Abstract
Neem is a perennial plant of family Meliaceae grown very commonly in India. During a survey in Rajasthan, India; a population of root-knot nematode was found in association with tender neem plants causing yellowing, stunting, and heavy root galling. Inspection of the perineal pattern morphology of the adult females, extracted from the galled roots, primarily led to identification of the species as Meloidogyne indica. Further, detailed morphological and morphometric illustrations of second-stage juveniles, males and females were carried out by light compound and scanning electron microscopy. Gross morphology and measurements were found consistent with the original description of M. indica infecting citrus by Whitehead (1968). The neem population was found to infect and reproduce on citrus. Additionally, evolutionary relationship was deduced by Maximum likelihood method using ITS rRNA, D2D3 expansion segment of 28S rRNA and mitochondrial COI sequences. Phylogenetic analyses based on these sequences showed sufficient divergence of M. indica to be differentiated as a unique species under the genus Meloidogyne.
Keywords: COI, D2D3, ITS, Meloidogyne indica, Neem
Neem or margosa or Indian lilac (Azadirachta indica A. Juss), a perennial plant of family Meliaceae, is well known for containing bioactive compounds against several insects and plant-parasitic nematodes (Kraus, 1995). Azadirachtin, the main chemical component responsible for the toxic effect has nematicidal potential with respect to reduced fecundity and hatching of various plant-parasitic nematodes (Alam, 1993). The final population density of root-knot nematodes (RKN) has been found to decline with the application of neem seed kernel extract as a soil drench, and use of neem cake as a soil amendment etc. (Riga and Lazarovits, 2001; Kumar and Khanna, 2006). In spite of the widespread use of neem-based botanicals in plant protection, it is still not free from disease and pest attack. In the Indian subcontinent, it is known to be infested by different insects and fungi (Beeson, 1953; Tewari, 1992; Cornezo, 1999), but any nematode species attacking neem is largely unknown to date.
RKN (Meloidogyne spp.) are the most economically important group of obligate sedentary plant-parasitic species distributed worldwide encompassing more than 3,000 plant hosts (Jones et al., 2013). Genus Meloidogyne is known to include more than 90 nominal species to date (Karssen, 2002; Karssen and Moens, 2006). The use of DNA-based diagnostics in combination with morphological and morphometric analyses aid in authoritative identification of the RKN species and also help in resolving phylogenetic relationships (Blok and Powers, 2009). Molecular markers that have proved useful in these studies include mitochondrial gene cytochrome c oxidase subunit I (COI) (Blok et al., 2002; Hebert et al., 2004; Xu et al., 2004), the small subunit 18S rRNA gene (De Ley et al., 2002; Tenente et al., 2004; Tigano et al., 2005), D2D3 expansion segment of the large subunit 28S rRNA (Chen et al., 2003; Palomares-Ruis et al., 2007) and ribosomal internal transcribed spacer (ITS) 1 and 2 (Powers and Harris, 1993; Powers et al., 1997). All these markers have proven useful for identification of Meloidogyne spp. (Castillo et al., 2003; Tigano et al., 2005; Powers et al., 2005).
During a survey in Jaipur district of Rajasthan, India a species of Meloidogyne was found in association with tender neem plants. The nematode species was initially considered to be M. indica (Whitehead, 1968) based on the perineal pattern morphology, when compared with the original species description (Whitehead, 1968). Accordingly, detailed morphological and morphometric illustrations were carried out for expanded and robust characterization of the species. Additionally information on molecular markers, viz., ITS rDNA, and D2D3 expansion segment of 28S rRNA gene and mitochondrial COI were also generated for the species that were not presented in its earlier description.
Materials and methods
Nematode population
Tender neem plants showing the symptoms of stunting and yellowing were collected along with the rhizosphere soil from Jaipur district, Rajasthan, India; located at N26°55′10″ and E75°47′16″. The soil type was found to be sandy to sandy loam with medium phosphorus and medium to high potassium content. Average annual rainfall in the area ranges from 500 to 600 mm (Anonymous, 1995). For diagnosis and identification purposes, females were directly dissected out of the galled roots for preparing perineal patterns and genomic DNA extraction. Eggmasses were hand-picked from the infected neem roots and were used for hatching the second-stage infective juveniles. Both second-stage juveniles (J2s) and males were collected by the modified Baermann’s technique (Whitehead and Hemming, 1965).
Morphological studies
For conducting taxonomic studies, the J2s and males were processed by the glycerol–ethanol method following standard procedures (Seinhorst, 1959). The morphological characters used for taxonomic identification were employed as described by Karssen (2002) and Carneiro et al. (2014). Morphometric illustrations and measurements (in μm) were made with a camera lucida and an ocular micrometer within an Olympus BX50 compound microscope. The posterior portion with cuticular markings surrounding the vulva and anus of stained-fixed females was cut using a fine-pointed scalpel to obtain the perineal pattern. The inner tissue was carefully removed using flexible bristle, trimmed and transferred to a drop of anhydrous glycerol on a clean glass slide (Southey, 1986) to study the perineal pattern morphology. Photomicrographs of perineal pattern, females, males, and J2s were taken with a Zeiss Axiocam M2m compound microscope equipped with differential interference contrast optics.
Scanning electron microscopy (SEM) was done using a JEOL (UK) JSM 6700 FEG scanning electron microscope equipped with a GATAN (UK) Alto 2500 Cryo unit. The instrument was prepared by cooling with liquid nitrogen and the temperature in the microscope chamber was maintained at −160°C throughout sample examination. The nematode specimens were pipetted onto a 595-grade WhatmanTM filter paper attached to a cryo stub with OCT mountant (Sakura Finetek, Europe NL) and frozen by plunging into pre-slushed liquid nitrogen. The sample was then transferred to the GATAN cryo chamber stage under vacuum and etched to remove contaminating ice by increasing the temperature to −95°C for 1 min. Once the temperature of the stage had returned to −160°C, the sample was coated with Au/Pd alloy for 1 min and transferred to the SEM chamber and mounted on the microscope stage for examination at −160°C. Images were recorded using the JEOL on board system and software.
Molecular characterization
For molecular analysis, single adult female was lysed in worm lysis buffer (Subbotin et al., 2000) and the molecular markers, viz., ITS, D2D3, and COI were PCR amplified using the primers listed in Table 1. The reaction condition for amplification of the ITS gene was followed as described in Subbotin et al. (2001, 2006). For D2D3 and COI genes, the thermal cycling program was used as described in Ye et al. (2007). Amplified products were separated by electrophoresis on 1% agarose gel and purified using QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). The gel purified PCR products were cloned into pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into E. coli DH5α competent cells (New England Biolabs). Recombinant plasmids were isolated from the positive clones (QIAGEN Plasmid Miniprep kit (Qiagen, Valencia, CA, USA)) and sequenced. The raw sequences were checked for quality using MEGA6 (Tamura et al., 2013), aligned, and a consensus sequence was generated for each gene using Bioedit software (Tom Hall; Ibis Biosciences, Carlsbad, CA).
Table 1.
List of primers used for polymerase chain reaction amplification in this study.
| Primer name | Gene | Sequence | References |
| V5367 | ITS | 5′-TTGATTACGTCCCTGCCCTTT-3′ | Vrain et al. (1992) |
| 26S | ITS | 5′-TTTCACTCGCCGTTACTAAGG-3′ | Vrain et al. (1992) |
| D2A | LSU | 5′-ACAAGTACCGTGAGGGAAAGTTG-3′ | Castillo et al. (2003) |
| D3B | LSU | 5′-TCGGAAGGAACCAGCTACTA-3′ | Castillo et al. (2003) |
| JB3 | COI | 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ | Bowles et al. (1992) |
| JB5 | COI | 5′-AGCACCTAAACTTAAAACATAATGAAAATG-3′ | Derycke et al. (2005) |
The obtained ITS, D2D3, and COI sequences were compared with the available sequences in the NCBI GenBank database using BLAST search. Corresponding published sequences (of Meloidogyne spp.) for each gene were retrieved (Table 2) and phylogenetic analyses were carried out using MEGA6 software (Tamura et al., 2013). The sequences were aligned using ClustalW and evolutionary history was deduced by the maximum likelihood (ML) method with selection of appropriate model using Modeltest (Posada and Crandall, 1998). The phylograms were bootstrapped 1,000 times (Felsenstein, 1985) to assess the degree of support for the phylogenetic branching as indicated in the consensus tree. Pratylenchus vulnus was used as out-group.
Table 2.
List of GenBank accession numbers used in phylogenetic analyses (** Not found in NCBI database).
| Species | ITS rRNA | D2D3 28S rRNA | COI mtDNA |
|---|---|---|---|
| Meloidogyne arabicida | ** | KF993624 | ** |
| Meloidogyne africana | ** | ** | KY433441 |
| Meloidogyne arenaria | AF387092 | JX987332 | JX683705 |
| Meloidogyne artiellia | KC545880 | AY150369 | KU517173 |
| Meloidogyne baetica | AY150366 | AY150367 | ** |
| Meloidogyne camelliae | JX912885 | KF542869 | KM887148 |
| Meloidogyne chitwoodi | AY281852 | AF435802 | KU517168 |
| Meloidogyne christiei | KR082319 | KR082317 | ** |
| Meloidogyne dunensis | EF612711 | EF612712 | ** |
| Meloidogyne duytsi | ** | ** | KU517177 |
| Meloidogyne enterolobii | KM046989 | KJ146862 | KT936633 |
| Meloidogyne ethiopica | KF482366 | KF482372 | ** |
| Meloidogyne exigua | ** | AF435795 | ** |
| Meloidogyne fallax | AY281853 | KC241969 | KU517182 |
| Meloidogyne graminicola | KM111531 | KJ728847 | KY250093 |
| Meloidogyne graminis | JN157866 | JN019326 | ** |
| Meloidogyne hapla | EU908052 | DQ145641 | JX683719 |
| Meloidogyne haplanaria | ** | ** | KU174206 |
| Meloidogyne hispanica | EU443613 | EU443607 | JX683712 |
| Meloidogyne ichinohei | ** | EF029862 | KY433448 |
| Meloidogyne incognita | KJ739707 | JX100425 | JX683696 |
| Meloidogyne indica | KC311146 | MF680038 | MF662179 |
| Meloidogyne inornata | KF482368 | KF482374 | ** |
| Meloidogyne izalcoensis | ** | KF993621 | ** |
| Meloidogyne javanica | KJ739709 | KC953092 | JX683711 |
| Meloidogyne konaensis | ** | AF435797 | ** |
| Meloidogyne lopezi | ** | KF993616 | ** |
| Meloidogyne luci | KF482365 | KF482371 | ** |
| Meloidogyne mali | JX978228 | KF880398 | KU517175 |
| Meloidogyne marylandi | JN157854 | JN019333 | |
| Meloidogyne minor | KC241953 | JN628436 | KU517178 |
| Meloidogyne naasi | KJ934132 | KC241979 | KU517170 |
| Meloidogyne panyuensis | ** | ** | ** |
| Meloidogyne paranaensis | ** | AF435799 | ** |
| Meloidogyne silvestris | EU570216 | EU570214 | ** |
| Meloidogyne spartelensis | KP896294 | KP895293 | KP997301 |
| Meloidogyne thailandica | AY858795 | EU364890 | ** |
| Meloidogyne trifoliophila | JX465593 | AF435801 | ** |
| Pratylenchus vulnus | FJ713011 | EU130885 | KX349427 |
| Hirschmanniella oryzae | DQ309588 | JX291142 | ** |
| Tylenchorhynchus leviterminalis | EF030984 | KJ475548 | ** |
| Heterodera glycines | HM370421 | GU595446 | ** |
| Radopholus similis | KJ845638 | JN091964 | KX349430 |
| Heterotheca mucronata | ** | ** | KR819278 |
| Tylenchorhynchus sp | ** | ** | KY639376 |
The sequences were deposited in the NCBI GenBank with the accessions KC311146, MF680038, and MF662179 for ITS, D2D3, and COI, respectively.
Inoculation of neem population on citrus
Inoculation assay was conducted in a soil system to test whether the nematode species infecting neem can parasitize and multiply on citrus. One-month-old citrus seedlings were planted in plastic pouches (height: 6 inch, diameter: 4 inch) filled with sterilized soil, and inoculated with 2 J2s/g soil. Freshly hatched J2s from the eggmasses, collected from neem roots, were used for inoculation. The experiment was conducted with five replicates.
Results
Plant symptoms and characterization of species
The infected neem seedlings showed the symptoms of yellowing of leaves coupled with stunted growth of the immature plants. Examination of the infected plants revealed the presence of heavy galling on the roots located mostly along the tap root axis with few lateral branches (Fig. 1A–C). For characterization and identification of the species, morphology of adult females, males and J2s was studied (Fig. 1D–K) including SEM photomicrographs (Fig. 2) with minute detailing.
Fig. 1.
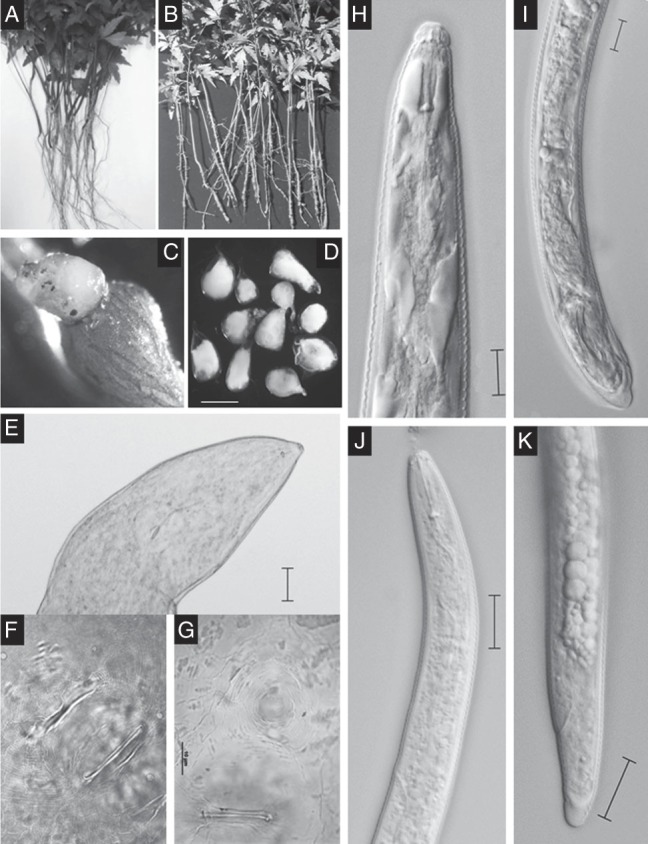
Plant symptoms (A–C) and morphology (D–K) of Meloidogyne indica infecting neem. A, Healthy neem seedlings; B, Infected neem seedlings devoid of lateral roots; C, Root gall with eggmass; D, Adult females; E, Anterior region of adult female; F and G, Perineal pattern morphology; H, Anterior region of male; I, Male tail; J, Anterior end of second-stage juvenile (J2); K, Second-stage juvenile tail (scale bar = D: 550 µm; E: 100 µm; H,I: 10 µm; J,K: 20 µm).
Fig. 2.
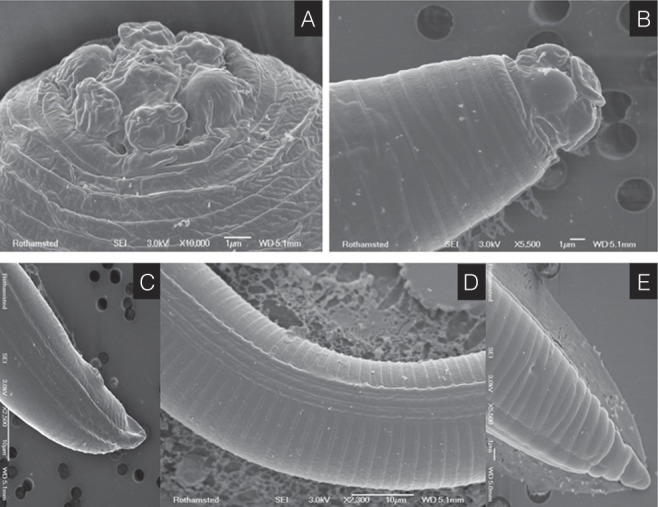
Scanning electron microscopy photomicrographs of Meloidogyne indica. A, Female lip region; B, Male lip region; C, Male tail; D, Lateral field with lateral lines; E, Second-stage juvenile tail (scale bar in µm).
The nematode species infecting neem was found to infect the citrus plants. The citrus roots showed the presence of galls with formation of eggmasses when observed at 35 days post inoculation (Fig. 3). The females, dissected out from the citrus roots, showed similar perineal pattern morphology with that of the neem population (data not shown).
Fig. 3.
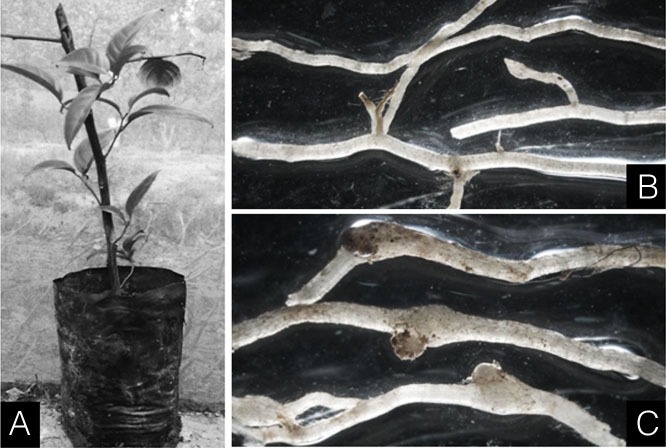
Citrus roots showing development of galls and eggmasses upon inoculation with the neem population of Meloidogyne indica. A, Citrus plant inoculated with neem population of M. indica; B, Healthy roots of citrus; C, Infected roots of citrus with galls and eggmasses.
General morphology
Female
Females are completely enclosed within root tissue. The body appeared translucent white, pyroid shaped, and variable in size. Neck prominent and short, bent at various angles. Body annuli smaller in anterior neck region. En face view with six prominent lips and stylet opening pore-like. Stylet small and strong, knobs well developed and ovoid. Position of Dorsal oesophageal gland orifice (DGO) is 2.90 ± 0.39 μm from stylet knob. Median bulb well developed, almost rounded. Vulva nearly terminal. The perineal cuticular pattern was rounded with low and rounded dorsal arch and devoid of lateral lines or any forking. Typically it is formed of very fine, closely spaced striae of the cuticle, forming a distinct tail whorl. Phasmid and anus were distinct. Commonly egg sac occurs outside the root.
Male
The body is vermiform, tapering anteriorly. Body annuli large and distinct. In lateral view, labial cap high and rounded. Lip annuli two behind the head cap. Labial disc rounded. Cephalic framework moderately developed. Stylet robust and thin with cone pointed, shaft cylindrical and knobs directed posteriorly. DGO 3.11 ± 0.17 μm from the stylet knob. Amphids very distinct. Lateral field with four lateral lines. Testes out-stretched. Tail is short conoid with narrow terminus and smooth end. Spicule arcuate, gubernaculum distinct.
Second-stage juvenile
The body is slender, vermiform, and finely annulated. Lip region not set off from the body. Cephalic framework very weakly developed, amphids distinct. Stylet relatively long, delicate with pointed conus and rounded knobs. DGO 2.86 ± 0.23 μm posterior to stylet knob. Metacorpus oval to rounded. Oesophageal gland lobe overlapping intestine ventrally. Excretory pore distinct, at the level of isthmus. Lateral field with four incisures running almost entire length of body and ending near hyaline tail terminus. Tail length short with very small hyaline portion, terminal blunt, and unstriated.
The morphometric measurements of females, males, and J2s for the present population of RKN species are presented in Table 3.
Table 3.
Morphometrics for Meloidogyne indica infecting neem and citrus. All linear measurements are in micrometer and in the form of mean ± SD.
| Neem population | Citrus population (After Whitehead, 1968) | ||||
|---|---|---|---|---|---|
| Character | J2 | Male | Female | J2 | Female |
| n | 20 | 15 | 30 | 25 | 8 |
| L | 484 ± 31.5 (430–520) | 1253 ± 80 (1180–1380) | 653 ± 92.2 (450–790) | 414 ± 4.5 (381–448) | – |
| Body width | 18 ± 1.5 (16.77–21.15) | 28 ± 4 (24.55–34.66) | 408 ± 75 (325–550) | – | – |
| A | 26.68 ± 1.9 (24.20–29.87) | 44.89 ± 3.5 (39.81–48.06) | 1.60 ± 0.3 (1.38–2.10) | – | – |
| Stylet length | 13.8 ± 0.1 (13.57–14.21) | 16.3 ± 0.4 (15.90–17.08) | 13.7 ± 0.4 (13.32–14.18) | 12 ± 0.9 (10–14) | 14 (12–16) |
| DGO | 2.8 ± 0.2 (2.45–3.25) | 3.1 ± 0.1 (2.92–3.30) | 2.9 ± 0.3 (2.49–3.67) | – | 3 (2–4) |
| Head-metacorpus | 50 ± 2.3 (46.45–53.02) | 73 ± 4.1 (68.70–78.14) | – | – | – |
| Head-oesophageal gland | 138 ± 4.8 (129.97–144.65) | – | – | – | – |
| b′ | 3.5 ± 0.1 (3.10–3.65) | – | – | – | – |
| c | 26.2 ± 1.2 (24.15–27.65) | – | – | 24.9 ± 1.36 (21.2–31) | – |
| c′ | 1.6 ± 0.1 (1.52–1.91) | – | – | 1.57 ± 0.012 (1.06–1.78) | – |
| Tail length | 18 ± 0.6 (17.50–19.50) | – | – | 16.8 ± 1.88 (13–20.1) | – |
| Anal body width | 11.1 ± 1.0 (9.85–12.45) | – | – | – | – |
| Spicule | – | 26 ± 0.6 (25.90–27.50) | – | – | – |
Differential diagnosis of the RKN species infecting neem showed maximum similarity with M. indica, originally described by Whitehead (1968) from New Delhi, India infecting Citrus aurantifolia. Minor variations were observed in the general morphology and morphometrics. The measurements originally taken by Whitehead (1968) for description of M. indica are also presented in Table 3 for comparison.
Molecular characterization
Phylogenetic analysis of the ITS rRNA and D2D3 fragment of 28S rRNA sequence revealed that the sequences were highly divergent from other Meloidogyne species described to date. Phylogenetic relationship as inferred from the ML method using ITS rRNA sequences (Fig. 4) showed that the species holds a basal position in the tree without clustering with other described species of Meloidogyne. The closest sequence-related neighbors were found to be M. panyuensis, M. camelliae, M. artiellia, M. mali and M. baetica with high bootstrap support. The ML tree based on the D2D3 expansion segment of the 28S rRNA gene (Fig. 5) showed that the species branched separately being close to the out-group taxon P. vulnus. Phylogenetic analysis of the mitochondrial COI sequence (Fig. 6) showed that the species branched out separately without clustering with any Meloidogyne species, having sequence similarity to M. artiellia. The dataset gave similar results across all tree reconstruction algorithms. Thus substantial sequence divergence for all the three genes tested distinguishes the species from other studied RKN.
Fig. 4.
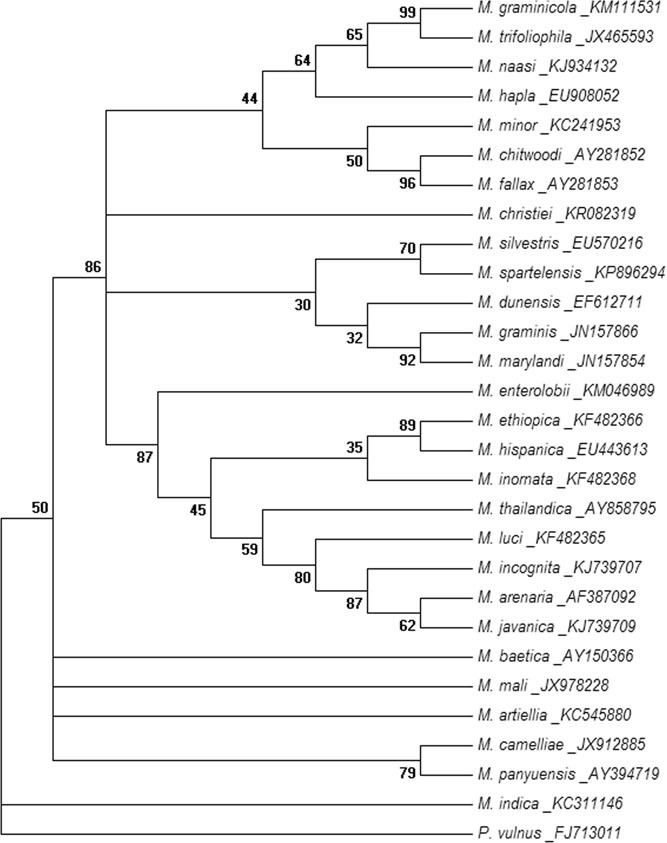
Evolutionary relationship of Meloidogyne indica using ITS rRNA sequence. The evolutionary history was inferred by using the maximum likelihood method based on Kimura 2-parameter model. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the analyzed taxa. Branches corresponding to partitions reproduced in less than 30% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value (–1869.0466). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 1.0929]). The analysis involved 29 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 170 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
Fig. 5.
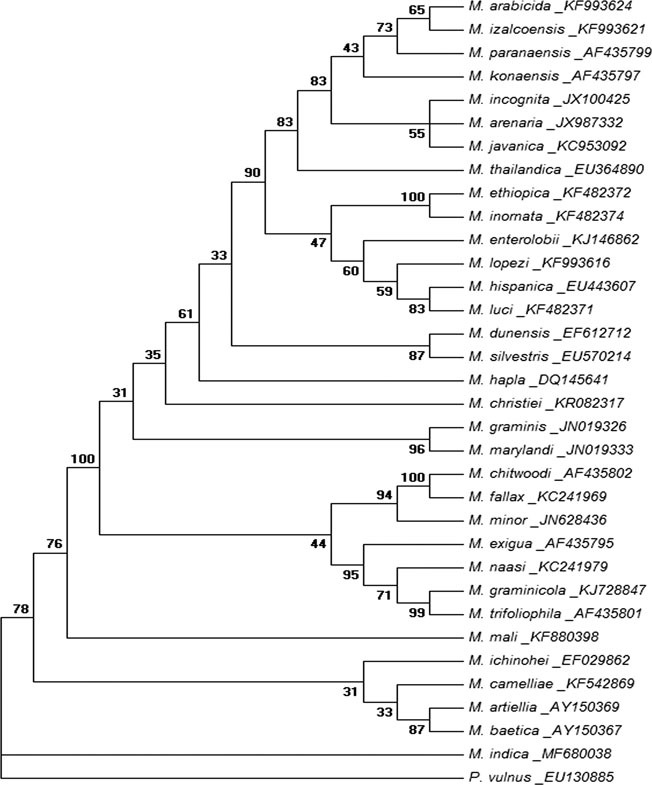
Evolutionary relationship of Meloidogyne indica using D2D3 expansion segment of 28S rRNA sequence. The evolutionary history was inferred by using the maximum likelihood method based on General Time Reversible model. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the analyzed taxa. Branches corresponding to partitions reproduced in less than 30% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach and then selecting the topology with superior log likelihood value (−4112.4125). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 0.4693]). The analysis involved 34 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 525 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
Fig. 6.
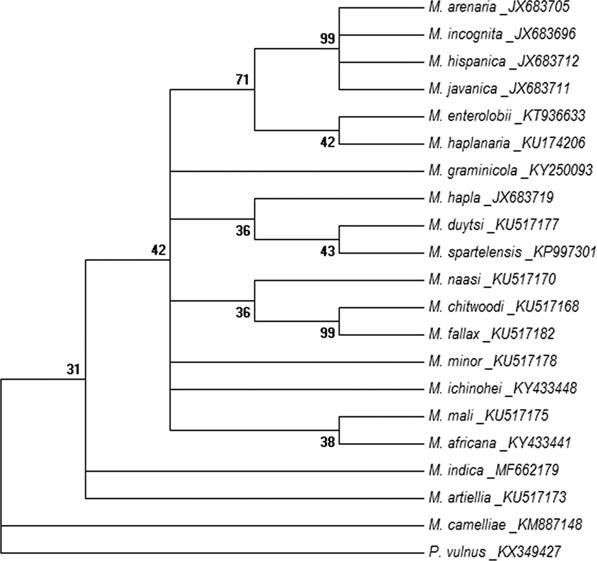
Evolutionary relationship of Meloidogyne indica using mitochondrial COI sequences. The evolutionary history was inferred by using the maximum likelihood method based on General Time Reversible model. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the analyzed taxa. Branches corresponding to partitions reproduced in less than 30% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value (−2471.1920). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 0.5728]). The analysis involved 21 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 309 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
Discussion
The present investigation includes identification and characterization of the RKN species parasitizing neem as M. indica (Whitehead, 1968) from Rajasthan, India. Meloidogyne indica was first described by Whitehead (1968) infecting Citrus aurantifolia from New Delhi, India. Whitehead (1968) described the species based on a limited number of specimens; and adequate morphometric data or any molecular phylogenetic analyses were neither presented in the original description nor any other publication to date. Hence, the present study provides additional and useful information on morphology, mophometrics, and molecular phylogeny of the species in detail. The morphometric illustrations are consistent with the description of Whitehead (1968) with slight deviations. The body length (483.70 ± 31.52 μm vs. 414 ± 4.5 µm) and tail length (18.41 ± 0.67 μm vs. 16.8 ± 1.88 µm) of the J2s of the neem population were found to be relatively larger than that of the citrus population. However, these differences may be considered as intra-specific variation for the species. Additional measurements for J2s include length of DGO; lip region-metacorpus length; length of glandular overlapping; a- and b′ ratio that were not presented in the earlier description. The J2s showed distinctiveness in the gross morphology of anterior and posterior ends, tail shape, and tail length typical for the species. The perineal pattern is characteristic for the species as described earlier. Morphometrics of males were not studied earlier and therefore the data generated in terms of body length, body width, stylet length, length of DGO, spicule length, and a- ratio could not be compared. Additionally, supplementary morphology has also been generated for males in this study. The scanning electron microscope photomicrographs showed minute morphological details of males, females, and J2s.
For molecular characterization, three molecular markers, viz., ITS rRNA, D2D3 expansion segment of 28S rRNA, and mitochondrial COI genes were PCR amplified and sequenced. Independent ML phylogenetic analyses for each of the three marker sequences of M. indica revealed sufficient divergence of this species demonstrating its novelty as a unique species under the genus Meloidogyne. Within evolutionary trees M. indica was a basal taxon relative to other Meloidogyne species when Pratylenchus was an out-group. This can probably establish the primitiveness of the species among the other RKN as they are presumably thought to arise from a Pratylenchus-like ancestor (Quist et al., 2015). Whitehead’s grouping of Meloidogyne spp. based on the tail shape of J2s and the number of head annuli of males, reflected the similarity of M. indica with M. artiellia in gross morphology (Whitehead, 1968). The similarity between these two species has also been validated in the present phylogenetic analyses based on ribosomal and mitochondrial genes.
So far M. indica has been documented from different species of citrus and Bt-cotton (Whitehead, 1968; Franklin, 1978; Patel et al., 1999; Vovlas and Inserra, 2000; Davis and Venette, 2004; Khan et al., 2017). In the present study the species was found to infect neem plants demonstrating the ability of the species to parasitize perennial hosts. The cross-inoculation of the neem population of M. indica to citrus plants revealed that it could successfully reproduce on citrus. Most Meloidogyne spp. are polyphagous and highly adapted towards their hosts. The ability of M. indica to infect neem may be a result of its co-evolution allowing the nematode to withstand the toxic compounds present in the neem plant. Detoxification of plant toxins is reported amongst insects where they can successfully use some enzymes for their own benefit (Dowd et al., 1983; Mello and Silva-Filho, 2002; Jeschke et al., 2016). Digestive enzymes in insects, viz., alpha-amylases, esterases and glutathione S-transferase were found to detoxify plant defence chemicals (Senthil-Nathan, 2013) and similar mechanism may be involved in the parasitism of M. indica on neem. One such example is Meloidogyne incognita infecting and multiplying on some Brassica spp., plants with nematicidal potential (Shivakumara et al., 2016).
Acknowledgments
This work was partly supported by a project looking at ‘Multitrophic Interactions in the Rhizosphere’ as part of the United Kingdom India Education and Research Initiative (UKIERI) funded by the British Council. The authors gratefully acknowledge Mrs. Kirstie Halsey, Rothamsted Bio-Imaging, Harpenden, UK and Mr. Purshotam, Division of Nematology, ICAR-IARI, New Delhi for their substantial help.
Literature Cited
- Alam M. M. 1993. Bioactivity against phytonematodes in Randhawa N.S., and Parmar B.S. (Eds), Neem Research and Development, New Delhi, Society of Pesticide Science, India, 123-143. [Google Scholar]
- Anonymous 1995. Soils of Rajasthan for optimizing land use (b: Executive summery) by National Bureau of Soil Survey and Land Use Planning, India http://krishikosh.egranth.ac.in/bitstream/1/2034146/1/94.pdf.
- Beeson C.F.C. 1953. The ecology and control of the forest insects of India and the neighboring countries, 2nd ed., Government of India Press, New Delhi. [Google Scholar]
- Blok V.C., and Powers T.O.. 2009. Biochemical and molecular identification, in Perry R.N., Moens M., and Starr J. (Eds), Root-Knot Nematodes, CAB International, UK, 98-112. [Google Scholar]
- Blok V.C., Wishart J., Fargette M., Berthier K., and Phillips M.S.. 2002. Mitochondrial DNA differences distinguishing Meloidogyne mayaguensis from the major species of tropical root-knot nematodes. Nematology 4: 773-781. [Google Scholar]
- Bowles J., Blair D., and McManus D.P.. 1992. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Molecular and Biochemical Parasitology 54: 165-174. [DOI] [PubMed] [Google Scholar]
- Carneiro R.M.D.G., Correa V.R., Almeida M.R.A., Gomes A.C.M.M., Deimi A.M., Castagnone-Sereno P., and Karssen G.. 2014. Meloidogyne luci n. sp. (Nematoda: Meloidogynidae), a root-knot nematode parasitising different crops in Brazil, Chile and Iran. Nematology 16: 289-301. [Google Scholar]
- Castillo P., Vovlas N., Subbotin S., and Troccoli A.. 2003. New root-knot nematode, Meloidogyne baetica n. sp. (Nematoda: Heteroderidae), parasitizing wild olive in southern Spain. Nematology 93: 1093-1102. [DOI] [PubMed] [Google Scholar]
- Chen P., Roberts P.A., Metcalf A.E., and Hyman B.C.. 2003. Nucleotide substitution patterning within the Meloidogyne rDNA D3 region and its evolutionary implications. Journal of Nematology 35: 404-410. [PMC free article] [PubMed] [Google Scholar]
- Cornezo A.T. 1999. Potentially destructive diseases of neem seedlings. Canopy International 25: 3. [Google Scholar]
- Davis E.E., and Venette R.C.. 2004. Mini risk assessment: Asian Citrus root knot nematodes, Meloidogyne citri Zhang, Gao and Weng; M. donghaiensis Zheng, Lin and Zheng; M. fujianenesis Pan; M. indica Whitehead; M. jianyangensis Yang, Hu, Chen and Zhu; M. kongi Yang, Wang and Feng; and M. mingnanica Zhang [Nematoda: Meloidogynidae]. Cooperative Agricultural Pest Survey, Animal and Plant Health Inspection Service, US Department of Agriculture.
- De Ley T.I., De Ley P., Vierstraete A., Karssen G., Moens M., and Vanfleteren J.. 2002. Phylogenetic analyses of Meloidogyne SSU rDNA. Journal of Nematology 34: 319-331. [PMC free article] [PubMed] [Google Scholar]
- Derycke S., Remerie T., Vierstraete A., Backeljau T., Van- fleteren J., Vincx M., and Moens T.. 2005. Mitochondrial DNA variation and cryptic speciation within the freeliving marine nematode Pellioditis marina. Marine Ecology Progress Series 300: 91-103. [Google Scholar]
- Dowd P.F., Smith C.M., and Sparks T.C.. 1983. Detoxification of plant toxins by insects. Insect Biochemistry 13: 453-468. [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791. [DOI] [PubMed] [Google Scholar]
- Franklin M.T. 1978. Meloidogyne, in Southey J.F. (Eds), Plant Nematology, Ministry of Agriculture Fisheries and Food; Her Majesty’s Stationery Office (HMSO), London, 98-124. [Google Scholar]
- Hebert P.D.N., Stoeckle M.Y., Zemlak T.S., and Francis C.M.. 2004. Identification of birds through DNA Barcodes. PLOS Biology 2: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke V., Gershenzon J., and Vassão D.G.. 2016. Insect detoxification of glucosinolates and their hydrolysis products. Advances in Botanical Research 80: 199-245. [Google Scholar]
- Jones J.T., Haegeman A., Danchin E.G.J., Gaur H.S., Helder J., Jones M.G.K., Kikuchi T., Manzanilla-López R., Palomares-Rius J.E., Wesemael W.M., and Perry R.N.. 2013. Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology 14: 946-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen G. 2002. The plant-Parasitic Nematode Genus Meloidogyne Göldi, 1892 (Tylenchida) in Europe, Brill, The Netherlands. [Google Scholar]
- Karssen G., and Moens M.. 2006. Root-knot nematodes, in Perry R.N., and Moens M. (Eds), Plant Nematology, CAB International, UK, 59-90. [Google Scholar]
- Khan M.R., Pal S., Patel A.D. Patel B.A., Ghule T.M., Singh A., and Phani V.. 2018. Further observation on Meloidogyne indica Whitehead, 1968 from India. Pakistan Journal of Zoology. [IN PRESS].
- Kraus W. 1995. Biologically active ingredients: azadirachtin and other triterpenoids in Schmutterer H., and Weinheim V.C.H. (Eds), The neem tree, Azadirachta indica A. Juss., and other meliaceous plants: Sources of unique natural products for integrated pest management, medicine, industry and other purposes, VCH, Germany, 35-74. [Google Scholar]
- Kumar S., and Khanna A.S.. 2006. Effect of neem-based products on the root-knot nematode, Meloidogyne incognita, and growth of tomato. Nematologia Mediterranea 34: 141-146. [Google Scholar]
- Mello M.O., and Silva-Filho M.C.. 2002. Plant-insect interactions: An evolutionary arms race between two distinct defense mechanisms. Brazilian Journal of Plant Physiology 14: 71-81. [Google Scholar]
- Palomares-Ruis J.E., Vovlas N., Troccoli A., LieBanas G., Landa B.B., and Castillo P.. 2007. A new root-knot nematode parasitizing sea rocket from Spanish Mediterranean coastal sand dunes: Meloidogyne dunesis n. sp. (Nematoda: Meloidogynidae). Journal of Nematology 39: 190-202. [PMC free article] [PubMed] [Google Scholar]
- Patel D.J., Patel B.A., Patel S.K., Patel R.L., and Patel R.G.. 1999. Root knot nematode, Meloidogyne indica on kagzi lime in north Gujarat. Indian Journal of Nematology 29: 197. [Google Scholar]
- Posada D., and Crandall K.A.. 1998. Modeltest: Testing the model of DNA substitution. Bioinformatics 14: 817-818. [DOI] [PubMed] [Google Scholar]
- Powers T.O., and Harris T.S.. 1993. A polymerase chain reaction method for identification of five major Meloidogyne species. Journal of Nematology 25: 1-6. [PMC free article] [PubMed] [Google Scholar]
- Powers T.O., Mullin P.G., Harris T.S., Sutton L.A., and Higgins R.S.. 2005. Incorporating molecular identification of Meloidogyne spp. into a large-scale regional nematode survey. Journal of Nematology 37: 226-235. [PMC free article] [PubMed] [Google Scholar]
- Powers T.O., Todd T.C., Burnell A.M., Murray P.C.B., Fleming C.C., Szalanski A.L., Adams B.J., and Harris T.S.. 1997. The rDNA internal transcribed spacer region as a taxonomic marker for nematodes. Journal of Nematology 29: 441-450. [PMC free article] [PubMed] [Google Scholar]
- Quist C.W., Smant G., and Helder J.. 2015. Evolution of plant parasitism in the phylum nematode. Annual Review of Phytopathology 53: 289-310. [DOI] [PubMed] [Google Scholar]
- Riga E., and Lazarovits G.. 2001. Development of an organic pesticide based on neem tree products. Journal of Nematology 33: 274, [Abstract]. [Google Scholar]
- Seinhorst J.W. 1959. A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica 15: 67-69. [Google Scholar]
- Senthil-Nathan S. 2013. Physiological and biochemical effect of neem and other Meliaceae plants secondary metabolites against Lepidopteran insects. Frontiers in Physiology 4: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumara T.N., Phani V., Yadava D.K., and Rao U.. 2016. Use of Brassicas as bio-denematizer can be questioned by root-knot nematodes. Indian Journal of Nematology 46: 189-193. [Google Scholar]
- Southey J.F. 1986. Laboratory methods for work with plant and soil nematodes, Ministry of Agriculture Fisheries and Food, Reference Book 402; Her Majesty’s Stationery Office (HMSO), London. [Google Scholar]
- Subbotin S.A., Sturhan D., Chizhov V.N., Vovlas N.V., and Baldwin J.G.. 2006. Phylogenetic analysis of Tylenchida Thorne, 1949 as inferred from D2 and D3 expansion fragments of the 28S rRNA gene sequences. Nematology 8: 455-474. [Google Scholar]
- Subbotin S.A., Vierstraete A., De Ley P., Rowe J., Waeyenberge L., Moens M., and Vanfleteren J.R.. 2001. Phylogenetic relationships within the cyst-forming nematodes (Nematoda: Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Molecular Phylogenetics and Evolution 21: 1-16. [DOI] [PubMed] [Google Scholar]
- Subbotin S.A., Waeyenberge A.L., and Moens M.. 2000. Identification of cyst-forming nematodes of the genus Heterodera (Nematoda: Heteroderidae) based on the ribosomal DNA-RFLPs. Nematology 2: 153-164. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S.. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenente G.C.M.V., De Ley P., De Ley T.I., Karssen G., and Vanfleteren J.R.. 2004. Sequence analysis of the D2/D3 region of the large subunit rDNA from different Meloidogyne isolates. Nematropica 34: 1-12. [Google Scholar]
- Tewari D.N. 1992. Monograph on neem, International Book Distributor, Dehra Dun, India. [Google Scholar]
- Tigano M.S., Carneiro R.M.D.G., Jeyaprakash A., Dickson D.W., and Adams B.J.. 2005. Phylogeny of Meloidogyne spp. based on 18S rDNA and the intergenic region of mitochondrial DNA sequences. Nematology 7: 851-862. [Google Scholar]
- Vovlas N., and Inserra R.N.. 2000. Root-knot nematodes as parasites of citrus. Proceedings of the International Society of Citriculture 2: 812-817. [Google Scholar]
- Vrain T.C., Wakarchuk D.A., Levesque A.C., and Hamilton R.I.. 1992. Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fundamental and Applied Nematology 15: 563-573. [Google Scholar]
- Whitehead A.G. 1968. Taxonomy of Meloidogyne (Nematodea: Heteroderidae) with descriptions of four new species. Transactions of the Zoological Society of London 31: 263-401. [Google Scholar]
- Whitehead A.G., and Hemming J.R.. 1965. A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Annals of Applied Biology 55: 25-38. [Google Scholar]
- Xu J., Liu P., Meng Q., and Long H.. 2004. Characterization of Meloidogyne species from China using isozyme, phenotypes and amplified mitochondrial DNA restriction fragment length polymorphism. European Journal of Plant Pathology 110: 309-315. [Google Scholar]
- Ye W., Giblin-Davis R.M., Davies K.A., Purcell M.F., Scheffer S.J., Taylor G.S., Center T.D., Morris K., and Thomas W.K.. 2007. Molecular phylogenetics and the evolution of host plant associations in the nematode genus Fergusobia (Tylenchida: Fergusobiinae). Molecular Phylogenetics and Evolution 45: 123-141. [DOI] [PubMed] [Google Scholar]


