Abstract
Metastasis can be a fatal step in breast cancer progression. Effective therapies are urgently required due to the limited therapeutic options clinically available. The aim of the present study was to investigate the effect of matrine (MAT), a traditional Chinese medicine, on the proliferation and migration of human breast cancer cells and its underlying mechanisms of action. The proliferation of MDA-MB-231 cells was inhibited and apoptosis was induced following treatment with MAT, as determined by MTT and Annexin-V-FITC/PI assays. Western blot analysis was used to detect the LC-3II/I levels and the results suggested that tumor autophagy is involved in the anti-tumor activity of MAT. To the best of our knowledge, this is the first study to report that MAT inhibits MDA-MB-231 and MCF-7 cell motility, potentially by targeting integrin β1 (ITGB1) and epithelial-to-mesenchymal transition (EMT), as indicated by Transwell® and siRNA interference assays. In conclusion, ITGB1 and EMT are involved in MAT-induced breast carcinoma cell death and the inhibition of metastasis. This may lead to the development of novel compounds for the treatment of breast cancer metastasis.
Keywords: matrine, breast cancer, integrin β1, epithelial- mesenchymal transition
Introduction
Breast cancer is the most invasive type of malignancy in females worldwide, leading to >39,000 deaths in the USA each year (1). Although a number of treatments have seen significant improvement over the years, breast cancer remains a paramount health issue and is at the forefront of medical research (2). It can be considered a heterogeneous disease segmented into five molecular subtypes: Luminal A, luminal B, HER2-enriched, basal-like and claudin-low (3). Treatment options for these cases include surgery, chemotherapy and/or radiotherapy (4). However, breast cancer remains a leading cause of cancer-associated mortality, especially among young women (5). Therefore, the treatments that currently available for patients with breast cancer require urgent improvement.
Chinese traditional herbs can kill tumor cells by acting on multiple targets with few adverse effects, making it an area of great research interest. Matrine (MAT), an alkaloid derived from Sophora Flavescens, is a traditional Chinese medicine used for the treatment of aggressive cancers (6). MAT was found to inhibit the progress of hepatic, cervical and gastric cancer (7), with a plethora of studies focusing on the pharmacological and clinical applications of MAT (8–10).
To the best of our knowledge, little attention has previously been paid to the effects of MAT on breast cancer metastasis. Migration is the driving process of cancer metastasis and corresponds to poor clinical symptoms, a deterioration in health and eventual death (11). A previous study compared different datasets and identified integrin β1 (ITGB1) as one of the crucial genes involved in breast cancer cell migration (12). In addition, ITGB1 is reportedly highly expressed in the claudin-low subtype of breast cancer (13). However, whether MAT inhibits the migration of breast cancer cells by mediating ITGB1 expression remains unclear.
In the present study, it was demonstrated that MAT dose-dependently inhibits proliferation and induces apoptosis in MDA-MB-231 cells. In addition, the present data provided novel evidence of MAT-induced inhibition of cell migration by targeting ITGB1 and the epithelial-to-mesenchymal transition (EMT) in breast cancer.
Materials and methods
Reagents
MAT was purchased from Sigma-Aldrich (Merck KGaA) and stored at 4°C. MAT was later dissolved in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) at a concentration of 20 mg/ml and stored at −20°C. Chloroquine diphosphate salt (CQ) was purchased from Sigma-Aldrich (Merck KGaA).
Cell culture
The human breast cancer cell lines MDA-MB-231 and MCF-7 (Shanghai Institute of Cell Biology, Chinese Academy of Sciences) were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 100 µg/ml penicillin/streptomycin (HyClone; GE Healthcare Life Sciences) in a humidified atmosphere containing 5% CO2 at 37°C.
Cell proliferation assay
To test the effect of MAT on MDA-MB-231 proliferation, 4×103 cells/well were seeded into 96-well culture plates (Nunc™; Thermo Fisher Scientific, Inc.) in 100 µl RPMI-1640 medium and then cultured in a 37°C 5% CO2 incubator overnight. The supernatant was then changed to one that contained different doses of MAT (0, 1 and 2 mg/ml) and cultured for 24 and 48 h, followed by another 2 h after 20 µl MTT (5 mg/ml; Promega Corporation) was added to each well. Optical density values were obtained using a plate reader at a wavelength of 490 nm.
Cell apoptosis assay
Annexin-V-FITC/PI double staining assays were performed to detect the effects of apoptosis on MDA-MB-231 cells. Cells were exposed to MAT (2 mg/ml) or the vehicle control for 48 h in a 24-well plate (3×105 cells/well), after which each group was washed with PBS three times followed by staining at room temperature with an Annexin-V-FITC Apoptosis Detection kit I (RT; BD Biosciences; Becton, Dickinson and Company). The number of apoptotic cells was counted using flow cytometry (FACSCanto™; BD Biosciences; Becton, Dickinson and Company) and the Flowjo Software (version 8.2.4; FlowJo LLC), according to the manufacturer's protocol.
Cell migration assay
Migratory abilities of MDA-MB-231 and MCF-7 cells were determined using a chemotaxis chamber (Corning Life Sciences) according to the manufacturer's protocol. In this assay, cell motility was assessed by migration through a membrane (24-well Transwell® plate, 8-µm pore size) towards a chemoattractant. Briefly, cells were seeded into the upper chambers of the Transwell® inserts (3×104 per well in serum-free medium). Medium with 10% FBS was used as the chemoattractant in the lower chambers. The medium contained tested substances or a vehicle. Following incubation at 37°C with 5% CO2 and 95% air for 16 h, cells were stained with Calcein-AM (0.2 µg/ml; cat. no. C3100MP; Invitrogen; Thermo Fisher Scientific, Inc.) at RT for 30 min. The migrated cells were counted using an eclipse Ti inverted microscope (Nikon Corporation). The number of cells that had migrated was determined using MetaMorph image analysis software (version 4.0; Molecular Devices, LLC) and the results are presented as the mean ± standard deviation (n=3).
RNA interference
Oligonucleotides for human ITGB1 siRNA kit was purchased from Guangzhou RiboBio Co., Ltd. The kit contains three predesigned duplexes targeting a specific ITGB1 gene. Cells were transfected with ITGB1 siRNA or NC at the concentration of 50 nmol/l using the opti-MEM plus X-treme GENE siRNA transfection reagent (Roche Diagnostics) according to the protocol of the manufacturer. The ITGB1 siRNA sequence was as follows: Forward, 5′-CCAUUCUGAUGAAUCUGAU-3′ and reverse, 5′-AUCAGAUUCAUCAGAAUGG-3′. After 48 h of post-transfection, western blot analyses were further performed.
RNA isolation and reverse transcription-quantitative (RT-q)PCR
Briefly, total cellular RNA was extracted using TRIzol® (Life Technologies; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Total RNA was extracted using TRIzol® and a Total RNA kit (Tiangen Biotech Co., Ltd.). cDNA was generated at 37°C using 1 mg total RNA and a QuantiTect Reverse Transcription kit (Qiagen GmbH). RT-qPCR was performed using the SYBR Green (Bio-Rad Laboratories, Inc.) method on an ABI Prism 7000 Sequence Detection System (Life Technologies; Thermo Fisher Scientific, Inc.). The primer for ITGB1 was 5′-CCTACTTCTGCACGATGTGATG-3′ (forward) and 5′-CCTTTGCTACGGTTGGTTACATT-3′ (reverse). The primer for β-actin (control) was 5′-CCACACCCGCCACCAGTTCG-3′ (forward) and 5′-TACAGCCCGGGGAGCATCGT-3′ (reverse). All primers were purchased from Sangon Biotech Co., Ltd., and diluted in DEPC water. The qPCR reaction was performed as follows: 95°C For 30 min, 40 cycles of 95°C for 15 sec and 56°C for 20 sec. To confirm the amplification specificity, the PCR products were subjected to melting curve analysis. The relative mRNA level of ITGB1 was normalized to the β-actin mRNA and analyzed by the comparative threshold (Cq) cycle method (2−ΔΔCq), according to previous research (14).
Western blot analysis
Protein concentration was measured using a bicinchoninic acid Protein Assay Reagent (Pierce; Thermo Fisher Scientific, Inc.). Total protein (20 µg/well) was separated via 10% SDS-PAGE and then transferred onto PVDF membranes at 250 mA for 1 h. Membranes were blocked at 37°C for 2 h with 5% non-fat milk in Tris-buffered saline/0.1% Tween-20 and then incubated at 4°C overnight with the following primary antibodies: Anti-ITGB1 (1:1,000; cat. no. ab183666; Abcam), anti-LC3 II/I (1:1,000; cat. no. ab128025; Abcam), anti-epithelial (E)-cadherin, anti-neural (N)-cadherin, anti-vimentin (1:1,000; cat. no. 9782T; EMT Antibody Sampler kit Cell; Cell Signaling Technology, Inc.), anti-GAPDH (1:5,000; cat. no. 10494-1-AP; ProteinTech Group, Inc.) and anti-β-actin (1:5,000; cat. no. 20536-1-AP; ProteinTech Group, Inc.). Subsequently, membranes were incubated at 37°C for 2 h with anti-rabbit IgG secondary antibodies (1:3,000; cat. no. 14708S; Cell Signaling Technology, Inc.) and the immunoblotted proteins were then detected using an Odyssey Western Blotting Detection System (Gene Tech Co., Ltd.) and Odyssey software (version 1.2).
Statistical analysis
All data are expressed as the mean ± standard deviation) based on experiments performed in triplicate, and were analyzed using SPSS 18.0 statistical analysis software (SPSS, Inc.). One-way analysis of variance followed by Student-Newman-Keuls post hoc test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
MAT inhibits MDA-MB-231 cell growth by inducing apoptosis
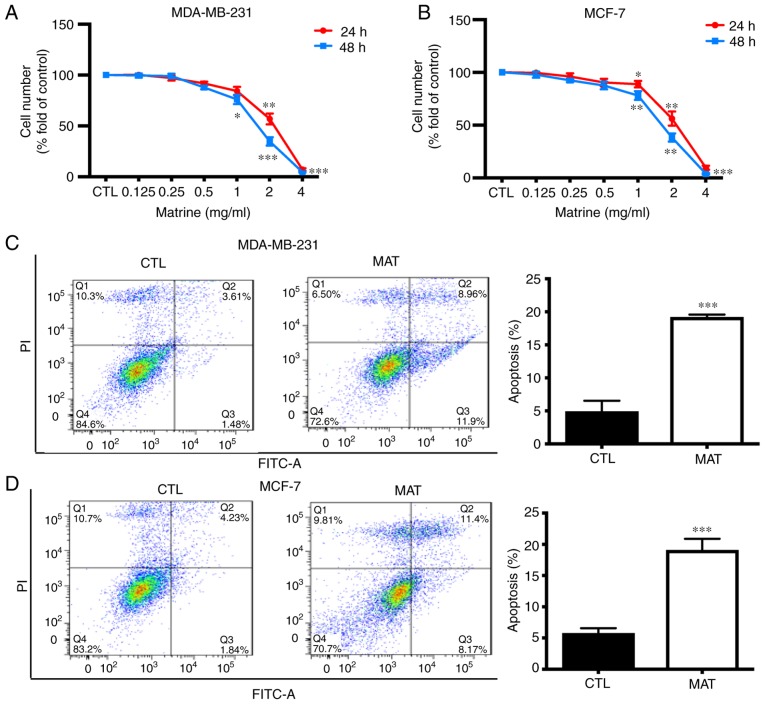
To determine the role of MAT in breast cancer, MDA-MB-231 cells were treated with various concentrations of MAT for 24 and 48 h, following which an MTT assay was performed to evaluate proliferation (Fig. 1A). The data demonstrated that MAT inhibited the proliferation of MDA-MB-231 cells in a dose- and time-dependent manner. Tumor growth was not only associated with abnormal proliferation, but was also dependent on a reduction in apoptosis. To confirm that the apoptosis observed in the cancer cells was induced by MAT, an Annexin-V-FITC/PI apoptosis assay was performed (Fig. 1B). For flow cytometry, MDA-MB-231 cells were treated with or without MAT (2 mg/ml) for 48 h. Cells in the late and early stages of apoptosis were observed in the upper and lower right quadrant of the plots (Q2 and Q4 areas), respectively. These results indicated that treatment with MAT was able to impair proliferation and induce apoptosis in breast cancer cells.
Figure 1.
MAT inhibits MDA-MB-231 and MCF-7 cell growth by inducing cell apoptosis. Following incubation with various concentrations of MAT (0–4 mg/ml) for 24 and 48 h, (A) MDA-MB-231 and (B) MCF-7 cell proliferation was measured by MTT assay. (C) MDA-MB-231 and (D) MCF-7 cells were treated with or without MAT (2 mg/ml) for 48 h and stained with Annexin V (5 µg/ml)/PI (10 µg/ml) prior to being analyzed by flow cytometry. Cells labeled with Annexin V(−) PI(+) are shown in the Q1 area, cells labeled with Annexin V(+) PI(+) in the Q2 area, cells labeled with Annexin V(−) PI(−) in the Q3 area and cells labeled with Annexin V(+) PI(−) in the Q4 area. *P<0.05, **P<0.01 and ***P<0.001 MAT vs. CTL by one-way analysis of variance followed by Student-Newman-Keuls post hoc test. MAT, matrine; NC, negative control; PI, propidium iodide; MAT, matrine; FITC, fluorescein isothiocyanate; CTL, control.
Role of autophagy in MAT-induced decrease in cell growth
Previous studies have demonstrated that both autophagy and apoptosis are involved in the effects observed in cancer cells treated with MAT, including acute myeloid leukemia (15) and osteosarcoma (16) cells. It was verified that MAT inhibits cell proliferation and induces apoptosis. In addition, the fact that apoptosis often occurs simultaneously with autophagy prompted the present study to investigate the association between MAT and autophagy. First, the autophagy inhibitor CQ, a small alkaline molecule that accumulates in lysosomes and reduces hydrolysis (17), was used. As shown in Fig. 2A and B, when MDA-MB-231 and MCF-7 cells were exposed to various doses of MAT with and without CQ, proliferation was significantly decreased in the MAT+CQ group, as compared with the MAT alone group (P<0.05).
Figure 2.
Effect of autophagy in MAT-reduced cell growth. (A) MDA-MB-231 and (B) MCF-7 cell growth analysis was performed following treatment with MAT (0, 1 and 2 mg/ml) for 24 and 48 h or pre-treatment with CQ (10 µM) for 1 h. Western blot analysis of LC3-II/I level in (C) MDA-MB-231 and (D) MCF-7 cells following treatment with vehicle CTL, MAT, CQ and MAT+CQ for 48 h. *P<0.05, **P<0.01 and ***P<0.001 MAT or MAT+CQ vs. CTL, #P<0.05 MAT+CQ vs. MAT by one-way analysis of variance followed by Student-Newman-Keuls post hoc test. MAT, matrine; CQ, chloroquine diphosphate salt; CTL, control.
Next the expression of LC3-II/I, one of the main autophagy regulatory proteins (18), was investigated following exposure to MAT in cells pre-treated with CQ for 1 h. The results showed that the expression of LC3-II/I was accumulated in MDA-MB-231 and MCF cells treated with MAT or with MAT+CQ. In addition, LC3-II/I in cells treated with MAT+CQ was significantly upregulated, as compared with cells treated with MAT alone (P<0.001; Fig. 2C and D). These results therefore suggested that autophagy is involved in MAT-induced breast cancer cell apoptosis.
MAT decreases the migratory capacity of MDA-MB-231 and MCF-7 cells potentially by targeting ITGB1
Metastasis is a primary cause of morbidity and mortality in patients with cancer (19), and cell migration and invasion are the most important steps in this complex process. Therefore, transwell assays were performed to detect the migratory capacity of breast cancer cells and found that MAT (2 mg/ml) significantly inhibited the migration of MDA-MB-231 and MCF-7 cells (P<0.05; Fig. 3A), indicating that MAT may be a promising anti-metastatic agent for breast cancer.
Figure 3.
MAT decreases the migratory capacity of MDA-MB-231 cells potentially by targeting ITGB1. (A) Migration was analyzed in MDA-MB-231 and MCF-7 cells with or without MAT treatment (1 and 2 mg/ml) for 48 h. (B) Reverse Transcription-quantitative PCR and (C) western blot analysis of ITGB1 mRNA and protein levels in MDA-MB-231 and MCF-7 cells following MAT treatment. *P<0.05 and **P<0.01 MAT vs. CTL by one-way ANOVA. The expression of ITGB1 was reduced in (D) MDA-MB-231 and (E) MCF-7 cells transfected with ITGB1 siRNA. β-actin was used as a loading control. (F) In the presence of siRNA targeting ITGB1, Transwell® assay was conducted to evaluate MDA-MB-231 and MCF-7 cell migration following transfection. Silencing ITGB1 results in decreased MDA-MB-231 and MCF-7 cell migration. *P<0.05, **P<0.01 and ***P<0.001 MAT vs. NC by one-way ANOVA followed by Student-Newman-Keuls post hoc test. MAT, matrine; ITGB1, integrin β1; ANOVA, analysis of variance; CTL, control; NC, negative control; si, small interfering.
ITGB1 is reportedly highly expressed in breast cancer and correlates with cell migration (20). Therefore the levels of ITGB1 in MDA-MB-231 and MCF-7 cells following treatment with MAT were investigated. As hypothesized, following incubation with MAT, the relative mRNA expression of ITGB1 was significantly decreased to 64.3 and 60.3% in MDA-MB-231 and MCF-7 cells, respectively (P<0.05). The protein activity of ITGB1 also decreased to 53 and 61.6% in MDA-MB-231 and MCF-7 cells, respectively compared with the control (CTL) group (Fig. 3B). To further confirm that ITGB1 is involved in MTA-impaired migration, siRNA was used to silence ITGB1 expression in MDA-MB-231 and MCF-7 cells. ITGB1-silencing was verified by RT-qPCR and western blot analysis (Fig. 3C and D). Furthermore, transfection with ITGB1 siRNA decreased the migratory capacity of MDA-MB-231 and MCF-7 cells (Fig. 3E). Overall, these data indicated that ITGB1 is involved in MAT-induced inhibition of breast cancer cell motility.
MAT regulates EMT in breast cancer cells
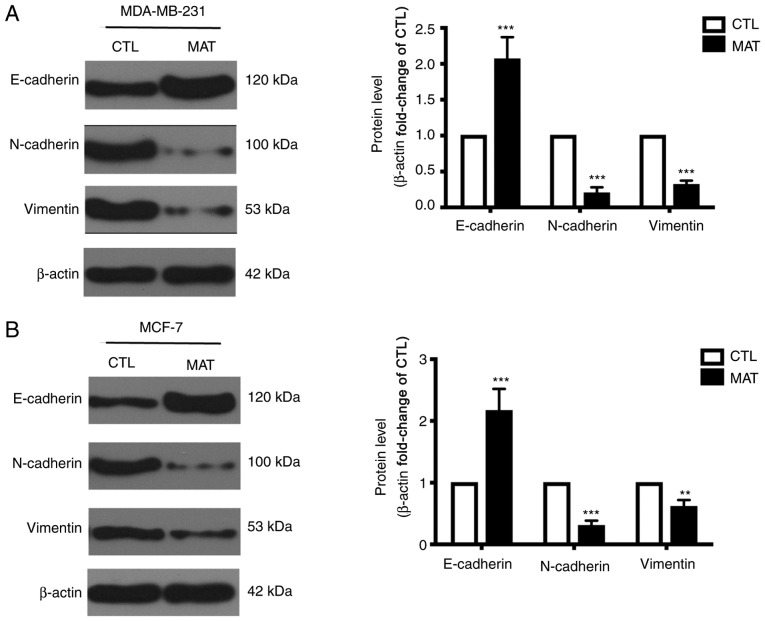
During EMT, cells lose epithelial characteristics and obtain mesenchymal properties, including decreased E-cadherin and increased N-cadherin and vimentin. Considering the significant effect of EMT on tumor cell migration, as well as that the process can be mediated by ITGB1 (21), the levels of certain EMT-associated markers were detected. MDA-MB-231 cells exhibited a mesenchymal phenotype, while MCF-7 cells exhibited the properties of epithelial cells. Thus, the expression of E-cadherin, N-cadherin and vimentin between MDA-MB-231 and MCF-7 cells was different in Fig. 4. In addition, the western blot assays of Fig. 4A and B were not performed on the same PVDF membranes, so the levels of these proteins in these two cells cannot be compared due to variation in experimental conditions. Incubation with MAT can markedly increase the expression of E-cadherin and reduce the levels of N-cadherin and vimentin compared with their CTL group. These changes demonstrated that EMT in MAT-treated MDA-MB-231 and MCF-7 breast cancer cells is blocked, reducing cell metastasis.
Figure 4.
MAT regulates the transition between epithelial and mesenchymal phenotypes in breast cancer cells. Western blot analysis of E-cadherin, N-cadherin and vimentin levels in (A) MDA-MB-231 and (B) MCF-7 cells following MAT treatment (2 mg/ml) for 48 h. **P<0.01 and ***P<0.001 MAT vs. CTL by one-way analysis of variance. MAT, matrine; CTL, control; E, endothelial; N, neural.
Discussion
Natural resources, especially traditional plant-based medicines, are being increasingly investigated as anti-tumor agents (22). MAT is a component of one such traditional plant (Sophora Flavescens), which has been shown to exert therapeutic effects on various types of solid tumors (23,24). In the present study, it was demonstrated that MAT exerts therapeutic effects on MDA-MB-231 and MCF-7 breast cancer cells through inhibiting proliferation and migration. Mechanistically, MAT induces apoptotic cell death, influences ITGB1 expression and blocks EMT to produce these anti-cancer effects.
Previous reports have demonstrated that MAT inhibits the growth of various types of tumors by inducing apoptosis and cell cycle arrest (25,26). The present study reinforced this by demonstrating the ability of MAT to inhibit MDA-MB-231 and MCF-7 cell growth and induce apoptosis. To investigate the mechanisms of MAT-induced cell growth inhibition, cell autophagy and LC3-II/I, two forms of LC3 were focused on. Cytoplasmic LC3-I is conjugated to phosphatidylethanolamine to form LC3-II, which is closely associated with autophagosome membranes and serves as a reliable marker for the monitoring of autophagy (27). First, it was found that cell growth inhibition was increased following the application of the autophagy inhibitor CQ, indicating that impaired autophagy aggravates MAT-induced cell growth inhibition. Secondly, the expression levels of LC3-II/I were further examined and the present data showed that the level of LC3-II/I was elevated following MAT treatment. In addition, its expression markedly increased with CQ co-treatment, suggesting that cancer cell autophagy and apoptosis could be targets for enhancing the anti-tumor effects of MAT.
Metastasis is a complex multistep process that involves cell growth, migration and transportation via the blood vessels. Therefore, the effects of MAT on cell migration, a crucial step in breast cancer metastasis, were detected. The Transwell® assay showed that MAT (1 mg/ml) may have inhibited cell migration, but no significant differences were observed. However, MAT (2 mg/ml) exhibited a marked ability to reduce cell migration in both MDA-MB-231 and MCF-7 cells. Consistent with present results, other studies reported that MAT is able to inhibit cell migration in multiple types of cancer (28–30), suggesting that MAT may be a promising anti-metastatic drug. Among metastasis-related genes, integrins are considered to mediate cell-cell crosstalk across the cellular membrane and play an important role in the maintenance of extracellular matrix (ECM) macromolecules (31). Furthermore, ITGB1 plays critical roles in breast cancer cell proliferation and motility, is highly expressed in aggressive breast tumors and drives metastasis (32,33). ITGB1 is a major adhesion receptor for various ECM components; therefore, the present study investigated the expression of ITGB1 in breast cancer cells following treatment with MAT. The mRNA and protein expression of ITGB1 was impaired in MDA-MB-231 and MCF-7 cells following MAT treatment. Based on siRNA analysis results, the present study hypothesized that ITGB1 and its downstream signaling network is regulated by MAT. The present data therefore provided a new target through which MAT can exert anti-cancer effects.
EMT, a phenotypic cellular process, leads to the loss of cell-cell adhesion. Consequently, cancer cell motility, migration and metastasis is triggered (34). Alterations in cadherin expression are typical in EMT, as is the downregulation of E-cadherin and upregulation of N-cadherin (35) and vimentin (as well as other mesenchymal proteins). A western blot experiment was performed in mesenchymal-like MDA-MB-231 cells and the effects of MAT on EMT were examined. The results showed that incubation of MAT in MDA-MB-231 cells increased the expression of epithelial markers and decreased the expression of mesenchymal marker. Collectively, these data showed that MAT interfered EMT in breast cancer cells. Previous studies also showed ZO1 (36) and E-cadherin (37) were upregulated during EMT process in MDA-MB-231 cells. In the present study, treatment with MAT resulted in the upregulation of E-cadherin and downregulation of N-cadherin and vimentin, strongly indicating that EMT is blocked by MAT. In addition, studies have demonstrated that ITGB1 and ITGB3 exhibit tumor-promoting effects via facilitating EMT in breast cancer and nasopharyngeal carcinoma (38,39). The knockdown of ITGB1 partly increased the expression of E-cadherin and decreased that of vimentin, fibronectin and N-cadherin in BT549 and Hs578T breast cancer cells (40). In the present study, it was shown that the induction and regulation of EMT by MAT may involve multiple molecular mechanisms, including the inhibition of ITGB1 expression.
The limitation of the present study is the lack of specific mechanism by which MAT regulates ITGB1. The authors will measure whether ITGB1 is transcriptionally regulated by MAT via luciferase reporter assays and explore the possibility of reversing MAT-inhibited cellular proliferation and migration by overexpressing ITGB1 in MDA-MB-231 and MCF-7 cells in the following study.
In conclusion, the present results revealed that MAT exerts modulatory effects on apoptotic cell death and that the inhibition of MAT-induced migration is potentially affected through the attenuation of ITGB1 and EMT. Therefore, MAT may serve as a novel suppressor of breast cancer.
Acknowledgements
The authors would like to thank Chief Attending Physician Dr Qinghua Yao, Zhejiang Cancer Hospital, for her durable support and constructive guidance.
Funding
The present study was supported by Zhejiang Medical and Health Science and Technology Plan (grant no. 2013KYB04).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LR wrote the manuscript and performed the experiment. WM and LW collected and interpreted the data. XW obtained funding and designed the study. All authors have read and approved the manuscript for publication.
Ethics approval and consent to participate
All experimental protocols were performed in accordance with the regulation of the Helsinki Declaration and were approved by Ethics Committee of our hospital. Written consent of the participants was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Smith RA, Andrews K, Brooks D, DeSantis CE, Fedewa SA, Lortet-Tieulent J, Manassaram-Baptiste D, Brawley OW, Wender RC. Cancer screening in the United States, 2016: A review of current American cancer society guidelines and current issues in cancer screening. CA Cancer J Clin. 2016;66:96–114. doi: 10.3322/caac.21336. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 3.Miller SM, Goulet DR, Johnson GL. Targeting the breast cancer kinome. J Cell Physiol. 2017;232:53–60. doi: 10.1002/jcp.25427. [DOI] [PubMed] [Google Scholar]
- 4.Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: An overview. Updates Surg. 2017;69:313–317. doi: 10.1007/s13304-017-0424-1. [DOI] [PubMed] [Google Scholar]
- 5.Simmons A, Burrage PM, Nicolau DV, Jr, Lakhani SR, Burrage K. Environmental factors in breast cancer invasion: A mathematical modelling review. Pathology. 2017;49:172–180. doi: 10.1016/j.pathol.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H, Xu M, Gao Y, Deng Z, Cao H, Zhang W, Wang Q, Zhang B, Song G, Zhan Y, Hu T. Matrine induces caspase-independent program cell death in hepatocellular carcinoma through bid-mediated nuclear translocation of apoptosis inducing factor. Mol Cancer. 2014;13:59. doi: 10.1186/1476-4598-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Zhang Y, Zhuang Y, Wang J, Ye J, Zhang S, Wu J, Yu K, Han Y. Matrine induces apoptosis in human acute myeloid leukemia cells via the mitochondrial pathway and Akt Inactivation. PLoS One. 2012;7:e46853. doi: 10.1371/journal.pone.0046853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng F, Wang J, Ding F, Xie Y, Zhang Y, Zhu J. Neuroprotective effect of matrine on MPTP-induced Parkinson's disease and on Nrf2 expression. Oncol Lett. 2017;13:296–300. doi: 10.3892/ol.2016.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang LP, Jiang JK, Tam JW, Zhang Y, Liu XS, Xu XR, Liu BZ, He YJ. Effects of matrine on proliferation and differentiation in K-562 cells. Leuk Res. 2001;25:793–800. doi: 10.1016/S0145-2126(00)00145-4. [DOI] [PubMed] [Google Scholar]
- 10.Long Y, Lin XT, Zeng KL, Zhang L. Efficacy of intramuscular matrine in the treatment of chronic hepatitis B. Hepatobiliary Pancreat Dis Int. 2004;3:69–72. [PubMed] [Google Scholar]
- 11.Sciacovelli M, Frezza C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 2017;284:3132–3144. doi: 10.1111/febs.14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klahan S, Huang WC, Chang CM, Wong HS, Huang CC, Wu MS, Lin YC, Lu HF, Hou MF, Chang WC. Gene expression profiling combined with functional analysis identify integrin beta1 (ITGB1) as a potential prognosis biomarker in triple negative breast cancer. Pharmacol Res. 2016;104:31–37. doi: 10.1016/j.phrs.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Zawistowski JS, Nakamura K, Parker JS, Granger DA, Golitz BT, Johnson GL. MicroRNA 9-3p targets β1 integrin to sensitize claudin-low breast cancer cells to MEK inhibition. Mol Cell Biol. 2013;33:2260–2274. doi: 10.1128/MCB.00269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Hu G, Dong Y, Ma R, Yu Z, Jiang S, Han Y, Yu K, Zhang S. Matrine induces Akt/mTOR signalling inhibition-mediated autophagy and apoptosis in acute myeloid leukaemia cells. J Cell Mol Med. 2017;21:1171–1181. doi: 10.1111/jcmm.13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma K, Huang MY, Guo YX, Hu GQ. Matrine-induced autophagy counteracts cell apoptosis via the ERK signaling pathway in osteosarcoma cells. Oncol Lett. 2016;12:1854–1860. doi: 10.3892/ol.2016.4848. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Qu X, Sheng J, Shen L, Su J, Xu Y, Xie Q, Wu Y, Zhang X, Sun L. Autophagy inhibitor chloroquine increases sensitivity to cisplatin in QBC939 cholangiocarcinoma cells by mitochondrial ROS. PLoS One. 2017;12:e0173712. doi: 10.1371/journal.pone.0173712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo S-H, Tang G, Ma K, Babij R, Cortes E, Vonsattel JP, Faust PL, Sulzer D, Louis ED. Macroautophagy abnormality in essential tremor. PLoS One. 2012;7:e53040. doi: 10.1371/journal.pone.0053040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talmadge JE, Fidler IJ. AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li WX, Sha RL, Bao JQ, Luan W, Su RL, Sun SR. Expression of long non-coding RNA linc-ITGB1 in breast cancer and its influence on prognosis and survival. Eur Rev Med Pharmacol Sci. 2017;21:3397–3401. [PubMed] [Google Scholar]
- 21.Xie G, Ji A, Yuan Q, Jin Z, Yuan Y, Ren C, Guo Z, Yao Q, Yang K, Lin X, Chen L. Tumour-initiating capacity is independent of epithelial-mesenchymal transition status in breast cancer cell lines. Br J Cancer. 2014;110:2514–2523. doi: 10.1038/bjc.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Li X, Bai M, Suo Y, Zhang G, Cao X. Matrine inhibited proliferation and increased apoptosis in human breast cancer MCF-7 cells via upregulation of Bax and downregulation of Bcl-2. Int J Clin Exp Pathol. 2015;8:14793–14799. [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, Huang K. Effects of the Chinese medicine matrine on experimental C. parvum infection in BALB/c mice and MDBK cells. Parasitol Res. 2012;111:1827–1832. doi: 10.1007/s00436-012-3030-7. [DOI] [PubMed] [Google Scholar]
- 24.Shao H, Yang B, Hu R, Wang Y. Matrine effectively inhibits the proliferation of breast cancer cells through a mechanism related to the NF-κB signaling pathway. Oncol Lett. 2013;6:517–520. doi: 10.3892/ol.2013.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li LQ, Li XL, Wang L, Du WJ, Guo R, Liang HH, Liu X, Liang DS, Lu YJ, Shan HL, Jiang HC. Matrine inhibits breast cancer growth via miR-21/PTEN/Akt pathway in MCF-7 cells. Cell Physiol Biochem. 2012;30:631–641. doi: 10.1159/000341444. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Hou C, Zhang S, Xie H, Zhou W, Jin Q, Cheng X, Qian R, Zhang X. Matrine upregulates the cell cycle protein E2F-1 and triggers apoptosis via the mitochondrial pathway in K562 cells. Eur J Pharmacol. 2007;559:98–108. doi: 10.1016/j.ejphar.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Yu KY, Wang YP, Wang LH, Jian Y, Zhao XD, Chen JW, Murao K, Zhu W, Dong L, Wang GQ, Zhang GX. Mitochondrial KATP channel involvement in angiotensin II-induced autophagy in vascular smooth muscle cells. Basic Res Cardiol. 2014;109:416. doi: 10.1007/s00395-014-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Zhou J, Cai D, Li M. Matrine inhibits the metastatic properties of human cervical cancer cells via downregulating the p38 signaling pathway. Oncol Rep. 2017;38:1312–1320. doi: 10.3892/or.2017.5787. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhang H, Yu P, Liu Q, Liu K, Duan H, Luan G, Yagasaki K, Zhang G. Effects of matrine against the growth of human lung cancer and hepatoma cells as well as lung cancer cell migration. Cytotechnology. 2009;59:191–200. doi: 10.1007/s10616-009-9211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Wang T, Wen X, Wei Y, Peng X, Li H, Wei L. Effect of matrine on HeLa cell adhesion and migration. Eur J Pharmacol. 2007;563:69–76. doi: 10.1016/j.ejphar.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 31.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 32.Xu Z, Zou L, Ma G, Wu X, Huang F, Feng T, Li S, Lin Q, He X, Liu Z, Cao X. Integrin β1 is a critical effector in promoting metastasis and chemo-resistance of esophageal squamous cell carcinoma. Am J Cancer Res. 2017;7:531–542. [PMC free article] [PubMed] [Google Scholar]
- 33.Jahangiri A, Nguyen A, Chandra A, Sidorov MK, Yagnik G, Rick J, Han SW, Chen W, Flanigan PM, Schneidman-Duhovny D, et al. Cross-activating c-Met/β1 integrin complex drives metastasis and invasive resistance in cancer. Proc Natl Acad Sci USA. 2017;114:E8685–E8694. doi: 10.1073/pnas.1701821114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang R, Zhang C, Liu G, Gu R, Wu H. MicroRNA-126 inhibits proliferation, migration, invasion and EMT in osteosarcoma by targeting ZEB1. J Cell Biochem. 2017;118:3765–3774. doi: 10.1002/jcb.26024. [DOI] [PubMed] [Google Scholar]
- 35.Araki K, Shimura T, Suzuki H, Tsutsumi S, Wada W, Yajima T, Kobayahi T, Kubo N, Kuwano H. E/N-cadherin switch mediates cancer progression via TGF-β-induced epithelial-to-mesenchymal transition in extrahepatic cholangiocarcinoma. Br J Cancer. 2011;105:1885–1893. doi: 10.1038/bjc.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Hao Y, Mao W, Xue X, Xu P, Liu L, Yuan J, Zhang D, Li N, Chen H, et al. LincK contributes to breast tumorigenesis by promoting proliferation and epithelial-to-mesenchymal transition. J Hematol Oncol. 2019;12:19. doi: 10.1186/s13045-019-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY, Xi QS. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012;18:BR299–BR308. doi: 10.12659/MSM.883262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y, Pan Y, Liu S, Jiang F, Jiao J. Elevation of MiR-9-3p suppresses the epithelial-mesenchymal transition of nasopharyngeal carcinoma cells via down-regulating FN1, ITGB1 and ITGAV. Cancer Biol Ther. 2017;18:414–424. doi: 10.1080/15384047.2017.1323585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YY, Kong LQ, Zhu XD, Cai H, Wang CH, Shi WK, Cao MQ, Li XL, Li KS, Zhang SZ, et al. CD31 regulates metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma via the ITGB1-FAK-Akt signaling pathway. Cancer Lett. 2018;429:29–40. doi: 10.1016/j.canlet.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Hou Y, Zhou M, Wen S, Zhou J, Xu L, Tang X, Du YE, Hu P, Liu M. Twist induces epithelial-mesenchymal transition and cell motility in breast cancer via ITGB1-FAK/ILK signaling axis and its associated downstream network. Int J Biochem Cell Biol. 2016;71:62–71. doi: 10.1016/j.biocel.2015.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.