BAT in obesity models [[50], [51], [52]], suggesting a potential basis for the effect on BAT thermogenesis and WAT beiging observed here. However, loss of TNF appears not to underpin the promotion of excessive weight gain [32], suggesting the contribution of additional ADAM17 substrates. It remains challenging to determine which additional ADAM17 targets are responsible, because ADAM17 cleaves more than 80 cellular substrates [35].
The impact of loss of iRhom2/ADAM17 can have potentially complex effects even on TNF signaling itself. ADAM17 can cleave not only TNF but also its receptors, TNFRI and TNFRII [36,53]. Therefore, deletion of iRhom2 could abolish TNF signaling in some tissues and potentiate it in others. Additionally, loss of iRhom2 could also alter the mode of TNF signaling (e.g. whether signaling occurs predominantly through TNFRI or TNFRII, which elicit distinct biological outcomes) [54,55]. Moreover, blocking soluble TNF (sTNF) production would abolish autocrine, paracrine, or endocrine TNF signaling, while increasing the relative abundance of membrane TNF (mTNF), which preferentially activates TNFRII [56,57]. In addition, loss of ADAM17 activity could increase the overall TNFRI or TNFRII levels, rendering the target cell more sensitive to TNF signaling [90].
Other substrates that may contribute to the ADAM17 KO metabolic phenotype include multiple members of the EGFR ligand family that are cleaved by ADAM17 to activate ErbB/EGFR family receptor tyrosine kinases [58]. Although studies on the role of EGFR ligands in metabolic regulation are sparse, the expression levels of the ErbB1 and ErbB4 ligand HB-EGF [59], which is highly expressed in human adipose tissue, correlates with the degree of adiposity. Increased plasma levels of HB-EGF in obese humans is positively associated with the incidence of coronary heart disease [60]. Moreover, neuregulin 4 (Nrg4), which is highly secreted from BAT, activates ErbB4 signaling in the liver, to downregulate hepatic lipogenesis [61]. Speculatively, because ErbB4 can be cleaved by ADAM17 [62,63], increased cell surface levels of ErbB4 may increase its signaling, potentially mediating some of the anti-obesogenic effects observed in iRhom2 KOs.
Interestingly, many of the phenotypes observed in our iRhom2 KO mice (e.g. increased BAT thermogenesis, improved insulin sensitivity, reduced weight gain, and decreased hepatic steatosis) are regulated by FGF21, a hormone secreted by the liver that acts on adipocytes. Activation of the FGF receptors by FGF21 requires a coreceptor, β-Klotho [64,65]. Several studies have reported that α-Klotho, a molecule closely related to β-Klotho, is cleaved by ADAM17 [66]. However, the role of ADAM17 in controlling the levels of FGF21 receptors or β-Klotho is not established.
It is interesting to note that there are no apparent metabolic differences between WT and iRhom2 KO mice when the animals are maintained on SD, whereas exposure of animals to HFD reveals enhanced thermogenesis in iRhom2 KOs. This raises the issue of why loss of iRhom2 enhances thermogenesis only within the context of HFD. Speculatively, one possibility is that conditions that license brown adipocytes to engage in thermogenesis (e.g. positive energy balance) may upregulate iRhom2 levels as part of a negative feedback loop to prevent uncontrolled thermogenesis. This may explain why the loss of iRhom2 results in elevated thermogenesis within the context of HFD.
An obvious question concerns what the implications of our work are for human metabolic disease. To the best of our knowledge, no publication has examined the impact of iRhom2 on obesity, diabetes, or BMI in human subjects. Studies in humans have focused on dominant mutations within the N-terminus of iRhom2 and their effect on hyperkeratosis [67] and esophageal cancer [68]. The proposed mechanistic basis of these disease-associated phenotypes is elevated EGFR activation [49].
Although ADAM17 null mutations in pediatric patients are associated with severe skin and gastrointestinal manifestations [69], there is no reported impact on metabolism. However, there is some evidence that, in adults, the expression of ADAM17 in skeletal muscle is positively associated with percentage body fat, type 2 diabetes, and lipid-induced insulin resistance and in the augmented inflammation associated with increased adiposity [70,71].
In spite of the extensive data showing that blocking TNF signaling improves insulin sensitivity in rodents, results in human studies have been mixed. Single doses of anti-TNF biologics have no effect in short term therapies, but some impact on insulin sensitivity has been observed in patients undergoing longer-term TNF inhibitor treatment for chronic inflammatory diseases [[72], [73], [74]] but not for metabolic syndrome [75]. However, as iRhom2 or ADAM17 KO mice have additional reported phenotypes (i.e., protection from obesity and increased energy consumption) compared to the TNF KOs, inhibition of the iRhom2/ADAM17 pathway could represent an alternative strategy for intervening in obesity/metabolic syndrome.
However, over the past two decades, pharmaceutical ADAM17 inhibitor programs have been closed down because of cytotoxicity associated with collateral targeting of other metalloproteases with similar active site architectures [76]. Even if ADAM17 could be inhibited specifically and chronically, there are additional concerns about unintended effects caused by systemic inhibition of ADAM17, particularly because its loss in humans is associated with inflammatory bowel and skin disease [69]. By contrast, iRhom2 KO mice lack most of the defects associated with loss of ADAM17, exhibit mature (active) ADAM17 in most somatic tissues, and do not exhibit epithelial defects. However, these mice are significantly protected from obesity and the range of other phenotypes reported for mice defective in TNF signaling. Given the relative absence of deleterious effects associated with deletion of iRhom2 compared to ADAM17, the targeting of iRhom2 could be an appealing prospect for future studies for the treatment of metabolic syndrome.
4. Experimental models
4.1. Experimental animals
iRhom2 KO mice were the kind gift of Matthew Freeman, the Dunn School, Oxford, UK, and have been previously described [21]. Mice were maintained in a SPF facility in a 12-hour light/dark cycle, at standard sub-thermoneutral conditions of 20–24 °C and an average of 50% humidity, in ventilated cages with corn cob as bedding. Four weeks-old iRhom2 KO or WT C57Bl/6 male mice were co-housed after weaning and throughout the experiments to homogenize differences in microbiota and other environmental conditions. They were given access to water ad libitum and standard (SD) or high-fat diet (HFD) (SNIFF diet D12492 60 kJ% fat, E15742: 60% of energy from fat). Typically, mice were sacrificed following up to 30 weeks of exposure to SD or HFD. Body weight was recorded weekly. Three independent HFD-induced obesity experiments, with n = 12 WT and 11 KO in two, and n = 11 WT and 9 KO in the third were performed. Typically, the analysis of the SD-fed mice was performed on 2–3 groups of 4–7 mice per genotype. In all experiments, mice were monitored twice per week to check for signs of discomfort or distress. A minimal interval of 1 week was maintained between metabolic measurements, to minimize stress on the mice. For high fat diet experiments, mice were housed in large cages [365 × 207 × 140 mm; floor area 530 cm2] containing a grid beneath the food repository that prevents the food from falling easily unto the floor of the cage. This avoids the stress associated with animals' fur becoming oily upon contact with the HFD. Each large cage has a defined capacity of 16 mice but we limited the numbers of mice to 9–12 per cage to provide sufficient space when the animals grow larger, to minimize stress.
Calorimetric measurements were performed using 6–8 mice per genotype and per diet and were conducted in temperature-controlled (24 °C) cages. After arrival at CiMUS, where the indirect calorimetry and metabolic cage experiments were performed, the mice were allowed to acclimate for 5 weeks before starting the experiments in the new facility. Both WT and iRhom2 KOs, fed on SD versus HFD, were exposed to identical conditions during transportation and acclimation.
4.2. FLAG-iRhom2 mice
FLAG-iRhom2 mice were generated via CRISPR/Cas9 as previously described [77]. In brief, one guide RNA (5′- GGTGCTACACTAACTGCAGA-3′) was designed to introduce a 3 x FLAG tag into Exon 19 of the mouse iRhom2 gene. An annealing oligo encoding the guide RNA was inserted into the plasmid pgRNAbasic, which contains a T7 promoter [77]. The linearized vector was used as a template for the production of sgRNA, produced by in vitro transcription using the MEGAshortscript T7 Kit (Life Technologies). RNA was cleaned using the MEGAclear kit (AM1908, Life Technologies). Cas9 mRNA was produced by in vitro transcription using the mMESSAGE mMACHINE T7 Ultra Kit (Life Technologies) and plasmid pT7-Cas9 as a template [77]. A 200bp homology template (5′-GCTTTCCCTTCACCAGCCGCTTCTGTGAGAAGTACGAGCTAGACCAGGTGCTACACGAGAATCTTTATTTTCAGGGCGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGATTACAAGGATGACGATGACAAGTAACTGCAGAGATTGTGTGTCTGCCCTGGGCCGTGTGTCTATGAACCGGTGGGGCCC-3′) was designed to contain 5′ and 3′ arms surrounding a TEV site and 3 x FLAG sequence. Homology template, Cas9 mRNA (10 ng/ml), and the sgRNA (10 ng/ml) were injected into the pronuclei of fertilized C57Bl/6 oocytes using standard procedures [78]. Genotyping from tail genomic DNA was performed using primers flanking the homology template listed in the Table of reagents (Table 1). We confirmed that the tagged form of iRhom2 retains its biological function, since macrophages isolated from mice containing one null allele of iRhom2 and one flag-tagged allele behave similarly to heterozygous animals in their capacity to secrete TNF (data not shown).
Table 1.
Key resource table.
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Mouse anti-Flag HRP | Sigma | A8592 |
| Mouse anti-p97 ATPase | Progen | 65278 |
| Rabbit anti-GAPDH | Cell Signaling Technology | 2118 |
| Mouse anti-HA-HRP | Roche | 12013819001 |
| Anti-rabbit HRP | Cell Signaling Technology | 1677074P2 |
| Anti-mouse HRP | Cell Signaling Technology | 1677076P2 |
| Rabbit anti-iRhom2 | Instituto Gulbenkian de Ciência | Produced in house |
| Rabbit anti-UCP1 | Abcam | Ab10983 |
| Anti-mouse CD45.2-PE | Instituto Gulbenkian de Ciência | Produced in house |
| Anti-mouse CD11b-FITC | Instituto Gulbenkian de Ciência | Produced in house |
| Anti-mouse F4/80-A647 | Biolegend | 123122 |
| Goat anti-TNF | R&D | AF-410 |
| Chemicals/Reagents | ||
| Albumin bovine serum fraction V fatty acid free | EMD Millipore | 126575 |
| Amphotericin B | Life Tech | 04195780D |
| Calcium Chloride (CaCl) | Sigma | 21114 |
| Collagenase | Fisher Bioreagents | BP2649-1 |
| Collagenase H Solution | Sigma | 11074059001 |
| Collagen I Rat Protein, Tail | Corning | 354249 |
| Chloroform | Acros | 10122190 |
| DAPI | ThermoFisher Scientific | D1306 |
| D-(+)Glucose | Sigma | G8270 |
| Dexamethasone | Cayman | CAYM11015-1 |
| Dispase II | Roche | 4942078001 |
| DMEM-F12 medium | Biowest | L0092-500 |
| Dulbecco’s Phosphate-Buffered Saline | Zen-Bio | DPBS-1000 |
| Eosin Y | Sigma | 230251 |
| Ethanol absolute | VWR | 20821.330 |
| Ethylenediaminetetraacetic acid (EDTA) | Fisher Chemical | 10080060 |
| Fc Block, clone 2.4G2 | Instituto Gulbenkian de Ciência | Produced in house |
| Fetal Bovine Serum | PAN Biotech | P30-3401 |
| Gentamicin Sulphate | Sigma | G1272 |
| Hematoxylin | Sigma | H3136 |
| Hanks’ Balanced Salt Solution | Biowest | L0611-500 |
| High-Fat Diet | SNIFF diet | D12492 |
| Hydrochloric acid 37% (HCl) | VWR | 20252.290 |
| HRP | Sigma | A8592 |
| Insulin Solution from Bovine Pancreas | Sigma | I0516 |
| New Born Calf Serum | Biowest | S075H-500 |
| (±)-Norepinephrine (+)-bitartrate salt | Sigma | A0937 |
| NZYol | NZYTech | MB18501 |
| Penicillin-Streptomycin Solution 100 x | Biowest | L0022-100 |
| Protease Inhibitor Cocktail | Roche | 11836153001 |
| Rosigliatozone | Cayman | CAYM71740-10 |
| Sodium Chloride (NaCl) | VWR | 27810.295 |
| Sodium Deoxycholate | Sigma | 30970 |
| Sodium Dodecyl Sulfate (SDS) | Fisher BioReagents | Cas151-21-3 |
| Sodium pyruvate | Sigma | P5280 |
| TRIS Base | VWR | PROL33621.260 |
| Triton X-100 | Fisher Chemical | T3751108 |
| 3-isobutyl-1-methylxanthine (IBMX) | AppliChem | A0695.0500 |
| 3,3’,5-Triiodo-L-thyronine sodium salt (T3) | Sigma | T6397 |
| Critical Commercial Kit Assays | ||
| Cholesterol CHOD-POD-Enz-Color | Spinreact | 201001092 |
| HDL c-D (HDL Cholesterol D) | Spinreact | 1001096 |
| LDL c-D (LDL Cholesterol D) | Spinreact | 41023 |
| High-Capacity cDNA reverse transcription kit | ThermoFisher Scientific | 4368814 |
| MEGAshortscript™ T7 Transcription Kit | Life Technologies | Am1354 |
| MEGAclear Kit | Life Technologies | AM1908 |
| mMESSAGE mMACHINE T7 Ultra Kit | Life Technologies | AMB1345 |
| Mouse Insulin ELISA | ALPCO | 80-INSMS-E01, E10 |
| RNA extraction Kit | NZYTech | MB13402 |
| Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies | 103015-100 |
| SuperScript® III First-Strand Synthesis SuperMix for q-RT-PCR | ThermoFisher Scientific | 11752-050 |
| Triglycerides GPO/POD-Enz-Color | Spinreact | 201001313 |
| Mouse recombinant TNF | R&D | 410-MT-010/CF |
| Mouse Strains | ||
| iRhom 2 KO | Matthew Freeman, Dunn School, Oxford, UK | [21] |
| C57BL/6 WT | Instituto Gulbenkian de Ciência | Produced in house |
| FLAG-iRhom2 | Instituto Gulbenkian de Ciência | Produced in house |
| Oligonucleotides | Sequences | |
| Cidea forward | 5’ TGCTCTTCTGTATCGCCCAGT 3’ | |
| Cidea reverse | 5’ GCCGTGTTAAGGAATCTGCTG 3’ | |
| Cox8b forward | 5’ GAACCATGAAGCCAACGACT 3’ | |
| Cox8b reverse | 5’ GCGAAGTTCACAGTGGTTCC 3’ | |
| Gapdh forward | 5’ AACTTTGGCATTGTGGAAGG 3’ | |
| Gapdh reverse | 5’ ACACATTGGGGGTAGGAACA 3’ | |
| Gnb1 forward | 5’ ccagttctggagacaccacat 3’ | |
| Gnb1 reverse | 5’ ggagcaagagacaggctca 3’ | |
| Gnb2 forward | 5’ ggcaaaaatctatgccatgc3 3’ | |
| Gnb2 reverse | 5’ tgatgagctttccgtcctg 3’ | |
| Il1 forward | 5’ GAAGAAGAGCCCATCCTCTG 3’ | |
| Il1 reverse | 5’ TCATCTCGGAGCCTGTAGTG 3’; | |
| Il6 forward | 5’ ACGGCCTTCCCTACTTCACA 3’ | |
| Il6 reverse | 5’ CATTTCCACGATTTCCCAGA 3’ | |
| Il10 forward | 5’ GCTCTTACTGACTGGCATGAG 3’ | |
| Il10 reverse | 5’ CGCAGCTCTAG GAGCATGTG 3’ | |
| Mcp1 forward | 5’ GGAAAAATGGATCCACACCTTGC 3’ | |
| Mcp1 reverse | 5’ TCTCTTCCTCCACCATGCAG-3’ | |
| Pgc1a forward | 5’ CCCTGCCATTGTTAAGACC 3’ | |
| Pgc1a reverse | 5’ TGCTGCTGTTCCTGTTTTC 3’ | |
| PRDM16 forward | 5’ CAGCACGGTGAAGCCATT 3’ | |
| PRDM16 reverse | 5’ GCGTGCATCCGCTTGTG 3’ | |
| Psmb2 forward | 5’ gagggcagtggagcttctta 3’ | |
| Psmb2 reverse | 5’ aggtgggcagattcaagatg 3’ | |
| Rhbdf2 forward | 5’ GCTCAACCGAAGCTATCGAC 3’ | |
| Rhbdf2 reverse | 5’ ACGAACGTCAGCCAGTAGGT 3’ | |
| Tbp forward | 5’ CAAACCCAGAATTGTTCTCCTT 3’ | |
| Tbp reverse | 5’ ATGTGGTCTTCCTGAATCCCT 3’ | |
| Tnf forward | 5’ ATGAGCACAGA AAGCATGATC 3’ | |
| Tnf reverse | 5’ TACAGGCTTGTCACTCGAATT 3’; | |
| Ucp1 forward | 5’ ACTGCCACACCTCCAGTCATT 3’ | |
| Ucp1 reverse | 5’ CTTTGCCTCACTCAGGATTGG 3’ | |
| Ywhaz forward | 5’ ttacttggccgaggttgct 3’ | |
| Ywhaz reverse | 5’ tgctgtgactggtccacaat 3’ | |
| FLAG-iRhom2 forward | 5’ ACATTACCTTCGGCACCAGCG 3’ | |
| FLAG-iRhom2 reverse | 5’ ATCTGGAGTCAGCCTTGAGT 3’ | |
| Taqman Probes | ||
| Actb | ThermoFisher Scientific-Applied Biosystems | ID-Mm00607939_s1 |
| Rhbdf2 | ThermoFisher Scientific-Applied Biosystems | ID-Mm00553469_m1 |
| Plasmids | ||
| pCL-Eco | Matthew Freeman, Dunn School, Oxford, UK | [84] |
| pT7-Cas9 | Instituto Gulbenkian de Ciência | [77] |
| pM6P.BLAST empty vector | F. Randow, Cambridge, UK | |
| pM6P.BLAST iRhom2-HA | Instituto Gulbenkian de Ciência | |
| SV40 Large T Ag | Matthew Freeman, Dunn School, Oxford, UK | |
4.3. Metabolic measurements
Following exposure to SD or HFD for 20–22 weeks, serum was collected after overnight fasting, and the levels of total cholesterol, triglycerides, HDL, and LDL were measured using an enzymatic-colorimetric kit (Spinreact). Insulin levels were calculated by ELISA (ALPCO). Blood glucose was measured using a gluco-analyzer (Roche Diagnostics). Oral glucose, insulin, and lipid tolerance tests (OGTT, ITT, and LTT, respectively) were performed on conscious mice following exposure to SD or HFD for 26–28 weeks. Glucose (1,5 g/Kg) (Sigma) was administrated per os to overnight fasted mice. Human recombinant insulin (0,9 U/Kg) (Lilly) was administrated intraperitoneally to 6 h-fasted mice. Blood glucose was measured at 0, 15, 30, 60, 90, and 120 min. The area under the curve (AUC) (%) was calculated using the trapezoid rule (the curve is divided into series of trapezoids and the sum of their area is the AUC). For the LTT, olive oil (0,01 μl/kg) was administrated per os to overnight fasted mice. Quantification of triglycerides in the serum was performed at 0, 1, 2, 3, and 4 h.
The quantitative insulin sensitivity check index (QUICKI) was calculated using the formula 1/log [fasting glucose] + log [fasting insulin]. Homeostatic model assessment of insulin resistance index (HOMA-IR) was calculated as ([fasting insulin] (mU/L) x [fasting glucose] (mmol/L))/14,1. Following exposure to SD or HFD for 24–25 weeks, mouse feces were collected after a 24 h period in order to analyze its fat content. Samples were then dried for 1 h at 70 °C. One gram of dried feces was then homogenized with a tissue grinder in PBS. Chloroform: methanol (2:1) was added to the homogenized samples and after centrifugation (1000G, 10 min) the lower liquid phase (containing the lipids) was collected and left to evaporate. Chloroform + 1% Triton-X-100 was added, and after evaporation, water was added and triglycerides and cholesterol were measured using an enzymatic-colorimetric kit (Spinreact).
After 30 weeks on HFD, total cholesterol and triglycerides in the livers was measured using the same kit described above. The result was normalized to the liver weight (mg/g). Briefly, a piece of the liver caudate lobe was homogenized in chloroform/methanol (2:1) using TissueLyser II (Qiagen) (30 Hz, 5 min). After overnight incubation, this mixture was centrifuged (800G, 15 min) and the supernatant was mixed with 1/5th of a volume of 0,9% NaCl. The lower phase was then dried out and butanol/(Triton X-100:methanol) (2:1) was added to perform the colorimetric analysis. Insulin extraction from the pancreas was performed using the method described by Rastogi et al. [79]. Pancreatic insulin content was measured using the kit described above.
4.4. Calorimetric measurements
Animals were analyzed for energy expenditure (EE), respiratory quotient (RQ), and locomotor activity (LA) using a calorimetric system (LabMaster; TSE Systems; Bad Homburg, Germany) as previously shown [[80], [81], [82]].
4.5. Macroscopic analysis
After exposure to HFD for 30 weeks, mice were sacrificed and blood was collected by cardiac puncture. After perfusion with PBS, brown adipose tissue (BAT), dorsal subcutaneous fat (sWAT), epididymal adipose tissue (eWAT), mesenteric fat, retroperitoneal fat, spleen, liver, pancreas, and limb muscle were collected. All the tissues were weighed, except for the muscle.
4.6. Western blotting
SVF-derived primary adipocytes were washed in PBS and lysed for 10 min on ice in TX-100 lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris–HCl, pH 7.4) containing complete protease inhibitor cocktail (Roche). Tissues from FLAG-iRhom2 and WT mice were lysed in a modified RIPA buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris–HCl, pH 7.4, 1 mM EDTA, 1% Sodium Deoxycholate, 0,1% SDS) containing complete protease inhibitor cocktail (Roche) in a TissueLyser II (QIAGEN). The samples were then quantified, normalized, and denatured with LDS at 65 °C for 15 min. The lysates were fractionated by SDS-PAGE and transferred onto PVDF membranes. After blocking with 5% milk in TBS-T for 30 min, the membranes were cut and incubated overnight with the primary antibodies mouse anti-Flag HRP (1:1000; A8592; Sigma) and rabbit anti-GAPDH (1:2000; 2118, Cell Signaling Technology), rabbit anti-iRhom2 (1:500; produced in house), mouse anti-p97 ATPase (1:1000; 65278, Progen), mouse anti-HA (1:5000; 12013819001 Roche). The following day, the membranes were washed and incubated with the secondary antibody anti-rabbit HRP (1:5000; 1677074P2, Cell Signaling Technology) or anti-mouse (1:5000; 1677076P2, Cell Signaling Technology). After washing, protein bands were detected using ECL.
4.7. Histopathological analysis
The collected samples were processed to paraffin-embedded sections (3 μm sections) and stained with H-E (Sigma). Histopathological analysis was performed by a pathologist in a blinded manner. Sections were examined under a Leica DMLB2 microscope and images were acquired with a Leica DFC320 camera. The extent of NAFLD was scored using the following classification: (0) no alterations, (1) mild hepatocyte swelling/tumefaction with even cytoplasm vacuolization, (2) moderate hepatocyte swelling/tumefaction with uneven hepatocyte vacuolization, (3) marked hepatocyte swelling/tumefaction with uneven hepatocyte vacuolization and mild to few ballooning, (4) equal to (3) plus moderate to marked hepatocyte ballooning. Oil red staining (Sigma) was performed on 8 μm liver and muscle cryosections, from samples fixed in 10% formalin solution for 48 h, dehydrated in 30% sucrose, and snap frozen in liquid nitrogen embedded in OCT. The liver and muscle fat content was scored using the following classification: (0) within normal limits, (1) mild to moderate, and (2) severe. WAT and BAT were scanned into digital images (NanoZoomer-SQ Digital slide scanner-Hamamatsu) and analyzed using Fiji software. The average WAT adipocyte size [average cross-sectional area per cell (μm2)] was determined using a macro on H-E staining of adipose tissue from 3 HFD-fed animals and 1–3 SD-fed mice per genotype per experiment (3 experiments). The applied macro can be provided under request. The average BAT adipocyte size was calculated as described previously [83]. Briefly, five low-power field images were acquired at regular spatial intervals, after which the diameters of a representative number of cells in each field were measured by an observer blinded to the conditions. Steatitis was scored using the following classification: (0) no alterations, (1) rare macrophage infiltration, (2) mild multifocal macrophage infiltration, (3) moderate multifocal macrophage infiltration, (4) moderate multifocal macrophage infiltration with steatonecrosis.
4.8. Adipocyte culture
Interscapular BAT and sWAT from 5 to 6 iRhom2 KO and WT 4-6-week-old mice fed with SD was combined, minced and digested in 2 ml of Hank's balanced salt solution (Biowest) supplemented with 1 mg/ml Collagenase (Fisher Bioreagents), 2.2 mg/ml Dispase II (Roche) and 10 mM CaCl2 at 37 °C in a shaker for 30 min. DMEM-F12 (Biowest) was added to stop digestion. Solution was filtered through a 100 μm strainer and the filtrate centrifuged at 4 °C, 500G for 10 min. The pellet was resuspended in DMEM-F12 and filtered through a 40 μm strainer and the filtrate centrifuged at 4 °C, 500G for 5 min. The resultant pellet was thereafter resuspended in complete medium (DMEM-F12 supplemented with 10% new born calf serum, 1% Penicillin-Streptomycin solution 100X, 10 μg/ml gentamicin sulphate (Sigma), and 2.5 μg/ml Amphotericin B (Life Tech)) and plated in 10 cm dish. The cells were incubated at 37 °C and 5% CO2. For adipocyte differentiation, plates were coated with collagen I, rat tail (Corning). Confluent cultures were exposed to induction medium (complete medium containing 10 μM dexamethasone, 2 μM rosiglitazone (Cayman), 1 μgml−1 insulin (Sigma), 1 mM 3-isobutyl methylxanthine (AppliChem), and 2 nM 3,3′,5-Triiodo-l-thyronine (Sigma)) on day 0 and post induction day 2. From post induction day 4 until day 6 cells were exposed to maintenance medium (complete medium supplemented with 1 μgml−1 insulin and 2 nM T3). Differentiated primary adipocytes were treated for 8 h with mouse TNF (10 ng/ml, 410-MT-010/CF, R&D) or with an anti-TNF antibody (1 μg/ml, AF-410, R&D) and compared to nontreated cells.
4.9. Retroviral transduction
HEK293 ET packaging cells (1 × 106) were transfected with pCL-Eco packaging plasmid [84] plus pM6P.BLAST empty vector (kind gift of F. Randow, Cambridge, UK), or pM6P containing mouse iRhom2 WT cDNA fused to a C-terminal HA tag, or SV40 Large T Ag. WT brown preadipocytes from a WT newborn mouse was immortalized as previously described [85]. In summary, isolated cells were split in two and seeded in a 35 mm plate at 40% confluence and one well was transduced with 2 ml of SV-40 supernatant supplemented with polybrene 8 μg/mL twice with a 24 h interval. The non-transduced cells were used as control. Then, the immortalized cells were transduced with the empty vector or iRhom2-HA viral supernatant supplemented with polybrene 8 μg/mL, and selected with blasticidin (8 μg/mL) for 2 days to generate stable cell lines. These cells were differentiated as mentioned above and incubated for 5 h with 0.4 μM or 0.04 μM of norepinephrine (Sigma) in DMEM F12 with 2% free fatty acid BSA (EMD Millipore) or only in the medium.
4.10. Seahorse assay
Oxygen consumption rate (OCR) was measured using a Seahorse XFe96 analyzer (Agilent Tech.) and the Seahorse XF Cell Mito Stress Test Kit according to instructions from the manufacturer. Specifically, primary brown preadipocytes pulled from 3 to 4 mice BAT were plated on collagen-coated Seahorse XF96 plates (10.000 cells per well) and differentiated as mentioned above. Cells were incubated for 5 h in medium alone (DMEM F12 with 2% free fatty acid BSA; EMD Millipore) or with medium containing 0.4 μM norepinephrine (Sigma). Then, they were washed twice with warm Seahorse XF Assay Medium (pH 7.4, 1 mM sodium pyruvate, 2 mM l-glutamine, 10 mM glucose), and XF Assay Medium (180 μL) was added per well. Cells were incubated in a non-CO2 incubator (37 °C; 1 h) prior to assay. The analyzer was programmed to calibrate and equalize samples, followed by 3 baseline measurements (3 min each) and mixing (3 min) between measurements prior to inhibitor injection. The inhibitors were injected in the following order: Oligomycin (2 μM); FCCP (1 μM); Rotenone/Antimycin A (0.5 μM). Three measurements (3 min each) were made following each injection with 3 min mixing between measurements. A volume of 20 μl of 10x TX-100 lysis buffer with protease inhibitor (described above) was added and the protein content of 2 replicates per condition was quantified for normalization.
4.11. Immunohistochemistry
Detection of UCP1 in WAT was performed using anti-UCP1 antibody (1:500; ab10983; Abcam) as previously reported [[86], [87], [88]].
4.12. Flow cytometry
Adipose tissue samples were digested in 1% collagenase H solution (Sigma) (1 h, 37 °C, with shaking). Spleens were mashed through a 70 μm cell strainer. The digested cells were centrifuged (5 min, 4 °C, 2000 rpm), and red blood lysis buffer was added (9 vol. NH4Cl and 1 vol. Tris-Cl). The cells were washed and filtered through a 70 μm strainer. Cells were incubated in Fc Block, clone 2.4G2 (1:100, produced in-house) and staining was then performed with anti-mouse CD45.2-PE, CD11b-FITC (1:100; produced in-house), and F4/80-A647 (1:100; 123122; Biolegend). After washing in PBS plus 2% new born calf serum, DAPI (0.2 μg/mL; Invitrogen) was added and the samples were analyzed using a Cyan ADP analyzer (Beckman Coulter) with FlowJo software, version 10.2. The gating strategy used is shown in Figure S4.
4.13. Quantitative transcriptional analysis
Interscapular BAT from iRhom2 KO and WT mice, and liver, hypothalamus, sWAT, and eWAT from WT mice fed with SD or HFD for 30 weeks were snap frozen in liquid nitrogen until RNA extraction (NZYTech). The same was applied to adipocytes differentiated in vitro. Interscapular BAT from 2 WT mice fed with SD and HFD for 30 weeks was combined, minced and digested as described above. The digested samples were filtered and centrifuged at 400G for 7 min at room temperature. The supernatant was collected, filtered, and after standing for 10 min, 1 ml was collected from the top layer to obtain mature brown adipocytes. The pellet (stromal vascular fraction or SVF) was washed twice. RNA was extracted from mature adipocytes, SVF and whole tissue using NZYol (NZYTech), followed by purification in columns (NZYTech). In the case of BAT from mice exposed to different temperatures, RNA was extracted from 50 mg of tissue using the Trizol reagent. First-strand cDNA was synthesized from total RNA using the SuperScript® III First-Strand Synthesis SuperMix or the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). Real-time PCR analysis was performed using the comparative CT method [89]. Gene expression was normalized to Gapdh, Actin or to the geometrical average of 5 different genes (Tbp, Ywhaz, Psmb2, Gnb2, and Gnb1). The primers sequence and Taqman probes used are listed in the Table of reagents (Table 1).
4.14. Statistics
To compare single measurements between control and test groups, unpaired t test with Welch's correction or the Mann–Whitney–Wilcoxon test were used. For repeated measurements, the two-way ANOVA was used. The statistical analysis was performed using GraphPad Prism, version 6. Results are presented as average ± SEM. P-values <0.05 were represented as (*), <0.01 as (**), <0.001 as (***), and <0.0001 as (****).
4.15. Study approval
Animal procedures carried out at Instituto Gulbenkian de Ciência were approved by the Portuguese national regulatory agency (DGAV – Direção Geral de Alimentação e Veterinária) and by the ethics and animal welfare committees of Instituto Gulbenkian de Ciência. Experiments performed at the University of Santiago de Compostela, Spain, were performed in agreement with the International Law on Animal Experimentation and were approved by the USC Ethical Committee (Project License 15010/14/006).
Author contributions
M.B., I.G.-G., L.F.M., M.L., and C.A. designed the research studies. M.B., A.A., I.F., E.B., E.S., E.C., M.C., D.P., A.N.C., S.C., and I.G.-G. carried out the experiments. P.F. performed the histopathological analysis. M.B., A.A., I.G.-G., A.D., M. L., A.V.P., F.J.O., J.M.F.-R., and C.A. interpreted the data. M.B. and C.A. prepared the manuscript. C.A. supervised the project. All authors read and approved the final manuscript.
Acknowledgements
The authors thank the Animal, Mouse Transgenics, Histopathology, Flow cytometry and the Antibody units of the Instituto Gulbenkian de Ciência. We thank Moises Mallo and Ana Nóvoa for advice and help in the generation of FLAG-iRhom2 mutant mice. We thank Jorge Ruas for advice concerning ex vivo primary adipocyte culture. C.A. acknowledges the support of Fundação Calouste Gulbenkian, Worldwide Cancer Research (14–1289), a Marie Curie Career Integration Grant (project no. 618769), Fundação para a Ciência e Tecnologia (FCT, SFRH/BCC/52507/2014; PTDC/BEX-BCM/3015/2014; LISBOA-01–0145-FEDER-031330). M.C. acknowledges the support of the FCT grant (SFRH/BPD/117216/2016). This work was developed with the support of the research infrastructure Congento, project LISBOA-01–0145-FEDER-022170, co-financed by Lisboa Regional Operational Programme (Lisboa 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF), and Foundation for Science and Technology (Portugal). M.L. was funded by Xunta de Galicia (2016-PG068), Ministerio de Economía y Competitividad (MINECO; SRTI2018-101840-B-I00) and Atresmedia. L.F.M. is an FCT Investigator and is supported by the European Community Horizon 2020 (ERC-2014-CoG 647888-iPROTECTION) and FCT (FCT: PTDC/BIM-MEC/4665/2014). S.C. and A.V.P. were funded by the Wellcome Trust strategic award (100574/Z/12/Z) and MRC MDU (MC_UU_12012/2).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.10.006.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
iRhom2 is expressed in metabolic tissues and increased in BAT during obesity, and its deletion protects mice from this disease. A Protein expression of iRhom2 in subcutaneous (sWAT), brown (BAT), retroperitoneal (Retrop.), mesenteric (Mesent.), and epididymal (eWAT) adipose tissues, pancreas, liver, and muscle from homozygous (iR2-FLAG) mice with an N-terminal FLAG tag incorporated into the endogenous iRhom2 gene. B Protein expression of iRhom2 in BAT from iR2-FLAG mice fed with SD versus HFD. In A, B We used iRhom2 WT BAT as a negative control. As a loading control we measured GAPDH protein level. Two experiments were performed. C RT-PCR analysis of Rhbdf2 (iRhom2) mRNA, normalized to a housekeeping control (beta Actin), in brown adipocytes differentiated in vitro from the stromal vascular fraction of 4-5 pooled WT or iRhom2 KO mice. The WT cells were treated for 8 h with TNF (10 ng/ml) or with an anti-TNF antibody (1 μg/ml) and compared to nontreated cells. The lack of iRhom2 gene expression in the iRhom2 KO brown adipocytes provides a specificity control. Three independent experiments. D Body weight over time of iRhom2 KO and WT mice fed with HFD from weeks 4 to 34 of age. In panels C,D error bars represent SEM; * represents p<0.05, **** represents p<0.0001. E-F Representative photographs of iRhom2 KO and WT fed with SD and HFD for 30 weeks.
HFD-fed iRhom2 KO mice do not exhibit significantly reduced accumulation of fat in the skeletal musculature. A-B Histopathological classification of skeletal muscle fat content (scores 0-2) (A), and representative photographs of skeletal muscle Oil red staining (100x magnification) (B) of iRhom2 KO and WT fed with HFD for 30 weeks. Two experiments with 5-6 mice per group. Error bars represent SEM; Scale bar = 100 μm.
Adipose tissues from HFD-fed iRhom2 KO mice have an increased frequency of smaller adipocytes. A Representative photographs of eWAT and sWAT from iRhom2 KO and WT mice fed with HFD for 30 weeks. B-F Histograms showing the adipocyte area frequency (%) from subcutaneous (sWAT) (B), mesenteric (C), epididymal (eWAT) (D), reptroperitoneal (E), and brown adipose tissue (BAT) (F) of iRhom2 KO and WT mice fed with HFD for 30 weeks. Three experiments with 3 mice per genotype. Error bars represent SEM.
Gating strategy used in the FACS analysis. A-E Cells were first gated on size (SS vs FS) (A) and then on singularity (Pulse Width vs FS) (B). This was followed by DAPI exclusion to gate for live cells (SS vs FL6) (C). Live cells were gated on expression of CD45.2 to select the myeloid cells (FL3 vs FL2) (D). Finally, myeloid cells were gated for co-expression of CD11b and F4/80 to identify the macrophages (FL8 vs FL1) (E).
HFD but not SD fed iRhom2 KO mice have increased cumulative energy expenditure. A-D Energy expenditure (EE) over time (A, B) and cumulative EE (C, D) of iRhom2 KO and WT mice fed with HFD (A, C) and SD (B, D) for 17 weeks. Results are representative of one experiment, with a total of n=8 and n=6 for SD and HFD experiments, respectively. Error bars represent SEM; * represents p<0.05.
iRhom2 loss leads to upregulation of thermogenic genes, whereas ambient temperature influences iRhom2 expression levels. A RT-PCR analysis of UCP1, PGC1a, Cidea, PRDM16, and Cox8b expression in RNA isolated from BAT samples isolated from WT versus iRhom2 KO mice fed on SD or HFD for 2 weeks. Two experiments with 3-4 replicates in each. B RT-PCR analysis of Rhbdf2 expression in the BAT of WT mice subjected to the following temperature conditions, relative to thermoneutrality (housed at 30°C for 4 weeks): (1) Mice housed at 21°C for the time of the study, (2) mice exposed to 5°C for 1 week, (3) mice acclimated for 4 weeks at 5°C, (4) mice acclimated for 4 weeks at 5°C and then exposed to 30°C for 1 week, (5) mice acclimated for 4 weeks at 5°C and then acclimated for 4 weeks at 30°C. One experiment with 4 independent samples per condition. C-H Basal (C) and maximal respiration (D), ATP production (E), coupling efficiency (F), spare respiratory capacity (G), and non-mitochondrial oxygen consumption (H) of brown adipocytes differentiated in vitro from the stromal vascular fraction of 4-5 pooled WT and iRhom2 KO mice and stimulated or not with norepinephrine. The results were normalized to the protein content. Two experiments with one or two independent samples per genotype, respectively. Error bars represent SEM; * represents p<0.05, ** represents p<0.01, *** represents p<0.001, **** represents p<0.0001.
References
- 1.van der Klaauw A., Farooqi I. The hunger genes: pathways to obesity. Cell. 2015;161:119–132. doi: 10.1016/j.cell.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz M.W., Seeley R.J., Zeltser L.M., Drewnowski A., Ravussin E., Redman L.M. Obesity pathogenesis: an endocrine society scientific statement. Endocrine Reviews. 2017;38:267–296. doi: 10.1210/er.2017-00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Després J.P., Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 4.Vidal-Puig A., Unger R.H. Special issue on lipotoxicity. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2010;1801:207–208. doi: 10.1016/j.bbalip.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 5.van Herpen N.A., Schrauwen-Hinderling V.B. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiology & Behavior. 2008;94:231–241. doi: 10.1016/j.physbeh.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg A.S., Obin M.S. Obesity and the role of adipose tissue in inflammation and metabolism. American Journal of Clinical Nutrition. 2006;83:461S–465S. doi: 10.1093/ajcn/83.2.461S. [DOI] [PubMed] [Google Scholar]
- 7.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 8.Choi C.H.J., Cohen P. Adipose crosstalk with other cell types in health and disease. Experimental Cell Research. 2017;360:6–11. doi: 10.1016/j.yexcr.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Stern J.H., Rutkowski J.M., Scherer P.E. Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metabolism. 2016;23:770–784. doi: 10.1016/j.cmet.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klingenberg M. UCP1 - a sophisticated energy valve. Biochimie. 2017;134:19–27. doi: 10.1016/j.biochi.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Hamann A., Flier J.S., Lowell B.B. Decreased brown fat markedly enhances susceptibility to diet-induced obesity, diabetes, and hyperlipidemia. Endocrinology. 1996;137:21–29. doi: 10.1210/endo.137.1.8536614. [DOI] [PubMed] [Google Scholar]
- 12.Bartelt A., Heeren J. Adipose tissue browning and metabolic health. Nature Reviews Endocrinology. 2014;10:24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- 13.Nedergaard J., Cannon B. The browning of white adipose tissue: some burning issues. Cell Metabolism. 2014;20:396–407. doi: 10.1016/j.cmet.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Wu J., Boström P., Sparks L.M., Ye L., Choi J.H., Giang A.H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thyagarajan B., Foster M.T. Beiging of white adipose tissue as a therapeutic strategy for weight loss in humans. Hormone Molecular Biology and Clinical Investigation. 2017;31 doi: 10.1515/hmbci-2017-0016. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava S., Veech R.L. Brown and brite: the fat soldiers in the anti-obesity fight. Frontiers in Physiology. 2019;10:38. doi: 10.3389/fphys.2019.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giralt M., Villarroya F. White, brown, beige/brite: different adipose cells for different functions. Endocrinology. 2013;154:2992–3000. doi: 10.1210/en.2013-1403. [DOI] [PubMed] [Google Scholar]
- 18.Giordano A., Frontini A., Cinti S. Convertible visceral fat as a therapeutic target to curb obesity. Nature Reviews Drug Discovery. 2016;15:405–424. doi: 10.1038/nrd.2016.31. [DOI] [PubMed] [Google Scholar]
- 19.Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annual Review of Immunology. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 20.Black R.A., Rauch C.T., Kozlosky C.J., Peschon J.J., Slack J.L., Wolfson M.F. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 21.Adrain C., Zettl M., Christova Y., Taylor N., Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIlwain D.R., Lang P.A., Maretzky T., Hamada K., Ohishi K., Maney S.K. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Maretzky T., Weskamp G., Monette S., Qing X., Issuree P.D. iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6080–6085. doi: 10.1073/pnas.1505649112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grieve A.G., Xu H., Künzel U., Bambrough P., Sieber B., Freeman M. Phosphorylation of iRhom2 at the plasma membrane controls mammalian TACE-dependent inflammatory and growth factor signalling. Elife. 2017;6 doi: 10.7554/eLife.23968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavadas M., Oikonomidi I., Gaspar C.J., Burbridge E., Badenes M., Félix I. Phosphorylation of iRhom2 controls stimulated proteolytic shedding by the metalloprotease ADAM17/TACE. Cell Reports. 2017;21:745–757. doi: 10.1016/j.celrep.2017.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maretzky T., McIlwain D.R., Issuree P.D., Li X., Malapeira J., Amin S. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11433–11438. doi: 10.1073/pnas.1302553110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cawthorn W.P., Sethi J.K. TNF-alpha and adipocyte biology. FEBS Letters. 2008;582:117–131. doi: 10.1016/j.febslet.2007.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotamisligil G.S. Foundations of immunometabolism and implications for metabolic health and disease. Immunity. 2017;47:406–420. doi: 10.1016/j.immuni.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hotamisligil G.S. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542:177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- 30.Paz K., Hemi R., LeRoith D., Karasik A., Elhanany E., Kanety H. A molecular basis for insulin resistance: elevated serine/threonine phosphorylation of IRS-1 and IRS-2 inhibits their binding to the juxtamembrane region of the insulin receptor and impairs their ability to undergo insulin-induced tyrosine phosphorylation. Journal of Biological Chemistry. 1997;272:29911–29918. doi: 10.1074/jbc.272.47.29911. [DOI] [PubMed] [Google Scholar]
- 31.Qatanani M., Lazar M.A. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Genes & Development. 2007;21:1443–1455. doi: 10.1101/gad.1550907. [DOI] [PubMed] [Google Scholar]
- 32.Uysal K.T., Wiesbrock S.M., Marino M.W., Hotamisligil G.S. Protection from obesity-induced insulin resistance in mice lacking TNF-α function. Nature. 1997;389:610–614. doi: 10.1038/39335. [DOI] [PubMed] [Google Scholar]
- 33.Gelling R.W., Yan W., Al-Noori S., Pardini A., Morton G.J., Ogimoto K. Deficiency of TNFalpha converting enzyme (TACE/ADAM17) causes a lean, hypermetabolic phenotype in mice. Endocrinology. 2008;149:6053–6064. doi: 10.1210/en.2008-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko H., Anzai T., Horiuchi K., Morimoto K., Anzai A., Nagai T. Tumor necrosis factor-α converting enzyme inactivation ameliorates high-fat diet-induced insulin resistance and altered energy homeostasis. Circulation Journal. 2011;75:2482–2490. doi: 10.1253/circj.cj-11-0182. [DOI] [PubMed] [Google Scholar]
- 35.Scheller J., Chalaris A., Garbers C., Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends in Immunology. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Peschon J.J., Slack J.L., Reddy P., Stocking K.L., Sunnarborg S.W., Lee D.C. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 37.Issuree P.D., Maretzky T., McIlwain D.R., Monette S., Qing X., Lang P.A. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. Journal of Clinical Investigation. 2013 doi: 10.1172/JCI66168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.H., Kim J., Chun J., Lee C., Im J.P., Kim J.S. Role of iRhom2 in intestinal ischemia-reperfusion-mediated acute lung injury. Scientific Reports. 2018;8:239. doi: 10.1038/s41598-018-22218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabbrini E., Sullivan S., Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilherme A., Virbasius J.V., Puri V., Czech M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nature Reviews Molecular Cell Biology. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lumeng C.N., Saltiel A.R. Inflammatory links between obesity and metabolic disease. Journal of Clinical Investigation. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qing X., Rogers L., Mortha A., Lavin Y., Redecha P., Issuree P.D. iRhom2 regulates CSF1R cell surface expression and non-steady state myelopoiesis in mice. European Journal of Immunology. 2016;46:2737–2748. doi: 10.1002/eji.201646482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siggs O.M., Xiao N., Wang Y., Shi H., Tomisato W., Li X. iRhom2 is required for the secretion of mouse TNFalpha. Blood. 2012;119:5769–5771. doi: 10.1182/blood-2012-03-417949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutens L., Stienstra R. Adipose tissue macrophages: going off track during obesity. Diabetologia. 2016;59:879–894. doi: 10.1007/s00125-016-3904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y., Tsai Y.-H., Xiao W., Ralls M.W., Stoeck A., Wilson C.L. Loss of ADAM17-mediated tumor necrosis factor Alpha signaling in intestinal cells attenuates mucosal atrophy in a mouse model of parenteral nutrition. Molecular and Cellular Biology. 2015;35:3604–3621. doi: 10.1128/MCB.00143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fenzl A., Kiefer F.W. Brown adipose tissue and thermogenesis. Hormone Molecular Biology and Clinical Investigation. 2014;19:25–37. doi: 10.1515/hmbci-2014-0022. [DOI] [PubMed] [Google Scholar]
- 47.Serino M., Menghini R., Fiorentino L., Amoruso R., Mauriello A., Lauro D. Mice heterozygous for tumor necrosis factor-alpha converting enzyme are protected from obesity-induced insulin resistance and diabetes. Diabetes. 2007;56:2541–2546. doi: 10.2337/db07-0360. [DOI] [PubMed] [Google Scholar]
- 48.Christova Y., Adrain C., Bambrough P., Ibrahim A., Freeman M. Mammalian iRhoms have distinct physiological functions including an essential role in TACE regulation. EMBO Reports. 2013;14:884–890. doi: 10.1038/embor.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosur V., Johnson K.R., Burzenski L.M., Stearns T.M., Maser R.S., Shultz L.D. Rhbdf2 mutations increase its protein stability and drive EGFR hyperactivation through enhanced secretion of amphiregulin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2200–E2209. doi: 10.1073/pnas.1323908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nisoli E., Briscini L., Giordano A., Tonello C., Wiesbrock S.M., Uysal K.T. Tumor necrosis factor alpha mediates apoptosis of brown adipocytes and defective brown adipocyte function in obesity. Proceedings of the National Academy of Sciences. 2000;97:8033–8038. doi: 10.1073/pnas.97.14.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porras A., Valladares A., Alvarez A.M., Roncero C., Benito M. Differential role of PPAR gamma in the regulation of UCP-1 and adipogenesis by TNF-alpha in brown adipocytes. FEBS Letters. 2002;520:58–62. doi: 10.1016/s0014-5793(02)02762-x. [DOI] [PubMed] [Google Scholar]
- 52.Valladares A., Roncero C., Benito M., Porras A. TNF-alpha inhibits UCP-1 expression in brown adipocytes via ERKs. Opposite effect of p38MAPK. FEBS Letters. 2001;493:6–11. doi: 10.1016/s0014-5793(01)02264-5. [DOI] [PubMed] [Google Scholar]
- 53.Reddy P., Slack J.L., Davis R., Cerretti D.P., Kozlosky C.J., Blanton R.A. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. Journal of Biological Chemistry. 2000;275:14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]
- 54.Hehlgans T., Pfeffer K. The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology. 2005;115:1–20. doi: 10.1111/j.1365-2567.2005.02143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmad S., Azid N.A., Boer J.C., Lim J., Chen X., Plebanski M. The key role of TNF-TNFR2 interactions in the modulation of allergic inflammation: a review. Frontiers in Immunology. 2018;9:761. doi: 10.3389/fimmu.2018.02572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grell M., Douni E., Wajant H., Löhden M., Clauss M., Maxeiner B. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 57.Rauert H., Wicovsky A., Müller N., Siegmund D., Spindler V., Waschke J. Membrane tumor necrosis factor (TNF) induces p100 processing via TNF receptor-2 (TNFR2) Journal of Biological Chemistry. 2010;285:7394–7404. doi: 10.1074/jbc.M109.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider M.R., Wolf E. The epidermal growth factor receptor ligands at a glance. Journal of Cellular Physiology. 2009;218:460–466. doi: 10.1002/jcp.21635. [DOI] [PubMed] [Google Scholar]
- 59.Raab G., Klagsbrun M. Heparin-binding EGF-like growth factor. Biochimica et Biophysica Acta. 1997;1333:F179–F199. doi: 10.1016/s0304-419x(97)00024-3. [DOI] [PubMed] [Google Scholar]
- 60.Matsumoto S., Kishida K., Shimomura I., Maeda N., Nagaretani H., Matsuda M. Increased plasma HB-EGF associated with obesity and coronary artery disease. Biochemical and Biophysical Research Communications. 2002;292:781–786. doi: 10.1006/bbrc.2002.6720. [DOI] [PubMed] [Google Scholar]
- 61.Wang G.X., Zhao X.Y., Meng Z.X., Kern M., Dietrich A., Chen Z. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nature Medicine. 2014;20:1436–1443. doi: 10.1038/nm.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rio C., Buxbaum J.D., Peschon J.J., Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. Journal of Biological Chemistry. 2000;275:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- 63.Cheng Q.C., Tikhomirov O., Zhou W., Carpenter G. Ectodomain cleavage of ErbB-4: characterization of the cleavage site and m80 fragment. Journal of Biological Chemistry. 2003;278:38421–38427. doi: 10.1074/jbc.M302111200. [DOI] [PubMed] [Google Scholar]
- 64.Ding X., Boney-Montoya J., Owen B.M., Bookout A.L., Coate K.C., Mangelsdorf D.J. βKlotho is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metabolism. 2012;16:387–393. doi: 10.1016/j.cmet.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams A.C., Cheng C.C., Coskun T., Kharitonenkov A. FGF21 requires βklotho to act in vivo. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049977. e49977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bloch L., Sineshchekova O., Reichenbach D., Reiss K., Saftig P., Kuro-o M. Klotho is a substrate for alpha-, beta- and gamma-secretase. FEBS Letters. 2009;583:3221–3224. doi: 10.1016/j.febslet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brooke M.A., Etheridge S.L., Kaplan N., Simpson C., O'Toole E.A., Ishida-Yamamoto A. iRHOM2-dependent regulation of ADAM17 in cutaneous disease and epidermal barrier function. Human Molecular Genetics. 2014;23:4064–4076. doi: 10.1093/hmg/ddu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blaydon D.C., Etheridge S.L., Risk J.M., Hennies H.C., Gay L.J., Carroll R. RHBDF2 mutations are associated with tylosis, a familial esophageal cancer syndrome. The American Journal of Human Genetics. 2012 doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blaydon D.C., Biancheri P., Di W.L., Plagnol V., Cabral R.M., Brooke M.A. Inflammatory skin and bowel disease linked to ADAM17 deletion. New England Journal of Medicine. 2011;365:1502–1508. doi: 10.1056/NEJMoa1100721. [DOI] [PubMed] [Google Scholar]
- 70.Monroy A., Kamath S., Chavez A.O., Centonze V.E., Veerasamy M., Barrentine A. Impaired regulation of the TNF-alpha converting enzyme/tissue inhibitor of metalloproteinase 3 proteolytic system in skeletal muscle of obese type 2 diabetic patients: a new mechanism of insulin resistance in humans. Diabetologia. 2009;52:2169–2181. doi: 10.1007/s00125-009-1451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Timmerman K.L., Connors I.D., Deal M.A., Mott R.E. Skeletal muscle TLR4 and TACE are associated with body fat percentage in older adults. Applied Physiology Nutrition and Metabolism. 2016;41:446–451. doi: 10.1139/apnm-2015-0567. [DOI] [PubMed] [Google Scholar]
- 72.Stagakis I., Bertsias G., Karvounaris S., Kavousanaki M., Virla D., Raptopoulou A. Anti-tumor necrosis factor therapy improves insulin resistance, beta cell function and insulin signaling in active rheumatoid arthritis patients with high insulin resistance. Arthritis Research and Therapy. 2012;14:R141. doi: 10.1186/ar3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paschou S.A., Kothonas F., Lafkas A., Myroforidis A., Loi V., Terzi T. Favorable effect of anti-TNF therapy on insulin sensitivity in nonobese, nondiabetic patients with inflammatory bowel disease. International Journal of Endocrinology. 2018;2018 doi: 10.1155/2018/6712901. 6712901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yazdani-Biuki B., Stelzl H., Brezinschek H.P., Hermann J., Mueller T., Krippl P. Improvement of insulin sensitivity in insulin resistant subjects during prolonged treatment with the anti-TNF-alpha antibody infliximab. European Journal of Clinical Investigation. 2004;34:641–642. doi: 10.1111/j.1365-2362.2004.01390.x. [DOI] [PubMed] [Google Scholar]
- 75.Wascher T.C., Lindeman J.H., Sourij H., Kooistra T., Pacini G., Roden M. Chronic TNF-α neutralization does not improve insulin resistance or endothelial function in “healthy” men with metabolic syndrome. Molecular Medicine. 2011;17:189–193. doi: 10.2119/molmed.2010.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murumkar P.R., DasGupta S., Chandani S.R., Giridhar R., Yadav M.R. Novel TACE inhibitors in drug discovery: a review of patented compounds. Expert Opinion on Therapeutic Patents. 2010;20:31–57. doi: 10.1517/13543770903465157. [DOI] [PubMed] [Google Scholar]
- 77.Casaca A., Nóvoa A., Mallo M. Hoxb6 can interfere with somitogenesis in the posterior embryo through a mechanism independent of its rib-promoting activity. Development. 2016;143:437–448. doi: 10.1242/dev.133074. [DOI] [PubMed] [Google Scholar]
- 78.Hogan B., Costantini F., Lacy E. 1986. Manipulating the mouse embryo: a laboratory manual.genesdev.cshlp.org [Google Scholar]
- 79.Rastogi G.K., Letarte J., Fraser T.R. Immunoreactive insulin content of 203 pancreases from foetuses of healthy mothers. Diabetologia. 1970;6:445–446. doi: 10.1007/BF01212080. [DOI] [PubMed] [Google Scholar]
- 80.Martins L., Seoane-Collazo P., Contreras C., González-García I., Martínez-Sánchez N., González F. A functional link between AMPK and orexin mediates the effect of BMP8B on energy balance. Cell Reports. 2016;16:2231–2242. doi: 10.1016/j.celrep.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martínez de Morentin P.B., González-García I., Martins L., Lage R., Fernández-Mallo D., Martínez-Sánchez N. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metabolism. 2014;20:41–53. doi: 10.1016/j.cmet.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martínez-Sánchez N., Seoane-Collazo P., Contreras C., Varela L., Villarroya J., Rial-Pensado E. Hypothalamic AMPK-ER stress-JNK1 Axis mediates the central actions of thyroid hormones on energy balance. Cell Metabolism. 2017;26:212–229. doi: 10.1016/j.cmet.2017.06.014. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nishimura S., Manabe I., Nagasaki M., Hosoya Y., Yamashita H., Fujita H. Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes. 2007;56:1517–1526. doi: 10.2337/db06-1749. [DOI] [PubMed] [Google Scholar]
- 84.Naviaux R.K., Costanzi E., Haas M., Verma I.M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. Journal of Virology. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klein J., Fasshauer M., Ito M., Lowell B.B., Benito M., Kahn C.R. β -adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in Brown adipocytes. Journal of Biological Chemistry. 1999;274:34795–34802. doi: 10.1074/jbc.274.49.34795. [DOI] [PubMed] [Google Scholar]
- 86.Martínez-Sánchez N., Moreno-Navarrete J.M., Contreras C., Rial-Pensado E., Fernø J., Nogueiras R. Thyroid hormones induce browning of white fat. Journal of Endocrinology. 2017;232:351–362. doi: 10.1530/JOE-16-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Contreras C., González-García I., Seoane-Collazo P., Martínez-Sánchez N., Liñares-Pose L., Rial-Pensado E. Reduction of hypothalamic endoplasmic reticulum stress activates browning of white fat and ameliorates obesity. Diabetes. 2017;66:87–99. doi: 10.2337/db15-1547. [DOI] [PubMed] [Google Scholar]
- 88.Alvarez-Crespo M., Csikasz R.I., Martínez-Sánchez N., Diéguez C., Cannon B., Nedergaard J. Essential role of UCP1 modulating the central effects of thyroid hormones on energy balance. Molecular Metabolism. 2016;5:271–282. doi: 10.1016/j.molmet.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 90.Badenes M., Adrain C. iRhom2 and TNF: Partners or enemies? Science Signaling. 29 Oct 2019;12(605):eaaz0444. doi: 10.1126/scisignal.aaz0444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
iRhom2 is expressed in metabolic tissues and increased in BAT during obesity, and its deletion protects mice from this disease. A Protein expression of iRhom2 in subcutaneous (sWAT), brown (BAT), retroperitoneal (Retrop.), mesenteric (Mesent.), and epididymal (eWAT) adipose tissues, pancreas, liver, and muscle from homozygous (iR2-FLAG) mice with an N-terminal FLAG tag incorporated into the endogenous iRhom2 gene. B Protein expression of iRhom2 in BAT from iR2-FLAG mice fed with SD versus HFD. In A, B We used iRhom2 WT BAT as a negative control. As a loading control we measured GAPDH protein level. Two experiments were performed. C RT-PCR analysis of Rhbdf2 (iRhom2) mRNA, normalized to a housekeeping control (beta Actin), in brown adipocytes differentiated in vitro from the stromal vascular fraction of 4-5 pooled WT or iRhom2 KO mice. The WT cells were treated for 8 h with TNF (10 ng/ml) or with an anti-TNF antibody (1 μg/ml) and compared to nontreated cells. The lack of iRhom2 gene expression in the iRhom2 KO brown adipocytes provides a specificity control. Three independent experiments. D Body weight over time of iRhom2 KO and WT mice fed with HFD from weeks 4 to 34 of age. In panels C,D error bars represent SEM; * represents p<0.05, **** represents p<0.0001. E-F Representative photographs of iRhom2 KO and WT fed with SD and HFD for 30 weeks.
HFD-fed iRhom2 KO mice do not exhibit significantly reduced accumulation of fat in the skeletal musculature. A-B Histopathological classification of skeletal muscle fat content (scores 0-2) (A), and representative photographs of skeletal muscle Oil red staining (100x magnification) (B) of iRhom2 KO and WT fed with HFD for 30 weeks. Two experiments with 5-6 mice per group. Error bars represent SEM; Scale bar = 100 μm.
Adipose tissues from HFD-fed iRhom2 KO mice have an increased frequency of smaller adipocytes. A Representative photographs of eWAT and sWAT from iRhom2 KO and WT mice fed with HFD for 30 weeks. B-F Histograms showing the adipocyte area frequency (%) from subcutaneous (sWAT) (B), mesenteric (C), epididymal (eWAT) (D), reptroperitoneal (E), and brown adipose tissue (BAT) (F) of iRhom2 KO and WT mice fed with HFD for 30 weeks. Three experiments with 3 mice per genotype. Error bars represent SEM.
Gating strategy used in the FACS analysis. A-E Cells were first gated on size (SS vs FS) (A) and then on singularity (Pulse Width vs FS) (B). This was followed by DAPI exclusion to gate for live cells (SS vs FL6) (C). Live cells were gated on expression of CD45.2 to select the myeloid cells (FL3 vs FL2) (D). Finally, myeloid cells were gated for co-expression of CD11b and F4/80 to identify the macrophages (FL8 vs FL1) (E).
HFD but not SD fed iRhom2 KO mice have increased cumulative energy expenditure. A-D Energy expenditure (EE) over time (A, B) and cumulative EE (C, D) of iRhom2 KO and WT mice fed with HFD (A, C) and SD (B, D) for 17 weeks. Results are representative of one experiment, with a total of n=8 and n=6 for SD and HFD experiments, respectively. Error bars represent SEM; * represents p<0.05.
iRhom2 loss leads to upregulation of thermogenic genes, whereas ambient temperature influences iRhom2 expression levels. A RT-PCR analysis of UCP1, PGC1a, Cidea, PRDM16, and Cox8b expression in RNA isolated from BAT samples isolated from WT versus iRhom2 KO mice fed on SD or HFD for 2 weeks. Two experiments with 3-4 replicates in each. B RT-PCR analysis of Rhbdf2 expression in the BAT of WT mice subjected to the following temperature conditions, relative to thermoneutrality (housed at 30°C for 4 weeks): (1) Mice housed at 21°C for the time of the study, (2) mice exposed to 5°C for 1 week, (3) mice acclimated for 4 weeks at 5°C, (4) mice acclimated for 4 weeks at 5°C and then exposed to 30°C for 1 week, (5) mice acclimated for 4 weeks at 5°C and then acclimated for 4 weeks at 30°C. One experiment with 4 independent samples per condition. C-H Basal (C) and maximal respiration (D), ATP production (E), coupling efficiency (F), spare respiratory capacity (G), and non-mitochondrial oxygen consumption (H) of brown adipocytes differentiated in vitro from the stromal vascular fraction of 4-5 pooled WT and iRhom2 KO mice and stimulated or not with norepinephrine. The results were normalized to the protein content. Two experiments with one or two independent samples per genotype, respectively. Error bars represent SEM; * represents p<0.05, ** represents p<0.01, *** represents p<0.001, **** represents p<0.0001.